Abstract
This study evaluated a PCR method for the rapid detection of clinically significant genotypes of vancomycin-resistant enterococci (VRE) in nosocomial surveillance specimens. Detection of the vanA and vanB genes by multiplex PCR using 657 specimens that showed presumptive growth of VRE on bile esculin azide agar containing 6 mg of vancomycin/liter was compared to the conventional method. The diagnostic values for the PCR compared to the phenotypic method were as follows: 99.8% specificity, 95.4% sensitivity, 98.8% positive predictive value, and 99.3% negative predictive value. The average cost per test for PCR is $8.26, compared to $9.45 for the phenotypic method. The average turnaround time for detecting a VRE is 48 h for PCR, compared to 96 h for the conventional method.
Since the late 1980s vancomycin-resistant enterococci (VRE) have emerged as an important cause of nosocomial infections. They are the third most common cause of hospital-acquired bacteremia (5). Outbreaks with VRE are also being reported with increasing frequency in many countries (1). The National Nosocomial Infection Surveillance (NNIS) system in the United States has reported an increase of VRE since 1989 (1). Recent data from The Surveillance Network database (TSN) in the United States for 1997 indicate that about 15% of all enterococci are resistant to vancomycin (4). The prevalence of VRE in Canadian hospitals has been substantially lower. A point prevalence study carried out by the Canadian Nosocomial Infection Surveillance Program (CNISP) in 1996 reported prevalences of 0.1 and 3.7% in hospitals in which VRE are not and are endemic, respectively (6).
Rapid identification of VRE-colonized patients is essential for the implementation of appropriate control measures to prevent the spread of VRE. Routine culture methods are time-consuming and expensive for the laboratory handling a large number of specimens. The microbiology lab at Hamilton Civic Hospitals receives an average of 1,200 VRE surveillance culture orders per month. PCR-based techniques that have been reported (7, 8) as alternative methods for detecting and identifying VRE directly from fecal samples are not cost-effective, especially when one is dealing with a large number of specimens and a low prevalence. The objective of the present study was to develop a simple, rapid, and cost-effective PCR method that could be used to detect vanA and vanB genes from the presumptive growth of VRE on bile esculin azide agar containing 6 mg of vancomycin/liter (BEAA-V6) as a routine method of screening nosocomial surveillance specimens. The new DNA-based method was compared with conventional culture and evaluated for its cost-effectiveness.
A total of 657 rectal swab specimens that produced black colonies on BEAA-V6 (PML Microbiologicals, Mississauga, Ontario, Canada) were used for the study. Cell suspensions of black colonies from BEAA-V6 in 0.5 N saline were prepared to a density equivalent to a McFarland standard of 1. Fifty microliters of the cell suspension was mixed with 50 μl of lysis buffer containing 15% Chelex-100 (Bio-Rad Laboratories, Mississauga, Ontario, Canada), 1% Brij 58 (Sigma Chemical Company, Mississuga, Ontario, Canada), and 1% Tween 20 (Bio-Rad), and the mixture was heated for 15 min at 95°C. It was then centrifuged for 2 min, and 2 μl of clear liquid was used as the template for PCR. Two in-frame DNA fragments of 732 and 635 bp, corresponding to vanA and vanB genes (2), respectively, were coamplified with a 370-bp universal eubacterial target (16S ribosomal DNA [rDNA]) (3) as an internal PCR control. After the conditions were optimized, the 25-μl PCR mix contained 1.5 mM MgCl2, 1× PCR buffer, 250 μM (each) dATP, dCTP, dGTP, and dTTP (Boehringer Mannheim, Laval, Quebec, Canada), 15 pmol of each vanA primer (forward, 175-GGGAAAACGACAATTGC-191; reverse, 907-GTACAATGCGCCGTTA-891), 30 pmol of each vanB primer (forward, 173-ATGGGAAGCCGATAGTC-189; reverse, 807-GATTTCGTTCCTCGACC-791), 3 pmol of each 16S rDNA universal target primer (forward, 1170-AACTGGAGGAAGGTGGGGAT-1189; reverse, 1522-AGGAGGTGATCCAACCGCA-1540) (Mobix, McMaster University, Hamilton, Ontario, Canada), and 1.25 U of AmpliTaq DNA polymerase (Roche Molecular Systems Inc., Branchburg, N.J.). Prefrozen master mixes (without the enzyme) were thawed, and aliquots were dispensed into PCR tubes after the enzyme was added. Two microliters of the DNA preparation was added to the reaction mixes and cycled with a temperature profile of 94°C for 30 s, 54°C for 30 s, and 72°C for 45 s in a GeneAmp PCR system 9700 (Perkin-Elmer Applied Biosystems, Mississauga, Ontario, Canada) for 30 cycles. A 5-min denaturation step at 94°C was included at the beginning of the cycling, and at the end a final 7-min extension step at 72°C was included. A vanA strain (Enterococcus faecalis), a vanB strain (Enterococcus faecium), and a vancomycin-susceptible E. faecalis strain (ATCC 29212) were run with each set of reactions as positive and negative controls.
PCR products were size separated by electrophoresis on 1% agarose (Promega Corporation, Madison, Wis.) gels containing 0.5 mg of ethidium bromide/liter to detect vanA, vanB, and 16S rDNA targets. A PCR ruler (100 bp) (Bio-Rad) was used as the molecular-size marker. Gel images were photodocumented by using a Gel Doc 1000 (Bio-Rad) for interpretation. Cell suspensions that did not show amplification of the universal target (370-bp product) were subcultured onto colistin-nalidixic acid agar (CNA), from which PCR was repeated the next day. Routine culture and phenotypic identification of VRE were carried out by Gram staining (Quelab Laboratories Inc., Montreal, Quebec, Canada) and with catalase (by using H2O2) (Regal Pharmaceuticals, Burlington, Ontario, Canada), pyrazinamidase (PML), leucine aminopeptidase (Key Scientific, Round Rock, Tex.), o-nitrophenyl-β-d-galactopyranoside (ONPG)-phenylalanine-Motility medium (Quelab), and brain heart infusion agar containing 6 mg of vancomycin/liter (PML).
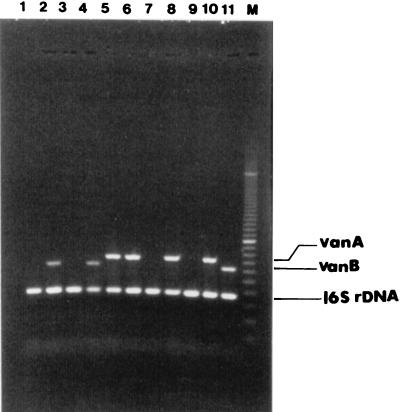
Amplification of vanA, vanB, and 16S rDNA targets produced distinct bands corresponding to their respective molecular sizes that were easily recognizable (Fig. 1). Thirteen percent (83 of 657) of presumptive growth was identified as VRE. Of these, 53 were vanA and 30 were vanB genotypes. Five percent of specimens (31 of 657) from BEAA-V6 were negative for the PCR amplification control. All PCR amplification control-negative specimens were repeated from the original BEAA-V6 plates and were negative for amplification. However, all 31 specimens produced a 370-bp band corresponding to the universal target when DNA was prepared from the growth on CNA. Four of these were identified as VRE (three vanA and one vanB). The PCR method produced one false-positive specimen. However, repeated testing by PCR showed it to be a true negative, and the initial positive result was attributed to a laboratory technologist’s error. The diagnostic values for the PCR from BEAA-V6 compared to the phenotypic method as the “gold standard” were as follows: 99.8% specificity, 95.4% sensitivity, 98.8% positive predictive value, and 99.3% negative predictive value. The use of CNA increased the sensitivity and negative predictive value to 100%. The average turnaround time for the PCR method is 48 h from the time of specimen setup, compared to 96 h for the routine phenotypic method. The estimated costs for both methods are given in Canadian dollars ($1.00 Canadian = $0.67 U.S.) in Table 1. The total cost per test for VRE was $8.26 for PCR compared to $9.45 for culture. However, the cost per specimen depends on the prevalence of black colonies that are gram-positive cocci on BEAA-V6. At present at the Hamilton Civic Hospitals, only 17.3% of all specimens produce black colonies on BEAA-V6, and only 9% require PCR or complete investigation by culture. At the current prevalence rate, reduced costs associated with negative specimens on BEAA-V6 bring the cost per specimen down to $3.10 and $2.95 for culture and PCR, respectively. The equipment cost for setting up PCR is $8,000 to $15,000 based on the workload. However, many laboratories already have the capacity to perform PCR.
FIG. 1.
Agarose gel electrophoresis of PCR-amplified vanA, vanB, and 16S rDNA gene targets from presumptive growth on BEAA-V6 medium. The negative control, E. faecalis ATCC 29212, was grown on CNA. Lanes 1 to 8, sample of routine nosocomial surveillance specimens; lane 9, E. faecalis ATCC 29212 (vancomycin susceptible); lane 10, an E. faecalis vanA strain; lane 11, an E. faecium vanB strain; lane 12, 100-bp PCR ruler (Bio-Rad).
TABLE 1.
Estimated costsa and turnaround times for screening VRE by the conventional and PCR methods
| Method | Cost per testb
|
Cost per specimenc
|
Turn-around time (h) | ||||
|---|---|---|---|---|---|---|---|
| Materials | Labord | Total | Materials | Labord | Total | ||
| PCR | 4.84 | 3.42 | 8.26 | 2.63 | 0.32 | 2.95 | 48 |
| Culture | 5.66 | 3.79 | 9.45 | 2.75 | 0.35 | 3.10 | 96 |
All costs are given in Canadian dollars ($1.00 Canadian = $0.67 U.S.).
Based on complete testing of a batch of 14 specimens and controls.
Based on 17.3% of all specimens producing black colonies on BEAA-V6 and only 9% being gram-positive cocci requiring PCR and full culture workup. All costs are based on handling a batch of 14 tests and controls.
Calculated at $30.32/h (in Canadian dollars) based on hands-on time.
This study examined whether PCR could be used in the clinical laboratory to replace traditional surveillance culture methods for detection of VRE in colonized patients. Since the prevalence of VRE is significantly low in our local setting (<10% of presumptive growth on BEAA-V6 is VRE), PCR methods using direct specimens are not cost-effective. In addition, such methods (7) are known to have relatively low sensitivities and do not allow for strain typing studies, an essential part of many infection control investigations. The possibility of culturing those specimens that are positive by PCR to recover enterococci for molecular typing would increase the overall cost of the surveillance system. Therefore, we examined the possibility of using presumptive growth of VRE on a screening medium (BEAA-V6) for the PCR method to overcome those limitations. Among screening media, BEAA-V6 has been proven effective and is widely used for surveillance culture of VRE (7). Since the significance of detecting vanC-containing VRE from rectal samples remains unclear (7), only vanA and vanB targets were used in the multiplex PCR. The 16S rDNA universal target is necessary to identify PCR inhibition. This is particularly important because only 13% of the specimens were VRE and 5% showed PCR inhibition when BEAA-V6 medium was used. Universal target amplification helps to identify specimens that might be considered false-negatives.
The PCR method has an added advantage in that it can reliably detect vanB-type VRE phenotypes, which are difficult to detect by commercially available susceptibility testing systems and automated susceptibility assays such as the MicroScan Rapid (Dade MicroScan Inc., West Sacramento, Calif.) and VITEK GPS-TA (bioMerieux Vitek Inc., Hazelwood, Mo.) tests.
In summary, the PCR method is an attractive alternative to culture and phenotypic methods for surveillance of VRE in hospitals with heavy workloads. Even though the PCR that uses presumptive growth of VRE on a screening medium is not as rapid as a PCR using direct specimens, it has a shorter turnaround time than the phenotypic method. It is a cost-effective method, particularly when the prevalence of VRE is low. It also provides isolates for strain typing without any additional costs.
Acknowledgments
This work was supported by the Hamilton Regional Laboratory Medicine Program.
REFERENCES
- 1.Centers for Disease Control and Prevention. Nosocomial enterococci resistant to vancomycin—United States, 1989–1993. Morbid Mortal Weekly Rep. 1993;42:597–599. [PubMed] [Google Scholar]
- 2.Dutka-Malen S, Evers S, Courvalin P. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J Clin Microbiol. 1995;33:24–27. doi: 10.1128/jcm.33.1.24-27.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Greisen K, Loeffelholz M, Purohit A, Leong D. PCR primers and probes for the 16S rRNA gene of most species of pathogenic bacteria, including bacteria found in cerebrospinal fluid. J Clin Microbiol. 1994;32:335–351. doi: 10.1128/jcm.32.2.335-351.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huycke M M, Sahm D F, Gilmore M S. Multiple-drug-resistant enterococci: the nature of the problem and an agenda for the future. Emerg Infect Dis. 1998;4:239–249. doi: 10.3201/eid0402.980211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Narmdari H, DelVecchio V. Application of PCR for the characterization of enterococci. Clin Microbiol Newsl. 1998;20:91–94. [Google Scholar]
- 6.Ofner-Agastin M E, Conly J, Paton S, Kureishi A, Nicolle L, Mulvey M, Johnson W, Johnston L The Canadian Hospital Epidemiology Committee. Vancomycin-resistant enterococci (VRE) in Canada—results of the Canadian Nosocomial Infection Surveillance Program 1996 VRE point prevalence surveillance project. Can J Infect Dis. 1997;8:73–78. doi: 10.1155/1997/297038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Satake S, Clark N, Rinland D, Nolte F S, Tenover F C. Detection of vancomycin-resistant enterococci in fecal samples by PCR. J Clin Microbiol. 1997;35:2325–2330. doi: 10.1128/jcm.35.9.2325-2330.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stosor V, Tornatore M A, Noskin G A, Tenover F C, Peterson L R. Abstracts of the 98th General Meeting of the American Society for Microbiology. Washington, D.C: American Society for Microbiology; 1998. Improved recovery of vancomycin-resistant enterococci (VRE) using a hot-start polymerase chain reaction (PCR) assay for the detection of vanA and vanB from rectal swabs, abstr. C-366; p. 192. [Google Scholar]