Key Words: astrocyte, bioinformatics analysis, central nervous system, exosomes, microglia, neurodegenerative disease, RNA transcriptomics, UBB
Abstract
Glial cells play an important role in signal transduction, energy metabolism, extracellular ion homeostasis and neuroprotection of the central nervous system. However, few studies have explained the potential effects of exosomes from glial cells on central nervous system health and disease. In this study, the genes expressed in exosomes from astrocytes and microglia were identified by deep RNA sequencing. Kyoto Encyclopedia of Genes and Genomes analysis indicated that several pathways in these exosomes are responsible for promoting neurodegenerative diseases, including Alzheimer’s disease, Parkinson’s disease and Huntington’s disease. Gene ontology analysis showed that extracellular exosome, mitochondrion and growth factor activity were enriched in exosomes from the unique astrocyte group, while extracellular exosome and mitochondrion were enriched in exosomes from the unique microglia group. Next, combined with the screening of hub genes, the protein-protein interaction network analysis showed that exosomes from astrocytes influence neurodegenerative diseases through metabolic balance and ubiquitin-dependent protein balance, whereas exosomes from microglia influence neurodegenerative diseases through immune inflammation and oxidative stress. Although there were differences in RNA expression between exosomes from astrocytes and microglia, the groups were related by the hub genes, ubiquitin B and heat shock protein family A (Hsp70) member 8. Ubiquitin B appeared to be involved in pleiotropic regulatory functions, including immune regulation, inflammation inhibition, protein catabolism, intracellular protein transport, exosomes and oxidative stress. The results revealed the clinical significance of exosomes from glia in neurodegenerative diseases. This study was approved by the Animal Ethics Committee of Nantong University, China (approval No. S20180102-152) on January 2, 2018.
Chinese Library Classification No. R446.1; R741.02; Q344+.14
Introduction
Central nervous system (CNS) injury is one of the leading causes of death or persistent disability in humans. One of the reasons for the inability to recover loss of human neurons is the limited regenerative ability of the CNS (Kyritsis et al., 2014). A growing body of evidence supports the importance of CNS resident glial cells such as microglia and astrocytes (Kwon and Koh, 2020) in neurodegenerative diseases such as Alzheimer’s disease, Parkinson’s disease and amyotrophic lateral sclerosis (Rothhammer and Quintana, 2015). On the one hand, there is increasing evidence that reactive astrocytes may play a facilitator role in CNS diseases by losing normal astrocyte functions or acquiring abnormal effects (Sofroniew and Vinters, 2010). On the other hand, reactive astrocytes communicate with almost all CNS cells, thereby promoting neurogenesis, angiogenesis and recovery of the CNS (Huang et al., 2019). Additionally, microglia are resident cells in the brain, regulating brain development, neural network formation and CNS damage (Colonna and Butovsky, 2017). For example, in a healthy human brain, microglia affect neural progenitor cell differentiation, astrocyte activation, neuronal homeostasis and synapse formation. In human brain diseases, they undergo substantial morphological, molecular and functional changes, presenting a new biological state related to the pathogenesis and progression of these diseases (Wright-Jin and Gutmann, 2019).
Recently, exosomes have also been demonstrated to play a role in the CNS, ranging from removing unwanted biomolecules to intercellular communication and transmission of pathogenic proteins (Hill, 2019). In clinical studies, exosomes are frequently proposed as therapeutic drug carriers. Moreover, because the release of exosomes and their molecular cargo are cell-type-specific, exosomes have also been proposed as potential biomarkers for detecting and diagnosing neurodegeneration (Barile and Vassalli, 2017). Furthermore, some reviews have emphasized the role of exosomes as messengers in the communication between neurons and glial cells in the brain under toxic or pathological conditions (Paolicelli et al., 2019), these messengers contribute to the occurrence of neuroinflammation and neurodegeneration (Guo et al., 2021).
However, the pathogenic or protective effects of exosomes on neurodegenerative diseases have not been conclusively established in vivo; hence it is necessary to explore the molecular mechanisms of exosomes participating in neurodegenerative diseases. Astrocytes and microglia are important sources of neurogenic exosomes in the process of neurodegeneration (You and Ikezu, 2019). This study used bioinformatic analysis to reveal the similarities and differences between exosomes from astrocytes and microglia in complex neurodegenerative diseases and the relationship between the similarities and differences at the exosome level. In summary, considering that neurodegenerative diseases may be the result of multiple factors, this study investigated whether there are target genes with multiple regulatory functions that control the occurrence and development of various neurodegenerative diseases. Therefore, we explored a unique approach, putting emphasis on the comparison between the similarities and differences and the connections between exosomes from microglia and astrocytes, to explore target genes and regulatory mechanisms that exert multipotent regulatory functions using biochemistry analysis, and to provide new ideas for the treatment of neurodegenerative diseases.
Materials and Methods
Workflow
Figure 1 shows the workflow for this study, including the databases, bioinformatics tools and corresponding tasks performed. To eliminate errors and omissions in the analysis process, biological replications and bioinformatics analyses of RNA sequencing data were conducted at least three times from May to November 2020. This study was approved by the Animal Ethics Committee of Nantong University (approval No. S20180102-152) on January 2, 2018.
Figure 1.
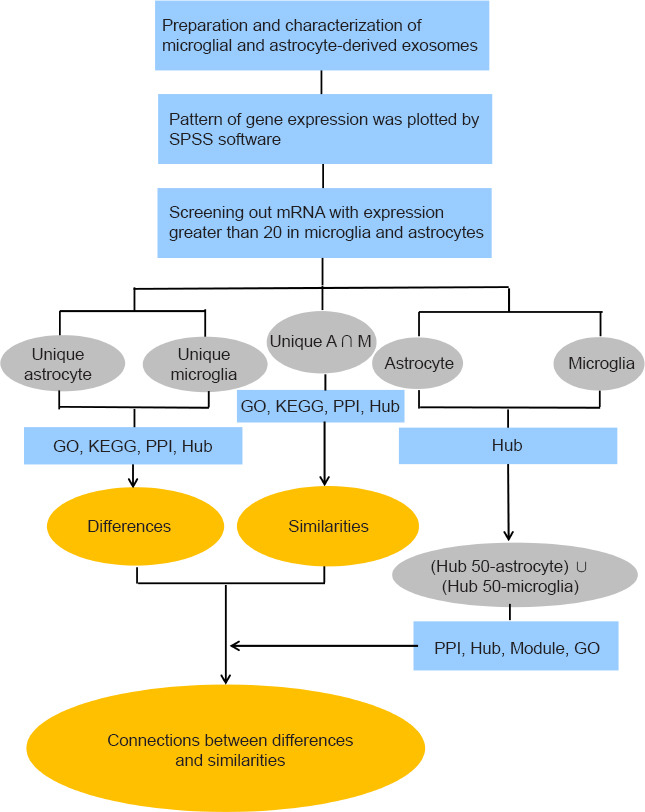
Workflow of the study.
The arrows represent the sequence of the analyses. The main results in the ellipse (in orange) were obtained by the analytical steps that are briefly described in the blue rectangles. The terms in the grey ellipses correspond to the group names. GO: Gene Ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes; PPI: protein-protein interaction network; SPSS: Statistical Product and Service Solutions software.
Astrocyte and microglia extraction and culture
Primary astrocytes and microglia were extracted from 40 1-day-old male Sprague Dawley rats provided by the Experimental Animal Center of Nantong University (license No. SCXK2014-0001). The rats (weight 3–5 g) were housed at 24°C and 55 ± 5% humidity with a 12-hour light/dark cycle. After they were anesthetized with isoflurane, the entire spine was excised, cut into pieces and incubated on ice in 10 mL of phosphate-buffered saline (PBS; Sigma-Aldrich, St. Louis, MO, USA) containing 1 mL of penicillin-streptomycin (Sigma-Aldrich). After discarding the supernatant and centrifuging at 300 × g for 3 minutes, 4 mL of trypsin (0.25%; Invitrogen, Carlsbad, CA, USA) was added to the precipitate, stirred well, placed at 37°C for a 15-minute digestion, and then 10% fetal bovine serum (complete medium; Invitrogen), penicillin (50 U/mL) and streptomycin (50 U/mL) were added. Centrifugation (179 × g, 3 minutes) was then performed and the precipitate was resuspended in medium containing 10% fetal bovine serum, penicillin (50 U/mL) and streptomycin (50 U/mL). The suspension was passed through a cell strainer and put into the incubator for differential attachment for 30 minutes to remove fibroblasts. Then, the supernatant was collected and placed in a flask (Sigma-Aldrich), which was placed on a shaking table at 180 r/min for 90 minutes. The supernatant was then collected to obtain microglia and the adhered cells, which were astrocytes, were kept (Hu et al., 2017).
Immunocytochemical staining
The purification of the astrocytes was confirmed by glial fibrillary acidic protein immunocytochemical staining, and the purification of the microglia was confirmed by ionized calcium-binding adapter molecule 1 immunocytochemical staining. Cultured astrocytes and microglia were first fixed with 4% paraformaldehyde (Sigma-Aldrich) for 30 minutes. Bovine serum albumin (5%; Invitrogen) was used as a blocking agent at room temperature for 1 hour. The cells were then incubated with anti-glial fibrillary acidic protein antibody (Cat# ab7260; mouse, 1:5000, Abcam, Cambridge, MA, USA) or anti-ionized calcium-binding adapter molecule 1 antibody (Cat# ab48004; mouse, 1:1000, Abcam) at 4°C for 12–16 hours. The secondary antibodies used were goat anti-mouse IgG H&L (Cy3®) preadsorbed (Cat# ab97035; 1:500, Abcam). Hoechst 33342 (Abcam) was used to label the cell nucleus. The reactions were visualized under a TCS SP2 confocal microscope or a ZEISS-AX10 fluorescence microscope (Carl Zeiss, Oberkochen, Germany). The morphology of the microglia and astrocytes was examined using ImageJ (Rawak Software Inc., Stuttgart, Germany).
Exosomes isolation and storage
When the density of the second-generation glial cells exceeded 80% and the cells were closely arranged and growing with cytoplasmic processes interlinked, they were exposed to exosome-depleted serum (Invitrogen) (Pusic et al., 2016) for approximately 24 hours. Then, the culture supernatant was cleared of cell debris and large vesicles by sequential centrifugation at 300 × g for 10 minutes, 1000 × g for 20 minutes and 10,000 × g for 30 minutes, followed by filtration with 0.2 μm syringe filters. Next, the sample was spun at 15,000 × g at 4°C for 2 hours to pellet the exosomes. Finally, the precipitates were resuspended in 200 μL of PBS and stored at –80°C (Miranda et al., 2018).
Transmission electron microscopy
The isolated exosomes were reseparated in PBS (Sigma-Aldrich), placed on a Formvar®-coated copper grid (150 Mesh, Ted Pella Inc., Redding, CA, USA) and fixed with 4% paraformaldehyde (Invitrogen) (Stuendl et al., 2016). The grids were negatively stained with 2% uranyl acetate (Invitrogen) containing 0.7 M oxalate (Invitrogen), pH 7.0, and visualized using a Philips 201 transmission electron microscope (Philips/FEI Inc., Briarcliff Manor, NY, USA).
Nanoparticle tracking analysis particle size analysis
The collected exosomes were diluted with PBS to 1 × 106/mL and injected into a NanoSight 500 (Version 2.2; NanoSight, Wiltshire, UK) with a 1-mL syringe. The Brownian motion of exosomes was captured, and the hydrodynamic diameters and the nanoparticle concentration were calculated by the Stocke-Einstein equation (Haney et al., 2015).
Western blot assay
Proteins from exosomes were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and electrotransferred to a nitrocellulose membrane (Bio-Rad, Hercules, CA, USA). The membrane was blocked with 5% non-fat dry milk (BBI Life Sciences, Shanghai, China) at room temperature for 1 hour and incubated overnight at 4°C with anti-CD63 antibody (Cat# ab59479; mouse, 1:1000, Abcam). According to the manufacturer’s guidelines, the observed molecular weight of CD63 varies in the range of 30–65 kDa. Antibody binding was detected by the secondary antibody, horseradish peroxidase-conjugated goat anti-mouse IgG H&L (ab205719, 1:20,000, Abcam). Antibody binding to the membranes was visualized by an enhanced chemiluminescence western detection system (Beyotime Biotechnology) and captured by a digital chemiluminescence scanner (C-Digit, LI-COR Biosciences, Lincoln, NE, USA). The intensities of the immunoblot bands were determined with Image Studio Version software (version 5.2, LI-COR Biosciences) (Zha et al., 2021).
RNA-sequencing analysis
To ensure the quality of the exosome samples, agarose gel electrophoresis was used to detect RNA degradation or genomic DNA pollution. Nanodrop (Ruibo, Guangzhou, China) was used to determine the purity of the total RNA. Furthermore, the RNA integrity was determined by Agilent 2200 Bioanalyzer (Ruibo) and the RNA integrity index calculated according to the RNA peak value, which requires an RNA integrity index > 7. Subsequently, RNA libraries were constructed in a series of procedures, including reverse transcription synthesis of double-stranded DNA, terminal repair, ligation of a sequencing linker, PCR amplification and library quality control. The sequencing was carried out by Guangzhou Ruibo Biotechnology Co., Ltd. (Guangzhou, China) and the sequencing mode was PE150 (Ruibo) (Gehring et al., 2020). Consequently, we obtained 41,191 genes, including long non-coding RNA (lncRNA), messenger RNA (mRNA) and circular RNA (cirRNA).
Screening of differentially expressed genes
Because astrocytes and microglia are the two primary glia cells in the brain, we preliminarily explored whether they lead to neurodegenerative changes through different biological functions. The R language (https://www.r-project.org/) was used in this study to identify differentially expressed genes in terms of fold-change (FC; microglia relative to astrocytes) and draw the corresponding heat maps and volcano maps, which helped lay the foundation for further study of the similarities, differences and relationships between the two. Log2(FC) < –2 or log2(FC) > 2 (Ambroise et al., 2011) and P < 0.05 (Perneger and Combescure, 2017) were considered statistically significant.
Analysis of biological processes and pathways
The Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were downloaded from the DAVID database (https://david.ncifcrf.gov/) (Huang da et al., 2009). The differentially expressed genes were grouped into genes unique to astrocyte exosomes, genes unique to microglial exosomes and genes unique to astrocyte and microglial exosomes (unique A ∩ M). In these groups the mRNA expression was higher than 20 fragments per kilobase of exon per million reads mapped (FPKM). We also generated and analyzed the following groups: (hub 50 astrocyte)∪ (hub 50 microglia), total RNA, mRNA and circRNA. In these analyses, the lower the false discovery rate was, the more significant the result of gene enrichment, and the higher the counts were, the more the genes contained in each GO or KEGG term were enriched. Subsequently, the corresponding bar charts, pie charts and bubble charts were drawn in R language. Among these, the pie charts especially contributed to describing the biological categories and their respective proportions. Regarding the KEGG bubble diagrams, the color darkness refers to the relevant P value of each functional pathway, and the circle size refers to the number of genes involved. In the mRNA analysis, the genes whose expression was more than 20 FPKM were selected and were divided into three groups by intersection analysis (http://bioinformatics.psb.ugent.be/webtools/Venn/) for further exploration of the similarities and the differences and the relationship between them. P < 0.05 was considered statistically significant and was consistent with the significance of functional enrichment results.
Protein-protein interaction
The genes of the different groups were uploaded to the String online database (https://string-db.org/), and the species was set as Rattus norvegicus. cytoHubba and MCODE in the Cytoscape software 3.6.1 (Smoot et al., 2011) were used to analyze the modules and hub genes with a high number of connections in the generated protein-protein interaction (PPI) networks. Among the 11 topology analysis methods included in cytoHubba, Botteleneck and Radiality (Chin et al., 2014) were chosen as the main topological analysis methods for scoring and ranking the genes. An interaction score > 0.9 (Szklarczyk et al., 2019) was considered statistically significant (Figure 1).
Statistical analysis
FPKM was used to normalize gene length and library size by R language. Consequently, the selection and grouping of the mRNAs were performed by SPSS 25.0 software (IBM Corp., Armonk, NY, USA). For example, the unique astrocyte group contained mRNAs showing expression of more than 20 FPKM in exosomes of astrocytes and less than 20 FPKM in exosomes of microglia. The unique astrocytes and microglia group contained mRNAs showing expression of more than 20 FPKM in the exosomes of both microglia and astrocytes, and the unique microglia group contained mRNAs showing expression of more than 20 FPKM in exosomes of microglia and less than 20 FPKM in exosomes of astrocytes. The quality of samples was confirmed and the parameters of RNA-sequencing (RNA-seq) data were set correctly, however, the same data were processed three times using SPSS 25.0, in an attempt to eliminate errors and omissions in the operation processes and to ensure P was statistically significant (P < 0.05).
Results
Characteristics of exosomes from astrocytes and microglia
The exosomes isolation process is shown in Figure 2A. Glial fibrillary acidic protein immunocytochemical staining showed that the astrocytes had large cell bodies, from which they sent out many long, branching processes (Additional Figure 1A (2.2MB, tif) ). The microglia, confirmed by immunocytochemical staining of ionized calcium-binding adapter molecule 1, had very small, short, club-shaped cell bodies. These club-like cell bodies protruded from several withered processes. The protrusion surface was rough, spiny and rarely branched (Additional Figure 1B (2.2MB, tif) ).
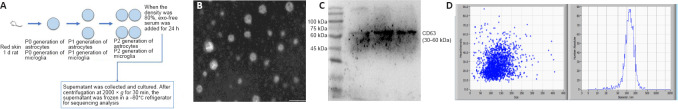
Figure 2.
Preparation and characterization of exosomes derived from astrocytes and microglia.
(A) A schematic diagram of the preparation of exosomes. (B) The morphological characteristics of exosomes were observed under an electron microscope. Scale bar: 200 nm. (C) The typical exosome marker, CD63, was detected by western blotting. (D) The Brownian motion and hydrodynamic diameter of exosomes were calculated by NTA analysis. NTA: Nanoparticle tracking analysis; P: passage.
As observed under the transmission electron microscope (Figure 2B), the exosomes had an obvious membrane boundary, showing a saucer or a cup structure of 30–100 nm. Furthermore, western blotting showed CD63 protein bands (Figure 2C), and the particles’ diameter was 50–200 nm, as assessed by nanoparticle tracking analysis particle size analysis (Figure 2D). The exosomes shown in Figure 2 were derived from astrocytes, and the quality assessment criteria for exosomes from microglia were the same as for those from astrocytes, which are disc-shaped or cup-shaped structures expressing CD63 with a particle diameter of 50–200 nm.
KEGG and GO enrichment analysis of different types of genes from the RNA-seq results
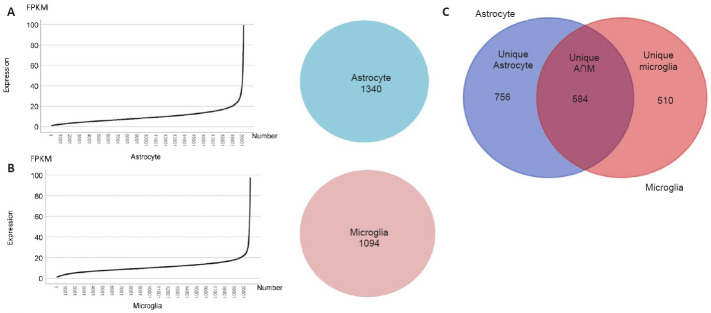
After normalizing the RNA-seq results, 41,191 genes were categorized into different types, e.g., lncRNA, mRNA and cirRNA. To understand the differences and similarities between exosomes from astrocytes and microglia, we selected from the RNA-seq results 20,840 mRNAs according to the proportion of various genes, which accounted for approximately half of all the categorized genes. In Figure 3, we visually showed the expression levels of all genes in the exosomes of astrocytes and microglia in a line graph, in which the x-axis indicates the number of each gene in order of gene expression from low to high. The distribution trends of the two types of exosomes were similar, with both showing a transition at the expression level of 20 FPKM (Figure 3A and B). Therefore, taking this turning point as a reference, we selected the mRNAs expressing more than 20 FPKM as genes that represent and define these two different cell types, and then divided them into three groups by intersection analysis (Figure 3C). The groups were termed unique astrocyte, unique microglia and unique astrocyte and microglia.
Figure 3.
Preliminary screening and grouping based on the distribution of gene expression.
(A) Left: Line diagram of gene expression in exosomes from astrocytes. The line graph shows all the genes from astrocyte exosomes sorted and numbered according to their expression level. The ordinate represents the expression level and the abscissa represents the gene number. Right: The number of mRNAs whose expression was greater than 20 FPKM in exosomes from astrocytes. (B) Left: Line diagram of gene expression in exosomes from microglia. The line graph shows all the genes from microglia exosomes sorted and numbered according to their expression level. The ordinate represents the expression level and the abscissa represents the gene number. Right: The number of mRNAs whose expression was greater than 20 FPKM in exosomes from microglia. (C) Grouping by intersection analysis. Left: The unique astrocyte group contained mRNAs with expression higher than 20 FPKM in exosomes from astrocytes and lower than 20 FPKM in exosomes from microglia. Middle: The unique astrocyte and microglia (unique A ∩ M) group contained mRNAs with expression higher than 20 FPKM in exosomes from microglia and astrocytes. Right: The unique microglia group contained mRNAs with expression higher than 20 FPKM in exosomes from microglia and lower than 20 FPKM in exosomes from astrocytes. FPKM: Fragments per kilobase of exon per million mapped fragments; mRNA: messenger RNA.
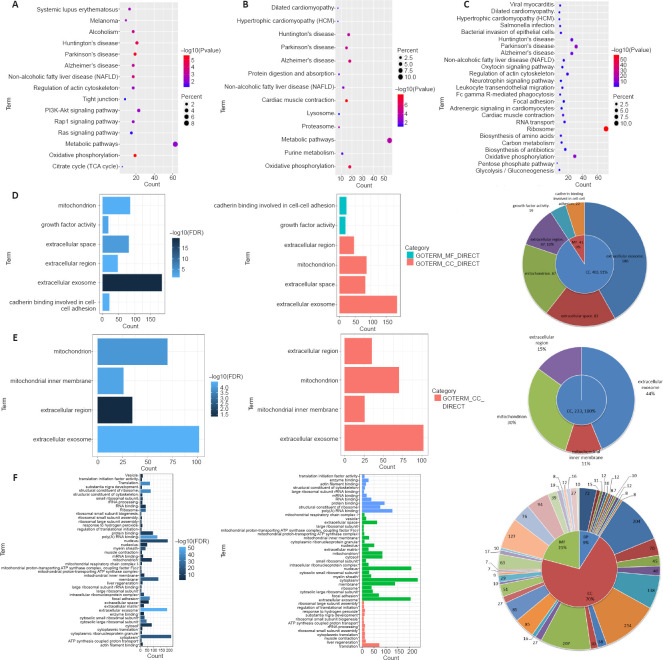
To elucidate what biological pathways or behaviors these genes are related to, we conducted KEGG pathway analysis using DAVID and found that the genes in the three groups were significantly enriched in typical neurodegenerative diseases, such as Alzheimer’s disease, Parkinson’s disease and Huntington’s disease (Figure 4A–C). These results suggest that exosomes from astrocytes and microglia are associated with neurodegenerative diseases.
Figure 4.
KEGG and GO analysis of the three mRNA groups based on the intersection analysis.
(A–C) The KEGG bubble chart was performed for (A) the unique astrocyte group, (B) the unique microglia group and (C) the unique astrocyte and microglia (unique A ∩ M) group. The P values indicated in red are more significant than those in blue. The dot size is directly proportional to the number of genes that are involved in the pathway. P < 0.05 was considered statistically significant. (D–F) GO was performed for (D) the unique astrocyte group, (E) the unique microglia group and (F) the unique A ∩ M group. Left panel: Bar charts where the bar length is proportional to the number of enriched genes. The darker the color, the more significant the term. Middle panel: Bar charts where terms were divided into CC, MF and BP. Right panel: Pie charts demonstrating the proportion of CC, MF and BP. The specific terms and the number of genes are shown in the outer circle. P < 0.05 was considered statistically significant. BP: Biological process; CC: cellular component; FDR: false discovery rate; GO: Gene Ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes; MF: molecular function.
To describe the functional information of molecular function (MF), cellular component (CC) and biological process (BP) involved in each gene group, we performed Gene Ontology analysis (Figure 4D–F). After comprehensive analysis based on false discovery rate and the number of genes in each GO term, it was observed that the differentially expressed genes were mainly enriched in terms of the cellular component, including extracellular exosome, mitochondrial and extracellular regions. Additionally, genes from the unique astrocytes were mainly enriched in growth factor activity and cadherin binding involved in cell-cell adhesion of molecular function. In contrast, genes from the unique microglia were mainly enriched in the mitochondrial inner membrane of the cellular component, indicating that the different exosomes are heterogenic in the pathogenesis of neurodegenerative diseases.
To further demonstrate that the gene expression in exosomes from astrocytes and microglia is different in neurodegenerative diseases, we performed a supplementary analysis on the 41,191 genes. There were 921 upregulated genes and 2952 downregulated genes (Additional Figure 2A (2.9MB, tif) and B (2.9MB, tif) ). GO analysis showed that most of these genes were enriched in G-protein coupled receptor activity, olfactory receptor activity, plasma membrane, integral component of membrane, G-protein coupled receptor signaling pathway and detection of chemical stimulus involved in sensory perception of smell (Additional Figure 2C (2.9MB, tif) ), of which BP accounted for the majority (Additional Figure 2D (2.9MB, tif) ). Furthermore, the KEGG analysis clearly showed that olfactory transduction was the most important pathway (Additional Figure 2E (2.9MB, tif) ). To further explore the specificity of the characteristics found, mRNAs, lncRNAs and cirRNAs were analyzed separately (Li et al., 2017). There were 363 upregulated and 1358 downregulated mRNAs (Additional Figure 3A (3MB, tif) and B (3MB, tif) ). Moreover, we observed that the GO terms including G-protein coupled receptor activity, olfactory receptor activity, plasma membrane, integral component of membrane, G-protein coupled receptor signaling pathway and detection of chemical stimulus involved in olfactory perception showed even distribution (Additional Figure 3C (3MB, tif) ) in BP, CC and MF (Additional Figure 3D (3MB, tif) ). Additionally, the differentially expressed mRNAs were closely related to the olfactory transduction pathway in the KEGG analysis (Additional Figure 3E (3MB, tif) ). Furthermore, there were 139 upregulated and 706 downregulated lncRNAs (Additional Figure 4A (2.2MB, tif) and B (2.2MB, tif) ) and 150 upregulated, and 89 downregulated cirRNAs (Additional Figure 4C (2.2MB, tif) and D (2.2MB, tif) ). There were specific differences that may be responsible for neurodegenerative diseases and some common mechanisms or functions among the identified genes that need to be further explored.
Differences between mRNAs in exosomes from astrocytes and microglia
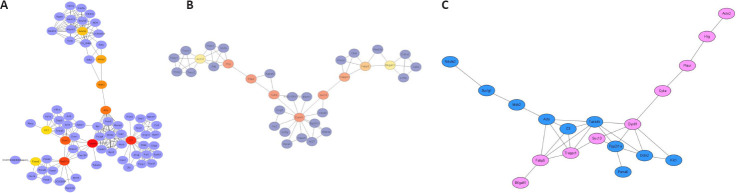
To further investigate the specific differences between the two types of exosomes, Cytoscape was used to construct PPI networks for the unique astrocyte and unique microglia groups. The top ten hub genes ranged from red to yellow based on their interaction score (Figure 5A and B). According to the Bottleneck analysis, the interaction scores of Tubb4b, C3, Ppp2r1a, Dctn2, Acly, Mdh2, Suclg1, Ndufs2, Klc1 and Psma6 in the unique astrocyte group were higher than the scores of the other genes. In the unique microglia group, the top 10 hub genes were Dynll1, Plaur, Sec13, Hrg, Cyba, Trappc1, Fabp5, Actn2, Actr3 and B4galt1. The hub genes in the unique astrocyte group were mainly related to the principal component of microtubules, modulating inflammation and possessing antimicrobial activity, signal transduction and synaptic formation during brain development (Table 1). However, the hub genes in the unique microglia group were mainly related to changing or maintaining cytoskeletal structures, localizing and promoting the formation of fibrinolytic enzyme, biogenesis of vesicles and mediating the elimination of necrotic cells (Table 2).
Figure 5.
Differences between exosomes from astrocytes and microglia based on PPI analysis of the unique astrocyte and the unique microglia groups.
(A, B) The top 10 hub genes in the unique astrocyte group (A) and the unique microglia group (B) are marked in red to yellow based on their interaction score in the PPI network analysis using cytoHubba (Bottleneck). The genes’ biological characteristics are listed in Tables 1 and 2. (C) The combined network based on the results in A and B. The blue nodes represent the top 10 hub genes from the unique astrocyte group and the pink nodes represent the top 10 hub genes from the unique microglia group. There was a pink gene node that was independent of the network map when drawing the network map, so this gene was not included in the results. Each node represents the protein indicated by the corresponding gene symbol, and each line represents an experimentally demonstrated protein-protein interaction. PPI: Protein-protein interaction network.
Table 1.
Functional roles of the top 10 hub genes in the unique astrocyte group ranked by the Bottleneck method
| Gene | Interaction score | Full name | Function |
|---|---|---|---|
| Tubb4b | 155 | Tubulin beta 4B class Ivb | The principal component of microtubules |
| C3 | 43 | Complement C3 | Modulating inflammation and possessing antimicrobial activity |
| Ppp2r1a | 40 | Protein phosphatase 2 scaffold subunit Aalpha | Signal transduction |
| Dctn2 | 35 | Dynactin subunit 2 | Synaptic formation during brain development |
| Acly | 33 | ATP citrate lyase | Important biosynthetic pathways |
| Mdh2 | 32 | Malate dehydrogenase 2 | Metabolic coordination |
| Suclg1 | 30 | Succinate-CoA ligase GDP/ADP-forming subunit alpha | Metabolic balance |
| Ndufs2 | 28 | Ubiquinone oxidoreductase core subunit S2 | Electron transport |
| Klc1 | 20 | Kinesin light chain 1 | Organelle transportation |
| Psma6 | 20 | Proteasome 20S subunit alpha 6 | The ATP-dependent degradation of ubiquitinated proteins |
Table 2.
Functional roles of the top 10 hub genes in the unique microglia group ranked by the Bottleneck method
| Gene | Interaction score | Full name | Function |
|---|---|---|---|
| Dynll1 | 43 | Dynein light chain LC8-type 1 | Changing or maintaining cytoskeletal structures |
| Plaur | 43 | Plasminogen activator, urokinase receptor | Localizing and promoting plasminformation |
| Sec13 | 43 | Nuclear pore and COPII coat complex component | Biogenesis of vesicles |
| Hrg | 43 | Histidine rich glycoprotein | Mediating the elimination of necrotic cells |
| Cyba | 43 | Cytochrome B-245 alpha chain | Membrane-bound oxidase |
| Trappc1 | 11 | Trafficking protein particle complex 1 | Vesicular transport |
| Fabp5 | 8 | Fatty acid binding protein 5 | Regulate the metabolism and actions of the ligands |
| Actn2 | 6 | Actinin alpha 2 | Anchor actin to a variety of intracellular structures. |
| Actr3 | 6 | Actin related protein 3 | Cell motility |
| B4galt1 | 6 | Beta-1,4-galactosyltransferase 1 | The biosynthesis of glycoconjugates |
Though it was clearly shown that the exosomes from astrocytes and microglia affect neurodegenerative diseases through different mechanisms, we also found connections among them (Figure 5C). Considering that each line in the figure is based on an experimentally demonstrated PPI, there be some associations between the two groups, however, whether these observations are significant remains to be verified.
To further confirm our conjecture about the possible connections, supplementary integrative analysis of the cirRNAs (Additional Figure 5 (2.6MB, tif) ) was conducted. The functional enrichment results demonstrated that cirRNAs in exosomes from astrocytes and microglia were associated with protein metabolism, such as metabolic and Ras signaling pathways. Because most of the measured lncRNAs were unannotated genes, they could not be analyzed. The associations among mRNAs require further research.
Similarities between mRNAs in exosomes from astrocytes and microglia
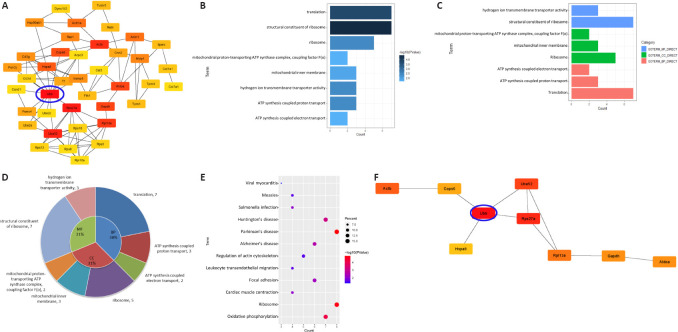
A PPI network for the unique A ∩ M group was constructed using String to find the similarities between exosomes from astrocytes and microglia. Fifty genes were identified as hub genes using the Cytoscape plug-in cytoHubba (Figure 6A). We found that Ubb had the largest number of interaction partners. Given that all cellular proteins and organelles are quality controlled, the ubiquitin-proteasome and autophagy systems help destroy and restore altered cellular components in situations where repairs are impossible. Ubb encodes ubiquitin, which plays a major role in the degradation of target cellular proteins, and abnormal expression of UBB has been detected in Alzheimer’s disease. Therefore, this research proposes that UBB may be a critical driving force in the occurrence and development of neurodegenerative diseases. The GO analysis demonstrated that genes of BP were significantly enriched in protein translation. Additionally, genes of CC were mainly enriched in ribosomes and genes of MF were mainly enriched in the structure of the ribosome (Figure 6B–D). These findings were associated with the abnormal accumulation of toxic proteins in neurodegenerative diseases, which were enriched in the KEGG analysis (Figure 6E).
Figure 6.
Similarities between exosomes from astrocytes and microglia determined by analysis of the unique astrocyte and microglia (unique A ∩ M) group.
(A) The network of the top 50 hub genes in the unique A ∩ M group. Genes are marked from red to yellow based on their interaction score in the PPI network analysis using cytoHubba (Radiality). (B) The length of the bars of the terms contained in the bar chart is directly proportional to the number of enriched genes in the GO analysis. The darker the color, the more significant the term. (C) Bar charts of GO terms divided into MF, BP and CC. (D) Pie chart demonstrating the proportion of CC, MF and BP. The specific terms and the number of genes are shown in the outer circle. (E) Bubble diagram showing the KEGG analysis. P indicated in red is significantly higher than that in blue. The dot size is proportional to the number of genes that are involved in the pathway. (F) The top 10 hub genes in the unique A ∩ M group are marked in red to yellow based on their interaction score in the PPI network analysis using cytoHubba (Bottleneck). Their biological characteristics are listed in Table 3. The Ubb node with the most direct interactions is highlighted in the brightest red and the blue circle. BP: biological process; CC: cellular component; GO: Gene Ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes; MF: molecular function; PPI: protein-protein interaction network; Ubb: ubiquitin B.
To reveal representative features further, we selected the top 10 hub genes from the unique A ∩ M group without retaining the corresponding nodes (Figure 6F). It was apparent that these proteins were mainly related to protein degradation, activation of protein kinases and signal transduction, damaged DNA repair and inhibition of inflammation (Table 3), which were associated with the differences we found in this study. We paid attention to heat shock protein family A (Hsp70) member 8 (HSPA8) because in our results it had an interaction with UBB, and it is a marker of almost all exosomes. Therefore, we speculated that UBB might be associated with multiple exosomes and help regulate different biological functions that lead to neurodegenerative changes.
Table 3.
Functional roles of the top 10 hub genes in the unique astrocyte and microglia group ranked by the Bottleneck method
| Gene | Interaction score | Full name | Function |
|---|---|---|---|
| Ubb | 185 | Ubiquitin B | Protein degradation |
| Rps27a | 125 | Ribosomal protein S27a | Protein degradation |
| Uba52 | 65 | Ubiquitin A-52 residue ribosomal protein fusion product1 | Activation of protein kinases and in signaling |
| Actb | 36 | Actin beta | Reparation of damaged DNA |
| Rpl13a | 35 | Ribosomal protein L13a | Inhibition of inflammation |
| Aldoa | 32 | Aldolase, fructose-bisphosphate A | Glycolysis and gluconeogenesis |
| Gapdh | 32 | Glyceraldehyde-3-phosphate dehydrogenase | The product catalyzes carbohydrate metabolism |
| Cops6 | 25 | COP9 signalosome subunit 6 | Phosphorylation |
| Hspa8 | 24 | Heat shock protein family A (Hsp70) member 8 | The disassembly of vesicles |
| ATP8 | 20 | Mitochondrially encoded ATP synthase membrane subunit 8 | Electron transport |
The relationship between the differences and similarities in the expression of exosomal mRNAs from astrocytes and microglia
Chronic and progressive neurodegeneration is the result of multiple factors. Even though Ubb was the most significant hub gene in all the groups, it was unclear whether there was a causal relationship between Ubb and genes we discovered previously. To find connections between the differences and the similarities, we merged the top 50 hub genes in the astrocytes (Additional Figure 6A (1.1MB, tif) ) and microglia groups (Additional Figure 6B (1.1MB, tif) ) to generate a PPI network (Figure 7A). We applied MCODE in Cytoscape to identify three biological function modules (Figure 7B) and showed the GO analysis based on each module in combination with a series of pie charts. The upper left module comprised genes from both exosome types and was most significantly enriched in exosome transport genes (Figure 7C), such as AP2M1 and FCHO1. In the lower-left module, genes from microglial exosomes were the majority and were significantly enriched in inflammatory response genes (Figure 7D). For example, Rpl13a, which encodes riboprotein, acts as a translation inhibitor of IFN-γ (Jia et al., 2012). Microglia-mediated chronic inflammatory responses can cause nerve damage (Subhramanyam et al., 2019). In the right module, genes from astrocyte exosomes were the majority and were significantly enriched in maintaining protein balance genes (Figure 7E), such as Psma6, which encodes a proteasome subunit and participates in the degradation of ATP-dependent ubiquitinated proteins by removing misfolded or damaged proteins that may impair cellular functions. Chung et al. (2013) have demonstrated that astrocytes are involved in cell phagocytosis. They bind to specific proteins like GULP PTB-domain-containing engulfment adaptor 1 (GULP1) and ATP binding cassette subfamily A member 1 (ABCA1) in target cells, triggering subsequent uptake and digestion of the target cells (Chung et al., 2013). It is worth noting that UBB was still the hub gene with the highest interaction score, suggesting that it may be the unifying step that plays an indispensable role in regulating the different developmental mechanisms of neurodegenerative diseases.
Figure 7.
The connections between the differences and similarities.
(A) Hub genes from the hub 50 astrocyte and hub 50 microglia are marked in red to yellow based on their interaction score using cytoHubba (Radiality). (B) The three most important modules obtained from the hub 50 astrocyte and hub 50 microglia. The blue nodes indicating genes from the hub 50 astrocyte (Additional Figure 6A (1.1MB, tif) ) are mainly concentrated in the right module. The yellow nodes indicating genes from the hub 50 microglia (Additional Figure 6B (1.1MB, tif) ) are mainly concentrated in the lower left module. The green nodes indicating genes from the hub 50 astrocyte and the hub 50 microglia are mainly concentrated in the upper left module. (C) Pie chart displaying the GO analysis of the genes enriched in the upper left module. (D) Pie chart displaying the GO analysis of the genes enriched in the bottom left module. (E) Pie chart displaying the GO analysis of the genes enriched in the right module. The Ubb node connecting the three modules has the most direct interactions and is highlighted by a blue circle. This gene encodes ubiquitin, which plays a major role in the degradation of target cellular proteins. Hub 50 astrocyte: the group containing the top 50 hub genes from astrocyte exosomes with expression levels greater than 20 FPKM. Hub 50 microglia: the group containing the top 50 hub genes from microglial exosomes with expression levels greater than 20 FPKM. GO: Gene Ontology; PPI: protein-protein interaction network; Ubb: ubiquitin B.
Discussion
In summary, we generated extensive transcriptome data on exosomes from microglia and astrocytes. Bioinformatics analyses demonstrated that these exosomes play an important role in the occurrence and development of neurodegenerative diseases, such as Huntington’s disease, Alzheimer’s disease and Parkinson’s disease. Exploring the differences in gene expression, we found that astrocyte exosomes affect neurodegenerative diseases mainly through metabolic balance and ubiquitin-dependent protein balance (Scrivo et al., 2018). In contrast, microglia exosomes affect neurodegenerative diseases mainly through immune inflammation and oxidative stress. Although there were apparent differences in gene expression between the exosome types, both were related to hub genes like UBB and HSPA8. It has been demonstrated that the UBB pool plays an indispensable role in determining the fate and self-renewal of neural stem cells (Park et al., 2020). Furthermore, studies have shown that neurodegenerative diseases are the clinical symptoms of neurological dysfunction caused by biochemical changes and protein accumulation inside or outside various anatomical sites such as neurons or glial cells (Kovacs, 2017). Disruption of UBB often leads to a protein balance disorder and neurogenesis inhibition (Ryu et al., 2014). Additionally, UBB always binds to HSPA8, which is a marker of almost all exosomes (Pratt et al., 2014). Therefore, through the connections between differences and similarities, it is easy to conclude that neurodegenerative diseases are the outcomes of multiple factors and UBB may play pleiotropic regulatory roles in neurodegeneration, such as immune regulation, inflammation inhibition, protein catabolism, intracellular protein transport, exosomes and oxidative stress. It was an interesting phenomenon that the three groups of genes were significantly enriched in typical neurodegenerative diseases. In the follow-up study, we will perform the corresponding comparative analysis of isolated exosomes from Alzheimer’s disease, Parkinson’s disease and Huntington’s disease and their matching controls.
Progressive neurodegenerative diseases in older people are the main cause of senile dementia, which has caused great suffering to patients and become a major burden on society. These diseases are characterized by memory loss and cognitive decline. Currently, there are no drugs or other therapeutic agents available to prevent disease progression (Tang and Le, 2016). A substantial amount of evidence has shown that the pathological processes of neurodegenerative diseases (Vaquer-Alicea and Diamond, 2019) may be associated with abnormalities in the ubiquitin-proteasome and autophagy/lysosomal systems (Nataf et al., 2019), the deposition of abnormal proteins and the degeneration of brain networks, as well as the subsequent protein toxicity (Guttenplan et al., 2020), oxidative stress (Tian et al., 2016), nerve inflammation (Kong et al., 2017) and programmed cell death (Aufschnaiter et al., 2017). However, it has been suggested that the formation of abnormal proteins in neurodegenerative diseases occurs before the clinical features appear, which indicates that early diagnosis and treatment are necessary. Until now, effective treatment strategies for neurodegenerative diseases mainly focused on preventing the formation of toxic proteins (Shrivastava et al., 2020), regulating accompanying brain inflammation (Simchovitz et al., 2017) and protecting neurons from cell death (Hwang and Zukin, 2018). However, we still lack sufficient understanding of the molecular mechanisms of the toxic protein formation and inflammatory responses.
Brain biopsies are performed under very limited circumstances, and neuroscientists have to deal with the absence of appropriate animal models for most neurological diseases and other limitations associated with human brain research. Most examinations can only be done through imaging techniques or postmortem dissection, leading to failure to detect and treat neurodegenerative diseases early. Therefore, effective early diagnosis and treatment are urgently needed to reduce poor prognosis. Several studies have demonstrated that the pathophysiological/biological processes leading to neurodegeneration involve neurons and glial cells, including astrocytes and microglia (Kam et al., 2020). For example, apoptosis is a widespread phenomenon that occurs in the brain under both physiological and pathological conditions. Dead cells must be quickly removed to avoid further toxic effects. Importantly, these processes are always carried out by microglia, which are defined as specialized phagocytes of the brain. However, unfortunately, they have long been overlooked (Diaz-Aparicio et al., 2016). Microarray technology has proven to be a useful method for identifying new biomarkers. Bioinformatics analysis of exosomes can reveal unknown cellular and molecular mechanisms of intercellular communication, organ homeostasis and diseases. Therefore, obtaining information about nerve cells from various exosomal genes would be very useful, which is why exosomes are now of great interest in the study of neurodegenerative diseases (Kalluri and LeBleu, 2020).
Exosomes were first described 30 years ago and have been explored as a means for drug or biomarker discovery, owing to their involvement in intercellular communication and the spread of disease states (Edgar, 2016). Studies have shown that the exosome is a type of endogenous extracellular vesicle (40–100 nm in diameter), which is considered as a new-generation natural nanoscale delivery system. Exosomes secreted by different types of cells carry different signaling molecules (such as RNA and proteins) and thus have great potential for targeted drug delivery and therapy (Liao et al., 2019). Moreover, Kumar et al. (2017) have proposed that some impaired micro RNA can be encapsulated by exosomes and play a key role in various human diseases, including neurodegenerative diseases such as Alzheimer’s disease, Huntington’s disease and Parkinson’s disease. Therefore, exosomes have opened up a new way for biomarker discovery. In this study, the complex regulatory mechanism of neurodegenerative diseases was functionally dissected, and the clinical significance of exosomes from astrocytes and microglia in neurodegenerative diseases was revealed. Our findings lay a foundation for revealing mechanisms of exosomes from astrocytes and microglia in complex neurodegenerative diseases, which will enable the discovery of exosome-mediated treatment strategies at the neurogliocyte level. However, it is worth noting that the current study was conducted using cell culture and transgenic animal models and needs to be verified by further research.
Currently, there are still limitations in the determination and analysis of exosomes. For example, it is difficult to distinguish different types of exosomes; hence it is impossible to optimize their analysis. Although they are potential biomarkers that are likely to be used to develop new, specific indicators of neuroinflammation or neurodegeneration, we do not fully understand their potential because they can reverse or promote neurodegenerative diseases. Furthermore, it is difficult to predict whether these potential biomarkers will be more clinically practical, convenient and cost-effective than traditional diagnostic methods such as magnetic resonance imaging. Although our data mining methods were limited to retrieving experimentally demonstrated PPIs, whether such interactions occur in exosomes from astrocytes and microglia remains to be determined.
In conclusion, our findings suggest that exosomes may be a valuable tool for the treatment of neurodegenerative diseases in the future. In this research, deep exosome RNA sequencing demonstrated that neurodegenerative diseases result from the combined action of multiple factors, and Ubb may play in neurodegenerative diseases multiple regulatory roles, such as immune regulation, inflammatory suppression, protein catabolism, intracellular protein transport, exosomes and oxidative stress. In this field, it is possible to reveal the pathogenic mechanisms and long-term biomarkers at the glial cell level and determine exosome-mediated treatment strategies. Although complex interventions are challenging, our research showed that exosome bioinformatics is a promising approach for the widespread use of genetically engineered and targeted therapies for CNS diseases.
Additional files:
Immunocytochemical staining of astrocytes and microglia.
(A) Astrocytes confirmed by GFAP (red, stained by Cy3) immunocytochemical staining. GFAP was shown in the cytoplasm of the astrocytes, and blue staining represents the nucleus. (B) Microglia confirmed by Iba1 (red, stained by Cy3) immunocytochemical staining. Iba1 was shown in the cytoplasm of microglia, and blue staining represents the nucleus. Scale bars: 100 μm. DAPI: 4’,6-Diamidino-2-phenylindole; GFAP: Glial fibrillary acidic protein; Iba1: ionize calcium binding adapt molecule 1.
GO analysis of all differential genes in exosomes from astrocytes and microglia.
(A) The heatmap was based on folding changes (microglia relative to astrocytes), where log2(FC) > 2 were highlighted in red while the log2 (FC) < -2 were highlighted in green. Preprocession included log2 normalization and data centralization. (B) The volcano images showed that the blue dots represented down-regulated genes, while the red dots represented up-regulated genes. (C) The left bar chart of GO analysis showed that the darker the color, the more significant the enrichment. In the right bar chart, the blue represented MF, the green represented the CC, and the pink represented BP. (D) The pie chart demonstrated the proportion of CC, MF and BP. Their specific terms and the number of genes were distributed in the outer circle. (E) In bubble diagram of KEGG analysis, P-values indicated in red were significantly higher than those in blue and the sizes of dots were proportional to the number of enriched genes. P-value < 0.05. BP: Biological process; CC: cellular component; GO: Gene Ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes; log2(FC): log2(fold change); MF: molecular function.
mRNA-level differential analysis between exosomes from astrocytes and microglia.
(A) The heatmap was based on mRNA-level folding changes (microglia relative to astrocytes), where log2(FC) > 2 were highlighted in red while log2 (FC) < -2 were highlighted in green. (B) The volcano figure showed that the blue dots represented down-regulated genes, while the red dots represented up-regulated genes. (C) The left bar chart of GO analysis showed that the darker the color, the more significant the enrichment. In the right bar chart, the blue represented MF, the green represented CC and the pink represented the BP. (D) The pie chart demonstrated the proportion of CC, MF and BP. Their specific terms and the number of genes were distributed in the outer circle. (E) The bubble diagram showed KEGG analysis. P values indicated in red were more significant than those in blue. The sizes of dots were directly proportional to the number of enriched genes. Pre-processing included log2 normalization and data centralization. P value < 0.05 was statistically significant. BP: Biological process; CC: cellular component; GO: Gene Ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes; log2(FC): log2(fold change); MF: molecular function; mRNA: messenger RNA.
ncRNA-level and cirRNA-level difference analysis between exosomes from astrocytes and microglia.
(A) The heatmap were based on ncRNA-level folding changes (microglia relative to astrocytes), where log2 (FC) > 2 were highlighted in red while log2 (FC) < -2 were highlighted in green. (B) In volcano figure, the blue dots represented down-regulated genes, while the red dots represented up-regulated genes. (C) The heatmap was based on cirRNA-level fold changes (microglia relative to astrocytes), where log2 (FC) > 2 were highlighted in red while log2 (FC) < -2 were highlighted in green. (D) The network diagram showed that the blue dots represented down-regulated genes, while the red dots represented up-regulated genes. Pre-processing included log2 normalization and data centralization. P-value < 0.05 was statistically significant.) cirRNA: Circular RNAs; log2 (FC): log2 (fold change); ncRNA: non-coding RNA.
GO and KEGG analysis based on cirRNAs in exosomes from astrocytes and microglia respectively.
(A) The specific terms of cirRNAs in exosomes from microglia. The upper bar chart showed that the darker the color, the more significant the enrichment. The bottom bar chart showed that the blue represented MF, the green represented CC and the pink represented BP. (B) The pie chart demonstrated the proportion of CC, MF and BP in the inner circle. Their specific terms and the number of genes were distributed in the outer circle. (C) The bubble diagram showed the KEGG analysis. P values indicated in red were more significant than those in blue. The dots size were proportional to the numbers of enrich genes. (D) The specific terms of cirRNAs in exosomes from microglia. The upper bar chart showed that the darker the color, the more significant the enrichment. The bottom bar chart showed that the blue represented MF, the green represented CC and the pink represented BP. (E) The pie chart demonstrated the proportion of CC, MF and BP. Their specific terms and the number of genes were distributed in the outer circle. (F) The bubble diagram showed the KEGG analysis. P values indicated in red were significantly higher than those in blue. The sizes of dots were proportional to the number of enriched genes. P value < 0.05 was statistically significant. BP: Biological process; CC: cellular component; cirRNA: circular RNAs; GO: Gene Ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes; MF: molecular function.
Mining the top 50 hub mRNA from the astrocyte and the microglia using Cytoscape.
(A) The PPI network of the hub 50 astrocyte showed that the top 50 hub genes from the astrocyte (Figure 3A) were marked from red to yellow using Cytohubba (Radiality). (B) The PPI network of the hub 50 microglia showed that the top fifty hub genes from microglia (Figure 3B) were marked from red to yellow using Cytohubba (Radiality). The “UBB” node with the most direct interactors was highlighted in the brightest red and selected by the blue circle. Hub 50 astrocyte: the group contained the top 50 hub mRNAs from astrocyte exosomes, and its expression levels were greater than 20 FPKM. Hub 50 microglia: the group contained the top 50 hub mRNAs from microglia exosomes, and its expression levels were greater than 20 FPKM. FPKM: Fragments per kilobase of exon model per million mapped fragments; PPI: protein protein interaction network; UBB: ubiquitin B.
Footnotes
Conflicts of interest: The authors declare no conflicts of interest.
Financial support: This study was supported by the National Natural Science Foundation of China, No. 81870975 (to SLZ); and Innovation and Entrepreneurship Training Program for College Students in Jiangsu Province of China, No. 202010304034Z (to FYZ). The funding sources had no role in study conception and design, data analysis or interpretation, paper writing or deciding to submit this paper for publication.
Institutional review board statement: This study was approved by the Animal Ethics Committee of Nantong University, China (approval No. S20180102-152) on January 2, 2018.
Copyright license agreement: The Copyright License Agreement has been signed by all authors before publication.
Data sharing statement: Datasets analyzed during the current study available from the corresponding author on reasonable request.
Plagiarism check: Checked twice by iThenticate.
Peer review: Externally peer reviewed.
Funding: This study was supported by the National Natural Science Foundation of China, No. 81870975 (to SLZ); and Innovation and Entrepreneurship Training Program for College Students in Jiangsu Province of China, No. 202010304034Z (to FYZ).
C-Editor: Zhao M; S-Editors: Yu J, Li CH; L-Editors: Yu J, Song LP; T-Editor: Jia Y
References
- 1.Ambroise J, Bearzatto B, Robert A, Govaerts B, Macq B, Gala JL. Impact of the spotted microarray preprocessing method on fold-change compression and variance stability. BMC Bioinformatics. 2011;12:413. doi: 10.1186/1471-2105-12-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barile L, Vassalli G. Exosomes: Therapy delivery tools and biomarkers of diseases. Pharmacol Ther. 2017;174:63–78. doi: 10.1016/j.pharmthera.2017.02.020. [DOI] [PubMed] [Google Scholar]
- 3.Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol 8 Suppl. 2014;4:S11. doi: 10.1186/1752-0509-8-S4-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chung WS, Clarke LE, Wang GX, Stafford BK, Sher A, Chakraborty C, Joung J, Foo LC, Thompson A, Chen C, Smith SJ, Barres BA. Astrocytes mediate synapse elimination through MEGF10 and MERTK pathways. Nature. 2013;504:394–400. doi: 10.1038/nature12776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colonna M, Butovsky O. Microglia function in the central nervous system during health and neurodegeneration. Annu Rev Immunol. 2017;35:441–468. doi: 10.1146/annurev-immunol-051116-052358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Edgar JR. Q&A: What are exosomes, exactly? BMC Biol. 2016;14:46. doi: 10.1186/s12915-016-0268-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gehring J, Hwee Park J, Chen S, Thomson M, Pachter L. Highly multiplexed single-cell RNA-seq by DNA oligonucleotide tagging of cellular proteins. Nat Biotechnol. 2020;38:35–38. doi: 10.1038/s41587-019-0372-z. [DOI] [PubMed] [Google Scholar]
- 8.Guo M, Hao Y, Feng Y, Li H, Mao Y, Dong Q, Cui M. Microglial exosomes in neurodegenerative disease. Front Mol Neurosci. 2021;14:630808. doi: 10.3389/fnmol.2021.630808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guttenplan KA, Weigel MK, Adler DI, Couthouis J, Liddelow SA, Gitler AD, Barres BA. Knockout of reactive astrocyte activating factors slows disease progression in an ALS mouse model. Nat Commun. 2020;11:3753. doi: 10.1038/s41467-020-17514-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haney MJ, Klyachko NL, Zhao Y, Gupta R, Plotnikova EG, He Z, Patel T, Piroyan A, Sokolsky M, Kabanov AV, Batrakova EV. Exosomes as drug delivery vehicles for Parkinson’s disease therapy. J Control Release. 2015;207:18–30. doi: 10.1016/j.jconrel.2015.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hill AF. Extracellular vesicles and neurodegenerative diseases. J Neurosci. 2019;39:9269–9273. doi: 10.1523/JNEUROSCI.0147-18.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu Z, Feng J, Bo W, Wu R, Dong Z, Liu Y, Qiang L, Liu M. Fidgetin regulates cultured astrocyte migration by severing tyrosinated microtubules at the leading edge. Mol Biol Cell. 2017;28:545–553. doi: 10.1091/mbc.E16-09-0628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang L, Nakamura Y, Lo EH, Hayakawa K. Astrocyte signaling in the neurovascular unit after central nervous system injury. Int J Mol Sci. 2019;20:282. doi: 10.3390/ijms20020282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hwang JY, Zukin RS. REST, a master transcriptional regulator in neurodegenerative disease. Curr Opin Neurobiol. 2018;48:193–200. doi: 10.1016/j.conb.2017.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jia J, Arif A, Willard B, Smith JD, Stuehr DJ, Hazen SL, Fox PL. Protection of extraribosomal RPL13a by GAPDH and dysregulation by S-nitrosylation. Mol Cell. 2012;47:656–663. doi: 10.1016/j.molcel.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science. 2020;367:eaau6977. doi: 10.1126/science.aau6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kam TI, Hinkle JT, Dawson TM, Dawson VL. Microglia and astrocyte dysfunction in parkinson’s disease. Neurobiol Dis. 2020;144:105028. doi: 10.1016/j.nbd.2020.105028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kong WL, Peng YY, Peng BW. Modulation of neuroinflammation: Role and therapeutic potential of TRPV1 in the neuro-immune axis. Brain Behav Immun. 2017;64:354–366. doi: 10.1016/j.bbi.2017.03.007. [DOI] [PubMed] [Google Scholar]
- 20.Kovacs GG. Concepts and classification of neurodegenerative diseases. Handb Clin Neurol. 2017;145:301–307. doi: 10.1016/B978-0-12-802395-2.00021-3. [DOI] [PubMed] [Google Scholar]
- 21.Kumar S, Vijayan M, Bhatti JS, Reddy PH. MicroRNAs as peripheral biomarkers in aging and age-related diseases. Prog Mol Biol Transl Sci. 2017;146:47–94. doi: 10.1016/bs.pmbts.2016.12.013. [DOI] [PubMed] [Google Scholar]
- 22.Kwon HS, Koh SH. Neuroinflammation in neurodegenerative disorders: the roles of microglia and astrocytes. Transl Neurodegener. 2020;9:42. doi: 10.1186/s40035-020-00221-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kyritsis N, Kizil C, Brand M. Neuroinflammation and central nervous system regeneration in vertebrates. Trends Cell Biol. 2014;24:128–135. doi: 10.1016/j.tcb.2013.08.004. [DOI] [PubMed] [Google Scholar]
- 24.Li S, Qian T, Wang X, Liu J, Gu X. Noncoding RNAs and their potential therapeutic applications in tissue engineering. Engineering. 2017;3:3–15. [Google Scholar]
- 25.Liao W, Du Y, Zhang C, Pan F, Yao Y, Zhang T, Peng Q. Exosomes: The next generation of endogenous nanomaterials for advanced drug delivery and therapy. Acta Biomater. 2019;86:1–14. doi: 10.1016/j.actbio.2018.12.045. [DOI] [PubMed] [Google Scholar]
- 26.Miranda AM, Lasiecka ZM, Xu Y, Neufeld J, Shahriar S, Simoes S, Chan RB, Oliveira TG, Small SA, Di Paolo G. Neuronal lysosomal dysfunction releases exosomes harboring APP C-terminal fragments and unique lipid signatures. Nat Commun. 2018;9:291. doi: 10.1038/s41467-017-02533-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nataf S, Guillen M, Pays L. Common neurodegeneration-associated proteins are physiologically expressed by human B lymphocytes and are interconnected via the inflammation/autophagy-related proteins TRAF6 and SQSTM1. Front Immunol. 2019;10:2704. doi: 10.3389/fimmu.2019.02704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paolicelli RC, Bergamini G, Rajendran L. Cell-to-cell communication by extracellular vesicles: focus on microglia. Neuroscience. 2019;405:148–157. doi: 10.1016/j.neuroscience.2018.04.003. [DOI] [PubMed] [Google Scholar]
- 29.Park CW, Jung BK, Ryu KY. Disruption of the polyubiquitin gene Ubb reduces the self-renewal capacity of neural stem cells. Biochem Biophys Res Commun. 2020;527:372–378. doi: 10.1016/j.bbrc.2020.04.042. [DOI] [PubMed] [Google Scholar]
- 30.Perneger TV, Combescure C. The distribution of P-values in medical research articles suggested selective reporting associated with statistical significance. J Clin Epidemiol. 2017;87:70–77. doi: 10.1016/j.jclinepi.2017.04.003. [DOI] [PubMed] [Google Scholar]
- 31.Pratt WB, Morishima Y, Gestwicki JE, Lieberman AP, Osawa Y. A model in which heat shock protein 90 targets protein-folding clefts: rationale for a new approach to neuroprotective treatment of protein folding diseases. Exp Biol Med (Maywood) 2014;239:1405–1413. doi: 10.1177/1535370214539444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pusic KM, Pusic AD, Kraig RP. Environmental enrichment stimulates immune cell secretion of exosomes that promote CNS myelination and may regulate inflammation. Cell Mol Neurobiol. 2016;36:313–325. doi: 10.1007/s10571-015-0269-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rothhammer V, Quintana FJ. Role of astrocytes and microglia in central nervous system inflammation. Introduction. Semin Immunopathol. 2015;37:575–576. doi: 10.1007/s00281-015-0521-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ryu HW, Park CW, Ryu KY. Disruption of polyubiquitin gene Ubb causes dysregulation of neural stem cell differentiation with premature gliogenesis. Sci Rep. 2014;4:7026. doi: 10.1038/srep07026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scrivo A, Bourdenx M, Pampliega O, Cuervo AM. Selective autophagy as a potential therapeutic target for neurodegenerative disorders. Lancet Neurol. 2018;17:802–815. doi: 10.1016/S1474-4422(18)30238-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shrivastava AN, Triller A, Melki R. Cell biology and dynamics of neuronal Na(+)/K(+)-ATPase in health and diseases. Neuropharmacology. 2020;169:107461. doi: 10.1016/j.neuropharm.2018.12.008. [DOI] [PubMed] [Google Scholar]
- 37.Simchovitz A, Heneka MT, Soreq H. Personalized genetics of the cholinergic blockade of neuroinflammation. J Neurochem 142 Suppl. 2017;2:178–187. doi: 10.1111/jnc.13928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sofroniew MV, Vinters HV. Astrocytes: biology and pathology. Acta Neuropathol. 2010;119:7–35. doi: 10.1007/s00401-009-0619-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stuendl A, Kunadt M, Kruse N, Bartels C, Moebius W, Danzer KM, Mollenhauer B, Schneider A. Induction of α-synuclein aggregate formation by CSF exosomes from patients with Parkinson’s disease and dementia with Lewy bodies. Brain. 2016;139:481–494. doi: 10.1093/brain/awv346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Subhramanyam CS, Wang C, Hu Q, Dheen ST. Microglia-mediated neuroinflammation in neurodegenerative diseases. Semin Cell Dev Biol. 2019;94:112–120. doi: 10.1016/j.semcdb.2019.05.004. [DOI] [PubMed] [Google Scholar]
- 42.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, Jensen LJ, Mering CV. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tang Y, Le W. Differential roles of M1 and M2 microglia in neurodegenerative diseases. Mol Neurobiol. 2016;53:1181–1194. doi: 10.1007/s12035-014-9070-5. [DOI] [PubMed] [Google Scholar]
- 44.Tian F, Yang W, Mordes DA, Wang JY, Salameh JS, Mok J, Chew J, Sharma A, Leno-Duran E, Suzuki-Uematsu S, Suzuki N, Han SS, Lu FK, Ji M, Zhang R, Liu Y, Strominger J, Shneider NA, Petrucelli L, Xie XS, et al. Monitoring peripheral nerve degeneration in ALS by label-free stimulated Raman scattering imaging. Nat Commun. 2016;7:13283. doi: 10.1038/ncomms13283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vaquer-Alicea J, Diamond MI. Propagation of protein aggregation in neurodegenerative diseases. Annu Rev Biochem. 2019;88:785–810. doi: 10.1146/annurev-biochem-061516-045049. [DOI] [PubMed] [Google Scholar]
- 46.Wright-Jin EC, Gutmann DH. Microglia as dynamic cellular mediators of brain function. Trends Mol Med. 2019;25:967–979. doi: 10.1016/j.molmed.2019.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.You Y, Ikezu T. Emerging roles of extracellular vesicles in neurodegenerative disorders. Neurobiol Dis. 2019;130:104512. doi: 10.1016/j.nbd.2019.104512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zha Z, Gao YF, Ji J, Sun YQ, Li JL, Qi F, Zhang N, Jin LY, Xue B, Yang T, Fan YP, Zhao H, Wang L. Bu Shen Yi Sui capsule alleviates neuroinflammation and demyelination by promoting microglia toward M2 polarization, which correlates with changes in miR-124 and miR-155 in experimental autoimmune encephalomyelitis. Oxid Med Cell Longev. 2021;2021:5521503. doi: 10.1155/2021/5521503. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Immunocytochemical staining of astrocytes and microglia.
(A) Astrocytes confirmed by GFAP (red, stained by Cy3) immunocytochemical staining. GFAP was shown in the cytoplasm of the astrocytes, and blue staining represents the nucleus. (B) Microglia confirmed by Iba1 (red, stained by Cy3) immunocytochemical staining. Iba1 was shown in the cytoplasm of microglia, and blue staining represents the nucleus. Scale bars: 100 μm. DAPI: 4’,6-Diamidino-2-phenylindole; GFAP: Glial fibrillary acidic protein; Iba1: ionize calcium binding adapt molecule 1.
GO analysis of all differential genes in exosomes from astrocytes and microglia.
(A) The heatmap was based on folding changes (microglia relative to astrocytes), where log2(FC) > 2 were highlighted in red while the log2 (FC) < -2 were highlighted in green. Preprocession included log2 normalization and data centralization. (B) The volcano images showed that the blue dots represented down-regulated genes, while the red dots represented up-regulated genes. (C) The left bar chart of GO analysis showed that the darker the color, the more significant the enrichment. In the right bar chart, the blue represented MF, the green represented the CC, and the pink represented BP. (D) The pie chart demonstrated the proportion of CC, MF and BP. Their specific terms and the number of genes were distributed in the outer circle. (E) In bubble diagram of KEGG analysis, P-values indicated in red were significantly higher than those in blue and the sizes of dots were proportional to the number of enriched genes. P-value < 0.05. BP: Biological process; CC: cellular component; GO: Gene Ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes; log2(FC): log2(fold change); MF: molecular function.
mRNA-level differential analysis between exosomes from astrocytes and microglia.
(A) The heatmap was based on mRNA-level folding changes (microglia relative to astrocytes), where log2(FC) > 2 were highlighted in red while log2 (FC) < -2 were highlighted in green. (B) The volcano figure showed that the blue dots represented down-regulated genes, while the red dots represented up-regulated genes. (C) The left bar chart of GO analysis showed that the darker the color, the more significant the enrichment. In the right bar chart, the blue represented MF, the green represented CC and the pink represented the BP. (D) The pie chart demonstrated the proportion of CC, MF and BP. Their specific terms and the number of genes were distributed in the outer circle. (E) The bubble diagram showed KEGG analysis. P values indicated in red were more significant than those in blue. The sizes of dots were directly proportional to the number of enriched genes. Pre-processing included log2 normalization and data centralization. P value < 0.05 was statistically significant. BP: Biological process; CC: cellular component; GO: Gene Ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes; log2(FC): log2(fold change); MF: molecular function; mRNA: messenger RNA.
ncRNA-level and cirRNA-level difference analysis between exosomes from astrocytes and microglia.
(A) The heatmap were based on ncRNA-level folding changes (microglia relative to astrocytes), where log2 (FC) > 2 were highlighted in red while log2 (FC) < -2 were highlighted in green. (B) In volcano figure, the blue dots represented down-regulated genes, while the red dots represented up-regulated genes. (C) The heatmap was based on cirRNA-level fold changes (microglia relative to astrocytes), where log2 (FC) > 2 were highlighted in red while log2 (FC) < -2 were highlighted in green. (D) The network diagram showed that the blue dots represented down-regulated genes, while the red dots represented up-regulated genes. Pre-processing included log2 normalization and data centralization. P-value < 0.05 was statistically significant.) cirRNA: Circular RNAs; log2 (FC): log2 (fold change); ncRNA: non-coding RNA.
GO and KEGG analysis based on cirRNAs in exosomes from astrocytes and microglia respectively.
(A) The specific terms of cirRNAs in exosomes from microglia. The upper bar chart showed that the darker the color, the more significant the enrichment. The bottom bar chart showed that the blue represented MF, the green represented CC and the pink represented BP. (B) The pie chart demonstrated the proportion of CC, MF and BP in the inner circle. Their specific terms and the number of genes were distributed in the outer circle. (C) The bubble diagram showed the KEGG analysis. P values indicated in red were more significant than those in blue. The dots size were proportional to the numbers of enrich genes. (D) The specific terms of cirRNAs in exosomes from microglia. The upper bar chart showed that the darker the color, the more significant the enrichment. The bottom bar chart showed that the blue represented MF, the green represented CC and the pink represented BP. (E) The pie chart demonstrated the proportion of CC, MF and BP. Their specific terms and the number of genes were distributed in the outer circle. (F) The bubble diagram showed the KEGG analysis. P values indicated in red were significantly higher than those in blue. The sizes of dots were proportional to the number of enriched genes. P value < 0.05 was statistically significant. BP: Biological process; CC: cellular component; cirRNA: circular RNAs; GO: Gene Ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes; MF: molecular function.
Mining the top 50 hub mRNA from the astrocyte and the microglia using Cytoscape.
(A) The PPI network of the hub 50 astrocyte showed that the top 50 hub genes from the astrocyte (Figure 3A) were marked from red to yellow using Cytohubba (Radiality). (B) The PPI network of the hub 50 microglia showed that the top fifty hub genes from microglia (Figure 3B) were marked from red to yellow using Cytohubba (Radiality). The “UBB” node with the most direct interactors was highlighted in the brightest red and selected by the blue circle. Hub 50 astrocyte: the group contained the top 50 hub mRNAs from astrocyte exosomes, and its expression levels were greater than 20 FPKM. Hub 50 microglia: the group contained the top 50 hub mRNAs from microglia exosomes, and its expression levels were greater than 20 FPKM. FPKM: Fragments per kilobase of exon model per million mapped fragments; PPI: protein protein interaction network; UBB: ubiquitin B.