Abstract
We are investigating the neuropathogenesis of Lyme disease caused by Borrelia burgdorferi in a nonhuman primate model. In the past, two separate pieces of tissue had to be used when both analyzing for the presence of the spirochete and examining the host response to infection. We have modified a procedure to purify DNA from the same sample after the extraction of RNA. The remaining material containing the DNA was precipitated, and residual organic reagent was removed prior to deproteinization and extraction of the DNA. This procedure now allows us to both assay for the presence of the Lyme microorganism and analyze the host response in the same tissue preparation.
In investigating the nonhuman primate (NHP) model of Lyme neuroborreliosis (5–7), the detection of the presence of the spirochete and assessment of the expression of host cellular genes in the same tissue sample were desirable. In the past, these two analyses were performed on separate tissue samples (3, 9) because of the difficulties we had in obtaining DNA and RNA preparations of adequate purity from the same tissue sample (2). This report describes a modified method for preparing DNA from the same tissue sample after the extraction of RNA to detect the presence of Borrelia burgdorferi and assess the expression of host cellular genes.
Tissue samples were from NHPs infected with B. burgdorferi, as previously described (5–7). Frozen tissue samples (15 to 30 mg) were placed into green-top FastRNA tubes (BIO-101, Vista, Calif.). While the samples were held at −80°C, 400 μl of TRIZOL (Life Technologies, Gaithersburg, Md.) was added to each, and tissues were homogenized with the Fast Prep FP120 tissue homogenizer (Savant Instruments, Inc., Holbrook, N.Y.) at a setting of 6 for 20 s; then the samples were placed on ice for 5 min. The homogenate was removed and transferred to a 1.5-ml screw-cap microcentrifuge tube. Approximately 85% of the input volume could be recovered from the FastRNA tubes. To each sample was added 80 μl of chloroform, and then the samples were shaken by hand for 15 s, left at room temperature for 2 to 3 min, and then centrifuged at 12,000 rpm and 4°C for 15 min. All centrifugation steps throughout the study were carried out in an Eppendorf microcentrifuge, model 5415C (Brinkmann Instruments, Inc., Westbury, N.Y.) with the standard rotor for 1.5-ml microcentrifuge tubes. As much of the upper aqueous layer was removed as possible and transferred to a 1.5-ml microcentrifuge tube, and RNA was prepared as per the manufacturer’s (Life Technologies) recommendations as described further below.
To the lower layer and interphase material, which contained the DNA of interest, 150 μl of 100% ethanol was added, and the samples were mixed by inversion and left at room temperature for 2 to 3 min before being centrifuged at 7,500 rpm and 4°C for 5 min. The supernatants were discarded, and the pellets were resuspended in 200 μl of 10 mM Tris-HCl (pH 7.4)–1 mM EDTA (TE). To each sample was added an equal volume (200 μl) of chloroform, and the samples were then vortexed prior to centrifugation at 7,500 rpm and 4°C for 5 min. The top (aqueous) portions were removed with as much of the interphase material as possible and transferred to 1.5-ml screw-cap microcentrifuge tubes. An equal volume (approximately 200 μl) of 2× digestion buffer (200 mM NaCl, 20 mM Tris-HCl [pH 8.0], 50 mM EDTA, 1% sodium dodecyl sulfate, and 0.2 mg of proteinase K/ml [added just before use]) (9) was added to each sample, and the samples were mixed and incubated at 50°C overnight. The samples were extracted twice with an equal volume of phenol-chloroform-isoamyl alcohol (50:48:2) (Life Technologies) and twice with an equal volume of chloroform. One hundred fifty microliters of 5 M ammonium acetate (approximately 0.7 volume) was added to each sample, along with 750 μl of 100% ethanol, and the samples placed on dry ice for 30 min before being centrifuged at 12,500 rpm and 4°C for 15 min. The supernatants were discarded, and the tubes were rinsed with cold 75% ethanol and tubes left to drain at room temperature for approximately 30 min. The precipitated DNA, which was sometimes not visible, was suspended in 25 to 30 μl of TE, and the tubes were placed at 50°C for 10 to 20 min to aid in dissolution of the DNA.
RNA was prepared from the upper, aqueous layer after addition of chloroform to the homogenate as per the manufacturer’s (Life Technologies) instructions. Briefly, 0.25 ml of isopropanol was added to each upper, aqueous phase, and the samples were vortexed and left at room temperature for at least 10 min before being centrifuged at 12,000 rpm and 4°C for 10 min. The supernatants were discarded, and the pellets were resuspended in 75% cold ethanol; then the samples were centrifuged at 7,500 rpm for 5 min. After the supernatants were decanted, the tubes were left to drain for not more than 10 min at room temperature, and RNAs were suspended in 12 to 20 μl of sterile, RNase-free water.
Expression of the genes encoding glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and the ubiquitously expressed cytokine transforming growth factor β1 (TGF-β1) (4) was assessed by reverse transcriptase (RT) PCR. cDNA was first prepared by using a Perkin-Elmer Gene Amp RNA PCR kit (Roche Molecular Systems, Inc., Branchburg, N.J.) with 1 μg of total RNA and random primers in a volume of 20 μl as per the manufacturer’s specifications. Before the addition of the RT, 10 U of RNase-free DNase I (Boehringer Mannheim Corp., Indianapolis, Ind.) was added to the RT mixture, and the reaction mixture was incubated for 30 min at 37°C and then 5 min at 95°C. For cDNA amplification, 3 μl of RT mixture was used for each 50 μl of PCR mixture. Each PCR mixture contained the standard components, as per the manufacturer’s specifications, along with 1.5 mM MgCl2 and 0.5 mM each GAPDH or 1.0 mM TGF-β1 primer. The human GAPDH and TGF-β1 primers were obtained from Genosys Biotechnologies, Inc. (The Woodlands, Tex.) and Stratagene (La Jolla, Calif.), respectively. PCR resulted in products of 398 and 187 bp for GAPDH and TGF-β1, respectively. After an initial 3-min incubation period at 95°C, the PCR mixtures were incubated in a PE9700 thermal cycler (Perkin-Elmer, Norwalk, Conn.) for 45 cycles consisting of 95°C for 1 min, 55°C for 1 min, and 72°C for 1 min, followed by a 7-min incubation at 72°C and cooling to 4°C.
To detect the presence of B. burgdorferi in the tissue, OspB PCR primers were used as previously described (7). A 50-μl PCR mixture, which contained the standard components as per the manufacturer’s (Perkin-Elmer) instructions, was used, along with 4 mM MgCl2, 0.5 mM each OspB primer, 300 ng of DNA, and 2.5 U of AmpliTaq Gold polymerase. To test for inhibition of the PCR by a DNA preparation, 300 ng of the suspected DNA was added, along with the same amount of a PCR-positive DNA sample. Before amplification of OspB was initiated, the PCR mixture was heated at 95°C for 9 min; then it was subjected to 45 cycles in a PE9700 thermal cycler as follows: 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s. After a final 7-min elongation step at 72°C, and the sample was cooled to 4°C. PCR products (15 μl) were analyzed by gel electrophoresis, using a 3% Nusieve (3:1) agarose gel (FMC, Rockland, Maine) with 0.5% ethidium bromide.
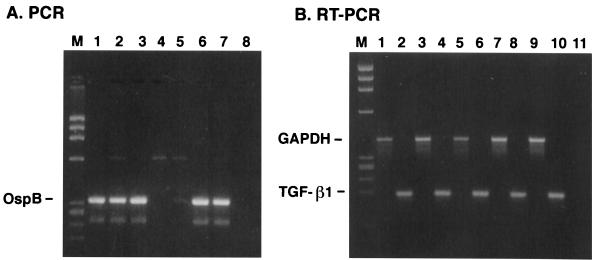
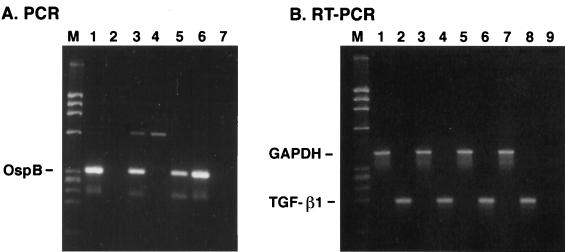
Table 1 shows the yields of RNA and DNA obtained from the same tissue sample in two separate experiments. One immediate result of the modified DNA extraction procedure was the consistently high 260 nm/280 nm absorbance ratios of the DNA preparations. These results were independent of the method (hand homogenization versus Fast Prep FP120) of DNA/RNA extraction (data not shown) and of the type of tissue used for extraction. The yield of DNA with the modified procedure was lower than that achieved when the DNA was extracted independently. However, some DNA preparations isolated in the latter manner inhibited the PCR, as demonstrated by a marked decrease in signal of a positive PCR DNA sample when the two DNA samples were mixed together (data not shown). This blocking inhibition of the PCR by the negative DNA sample, presumably due to the presence of Taq polymerase inhibitors, was either eliminated or not as severe when the DNA was prepared by the modified procedure (Fig. 1 and 2). The lower yield of DNA by this method is most likely the result of the less efficient recovery of DNA fragments, generated by the homogenization process, and the loss of DNA in the upper RNA fraction after treatment of the tissue homogenate with chloroform. Some tissues, such as the bladder, gave consistently higher yields of DNA than other tissues, whereas DNA yields from the spinal cord (lumbar) were generally lower. Nevertheless, we were able to detect the presence of B. burgdorferi in some DNA preparations from the central nervous system, bladder, and heart tissue (Fig. 1 and 2). The yield of RNA was consistently higher (2- to 24-fold [Table 1]) than the amount of DNA recovered from the same tissue, but the 260 nm/280 nm absorbance ratios of the RNA preparations were generally lower (1.7 versus 1.9) than those of the DNAs from the same samples. Expression of GAPDH and the cytokine TGF-β1 could be assessed in the different RNA preparations (Fig. 1 and 2).
TABLE 1.
Yields of RNA and DNA from the same tissue sample
| Expta | Tissue | RNA prepn
|
DNA prepn
|
||
|---|---|---|---|---|---|
| Yield (μg) | A260/A280 ratio | Yield (μg) | A260/A280 ratio | ||
| 1 | Heart (apex) | 18.8 | 1.74 | 5.49 | 1.95 |
| Spinal cord (lumbar) | 15.7 | 1.75 | 1.69 | 2.16 | |
| Bladder | 32.2 | 1.75 | 10.9 | 1.99 | |
| Temporal lobe | 17.7 | 1.76 | 6.56 | 1.97 | |
| Pons | 17.4 | 1.75 | 4.31 | 1.89 | |
| 2 | Heart | 37.2 | 1.78 | 5.40 | 1.97 |
| Midbrain | 38.4 | 1.77 | 3.10 | 1.90 | |
| Spinal cord (thoracic) | 7.0 | 1.70 | 3.15 | 2.00 | |
| Cerebellum | 51 | 1.75 | 9.12 | 1.94 | |
Tissues in experiments 1 and 2 are from different NHPs.
FIG. 1.
Gel analysis of PCR and RT PCR products prepared from DNA and RNA, respectively, from tissues in experiment 1. (A) PCR of DNAs for OspB. Lanes: 1, heart (apex); 2, spinal cord (lumbar); 3, bladder; 4, temporal lobe; 5, pons; 6, heart (apex) plus temporal lobe; 7, heart (apex) plus pons; 8, no-DNA control. (B) RT PCR of RNAs. Lanes: 1, heart (apex), GAPDH; 2, heart (apex), TGF-β1; 3, spinal cord (lumbar), GAPDH; 4, spinal cord (lumbar), TGF-β1; 5, bladder, GAPDH; 6, bladder, TGF-β1; 7, temporal lobe, GAPDH; 8, temporal lobe, TGF-β1; 9, pons, GAPDH; 10, pons, TGF-β1; 11, no-cDNA control; M, DNA size markers.
FIG. 2.
Gel analysis of PCR and RT PCR products prepared from DNA and RNA, respectively, from tissues in experiment 2. (A) PCR of DNAs for OspB. Lanes: 1, heart; 2, midbrain; 3, spinal cord (thoracic); 4, cerebellum; 5, heart plus midbrain; 6, heart plus cerebellum; 7, no-DNA control. (B) RT PCR of RNAs. Lanes: 1, heart, GAPDH; 2, heart, TGF-β1; 3, midbrain, GAPDH; 4, midbrain, TGF-β1; 5, spinal cord (thoracic), GAPDH; 6, spinal cord (thoracic), TGF-β1; 7, cerebellum, GAPDH; 8, cerebellum, TGF-β1; 9, no-cDNA control; M, DNA size markers.
In summary, we have modified a method for the isolation of DNA of high purity from samples in which the RNA has already been extracted. This procedure can be used to prepare DNA from most types of tissues or cells after extraction of the RNA portion. Using this procedure, we have demonstrated that we can detect the presence of B. burgdorferi and the expression of host cellular genes in the same tissue samples obtained from NHPs, which are serving as a model for Lyme neuroborreliosis.
Acknowledgments
This research was supported by grant RO1 NS34715 from the National Institutes of Health (to A.R.P.).
REFERENCES
- 1.Bergstrom S, Bundoc V, Barbour A. Molecular analysis of linear plasmid-encoded major surface proteins, OspA and OspB, of the Lyme disease spirochaete Borrelia burgdorferi. Mol Microbiol. 1989;3:479–486. doi: 10.1111/j.1365-2958.1989.tb00194.x. [DOI] [PubMed] [Google Scholar]
- 2.Chmczynski P. A reagent for the single-step simultaneous isolation of RNA, DNA, and proteins from cell and tissue samples. BioTechniques. 1993;15:532–536. [PubMed] [Google Scholar]
- 3.Chmczynski P, Sacchi N. Single step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 4.Husmann I, Soulet L, Gautron J, Martelly I, Barritault D. Growth factors in skeletal muscle regeneration. Cytokine Growth Factor Rev. 1996;7:249–258. doi: 10.1016/s1359-6101(96)00029-9. [DOI] [PubMed] [Google Scholar]
- 5.Pachner A R, Amemiya K, Delaney E, O’Neill T, Hughes C A N, Zhang W-F. Interleukin-6 expressed at high levels in the CNS in Lyme neuroborreliosis. Neurology. 1997;49:147–152. doi: 10.1212/wnl.49.1.147. [DOI] [PubMed] [Google Scholar]
- 6.Pachner A R, Delaney E, O’Neill T. Neuroborreliosis in the nonhuman primate: Borrelia burgdorferi persists in the central nervous system. Ann Neurol. 1995;38:667–669. doi: 10.1002/ana.410380417. [DOI] [PubMed] [Google Scholar]
- 7.Pachner A R, Delaney E, O’Neill T, Major E. Inoculation of nonhuman primates with the N40 strain of Borrelia burgdorferi leads to a model of Lyme neuroborreliosis faithful to the human disease. Neurology. 1995;45:165–172. doi: 10.1212/wnl.45.1.165. [DOI] [PubMed] [Google Scholar]
- 8.Persing D H, Rutledge B J, Rys P N, Podzorski D S, Mitchell P D, Reed K D, Liu B, Fikrig E, Malawista S E. Target imbalance: disparity of Borrelia burgdorferi genetic material in synovial fluid from Lyme arthritis patients. J Infect Dis. 1994;169:668–672. doi: 10.1093/infdis/169.3.668. [DOI] [PubMed] [Google Scholar]
- 9.Strauss W M. Preparation of genomic DNA from mammalian tissue. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 2. New York, N.Y: John Wiley & Sons; 1998. pp. 2.2.1–2.2.3. [Google Scholar]