Abstract
Antigen-bearing proteins become progressively unavailable to immunodetection after prolonged storage of routine sections, exposed to a variety of agents, such as moisture, oxygen, and temperature. By proteomic analysis, the antigens are retained in the sections and definitely in the tissue block, pointing to fixation-independent, storage time–dependent protein modifications. Based on previous experience, we hypothesized that a combined exposure to a reducing agent and to chemicals favoring protein conformation changes would reverse the masking in aged sections. Disaccharides, lactose and sucrose, and a surfactant, added to a standard antigen retrieval buffer, reverse the negative changes in aged sections. Furthermore, they provide enhanced access to antigens in freshly cut sections, but not universally, revealing additional factors, besides heat and calcium chelation, required for antigen retrieval of individual proteins:
Keywords: antigen retrieval, antigenic variation, fluorescent image analysis, formalin-fixed paraffin-embedded tissue section, immunohistochemistry, long-term storage of FFPE tissue section, quantitative methods
Introduction
Prolonged storage of routinely cut and processed sections affixed to glass slides reproducibly results in loss of stainability. There is ample literature published about this phenomenon; according to a recent and most comprehensive investigation about the causes of this unwanted effect,1 there is loss of antigens from the sections caused by exposure to moisture during the embedding and/or the storage, and to a multiplicity of concurring causes, reviewed in Haragan et al.2 Interestingly, this phenomenon affects some antigens (e.g., membrane antigens) but not others.3,4
To further confound the comprehension of the problem, it is well known that exposure to high heat in the presence of calcium chelators, the antigen retrieval (AR) technique,5 is able to restore immunostainability in freshly cut sections from fixed and processed tissue [formalin-fixed paraffin-embedded (FFPE)], but not on aged sections.1,3,4,6 It is thus difficult to understand why aging may progressively limit this ability in a time- and/or moisture-dependent fashion, once the fixative is long gone from the tissue.
Hence, the conclusion is that the antigen is lost from the section, possibly by degradation, hydrolysis of protein–protein cross-links, changes in cross-linked protein conformation, loss of discontinuous epitopes, or masking of linear ones.2
However, a dual investigation by immunostaining and proteomic assay combined,2 although focused on one single antigen, programmed death-ligand 1 (PD-L1), disputes that the antigen is lost: Despite a failure to stain aged sections, the protein can be biochemically demonstrated in the tissue sections.2 Resectioning the block and staining fresh sections produces the expected staining, proofing that the antigens are retained in the tissue block.7
Furthermore, a tissue section which has been dewaxed, antigen retrieved once, repeatedly exposed (~30× for a total of 15 hr) to beta-mercaptoethanol and sodium dodecyl sulfate (SDS) at 56C, and stored in a glycerol–water–sucrose medium at −20C, for a total of about 10 months since the beginning, shows a minimal variation of immunostainability (see Supplemental Fig. 1 in Manzoni et al.)8.
During our previous investigations about the mechanism of antigen fixation and antibody removal methods,9–11 we observed that antigen masking may occur independently of cross-linking, for example, upon removal of the protein-associated water.9 Furthermore, once a protein is restored to immunoavailability after AR, it can be brought back to an immunoexcluded state by drying9 or by the application of denaturants such as guanidine HCl.11 Disaccharides such as lactose or sucrose can help in avoiding these negative effects by acting as molecular water substitutes and/or protein conformation facilitators.9
Finally, formalin-induced bonds formed after 48 hr of fixation of frozen sections from fresh tissue can be reversed completely by as little as 20 min of AR, leading to tissue loss,12 which do not occur in routinely processed tissue (FFPE) fixed in formalin for shorter time.
It is thus possible that upon storage, molecular changes occur which further mask the epitope availability because of protein conformation changes caused by time, moisture, atmospheric agents, but not dependent on fixative-induced cross-links. These changes may affect the epitope itself, neighboring proteins, or both.
To investigate these factors and provide a solution, we tested a surfactant (SDS), a protein folding facilitator,11,13 and a reductive agent,9 these latter two combined in a single reducing disaccharide, lactose.
Materials and Methods
Human Specimens
Human surgical pathology specimen leftovers (pediatric tonsils, discarded serial sections from routinely processed FFPE) were used; fresh specimens were fixed overnight at RT in buffered 4% formaldehyde (Bio-Optica Milano Spa; Milano, Italy), processed through a graded ethanol gradient, then in xylene, and embedded in molten paraffin for sectioning.
The study has been approved by the Institutional Review Board Comitato Etico Brianza, N. 3204, “High-dimensional single cell classification of pathology (HDSSCP),” October 2019. Patients consent was obtained or waived according to article 89 of the EU general data protection regulation 2016/679 (GDPR) and decree N. 515, 12/19/2018 of the Italian Privacy Authority.
Tissue Microarrays
Tissue microarrays (TMAs) were prepared as previously published14 on a Tissue Microarrayer Galileo model TMA CK4600 (Integrated System Engineering srl; Milan, Italy). Cores of 0.6 and 1 mm were used. The following TMAs were prepared and multiple serial sections obtained at the time of preparation and again in April 2021:
11T1: colonic polyps from 29 patients, resected from 2005 to 2008. Two 1-mm cores each. Prepared in August 2011.
11T14: colon cancer from 46 patients, resected in 2011. Three 0.6-mm cores each. Prepared in December 2011.
12T30: colon cancer from 34 patients, resected in 2011. Three 1-mm cores each. Prepared in February 2012.
11T4: normal placentas from 8 patients, delivered in 2011. Three 2-mm cores each. Prepared in December 2011.
After sectioning, slides were dried at 40C overnight in a dry oven and stored in plastic boxes (Kartell; Noviglio, Italy) at room temperature in a lab room. No paraffin coating on slides was performed. No desiccant was added to the boxes. Air conditioning to the room was provided by a poorly functioning wall unit.
AR
AR was performed placing the dewaxed, rehydrated sections10 in a 800-ml glass container filled with the retrieval solutions (see below), irradiated in a household microwave oven at full speed for 8 min, followed by intermittent electromagnetic radiation to maintain constant boiling for 30 min, and cooling the sections to about 50C before use.
AR Solutions
EDTA pH 8 (1-mM EDTA in 10-mM Tris buffer pH 8, cat. no. T9285; Merck Life Science S.r.l., Milano, Italy) was the reference buffer, henceforth named “EDTA.” 1% SDS (cat. no. 74255; Merck) was added to the EDTA buffer (“SDS”). Near saturation lactose (10%; ACEF, Piacenza, Italy) or sucrose (10% w/v; refined granulated sugar) was dissolved in the EDTA buffer (“Lact”; “Sucr”). A combination of EDTA buffer with 1% SDS and 10% lactose was prepared (“FULL”). No other AR with a different pH was used, nor an enzymatic retrieval solution.
Immunohistochemistry and Immunofluorescence
Primary antibodies, optimally diluted, (Table 1) were applied overnight, washed in 50-mM Tris-HCl buffer (pH 7.5) containing 0.01% Tween-20 (Merck) and 100-mM sucrose (TBS-Ts),15 counterstained with a horseradish peroxidase–conjugated polymer (Vector Laboratories; Burlingame, CA), washed, developed in 3,3′-diaminobenzidine (Dako; Glostrup, Denmark), lightly counterstained, and mounted.
Table 1.
Primary and Secondary Antibodies.
| Primary Antibodies | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Name | Clone | Isotype | Cat. No. | Source | Concentration | Dilution | Lot | RRID_AB | AR Preferred pH |
| BCL-2 | Bcl-2-100 | Mo IgG1 | B3170-.2ML | Merck Sigma Aldrich | 5.1 mg/ml | 1 µg/ml | 022M4805 | 258541 | High |
| Blimp-1/PRDM1 | 6D3 | Rat IgG2a | sc-47732 | SCBT | 200 µg/ml | 1 µg/ml | NA | 628168 | High |
| CD4 | EPR6855 | Rb Mab | ab133616 | Abcam/Epitomics | 166 µg/ml | 1 µg/ml | GR3215096 | 2819211 | High |
| CD14 | HPA002127 | Rb Ab | HPA002127 | Merck Sigma Aldrich | 400 µg/ml | 1 µg/ml | A113974 | 1078440 | Low |
| CD34 | 43A1 | Mo IgG3 | sc-65261 | SCBT | 100 µg/m1 | 1 µg/ml | A1212 | 631133 | High |
| Cytokeratin 19 | A53-B/A2 | Mo IgG2a | sc-6278 | SCBT | 200 µg/ml | 1 µg/ml | H0614 | 627851 | High |
| HLA-DR | SPM288 | Mo IgG2b | sc-56545 | SCBT | 200 µg/ml | 1 µg/ml | B0909 | 1125217 | High |
| IRF4 | IRF-4 (M-17) | Goat | sc-6059 | SCBT | 100 µg/ml | 1 µg/ml | J2015 | 2127145 | High |
| Ki-67 | 4A1 | Mo IgG2b | SAB5300423 | Merck Sigma Aldrich | NA | 1:200 | 12110662 | 10980978 | High |
| MLH1 | ES05 | Mo IgG1 | M364029 | Dako | 91.6 µg/ml | 1 µg/ml | 10053229 | 2631352 | High |
| SOX9 | CL0639 | Mo IgG2a | AMAB90795 | Merck Sigma Aldrich | 1000 µg/ml | 1 µg/ml | 2712 | 2665670 | NA |
| Vimentin | V9 | Mo IgG1 | M0725 | Dako | 156 µg/ml | 1 µg/ml | 83947 | 83947 | High |
| Secondary Antibodies | |||||||||
| Name | Notes | Cat. No. | Source | Concentration | Dilution | Lot | RRID_AB | ||
| Brilliant Violet 480 AffiniPure Goat Anti-Mouse IgG, Fcγ Subclass 2a Specific* | Fc-specific | 115-685-206 | Jackson Immunoresearch | 0.2 mg/ml | 1:100 | 130919 | 2651097 | ||
| Alexa Fluor 488 Goat Anti-Mouse, Fcγ Subclass IgG1 Specific* | Fc-specific | 115-545-205 | Jackson Immunoresearch | 1.6 mg/ml | 1:200 | 145537 | 2338854 | ||
| Rhodamine Red-X (RRX) AffiniPure Goat Anti-Mouse IgG, Fcγ Subclass 2b Specific* | Fc-specific | 115-295-207 | Jackson Immunoresearch | 1.8 mg/ml | 1:200 | 118791 | 2338771 | ||
| Alexa Fluor 647 AffiniPure Donkey Anti-Rabbit IgG** | H+L | 711-605-152 | Jackson Immunoresearch | 1.5 mg/ml | 1:200 | 138314 | 2492288 | ||
| PerCp AffiniPure Goat Anti-Mouse IgG, Fcγ Subclass 3 Specific* | Fc-specific | 115-125-209 | Jackson Immunoresearch | 0.5 mg/ml | 1:200 | 88606 | 2338635 | ||
| Brilliant Violet 480 AffiniPure Donkey Anti-Rabbit IgG (H+L)** | H+L | 711-685-152 | Jackson Immunoresearch | 0.2 mg/ml | 1:100 | 136043 | 2651109 | ||
| Alexa Fluor 488-AffiniPure Donkey Anti-Goat IgG (H+L)** | H+L | 705-545-147 | Jackson Immunoresearch | 1.5 mg/ml | 1:200 | 139173 | 2336933 | ||
| Rhodamine Red-X (RRX) AffiniPure Donkey Anti-Mouse IgG(H+L)* | H+L | 715-295-151 | Jackson Immunoresearch | 1.5 mg/ml | 1:200 | 117959 | 2340832 | ||
| Alexa Fluor 647 AffiniPure Donkey Anti-Rat IgG (H+L)** | H+L | 712-605-153 | Jackson Immunoresearch | 1.5 mg/ml | 1:200 | 133374 | 2340694 | ||
Secondary antibody absorption
(min X Hu, Bov, Rb Sr Prot).
(min X Bov, Ck, Gt, GP, Sy Hms, Hrs, Hu, Ms, Rat, Shp Sr Prot).
Multiple immunofluorescent labeling was previously described in detail.11
Briefly, the sections were incubated overnight with optimally diluted primary antibodies in combination of up to five, washed, and counterstained with specific distinct fluorochrome-tagged secondary antibodies (Table 1).11 The slides, counterstained with 4′,6-diamidino-2-phenylindole (DAPI) and mounted, were scanned on an S60 Hamamatsu scanner (Nikon; Italia) at 20× magnification.
The filter setup for seven color acquisition [DAPI, BV480, FITC, tetramethyl rhodamine isothiocyanate (TRITC), Cy5, PerCp, autofluorescence (AF)] was as shown in Supplemental Fig. 1; all filters were from Semrock (Optoprim srl; Monza, Italy). The seven color multiplex, not previously published, was implemented to minimize section requirement and maximize the number of parameters to be assessed on the very same section. The scanning time, once set for each marker at the beginning of the experiments, was maintained unchanged throughout the study.
Single-patient (two cores) assessment of antigenicity changes over treatments was tested with a five antibody panel (Table 1) comprising CD14, Ki-67, cytokeratin 19 (KRT19), vimentin, and CD34.
Preparation of Immunofluorescent Images for Image Analysis
Single .ndpi images for each case were saved as .tiff files and AF was subtracted.11
Data Analysis
Fluorescence was quantified as published in Scalia et al.10 by selecting each case with the Fiji “rectangle” tool from the TMA image and exporting the histogram pixel values in a dedicated Excel file (Microsoft; Redmond, WA) (Supplemental Table 1: FluoQuant-fromHistogr.xlsx),16 available as a template at Bicocca Open Archive Research Data (BOARD). DOI: http://dx.doi.org/10.17632/z65mr2yfc7.1. Care was taken to select contiguous serial sections where duplicate cores per specimen, represented in serial sections, were chosen for analysis. Seven to 14 individual specimens (patients) were used for the analysis across the experiments. The fluorescence channel collecting 90% of the positive pixels was used for the analysis, after gating out pixels in the negative range (channels 0 to ~15/255).
Data were analyzed and graphically represented with StatPlus:mac, AnalystSoft Inc.—statistical analysis program for Mac OS. vv6. (http://www.analystsoft.com/en/).
Fiji, Adobe Photoshop and Adobe Illustrator were used to prepare the iconography.
Results
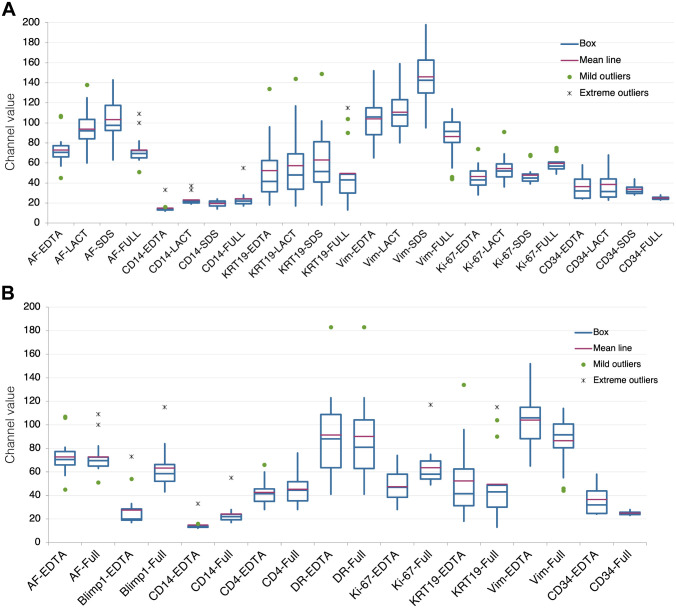
To test the contribution of a surfactant agent (SDS) to the antigen unmasking on aged sections, a comparison between plain EDTA buffer and an EDTA + SDS buffer was made with an antibody panel (Table 1). Improved staining was obtained for CD14 and vimentin, but not for other markers (Fig. 1A). AF was enhanced (Fig. 1A). The unmasking power of SDS alone has been discounted previously,17 thus was not further investigated.
Figure 1.
Effect of antigen retrieval modifications on immunodetection. A: Variation in fluorescence intensity (expressed as channel value for 90% of the positive pixels) for each marker according to the four AR treatments: EDTA, EDTA + Lactose (LACT), EDTA + SDS (SDS), and EDTA + Lactose + SDS (FULL). Fourteen samples tested for each marker, except Ki-67 (13 samples) and CD34 (4 samples). B: Comparison of variation in fluorescence intensity (expressed as channel value for 90% of the positive pixels) between EDTA and FULL AR for each marker. Fourteen samples tested for each marker except CD34 (4 samples). Abbreviations: AR, antigen retrieval; SDS, sodium dodecyl sulfate; AF, autofluorescence.
Oxidation may be a factor for the antigen masking over time2; thus, we added to the AR buffer lactose, a reducing disaccharide, which has the advantage also to be a protein folding facilitator.11,13 The comparison of the EDTA AR buffer with an EDTA + Lactose AR buffer showed an improved staining with the latter for CD14, vimentin, and Ki-67 (Fig. 1A); 10% lactose alone as an AR buffer did not retrieve the antigens tested (not shown).
When we tested the addition of both SDS and lactose to the AR EDTA buffer (FULL), we obtained a substantial improvement for CD14, Ki-67 but not for KRT19, vimentin, and CD34; these latter two had a reduced immunoreactivity, compared with plain EDTA or the addition of lactose or SDS. AF was reduced (Fig. 1A).
A larger number of markers was tested (Table 1), comparing directly the EDTA and the full AR buffers (Fig. 1B and Supplemental Fig. 2); PRDM1/Blimp1, CD14, and Ki-67 detection was enhanced but not for the rest of the panel. We failed to detect Sox9; MLH1 and bcl2 were detected at low levels by IHC only and could not produce quantifiable results by immunofluorescence (not shown).
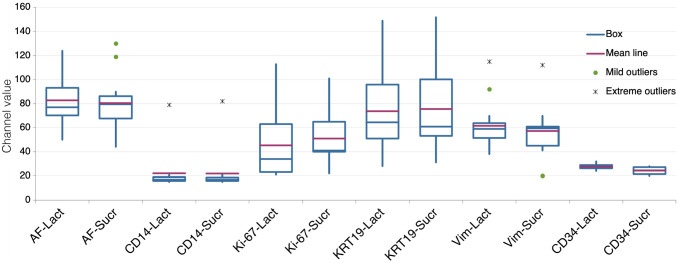
To test for the capacity of a reductive agent to enhance the immunodetection in vintage sections, we compared EDTA + Lactose with EDTA to which sucrose, a non-reducing disaccharide, was added. The results show that both the reducing and the non-reducing disaccharides are equally able to rejuvenate vintage sections; thus, the role of a reducing agent is mild or irrelevant (Fig. 2).
Figure 2.
Comparison of the effect of lactose or sucrose, added to the EDTA in the AR buffer. Variation in fluorescence intensity (expressed as channel value for 90% of the positive pixels) for each marker according to the two AR treatments: EDTA + Lactose (LACT) and EDTA + Sucrose (SUCR). Fourteen samples tested for each marker, except Ki-67 (13 samples) and CD34 (4 samples). Abbreviations: AR, antigen retrieval; AF, autofluorescence.
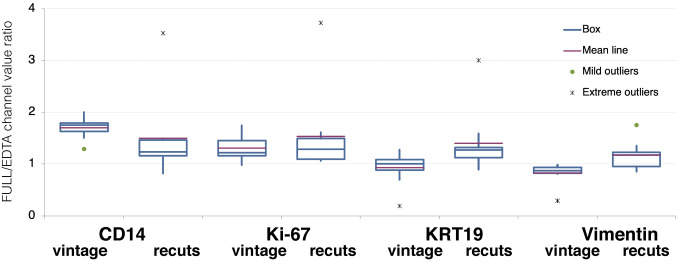
To test whether providing protein conformation facilitators during AR would benefit vintage sections only or freshly cut sections as well, a comparison was made between vintage sections and recuts: A significant differential increase was obtained for CD14 in vintage sections and a small decrease in aged sections stained for vimentin (Fig. 3 and Supplemental Fig. 3). For the remaining markers, the variation across the two AR buffers was not significant.
Figure 3.
Relative variation of fluorescence intensity by comparing EDTA vs FULL AR on vintage sections and recuts. The channel value for each marker shown after AR FULL, divided by the same value for EDTA, was calculated and plotted for vintage sections and for recuts. Analysis of variance shows p=0.006 significance overall, with the contribution of CD14 (p=0.04) and vimentin (p=0.03) on vintage sections. Between 8 and 10 samples per experimental point. Abbreviation: AR, antigen retrieval.
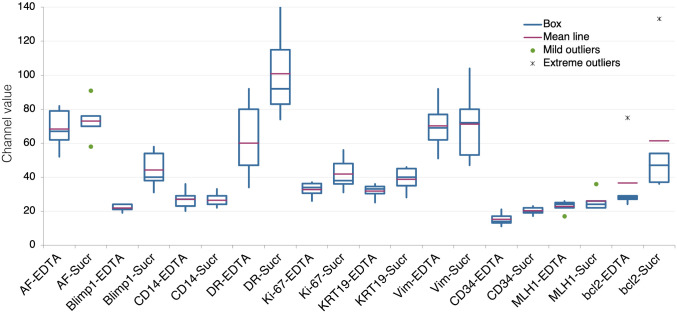
The amount of tissue remaining in the block may condition the depth at which recuts can be made, which may influence the recovery of immunogenicity.7 To assess whether adding a disaccharide during AR would influence fresh sections from recent tissue blocks, we tested a panel of antibodies on five large and small intestine surgical specimens, processed from 2013 to 2017, from which fresh sections were obtained by deep sectioning. Individual antigens were enhanced, some considerably (Blimp-1/PRDM1, bcl-2), others remained unchanged (Fig. 4). At variance with recuts from vintage TMA blocks, some antigens (e.g., CD14) did not benefit from the AR + Sucrose buffer, suggesting that recuts from vintage blocks, because of shallow resectioning, contain time-dependent conformational changes, amenable to rejuvenation. This was supported by the low levels of antigenicity for bcl-2, MLH1, and Sox9 in vintage blocks recuts, but immunodetection on recent material (Fig. 4). The combination of EDTA, sucrose, and SDS (FULL) reduced instead of improving the detection of Ki-67 and CD34 (Supplemental Fig. 4) and was not further pursued on fresh sections.
Figure 4.
Comparison of variation in fluorescence intensity (expressed as channel value for 90% of the positive pixels) between EDTA and EDTA + Sucrose (SUCR) AR for each marker on fresh, recent sections. Five samples per marker. Abbreviation: AR, antigen retrieval.
Discussion
An unknown fraction of antigens become progressively unavailable for detection during tissue section aging, for unknown reasons. We hypothesize that factors not linked to fixation or degradation were the cause and indeed countering protein misfolding via heat-mediated delivery of protein folding facilitators (disaccharides, SDS) restores, albeit partially, immunodetection, at least for a limited number of antigens and tissue type tested.
We also show that this rejuvenating effect is more pronounced in aged sections, mild in superficial recuts, and nil on fresh sections, at least for one of the markers used, CD14. Limitations in the availability of 10-year-old sections for testing did not allow a more expanded panel, despite an improved throughput in multiplex immunofluorescence.
Remarkably, some proteins (HLA-DR, CD4, KRT19) in aged sections are not or minimally affected by the procedure, pointing to a protein-specific or at least an epitope-specific effect of aging. Persistence of immunoavailability of some epitopes over the years has been noted previously.4,18 The causes for this distinct behavior are unknown.
In summary, we show that most if not all antigens remain in the tissue, as predicted by the proteomic data,2 but in various states of inaccessibility; to this regard, indeed, our method can demonstrate PD-L1 in vintage sections (Supplemental Fig. 5).
Recent work7 shows that the process which leads to antigenic diminution can affect the tissue block as well, starting from the most superficial layers and decreasing along deeper cuts in the block. Our section recuts, which have been very superficial because of the amount of residual material, may represent an intermediate phase of the process.
The fact that antigens can be detected again by adding conformational modifiers to a standard AR buffer seems to counter the argument that some antigens undergo moisture-dependent hydrolysis in stored sections,1 and thus loss from the tissue. The two hypotheses however are not mutually exclusive. Protein conformation changes may facilitate a non-enzymatic hydrolysis,19 either in situ or upon extraction from the section. This hypothesis can be tested by adding sucrose to the AR buffer before protein extraction and quality evaluation.
We obtained these results by (1) devising an improved multiplex immunofluorescence approach (seven colors) suitable to common IF filters, microscopes, and scanners, (2) using the TMA technology,14 which allowed the analysis of more than a dozen separate tissue samples consistently across the experimental panel over each experimental point, and (3) relying on an expert technical help which allows the analysis of virtually identical samples over serial TMA tissue sections (Supplemental Fig. 6). On top of that, a robust IF method11 and a simple quantitative tool (see Supplemental Table 1 FluoQuant-fromHistogr.xlsx also available as a template at BOARD. DOI: http://dx.doi.org/10.17632/z65mr2yfc7.1)16 allows very precise estimation of antigenic variations over samples, procedures, and antigens.
Previously published methods to rejuvenate vintage sections involve extending AR time,7 changing AR buffer,6 and deep sectioning,7 which is not applicable if the tissue block and/or residual tissue is not available; all these methods are nonspecific in nature and do not address the mechanism of antigen disappearance in vintage sections. AR solutions which may alter the protein conformation by changing ionic strength and pH have been tested before,20 but never to rescue antigens in aged sections.
We could not fully reconstitute immunostainability: This may has to do with irreversible protein changes upon storage, similar to what we have already observed upon storing disaccharide-protected dewaxed sections.9
SDS has been used for protein extraction from routinely processed sections, together with high temperature and an alkaline buffer.21,22 In the context of rejuvenation as a component of an AR buffer, on one side, it seems to synergize with the disaccharides in re-exposing a subgroup of antigens on vintage sections (Fig. 1), on the other side is blunting the re-exposure of the same antigens on fresh sections (Fig. 4 and Supplemental Fig. 4). We have previously documented analogous properties of SDS, where, together with beta-mercaptoethanol and for a brief exposure to heat, did enhanced the antigen detection on FFPE sections.23 Longer exposures to the mixture caused tissue disaggregation. SDS in a buffer containing EDTA and disaccharides may work a fine balance between antigen enhancement and extraction of the protein from the tissue, being the storage time of the sections and the associated protein changes the balance between the two. In other words, time-dependent regressing changes which affect antigen detection upon storage may make the tissue more resistant to extraction.
Countering the age-dependent tissue oxidation seems not helpful. Lactose, the only reducing disaccharide tested, is very inconvenient to use because of its poor solubility in water. To further pursue the addition of a reducing agent, a better alternative may be combining a disaccharide such as sucrose with a reducing monosaccharide such as glucose, both very soluble and which can be prepared as a concentrated solution to be added to the AR buffer. Stronger agents such as beta-mercaptoethanol are affecting the tissue integrity when used at a temperature above 56C.23
A chelating agent, in our experiments EDTA, seems essential to produce the retrieval and to allow rejuvenation of tissue antigens. In the absence of it, SDS or a disaccharide do not produce retrieval, pointing to two separate effects working in concert to produce the desired epitope conformation. Which may result in a favorable, neutral, or unfavorable conformation, as shown in this work and in additional ongoing experiments, in which, for example, CD20 detection by the L26 antibody on fresh sections is halved by the addition of sucrose to the AR solution (not shown).
A dual AR buffer condition has been established over the years, following the observation that each antibody for routine material best performs at low (pH 6) or high pH (pH 8–10).24 Worth noting is that some of the previous manuscripts concerning vintage section staining have used an AR buffer with the inappropriate pH for the antibody/ies in use.18 The panel of antibodies we used required a pH 8–10 AR buffer (Table 1); thus, we did not investigate the effect of pH solely for vintage sections rejuvenation. The rationale for a selective pH requirement is also obscure. What our results highlight, however, is that AR buffer requirements are much more complex and variegated than previously thought and most likely involve protein conformation in an unique protein- or epitope-specific fashion. Aging-associated changes do modify the epitope accessibility and in some cases are reversed by enhanced conformational changes.
Supplemental Material
Supplemental material, sj-pdf-1-jhc-10.1369_00221554211047287 for Rejuvenated Vintage Tissue Sections Highlight Individual Antigen Fate During Processing and Long-term Storage by Francesco Mascadri, Maddalena M. Bolognesi, Daniela Pilla and Giorgio Cattoretti in Journal of Histochemistry & Cytochemistry
Acknowledgments
The authors thank Lorenza “Loredana” Tusa, Antonella Musaro, and Lorella Riva for immunohistochemistry help.
Footnotes
Competing Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: GC and FM equally designed the experiments. MMB devised the image analysis algorithms and performed visual and digital image analysis. FM and DP performed immunostaining experiments. GC and MMB wrote the article. All authors have read and approved the final article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work has been supported by the Departmental University of Milano-Bicocca funds, by Regione Lombardia POR FESR 2014–2020, Call HUB Ricerca ed Innovazione: ImmunHUB to Giorgio Cattoretti. M.M.B. is a PhD student in the DIMET PhD Program call XXXV of the Department of Medicine and Surgery of the University of Milano-Bicocca since November 2019.
Contributor Information
Francesco Mascadri, Pathology, Department of Medicine and Surgery, Università di Milano-Bicocca, Monza, Italy.
Maddalena M. Bolognesi, Pathology, Department of Medicine and Surgery, Università di Milano-Bicocca, Monza, Italy.
Daniela Pilla, Department of Pathology, ASST Monza, Ospedale San Gerardo, Monza, Italy.
Giorgio Cattoretti, Pathology, Department of Medicine and Surgery, Università di Milano-Bicocca, Monza, Italy; Department of Pathology, ASST Monza, Ospedale San Gerardo, Monza, Italy.
Literature Cited
- 1. Xie R, Chung J-Y, Ylaya K, Williams RL, Guerrero N, Nakatsuka N, Badie C, Hewitt SM. Factors influencing the degradation of archival formalin-fixed paraffin-embedded tissue sections. J Histochem Cytochem. 2011;59(4):356–65. doi: 10.1369/0022155411398488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Haragan A, Liebler DC, Das DM, Soper MD, Morrison RD, Slebos RJC, Ackermann BL, Fill JA, Schade AE, Gosney JR, Gruver AM. Accelerated instability testing reveals quantitative mass spectrometry overcomes specimen storage limitations associated with PD-L1 immunohistochemistry. Lab Invest. 2020;100:874–86. doi: 10.1038/s41374-019-0366-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Grillo F, Pigozzi S, Ceriolo P, Calamaro P, Fiocca R, Mastracci L. Factors affecting immunoreactivity in long-term storage of formalin-fixed paraffin-embedded tissue sections. Histochem Cell Biol. 2015;144(1):93–9. doi: 10.1007/s00418-015-1316-4. [DOI] [PubMed] [Google Scholar]
- 4. van den Broek LJ, van de Vijver MJ. Assessment of problems in diagnostic and research immunohistochemistry associated with epitope instability in stored paraffin sections. Appl Immunohistochem Mol Morphol. 2000;8(4):316–21. [PubMed] [Google Scholar]
- 5. Shi S-R, Shi Y, Taylor CR. Antigen retrieval immunohistochemistry: review and future prospects in research and diagnosis over two decades. J Histochem Cytochem. 2011;59(1):13–32. doi: 10.1369/jhc.2010.957191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wester K, Wahlund E, Sundstrom C, Ranefall P, Bengtsson E, Russell PJ, Ow KT, Malmstrom PU, Busch C. Paraffin section storage and immunohistochemistry. Effects of time, temperature, fixation, and retrieval protocol with emphasis on p53 protein and MIB1 antigen. Appl Immunohistochem Mol Morphol. 2000;8(1):61–70. [PubMed] [Google Scholar]
- 7. Grillo F, Campora M, Pigozzi S, Bonadio S, Valle L, Ferro J, Paudice M, Dose B, Mastracci L. Methods for restoration of ki67 antigenicity in aged paraffin tissue blocks. Histochem Cell Biol. 2021;156:183–90. doi: 10.1007/s00418-021-01987-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Manzoni M, Bolognesi MM, Antoranz A, Mancari R, Carinelli S, Faretta M, Bosisio FM, Cattoretti G. The adaptive and innate immune cell landscape of uterine leiomyosarcomas. Sci Rep. 2020;10(1):702–10. doi: 10.1038/s41598-020-57627-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Boi G, Scalia CR, Gendusa R, Ronchi S, Cattoretti G. Disaccharides protect antigens from drying-induced damage in routinely processed tissue sections. J Histochem Cytochem. 2016;64(1):18–31. doi: 10.1369/0022155415616162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Scalia CR, Boi G, Bolognesi MM, Riva L, Manzoni M, DeSmedt L, Bosisio FM, Ronchi S, Leone BE, Cattoretti G. Antigen masking during fixation and embedding, dissected. J Histochem Cytochem. 2017;65(1):5–20. doi: 10.1369/0022155416673995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bolognesi MM, Manzoni M, Scalia CR, Zannella S, Bosisio FM, Faretta M, Cattoretti G. Multiplex staining by sequential immunostaining and antibody removal on routine tissue sections. J Histochem Cytochem. 2017;65(8):431–44. doi: 10.1369/0022155417719419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bolognesi MM, Mascadri F, Furia L, Faretta M, Bosisio FM, Cattoretti G. Antibodies validated for routinely processed tissues stain frozen sections unpredictably. Biotechniques. 2021;70(3):137–48. doi: 10.2144/btn-2020-0149. [DOI] [PubMed] [Google Scholar]
- 13. Tsumoto K, Ejima D, Kumagai I, ARAKAWA T. Practical considerations in refolding proteins from inclusion bodies. Protein Expr Purif. 2003;28(1):1–8. doi: 10.1016/S1046-5928(02)00641-1. [DOI] [PubMed] [Google Scholar]
- 14. Pilla D, Bosisio FM, Marotta R, Faggi S, Forlani P, Falavigna M, Biunno I, Martella E, De Blasio P, Borghesi S, Cattoretti G. Tissue microarray design and construction for scientific, industrial and diagnostic use. J Pathol Inform. 2012;3:42. doi: 10.4103/2153-3539.104904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cattoretti G, Bosisio F, Marcelis L, Bolognesi MM. Multiple iterative labeling by antibody neodeposition (MILAN). Nat Res 2019. [cited 2019 September 25]. Available from: https://protocolexchange.researchsquare.com/article/nprot-7017/v5
- 16. Cattoretti G, Bolognesi MM, Pilla D, Mascadri F. Rejuvenated vintage tissue sections highlight individual antigen fate during processing and long term storage. V1 Database Bicocca Open Archive Research Data (BOARD) Mendeley Data, UNIMIB; 2021. Available from: https://board.unimib.it/datasets/z65mr2yfc7/1 [DOI] [PMC free article] [PubMed]
- 17. Morgan JM, Navabi H, Jasani B. Role of calcium chelation in high-temperature antigen retrieval at different pH values. J Pathol. 1997;182(2):233–7. doi: [DOI] [PubMed] [Google Scholar]
- 18. Bertheau P, Cazals-Hatem D, Meignin V, de Roquancourt A, Verola O, Lesourd A, Sene C, Brocheriou C, Janin A. Variability of immunohistochemical reactivity on stored paraffin slides. J Clin Pathol. 1998;51(5):370–4. doi: 10.1136/jcp.51.5.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lauer TM, Wood GP, Farkas D, Sathish HA, Samra HS, Trout BL. Molecular investigation of the mechanism of non-enzymatic hydrolysis of proteins and the predictive algorithm for susceptibility. Biochemistry. 2016;55(23):3315–28. [DOI] [PubMed] [Google Scholar]
- 20. Emoto K, Yamashita S, Okada Y. Mechanisms of heat-induced antigen retrieval: does pH or ionic strength of the solution play a role for refolding antigens? J Histochem Cytochem. 2005;53(11):1311–21. doi: 10.1369/jhc.5C6627.2005. [DOI] [PubMed] [Google Scholar]
- 21. Chung JY, Lee SJ, Kris Y, Braunschweig T, Traicoff JL, Hewitt SM. A well-based reverse-phase protein array applicable to extracts from formalin-fixed paraffin-embedded tissue. Proteomics Clin Appl. 2008;2(10-11):1539–47. doi: 10.1002/prca.200800005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shi S-R, Datar R, Liu C, Wu L, Zhang Z, Cote RJ, Taylor CR. DNA extraction from archival formalin-fixed, paraffin-embedded tissues: heat-induced retrieval in alkaline solution. Histochem Cell Biol. 2004;122(3):211–8. doi: 10.1007/s00418-004-0693-x. [DOI] [PubMed] [Google Scholar]
- 23. Scalia CR, Gendusa R, Cattoretti G. A 2-step laemmli and antigen retrieval method improves immunodetection. Appl Immunohistochem Mol Morphol. 2016;24(6):436–46. doi: 10.1097/PAI.0000000000000203. [DOI] [PubMed] [Google Scholar]
- 24. Shi SR, Imam SA, Young L, Cote RJ, Taylor CR. Antigen retrieval immunohistochemistry under the influence of pH using monoclonal antibodies. J Histochem Cytochem. 1995;43(2):193–201. doi: 10.1177/43.2.7822775. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-jhc-10.1369_00221554211047287 for Rejuvenated Vintage Tissue Sections Highlight Individual Antigen Fate During Processing and Long-term Storage by Francesco Mascadri, Maddalena M. Bolognesi, Daniela Pilla and Giorgio Cattoretti in Journal of Histochemistry & Cytochemistry