Abstract
Antibiotic-resistant Campylobacter constitutes a serious threat to public health. The clonal expansion of resistant strains and/or the horizontal spread of resistance genes to other strains and species can hinder the clinical effectiveness of antibiotics to treat severe campylobacteriosis. Still, gaps exist in our understanding of the risks of acquisition and spread of antibiotic resistance in Campylobacter. While the in vitro transfer of antimicrobial resistance genes between Campylobacter species via natural transformation has been extensively demonstrated, experimental studies have favored the use of naked DNA to obtain transformants. In this study, we used experimental designs closer to real-world conditions to evaluate the possible transfer of antimicrobial resistance genes between Campylobacter strains of the same or different species (Campylobacter coli or Campylobacter jejuni) and originating from different animal hosts (swine or turkeys). This was evaluated in vitro through co-culture experiments and in vivo with dual-strain inoculation of turkeys, followed by whole genome sequencing of parental and newly emerged strains. In vitro, we observed four independent horizontal gene transfer events leading to the acquisition of resistance to beta-lactams (blaOXA), aminoglycosides [aph(2′′)-If and rpsL] and tetracycline [tet(O)]. Observed events involved the displacement of resistance-associated genes by a mutated version, or the acquisition of genomic islands harboring a resistance determinant by homologous recombination; we did not detect the transfer of resistance-carrying plasmids even though they were present in some strains. In vivo, we recovered a newly emerged strain with dual-resistance pattern and identified the replacement of an existing non-functional tet(O) by a functional tet(O) in the recipient strain. Whole genome comparisons allowed characterization of the events involved in the horizontal spread of resistance genes between Campylobacter following in vitro co-culture and in vivo dual inoculation. Our study also highlights the potential for antimicrobial resistance transfer across Campylobacter species originating from turkeys and swine, which may have implications for farms hosting both species in close proximity.
Keywords: antimicrobial resistance, Campylobacter, transformation, turkey, swine, animal experiment, co-culture experiment, horizontal gene transfer
Introduction
With an estimated 9.4 million yearly cases in the United States alone, human foodborne diseases are an important public health concern (Scallan et al., 2011). Among bacterial foodborne pathogens, Campylobacter and Salmonella, cause the largest proportion of bacterial illnesses in the United States and other industrialized nations. In 2019, among the eight bacterial pathogens monitored by the Foodborne Diseases Active Surveillance Network (FoodNet), the overall incidence per 100,000 population was highest for Campylobacter (19.5 cases; Tack et al., 2020). While campylobacteriosis is largely a self-limiting illness, it is accompanied by acute gastroenteritis and is a leading antecedent for the severe autoimmune complication Guillain-Barré syndrome (Allos, 1997; Nachamkin, 2002; Scallan Walter et al., 2020). This, together with additional autoimmune sequelae, such as post-infectious irritable bowel syndrome and reactive arthritis, confers a high disease burden to human campylobacteriosis in the United States and worldwide (Batz et al., 2013; Kirk et al., 2015; Scallan et al., 2015; Scallan Walter et al., 2019).
Campylobacter spp. are Gram-negative bacteria that colonize the gastrointestinal tract of various animals. Since its first description in 1963, the genus has grown to include several important human and animal pathogens (Man, 2011). Two species, Campylobacter coli and Campylobacter jejuni, are the primary contributors to human campylobacteriosis (>Nachamkin et al., 2008). Campylobacter gastroenteritis usually occurs after contact with raw poultry, ingestion of undercooked animal products, raw milk/vegetables/fruits, or contaminated water, or direct transmission from pets and other animals (Facciola et al., 2017). Poultry are recognized as the most important animal reservoir for infection to humans, with 60–80% of human cases in Europe attributable to exposure to raw or undercooked poultry products [European Food Safety Authority (EFSA) and European Centre for Disease Prevention and Control (ECDC), 2015].
Ciprofloxacin (fluoroquinolone) and azithromycin (macrolide) are the drugs of choice to treat Campylobacter enteritis, while intravenous aminoglycosides are used for serious bacteremia and other systemic infections due to Campylobacter (Aarestrup and Engberg, 2001). Recent years have seen a worldwide rise in Campylobacter antimicrobial resistance (AMR) to fluoroquinolones (Gupta et al., 2004; Nelson et al., 2007) and, to a lesser extent, macrolides and other classes of antibiotics (Padungton and Kaneene, 2003; Moore et al., 2006; Luangtongkum et al., 2009). As Campylobacter spp. are zoonotic pathogens, the development and spread of AMR in the animal reservoir may significantly limit the options for clinical treatment of campylobacteriosis in humans (Moore et al., 2006). Every year, an estimated 700,000 deaths are attributable to antimicrobial-resistant infections worldwide, a number projected to rise to 10 million deaths per year by 2050 in the absence of additional control measures (O’Neill, 2014). In 2019, the CDC listed drug-resistant Campylobacter as a serious threat, causing twice as many infections (448,400) in the United States each year as drug-resistant Salmonella [Centers for Disease Control and Prevention (CDC), 2019], and the World Health Organization has listed fluoroquinolone-resistant Campylobacter as one of six high priority antimicrobial-resistant bacteria (Tacconelli et al., 2018).
Several studies have linked the use of antimicrobials – in particular fluoroquinolones – in food animal production and veterinary medicine with the emergence and spread of resistant Campylobacter (Alfredson and Korolik, 2007). In the United States, the introduction of the fluoroquinolones, sarafloxacin, and enrofloxacin in the 1990s as growth promoters in poultry has been associated with an increase of fluoroquinolone-resistant Campylobacter in human cases (Anderson et al., 2003).
Campylobacter spp. are intestinal commensals shared among livestock species, thus providing a possible pathway for AMR to spread between different host species. Campylobacter jejuni is the dominant Campylobacter species in poultry while C. coli is the most common species recovered from swine (Horrocks et al., 2009), but this host specificity is not strict. A study conducted in China between 2008 and 2014 observed an apparent shift of the dominant species from C. jejuni to C. coli in chickens, with this species shift coinciding with an increased prevalence of macrolide-resistant C. coli (Wang et al., 2016). Interestingly, both species colonize turkeys at high rates (Wright et al., 2008; Wesley et al., 2009).
The in vitro transfer of antimicrobial resistance genes (ARGs) between different Campylobacter species via natural transformation has been extensively demonstrated. For instance, erm(B) and aph(2″)-If (conferring resistance to macrolides and aminoglycosides, respectively) could be transferred from C. coli to C. jejuni via natural transformation (Qin et al., 2014; Wang et al., 2014). However, previous experimental studies have favored the use of extracted DNA in transformations. The aim of this study was to evaluate the possible transfer of ARGs between different Campylobacter strains of the same or different species (C. coli and C. jejuni) and originating from different host species (swine and turkeys) using experimental designs closer to real-world conditions. This was evaluated both in vitro, using co-culture experiments, and in vivo, using experimental dual-strain infections of turkeys.
Materials and Methods
Campylobacter Strains and Growth Conditions
The C. jejuni and C. coli strains used in this study were isolated from turkey feces or ceca, from swine feces, or from the environment at different farms in eastern North Carolina, South Carolina, or Virginia between 2003 and 2014 (Table 1). Strains were first isolated using direct plating on modified charcoal cefoperazone deoxycholate agar (mCCDA; Oxoid, Basingstoke, Hampshire, England) and incubated in anaerobic jars containing a CampyPak Plus microaerobic system (Becton Dickinson, Sparks, MD) at 42°C for 48h (Smith et al., 2004). Strains were then subcultured on different media: (i) non-selective Mueller-Hinton agar (MHA; Becton Dickinson, United States), (ii) modified selective Campy Line agar (CLA-S; Line, 2001) containing 25μg/ml sulfamethoxazole in addition to the original antibiotics (trimethoprim, vancomycin, and polymyxin B; Line et al., 2008), and (iii) selective and differential CHROMagar™ Campylobacter (CAC; CHROMagar, Paris, France), and incubated at 42°C in an 10% CO2 incubator.
Table 1.
Campylobacter jejuni and Campylobacter coli strains used in this study.
| Species and strain1 | Sample | Source | Sampling date |
|---|---|---|---|
| Cc 6461 | feces | swine | 04/2004 |
| Cc 6067 | drinker water* | turkey house | 11/2003 |
| Cc 13150 | feces | turkey | 09/2012 |
| Cj 15152A | cecum | turkey | 11/2004 |
| Cj 15152B | cecum | turkey | 11/2004 |
| Cj 14229–5 | feces | turkey | 08/2013 |
| Cj 14398–5 | feces | turkey | 09/2013 |
| Cj 11601MD | jejunum | turkey | 09/2006 |
| Cj 6631 | feces | turkey | 05/2004 |
| Cj 6535 | feces | turkey | 04/2004 |
| Cj JCC | cecum | turkey (processing plant) | 10/2014 |
Strains highlighted in gray have been sequenced previously (Dutta et al., 2016).
Water sampled from the drinker in the turkey house.
Cc, Campylobacter coli and Cj, Campylobacter jejuni.
Antibiotic Susceptibility Profiles
Campylobacter spp. strains were selected on the basis of their AMR profiles. They were initially tested on MHA plates with selected antibiotics at the following concentrations: gentamicin (50μg/ml), kanamycin (50μg/ml), streptomycin (50μg/ml), ampicillin (50μg/ml), erythromycin (20μg/ml), and tetracycline (15μg/ml). The same concentrations of antibiotics were used in selective MHA used to recover “newly emerged strains” (see next paragraph) after in vitro and in vivo experiments. Antibiotics were purchased from Sigma-Aldrich (Saint Louis, Missouri, United States). Selected strains and “newly emerged strains” (see experiments below) were further tested by the Veterinary Diagnostic Laboratory at Iowa State University (ISUVDL) using CAMPY2 Sensititre® plates (VersaTREK Diagnostics/Thermo Fisher, United States) to determine minimal inhibitory concentrations to azithromycin (AZI), ciprofloxacin (CIP), clindamycin (CLI), erythromycin (ERY), florfenicol (FFN), gentamicin (GEN), nalidixic acid (NAL), telithromycin (TEL), and tetracycline (TET) as per the manufacturer’s guidelines. Antimicrobial resistance was determined according to the interpretative criteria recommended by the Clinical and Laboratory Standards Institute [Clinical and Laboratory Standards Institute (CLSI), 2017].
In vitro Transfer of ARGs
In vitro experiments were conducted with 11 strain pairs to investigate the possible transfer of ARGs conferring resistance to erythromycin, kanamycin, gentamicin, streptomycin, and tetracycline. Experiments involved a single species (Two pairs with C. coli×C. coli; 5 pairs with C. jejuni×C. jejuni) or two species (Four pairs with C. coli×C. jejuni) and utilized two strains with mutually exclusive resistance for selection. The donor and the recipient strains were identified a posteriori. Campylobacter colonies showing a dual-resistance pattern, i.e., resistance markers of both parental strains, are hereafter referred to as newly emerged strains (NES) for “newly emerged strain.”
Whole genome sequencing was performed to identify if the newly acquired phenotypic resistance was due to a resistance-associated point mutation, a displacement of a preexisting non-functional resistance gene by a functional version, or a new acquisition of a resistance gene. The experiments utilized three different techniques:
Transformation With Naked DNA on Solid Media
Donor DNA was extracted from a Campylobacter liquid culture grown in Mueller-Hinton broth using the PureLink™ Genomic DNA kit (Invitrogen, Carlsbad, CA) following the manufacturer’s instructions. A loopful from a 24-h-old culture of one strain grown on CLA-S microaerobically at 42°C was spotted into MHA plates (five spots per plate) and 4μl genomic DNA (80μg/μl) of the other strain (this was only tested with strain 6461 DNA) was added to each spot and mixed, with the diameter of each spot after mixing being ca. 0.5cm. The plates were incubated up to 24h microaerobically at 42°C.
Co-culture of Two Strains on Solid and Biphasic Media
A loopful of the two strains was spotted into MHA solid media (five spots per plate) on the same spot and mixed, with the diameter of each spot after mixing being ca. 0.5cm. The plates were incubated for up to 24h microaerobically at 42°C. The biphasic medium consisted of 5ml of MHA overlaid with 5ml of Mueller-Hinton broth in a sterile vented CELLSTAR® 25-cm2 tissue culture flask (Greiner Bio-One North America Inc., United States). A loopful of both strains was added to the media in duplicate and co-cultured for up to 24h microaerobically at 42°C. After incubation, either all of the material from a spot (solid media) or 100μl of broth (biphasic media) was spread on 12-cm-diameter MHA plates containing either individual antibiotics (for the recovery and enumeration of parental strains) or an appropriate combination of antibiotics (for the recovery and enumeration of NES). This added up to three selective plates per replicate, for a total of seven replicates (five spots from solid media and two replicates from biphasic media). Colonies were further plated on selective CAC to confirm Campylobacter.
Experimental Design of the Animal Study
This experiment was conducted according to National Animal Disease Center Institutional Animal Care and Use Committee approved protocol. Male day-of-hatch Nicolas turkey poults (n=135) were obtained from a commercial hatchery and co-housed upon delivery and the following day to acclimate prior to the start of the study. The rooms were on a 12-h light/dark cycle with positive air pressure (> 10 air changes per hour). Turkeys were fed a turkey starter ration (Damron and Sloan, 1995) ad libitum for the entire 6-week study. After the acclimation, poults were randomly split into four rooms: three rooms (n=35 each) for inoculated birds and one (n=30) for controls. In the control group, (n=30) birds were gavaged with 1ml of sterile 1×phosphate-buffered saline (PBS) at Day 2 and euthanized for necropsy in groups of 10 on Days 8, 15, and 22 (Table 2). In the inoculation groups, birds were inoculated by oral gavage with 1ml of a Campylobacter suspension at Day 2 and a subset (n=10 per room) was euthanized for necropsy on Day 8. The remaining birds (n=25) were inoculated at Day 9 (with a strain different to the one inoculated at Day 2) with some (n=10) euthanized for necropsy on Day 15 and the remaining birds (n=15) euthanized for necropsy on Day 22 (Table 2). Birds were euthanized by CO2 asphyxiation followed by cervical dislocation and the cecal contents of each poult were harvested and placed in 50-ml tubes which were stored on ice until analysis for Campylobacter, typically within 2h.
Table 2.
Design and timeline of turkey infections.
| Bird age (days) | Day1 | Room 7 (control group) | Room 9 (inoculation group) | Room 10 (inoculation group) | Room 11 (inoculation group) |
|---|---|---|---|---|---|
| Hatch | D0 | delivery/co-mingling (n=135) | – | – | |
| 1 | D1 | co-mingling (n=135) | – | – | |
| 2 | D2 | n=30 PBS | n=35 Inoculation Cc6461 | n=35 Inoculation Cc13150 | n=35 Inoculation Cc6067 |
| 8 | D8 | Necropsy n=10 | Necropsy n=10 | Necropsy n=10 | Necropsy n=10 |
| 9 | D9 | – | n=25 Inoculation Cj14229-5 | n=25 Inoculation Cc6461 | n=25 Inoculation Cc6461 |
| 15 | D15 | Necropsy n=10 | Necropsy n=10 | Necropsy n=10 | Necropsy n=10 |
| 22 | D22 | Necropsy n=10 | Necropsy n=15 | Necropsy n=15 | Necropsy n=15 |
Day pertains to experimental time points corresponding to the indicated bird ages. PBS: birds were gavaged with sterile 1× phosphate-buffered saline. Cc and Cj preceding the strain number indicate C. coli and C. jejuni, respectively.
One room housed the control birds (gavaged with PBS) and three rooms housed the turkeys dual inoculated with Campylobacter spp.
Campylobacter inocula were prepared as follows: Each strain was cultured at 42°C in a 10% CO2 incubator on selective CLA-S medium for 48h and a loopful (approx. five colonies) was transferred into biphasic media in CELLSTAR® 25-cm2 tissue culture flasks (Greiner Bio-One North America Inc., United States) including CLA-S and Bolton broth (5ml/5ml) and cultured without shaking under the same conditions. The next day, a loopful (10μl) of the inoculated Bolton broth was used to inoculate two CELLSTAR® 75-cm2 tissue culture flasks (Greiner Bio-One North America Inc., United States) with the same biphasic media (10ml/10ml). Cultures were grown without shaking at 42°C microaerobically overnight. On the day of challenge, broth cultures were adjusted to an OD600 value of 0.4, centrifuged at maximum speed (5,251×g) for 10min, and resuspended in an identical volume of PBS. Campylobacter motility was assessed using dark-field microscopy as previously described (Sylte et al., 2019) and confirmed for all inocula.
Enumeration of Campylobacter in Inoculum and Cecal Contents
Colony forming units (CFU) in suspensions utilized as inoculum were enumerated by plating appropriate dilutions on CAC and enumerating colonies following incubation at 42°C in a 10% CO2 incubator. To enumerate Campylobacter in cecal contents, 1g of cecal material was diluted in 9ml of sterile PBS (making the 10−1 dilution), vortexed for 5s, and serially diluted to 10−6. Using the track-plating dilution method (Jett et al., 1997; Siragusa, 1999), we plated 10μl of each dilution in duplicate on CAC to which appropriate antibiotics were added as needed (see Supplementary Table S1). The plated volume was increased from 10μl to 100μl for the recovery of NES on selective media (Day 15 and 22). Plates were incubated for up to 72h at 42°C in a 10% CO2 incubator and magenta colonies (Campylobacter) were enumerated to calculate CFU/g of cecal contents. For statistical purposes, if no putative Campylobacter colonies were obtained on selective media, the CFU was assigned the value of 103CFU/g of cecal contents, i.e., the limit of detection (Sylte et al., 2018).
DNA Extraction, Library Preparation, and Sequencing
From selective media showing NES, a single colony was picked and subcultured on biphasic media (MH) overnight at 42°C in a 10% CO2 incubator and used for DNA extraction using the PureLink™ Genomic DNA kit (Invitrogen, Carlsbad, CA) following the manufacturer’s instructions. DNA concentrations were determined using a NanoDrop™ spectrophotometer (Thermo Fisher Scientific) and the Qubit® dsDNA Broad Range Assay with the Qubit Fluorimeter (Qiagen), and normalized to 200ng input DNA in 30μl final volume for Illumina MiSeq sequencing; 400ng DNA in 7.5μl final volume for sequencing using an Oxford Nanopore MinION standard flow cell; and 200ng DNA in 3.75μl final volume for sequencing using an Oxford Nanopore MinION Flongle flow cell.
DNA libraries were generated with the Nextera™ DNA Flex Library Prep kit (Illumina, San Diego, CA) according to the manufacturer’s instructions for MiSeq sequencing. For MinION sequencing, DNA libraries were generated with the rapid barcoding kit (SQK-RBK004; Oxford Nanopore, Oxford, United Kingdom) according to the manufacturer’s instructions.
Genome sequencing was performed at USDA, NADC, and Ames on a paired-end 300PE MiSeq (Illumina) instrument using MiSeq Reagent kit v3 and on a MinION instrument (Oxford Nanopore) using a standard flow cell (FLO-MIN106 R9.4.1) or a Flongle flow cell (FLO-FLG106). The MinION flow cell was run for 48–72h, and the resultant reads with a quality score > Q7 were either base called live in MinKNOW or afterward with Guppy v. 3.1.5 (Wick et al., 2019).
Bioinformatic Analysis
Assembly
Multiple web-based bioinformatics tools available on the online Galaxy platform1 (Afgan et al., 2018) were used to generate genome assemblies. FastQC Galaxy Version 0.72+galaxy1 (Andrews, 2010) was used to inspect the quality of the Illumina reads. Short-read (MiSeq, Illumina Inc.) and long-read (MinION™ Nanopore Inc.) sequences were assembled using Unicycler Galaxy Version 0.4.8.0 (Wick et al., 2017). For one isolate, Unicycler was unable to produce a complete genome and we generated a complete consensus assembly using the Trycycler pipeline2 – from assemblies produced by flye (Kolmogorov et al., 2019), miniasm (Wick and Holt, 2019), and raven3 – which was then polished with pilon (Walker et al., 2014). Assembled genomes were annotated using Prokka Galaxy Version 1.14.5 (Seemann, 2014). Default parameters were used for all software, except for the parameter – min_fasta_length which was set to 1,000 in Unicycler.
Pangenome Analysis
To determine the parental strain that served as a recipient or a donor for a specific NES (i.e., carrying resistance markers of both parental strains), the average nucleotide identity (ANI) between each of the parental genomes and the relevant NES was calculated with pyani (Pritchard et al., 2016). The parental genome with the greatest ANI relative to the NES was identified as the recipient. Next, for each in vitro experiment, three genomes (two parental and one NES) were included in a pangenome analysis using Roary (Page et al., 2015). The stringency for assigning coding sequences to a gene was set to 99% identity. The tool gifrop4 (v0.0.6) was used to help identify transfer of genes or consecutive blocks of genes (hereafter referred to as a “genomic island”) absent in the recipient strain but present in the NES. The workflow used to complete this analysis is available online.5
Genotypic Analysis for AMR and Virulence
The resistance determinants of Campylobacter parental strains were determined using the web-based tool ResFinder 3.16 (Zankari et al., 2012). Contigs (FASTA files) from each sample were screened for (i) chromosomal point mutations (both known and unknown) and (ii) acquired resistance genes with the default settings of 90% minimum identity and 20% minimum template length. For successful co-culture experiments, genomes were further screened for AMR and virulence genes using ABRicate version 1.0.1, which includes the databases ResFinder, CARD, ARG-ANNOT, and NCBI (Zankari et al., 2012; Gupta et al., 2014; Jia et al., 2017; Feldgarden et al., 2019). For ABRicate hits, the Prokka annotations were replaced with those from ABRicate. The NCBI AMRfinder tool (v3.9.8; Feldgarden et al., 2019) was also used to validate point mutation-based resistances.
Results
Several types of selective media were tested to recover parental strains and NES. Parental strain Cc 6461, resistant to streptomycin but sensitive to ampicillin (see resistance patterns below), did not grow on CAC or MHA supplemented with streptomycin and ampicillin but grew on CLA-S with the same added antibiotics, i.e., showing antagonistic interactions between the antibiotics already present in CLA-S and the additional antibiotics. For this reason, CLA-S usage was discontinued during the course of the study and CAC was exclusively employed.
Resistance Patterns of Wild-Type Campylobacter Strains
Phenotypic and genotypic resistance patterns of Campylobacter strains used in the co-culture experiments are summarized in Table 3, and minimum inhibitory concentration (MIC) results are provided in Supplementary Table S2. All strains exhibited resistance to one or more antimicrobials. Our genomic analyses detected streptomycin resistance either via the previously described rpsL K88R substitution (Olkkola et al., 2010) in Cj 6535 and Cj 6631, or via aadE (Pinto-Alphandary et al., 1990) in Cc 6461 (Table 3). Our analyses also identified two genes associated with aminoglycoside resistance in different strains: aph(2″)-If associated with both kanamycin and gentamicin resistance (Yao et al., 2017), and aph(3′)-IIIa associated with resistance to kanamycin but not to gentamicin (Gibreel et al., 2004; Crespo et al., 2016; Table 3). Erythromycin-resistant strain Cc 6461 showed the known substitution A2075G in all three copies of the 23S rRNA gene (Vacher et al., 2003); erm(B) associated with erythromycin resistance in certain Campylobacter strains (Qin et al., 2014) was not identified. The L22 A103V substitution associated with low-level macrolide resistance (Payot et al., 2006) was in the genome of two strains, Cc13150 and CjJCC, even though phenotypic macrolide resistance was not detected (erythromycin MICs 0.5μg/ml and 0.12μg/ml, respectively). Nalidixic acid (quinolone) resistance was associated with the gyrA T86I substitution (Wang et al., 1993) in the genome of three strains. Of note, this was identified using the AMRFinder tool (Feldgarden et al., 2019) but not ResFinder in the genome of two strains (Table 3). A closer analysis revealed that the T86I substitution was mediated by T86I ACA>ATT instead of the typically observed T86I ACA>ATA.
Table 3.
Antimicrobial resistance patterns of Campylobacter strains (n=11).
| Species and strain 1 | Phenotypic resistance | Chromosomal point mutations | Acquired antimicrobial resistance gene | ||
|---|---|---|---|---|---|
| Mutation | Predicted resistance phenotype | Resistance gene | Predicted resistance phenotype | ||
| Cj 15152A | AKG | aph(2′)-If | KG (aminoglycosides) | ||
| blaOXA |
|
||||
| Cj 15152B | AKG | aph(2″)-If | KG (aminoglycosides) | ||
| blaOXA | A (beta-lactams) | ||||
| Cc 13150a | AKQ | L22 A103Vb | Macrolides (low) | aph(3′)-IIIa | K (aminoglycosides) |
| gyrA T86Ic | Q (quinolones) | blaOXA | A (beta-lactams) | ||
| tet(O) p | T (tetracycline) | ||||
| Cj 14398–5 | AKG | aph(2″)-If | KG (aminoglycosides) | ||
| blaOXA | A (beta-lactams) | ||||
| Cj 11601MD | AKQT | gyrA T86I | Q (quinolones) | aph(3′)-IIIa | K (aminoglycosides) |
| blaOXA | A (beta-lactams) | ||||
| tet(O) p | T (tetracycline) | ||||
| Cc 6461 | TSE | 23S A2075G | E (macrolides) | aadE-Cc | S (aminoglycosides) |
| tet(O) | T (tetracycline) | ||||
| Cj 14229–5 | AKGT | aph(2″)-If | KG (aminoglycosides) | ||
| blaOXA | A (beta-lactams) | ||||
| tet(O) | T (tetracycline) | ||||
| Cj JCC | T | L22 A103Vb | Macrolides (low) | tet(O) p | T (tetracycline) |
| Cj 6535 | ATS | rpsL K88R | S (aminoglycoside) | blaOXA | A (beta-lactams) |
| tet(O/32/O) p | T (tetracycline) | ||||
| Cj 6631 | ATS | rpsL K88R | S (aminoglycoside) | blaOXA | A (beta-lactams) |
| tet(O/32/O) p | T (tetracycline) | ||||
| Cc 6067 | AQT | gyrA T86Ic | Q (quinolones) | blaOXA | A (beta-lactams) |
| tet(O) | T (tetracycline) | ||||
Strains highlighted in gray have been sequenced previously (Dutta et al., 2016).
Strain 13150 is phenotypically susceptible to tetracycline but carries a plasmid-borne tet(O).
L22 mutation was identified using the NCBI AMRfinder tool.
gyrA T86I substitution was identified using the AMRFinder tool but not ResFinder.
Plasmid-borne tet(O).
Cc, Campylobacter coli; Cj, Campylobacter jejuni; A, ampicillin/carbenicillin; E, erythromycin; G, gentamicin; K, kanamycin; Q, nalidixic acid; S, streptomycin; and T, tetracycline. Phenotypic resistance patterns were identified by plating the different strains on Mueller-Hinton agar supplemented with antibiotics. Genotypic resistance patterns – chromosomal point mutations and acquired antimicrobial resistance genes – were identified by sequencing the genomes of each strain (Illumina MiSeq and MinION Nanopore) and analyzing the contigs (obtained with Unicycler) using the ResFinder 3.1 web-based tool (available at https://cge.cbs.dtu.dk/services/ResFinder/), ABRicate, and the NCBI AMRfinder tool. “Chromosomal point mutations” refer to known mutations conferring a phenotypic resistance described under “predicted resistance phenotype.” Unknown mutations identified with ResFinder (in genes where known mutations occur; no predicted phenotype) are summarized in Supplementary Table S3. For the three previously sequenced strains highlighted in gray (Dutta et al., 2016), the reported AMR patterns were confirmed and additional information was obtained about beta-lactam resistance.
We obtained certain unexpected findings in the determination of AMR determinants. Specifically, Cc 13150 was phenotypically susceptible to tetracycline but harbored tet(O), typically associated with tetracycline resistance in Campylobacter spp. (Taylor et al., 1987; Zhao et al., 2016).
Resistance Patterns of “Newly Emerged Strains” (NES) Recovered From in vitro Experiments
Results from the 12 co-culture experiments are summarized in Table 4. In all co-culture experiments, we recovered both parental strains (data not shown). In six experiments, we also recovered isolates with a dual-resistance pattern, i.e., resistance markers of both parental strains (Figure 1). Putative NES had their genomes sequenced to characterize the resistance determinants.
Table 4.
Campylobacter co-culture in vitro.
| Parental strains (resistance pattern) | Antibiotics added to CAC | Putative NES | Recipient strain (NES pattern) | Acquired AMR | |
|---|---|---|---|---|---|
| Cj 6631 (TS) | Cj 15152A (AKG) | TK | no | ||
| Cj 6631 (TS) | Cj 15152B (AKG) | TK | no | ||
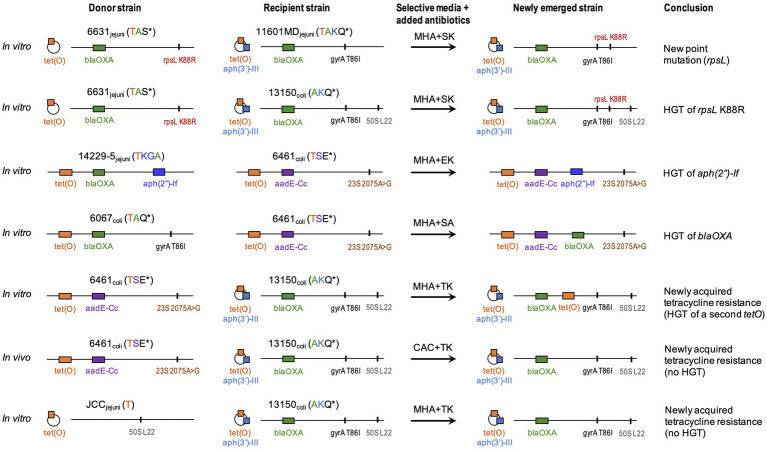
| Cj 6631 (ATS*) | Cj 11601MD (AKTQ*) | SK | yes | Cj 11601MD (AKTS*Q*) | Point mutation rpsL (S resistance) |
| Cj 6631 (ATS*) | Cc 13150 (AKQ*)+tet(O) | SK | yes | Cc 13150 (AKS*Q*)+tet(O) | HGT of mutated rpsL (S resistance) |
| SKQ | yes | ||||
| Cc 6461 (TE*S) | Cc 13150 (AKQ*)+tet(O) | TK | yes | Cc 13150 (ATKQ*)+tet(O) | Newly acquired phenotypic T / HGT tet(O) |
| SA | no | ||||
| SK | no | ||||
| SKQ | no | ||||
| Cc 6067 (ATQ*) | Cc 6461 (TE*S) | SA | yes | Cc 6461 (ATE*S) | HGT blaOXA (A resistance) |
| Cj 6535 (ATS*) | Cj 11601MD (AKTQ*) | SK | no | ||
| SKQ | no | ||||
| Cj JCC (T) | Cj 14398–5 (AKG) | TK | no | ||
| Cj JCC (T) | Cc 13150 (AKQ*)+tet(O) | TK | yes | Cc 13150 (AKTQ*)+tet(O) | Newly acquired phenotypic T / no HGT |
| Cj 14229–5 (AKGT) | Cc 6461 (TE*S) | SG | yes | Cc 6461 (TE*SKG) | HGT aph(2″)-If (KG resistance) |
| EK | yes | ||||
| SEK | yes | ||||
| SKG | yes | ||||
| Cc 6461 (TE*S) | Cj 14398–5 (AKG) | SKG | no | ||
| Cc 6461 (TE*S) | Cj 11601MD (AKTQ*) | SK | no | ||
| SKQ | no | ||||
indicates resistance due to a point mutation. Newly acquired resistance in NES compared to the parental recipient is in bold.
Cc, Campylobacter coli; Cj, Campylobacter jejuni; A, ampicillin/carbenicillin; E, erythromycin; G, gentamicin; K, kanamycin; Q, nalidixic acid; S, streptomycin; and T, tetracycline. The two parental strains were (i) co-cultured on Mueller-Hinton Agar (MHA), (ii) co-cultured on biphasic medium (5ml MHA+5ml Mueller-Hinton broth), or (iii) treated so that genomic DNA of one parental strain was mixed into colonies of the second parental strain on MHA. Resulting cultures were plated on selective media to recover newly emerged strains (NES). Whole genome sequence data from NES and parental strains were compared to identify donor vs recipient. The phenotypic resistance of each strain is provided in parentheses; Cc 13,150 which harbors tet(O) but does not express resistance to tetracycline is not indicated as “T.”
Figure 1.
Summary of in vitro and in vivo experiments. Strains names are followed by Campylobacter species and phenotypic resistance. Acronyms include A, ampicillin/carbenicillin; E, erythromycin; G, gentamicin; K, kanamycin; Q, nalidixic acid; S, streptomycin; and T, tetracycline. *indicates resistances due to a point mutation. Genotypic resistance can be carried on a chromosome (horizontal line) or a plasmid (circle). Resistance genes (colored blocks) are as: tet(O) (T resistance); aph(3′)-IIIa (K resistance); blaOXA (A resistance); aadE-Cc (S resistance); and aph(2′)-If (KG resistance). Point mutations (vertical lines) are as: gyrA T86I (nalidixic acid/ciprofloxacin: Q resistance); 23S A2075G (E resistance); rpsL K88R (S resistance); and L22 A103V (low macrolide resistance). Whole genome sequence data from newly emerged strain and parental strains were compared to identify donor vs recipient. CAC, CHROMagar™ Campylobacter; MHA, Mueller-Hinton agar; and HGT, horizontal gene transfer. Strain 13150 (phenotypically susceptible to tetracycline) carries a plasmid-borne non-functional tet(O) represented as a striped orange block.
Assessment of in vitro HGT via Experimental Dual Inoculation in Turkeys
Based on the results from the in vitro experiments, three Campylobacter pairs were chosen for further work in vivo. Turkeys were inoculated with the following strain pairs: Cc 6461 and Cj 14229–5 (room 9); Cc 6461 and Cc 13150 (room 10); and Cc 6461 and Cc 6067 (room 11; Table 2). The poults were free of Campylobacter prior to oral gavage and the control room (gavaged with PBS) remained free of Campylobacter throughout the experiment.
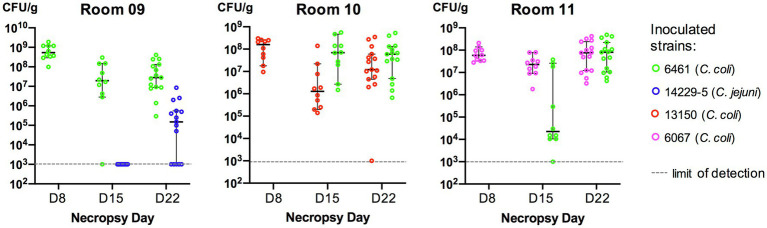
On the first inoculation (Day 2), inoculum concentrations of the strains at the day of challenge were as: 3.8×1010CFU/ml (strain 6461, room 9); 4.7×109CFU/ml (strain 13150, room 10); and 1.0×109CFU/ml (strain 6067, room 11). In birds necropsied at Day 8, all three inoculated strains could be recovered from rooms 9 to 11 (Figure 2). On the second inoculation (Day 9), inoculum concentrations of the strains at the day of challenge were as: 10×108CFU/ml (strain 14229–5, room 9); 5.5×108CFU/ml (strain 6461, room 10); and 9.5×108CFU/ml (strain 6461, room 11). In birds necropsied at Day 15, five of the six strains could be recovered from rooms 9 to 11, with strain 14229–5 not being recovered from room 9 on selective media (CAC with kanamycin; Figure 2). However, 1week later in birds necropsied at Day 22, both parental strains could be recovered from all necropsied birds (Figure 2).
Figure 2.
Cecal colonization levels (CFU/g cecal content) of Campylobacter in inoculated turkeys. Each bird was inoculated with two Campylobacter strains (one at Day 2 and the other at Day 9). Cc, Campylobacter coli and Cj, Campylobacter jejuni. In each room, 10–15 birds were euthanized at each necropsy and Campylobacter was recovered from cecal contents (see Methods). If no colonies resembling Campylobacter grew on selective media, the sample was assigned the value of 103CFU/g of cecal contents (limit of detection). Mean CFU and 95% confidence intervals are represented (as bars) for each set.
On dual-selective CAC media designed to recover NES, we recovered a single colony from the cecal content of bird N3-R10-137 infected with Cc 6461 and Cc 13150 on CAC with tetracycline and kanamycin (TK). Cc 6461 was tetracycline resistant and carried a chromosomal tet(O), while Cc 13150 was, as indicated above, phenotypically susceptible to tetracycline, despite carrying a plasmid-borne tet(O). The hybrid resistance pattern (TK) in the NES was compatible with the acquisition of tetracycline resistance (T) from donor Cc 6461. The pangenome analysis indicated that the colonies recovered on TK were highly similar to the parental strain Cc 13150. A close comparison of the tetO regions in the parental strains and the NES suggested a homologous recombination event via which the non-functional tet(O) of Cc13150 was replaced by a functional tet(O) from donor strain Cc 6461 (Table 5; see details in “Pangenome analysis results”). The hypothesis that the defective tet(O) function was restored by a gene displacement is further supported by the detection of other homologous recombination events in other regions of the NES genome.
Table 5.
Animal experiment results.
| Parental strains (resistance pattern) | Antibiotics added to CAC | Putative NES | Recipient strain (NES pattern) | Acquired AMR | |
|---|---|---|---|---|---|
| Cj 14229–5 (AKGT) | Cc 6461 (TE*S) | SG | no | ||
| Cc 6461 (TE*S) | Cc 13150 (AKQ*)+tet(O) | TK | yes | Cc 13150 (ATKQ*)+tet(O) | Newly acquired phenotypic T/HGT tet(O) |
| Cc 6067 (ATQ*) | Cc 6461 (TE*S) | SA | no | ||
indicates resistances due to a point mutation. New resistance in NES as compared to recipient is in bold.
Cc, Campylobacter coli; Cj, Campylobacter jejuni; A, ampicillin/carbenicillin; E, erythromycin; K, kanamycin; Q, nalidixic acid; S, streptomycin; T, tetracycline; and HGT, horizontal gene transfer. Phenotypic T: The NES is genetically similar to recipient strain 13150 (tetracycline susceptible) except it has a newly acquired tetracycline resistance. Turkeys were inoculated with two Campylobacter strains 7days apart. Cecal contents were plated after necropsy (Days 8, 15, and 22) on selective CHROMagar™ Campylobacter (CAC) plus additional antibiotics. The genome of recovered newly emerged strains (NES) – showing resistance markers of both parental strains – was sequenced and compared with both parental strains to identify donor vs recipient. The phenotypic resistance of each strain is provided in parentheses. Strain Cc 6461 carries tet(O) and is tetracycline resistant; strain Cc 13150 carries tet(O) but is tetracycline susceptible and thus not noted “T.”
Pangenome Analysis Results
Overall, the emergence of a novel resistance trait in the recipient strains occurred via a spontaneous point mutation or via a recombination-mediated horizontal gene transfer. We observed different HGT events: (i) the transfer of a genomic island including an ARG that was previously absent or (ii) the allelic transfer of an ARG, i.e., the replacement of an ARG already present by a mutated version of the same gene via homologous recombination.
The pangenome analysis was used to determine which of the two strains in the co-culture were the source of the ARG vs the recipient of the acquired resistance and to highlight genomic differences between parental strains and the NES, i.e., gene or genomic island transfers (genes absent in the recipient but present in both donor and NES) and deletions (genes present in the recipient but not the NES).
Mutation-Mediated Novel Resistance
In the in vitro co-cultures of Cj 6631 and Cj 11601MD, the NES recovered on KS was highly similar to the parental strain Cj 11601MD with an additional mutated version of gene rpsL (K88R) conferring resistance to streptomycin (Figure 1). To differentiate between the horizontal transfer of K88R rpsL from Cj 6631and a novel spontaneous mutation in Cj 11601MD, we compared the rpsL sequences between the strains. Aside from the K88R mutation, Cj 6631 rpsL sequence included two single-nucleotide polymorphisms that were absent in the Cj 11601MD and the NES genomes. Because the rest of the NES genome was also highly similar to Cj 11601MD, this suggest a spontaneous point mutation in Cj 11601MD, rather than the displacement of a mutated rpsL from Cj 6631.
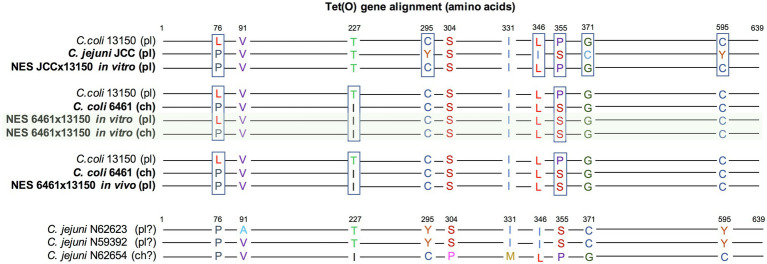
In in vitro co-cultures of Cj JCC and Cc 13150, the NES recovered on TK (compatible with the acquisition of tetracycline resistance from donor Cj JCC) was highly similar to parental strain Cc 13150 but expressing resistance to tetracycline (Figure 1). Sequencing revealed that all three strains (parental and NES) carried tet(O) commonly associated with tetracycline resistance in Campylobacter (Sougakoff et al., 1987), despite Cc 13150 being phenotypically susceptible to tetracycline (MIC=0.25μg/ml). We compared the “functional” and the “nonfunctional” versions of tet(O) and found that the “nonfunctional” version (in Cc 13150) had a leucine at residue 76, whereas the “functional” versions had a proline at this position (Figure 3). The observed tet(O) L76P substitution seems to restore the functionality of tet(O) (i.e., tetracycline resistance) in the NES. Of note, when adding three published tetracycline-susceptible strains carrying a “nonfunctional” tet(O) to the comparison (Whitehouse et al., 2018), a proline was identified at residue 76 of the tet(O) gene, and tet(O) in one of these strains (N59392) was identical to tet(O) of the tetracycline-resistant strain Cj JCC (Figure 3); hence, an explanation other than the L76P mutation must be found for these three published tetracycline-susceptible strains that harbor tet(O).
Figure 3.
Alignment of the tet(O) genes of Campylobacter strains. Parental strains Cc 6461, Cc 13150, and Cj JCC were compared to newly emerged strains (NES) recovered from in vitro co-culture experiments and the experimental infections of turkeys. Three previously published tetracycline-susceptible strains carrying tet(O) (Whitehouse et al., 2018) were included for comparison (BioProject PRJNA292664; accession numbers N59392, N62623, and N62654). The tet(O) gene was either plasmid-borne (pl) or chromosomal (ch; there is some uncertainty noted “?” for the public strains as the draft genomes are broken into many contigs). The tetracycline-resistant NES recovered from the in vitro co-culture of Cc 6461 and Cc 13150 carried two copies of the tet(O) gene, one chromosomal [corresponding to the chromosomal tet(O) of Cc 6461 (Crespo et al., 2012)] and one plasmid-borne (highlighted in green). The “nonfunctional” version of Cc 13150 has a leucine at residue 76, whereas the “functional” versions have a proline at residue 76.
Horizontal Transfer of Resistance Genes
In co-cultures of Cc 6461 and Cc 13150, the pangenome analysis detected the transfer of a chromosomal genomic island including tet(O) (2661bp) from donor Cc 6461 to the chromosome of the recipient Cc 13150 (chromosomal), leading to two copies of tet(O) in the NES, one chromosomal, newly acquired, and the original plasmid-borne copy (Figures 1, 3; Table 6). Of note, the plasmid-borne tet(O) in the NES differed from the plasmid-borne tet(O) in strain Cc 13150 by two amino acids. It was not possible to determine if this was due to a double mutation in the original gene or a displacement with a tet(O) from donor Cc 6461 with an additional point mutation (Figure 3).
Table 6.
Detailed information regarding genomic islands transfers.
| Experiment | Start | End | Island length | Number genes | Genes transferred | Annotation |
|---|---|---|---|---|---|---|
| 6631×13150 in vitro | 433,527 | 439,905 | 6,378 | 8 | rpsL rpsG fusA group_1637 group_1638 group_1639 group_1640 trpC | 30S ribosomal protein S12 (K88R-mutated) 30S ribosomal protein S7 elongation factor G exporting protein methyltransferase domain-containing protein hypothetical protein lipoprotein indole-3-glycerol phosphate synthase |
| 14229–5×6461 in vitro | 488,562 | 493,264 | 4,703 | 6 | group_363 group_364 group_368 group_367 group_366 group_365 | IS200/IS605 family transposase IS605 aminoglycoside O-phosphotransferase APH(2″)-If hypothetical protein IS1595 family transposase ISAcsp6 hypothetical protein hypothetical protein |
| 6067×6461 in vitro | 1,502,456 | 1,504,198 | 1,743 | 2 | group_177 btuD | beta-lactamase OXA-133 vitamin B12 import ATP-binding protein BtuD |
| 6461×13150 in vitro | 140,166 | 142,826 | 2,661 | 2 | tet(O) group_901 | tetracycline resistance ribosomal protection protein Tet(O) hypothetical protein |
For each experiment, three genomes (from the two parental strains and one “newly emerged strain”) were included in a pangenome analysis using Roary (Page et al., 2015). Antimicrobial resistance genes and resistance-conferring mutated genes are in bold. Of note, genes present in both parental strains and the newly emerged strain, and contiguous to the transferred genes might have also been transferred as part of the AMR genomic island – but undetectable by the analysis because they are 100% identical, suggesting that the “island length” below is underestimated.
A different outcome was observed in the in vivo dual inoculation of the same two strains, Cc 6461 and Cc 13150. Instead of the acquisition by Cc 13150 of a second copy of tet(O) detected in vitro, we observed in vivo the displacement of the non-functional tet(O) present in the plasmid of recipient strain Cc 13150 by the functional tet(O) from donor strain Cc 6461 by homologous recombination.
Additional instances of HGT of resistance genes were noted. In co-cultures of Cc 6461 and Cc 6067, the NES recovered on SA (compatible with either the acquisition of streptomycin resistance from Cc 6461 or the acquisition of ampicillin resistance from Cc 6067) showed the acquisition of ampicillin resistance via transfer of a genomic island including blaOXA (1743bp) from donor Cc 6067 to recipient Cc 6461 (Figure 1; Table 6). In co-cultures of Cc 6461 and Cj 14229–5, the NES recovered on EK showed the acquisition of kanamycin/gentamicin resistance via the transfer of a genomic island (4703bp) including aph(2″)-If from donor 14229–5 to recipient 6461 (Figure 1; Table 6).
In addition to acquisition of new ARGs, we noted HGT-mediated transfer of genes harboring specific substitutions associated with resistance. For instance, in co-cultures of Cj 6631 and Cc 13150, the NES recovered on KS was highly similar to the parental strain Cc 13150 with an additional K88R-mutated version of rpsL (Figure 1). The pangenome analysis confirmed the acquisition of streptomycin resistance via the transfer of a genomic island (6378bp) including a K88R-mutated version of rpsL from donor Cj 6631 to recipient Cc 13150, and not a novel spontaneous mutation of rpsL (Table 6).
In the co-culture of Cc 6461 and Cc 6067, several virulence-associated genes (cheW, cheA, cheV, fliF, and fliG) were also identified in close proximity but not contiguous to the transferred genomic island including the resistance gene blaOXA, while in the co-culture of Cc 6461 and Cc 13150, no virulence genes were identified in close proximity to the genomic island that harbored the resistance gene tet(O) (Supplementary Figure 1).
Of note, aside from the resistance genes transfers, each NES showed various other transfer or deletion events that are not fully detailed here (see Supplementary Table S4 for details).
Reproducibility of the Horizontal Transfer of Resistance Genes
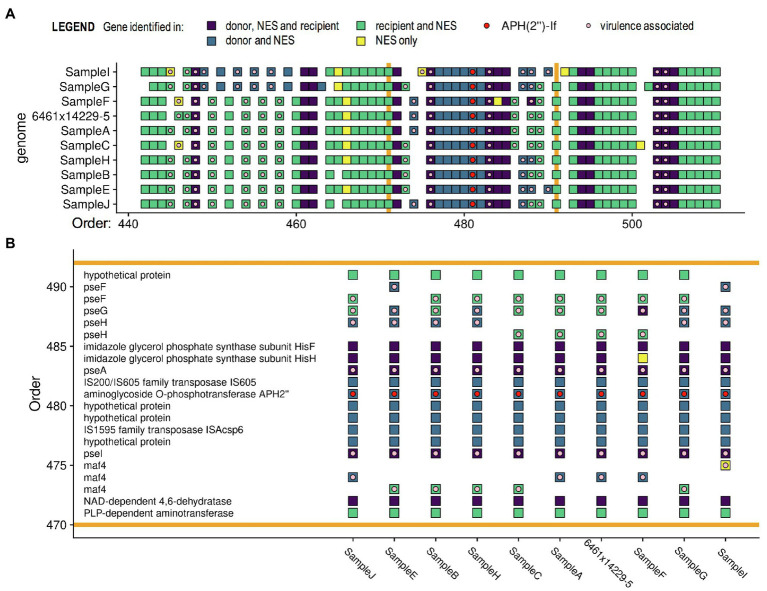
To determine reproducibility of the observed HGT events, we replicated the co-culture of Cc 6461 and Cj 14229–5, and subcultured and sequenced 10colonies (A–J) showing a dual-resistance pattern (SG). The NES originally sequenced and the nine newly sequenced NES from the replicate experiment (A–C and E–J) showed the same transfer of a genomic island including the resistance gene aph(2″)-If and five other genes at the same location in the recipient genome, but some variability in other transfer and deletion events elsewhere in the genome (Figure 4 and Supplementary Table S4). We detected the presence of genes associated with Campylobacter motility (such as pseA and pseI) in close proximity to aph(2″)-If in all sequences, with additional virulence-associated genes (e.g., maf4) being transferred in six out of 10 isolates (Figure 4). Of note, genes present in both parental strains and the NES (in dark purple in Figure 4) and contiguous to the genomic island carrying the ARG (in blue in Figure 4) might have been transferred concomitantly with it but were invisible to the analysis because these genes were 100% identical in the parental strains and the NES. Such findings suggest that contiguous pseA, pseI, and maf4 (involved in Campylobacter motility) were likely transferred with AMR as part of a single HGT, rather than multiple HGTs (Figure 4).
Figure 4.
Pangenome comparisons of 10 isolates of in vitro co-cultures of C. coli 6461 and C. jejuni 14229–5. (A) Only a section of the genomes is presented, highlighting (B) the transferred genomic island (between the orange bars) including resistance gene aph(2′)-If associated with kanamycin/gentamicin resistance. Each square represents a gene (annotated using PROKKA). The pangenome analysis was done using Roary, the virulence (light pink dot) and resistance genes (red dot) were annotated using ABRicate, and the figure was made with ggplot2 in “R.” Genes annotated as virulence genes are associated with Campylobacter motility. Of note, genes classified as the same gene in two strains have sequences ≥ 99% identical. Genes in yellow, i.e., found only in the newly emerged strain (NES), are genes that are less than 99% identical to the two parental genes, which might be due to chimeras of donor and recipient versions of the gene.
Discussion
Despite the increasing prevalence of AMR in C. jejuni and C. coli in food animals, such as swine and poultry, as well as the potential impact to human campylobacteriosis treatment, there is limited information on Campylobacter AMR acquisition in animals co-infected with different Campylobacter strains. This is especially true for turkeys, which are known for carrying multidrug-resistant Campylobacter strains of both C. coli and C. jejuni (Wright et al., 2008; Wesley et al., 2009). Here, we used turkey dual inoculations to investigate AMR transfer between different Campylobacter strains and species (originating from swine and turkeys), after also investigating the process in simpler models involving co-cultures in vitro.
Using in vitro co-cultures, we identified the horizontal transfer of chromosomal genes conferring resistance to beta-lactam (blaOXA), aminoglycoside [aph(2″)-If; rpsL K88R], and tetracycline [tet(O)], as well as a spontaneous mutation (in rpsL) conferring a novel resistance to streptomycin. Our findings suggest that even though barriers exist to keep exogenous DNA (e.g., from gut commensals) from being incorporated in Campylobacter species – through CRISPR-Cas and restriction-modification systems (Gardner and Olson, 2012) – transformation can mediate the horizontal transfer of resistance determinants between C. coli strains, as well and between C. jejuni and C. coli, including between strains originating from different animal hosts (swine and turkey).
Both C. coli and C. jejuni are naturally competent, i.e., can take up naked DNA without any special treatment (Wang and Taylor, 1990). Interestingly, the HGT-mediated acquisition of previously absent resistance genes always occurred through the transfer of consecutive blocks of genes – genomic islands – via homologous recombination (i.e., in regions of the genome showing high similarity between the recipient and donor strains). Our study also found two occurrences of resistance-associated genes being displaced by a mutated version of the same gene through homologous recombination (HGT of a single gene). Interestingly, even though we detected tet(O)-carrying plasmids in several parental strains (Figure 1), we did not observe any transfer of such plasmids. This supports previous reports that transformation of Campylobacter with plasmid DNA is much less efficient than with chromosomal DNA (Wang and Taylor, 1990; Taylor, 1992). In vitro studies on the natural transformation of Streptococcus pneumoniae (Prudhomme et al., 2002) and Acinetobacter spp. (De Vries and Wackernagel, 2002) identified short (3–10 nucleotides) stretches of sequence identity (microhomology) between donor and recipient DNA serving as recombinational anchors, leading to homology-facilitated illegitimate recombination. For S. pneumoniae, transformation occurred preferentially within segments of high GC content (Prudhomme et al., 2002). Further experimental studies will be needed to identify the mechanisms involved in the homologous recombination of Campylobacter spp.
In our experiments, streptomycin resistance was acquired once through a K88R mutation of rpsL, and once through a horizontal transfer of a K88R-mutated rpsL. The horizontal transfer of mutated genes was expected as previous studies have suggested that, in the absence of selective pressure, the frequency of transformation to resistance greatly exceeds the frequency of spontaneous mutations to resistance (transformation to resistance was observed with mutated rpsL (Oyarzabal et al., 2007), 23S rRNA (Kim et al., 2006), and gyrA (Wilson et al., 2003; Kim et al., 2008).
Of note, the acquisition of a genomic island including a K88R-mutated version of rpsL in the NES recovered from the co-culture of Cj 6631 and Cc 13150 was only detected after manually checking the rpsL region in order to confirm the occurrence of a spontaneous point mutation K88R. From the pangenome perspective, rpsL was more than 99% identical in all genomes (one nucleotide difference between the mutated and non-mutated gene) and hence was excluded from the list of possibly transferred genes. This also happened when trying to detect a mutated version of tet(O) (which restored the gene functionality) in the NES recovered in vivo (6461×13150). The pangenome framework can struggle to identify allelic transfers, such as these, where allelic versions of the gene conferring resistance are highly similar.
In vitro results were highly reproducible – co-culture of specific pairs consistently yielded NES recovered on selective media while other pairs consistently yielded no NES recovered – suggesting that HGT events are not random and/or that some strains might be more prone to horizontal gene transfers. In NES sequenced from co-cultures of Cc 6461 and Cj 14229–5, the horizontal transfer of a resistance gene consistently occurred through the transfer of the same genomic island, and at the same place on the recipient genome, showing that transformation is not random, and possibly indicating “recombination hotspots” (Yahara et al., 2014).
Of note, Cc 6461, the only swine-derived strain in our panel, was involved in four HGT events (including in a turkey host) as a donor and as a recipient, which suggests this strain may be more prone to genetic exchanges with other strains. The ability of turkeys to be colonized by both C. coli and C. jejuni has triggered interest as well as concern because swine and turkey farms can be located in close proximity, which is the case in the southeastern United States where our Campylobacter strains were isolated (Wright et al., 2008; Bolinger et al., 2018). In our dual-inoculation experiment, co-colonization with different strains of C. coli (assessed by plating cecal content) was successful with no apparent dominance between the two strains; co-colonization with C. jejuni and C. coli also occurred even though the C. jejuni strain (inoculated second) was not recovered the first week after inoculation but was recovered 2weeks after inoculation. Turkeys, if co-colonized by C. coli and C. jejuni, could be a host species where resistance transfers between the two species are more likely to occur and be further transferred to other animal host species. The possible transfer of resistance between bacteria in swine and turkeys warrants further investigation.
Interestingly, in two out of five experiments that showed the acquisition of resistance through HGT, the transfer occurred in close proximity to genes related to chemotaxis or the biosynthesis of functional flagella, and thereby to motility and pathogenesis of Campylobacter (Guerry et al., 2006; Salah Ud-Din and Roujeinikova, 2018). Genes associated with motility/colonization are frequently recombining genes and are considered to contribute to the pathogen’s adaptive potential (Park et al., 2020). Hence, the co-localization of ARGs near such motility genes on recombination hotspots could impact the transmission rate of ARGs through the population.
Sequencing strain Cc 13150, phenotypically susceptible to tetracycline, revealed a plasmid-borne tet(O). Such a discrepancy between phenotypic and genotypic tetracycline resistance in Campylobacter is uncommon but has been reported before, being noted in four out of 327 tet(O)-harboring isolates (Whitehouse et al., 2018). In three experiments involving strain Cc 13150, a newly acquired phenotypic resistance to tetracycline was observed in NES. Different mechanisms were involved as: a tet(O) P76L mutation that restored the function of the gene present in a plasmid in Cc 13150, or the acquisition of a new (functional) tet(O) gene by HGT. In one HGT event, a second (chromosomal) tet(O) was acquired, while in the other HGT, the non-functional tet(O) present in Cc 13150 was replaced by a functional tet(O) originating from donor Cc 6461 by homologous recombination.
Our study provides evidence for the natural horizontal transfer of a chromosomal tet(O) between different C. coli strains in the turkey intestine. During the in vivo experiment, birds were not inoculated with both strains at the same time to ensure that any resistance gene transfer would occur in the animal intestine (post-colonization) rather than during a co-inoculation. From the cecal contents of dual-strain inoculated birds, both parental strains were recovered, as well as a single NES from a bird that was inoculated with strains Cc 6461 and Cc 13150. In the recovered NES – with a newly acquired tetracycline resistance gene compared to the recipient strain – we detected a horizontal gene transfer on a plasmid, leading to the displacement of a non-functional tet(O) by a functional tet(O) by homologous recombination. This result was slightly different from the in vitro co-culture result where a second (chromosomal) tet(O) was acquired.
Several hypotheses could explain why other horizontal transfers of ARGs observed in vitro were not detected in vivo. Firstly, transformation between different Campylobacter strains and species is likely a rare event and it might be more difficult to recover recombinant strains from complex environments, such as cecal contents. Secondly, laboratory conditions are markedly different from the environment encountered by Campylobacter colonizing a turkey cecum. In vivo, naked DNA may become degraded by extracellular nucleases of other cecal microbes, reducing the frequency of transformation. Finally, transmission and adaptation of a new phenotype may take longer than our study – turkeys are reared up to 21weeks of age before slaughter – or transformation frequency might be lower in vivo.
This work provides further evidence that transformation can mediate the in vitro and in vivo HGT of antimicrobial resistance genes in Campylobacter, within and between species (C. coli and C. jejuni). In vitro, resistance gene transfer was not a rare event and was highly reproducible between specific strains, and homologous recombination seemed to be a mechanism of choice for resistance spread. Finally, some strains seem more prone to the acquisition or transfer of resistance genes. Being able to identify such strains could be important for prevention of antimicrobial resistance transmission.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The animal study was reviewed and approved by the National Animal Disease Center (NADC) Institutional Animal Care and Use Committee (IACUC).
Author Contributions
Project design and coordination were performed by TL. ZH isolated the Campylobacter strains used in the study and tested them for phenotypic resistance. VG-C performed the in vitro experiments, DNA extraction, sample library preparation, and sequencing. TL, VG-C, JM, JQ, and MS performed the animal experiment. JT and VG-C performed the bioinformatics analyses. VG-C wrote the initial manuscript and later versions were based on input and suggestions from all. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the United States Department of Agriculture-Agricultural Research Service (USDA-ARS) funds under project 5030-31320-004-00D. Partial support was also provided by the USDA-NIFA award 2018-67017-27927. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the USDA. USDA is an equal opportunity provider and employer. The Galaxy server that was used for some bioinformatics calculations is in part funded by the Collaborative Research Centre 992 Medical Epigenetics (DFG grant SFB 992/1 2012) and the German Federal Ministry of Education and Research [BMBF grants 031 A538A/A538C RBC, 031L0101B/031L0101C de.NBI-epi, and 031 L0106 de.STAIR (de.NBI)].
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Matt H. Inbody, Brandon Ritland, and Jennifer A. Jones at USDA for their technical support in the laboratory and during the animal experiment. We thank Dr. David Alt for performing the MiSeq Illumina sequencing and Dr. Daniel Nielsen for helping on the MinION Nanopore sequencing. We also thank Dr. Shaohua Zhao from the FDA, Division of Animal and Food Microbiology, for providing the accession numbers of three tetracycline-susceptible Campylobacter strains carrying tet(O). This research was supported by an appointment to the Agricultural Research Service (ARS) Research Participation Program administered by the Oak Ridge Institute for Science and Education (ORISE) through an interagency agreement between the U.S. Department of Energy (DOE) and the U.S. Department of Agriculture (USDA). ORISE is managed by ORAU under DOE contract number DE- SC0014664. All opinions expressed in this paper are the authors’ and do not necessarily reflect the policies and views of USDA, ARS, DOE, or ORAU/ORISE. USDA is an equal opportunity provider and employer. This research used resources provided by the SCINet project of the USDA Agricultural Research Service, ARS project number 0500-00093-001-00-D.
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at https://www.frontiersin.org/articles/10.3389/fmicb.2021.732969/full#supplementary-material
SUPPLEMENTARY FIGURE S1 | Results of the pangenome analysis of in vitro co-culture of Campylobacter strains. Only a section of the genome is presented, highlighting the region surrounding the resistance gene of interest. (S1A) Co-culture of 6461 and 6067; (S1B and S1C) Co-culture of 6461 and 13150. Of note, the newly emerged strain carries two copies of tet(O), one chromosomal (S1B) and one plasmidic (S1C).
SUPPLEMENTARY TABLE S1 | List of media with combination of antibiotics used for the plating of turkey cecal content by room and necropsy day. Different combinations were used to recover one (at Day 8) or two parental strains (Day 15 and Day22) as well as a possible newly emerged strain with dual resistance.
SUPPLEMENTARY TABLE S2 | Minimal inhibitory concentrations (MICs) from CAMPY2 Sensititre® plates (VersaTREK Diagnostics/Thermo Fisher, United States). Antimicrobial levels are given in μg/ml. Eight antimicrobials were tested. S, susceptible; R, resistant; and NI, no interpretation because guidelines have not been established by the Clinical and Laboratory Standards Institute (CLSI) for the bacteria/drug combination being tested. Testing was conducted by the Veterinary Diagnostic Laboratory at ISUVDL. Sensitivity testing failed for strain 14229–5, the result provided being “Isolate was unable to grow for sensitivity (two attempts).”
SUPPLEMENTARY TABLE S3 | Unknown mutations (in genes where specific known mutations confer resistance) in Campylobacter sp. as identified using the web-based tool ResFinder 4.17 (Zankari et al., 2012). Contigs (FASTA files) from each sample were screened for “Chromosomal point mutations - Unknown mutations” with the default settings of 90% minimum identity and 20% minimum template length.
SUPPLEMENTARY TABLE S4 | Detailed horizontal gene transfers observed in the newly emerged strains (NES). For each in vitro transfer experiment, three genomes (from two parental and one NES strain) were included in the pangenome analysis using Roary (Page et al., 2015). The stringency for assigning coding sequences to a gene was set to 99% identity. The tool gifrop8 (v0.0.6) was used to help identify transfer of genes or consecutive blocks of genes – hereafter referred to as a “genomic island” – absent in the recipient strain but present in the NES. The workflow used to complete this analysis is available online.9
References
- Aarestrup F. M., Engberg J. (2001). Antimicrobial resistance of thermophilic campylobacter. Vet. Res. 32, 311–321. doi: 10.1051/vetres:2001127, PMID: [DOI] [PubMed] [Google Scholar]
- Afgan E., Baker D., Batut B., van Den Beek M., Bouvier D., Cech M., et al. (2018). The galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 46, W537–W544. doi: 10.1093/nar/gky379, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfredson D. A., Korolik V. (2007). Antibiotic resistance and resistance mechanisms in campylobacter jejuni and campylobacter coli. FEMS Microbiol. Lett. 277, 123–132. doi: 10.1111/j.1574-6968.2007.00935.x, PMID: [DOI] [PubMed] [Google Scholar]
- Allos B. M. (1997). Association between campylobacter infection and Guillain-Barre syndrome. J. Infect. Dis. 176, S125–S128. doi: 10.1086/513783, PMID: [DOI] [PubMed] [Google Scholar]
- Anderson A. D., Nelson J. M., Rossiter S., Angulo F. J. (2003). Public health consequences of use of antimicrobial agents in food animals in the United States. Microb. Drug Resist. 9, 373–379. doi: 10.1089/107662903322762815, PMID: [DOI] [PubMed] [Google Scholar]
- Andrews S. (2010). FastQC: a quality control tool for high throughput sequence data. Available at: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- Batz M. B., Henke E., Kowalcyk B. (2013). Long-term consequences of foodborne infections. Infect. Dis. Clin. N. Am. 27, 599–616. doi: 10.1016/j.idc.2013.05.003, PMID: [DOI] [PubMed] [Google Scholar]
- Bolinger H. K., Zhang Q., Miller W. G., Kathariou S. (2018). Lack of evidence for erm(B) infiltration Into erythromycin-resistant campylobacter coli and campylobacter jejuni from commercial Turkey production in eastern North Carolina: a Major Turkey-growing region in the United States. Foodborne Pathog. Dis. 15, 698–700. doi: 10.1089/fpd.2018.2477, PMID: [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (CDC) . (2019). “Antibiotic resistance threats in the United States, 2019”. (Atlanta, GA: U.S. Department of Health and Human Services, CDC).
- Clinical and Laboratory Standards Institute (CLSI) . (2017). “Methods for antimicrobial susceptibility testing of infrequently isolated or fastidious bacteria isolated from animals” in CLSI supplement VET06. 1st Edn. (Wayne, PA, USA: Clinical and Laboratory Standards Institute; ). [Google Scholar]
- Crespo M. D., Altermann E., Olson J., Miller W. G., Chandrashekhar K., Kathariou S. (2016). Novel plasmid conferring kanamycin and tetracycline resistance in the Turkey-derived campylobacter jejuni strain 11601MD. Plasmid 86, 32–37. doi: 10.1016/j.plasmid.2016.06.001, PMID: [DOI] [PubMed] [Google Scholar]
- Crespo M. D., Olson J. W., Altermann E., Siletzky R. M., Kathariou S. (2012). Chromosomal tet(O)-harboring regions in campylobacter coli isolates from turkeys and swine. Appl. Environ. Microbiol. 78, 8488–8491. doi: 10.1128/AEM.02258-12, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damron B. L., Sloan D. R. (1995). Poultry Diets for Small Flocks. IFAS Extension: University of Florida. [Google Scholar]
- De Vries J., Wackernagel W. (2002). Integration of foreign DNA during natural transformation of Acinetobacter sp. by homology-facilitated illegitimate recombination. Proc. Natl. Acad. Sci. U. S. A. 99, 2094–2099. doi: 10.1073/pnas.042263399, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta V., Altermann E., Olson J., Wray G. A., Siletzky R. M., Kathariou S. (2016). Whole-genome sequences of agricultural, host-associated campylobacter coli and campylobacter jejuni strains. Genome Announc. 4:e00833. doi: 10.1128/genomeA.00833-16, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Food Safety Authority (EFSA) and European Centre for Disease Prevention and Control (ECDC) (2015). The European union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2013. EFSA J. 13:3991. doi: 10.2903/j.efsa.2015.3991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facciola A., Riso R., Avventuroso E., Visalli G., Delia S. A., Lagana P. (2017). Campylobacter: from microbiology to prevention. J. Prev. Med. Hyg. 58, E79–E92. PMID: [PMC free article] [PubMed] [Google Scholar]
- Feldgarden M., Brover V., Haft D. H., Prasad A. B., Slotta D. J., Tolstoy I., et al. (2019). Validating the AMRFinder tool and resistance gene database by using antimicrobial resistance genotype-phenotype correlations in a collection of isolates. Antimicrob. Agents Chemother. 63:e00483. doi: 10.1128/AAC.00483-19, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner S. P., Olson J. W. (2012). Barriers to horizontal gene transfer in Campylobacter jejuni. Adv. Appl. Microbiol. 79, 19–42. doi: 10.1016/B978-0-12-394318-7.00002-4, PMID: [DOI] [PubMed] [Google Scholar]
- Gibreel A., Skold O., Taylor D. E. (2004). Characterization of plasmid-mediated aphA-3 kanamycin resistance in Campylobacter jejuni. Microb. Drug Resist. 10, 98–105. doi: 10.1089/1076629041310127, PMID: [DOI] [PubMed] [Google Scholar]
- Guerry P., Ewing C. P., Schirm M., Lorenzo M., Kelly J., Pattarini D., et al. (2006). Changes in flagellin glycosylation affect Campylobacter autoagglutination and virulence. Mol. Microbiol. 60, 299–311. doi: 10.1111/j.1365-2958.2006.05100.x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A., Nelson J. M., Barrett T. J., Tauxe R. V., Rossiter S. P., Friedman C. R., et al. (2004). Antimicrobial resistance among Campylobacter strains, United States, 1997-2001. Emerg. Infect. Dis. 10, 1102–1109. doi: 10.3201/eid1006.030635, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S. K., Padmanabhan B. R., Diene S. M., Lopez-Rojas R., Kempf M., Landraud L., et al. (2014). ARG-ANNOT, a new Bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob. Agents Chemother. 58, 212–220. doi: 10.1128/AAC.01310-13, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horrocks S. M., Anderson R. C., Nisbet D. J., Ricke S. C. (2009). Incidence and ecology of Campylobacter jejuni and coli in animals. Anaerobe 15, 18–25. doi: 10.1016/j.anaerobe.2008.09.001, PMID: [DOI] [PubMed] [Google Scholar]
- Jett B. D., Hatter K. L., Huycke M. M., Gilmore M. S. (1997). Simplified agar plate method for quantifying viable bacteria. BioTechniques 23, 648–650. doi: 10.2144/97234bm22, PMID: [DOI] [PubMed] [Google Scholar]
- Jia B. F., Raphenya A. R., Alcock B., Waglechner N., Guo P. Y., Tsang K. K., et al. (2017). CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 45, D566–D573. doi: 10.1093/nar/gkw1004, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. S., Carver D. K., Kathariou S. (2006). Natural transformation-mediated transfer of erythromycin resistance in Campylobacter coli strains from turkeys and swine. Appl. Environ. Microbiol. 72, 1316–1321. doi: 10.1128/AEM.72.2.1316-1321.2006, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. S., Kim J. W., Kathariou S. (2008). Differential effects of temperature on natural transformation to erythromycin and nalidixic acid resistance in Campylobacter coli. Appl. Environ. Microbiol. 74, 6121–6125. doi: 10.1128/AEM.01075-08, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk M. D., Pires S. M., Black R. E., Caipo M., Crump J. A., Devleesschauwer B., et al. (2015). World Health Organization estimates of the global and regional disease burden of 22 foodborne bacterial, protozoal, and viral diseases, 2010: a data synthesis. PLoS Med. 12:e1001921. doi: 10.1371/journal.pmed.1001940, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolmogorov M., Yuan J., Lin Y., Pevzner P. A. (2019). Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 37, 540–546. doi: 10.1038/s41587-019-0072-8, PMID: [DOI] [PubMed] [Google Scholar]
- Line J. E. (2001). Development of a selective differential agar for isolation and enumeration of Campylobacter spp. J. Food Prot. 64, 1711–1715. doi: 10.4315/0362-028X-64.11.1711, PMID: [DOI] [PubMed] [Google Scholar]
- Line J. E., Bailey J. S., Berrang M. E. (2008). Addition of sulfamethoxazole to selective media aids in the recovery of Campylobacter spp. from broiler rinses. J Rapid Methods Autom Microbiol 16, 2–12. doi: 10.1111/j.1745-4581.2008.00111.x [DOI] [Google Scholar]
- Luangtongkum T., Jeon B., Han J., Plummer P., Logue C. M., Zhang Q. (2009). Antibiotic resistance in Campylobacter: emergence, transmission and persistence. Future Microbiol. 4, 189–200. doi: 10.2217/17460913.4.2.189, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man S. M. (2011). The clinical importance of emerging Campylobacter species. Nat. Rev. Gastroenterol. Hepatol. 8, 669–685. doi: 10.1038/nrgastro.2011.191, PMID: [DOI] [PubMed] [Google Scholar]
- Moore J. E., Barton M. D., Blair I. S., Corcoran D., Dooley J. S., Fanning S., et al. (2006). The epidemiology of antibiotic resistance in Campylobacter. Microbes Infect. 8, 1955–1966. doi: 10.1016/j.micinf.2005.12.030, PMID: [DOI] [PubMed] [Google Scholar]
- Nachamkin I. (2002). Chronic effects of Campylobacter infection. Microbes Infect. 4, 399–403. doi: 10.1016/S1286-4579(02)01553-8, PMID: [DOI] [PubMed] [Google Scholar]
- Nachamkin I., Szymanski C., Blaser M. eds. (2008). Campylobacter. 3rd Edn. Washington, DC: ASM Press. [Google Scholar]
- Nelson J. M., Chiller T. M., Powers J. H., Angulo F. J. (2007). Fluoroquinolone-resistant Campylobacter species and the withdrawal of fluoroquinolones from use in poultry: a public health success story. Clin. Infect. Dis. 44, 977–980. doi: 10.1086/512369, PMID: [DOI] [PubMed] [Google Scholar]
- Olkkola S., Juntunen P., Heiska H., Hyytiäinen H., Hänninen M.-L. (2010). Mutations in the rpsL gene are involved in streptomycin resistance in. Campylobacter coli. Microb. Drug Resist. 16, 105–110. doi: 10.1089/mdr.2009.0128 [DOI] [PubMed] [Google Scholar]
- O,Neill J. (2014). Review on Antimicrobial Resistance. Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations. Available at: https://amr-review.org/sites/default/files/AMR%20Review%20Paper%20-%20Tackling%20a%20crisis%20for%20the%20health%20and%20wealth%20of%20nations_1.pdf
- Oyarzabal O. A., Rad R., Backert S. (2007). Conjugative transfer of chromosomally encoded antibiotic resistance from helicobacter pylori to Campylobacter jejuni. J. Clin. Microbiol. 45, 402–408. doi: 10.1128/JCM.01456-06, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padungton P., Kaneene J. B. (2003). Campylobacter spp in human, chickens, pigs and their antimicrobial resistance. J. Vet. Med. Sci. 65, 161–170. doi: 10.1292/jvms.65.161, PMID: [DOI] [PubMed] [Google Scholar]
- Page A. J., Cummins C. A., Hunt M., Wong V. K., Reuter S., Holden M. T., et al. (2015). Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31, 3691–3693. doi: 10.1093/bioinformatics/btv421, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C. J., Li J., Zhang X., Gao F., Benton C. S., Andam C. P. (2020). Genomic epidemiology and evolution of diverse lineages of clinical Campylobacter jejuni Cocirculating in New Hampshire, USA, 2017. J. Clin. Microbiol. 58, e02070–e020019. doi: 10.1128/JCM.02070-19, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payot S., Bolla J. M., Corcoran D., Fanning S., Megraud F., Zhang Q. (2006). Mechanisms of fluoroquinolone and macrolide resistance in Campylobacter spp. Microbes Infect. 8, 1967–1971. doi: 10.1016/j.micinf.2005.12.032, PMID: [DOI] [PubMed] [Google Scholar]
- Pinto-Alphandary H., Mabilat C., Courvalin P. (1990). Emergence of aminoglycoside resistance genes aadA and aadE in the genus Campylobacter. Antimicrob. Agents Chemother. 34, 1294–1296. doi: 10.1128/AAC.34.6.1294, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard L., Glover R. H., Humphris S., Elphinstone J. G., Toth I. K. (2016). Genomics and taxonomy in diagnostics for food security: soft-rotting enterobacterial plant pathogens. Anal. Methods 8, 12–24. doi: 10.1039/C5AY02550H [DOI] [Google Scholar]
- Prudhomme M., Libante V., Claverys J. P. (2002). Homologous recombination at the border: insertion-deletions and the trapping of foreign DNA in Streptococcus pneumoniae. Proc. Natl. Acad. Sci. U. S. A. 99, 2100–2105. doi: 10.1073/pnas.032262999, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin S., Wang Y., Zhang Q., Zhang M., Deng F., Shen Z., et al. (2014). Report of ribosomal RNA methylase gene erm(B) in multidrug-resistant Campylobacter coli. J. Antimicrob. Chemother. 69, 964–968. doi: 10.1093/jac/dkt492, PMID: [DOI] [PubMed] [Google Scholar]
- Salah Ud-Din A. I. M., Roujeinikova A. (2018). Flagellin glycosylation with pseudaminic acid in Campylobacter and helicobacter: prospects for development of novel therapeutics. Cell. Mol. Life Sci. 75, 1163–1178. doi: 10.1007/s00018-017-2696-5, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scallan E., Hoekstra R. M., Angulo F. J., Tauxe R. V., Widdowson M. A., Roy S. L., et al. (2011). Foodborne illness acquired in the United States--major pathogens. Emerg. Infect. Dis. 17, 7–15. doi: 10.3201/eid1701.P11101, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scallan E., Hoekstra R. M., Mahon B. E., Jones T. F., Griffin P. M. (2015). An assessment of the human health impact of seven leading foodborne pathogens in the United States using disability adjusted life years. Epidemiol. Infect. 143, 2795–2804. doi: 10.1017/S0950268814003185, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scallan Walter E. J., Crim S. M., Bruce B. B., Griffin P. M. (2019). Postinfectious irritable bowel syndrome After Campylobacter infection. Am. J. Gastroenterol. 114, 1649–1656. doi: 10.14309/ajg.0000000000000408, PMID: [DOI] [PubMed] [Google Scholar]
- Scallan Walter E. J., Crim S. M., Bruce B. B., Griffin P. M. (2020). Incidence of Campylobacter-associated Guillain-Barre syndrome estimated from health insurance data. Foodborne Pathog. Dis. 17, 23–28. doi: 10.1089/fpd.2019.2652, PMID: [DOI] [PubMed] [Google Scholar]
- Seemann T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153, PMID: [DOI] [PubMed] [Google Scholar]
- Siragusa G. R. (1999). Statistical validation of the track-dilution plating method from ground beef and carcass surface samples. J. Rapid Methods Automa. Microbiol. 7, 155–161. doi: 10.1111/j.1745-4581.1999.tb00385.x [DOI] [Google Scholar]
- Smith K., Reimers N., Barnes H. J., Lee B. C., Siletzky R., Kathariou S. (2004). Campylobacter colonization of sibling Turkey flocks reared under different management conditions. J. Food Prot. 67, 1463–1468. doi: 10.4315/0362-028X-67.7.1463, PMID: [DOI] [PubMed] [Google Scholar]
- Sougakoff W., Papadopoulou B., Nordmann P., Courvalin P. (1987). Nucleotide sequence and distribution of gene tetO encoding tetracycline resistance in Campylobacter coli. FEMS Microbiol. Lett. 44, 153–159. doi: 10.1111/j.1574-6968.1987.tb02260.x [DOI] [Google Scholar]
- Sylte M. J., Inbody M. H., Johnson T. A., Looft T., Line J. E. (2018). Evaluation of different Campylobacter jejuni isolates to colonize the intestinal tract of commercial Turkey poults and selective media for enumeration. Poult. Sci. 97, 1689–1698. doi: 10.3382/ps/pex384, PMID: [DOI] [PubMed] [Google Scholar]
- Sylte M. J., Johnson T. A., Meyer E. L., Inbody M. H., Trachsel J., Looft T., et al. (2019). Intestinal colonization and acute immune response in commercial turkeys following inoculation with Campylobacter jejuni constructs encoding antibiotic-resistance markers. Vet. Immunol. Immunopathol. 210, 6–14. doi: 10.1016/j.vetimm.2019.02.003, PMID: [DOI] [PubMed] [Google Scholar]
- Tacconelli E., Carrara E., Savoldi A., Harbarth S., Mendelson M., Monnet D. L., et al. (2018). Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 18, 318–327. doi: 10.1016/S1473-3099(17)30753-3, PMID: [DOI] [PubMed] [Google Scholar]
- Tack D. M., Ray L., Griffin P. M., Cieslak P. R., Dunn J., Rissman T., et al. (2020). Preliminary incidence and trends of infections with pathogens transmitted commonly Through food – foodborne diseases active surveillance network, 10 U.S. sites, 2016-2019. MMWR Morb. Mortal. Wkly Rep. 69, 509–514. doi: 10.15585/mmwr.mm6917a1, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor D. E. (1992). Genetics of Campylobacter and Helicobacter. Annu. Rev. Microbiol. 46, 35–64. doi: 10.1146/annurev.mi.46.100192.000343, PMID: [DOI] [PubMed] [Google Scholar]
- Taylor D. E., Hiratsuka K., Ray H., Manavathu E. K. (1987). Characterization and expression of a cloned tetracycline resistance determinant from Campylobacter jejuni plasmid pUA466. J. Bacteriol. 169, 2984–2989. doi: 10.1128/jb.169.7.2984-2989.1987, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacher S., Menard A., Bernard E., Megraud F. (2003). PCR-restriction fragment length polymorphism analysis for detection of point mutations associated with macrolide resistance in Campylobacter spp. Antimicrob. Agents Chemother. 47, 1125–1128. doi: 10.1128/AAC.47.3.1125-1128.2003, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker B. J., Abeel T., Shea T., Priest M., Abouelliel A., Sakthikumar S., et al. (2014). Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Dong Y., Deng F., Liu D., Yao H., Zhang Q., et al. (2016). Species shift and multidrug resistance of Campylobacter from chicken and swine, China, 2008-14. J. Antimicrob. Chemother. 71, 666–669. doi: 10.1093/jac/dkv382, PMID: [DOI] [PubMed] [Google Scholar]
- Wang Y., Huang W. M., Taylor D. E. (1993). Cloning and nucleotide sequence of the Campylobacter jejuni gyrA gene and characterization of quinolone resistance mutations. Antimicrob. Agents Chemother. 37, 457–463. doi: 10.1128/AAC.37.3.457, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Taylor D. E. (1990). Natural transformation in Campylobacter species. J. Bacteriol. 172, 949–955. doi: 10.1128/jb.172.2.949-955.1990, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhang M., Deng F., Shen Z., Wu C., Zhang J., et al. (2014). Emergence of multidrug-resistant Campylobacter species isolates with a horizontally acquired rRNA methylase. Antimicrob. Agents Chemother. 58, 5405–5412. doi: 10.1128/AAC.03039-14, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesley I. V., Rostagno M., Hurd H. S., Trampel D. W. (2009). Prevalence of Campylobacter jejuni and Campylobacter coli in market-weight turkeys on-farm and at slaughter. J. Food Prot. 72, 43–48. doi: 10.4315/0362-028X-72.1.43, PMID: [DOI] [PubMed] [Google Scholar]
- Whitehouse C. A., Young S., Li C., Hsu C. H., Martin G., Zhao S. (2018). Use of whole-genome sequencing for Campylobacter surveillance from NARMS retail poultry in the United States in 2015. Food Microbiol. 73, 122–128. doi: 10.1016/j.fm.2018.01.018, PMID: [DOI] [PubMed] [Google Scholar]
- Wick R. R., Holt K. E. (2019). Benchmarking of long-read assemblers for prokaryote whole genome sequencing. F1000Res 8:2138. doi: 10.12688/f1000research.21782.4, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick R. R., Judd L. M., Gorrie C. L., Holt K. E. (2017). Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13:e1005595. doi: 10.1371/journal.pcbi.1005595, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick R. R., Judd L. M., Holt K. E. (2019). Performance of neural network basecalling tools for Oxford Nanopore sequencing. Genome Biol. 20:129. doi: 10.1186/s13059-019-1727-y, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson D. L., Bell J. A., Young V. B., Wilder S. R., Mansfield L. S., Linz J. E. (2003). Variation of the natural transformation frequency of Campylobacter jejuni in liquid shake culture. Microbiology 149, 3603–3615. doi: 10.1099/mic.0.26531-0, PMID: [DOI] [PubMed] [Google Scholar]
- Wright S. L., Carver D. K., Siletzky R. M., Romine S., Morrow W. E., Kathariou S. (2008). Longitudinal study of prevalence of Campylobacter jejuni and Campylobacter coli from turkeys and swine grown in close proximity. J. Food Prot. 71, 1791–1796. doi: 10.4315/0362-028X-71.9.1791, PMID: [DOI] [PubMed] [Google Scholar]
- Yahara K., Didelot X., Ansari M. A., Sheppard S. K., Falush D. (2014). Efficient inference of recombination hot regions in bacterial genomes. Mol. Biol. Evol. 31, 1593–1605. doi: 10.1093/molbev/msu082, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao H., Liu D., Wang Y., Zhang Q., Shen Z. (2017). High prevalence and predominance of the aph(2″)-If gene conferring aminoglycoside resistance in Campylobacter. Antimicrob. Agents Chemother. 61, e00112–e00117. doi: 10.1128/AAC.00112-17, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zankari E., Hasman H., Cosentino S., Vestergaard M., Rasmussen S., Lund O., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67, 2640–2644. doi: 10.1093/jac/dks261, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S., Tyson G. H., Chen Y., Li C., Mukherjee S., Young S., et al. (2016). Whole-genome sequencing analysis accurately predicts antimicrobial resistance phenotypes in Campylobacter spp. Appl. Environ. Microbiol. 82, 459–466. doi: 10.1128/AEM.02873-15, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SUPPLEMENTARY FIGURE S1 | Results of the pangenome analysis of in vitro co-culture of Campylobacter strains. Only a section of the genome is presented, highlighting the region surrounding the resistance gene of interest. (S1A) Co-culture of 6461 and 6067; (S1B and S1C) Co-culture of 6461 and 13150. Of note, the newly emerged strain carries two copies of tet(O), one chromosomal (S1B) and one plasmidic (S1C).
SUPPLEMENTARY TABLE S1 | List of media with combination of antibiotics used for the plating of turkey cecal content by room and necropsy day. Different combinations were used to recover one (at Day 8) or two parental strains (Day 15 and Day22) as well as a possible newly emerged strain with dual resistance.
SUPPLEMENTARY TABLE S2 | Minimal inhibitory concentrations (MICs) from CAMPY2 Sensititre® plates (VersaTREK Diagnostics/Thermo Fisher, United States). Antimicrobial levels are given in μg/ml. Eight antimicrobials were tested. S, susceptible; R, resistant; and NI, no interpretation because guidelines have not been established by the Clinical and Laboratory Standards Institute (CLSI) for the bacteria/drug combination being tested. Testing was conducted by the Veterinary Diagnostic Laboratory at ISUVDL. Sensitivity testing failed for strain 14229–5, the result provided being “Isolate was unable to grow for sensitivity (two attempts).”
SUPPLEMENTARY TABLE S3 | Unknown mutations (in genes where specific known mutations confer resistance) in Campylobacter sp. as identified using the web-based tool ResFinder 4.17 (Zankari et al., 2012). Contigs (FASTA files) from each sample were screened for “Chromosomal point mutations - Unknown mutations” with the default settings of 90% minimum identity and 20% minimum template length.
SUPPLEMENTARY TABLE S4 | Detailed horizontal gene transfers observed in the newly emerged strains (NES). For each in vitro transfer experiment, three genomes (from two parental and one NES strain) were included in the pangenome analysis using Roary (Page et al., 2015). The stringency for assigning coding sequences to a gene was set to 99% identity. The tool gifrop8 (v0.0.6) was used to help identify transfer of genes or consecutive blocks of genes – hereafter referred to as a “genomic island” – absent in the recipient strain but present in the NES. The workflow used to complete this analysis is available online.9
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.