Abstract
The aim of this work is to study the dosimetric effect from generated synthetic computed tomography (sCT) from magnetic resonance (MR) images using a deep learning algorithm for Gamma Knife (GK) stereotactic radiosurgery (SRS). The Monte Carlo (MC) method is used for dose calculations. Thirty patients were retrospectively selected with our institution IRB’s approval. All patients were treated with GK SRS based on T1-weighted MR images and also underwent conventional external beam treatment with a CT scan. Image datasets were preprocessed with registration and were normalized to obtain similar intensity for the pairs of MR and CT images. A deep convolutional neural network arranged in an encoder–decoder fashion was used to learn the direct mapping from MR to the corresponding CT. A number of metrics including the voxel-wise mean error (ME) and mean absolute error (MAE) were used for evaluating the difference between generated sCT and the true CT. To study the dosimetric accuracy, MC simulations were performed based on the true CT and sCT using the same treatment parameters. The method produced an MAE of 86.6 ± 34.1 Hundsfield units (HU) and a mean squared error (MSE) of 160.9 ± 32.8. The mean Dice similarity coefficient was 0.82 ± 0.05 for HU > 200. The difference for dose-volume parameter D95 between the ground true dose and the dose calculated with sCT was 1.1% if a synthetic CT-to-density table was used, and 4.9% compared with the calculations based on the water-brain phantom.
Keywords: Gamma knife, radiosurgery, synthetic CT, deep learning
Introduction
Accurate dose calculation is crucial to the quality of radiation treatment planning. Successful prediction of the delivered dose in radiosurgery can increase survival and reduce side effects for cancer patients. 1 However, there are institutions that treat brain tumors in Gamma Knife stereotactic radiosurgery (GK SRS) solely based on magnetic resonance (MR) images with a crude dose algorithm which assumes that the brain consists of water. The commonly used algorithm in the treatment planning system (TPS) is called TMR (tissue maximum ratio) classic or TMR 10, which is based on the assumption of a brain consisting of water. This assumption introduces error in the prediction of radiation dosage due to its ignorance of the heterogeneity in a human brain. Researchers and clinicians are aware of this issue and efforts have been made to improve the accuracy of calculations.2‐10 For example, a CT-based convolution dose algorithm, which considers approximately the effect of heterogeneity, has been provided in newer TPS versions. 11 Recently, a Monte Carlo (MC) dose algorithm based on a virtual source model has shown6‐8 that the CT-based dose algorithm may yield dose difference up to 11% to 20% compared with the TMR classic dose algorithm.8‐10 To take advantage of these accurate dose algorithms, the brain needs to be represented with close to real density and material information derived from computerized tomography (CT). For institutions that treat brain tumors in GK SRS solely based on MR images, there is a need to generate a synthetic computed tomography (sCT) from an MR image to create the calculation volume of the brain in order to take advantage of these dose algorithms.
Existing methods to generate sCT from MR images are mainly categorized into 3 types: tissue segmentation-based, atlas-based, and learning-based approach. In the tissue segmentation-based method, the MR image is segmented into different tissues and then the predefined HUs are assigned to each tissue class. 12 Multiple MR sequences are generally needed to distinguish ambiguous tissues, such as air and bone, which can both appear dark (low intensity) in certain MR images. This method is obviously highly dependent on the accuracy of tissue segmentation. The atlas-based method registers and aligns MR images to atlas MR images, and then deforms the corresponding atlas CT images according to the MR registered transformation to generate estimated CT images, and thus this method relies on the accuracy of image registration. Several approaches to improve the registration accuracy are proposed,13,14 such as finding the most similar atlas MR, creating an average atlas from multiple atlases. In the learning-based method, the nonlinear relationship between the MR and CT image is learned from a training dataset and then the learned knowledge is used to create an sCT from a given MR image. A number of methods have been developed in this category, such as structured random forests and convolutional neural networks (CNNs).15‐18 In the structured random forest method, 16 the input MR image is first represented by features and then the features are mapped to the estimated CT image. The quality of feature extraction is critical to the performance of this method. The other learning-based approach, the CNN method,15‐17 uses multilayer, fully trainable models that learn features by itself in the form of convolutional kernels. It has been widely used in both computer vision and medical image fields.
Several studies19‐22 have been carried out to investigate the dosimetric effect of using MR-generated sCT for treatment planning in external beam radiotherapy. However, very few works have been done for GK radiosurgery. 23 In this work, we employ the deep learning network ResU-Net,24,25 one of the CNN architectures, to learn the direct mapping from MR images to the corresponding CT images. The network consists of 10 ResNet 25 blocks that are arranged in an encoder–decoder fashion. We collected 30 pairs of MR and CT images from 30 patients with our institution IRB's approval for the network training. Each patient underwent both GK SRS and whole brain or brain stereotactic body radiotherapy (SBRT) treatment. The T1-weighted MR image was used for SRS treatment, and the CT image was used for whole brain or SBRT. A fivefold cross-validation26‐28 procedure was performed in which the image data of 24 patients were used as the training dataset and the remaining 6 patients were used as the test set. Cross-validation is a resampling procedure used to evaluate machine learning models on a limited data sample. In the procedure, the given data sample is split into several groups to estimate the skill of a model on unseen data. The approach has also been used in Han's work 15 to evaluate the performance of the proposed CNN method for MR-based sCT generation. In our study, we split 30 patient data into 5 groups. For each group, we took the group as the test data set and took the remaining 4 groups as the training dataset and the evaluation statistic was performed on the test data for the group. The performance of the model is a summary from all 5 groups. The goals of our work are twofold: to investigate the possibility of generating clinically applicable sCT using the ResU-Net and study the dosimetric accuracy of applying the generated sCT in MC simulations for GK SRS treatments.
Materials and Methods
Dataset and Preprocessing
A total of 30 subjects were retrospectively selected for this study from a collection of patients between 2017 and 2019 that underwent both GK radiosurgery and conventional external beam treatment. Our study was approved by University Hospitals (UH, Cleveland Medical Center, Cleveland OH) IRB administration offices under Chart Review Data Protocol 060519 (approval no. STUDY20200313). The request for a full waiver of consent was also approved by UH IRB.
For GK radiosurgery, A T1-weighted MR image was obtained for treatment planning. For external beam treatment of the whole brain or SBRT, a CT image was obtained. The MR image-based GK treatment plans were created with Leksell GammaPlan (LGP) TPS (version 10). The MR image was obtained using a 1.5T Siemens Espree scanner (Erlangen, Germany) with TE 3.28 ms, TR 2160 ms, and flip angle 15° in our hospital. The size of the image was 256 × 256 and the slice thickness was 1 mm. No additional special procedures were taken to consider the artifacts in MR images, and the GK frames were in the MR training image dataset. The LGP plan in the format of RTPlan was retrieved from the planning system to provide necessary input information for MC dose calculations. The CT images were acquired with a Siemens Somatom scanner with a slice thickness of 1 mm for SBRT or 3 mm for whole-brain treatment. The range of the days between the acquisition of the MR and CT images were from 3 days to 782 days (median: 195 day, meanL 194 days, std: 220 days). Image preprocessing was performed on the CT in order to match the image size and spacing of the MRI for data training using MIM software v6.1 (Cleveland, OH). The CT image was aligned to its corresponding MR image using the rigid registration method based on mutual information and saved with the same image size and resolution as the MR image using interpolation. The final preprocessed images had a size of 256 × 256 × 192 and a resolution of 1 × 1 × 1 mm3. To reduce the computational load, image patches of size 64 × 64 × 5 were extracted from the MR images and corresponding CT images for the model training. The subvolumes were generated by slicing through the whole volume with a stride size of 16 × 16 × 1 in the x, y, and z directions.
Deep Learning Network Architecture
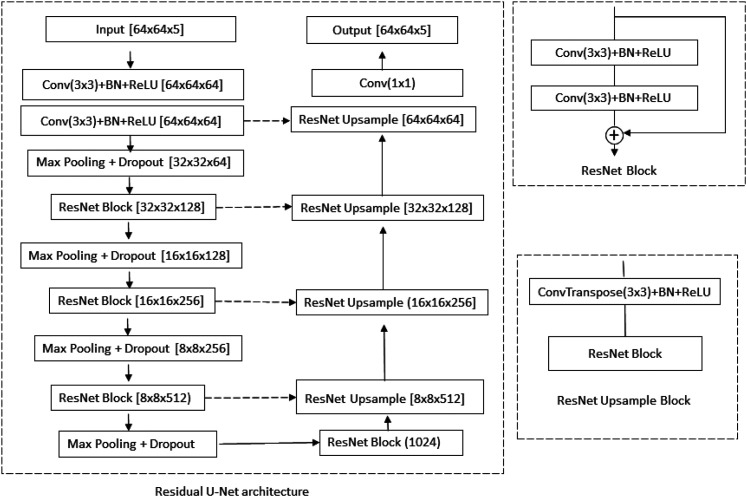
An artificial neural network consists of neurons that have learnable weights and biases, arranged as layers in the network. As more layers are stacked, the network has better learning capacities. The underlying idea of a deep learning network is to learn multilayer hierarchical feature representation so that filters used by deeper layers have much broader receptive fields to capture global information. The ResU-Net method24,25 was employed (details listed in Figure 1) in our work. All convolutions use 3 × 3 kernels except the final one and the identity filter which uses 1 × 1 kernel. Each convolution is followed by batch normalization to increase the network stability by subtracting the batch mean from the output of the previous activation layer and dividing by the batch standard deviation. The nonlinear rectified linear unit (ReLU) is used after batch normalization to introduce nonlinearity to the model and helps to alleviate the vanishing gradient problem. The convolutional block is then followed by a 2 × 2 average pooling to reduce the resolution in the encoder and a bilinear up-sampling to double the resolution in the decoder. Dropout, batch normalization, and data augmentation (image flipping) were used in the network to reduce over-fitting.
Figure 1.
Overall architecture of the residual U-Net network. Conv: convolution; BN: batch normalization
The model was implemented based on the deep learning library PyTorch. It runs on a single GPU (NVIDIA Tesla K40c) machine. We used the default weight initialization for each layer provided by PyTroch, which is a uniform distribution bounded by the inverse of square root of the number of input features. We used the library included function L1Loss for the cost function and used Adam optimizer for optimization. We have run 12 epochs about 200 000 iterations for each fold for training. It took about 8 h so that about 40 h were needed to generate 5 evaluation models. After a model is obtained, it took about 1 min and 20 s to generate a set of sCT (about 150 slices) for each test case in the hold-out fold.
Evaluation of Predict sCT
The accuracy of the predicted sCT for each test case was evaluated by comparing it with the true CT using a voxel-wise MAE,
where N is the total number of voxels inside the region of interest which excludes surrounding air. Similarly, the voxel-wise mean error (ME) and mean squared error (MSE) were also calculated to determine if the generated sCT was biased toward an over- or under-estimation of the CT number and the variation of the difference, respectively. We call the aligned CT as the true CT although the treatment GK plan was actually based on the MR image.
The other metric used in this work is the DSC. It was used to evaluate the correctness of the sCT bone geometry, that is,
where and are the volumes of bones in the sCT and true CT, respectively. From 0 to 1, DSC measures the degree of overlap of bony structures between the volumes. DSC = 0 means no overlap and DSC = 1 means complete overlap. Voxels with a CT number greater than 200 HU were identified as bones.
Dosimetric Evaluation
We carried out MC dose calculations based on the true CT and the corresponding sCT using the same beam configuration and total beam-on time in the clinical treatment plans. The transport of the particles was carried out using our in-house developed MC code, which has been described in our previous work.7,8 Briefly, the 3 physics processes, that is, Compton scattering, photoelectric ionization, and pair production were considered in the code for photon transport and the class II condensed history method was used for electron transport. The hard interactions such as inelastic collision and bremsstrahlung were simulated explicitly for energies above certain cutoffs. The continuous slowing down approximation was employed for energies below the thresholds. The cut-off energy for absorption was set to 50 keV for photons and 200 keV for electrons in our simulations, which is corresponding approximately to a mean free path of 1 mm (the voxel size in our calculations was 1 mm). The simulation statistical uncertainty in our work is defined as the maximum uncertainty for the voxels of the region of 50% of the maximum dose. A virtual source model was employed.7,8 In the model, the source locations were shifted and the calculation volume was kept fixed in the frame coordinate system, not like in the GK treatments the irradiated volume is shifted to the focus point. In addition, we designed an efficient method for sampling particles from multitarget and multishot beam configurations. The code has been used to study the dose difference between the GK TPS algorithms and the MC method in extreme heterogeneous cases resulting from the inhomogeneous effect using a designed heterogeneous phantom with multilayer materials, including air, bone, and tissue. More than 10% discrepancy in dose was reported 8 for the targets adjacent to the interfaces. The number of targets and shots for the test cases in the current work ranged from 2 to 13 targets and 3 to 105 shots with mixed collimators and gamma angles (70°, 90°, and 110°). The prescriptions ranged from 12 to 20 Gy prescribed to the 50% isodose line. The dose algorithm used in the treatment was TMR10 based on the MR image. The coordinates, collimator configurations, and irradiate time of the shots were used as inputs to the MC simulations. For a fair comparison between the true CT and the corresponding sCT, the same CT-to-density table was used to convert the CT number to voxel-wise density and material. A total number of simulated particles (1 × 109) was used to reduce the statistical uncertainty for the region of 50% maximum dose. The MC calculated dose was then converted into the RTDose format and was imported into a third-party software (MIM Software, Cleveland OH) for display and dose-volume histogram (DVH) calculations. The differences of DVH parameters, such as D95 (dose received by 95% of the target volume) for the targeted lesion and V12Gy (percentage volume received a dose greater than 12 Gy) for the whole brain, were calculated for comparison. An additional comparison method, the γ-index,29,30 was used to quantitatively compare the 3D dose distributions for the dose points greater than 10% of the maximum value. The passing rate was computed using the criterion of 1 mm/1% in this method. To study the influence of dose distributions using sCT, we also carried out MC calculations using the water-based brain phantom. The water phantom was created by assigning water to the skull structure exported from the TPS.
For radiotherapy purposes, one of the goals of generating sCT is to apply the sCT in treatment dose planning. The patient CT is converted to a density volume using the CT-to-density table (CDT) for a specific CT scanner. The CDT is precalibrated using a phantom with materials of known densities. We call it the clinical CDT. Practically, a DCCN model can be built directly by mapping the MR images to the density volumes by simply replacing the CT image with the density volume as input data. In doing so, however, the resulting model will be specific to that particular CT scanner. For a different CT scanner, the model must be reconstructed using the density volume generated from the scanner-specific CDT. In another approach, we may create a synthetic CDT to convert the synthetic CT number to the density volume by mapping the CT number of an individual voxel of the sCT to the true density generated from the CDT using the true CT for each case. The synthetic CDT can be viewed as a supplemental correction step for the generation of the density volume for a specific scanner. To create a synthetic CDT, an array of 512 elements starting from −1024 to 3072 HU (each bin has a range of 8 HU) was used to store the density values. For each bin, multiple density values may be corresponding to this synthetic CT bin. The averaged density value was used in the synthetic CDT. The final synthetic CDT was the average of 30 individual tables and it was used to study the effect on the dose calculations.
Results
Evaluation of sCT Images
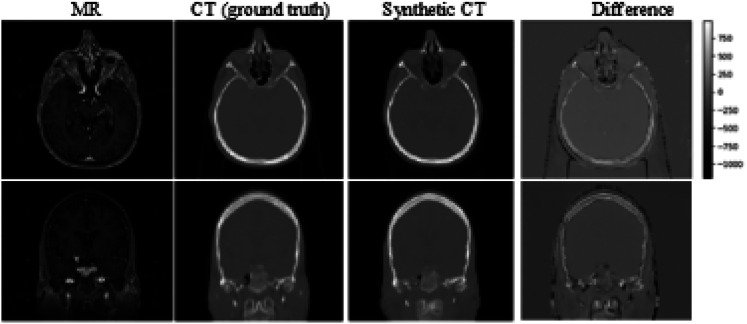
Example views of generated sCT (for subject #12) and the difference between the sCT and the true CT were shown in Figure 2 with the corresponding MR image. We can see that the deep learning network predicts CT values accurately for most of all soft tissues in the head region, except interfaces between tissues and air. The possible sources of large discrepancy are due to high-intensity gradients at these areas and imperfect registration between the input MRI and CT image. This similar observations were reported in other studies as well.15,19 Note that the headrest and the immobilization mask were shown in the true CT for external beam treatment (the couch table was not shown in the figure). It can be seen in the different images because neither headrest nor immobilization mask was used in MR scans for GK treatments. Instead, an MRI-compatible stereotactic head frame was used, as shown in Figure 2 (3 bright dots at each side of the axis and coronal slice view). Our synthetic CT model did predict the head frame, however, in low HU values (about −1000), and they were invisible in the sCT image. The frame in the sCT has no adverse impact on dose calculations because the purpose of the head frame is only used for the definition of the coordinate system. It can be neglected in the dose calculations (as in the commercial TPS).
Figure 2.
An example of MR image and corresponding aligned CT (designated as ground truth). The difference map is shown in the fourth column.
Abbreviations: MR, magnetic resonance; CT, computerized tomography.
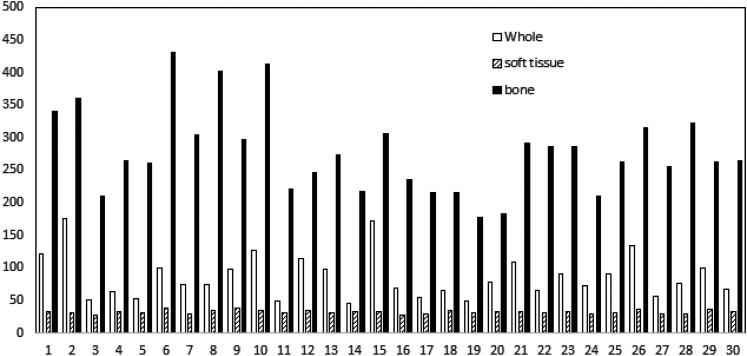
Three metrics (ME, MAE, and MSE) and the standard deviation were listed in Table 1. The ME was −14.6 ± 28.5 HU and −50.5 ± 98.6 HU for soft tissue and bones, respectively. The HU ranges for soft tissue and bone are defined differently in the literature.17,25,31,32 In our work, we define the HU range for soft tissue as greater than −200 HU and less than 200 HU, and the HU range for bone as greater than 200 HU. The MAE was 32.2 ± 2.9 HU and 277.7 ± 64.4 for soft tissue and bones, respectively. For the region of whole brain including air, the MAE was 86.5 ± 34.1 HU and the MSE was 160.9 ± 32.8. The MAE values for soft tissue, bone, and the whole brain for all 30 subjects were plotted in Figure 3. We can see that the MAE was less than 20 HU for bones for all cases.
Table 1.
Summary of HU difference for different tissue types (soft tissue: HU < 200 and HU > −200, bone: HU > 200) between the true CT and sCT measured by ME, MAE, and MSE.
| Whole | Tissue | Bone | |||||||
|---|---|---|---|---|---|---|---|---|---|
| ME | MAE | MSE | ME | MAE | MSE | ME | MAE | MSE | |
| Median | −11.4 | 75.6 | 154.5 | −9.5 | 32.5 | 54.0 | −34.0 | 264.6 | 354.8 |
| Mean | −4.7 | 86.5 | 160.9 | −14.6 | 32.2 | 54.6 | −50.5 | 277.7 | 360.2 |
| Std | 49.8 | 34.1 | 32.8 | 28.5 | 2.9 | 2.7 | 98.6 | 64.4 | 75.3 |
Abbreviations: HU, Hounsfield unit; CT, computerized tomography; sCT, synthetic computed tomography; ME, mean error; MAE, mean absolute error; MSE, mean squared error.
Figure 3.
The mean absolute error (MAE) values for all 30 subjects for different tissue types. Whole stands for whole brain.
To measure the accuracy of the generated sCT for bones, we calculated the DSC for all 30 cases. The median, mean, and standard derivation for DSC are 0.87, 0.86, and 0.04, respectively. For peak signal-to-noise ratio (PSNR) between the true CT and sCT, the calculated statistics are 24.3, 24.3, and 1.3 for median, mean, and standard derivation, respectively.
Evaluation of Dosimetric Effects Using sCT for GK Dose Calculations in the Six-Patient Test Set
Within this cohort, the total simulated targets and shots were 41 and 218. The shots were a mix of 4, 8, and 16 mm collimators. The treated plans were originally calculated with the TPS TMR dose algorithm. We excluded the TPS calculated dose in comparison due to the different algorithms used in the TPS. We focus on the difference that comes from the different phantoms of the brain, that is, the water-based phantom, the phantom generated from the true CT and the sCT. The calculation time was approximately 10 min for each case in the MC simulation and the maximum statistical uncertainty for the region of 50% maximum dose was less than 1.0%.
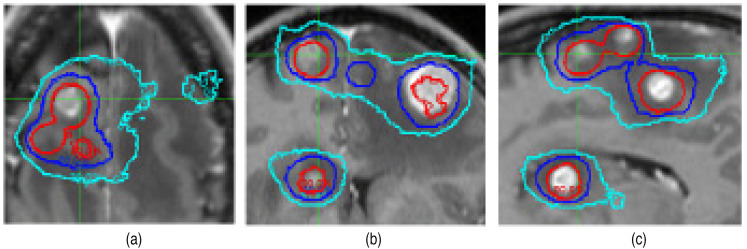
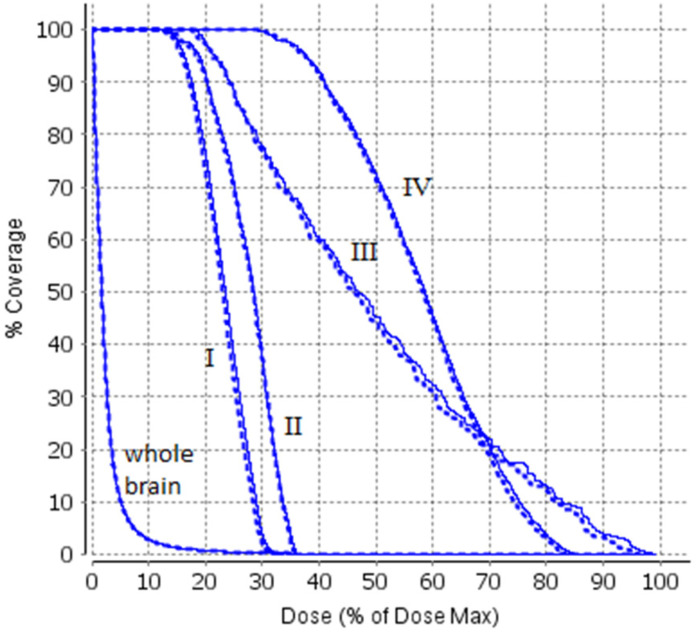
We first compare the dose distribution between the sCT and true CT-based calculations. Figure 4 shows an example of comparison of the dose contours at the axial (a), coronal (b), and sagittal (c) plane (for case #1), and the example of comparison of DVH is shown in Figure 5. The passing rates of comparison between the sCT and true CT-based calculations were 96.5%, 98.6%, 96.9%, 97.4%, 96.2%, and 95.4%, respectively, for the 6 cases using 1 mm/1% criterion for the dose points greater than 10% of the maximum dose value.
Figure 4.
Comparison of Monte Carlo dose contours based on the true CT and sCT (case #1). The axial (a), coronal (b), and sagittal (c) planes are shown. Three iso dose lines are demonstrated: red: 80%, blue: 50%, and cyan: 30%. The thin lines are the results calculated from the true CT and the thick lines are the results calculated from the sCT.
Abbreviations: CT, computerized tomography; sCT, synthetic computed tomography.
Figure 5.
An illustration of comparison of DVH for 4 treatment targets (case #1). The first curve is for the whole brain. The dotted lines were calculated based on the true CT and the sold lines were calculated based on the sCT.
Abbreviations: DVH, dose-volume histogram; CT, computerized tomography; sCT, synthetic computed tomography.
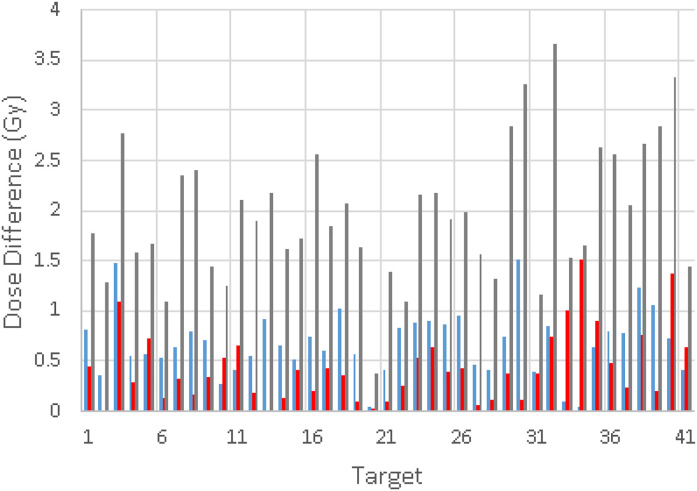
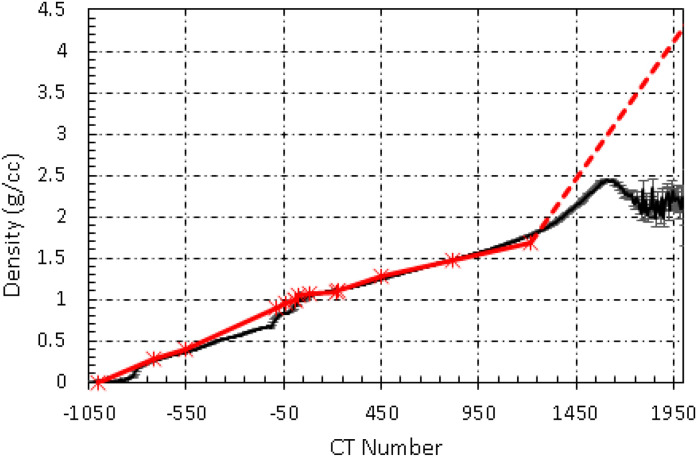
The comparison of dose distributions among different phantom volumes is shown in Figure 6. The dose calculation using the phantom created with the true CT and clinical CDT was used as a benchmark. The other phantoms used in the comparison were (1) the phantom (phantom_sCT_1) created using the sCT with clinical CDT; (2) the phantom (phatom_sCT_2) created using the sCT with synthetic CDT and (3) the water brain phantom (phantom_water) created with the skull structure from the treatment plan. The mean dose difference of D95 was 0.68 Gy based on phantom_sCT_1, 0.44 Gy based on phatom_sCT_2, and 1.97 Gy based on phantom_water compared with the benchmark. The differences were corresponding to 1.7%, 1.1%, and 4.9% of the prescribed dose, respectively if 20 Gy to 50% isodose line is used as the prescription. The mean differences of V12Gy were 0.78, 0.43, and 2.22 cc for the calculation based on phantom_sCT_1, phantom_sCT_2, and phantom_water, respectively. These results showed that the calculations based on the phantom created with the sCT and synthetic CDT were closer to the benchmark. Figure 7 shows the curve of the synthetic CDT and the clinical CDT. Statistical error bars were also shown for the curve of the synthetic CDT. The error bars were small for HU < 1200 HU and became larger for higher HU. The synthetic CDT agreed with the clinical CDT except for HU greater than 1200.
Figure 6.
Dose differences of D95 for all 41 lesions from the MC calculations based on (a) the water-based phantom (gray), (b) the sCT with the clinical CDT (blue), and (c) the sCT with the synthetic CDT (red), compared with the calculation based on the true CT and the clinical CDT.
Abbreviations: CT, computerized tomography; sCT, synthetic computed tomography; MC, Monte Carlo; CDT, CT-to-density table.
Figure 7.
The synthetic CDT (black) generated from sCT. The clinical CDT (red) is also shown for comparison.
Abbreviations: sCT, synthetic computed tomography; CDT, CT-to-density table.
Discussion
Using MR-generated sCT for treatment planning has been investigated for external beam radiotherapy in the brain, head, and neck, lung, prostate, and pelvis.12,20‐22,33 The main advantage of MR imaging is its superior soft-tissue contrast for accurate target delineation. It is true for GK stereotactic radiosurgery as well. In this work, we applied a deep learning CNN to generate sCTs from T1-weighted MR images for GK SRS. Our method produced an overall average MAE of 86.5 HU and an MSE of 160.9 using 30 test cases, which is similar to the results from Han15 (84.8 HU and 188.6, respectively) based on 18 test subjects. Other examples of reported MAE results using CNN in the literature for the brain sCT are 131 HU 31 based on 10 test patients and 108.1 HU 32 using 15 patients.
To evaluate the dosimetric effect in GK SRS planning using the sCT, we carried out MC dose calculations based on different phantom volumes combined with a clinical and a virtual CDT. The same beam configurations and parameters were used in the dose calculations except that the phantom was different for the sake of fair comparison. We found that the result based on the phantom created with the sCT and synthetic CDT was closer to the ground truth dose distribution. The result based on the water-brain assumption showed that the D95 can be 4.9% and the V12Gy can be 2.2 cc different from the ground truth. It indicates that heterogeneity is important and should be taken into account in the GK SRS planning. The reason that the results based on the sCT with synthetic CDT were better than with CDT is that the synthetic CDT was provided as a supplemental correction step for the generation of the density volume. The synthetic CDT was built by mapping the CT number in the sCT to the true density. It agreed with the clinical CDT except for the range around HU of 150 and HU > 1200. We noticed that the statistic error bars were larger, and our investigation found that it was due to the number of voxels with high HU at the denture region was small comparing with the whole brain region, and therefore the statistical noise is larger. This inaccuracy will not affect our dose calculations as no beams will pass through those regions.
The factor that affects the accuracy of the model can be imperfect image registration between the MR and CT pair. Inaccurate alignment between ground truth MR and CT in the training dataset can cause inaccuracy of the model. The various artifacts such as dental implants, geometrical distortion in MR images, and anatomy changes from brain surgery between the time of acquisition can also directly affect the mapping. The other difficulty is from the indistinguishable intensity between air and bone since both are shown black in the T1 MR image (however, not uniformly black for bone). As also shown in other works,13,22,33 our model predicted accurate CT values except for interfaces between bones and tissue/air. We showed that the error could lead to a discrepancy of 1.1% for D95 if compared with the true dose distribution.
Recently, another kind of deep machine learning algorithm, 34 generative adversarial networks (GANs), has been used in sCT generations from MR images.34‐37 A GAN consists of 2 CNNs: a generative network generating candidates and a discriminative network evaluating them. Emami et al 37 achieved an MAE of 89.3 HU using 15 patient data and Kazemifar et al 38 achieved an MAE of 47.2 HU with training dataset of 77 patients for sCT generation from T1-weighted MR images, while Koike et al 32 reported an MAE of 108.1 HU for multisequence (T1- and T2-weighted, fluid-attenuated inversion recovery) MR images. The results demonstrate that the performance metrics not only depends on the model but also on underlying training data. The operation of a CNN is generally opaque. Adjustable components are usually the loss functions used in the model. The quality of training data, such as good alignment, uniformity of scan protocol usage, and minimization of imaging artifacts, is also critical to accurate predictions by the trained model. It appears that the GAN is a very promising approach for sCT generation. Our future work will focus on this approach and also on improving the quality of our training data for GK SRS planning.
Conclusions
We have developed a deep learning algorithm based on convolutional neural networks to convert MRI to sCT for Gamma Knife treatment planning. The performance metrics such as MAE and ME show acceptable accuracy when comparing the pixel values of the true CT to those of the sCT. To evaluate the dosimetric effect from generated sCT, we employ the MC algorithm in the dose calculations based on the true CT and the sCT. To ensure a fair comparison, all the input parameters including shot coordinates, times are the same in both calculations. A difference of 1.1% in dose-volume parameter D95 was found compared with the calculations using the true CT. Since the treatment planning for most of GK treatments is MRI-based, there is more of a need for generating sCT from MR in order to take advantage of a more accurate dose algorithms such as the convolution or MC method. In this work, we have demonstrated the feasibility of using sCT for MR-only GK treatment planning.
Acknowledgments
This work was partially supported by Elekta Limited.
Abbreviations
- CDT
CT-to-density table
- CNNs
convolutional neural networks
- DSC
Dice similarity coefficient
- MAE
mean absolute error
- MC
Monte Carlo
- ME
mean error
- MR
magnetic resonance
- MSE
mean squared error
- GAN
generative adversarial networks
- GK
Gamma Knife
- HU
Hounsfield unit
- LGP
Leksell GammaPlan
- ReLU
rectified linear unit
- SBRT
stereotactic body radiotherapy
- sCT
synthetic computed tomography
- SRS
stereotactic radiosurgery
- TMR
tissue maximum ratio
Footnotes
Declaration of Conflicting Interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Ethical Approval: Our study was approved by University Hospitals (UH) IRB administration offices under Chart Review Data Protocol 060519 (approval no STUDY20200313). The request for a full waiver of consent was also approved by UH IRB.
ORCID iD: Jiankui Yuan https://orcid.org/0000-0001-8352-9206
References
- 1.Regis J, Tamura M, Guillot Cet al. et al. Radiosurgery with the world’s first fully robotized Leksell gamma knife PerfeXion in clinical use: a 200-patient prospective, randomized, controlled comparison with the gamma knife 4C. Neurosurgery. 2009;64(2):346‐355; discussion 355-346. [DOI] [PubMed] [Google Scholar]
- 2.Best R, Gersh J, Wiant D, Bourland J. Gamma knife perfexion dosimetry: a Monte Carlo model of one sector [abstract]. Med Phys.2012;39(6):3812-3813. [DOI] [PubMed] [Google Scholar]
- 3.Pipek J, Novotny J, Kozubikova P. A modular Geant4 model of Leksell gamma knife perfexion. Phys Med Biol. 2014;59(24):7609‐7623. [DOI] [PubMed] [Google Scholar]
- 4.Moskvin V, DesRosiers C, Papiez L, Timmerman R, Randall M, DesRosiers P. Monte Carlo simulation of the Leksell gamma knife: I. Source modelling and calculations in homogeneous media. Phys Med Biol. 2002;47(12):1995‐2011. [DOI] [PubMed] [Google Scholar]
- 5.Moskvin V, Timmerman R, DesRosiers Cet al. et al. Monte Carlo simulation of the Leksell gamma knife: II. Effects of heterogeneous versus homogeneous media for stereotactic radiosurgery. Phys Med Biol. 2004;49(21):4879‐4895. [DOI] [PubMed] [Google Scholar]
- 6.Novotny J, Bhatnagar JP, Niranjan Aet al. Dosimetric comparison of the Leksell gamma knife perfexion and 4C. J Neurosurg 2008;109:8‐14. [DOI] [PubMed] [Google Scholar]
- 7.Yuan J, Lo SS, Zheng Yet al. et al. Development of a Monte Carlo model for treatment planning dose verification of the Leksell gamma knife perfexion radiosurgery system. J Appl Clin Med Phys. 2016;17(4):6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yuan J, Machtay M. A Monte Carlo model and its commissioning for the Leksell gamma knife perfexion radiosurgery system. Med Phys. 2017;44(9):L4910-4918. [DOI] [PubMed] [Google Scholar]
- 9.Xu AY, Bhatnagar J, Bednarz G, et al. Dose differences between the three dose calculation algorithms in Leksell GammaPlan. J Appl Clin Med Phys. 2014;15(5):4844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fallows P, Wright G, Harrold N, Bownes P. A comparison of the convolution and TMR10 treatment planning algorithms for gamma knife([R]) radiosurgery. J Radiosurg SBRT. 2018;5(2):157‐167. [PMC free article] [PubMed] [Google Scholar]
- 11.Elekta Instrument AB 2010 The convolution algorithm in Leksell Gammaplan 10 Technical Report no. 018881.01.
- 12.Hsu S-H, Cao Y, Huang K, Feng M, Balter JM. Investigation of a method for generating synthetic CT models from MRI scans of the head and neck for radiation therapy. Phys Med Biol. 2013;58(23):8419‐8435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dowling JA, Lambert J, Parker Jet al. An atlas-based electron density mapping method for magnetic resonance imaging (MRI)-alone treatment planning and adaptive MRI-based prostate radiation therapy. Int J Radiat Oncol Biol Phys. 2012;83:e5. [DOI] [PubMed] [Google Scholar]
- 14.Uh J, Merchant TE, Li Y, Li X, Hua C. MRI-based treatment planning with pseudo CT generated through atlas registration. Med Phys. 2014;41:051711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han X. MR-based synthetic CT generation using a deep convolutional neural network method. Med Phys. 2017;44:1408. [DOI] [PubMed] [Google Scholar]
- 16.Nie D, Cao X, Gao Y, Wang L, Shen D. Estimating CT image from MRI data using 3D fully convolutional networks. Deep Learn Data Label Med Appl. 2016;170-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiang L, Wang Q, Nie Det al. et al. Deep embedding convolutional neutral network for synthesizing CT image from T1-weighted MR image. Med Image Anal. 2018;47:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huynh T, Gao Y, Kang Jet al. et al. Estimating CT image from MRI data using structured random forest and auto-context model. IEEE Trans Med Imaging. 2016;35:174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andreasen D, Leemput KV, Hansen RH, Anderson JA, Edmund JM. Patch-based generation of a pseudo CT from conventional MRI sequences for MRI-only radiotherapy of the brain. Med Phys. 2015;42(4):1596‐1605. [DOI] [PubMed] [Google Scholar]
- 20.Kim J, Garbarino K, Schultz L, et al. Dosimetric evaluation of synthetic CT relative to bulk density assignment-based magnetic resonance-only approaches for prostate radiotherapy. Radiat Oncol. 2015;10:239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paradis E, Cao Y, Lawrence TSet al. et al. Assessing the dosimetric accuracy of magnetic resonance-generated synthetic CT images for focal brain VMAT radiation therapy. Int J Radiat Oncol Biol Phys. 2015;93(5):1154‐1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng W, Kim JP, Kadbi M, Movsas B, Chetty IJ, Glide-Hurst CK. Magnetic resonance-based automatic air segmentation for generation of synthetic computed tomography scans in the head region. Int J Radiat Oncol Biol Phys. 2015;93(3):497‐506. [DOI] [PubMed] [Google Scholar]
- 23.Fatemi A, Kanakamedala MR, Chunli Yang C, Morris B, Duggar WN, Vijayakumar S. Evaluation of the geometric and dosimetric accuracy of synthetic computed tomography images for magnetic resonance imaging-only stereotactic radiosurgery. Cureus J Med Sci. 2019;11(4):e4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ronneberger O, Fischer P, Brox T. U-net: convolutional networks for biomedical image segmentation. 2015, arXiv:1505.04597. [Google Scholar]
- 25.He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. 2015, arXiv: 1512.03385. [Google Scholar]
- 26.Allen D. The relationship between variable selection and data augmentation and a method for prediction. Technometrics. 1974;16(1):125‐127. [Google Scholar]
- 27.Stone M. Cross-validatory choice and assessment of statistical predictions. J R Stat Soc: Series B (Methodol). 1974;36(2):111‐147. [Google Scholar]
- 28.Stone M. An asymptotic equivalence of choice of model by cross-validation and Akaike’s criterion. J R Stat Soc: Series B (Methodol) 1977;39(1), 44-47 [Google Scholar]
- 29.Low DA, Harms WB, Mutic S, Purdy JA. A technique for the quantitative evaluation of dose distributions. Med Phys. 1998;25(5):656‐661. [DOI] [PubMed] [Google Scholar]
- 30.Yuan J, Chen W. A gamma dose distribution evaluation technique using the k-d tree for nearest neighbor searching. Med Phys. 2010;37(9):4868‐4873. [DOI] [PubMed] [Google Scholar]
- 31.Wang Y, Liu C, Zhang X, Deng W. Synthetic CT generation based on T2 weighted MRI of nasopharyngeal carcinoma (NPC) using a deep convolutional neural network (DCNN). Front Oncol. 2019;9:1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koike Y, Akino Y, Sumida Iet al. Feasibility of synthetic computed tomography generated with an adversarial network for multi-sequence magnetic resonance-based brain radiotherapy. J Radiat Res. 2020;61:92‐103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang H, Chandarana H, Block KT, Vahle T, Fenchel M, Das IJ. Dosimetric evaluation of synthetic CT for magnetic resonance-only based radiotherapy planning of lung cancer. Radiat Oncol. 2017;12:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goodfellow I, Pouget-Abadie J, Mirza Met al. et al. Generative adversarial networks. In: Proceedings of the international conference on neural information processing systems (NIPS 2014), pp.2672‐2680. [Google Scholar]
- 35.Wolterink JM, Dinkla AM, Savenije MHF, Seevinck PR, van den Berg CAT, Isgum I. Deep MR to CT synthesis using unpaired data. 2017, arXiv.org. [Google Scholar]
- 36.Nie D, Trullo R, Lian Jet al. et al. Medical image synthesis with context-aware generative adversarial networks. Med Image Comput Comput Assist Interv. 2017;10435:417‐425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Emami HH, Dong M, Nejad-Davarani SP, Glide-Hurst CK. Generating synthetic CTs from magnetic resonance images using generative adversarial networks. Med Phys. 2018;45(8):3627‐3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kazemifar S, McGuire S, Timmerman Ret al. et al. MRI-only brain radiotherapy: assessing the dosimetric accuracy of synthetic CT images generated using a deep learning approach. Radiother Oncol. 2019;13:56. [DOI] [PubMed] [Google Scholar]