Abstract
Sulfonylurea receptor (SUR) belongs to the adenosine 5′-triphosphate (ATP)-binding cassette (ABC) transporter family; however, SUR is associated with ion channels and acts as a regulatory subunit determining the opening or closing of the pore. Abcc8 and Abcc9 genes code for the proteins SUR1 and SUR2, respectively. The SUR1 transcript encodes a protein of 1582 amino acids with a mass around 140–177 kDa expressed in the pancreas, brain, heart, and other tissues. It is well known that SUR1 assembles with Kir6.2 and TRPM4 to establish KATP channels and non-selective cation channels, respectively. Abbc8 and 9 are alternatively spliced, and the resulting transcripts encode different isoforms of SUR1 and SUR2, which have been detected by different experimental strategies. Interestingly, the use of binding assays to sulfonylureas and Western blotting has allowed the detection of shorter forms of SUR (~65 kDa). Identity of the SUR1 variants has not been clarified, and some authors have suggested that the shorter forms are unspecific. However, immunoprecipitation assays have shown that SUR2 short forms are part of a functional channel even coexisting with the typical forms of the receptor in the heart. This evidence confirms that the structure of the short forms of the SURs is fully functional and does not lose the ability to interact with the channels. Since structural changes in short forms of SUR modify its affinity to ATP, regulation of its expression might represent an advantage in pathologies where ATP concentrations decrease and a therapeutic target to induce neuroprotection. Remarkably, the expression of SUR1 variants might be induced by conditions associated to the decrease of energetic substrates in the brain (e.g. during stroke and epilepsy). In this review, we want to contribute to the knowledge of SUR1 complexity by analyzing evidence that shows the existence of short SUR1 variants and its possible implications in brain function.
Key Words: brain edema, epilepsy, Parkinson's disease, stroke, sulfonylurea receptor 1, SUR1, traumatic brain injury, TRPM4
Introduction
The adenosine 5′-triphosphate (ATP)-binding cassette (ABC) transporters is a superfamily of membrane proteins that involve ATPase activity and the subsequent ATP hydrolysis to transport a diversity of substrates across the cell membrane. These proteins are evolutionarily conserved and widely distributed in different phyla. In the human genome, there are 48 genes coding for proteins of the ABC superfamily. The ABC transporters have been classified according to their function as exporters, importers, and non-transporters. In mammals, sequence homology and gene structure have also been used to group them into seven subfamilies (ABCA to ABCG) (Thomas et al., 2020). Under physiological conditions, most ABC transporters function either importing or exporting substances; however, there are exceptions, such as the sulfonylurea receptor (SUR), which is associated with ion channels and acts as a regulatory subunit determining the opening or closing of the channel (Tinker et al., 2018).
SUR is a paradoxical ABC transporter due to its function as a non-transporter. It has been widely described that SUR assembles with Kir6.2, an inward rectifier K+ channel, to establish KATP channels (Puljung, 2018). KATP channels are expressed in many cell types and tissues, including the brain, ovary, heart, kidney, skeletal, and smooth muscles, where synchronize cell metabolism with electrical activity regulating transmembrane potassium fluxes (Bal et al., 2018; Filipets et al., 2019; Kaya et al., 2019; Kim et al., 2020). Early stoichiometry studies indicated that a 1:1 ratio between Kir6.2 and SUR subunits is necessary for channel assembly. Therefore, the active KATP channel complex is a hetero-octamer of four Kir6.2 subunits arranged at the center of the complex, forming the channel pore and surrounded by four regulatory SUR subunits (Li et al., 2017) (Figure 1A). Intracellular nucleotides regulate the KATP channels, thus increasing ADP concentrations open the channel eliciting K+ efflux, membrane hyperpolarization, and inhibition of electrical activity. Conversely, ATP closes the channel by coupling in one of the four binding sites located in the Kir6.2 subunits. Although the mechanism is not yet fully elucidated, it has been proposed that SUR determines the KATP channel opening by Mg-ADP binding where Mg2+ is necessary to trigger a conformation change in SUR1 (Puljung et al., 2019) (Figures 1B and 2). On the other hand, sulfonylureas are compounds that inhibit the activity of the KATP channel expressed in pancreatic β cells, and according to their ability to stimulate insulin secretion, they have been used in the management of diabetes mellitus type 2. Second-generation drugs called glyburide or glibenclamide are 100 to 1000 more potent stimulators of insulin secretion. Regarding the specific mechanism of action, it has been suggested that sulfonylurea inhibition of KATP channels provokes membrane depolarization, activation of voltage-dependent Ca2+ channels, Ca2+ influx, and finally Ca2+-dependent insulin granule exocytosis (de Wet and Proks, 2015).
Figure 1.
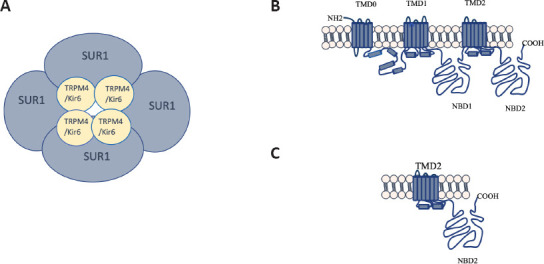
Bipartite structure of the SUR1 channels.
(A) Schematic representation of the SUR1 with the ATP dependent inwardly rectifying ion channel Kir6 and the TRPM4. The complex is integrated by four subunits of the ionic channel and four regulatory subunits SUR1. (B) Typical structure of the SUR1 protein with a molecular weight of 140 to 180 kDa that contains three TMD and two NBD. (C) Hypothetical structure of the SUR1 short form of 65 kDa, which conserve the NBD2. COOH: Carboxy-terminus; Kir6: inward rectifier potassium ion channel; NBD: nucleotide binding domains; NH2: amine-terminus; SUR1: sulfonylurea receptor 1; TMD: transmembrane domains; TRPM4: Transient receptor potential melastatin 4.
Figure 2.
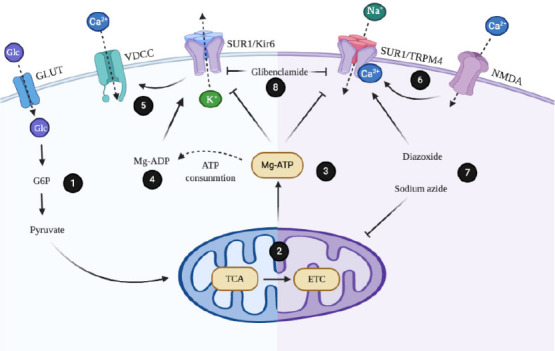
Regulation mechanisms of sulfonylurea receptor 1 (SUR1) channels.
(1) Glucose (Glc) capture by glucose transporters (GLUT) allows its conversion to glucose-6-phosphate (G6P) and pryruvate. (2) Metabolites that are products of glycolysis feed mitochondrial metabolism by entering to the cycle of the tricarboxylic acids (TCA) and the mitochondrial electron transport chain (ETC). (3) Mg-ATP generated during mitochondrial metabolism binds to SUR1 and blocks channels activity. (4) Generation of Mg-ADP displace Mg-ATP from its binding site and favors opening the potassium channel formed by the inwardly rectifying ion channel (Kir6) and the regulatory subunit SUR1. (5) Efflux of K+ favors cellular depolarization and voltage-gated Ca2+ channels (VDCC) opening. (6) Activity of the glutamate receptor N-methyl-D-aspartate (NMDA) in neurons favors internalization of Ca2+. This event activates the transient receptor potential melastatin 4 (TRPM4) and the regulatory subunit SUR1. (7) Diazoxide directly activates SUR1-TRPM4 channel, while sodium azide interferes with mitochondrial metabolism, favoring drop of Mg-ATP. (8) Sulfonylureas such as glibenclamide block the activity of SUR1 channels.
Interestingly, reports have described that SUR is associated with and regulates non-selective channels permeable to monovalent cations (e.g., Na+, K+) such as the transient receptor potential melastatin-4 (TRPM4) (Woo et al., 2020). Similar to KATP channels, TRPM4 is a hetero-multimeric composed of the co-assembly of four pore-forming subunits and four SUR regulatory subunits (Figure 1A). Electrophysiological analysis has revealed that SUR-TRPM4 is a Ca2+-activated cation channel; therefore, intracellular Ca2+ opens the channel and consequently elicits Na+ influx. Also, it is reported that binding of intracellular ATP can modulate the TRPM4 activity and probably this modulation could be regulated by SUR. Importantly, TRPM4 dysfunction has been linked to pathological processes (Amarouch et al., 2020) (Figure 2).
There are two SUR isoforms named SUR1 and SUR2, which are encoded by the ABC subfamily C 8 (Abcc8) and 9 (Abcc9) genes, respectively. Two alternative splicing of the last exon of SUR2 gives rise to the SUR2A and SUR2B isoforms. These two peptides differ only in the last 42 amino acids and consequently are considered highly homologous (Yamada and Kurachi, 2005). SUR1 also has splicing isoforms; however, little is known about their identity. Therefore, this review analyzes current information about isoforms and the alternative splice variants for SUR1 to understand the relationship between diversity, complexity, and function. Importantly, SUR1 function is relevant in the central nervous system in some process of neurodegeneration and in some diseases, therefore it can be a target to induce neuroprotection. The purpose of the review was to show the evidence of the possible expression of short forms of SUR1 in the brain. This information might be relevant to understand SUR1 function in the damaged brain and help to resolve problems in forthcoming investigations.
Search Strategy and Selection Criteria
The literature selection in this review was done by exploring terms associated to SUR1 expression, its isoforms, and its function in the brain. An electronic search was performed using the MeSH terms: SUR, SUR1, SUR2, Sulfonylurea Receptors, Abcc8, ATP-binding cassette transporters, glyburide, glibenclamide, TRPM4, K(ATP), ion channel, protein isoforms, splice variant, expression, brain, heart, pancreas, metabolism, ischemic stroke, cerebral hemorrhage, intracerebral hemorrhage, traumatic brain injury, epilepsy, status epilepticus, Parkinson’s, Alzheimer’s, neurodegenerative disease, blood brain barrier, and its combination in the Google scholar and PubMed databases. The results were further screened by title and abstract. Because the first part of the review focused on the description of evidence associated to the identity of the 65 kDa SUR1 isoform, we included references published between January 1, 1980 and December 16, 2020. Additionally, a search for the association of SUR1 function with neurodegeneration and brain injury was performed between January 1, 2015 and December 16, 2020. The included articles were limited to those published in English and no books, symposiums, and conferences were included.
Sulfonylurea Receptor 1 Variants
The ABCC8 human gene comprises 39 exons distributed in 84,348 bp (Genebank NG_008867.1) which is comparable to the Abcc8 rat gene (Figure 3). The Abcc8 promoter contains G + C rich region with no TATA box; the first 173 base pairs of the 5’flanking sequence is sufficient for maximal promoter activity (Ashfield and Ashcroft, 1998). Sequences of the 5’ untranslated region of the SUR1 transcripts are identical between mouse insulinoma and mouse brain, indicating that at least pancreatic β-cells and the nervous tissue use the same transcription start site, suggesting a similar situation for normal tissue (Kim et al., 2002). The 4749-nucleotide transcript from Rattus norvegicus (NM_013039.2) is present in total RNA obtained from different tissues (Figure 3). Accordingly, a hybridization/RNase protection assay showed that in mouse many tissues such as the brain, heart, skeletal muscle, and pancreas, with exception of the liver, express abundantly a main transcript with the same extension. This SUR mRNA of complete size is expressed all over the brain, but the hippocampus and cerebellum showed the highest levels of hybridization in the adult brain (Hernández-Sánchez et al., 1997).
Figure 3.
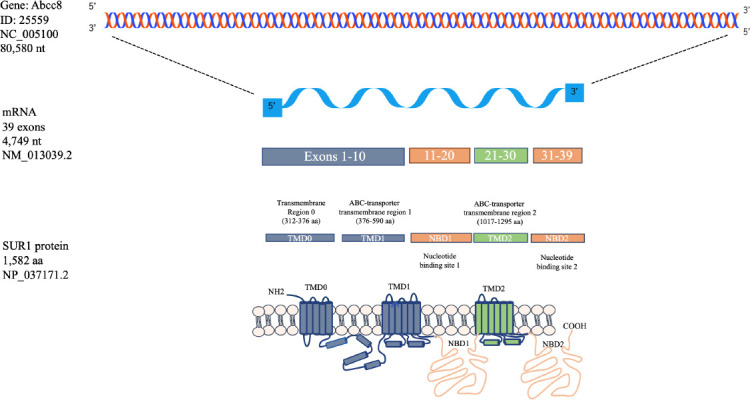
Graphic representation of the Abcc8 gene that encodes for the mRNA and protein of the sulfonylurea receptor 1.
The gene Abcc8 codes for an mRNA of 39 exons. Exons 1 to 10 code the two first TMD. The first NBD1 is included in the exons 11 to 20 while the NBD2 is included in the exons 31 to 39. The third TMD is coded in the exons 21 to 30. Abcc8: ATP-binding cassette transporter subfamily C member 8; COOH: carboxy-terminus; NBD1: nucleotide-binding domain; NH2: amine-terminus; TMD: Transmembrane domains.
The open reading frame of rat SUR1 cDNA encodes a protein of 1582 amino acids with a mass of 177,102 Da (Figure 3). This is consistent with the size of the complete SUR1 protein obtained from native β-cells although larger than expected (Aguilar-Bryan et al., 1995). In the β-cell line MIN6, gel filtration chromatography and SDS-PAGE displayed a molecular weight of approximately 140–170 kDa which are differentially glycosylated forms (Nelson et al., 1996). Furthermore, several SUR1 spliced variants with around 4,720 nucleotides in size have been identified (Additional Table 1). Interestingly, many tissues express shorter species of the protein which identity is unclear. Some tissues like the heart and pancreas express the mRNA and protein of the complete size, and smaller proteins; however, the variants function has not been characterized (Huang et al., 2019). Therefore, sometimes it has been considering that unconventional bands detected by antibodies or other techniques represent unspecific signals.
Additional Table 1.
Variants of SUR1
| Variant name | Tissues expressing SUR1 | Modification | Isoform size (Nucleotides) | Reference |
|---|---|---|---|---|
| SUR1 | Brain, heart, lung, skeletal muscle, kidney, stomach, pancreas, and liver | None | 4749 | Hernández-Sánchez et al. , 1999 |
| SUR1A2 | Brain, heart, skeletal muscle, pancreas, thymus, and testis | Single amino acid substitution in the NBD1 (Thr699Ile) | 4749 | Gros et al., 2002 |
| SUR1BΔ31 | Brain, heart, kidney, pancreas, spleen, thymus, testis, and intestine | Deletion of TM 16 – 17 | 4635 | Gros et al., 2002 |
| mSUR1Δ33 | Heart, midbrain, and hypothalamus | Frame shift and deletion at NBD2 | 4619 | Sakura et al., 1999 |
| rSUR1Δ17 | Brain, heart, lung, skeletal muscle, kidney, stomach, pancreas, and liver | Deletion at NBD1 | 4720 | Hambrock et al., 2002 |
| rSUR1Δ19 | Brain, heart, lung, pancreas, and liver | Deletion at NBD1 | 4651 | Hambrock et al., 2002 |
| rSUR1Δ17/Δ19 | Capillaries and cardiomyocytes | Deletion at NBD1 | 4622 | Hambrock et al., 2002 |
| rSUR1C | Brain, atrium, ventricle, lung, liver, kidney, and skeletal muscle | A truncated C-terminal fragment | 984 | Hambrock et al., 2002 |
| hSUR1Δ2 | Pancreas, brain, kidney, heart, skeletal muscle, testis, uterus, colon, spinal cord, and stomach | Lacks NBDs and sulfonylurea binding sites | 4608 | Schmid et al., 2012 |
NBD: Nucleotide-binding domain; SUR1: sulfonylurea receptor 1; TM: transmembrane segments.
The simpler splice variant of SUR1 was obtained from total RNA from pancreas/islet of Langerhans of rat (RINm5F cells). This novel isoform, named SUR1A2 (Genbank X97279), displays a single nucleotide polymorphism and the protein differs from SUR1 by the substitution of an isoleucine for the threonine at position 699. This mutation is in the NBD1, just before the Walker A motif. Remarkably, the human SUR1 has an isoleucine at position 699. Co-expression of Kir6.2 and SUR1A2 allowed to demonstrate that the variant can form functional KATP channels (Gros et al., 2002). Similarly, a SUR1 sequence (GenBank X97279) with four aminoacids substitutions (T487S, P835Q, G836R and G1313R) and the insertion of a serine after S741 was described in the rat hypothalamus but was not considered a true variant because it has the functional properties to SUR1 (Sakura et al., 1999).
Of the 39 exons that integrate the Abcc8 gene, only those found within the coding region of the NBD1 and 2 can be spliced out without generating a frame shift. Three splice forms originated from alternative splicing of the region coding for the NBD1 (exons 16–20) have been found in RNA obtained from guinea pig tissues including ventricular cardiomyocytes and capillary endothelial cells. These are the splice forms lacking exons 17 (SUR1-17, in which 36 nucleotides are spliced out), 19 (SUR1-19, in which 99 nucleotides are spliced out), or both (SUR1-17, 19). Transcripts of SUR1-17 and SUR1-19 showed a widespread distribution, whereas SUR1-17, 19 was found exclusively in cardiomyocytes. To evaluate the binding activity to glibenclamide, the previously characterized cassettes Δ17 and Δ19 were subcloned in the rat SUR1/pBF1 vector. Only rat SUR1 and rat SUR1 17 exhibited high affinity for glibenclamide suggesting the importance of exon 19 for the binding. No currents were detected in path clamp assays performed in cells transfected with Kir6.2/rSUR1 Δ19, demonstrating the role of this exon in the functionality of the channel (Hambrock et al., 2002).
A variant named SUR1BΔ31 lacks exon 31 (113 nucleotides) which does not affect the open reading frame and encodes a 1544 amino acid protein with a calculated molecular weight of 173.3 kDa (Genbank AF039595). The hydropathic analysis revealed that the deletion of exon 31 provokes the loss of the transmembrane spanning helices (TM) 16 and 17 without affecting the NBD2. However, SUR1BΔ31 co-expressed with Kir6.2 in Xenopus oocytes or HEK-293 cells does not induce KATP currents. This fact is probably associated to an impairment in the membrane trafficking or interaction between subunits. The deletion changes significantly the glibenclamide binding reducing the affinity ~500 times (Gros et al., 2002). A similar protein named SUR1bΔ33 lacking exon 33 (131 nucleotides) was isolated from rat and mouse ventromedial hypothalamus. The deletion is located before the NBD2 and introduces a frameshif at aminoacid 1330 originating a truncated protein with the complete elimination of the NBD2. This modification does not inhibit the interaction with Kir6.2 but reduces the sensitivity to the inhibitory effect of ADP and diazoxide. Furthermore, the framshift adds 25 residues to the C-terminus that reduces the ATP sensitivity (Sakura et al., 1999).
On the other hand, a cardiac ventricle cDNA library was screened with a fragment of the COOH-terminal of the guinea pig SUR1 (gpSUR1) and a modified splice variant named SUR1c (COOH-terminal SUR1-fragment) was found. The gpSUR1 cDNA contains 1663 nucleotides in length (GenBank AF183921). The 5’ noncoding region (1–556) is an unknown sequence followed with the known sequence of exon 28. The putative coding region includes exons 31 to 39 (557–1552 nucleotides) with a putative start-codon in exon 31. The gpSUR1 is transcribed in a short mRNA of 984 nucleotides that allows synthesis of a predicted molecular weight protein of 36 kDa. SUR1c is highly expressed in the atria and ventricles of guinea pig heart. The sequence probably contains the last transmembrane regions and the COOH-terminal domain of gpSUR1. Experiments have showed that channels transfected with Kir6.2/gpSUR1C did not display currents (Hambrock et al., 2002).
Likewise, an alternatively spliced form of SUR1 lacking exon 2 (SUR1Δ2) has a size of 4608 nucleotides. Omission of exon 2 causes a frame shift and an immediate stop codon in exon 3 leading to translation of a 5.6 kDa peptide that comprises the N-terminal extracellular domain and the first transmembrane helix of SUR1 (Schmid et al., 2012).
In summary, the SUR1 variants have been characterized at genomic level and described in diverse tissues including the brain and heart (Additional Table 1). SUR1 represents a small group of proteins with molecular weights oscillating around 140 kDa (the immature glycosylated form) except for the 5.6 and 30 kDa isoforms. Remarkably, the spliced forms in the NBDs have been analyzed because the sequence of the loops is involved in the pharmacological properties of the channels. These studies described the electrophysiological properties of the spliced forms using cloning techniques but avoiding the detection of the endogenous protein; therefore, some information is missing. The use of technics such as binding assays to sulfonylureas and Western blotting has allowed the detection of shorter forms of the protein SUR1 which has not been described.
The 65 kDa Short Form of Sulfonylurea Receptor
Results obtained from binding analysis of glibenclamide analogues have shown the existence of diverse putative SUR binding sites in β-cell membranes (Additional Table 2). Exposure of insulinoma β-cell membranes to [³H] glibenclamide followed by long-wavelength UV light irradiation results in the covalent labeling of two proteins with an apparent molecular mass of 30 and 140 kDa on SDS/PAGE. The radioactivity was measured in liquid scintillation counting after the protein contained in the sliced pieces of gels was digested. The 140 kDa was the minor component but the binding to [³H] glibenclamide was saturable and could be displaced in a concentration-dependent manner by other sulfonylureas (i.e., unlabeled glibenclamide or tolbutamide). The photolabeling of the 33 kDa protein was not significantly affected (Kramer et al., 1988).
Additional Table 2.
Commercial antibodies directed to SUR1
| Catalogue number | Trademark | Description | Specificity | Biological sample expressing SUR1 (used as positive control) | Size of the peptides identified (kDa) | Inmunogen and epitopes |
|---|---|---|---|---|---|---|
| ab134292 | Abcam, Cambridge, United Kingdom | Mouse monoclonal | SUR1, does not cross react with SUR2B | Rat brain membrane lysate | 177 | Cytoplasmic C terminal amino acids 1548- 1582 of rat SUR1 |
| ab217633 | Abcam, Cambridge, United Kingdom | Rabbit policlonal | SUR1 | Rat hippocampus lysate | 177 | C-terminal of the human protein sequence |
| ab77478 (discontinued) | Abcam, Cambridge, United Kingdom Merck Millipore, | Goat polyclonal | SUR1 | Human cerebellum lysate | 177 and 23 | NP_000343.2 |
| AB2241 (discontinued) | Burlington, Massachusetts, United States Merck Millipore, Burlington, | Rabbit polyclonal | SUR1 | Rat P1 Heart lysate | 60, 70, and 90 | Cytoplasmic domain |
| AB2242 (discontinued) | Massachusetts, United States Abgent, San Diego, California, United | Rabbit polyclonal | SUR1/2 | Rat pancreas lysate | 70 | Cytoplasmic domain |
| ASM10266 | States FabGennix, Frisco, | Mouse monoclonal | SUR1/2 | Rat Brain | 175,75, and 65 | Cytoplasmic C-terminus of rat SUR2B Synthetic peptide corresponding to |
| sur1-101AP | Texas, United States Merck Millipore, Burlington, | Rabbit polyclonal | SUR1 | Purified Protein | 100 and 180 | positions 1560-1582 |
| MABN501 (discontinued) | Massachusetts, United States Santa cruz Biotechnology, Santa Cruz, California, | Mouse monoclonal clone N289/16 | SUR1 | Mouse hypothalamus tissue lysate | 180 and ~68 | C-terminus of SUR1 |
| sc-293436 | United States StressMarq Biosciences, Victoria, | Mouse monoclonal | SUR1 | Recombinant protein | 40 | Amino acids 611-710 |
| PerCP | Canada Thermo Fisher Scientific, Waltham, Massachusetts, United | Mouse monoclonal | SUR1 | Rat brain | 160 | Cytoplasmic C-terminus amino acids 1548-1582 of a fusion protein of rat SUR1 |
| PA5-42392 | States Thermo Fisher Scientific, Waltham, Massachusetts, United | Rabbit polyclonal | SUR1 | Human 293T cell lysate Human placenta tissue, rat brain | 150, 45, and 33 | Synthetic peptide directed towards the N- terminal of human Abcc8 |
| PA5-78696 | States | Rabbit Polyclonal | SUR1 | tissue, and mouse brain tissue lysate | 180 | Synthetic peptide corresponding to a sequence of human SUR1 |
SUR1: Sulfonylurea receptor 1.
Furthermore, a sulfonylurea binding protein of approximately 65 kDa was first detected in the pancreatic β-cell line obtained from the hamster insulin-secreting tumor (HIT) T15 and normal rat pancreatic islets. The binding of [3H] glibenclamide to solubilized membranes was measured after separation of the bound and free glibenclamide by rapid gel filtration column (Sephadex G-50). The recovered in the eluate was superior compared to that obtained with the conventional gel filtration method. The binding of [125I]-glibenclamide to the microsomal preparations or solubilized membranes allowed the detection of a unique protein of 65 kDa that showed dose-dependence and a saturable binding site with a Kd value of 3.3 nM and Bmax of 90 fmol/mg of protein. The unlabeled glibenclamide, tolbutamide, and meglitinide compete with the binding of [125I]-glibenclamide and glibenclamide decreased the photolabeling of the 65 kDa protein; moreover, albumin was not associated to glibenclamide under the conditions used, all data indicative of a specific binding (Niki et al., 1991). This finding suggested that the 65 kDa protein constitutes the sulfonylurea receptor; however, this proposal differs from the initial idea that the receptor consisted of a 140 kDa protein. Therefore, authors proposed that 140 kDa protein could be a dimer of the 65 kDa receptor; although it is a possibility, subsequent analysis demonstrated that the complete protein is indeed expressed (Skeer et al., 1994).
However, it appears that the specific conditions of the assays change the protein species that can be detected. Remarkably, the quantitative photolabeling method with some modifications in the wavelength energy (i.e., length of time) and electrophoresis conditions allow the detection of four major proteins with a relative molecular weight of 140, 65, 55, and 33 kDa with equal abundancy (0.5 pmol/mg of membrane protein) in cell membranes of HIT cells. Glyburide specifically associates with high or low affinity to these proteins (Kd = 7 nM and 16 μM) (Nelson et al., 1992). According to a previous study, the highest affinity receptor corresponds to the 140 kDa species (Aguilar-Bryan et al., 1990). Interestingly, the amount of this protein was reduced significantly after boiling the samples in standard Laemmli SDS buffer at pH 6.8; this effect is avoided by eliminating the use of HCl to adjust the pH. Additionally, high energy (> 0.4 to 0.8 J/cm2) also occasioned disappearence of the 140 kDa species (Nelson et al., 1992). These results suggested that the 65 kDa species could be product from proteolysis of the 140 kDa form; nevertheless, this idea was discarded by posterior findings. Even though the amount of the 140 kDa protein varies significantly depending on photocoupling and the electrophoresis conditions, the appearance of the ~65 kDa band did not correlate with the disappearance of the ~140 kDa band; additionally, protease inhibitors do not inhibit the appearance of the ~65 kDa band (Aguilar-Bryan et al., 1990; Kramer et al., 1994). Importantly, experiments performed by enzymatic digestion (Matsuo et al., 2000) suggest that if a SUR1 protein of 65 kDa that contains the COOH terminal is expressed, it will form functional channels.
Since sulfonylureas have different binding sites in the receptor or different binding affinities, the photolabeling of β-cell membrane proteins with [3H]-glibenclamide reveled binding to two polypeptides of 140 and 33 kDa. In contrast, [3H]-glimepiride exclusively bind to a 65 kDa polypeptide, while two photolabile sulfonylureas N3-[3H]33055 and [125I]-35623 recognizing a 33 and 65 kDa proteins, respectively. The specificity was corroborated by competitive assays in the presence of excess of unlabeled molecules. Interestingly, solubilization of membranes with non-ionic detergents (i.e., Triton X-100 or CHAPS) increased photoaffinity labeling of the 65 kDa by [3H]-glimepiride. In contrast, the 140 kDa protein was no longer labeled with [3H]-glibenclamide in solubilized membranes, instead the 65 kDa protein was labeled. Competitive binding assays suggested that solubilization dissociate the putative sulfonylurea receptor into monomeric 65 and 140 kDa proteins and support the hypothesis that the functional receptor is composed of subunits (Kramer et al., 1994).
In solubilized proteins obtained from microsomes of the pig cerebral cortex, the photolabeling technique allowed the finding of at least six membrane proteins. Quantitative analysis showed that 9.2 ± 0.6% of the binding sites occupied by the glibenclamide analogue N-[4-[2-(4-azido-2-hydroxy-5-iodobenzamido)ethyl] benzenesulfonyl]-N′-cyclohexylurea ([125I]-N3-GA) were incorporated into the ~175 kDa protein. This polypeptide could represent the high-affinity sulfonylurea receptor, because photoincorporation was inhibited by other sulfonylureas: 0.1 μM glibenclamide, 1 μM glipizide, and 1 mM toltutamide. Dissociation experiments also showed that 24 to 28% of binding of [125I]-N3-GA is displaced by 1 µM glibenclamide. However, binding inhibition was not observed with the proteins of 94, 66, 55, 43, and 33 kDa which might represent nonspecific photoincorporation of the radioligand, because they were still labeled in the presence of 100 nM glicenclamide. These results again suggested that 175 kDa protein was the specific receptor to sulfonylurea (Schwanstecher et al., 1994). However, results could be associated to the different binding affinities of the SUR species to sulfonylureas.
A decrease in the expression of the 100 and 140 kDa peptides has been demonstrated in the double-knockout (SUR1–/–) mice. This strategy did not abolish the expression of the 65 kDa protein, suggesting lack of specificity for it; though, its expression was decreased. A region of approximate 1000 bp including the proximal promoter and the exon 1 of the Abcc8 gene was removed in the knockdown model. This strategy eliminates the transcription start site; therefore, the mRNA should not be detected, nevertheless the SUR1 mRNA levels remain unchanged. To explain the persisted mRNA, it was proposed the existence of an alternative start site for transcription, but this still needs confirmation. Moreover, in a transgenic mouse overexpressing SUR1 was found that bands of 140 and 100 but also the 65 kDa increased its expression (Flagg et al., 2009). These results suggest that expression of the 65 kDa protein depends on Abcc8 gene transcription.
Several commercial SUR1 antibodies detect bands of different molecular weights; however, these bands have been suggested to be non-specific (Additional Table 3). With this argument, antibodies have been extensively verified for specificity (Flagg et al., 2009). Despite the efforts, some antibodies continue recognizing bands of 140, 100, and 65 kDa in diverse tissues. On the other hand, most recently published studies mention the presence of a 150 to 177 kDa SUR1 protein; however, the blot is generally trimmed (Additional Table 4). This situation likely accounts for our failure to exhibit SUR1 short form expression. All this information suggests the existence of sulfonylurea receptors of different sizes which function has not been described but represent an issue for future research.
Additional Table 3.
Union of radioactive compounds to SUR1 variants
| Compound | Photolabeled proteins (kDa) | Reference | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| 140–180 | 90–100 | 65–70 | 50–55 | 33–40 | ||
| [γ-32P]-ATP | + | + | + | + | + | Matsuo et al., 2000 |
| [3H]-Glibenclamide | + | + | - | + | Nikiki et al., 1991; Kramer et al., 1994; Braun et al., 1997; Barg et al., 1999 | |
| [3H]-Glimepiride | - | + | - | - | Kramer et al., 1994 | |
| [125I]-35623 | - | + | - | Kramer et al., 1994 | ||
| N3-[3H]-33055 | - | - | - | + | Kramer et al., 1994 | |
| 5-[125I]-Iodo-2-hydroxyglybenclamide | + | + | + | + | + | Aguilar-Bryan et al., 1990; Nelson et al., 1992 |
| [251]-N3GA | + | + | + | + | + | Schwanstecher et al., 1994 |
Additional Table 4.
SUR1 detected in diverse experimental models
| Experimental model | Method of detection | Antibody anti-SUR1 | Size of SUR1 (kDa) | Observations | Reference |
|---|---|---|---|---|---|
| Middle cerebral artery occlusion | qPCR, WB, IH | SantaCruz Biotechnology, SantaCruz, California, UnitedStates (SC-5789) | 65* | Resveratrol prevents the upregulation of the mRNA and protein of SUR1which is induced bymiddle cerebral artery occlusion | Alquisiras-Burgos et al., 2020 |
| Malignant infarction of the middle cerebral artery in a porcine model | IH | SantaCruz Biotechnology, SantaCruz, California, UnitedStates | N.A. | SUR1was expressed in astrocytes, neurons and capillary endothelial cells and was inducedafter occlusion in the penumbra and core regions. TRPM-4showed a moderate expression | Arikan et al., 2017 |
| Brainstem coronalslices containingthe ventral cochlear nucleus from mice BALB/c strain | qPCR, WB, and IH | SantaCruz Biotechnology, SantaCruz, California, UnitedStates (SC-5789) | 160* | Strongstraining in cell bodies involving cytoplasm and cellular membrane. Majority of stellate neurons of the ventral cochlear nucleus express functional KATP channels | Bal et al., 2017 |
| Human (diabetic patient), african green monkey, cynomolgus macaque, and rodent retina. Neonatal hyperglycemia | IH | N.A. | N.A. | SUR1was expressed in retina, enriched inthe macula and colocalized withTRPM4 andKir6.2. Glibenclamide acts as a neuroprotectant usingvarious experimentalmodels thatduplicate neurodegeneration inretina | Berdugo et al., 2021 |
| Brain mitochondria from Sprague-Dawley rats | WB | SantaCruz Biotechnology, SantaCruz, California, UnitedStates (SC-5789) | 13, 30, 45, and 97 | Anti-SUR1antibody recognized several bands inbrain homogenate and mitochondrialsample. The 45and 97kDa bands were eliminated withthe blocking peptide. The methods used could not confirm the presenceof anmitoKATP channel | Brustovetsky et al., 2005 |
| Brain edema induced after subarachnoid hemorrhage in rats | WB, IH | Abcam,Cambridge, UnitedKingdom (ab134292) | 177* | Pituitaryadenylate cyclase-activating polypeptide knockout-induced brainedema aggravated the increase in SUR1and AQP4. SUR1 was barely expressed in the brain of sham groups, increased after 3 h, peaked at 24h and return to basalafter 72h of subarachnoid hemorrhage. Its expression increased on neurons,astrocytes and endothelialcells | Fang et al., 2020 |
| Human tissue from multiple sclerosis patients. Experimental autoimmune encephalomyelitis in murine model. TNF+ IFNγ stimulationin primary astrocyte cultures | IH, qPCR | Custom | N.A. | Quiescent astrocytes donot express SUR1 butthe mRNA of SUR1 and TRPM4were upregulated by TNF+ IFNγ treatment. Similarly, activewhite matter multiple sclerosis lesions showed SUR1 expression astrocytes but not microglia/macrophage cells | Gerzanichet al., 2017 |
| Immortalized murine and humanbrain endothelial cells. Activation by TNF exposure | IH, qPCR, WB | Custom | 150 -170* | Activation induced the upregulation of SUR1 and TRPM4. SUR1 inhibition didnot affect secretion of MMP-9 | Gerzanichet al., 2018 |
| Spinal cord injury | RT-PCR, WB | SantaCruz Biotechnology, SantaCruz, California, UnitedStates | Not indicated* | Spinal cord injury increased mRNA of SUR1and TRPM4 andpeaked after 1 and 8 h, respectively. Two forms of SUR1 andTRPM4 proteins were detected (mature and immature associated tothe glycosylated state). Ghrelin treatment inhibitedincreases | Lee et al., 2014 |
| MPTP-induced Parkinson disease in C57BL/6 mice | qPCR, WB, IH | Abcam,Cambridge, UnitedKingdom (ab32844) | 170* | MPTP increased 31% the mRNA and protein level of SUR1 | Gonget al., 2014 |
| Asphyxial cardiac arrest/ cardiopulmonary resuscitation (ACA/CRP) inSprague- Dawley rats | WB, IH | CWBIO, Beijing, China | 170* | SUR1and TRPM4 were upregulated after 24 h of ACA/CPR incortex and hippocampus. Target temperature management reduced SUR1 and TRPM positive cells (neurons, astrocytes, microglia and endothelial cells) | Huanget al., 2016 |
| Intracerebral hemorrhage in the autoblood-induced rat model | WB, IH, qPCR | Abcam,Cambridge, UnitedKingdom | 177* | SUR1and TRPM4 expression was increased in neurons and endothelialcells surroundingthe hematoma 24h after intracerebral hemorrhage, butnot Kir6.2 | Jianget al., 2017 |
| Cultured primary cholinergic neurons from cortex and hippocampus stimulated withAβ1-42 as model of Alzheimer’s disease | WB | N.A | 170* | Aβ1-42 upregulated the expression of kir6.2/SUR1through activation of NF-κB, p38MAPK andPKC after 72h. The NF-κB inhibitor SN50 decreased SUR1 expression | Li et al., 2019 |
| Renovascular hypertension in male Sprague-Dawley rats | WB | Abcam,Cambridge, UnitedKingdom (ab32844) | 170* | Baroreflex sensitivitywas increased byacute intravenous NaHS (a donor of Hydrogen sulfide) administrationto renovascular hypertensive rats. The increase was associated to upregulation of Kir6.2 and SUR1 | Li et al., 2020 |
| Model of status epilepticus inmale Sprague-Dawley rats | WB, qPCR, IH | CWBIO, Beijing, China | 170* | Protein and mRNA levels of SUR1 and TRPM4were increased in hippocampus and piriform cortex after 6 h of status epilepticus and lasted for 3 and 7 days, respectively. SUR1 was expressed in neurons and endothelialcells. Glibenclamide prevented the upregulation | Lin et al., 2017 |
| Rat model of diabetes mellitus induced by streptozotocin and subjected topermanent middle cerebral artery occlusion | WB | SantaCruz Biotechnology, SantaCruz, California, UnitedStates (SC-5789) SantaCruz Biotechnology, | Not indicated* | Liraglutidereduced neurological deficits induced bycerebral ischemia and decreased the expression of SUR1 and Kir6.2 inmitochondria | Shiet al., 2019 |
| Hypoxia inPC12 cells | IH | SantaCruz, California, UnitedStates | N.A. | Hypoxia increased Kir6.2 and SUR1 mRNA and protein expression | Singh et al., 2019 |
| SH-SY5Y cell line exposed to manganese to simulatethe effect observed in Parkinson disease | WB | N.A | Not indicated* | Manganese repressed GABA A receptors and induced GABA B receptors. Additionally, Kir6.1 andKir6.2 decreased, but SUR1 expression gradually increased and SUR2 did notchange. These changes were foundas potential factors for the secretion of α- synuclein | Sunet al., 2020 |
| Tissue from Hirschsprung’s Disease patients | WB, IH | Abcam,Cambridge, UnitedKingdom | 175* | Kir6.1,Kir6.2, SUR1, and SUR2 were expressed in the humancolon. Their expression decreased inthe ganglionic bowel | Tomuschatet al., 2016 |
| Middle cerebral artery occlusion in Sprague-Dawley rats | WB | Abcam,Cambridge, UnitedKingdom | 178* | Middle cerebral artery occlusion increased SUR1protein levels but neither delayed hypothermia nor glibenclamide prevented this up-regulation | Wu et al., 2016 |
| Global brain ischemia produced by4-vessel occlusion | WB | Eter Life, Birmingham, UnitedKingdom | 175* | The expression of the two subunits of mitoKATP, SUR1and Kir 6.2 were increased by the pretreatment withthe hypobaric hypoxia that induced neuronal protection | Zhang et al., 2016 |
| Intracerebral hemorrhage | WB | Abcam,Cambridge, UnitedKingdom (ab32844) | Not indicated* | Expression of SUR1was upregulated in the perihematomaltissue 24 and 72h after intracerebralhemorrhage | Zhou et al., 2018 |
*Incomplete image of the blot that shows a band of around 150–170 kDa. AQP4: aquaporin; IF: Immunohistochemistry; IFNγ: interferon gamma; MAPK: mitogen activated protein kinase; MPTP: 1- methyl-4-phenyl-1,2,3,6-tetrahydropyridine; PKC: protein kinase C; qPCR: quantitative real-time reverse transcriptase PCR; SUR1: sulfonylurea receptor 1; TNF: tumor necrosis factor; TRPM4: transient receptor potential melastatin 4; WB: western blot.
Functional Implications of Sulfonylurea Receptor Short Forms
Experiments of enzymatic digestion of the [32P]-ATP labeled SUR1 allowed the functional analysis of the mutant SUR1-R1420C in hyperinsulinemic hypoglycemia (Matsuo et al., 2000). Based on this study, we hypothesized that a 65 kDa SUR1 peptide that conserves the COOH terminal will retain the nucleotide binding site, the ATPase activity and the sulfonylurea binding site, regions that are essential to form functional channels (Figure 1C). Although these data suggest the ability of the short form of SUR1 to interact with Kir6, the structural changes necessarily impact the protein function and drug susceptibility. For instance, glibenclamide interacts with the NBD1/TM12 located in the N-terminal and with the TM1 – 2 (Martin et al., 2017). Therefore, the 65 kDa that lacks the region proximal to the N-terminal (TM0, TM1, and NBD1) probably have different sensitivity to glibenclamide binding. Interestingly, this hypothesis is reproducible in a non-conventional form of 68 kDa of SUR2 which has been functionally characterized showing insensitivity to glibenclamide and increased sensitivity ATP in the KATP current (Pu et al., 2009). Remarkably, the 65 kDa SUR1 protein have been observed in brain pathologies where ATP decreases; for example, brain ischemia induces the overexpression of SUR1 short form of 65 kDa (Alquisiras-Burgos et al., 2020). This situation could represent an adaptation mechanism to regulate ionic channels activity under conditions of metabolic stress with decreased ATP concentrations. This evidence coincides with studies performed in cellular cultures where the expression of the SUR1 variants with high (140 kDa) or low affinity (65 kDa) to sulfonylurea depend largely on the culture conditions. Under high glucose (25 mM) HIT cells express both sulfonylurea receptors, with high (Kd = 1 nM) and low-affinity to glibenclamide (Kd = 100 nM–1 μM). While cells cultured in a low glucose medium (11 mM) express the low-affinity receptor and exhibit loss of the high-affinity sulfonylurea binding sites (Aguilar-Bryan et al., 1990; Rabalet et al., 1996). These results suggest that short forms variants of SUR1 might come from alternative splicing and could be depend on an exogenous stimulus.
On the other hand, immunoprecipitation assays have showed that the SUR2 short forms (65 and 28 kDa) are part of a functional channel even coexisting with the typical forms of the 140 kDa receptor (Pu et al., 2009). Likewise, different SUR forms can co-assemble into KATP channels and generate distinct metabolic sensitivities and pharmacological profiles. For example, under conditions in which different SUR subtypes (i.e., SUR1 and SUR2) are co-expressed; the proteins retain the ability to co-assemble in a functional channel complex that shows increased KATP currents (Chan et al., 2008). Similarly, an intra-exonic splice variant of SUR2A (55 kDa) expressed in the mitochondria of the heart and brain was found to be more resistant to Ca2+ inhibition and insensitive to glibenclamide (Aggarwal et al., 2013). These data indicate that co-assemble of the short forms of SUR1 might generate a channel with greater sensitivity to ATP but also a channel with a greater current capacity. This situation has important implications in pathologies where the short forms of SUR could participate in the ionic imbalance as occur in the brain subjected to damage (Alquisiras-Burgos et al., 2020).
Consequences Associated with Sulfonylurea Receptor 1 Expression in the Brain
Ionic imbalance is the beginning of a series of secondary damage events that result in edema formation in multiple brain diseases (Stokum et al., 2016). Ion channels and transporters (i.e., aquaporins, SUR1-TRPM4, chloride channels, glucose transporters, and proton-sensitive channels) mediate cerebral edema formation and have been investigated as potential therapeutic targets (Jha et al., 2020; Luo et al., 2020). KATP channels (e.g., SUR1-KIR6.2) or SUR1-TRPM4 channels have opposite effects on membrane potential. Therefore, edema formation is mediated by SUR1-TRPM4 activation and clearly associated to a high morbidity and mortality in large hemispheric infarction and severe traumatic brain injury (TBI) but also with other central nervous system conditions (Caffes et al., 2015; Bianchi et al., 2018). The only treatment for severe brain swelling is decompressive craniectomy, which has significant disadvantages for patients; consequently, finding compounds for prevention of cerebral edema is essential. Importantly, glibenclamide has shown effective results in animal models but also in phase 2 clinical studies in patients suffering from large hemispheric infarction (Pergakis et al., 2019).
Cerebral edema development results from activation of multiple pathways some of which involve SUR1 activity (Figure 4). SUR1 is not normally expressed in the central nervous system, although it is upregulated after injury and lead activation of the complex SUR1-TRPM4-AQP4 (Stokum et al., 2018; Gerzanich et al., 2019; Alquisiras-Burgos et al., 2020; Figure 4). In patients, changes in SUR1 expression are observed 48 to 72 hours after stroke and are linked with negative outcomes, while decreasing SUR1 is associated with negligible intracranial hypertension and positive outcomes. Interestingly, cellular edema can be detected as soon as 60 minutes post injury and remain up to 14 days. The peak of intracranial pressure is preceded by the SUR1 overexpression in 91.7% of patients. Thus, early SUR1 increases observed in cerebrospinal fluid may indicate intracranial hypertension and represent and perfect biomarker for cerebral edema formation (Jha et al., 2017). Additionally, serum SUR1 and TRPM4 levels are upregulated in the peripheral blood samples of patients with subarachnoid hemorrhage, supporting their utility as a therapeutic target (Dundar et al., 2020).
Figure 4.
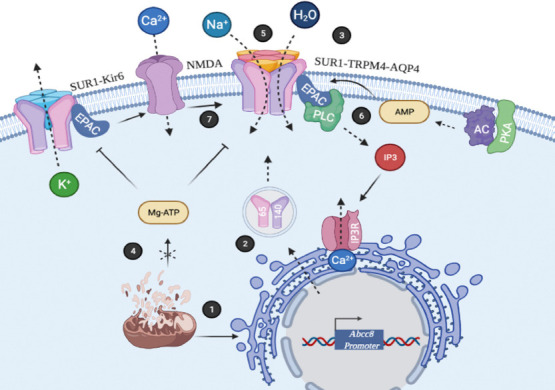
SUR1 overexpression in the brain.
(1) Metabolic stress stimulates Abcc8 gene expression. (2) Translation of the mRNA coding for SUR1 protein isoforms (140 and 65 kDa). (3) Assemble of SUR1-TRPM4-AQP4 complex. (4) Reduced levels of Mg-ATP of mitochondrial origin induce activation of SUR1. (5) The complex SUR1-TRPM4-AQP4 participates in the ionic imbalance and favors influx of water. (6) PKA/AC/EPAC/PLC axis induces IP3 production. Ca2+ release from the endoplasmic reticulum after IP3R stimulation regulates TRPM4 and Kir6 channels. (7) SUR1-Kir6 and EPAC modulates NMDAR activity. Abcc8: ATP-binding cassette transporter subfamily C member 8; AC: adenylate cyclase; AQP4: aquaporin 4; EPAC: exchange proteins directly activated by cAMP; IP3: inositol 1,4,5-trisphosphate; IP3R: inositol 1,4,5-trisphosphate receptor; Kir6: inward rectifier K+ channel; Mg-ATP: adenosine triphosphate; PKA: protein kinase A; PLC: phospholipase C; SUR1: sulfonylurea receptor 1; TRPM4: transient receptor potential melastatin-4.
Under physiological conditions, TRPM4 is constitutively expressed and involved in the regulation of Ca2+ entry. In neurodegenerative pathologies it is likely that the increase in its expression exist as an endogen mechanism to protect cells from the massive internalization of Ca2+ (Yan et al., 2020); however, the protective function is inadvertently, transformed into a mechanism of cellular damage. TRPM4 has been used as target to reduce edema and develop blockers for stroke management. TRPM4 upregulation in brain endothelium begins as soon as as soon as 2 hours post stroke-reperfusion in a rat model. Suppression of TRPM4 by treatment with siRNA reduced the infarct volume cerebral and edema suggesting that the blood brain barrier integrity is preserved (Chen et al., 2019a). Also, a TRPM4-specific antibody which inhibits the channel current by binding to the pore, reduces the TRPM4 surface location. This treatment prevents hypoxia-induced cell swelling and preservers blood-brain barrier integrity (Chen et al., 2019b). Similarly, in a model of status epilepticus the knockout of TRPM4 reduces cerebral edema (Chen et al., 2020).
Emerging evidence shows that astrocytes play important roles in the development of edema in cerebral ischemia. Astrocytes are responsible for the regulation of brain homeostasis and are capable to adjust energy production to the demand of neuronal activity (Felix et al., 2020). Basal level of TRPM4 expression is found in neurons and endothelial cells but not in astrocytes. Therefore, TRPM4 is a major responsible of cell swelling in these cells immediately after hypoxia even before the upregulation of TRPM4 induced by the middle cerebral artery occlusion model (MCAO) (Wei et al., 2020). However, TRPM4 overexpress in reactive astrocytes, which also contribute to cell swelling, showing its major resistance to initial conditions of ischemia (Stokum et al., 2018).
On the other hand, Kir6.1 is highly expressed in astrocytes in physiological conditions and the astrocytic Kir6.1 knockout exhibited larger infarct areas and more severe brain edema and neurological deficits in the MCAO (Zhong et al., 2019). Similarly, the use of antisense oligonucleotides directed against Abcc8 and Trpm4 on hemispheric swelling after permanent MCAO demonstrated that in post-ischemic tissues the infarct volume and swelling is reduced. In contrast, the antisense oligonucleotides directed against Kcnj8 (KIR6.1) and Kcnj11 (KIR6.2) have no effect on swelling (Woo et al., 2020). Likewise, after contusion expansion induced by TBI, TRPM4 and KIR6.2 are upregulated in astrocytes but only inhibition of SUR1 and TRPM4 reduces hemorrhagic progression of contusion (Gerzanich et al., 2019). These results clearly show that SUR1 and TRPM4 participate in edema progression and emphasize the importance to identify the SUR1 isoforms induced in brain subjected to damage.
Dopamine-releasing neurons in the Substantia nigra are susceptible to neurodegeneration, being the pathological hallmark of Parkinson’s disease. Interestingly, impaired function of ion channels contributes to their vulnerability. The KATP channels expressed in dopaminergic neurons of the Substantia nigra inhibit energy-demanding electrical activity and protect cells from overexcitability, particularly in conditions of metabolic stress. However, KATP channels have bidirectional effects that avoid chronic inhibition of dopamine release and facilitate the switch to N-methyl-D-aspartate (NMDA) glutamate receptor-mediated burst activity (Knowlton et al., 2018). Nevertheless, sustained activity in response to metabolic stress seems to trigger degeneration, since KATP knockout rescued dopaminergic neurons from degeneration (Duda et al., 2016). Interestingly, in human Parkinson’s disease, dopaminergic neurons express 2-fold higher levels of SUR1 and 10-fold the NMDA receptor subunit NR1 than in normal condition, consistent with high levels of burst activity in patients (Duda et al., 2016).
Additionally, iron metabolism is associated with dopaminergic neurons damage in Parkinson’s disease. Recently, it was demonstrated that Kir6.2 knockout suppressed the excessive iron accumulation in MPTP-treated mouse midbrain. This effect was associated to reduction in the expression of one of the components of ferritin, the main iron storage. Nevertheless, it was also found that glibenclamide inhibited the release of lactate dehydrogenase induced by MPP+ suggesting the involvement of SUR1 in the neuronal degeneration (Zhang et al., 2018). Interestingly, astrocytic Kir6.1 knockout mouse showed increased dopaminergic neuron loss in Substantia nigra compacta revealing that this subunit channel protects against neurodegeneration (Hu et al., 2019). These results suggest that KATP channels and SUR1 provide a targeting protective strategy for prevention of neurodegeneration in Parkinson’s disease and again expose the importance to identify the isoforms.
TRPM4 is an important regulator of membrane potential in excitable and non-excitable cell types and is an essential co-activator of the NMDA receptors (Menigoz et al., 2016). Recently, it was demonstrated that the NMDA receptor co-immunoprecipitated with TRPM4 in lysates from cultured mouse hippocampal neurons and brain lysates from the mouse hippocampus and cortex. Importantly, TRPM4-derived peptides that contain the cytoplasmatic N-terminal region of the TRPM4 fused to a glycosylphosphatidylinositol linker that mimic the native localization of the receptor achieve a strong neuroprotection in different models of acute neurodegeneration. These data revel that physical coupling of the receptors is associated to excitotoxicity and mediate the cellular death in the oxygen glucose deprivation model in cultured hippocampal neurons, in ischemic stroke induced by MCAO and in retinal ganglion cell degeneration induced by the intravitreal injection of NMDA (Yan et al., 2020). This information is relevant because TRPM4 is modulated by SUR1, which overexpression is induced under energy stress conditions; therefore, the sensibility of cells to damage could be associated to expression of the SUR1 isoforms (Figure 4).
SUR1 is also an active participant in epilepsy manifestation in different interesting ways. Intracellular cAMP is generated from ATP by the action of adenylate cyclase. The exchange proteins directly activated by cAMP (EPAC1 and EPAC2) are one type of the cAMP effector proteins expressed in mammals. Epac1 cDNA reveals two glucose responsive elements and hyperglycemia stimulates transcription and translation of Epac1. Additionally, the human EPAC1 promoter also contains a hypoxia responsive element and hypoxia enhances Epac1 expression in mouse primary cortical cells (Robichaux and Cheng, 2018). EPACs are expressed in pancreatic β-cells, where are associated to KATP channels and modulate Ca2+-dependent secretion of insulin; in these cells, SUR1 selectively activates Epac2 isoform (Herbst et al., 2011). Similarly, EPACs regulate ion channels to depolarize or hyperpolarize neurons. For example, EPAC enhances the activity of the TRPM4 through Ca2+ released from inositol 3-phosphate stores (Robichaux and Cheng, 2018). Accordingly, in the hippocampus and prefrontal cortex, EPAC2 regulates KATP channel open probability via a direct inhibition of the SUR1 receptor. Furthermore, ablation of the SUR1 receptor that intercepts EPAC binding inhibits glutamate release and reduces seizure vulnerability in mice (Zhao et al., 2013). In a different experimental model, it was observed that picrotoxin-kindling convulsions decrease amount of Kir6.2 and SUR1 mRNAs in the dentate gyrus. In contrast, when seizures are re-induced, both subunits were transiently up regulated indicating that KATP channels in brain enhance seizure susceptibility and alter seizure propagation of chronic epilepsy (Köhling et al., 2016). All these data exhibit the importance of SUR1 function in the regulation of epilepsy, a severe neurological disorder associated with an increased glutamate release (Figure 4). Furthermore, there are other inducers, such as post-traumatic epilepsy, which is caused by TBI. This evidence suggests involvement of SUR1 and TRPM4 in a complicated manner. For instance, in a mouse model of status epilepticus (behavioral seizures induced by lithium and pilocarpine), the knockout of TRPM4 preserve blood-brain barrier integrity and reduces cerebral edema having as consequence the improvement of neurologic outcome and reduction of mortality. At cellular level TRPM4 absence diminished neuronal loss and astrocytosis in the hippocampus and piriform cortex (Chen et al., 2020).
Besides, metabolic abnormalities are linked with an augmented risk of epilepsy development. Combination of diverse factors associated to mitochondrial dysfunction and metabolic alterations elicits a compromised supply of energy in the brain that precede to epileptic seizures. Glucocorticoid receptors regulate hypothalamus-pituitary-adrenal axis and mechanisms related to metabolic function; interestingly, glucocorticoid metabolism is altered during epileptogenesis. After pilocarpine administration, a model of temporal lobe epilepsy, rats become rigorously obese and showed substantial modifications in the hippocampal expression level of genes that are involved in energy metabolism and glucocorticoid regulation. Among the genes altered, the Abcc8 mRNA levels are downregulated and Kcnj11 levels tend to increase (Kundap et al., 2020). This situation is supported by the observation that epilepsy is positively modulated with diets such as the ketogenic diet or caloric restriction which produce a hyperpolarization mediated by the KATP channels (Rubio et al., 2020). These data show the complex interaction of SUR1 proteins with ion channels and the importance of their study during brain disease.
Conclusion
SUR1 function in the central nervous system has not been described in detail. Although evidence indicates that the short forms of SUR integrate functional channels, no proofs for the short SUR1 protein function exist. Multiple attempts have been made to demonstrate the identity of the 65 kDa protein but further experiments are necessary to clarify SUR1 isoforms participation in the brain. Based on the behavior of short forms of SUR2 and the detection of SUR1 overexpression in cerebral stroke, we propose that short variants of SUR1 have functional consequences in the brain during development of disease.
Additional files:
Additional file 1: Open peer review report 1 (95.6KB, pdf) .
Additional Table 1: Variants of SUR1.
Additional Table 2: Commercial antibodies directed to SUR1.
Additional Table 3: Union of radioactive compounds to SUR1 variants.
Additional Table 4: SUR1 detected in diverse experimental models.
Footnotes
Conflicts of interest: The authors declare no conflicts of interest.
Financial support: This work was supported by the CONACYT (FORDECYT-PRONACES/170733/2020 to PA and CB-2016-287959 to MRO). IAB is a doctoral student from Programa de Doctorado en Ciencias Biomédicas, Universidad Nacional Autónoma de México (UNAM) and beneficiary of scholarship No. 275610 from CONACYT.
Copyright license agreement: The Copyright License Agreement has been signed by all authors before publication.
Plagiarism check: Checked twice by iThenticate.
Peer review: Externally peer reviewed.
Open peer reviewer: Noah Goshi, University of California Davis, USA.
Funding: This work was supported by the CONACYT (FORDECYT-PRONACES/170733/2020 to PA and CB-2016-287959 to MRO). IAB is a doctoral student from Programa de Doctorado en Ciencias Biomédicas, Universidad Nacional Autónoma de México (UNAM) and beneficiary of scholarship No. 275610 from CONACYT.
P-Reviewer: Goshi N; C-Editors: Zhao M, Zhao LJ, Qiu Y; T-Editor: Jia Y
References
- 1.Aggarwal NT, Shi NQ, Makielski JC. ATP-sensitive potassium currents from channels formed by Kir6 and a modified cardiac mitochondrial SUR2 variant. Channels (Austin) 2013;7:493–502. doi: 10.4161/chan.26181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aguilar-Bryan L, Iii AEB, Nelson A. Photoaffinity labeling and partial purification of the beta cell sulfonylurea receptor using a novel, biologically active glyburide analog. J Biol Chem. 1990;265:8218–8224. [PubMed] [Google Scholar]
- 3.Aguilar-Bryan L, Nichols CG, Wechsler SW, Iv JPC, Ill AEB, Gonzalez G, Herrera-sosa H, Nguy K, Bryan J, Nelsont DA. Cloning of the P cell high-affinity sulfonylurea receptor: a regulator of insulin secretion. Science. 1995;268:423–426. doi: 10.1126/science.7716547. [DOI] [PubMed] [Google Scholar]
- 4.Alquisiras-Burgos I, Ortiz-Plata A, Franco-Pérez J, Millán A, Aguilera P. Resveratrol reduces cerebral edema through inhibition of de novo SUR1 expression induced after focal ischemia. Exp Neurol. 2020;330:113353. doi: 10.1016/j.expneurol.2020.113353. [DOI] [PubMed] [Google Scholar]
- 5.Amarouch MY, El Hilaly J. Inherited cardiac arrhythmia syndromes: focus on molecular mechanisms underlying TRPM4 channelopathies. Cardiovasc Ther. 2020;2020:6615038. doi: 10.1155/2020/6615038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arikan F, Martínez-Valverde T, Sánchez-Guerrero A, Campos M, Esteves M, Gandara D, Torné R, Castro L, Dalmau A, Tibau J, Sahuquillo J. Malignant infarction of the middle cerebral artery in a porcine model. A pilot study. PLoS One. 2017;12:e0172637. doi: 10.1371/journal.pone.0172637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ashfield R, Ashcroft SJ. Cloning of the promoters for the beta-cell ATP-sensitive K-channel subunits Kir6.2 and SUR1. Diabetes. 1998;7:1274–1280. doi: 10.2337/diab.47.8.1274. [DOI] [PubMed] [Google Scholar]
- 8.Bal R, Ozturk G, Etem EO, Him A, Cengiz N, Kuloglu T, Tuzcu M, Yildirim C, Tektemur A. Modulation of excitability of stellate neurons in the ventral cochlear nucleus of mice by ATP-sensitive potassium channels. J Membr Biol. 2018;251:163–178. doi: 10.1007/s00232-017-0011-x. [DOI] [PubMed] [Google Scholar]
- 9.Berdugo M, Delaunay K, Naud MC, Guegan J, Moulin A, Savoldelli M, Picard E, Radet L, Jonet L, Djerada L, Gozalo C, Daruich A, Beltrand J, Jeanny JC, Kermorvant-Duchemin E, Crisanti P, Polak M, Behar-Cohen F. The antidiabetic drug glibenclamide exerts direct retinal neuroprotection. Transl Res. 2021;229:83–99. doi: 10.1016/j.trsl.2020.10.003. [DOI] [PubMed] [Google Scholar]
- 10.Bianchi B, Smith PA, Abriel H. The ion channel TRPM4 in murine experimental autoimmune encephalomyelitis and in a model of glutamate-induced neuronal degeneration. Mol Brain. 2018;11:41. doi: 10.1186/s13041-018-0385-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Braun M, Anderie I, The F. Evidence for a 65 kDa sulfonylurea receptor in rat pancreatic zymogen granule membranes. FEBS Lett. 1997;411:255–259. doi: 10.1016/s0014-5793(97)00711-4. [DOI] [PubMed] [Google Scholar]
- 12.Brustovetsky T, Shalbuyeva N, Brustovetsky N. Lack of manifestations of diazoxide/5-hydroxydecanoate-sensitive KATP channel in rat brain nonsynaptosomal mitochondria. J Physiol. 2005;568:47–59. doi: 10.1113/jphysiol.2005.091199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caffes N, Kurland DB, Gerzanich V, Simard JM. Glibenclamide for the treatment of ischemic and hemorrhagic stroke. Int J Mol Sci. 2015;16:4973–4984. doi: 10.3390/ijms16034973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan KW, Wheeler A, Csanády L. Sulfonylurea receptors type 1 and 2A randomly assemble to form heteromeric KATP channels of mixed subunit composition. J Gen Physiol. 2008;131:43–58. doi: 10.1085/jgp.200709894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen B, Ng G, Gao Y, Low SW, Sandanaraj E, Ramasamy B, Sekar S, Bhakoo K, Soong TW, Nilius B, Tang C, Robins EG, Goggi J, Liao P. Non-invasive multimodality imaging directly shows TRPM4 inhibition ameliorates stroke reperfusion injury. Transl Stroke Res. 2019a;10:91–103. doi: 10.1007/s12975-018-0621-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen B, Gao Y, Wei S, Low SW, Ng G, Yu D, Tu TM, Soong TW, Nilius B, Liao P. TRPM4-specific blocking antibody attenuates reperfusion injury in a rat model of stroke. Pflugers Arch. 2019b;471:1455–1466. doi: 10.1007/s00424-019-02326-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen X, Liu K, Lin Z, Huang K, Pan S. Knockout of transient receptor potential melastatin 4 channel mitigates cerebral edema and neuronal injury after status epilepticus in mice. J Neuropathol Exp Neurol. 2020;79:1354–1364. doi: 10.1093/jnen/nlaa134. [DOI] [PubMed] [Google Scholar]
- 18.de Wet H, Proks P. Molecular action of sulphonylureas on KATP channels: a real partnership between drugs and nucleotides. Biochem Soc Trans. 2015;43:901–907. doi: 10.1042/BST20150096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duda J, Pötschke C, Liss B. Converging roles of ion channels, calcium, metabolic stress, and activity pattern of Substantia nigra dopaminergic neurons in health and Parkinson’s disease. J Neurochem 139 Suppl. 2016;1:156–178. doi: 10.1111/jnc.13572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dundar TT, Abdallah A, Yurtsever I, Guler EM, Ozer OF, Uysal O. Serum SUR1 and TRPM4 in patients with subarachnoid hemorrhage. Neurosurg Rev. 2020;43:1595–1603. doi: 10.1007/s10143-019-01200-6. [DOI] [PubMed] [Google Scholar]
- 21.Fang Y, Shi H, Ren R, Huang L, Okada T, Lenahan C, Gamdzyk M, Travis ZD, Lu Q, Tang L, Huang Y, Zhou K, Tang J, Zhang J, Zhang JH. Pituitary adenylate cyclase-activating polypeptide attenuates brain edema by protecting blood-brain barrier and glymphatic system after subarachnoid hemorrhage in rats. Neurotherapeutics. 2020;17:1954–1972. doi: 10.1007/s13311-020-00925-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Felix L, Delekate A, Petzold GC, Rose CR. Sodium fluctuations in astroglia and their potential impact on astrocyte function. Front Physiol. 2020;11:871. doi: 10.3389/fphys.2020.00871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Filipets N, Filipets О, Slobodian K, Gabunia L, Gvishiani M, Maxaradze T. Pharmaceutical activators of atp-dependent potassium channels as potential nephroprotectors against glomerular and tubular damage to the nephron. Georgian Med News. 2019:163–166. [PubMed] [Google Scholar]
- 24.Flagg TP, Kurata HT, Masia R, Caputa G, Nichols CG. Differential structure of atrial and ventricular KATP: atrial KATP channels require SUR1. Circ Res. 2009;103:1458–1465. doi: 10.1161/CIRCRESAHA.108.178186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gerzanich V, Makar KT, Reddy GP, Kwon S, Stokum JA, Kyoon SW, Ivanova S, Ivanov A, Mehta RI, Brooke AM, Bryan J, Bever CT, Simard JM. Salutary effects of glibenclamide during the chronic phase of murine experimental autoimmune encephalomyelitis. J Neuroinflammation. 2017;14:177. doi: 10.1186/s12974-017-0953-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gerzanich V, Seong MK, Kyoon SW, Ivanov A, Simard JM. SUR1-TRPM4 channel activation and phasic secretion of MMP-9 induced by tPA in brain endothelial cells. PLoS One. 2018;13:e0195526. doi: 10.1371/journal.pone.0195526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gerzanich V, Stokum JA, Ivanova S, Woo SK, Tsymbalyuk O, Sharma A, Akkentli F, Imran Z, Aarabi B, Sahuquillo J, Simard JM. Sulfonylurea receptor 1, transient receptor potential cation channel subfamily M member 4, and KIR6.2: role in hemorrhagic progression of contusion. J Neurotrauma. 2019;36:1060–1079. doi: 10.1089/neu.2018.5986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gong XG, Sun HM, Zhang Y, Zhang SJ, Gao YS, Feng J, Hu JH, Gai C, Guo ZY, Xu H, Ma L. Da-bu-yin-wan and qian-zheng-san to neuroprotect the mouse model of Parkinson’s disease. Evid Based Complement Alternat Med. 2014;2014:729195. doi: 10.1155/2014/729195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gros L, Trapp S, Dabrowski M, Ashcroft FM, Bataille D, Blache P. Characterization of two novel forms of the rat sulphonylurea receptor SUR1A2 and SUR1B D 31. Br J Pharmacol. 2002;137:98–106. doi: 10.1038/sj.bjp.0704836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hambrock A, Preisig-mu R, Russ U, Daut R, Piehl A, Hanley PJ, Ray J, Quast U, Derst C, Preisig-mu R, Piehl A, Hanley PJ, Ray J, Quast U, Derst C. Four novel splice variants of sulfonylurea receptor 1. Am J Physiol Cell Physiol. 2002;283:C587–598. doi: 10.1152/ajpcell.00083.2002. [DOI] [PubMed] [Google Scholar]
- 31.Herbst KJ, Coltharp C, Amzel LM, Zhang J. Direct activation of Epac by sulfonylurea is isoform selective. Chem Biol. 2011;18:243–251. doi: 10.1016/j.chembiol.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hernández-Sánchez C, Wood TL, LeRoith D. Developmental and tissue-specific sulfonylurea receptor gene expression. Endocrinology. 1997;138:705–711. doi: 10.1210/endo.138.2.4954. [DOI] [PubMed] [Google Scholar]
- 33.Hu ZL, Sun T, Lu M, Ding JH, Du RH, Hu G. Kir6.1/K-ATP channel on astrocytes protects against dopaminergic neurodegeneration in the MPTP mouse model of Parkinson’s disease via promoting mitophagy. Brain Behav Immun. 2019;81:509–522. doi: 10.1016/j.bbi.2019.07.009. [DOI] [PubMed] [Google Scholar]
- 34.Huang K, Wang Z, Gu Y, Hu Y, Ji Z, Wang S, Lin Z, Li X, Xie Z, Pan S. Glibenclamide is comparable to target temperature management in improving survival and neurological outcome after asphyxial cardiac arrest in rats. J Am Heart Assoc. 2016;5:e003465. doi: 10.1161/JAHA.116.003465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang Y, Hu D, Huang C, Nichols CG. Genetic discovery of ATP-sensitive K+ channels in cardiovascular diseases. Circ Arrhythm Electrophysiol. 2019;12:e007322. doi: 10.1161/CIRCEP.119.007322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jha RM, Puccio AM, Chou SH, Chang CH, Wallisch JS, Molyneaux BJ, Zusman BE, Shutter LA, Poloyac SM, Janesko-Feldman KL, Okonkwo DO, Kochanek PM. Sulfonylurea receptor-1: a novel biomarker for cerebral edema in severe traumatic brain injury. Crit Care Med. 2017;45:e255–264. doi: 10.1097/CCM.0000000000002079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jha RM, Bell J, Citerio G, Hemphill JC, Kimberly WT, Narayan RK, Sahuquillo J, Sheth KN, Simard JM. Role of sulfonylurea receptor 1 and glibenclamide in traumatic brain injury: a review of the evidence. Int J Mol Sci. 2020;21:409. doi: 10.3390/ijms21020409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang B, Li L, Chen Q, Tao Y, Yang L, Zhang B, Zhang JH, Feng H, Chen Z, Tang J, Zhu G. Role of glibenclamide in brain injury after intracerebral hemorrhage. Transl Stroke Res. 2017;8:183–193. doi: 10.1007/s12975-016-0506-2. [DOI] [PubMed] [Google Scholar]
- 39.Kaya ST, Bozdogan O, Ozarslan TO, Taskin E, Eksioglu D, Erim F, Firat T, Yasar S. The protection of resveratrol and its combination with glibenclamide, but not berberine on the diabetic hearts against reperfusion-induced arrhythmias: the role of myocardial KATP channel. Arch Physiol Biochem. 2019;125:114–121. doi: 10.1080/13813455.2018.1440409. [DOI] [PubMed] [Google Scholar]
- 40.Kim JW, Seghers V, Cho JH, Kang Y, Kim S, Ryu Y, Baek K, Aguilar-Bryan L, Lee YD, Bryan J, Suh-Kim H. Transactivation of the mouse sulfonylurea receptor I gene by BETA2/NeuroD. Mol Endocrinol. 2002;16:1097–1107. doi: 10.1210/mend.16.5.0934. [DOI] [PubMed] [Google Scholar]
- 41.Kim JM, Song KS, Xu B, Wang T. Role of potassium channels in female reproductive system. Obstet Gynecol Sci. 2020;63:565–576. doi: 10.5468/ogs.20064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Knowlton C, Kutterer S, Roeper J, Canavier CC. Calcium dynamics control K-ATP channel-mediated bursting in substantia nigra dopamine neurons: a combined experimental and modeling study. J Neurophysiol. 2018;119:84–95. doi: 10.1152/jn.00351.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Köhling R, Wolfart J. Potassium channels in epilepsy. Cold Spring Harb Perspect Med. 2016;6:a022871. doi: 10.1101/cshperspect.a022871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kramer W, Oekonomopulos R, Pünter J, Summ HD. Direct photoaffinity labeling of the putative sulfonylurea receptor in rat beta-cell tumor membranes by [3H]glibenclamide. FEBS Lett. 1988;229:355–359. doi: 10.1016/0014-5793(88)81155-4. [DOI] [PubMed] [Google Scholar]
- 45.Kramer W, Müller G, Girbig F, Gutjahr U, Kowalewski S, Hartz D, Summ H. Differential interaction of glimepiride and glibenclamide with the fl-cell sulfonylurea receptor II. Photoaffinity labeling of a 65 kDa protein by [3H]glimepiride. Biochim Biophys Acta. 1994;1191:278–290. doi: 10.1016/0005-2736(94)90178-3. [DOI] [PubMed] [Google Scholar]
- 46.Kundap UP, Paudel YN, Shaikh MF. Animal models of metabolic epilepsy and epilepsy associated metabolic dysfunction: a systematic review. Pharmaceuticals (Basel) 2020;13:106. doi: 10.3390/ph13060106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee JY, Choi HY, Na WH, Ju BG, Yune TY. Ghrelin inhibits BSCB disruption/hemorrhage by attenuating MMP-9 and SUR1/TrpM4 expression and activation after spinal cord injury. Biochim Biophys Acta. 2014;1842:2403–2412. doi: 10.1016/j.bbadis.2014.09.006. [DOI] [PubMed] [Google Scholar]
- 48.Li N, Wu JX, Ding D, Cheng J, Gao N, Chen L. Structure of a pancreatic ATP-sensitive potassium channel. Cell. 2017;168:101–110. doi: 10.1016/j.cell.2016.12.028. [DOI] [PubMed] [Google Scholar]
- 49.Li Y, Ba M, Du Y, Xia C, Tan S, Ng KP, Ma G. Aβ1-42 increases the expression of neural KATP subunits Kir6. 2/SUR1 via the NF-κB, p38 MAPK and PKC signal pathways in rat primary cholinergic neurons. Hum Exp Toxicol. 2019;38:665–674. doi: 10.1177/0960327119833742. [DOI] [PubMed] [Google Scholar]
- 50.Li Y, Feng Y, Liu L, Li X, Li XY, Sun X, Li KX, Zha RR, Wang HD, Zhang MD, Fan XX, Wu D, Fan Y, Zhang HC, Qiao GF, Li BY. The baroreflex afferent pathway plays a critical role in H2S-mediated autonomic control of blood pressure regulation under physiological and hypertensive conditions. Acta Pharmacol Sin. 2020 doi: 10.1038/s41401-020-00549-5. doi: 10.1038/s41401-020-00549-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin Z, Huang H, Gu Y, Huang K, Hu Y, Ji Z, Wu Y, Wang S, Yang T, Pan S. Glibenclamide ameliorates cerebral edema and improves outcomes in a rat model of status epilepticus. Neuropharmacology. 2017;121:1–11. doi: 10.1016/j.neuropharm.2017.04.016. [DOI] [PubMed] [Google Scholar]
- 52.Luo ZW, Ovcjak A, Wong R, Yang BX, Feng ZP, Sun HS. Drug development in targeting ion channels for brain edema. Acta Pharmacol Sin. 2020;41:1272–1288. doi: 10.1038/s41401-020-00503-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martin GM, Yoshioka C, Rex EA, Fay JF, Xie Q, Whorton MR, Chen JZ, Shyng SL. Cryo-EM structure of the ATP-sensitive potassium channel illuminates mechanisms of assembly and gating. eLife. 2017;6:e24149. doi: 10.7554/eLife.24149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Matsuo M, Tanabe K, Kioka N, Amachi T, Ueda K. Different binding properties and affinities for ATP and ADP among sulfonylurea receptor subtypes, SUR1, SUR2A, and SUR2B. J Biol Chem. 2000;275:28757–28763. doi: 10.1074/jbc.M004818200. [DOI] [PubMed] [Google Scholar]
- 55.Menigoz A, Ahmed T, Sabanov V, Philippaert K, Pinto S, Kerselaers S, Segal A, Freichel M, Voets T, Nilius B, Vennekens R, Balschun D. TRPM4-dependent post-synaptic depolarization is essential for the induction of NMDA receptor-dependent LTP in CA1 hippocampal neurons. Pflugers Arch. 2016;468:593–607. doi: 10.1007/s00424-015-1764-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nelson DA, Aguilar-Bryan L, Bryan J. Specificity of photolabeling of beta-cell membrane proteins with an 125I-labeled glyburide analog. J Biol Chem. 1992;267:14928–14933. [PubMed] [Google Scholar]
- 57.Nelson DA, Bryan J, Wechsler S, Clement JP, 4th, Aguilar-Bryan L. The high-affinity sulfonylurea receptor: distribution, glycosylation, purification, and immunoprecipitation of two forms from endocrine and neuroendocrine cell lines. Biochemistry. 1996;35:14793–14799. doi: 10.1021/bi960777y. [DOI] [PubMed] [Google Scholar]
- 58.Niki I, Welsh M, Berggren PO, Hubbrard P, Ashcroft SJH. Characterization of the solubilized glibenclamide receptor in hamster pancreatic fl-cell line, HIT T15. Biochem J. 1991;277:619–624. doi: 10.1042/bj2770619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pergakis M, Badjatia N, Chaturvedi S, Cronin CA, Kimberly WT, Sheth KN, Simard JM. BIIB093 (IV glibenclamide): an investigational compound for the prevention and treatment of severe cerebral edema. Expert Opin Investig Drugs. 2019;28:1031–1040. doi: 10.1080/13543784.2019.1681967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pu JL, Ye B, Kroboth SL, McNally E, Makielski JC, Shi NQ. Cardiac sulfonylurea receptor short form-based channels confer a glibenclamide-insensitive KATP activity. J Mol Cell Cardiol. 2009;44:188–200. doi: 10.1016/j.yjmcc.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Puljung MC. Cryo-electron microscopy structures and progress toward a dynamic understanding of KATP channels. J Gen Physiol. 2018;150:653–669. doi: 10.1085/jgp.201711978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Puljung M, Vedovato N, Usher S, Ashcroft F. Activation mechanism of ATP-sensitive K+ channels explored with real-time nucleotide binding. Elife. 2019;8:e41103. doi: 10.7554/eLife.41103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rabalet B, Mmell CJ, Johnson DG, Levinz SR. Sulfonylurea binding to a low-affinity site inhibits the Na/K-ATPase and the KATP channel in insulin-secreting cells. J Gen Physiol. 1996;107:231–241. doi: 10.1085/jgp.107.2.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Robichaux WG, 3rd, Cheng X. Intracellular cAMP sensor EPAC: physiology, pathophysiology, and therapeutics development. Physiol Rev. 2018;98:919–1053. doi: 10.1152/physrev.00025.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rubio C, Luna R, Rosiles A, Rubio-Osornio M. Caloric restriction and ketogenic diet therapy for epilepsy: a molecular approach involving Wnt pathway and KATP channels. Front Neurol. 2020;11:584298. doi: 10.3389/fneur.2020.584298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sakura H, Trapp S, Liss B, Ashcroft FM. Altered functional properties of KATP channel conferred by a novel splice variant of SUR1. J Physiol. 1999;521:337–350. doi: 10.1111/j.1469-7793.1999.00337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schmid D, Stolzlechner M, Sorgner A, Bentele C, Assinger A, Chiba P, Moeslinger T. An abundant, truncated human sulfonylurea receptor 1 splice variant has prodiabetic properties and impairs sulfonylurea action. Cell Mol Life Sci. 2012;69:129–148. doi: 10.1007/s00018-011-0739-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schwanstecher M, Lser S, Chudziak F, Bachmann C, Panten U. Photoaffinity labeling of the cerebral sulfonylurea receptor using a novel radioiodinated azidoglibenclamide analogue. J Neurochem. 1994;63:698–708. doi: 10.1046/j.1471-4159.1994.63020698.x. [DOI] [PubMed] [Google Scholar]
- 69.Shi N, He J, Guo Q, Liu T, Han J. Liraglutide protects against diabetes mellitus complicated with focal cerebral ischemic injury by activating mitochondrial ATP-sensitive potassium channels. Neuroreport. 2019;30:479–484. doi: 10.1097/WNR.0000000000001225. [DOI] [PubMed] [Google Scholar]
- 70.Singh BL, Chen L, Cai H, Shi H, Wang Y, Yu Ch, Chen X, Han X, Cai X. Activation of adenosine A2a receptor accelerates and A2a receptor antagonist reduces intermittent hypoxia induced PC12 cell injury via PKC-KATP pathway. Brain Res Bull. 2019;150:118–126. doi: 10.1016/j.brainresbull.2019.05.015. [DOI] [PubMed] [Google Scholar]
- 71.Skeer JM, Dégano P, Coles B, Potier M, Ashcroft FM, Ashcroft SJ. Determination of the molecular mass of the native beta-cell sulfonylurea receptor. FEBS Lett. 1994;338:98–102. doi: 10.1016/0014-5793(94)80124-x. [DOI] [PubMed] [Google Scholar]
- 72.Stokum JA, Gerzanich V, Simard JM. Molecular pathophysiology of cerebral edema. J Cereb Blood Flow Metab. 2016;36:513–538. doi: 10.1177/0271678X15617172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stokum JA, Kwon MS, Woo SK, Tsymbalyuk O, Vennekens R, Volodymyr G, Simard JM. SUR1-TRPM4 and AQP4 form a heteromultimeric complex that amplifies ion/water osmotic coupling and drives astrocyte swelling. Glia. 2018;66:108–125. doi: 10.1002/glia.23231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sun Y, He Y, Yang L, Liang D, Shi W, Zhu X, Jiang Y, Ou C. Manganese induced nervous injury by α-synuclein accumulation via ATP-sensitive K(+) channels and GABA receptors. Toxicol Lett. 2020;10:164–170. doi: 10.1016/j.toxlet.2020.07.008. [DOI] [PubMed] [Google Scholar]
- 75.Thomas C, Aller SG, Beis K, Carpenter EP, Chang G, Chen L, Dassa E, Dean M, Duong Van Hoa F, Ekiert D, Ford R, Gaudet R, Gong X, Holland IB, Huang Y, Kahne DK, Kato H, Koronakis V, Koth CM, Lee Y, et al. Structural and functional diversity calls for a new classification of ABC transporters. FEBS Lett. 2020;594:3767–3775. doi: 10.1002/1873-3468.13935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tinker A, Aziz Q, Li Y, Specterman M. ATP-sensitive potassium channels and their physiological and pathophysiological roles. Compr Physiol. 2018;8:1463–1511. doi: 10.1002/cphy.c170048. [DOI] [PubMed] [Google Scholar]
- 77.Tomuschat Ch, O’Donnell AM, Coyle D, Dreher N, Kelly D, Puri P. Altered expression of ATP-sensitive K(+) channels in Hirschsprung’s disease. J Pediatr Surg. 2016;51:948–952. doi: 10.1016/j.jpedsurg.2016.02.060. [DOI] [PubMed] [Google Scholar]
- 78.Wei S, Low SW, Poore CP, Chen B, Gao Y, Nilius B, Liao P. Comparison of anti-oncotic effect of TRPM4 blocking antibody in neuron, astrocyte and vascular endothelial cell under hypoxia. Front Cell Dev Biol. 2020;8:562584. doi: 10.3389/fcell.2020.562584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Woo SK, Tsymbalyuk N, Tsymbalyuk O, Ivanova S, Gerzanich V, Simard JM. SUR1-TRPM4 channels, not K(ATP), mediate brain swelling following cerebral ischemia. Neurosci Lett. 2020;718:134729. doi: 10.1016/j.neulet.2019.134729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wu Z, Zhu SZ, Hu YF, Gu Y, Wang SN, Lin ZZ, Xie ZS, Pan SY. Glibenclamide enhances the effects of delayed hypothermia after experimental stroke in rats. Brain Res. 2016;15:113–122. doi: 10.1016/j.brainres.2016.04.067. [DOI] [PubMed] [Google Scholar]
- 81.Yamada M, Kurachi Y. Focused issue on K ATP channels A functional role of the C-terminal 42 amino acids of SUR2A and SUR2B in the physiology and pharmacology of cardiovascular ATP-sensitive K+ channels. J Mol Cell Cardiol. 2005;39:1–6. doi: 10.1016/j.yjmcc.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 82.Yan J, Bengtson CP, Buchthal B, Hagenston AM, Bading H. Coupling of NMDA receptors and TRPM4 guides discovery of unconventional neuroprotectants. Science. 2020;370:eaay3302. doi: 10.1126/science.aay3302. [DOI] [PubMed] [Google Scholar]
- 83.Zhang Q, Li C, Zhang T, Ge Y, Han X, Sun S, Ding J, Lu M, Hu G. Deletion of Kir6.2/SUR1 potassium channels rescues diminishing of DA neurons via decreasing iron accumulation in PD. Mol Cell Neurosci. 2018;92:164–176. doi: 10.1016/j.mcn.2018.08.006. [DOI] [PubMed] [Google Scholar]
- 84.Zhang S, Guo Z, Yang S, Ma H, Fu C, Wang S, Zhang Y, Liu Y, Hu J. Chronic intermittent hybobaric hypoxia protects against cerebral ischemia via modulation of mitoK ATP. Neurosci Lett. 2016;2:8–16. doi: 10.1016/j.neulet.2016.10.025. [DOI] [PubMed] [Google Scholar]
- 85.Zhao K, Wen R, Wang X, Pei L, Yang Y, Shang Y, Bazan N, Zhu LQ, Tian Q, Lu Y. EPAC inhibition of SUR1 receptor increases glutamate release and seizure vulnerability. J Neurosci. 2013;33:8861–8865. doi: 10.1523/JNEUROSCI.5686-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhong CJ, Chen MM, Lu M, Ding JH, Du RH, Hu G. Astrocyte-specific deletion of Kir6.1/K-ATP channel aggravates cerebral ischemia/reperfusion injury through endoplasmic reticulum stress in mice. Exp Neurol. 2019;311:225–233. doi: 10.1016/j.expneurol.2018.10.005. [DOI] [PubMed] [Google Scholar]
- 87.Zhou F, Liu Y, Yang B, Hu Z. Neuroprotective potential of glibenclamide is mediated by antioxidant and anti-apoptotic pathways in intracerebral hemorrhage. Brain Res Bull. 2018;142:18–24. doi: 10.1016/j.brainresbull.2018.06.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


