Abstract
Triple negative breast cancer (TNBC) is an aggressive type of breast cancer with very little treatment options. TNBC is very heterogeneous with large alterations in the genomic, transcriptomic, and proteomic landscapes leading to various subtypes with differing responses to therapeutic treatments. We applied a multi-omics data integration method to evaluate the correlation of important regulatory features in TNBC BRCA1 wild-type MDA-MB-231 and TNBC BRCA1 5382insC mutated HCC1937 cells compared with non-tumorigenic epithelial breast MCF10A cells. The data includes DNA methylation, RNAseq, protein, phosphoproteomics, and histone post-translational modification. Data integration methods identified regulatory features from each omics method that had greater than 80% positive correlation within each TNBC subtype. Key regulatory features at each omics level were identified distinguishing the three cell lines and were involved in important cancer related pathways such as TGFβ signaling, PI3K/AKT/mTOR, and Wnt/beta-catenin signaling. We observed overexpression of PTEN, which antagonizes the PI3K/AKT/mTOR pathway, and MYC, which downregulates the same pathway in the HCC1937 cells relative to the MDA-MB-231 cells. The PI3K/AKT/mTOR and Wnt/beta-catenin pathways are both downregulated in HCC1937 cells relative to MDA-MB-231 cells, which likely explains the divergent sensitivities of these cell lines to inhibitors of downstream signaling pathways. The DNA methylation and RNAseq data is freely available via GEO GSE171958 and the proteomics data is available via the ProteomeXchange PXD025238.
Multi-omics data integration of triple negative breast cancer (TNBC) provides insight into biological pathways.
Introduction
Breast cancer is a heterogeneous type of cancer with various degrees of aggressiveness and patient outcome and is the second leading cause of cancer-related deaths in women in the United States.1,2 Breast cancer includes five molecular subtypes: luminal A (estrogen–receptor (ER) and/or progesterone–receptor (PR) positive), luminal B (ER and/or PR positive and human epidermal growth factor receptor2 (HER2) positive or negative), normal-like (ER and/or PR positive, HER2 negative), HER2-enriched (ER and PR negative, HER2 positive), and triple-negative/basal like (ER/PR/HER2 negative)1,3 Triple negative breast cancer (TNBC) accounts for 10–15% of all breast carcinomas3 and affects mostly younger women and women of African American descent.1
TNBC is associated with worse prognoses than other types of breast cancer, which is attributed to its aggressiveness and heterogeneity.4 The various subtypes and heterogeneity of breast cancer are largely due to altered genomic, transcriptomic, and proteomic molecular signatures that impact various signaling pathways. About 5% of breast cancer tumors carry heritable gene mutations, such as the most common mutation in the breast cancer gene1 (BRCA1). Of these BRCA1 mutant tumors, the BRCA1 5382insC mutation is one of the most common.5 This study by Pogoda et al. showed that out of 124 TNBC patients that underwent genetic counselling with BRCA1/2 mutation tests, 30 had a mutation detected. Of those 30 patients, 18 patients had a BRCA1 5382insC mutation.5
Despite their established diversity, TNBC subtypes are all treated with chemotherapy, which too often results in recurrence.6 This treatment strategy is clearly inadequate as genetic alterations in key genes that seemingly define the TNBC aggressiveness (e.g. PIK3CA and AKT) have been characterized – rendering treatments which target these proteins ineffective in a subset of TNBC patients.7 A study by Gu et al. 2016 demonstrated TNBC tumors with a BRCA1 mutant are insensitive to mitogen-activated protein kinase 1 and 2 (MEK 1/2) inhibitors, limiting the therapeutic potential against these cancer subtypes.8 MEK inhibitors have been used to successfully treat other genomic mutated cancers, such as melanoma.8 Therefore, understanding the molecular complexity of various subtypes of breast cancer is of primary importance.
The advent of personalized medicine brings great promise to those suffering from this challenging disease as distinct, potentially druggable, molecular targets with unique alterations have been identified. Development of treatment strategies that would broadly target TNBC patients necessitates a comprehensive understanding of the underpinnings of this disease. To achieve such understanding, we characterized two subtypes of TNBC by examining and integrating information on their epigenetic, transcriptomic, proteomic and phospho-proteomic profiles. Specifically, we characterized the BRCA1-wild-type MDA-MB-231 TNBC, and the BRCA15382insC HCC1937 TNBC cell lines as well as the MCF10A cell line as a normal breast epithelial control. Our multi-omics approach highlights the diversity of the different TNBC subtypes, and sharpens our understanding of the molecular pathways that govern this challenging breast cancer.
Methods
Cell culture
MCF10A cells were derived from benign proliferative breast tissue and are the most commonly used cell line to study normal breast cells.9 MDA-MB-231 cell line was isolated from a pleural effusion of a patient with invasive ductal carcinoma. This cell line is used to model late-stage breast cancer.10,11 HCC1937 cell line (Hamon Cancer Center) was established from a grade III infiltrating ductal primary breast tumor from a patient with a germ-line BRCA1 mutation. The primary tumor showed large vacuoles in many of the cells suggestive of secretory variant of infiltrating intraductal carcinoma. HCC1937 cell line is homozygous for a BRCA1 5382insC mutation.12 Each cell line was purchased from American Type Culture Collection (ATCC; CRL-2336, HTB-26, CRL-10317). The MCF10A cells were cultured in DMEM/F12 supplemented with 20 ng mL−1 hEGF, 100 ng mL−1 cholera toxin, 10 μg mL−1 bovine insulin, 500 ng mL−1 hydrocortisone, 5% horse serum, and 1% penicillin/streptomycin. MDA-MB-231 cells were cultured in DMEM and supplemented with 10% FBS and 1% penicillin/streptomycin. HCC1937 cells were cultured in RPMI supplemented with 10% FBS and 1% penicillin/streptomycin.
Experimental design
Biological replicates of MCF10A, MDA-MB-231, and HCC1937 cell lines were grown in culture and harvested for each omics method. In the DNA methylation experiment an Illumina Infinium Methylation EPIC Bead Chip 16 sample array was utilized and therefore, we sequenced 4 biological replicates of MCF10A, and 6 biological replicates each for MDA-MB-231 and HCC1937 cell lines to maximize the 16 sample array. In the RNAseq experiment, we sequenced a total of 11 samples (3 biological replicates of MCF10A, 4 biological replicates for MDA-MB-231 and HCC1937 cell lines) in order to maximize sequencing capacity on an Illumina NextSeq 500 high output flow cell. In the protein, phosphopeptide, and histone post-translational data sets, we used biological triplicates for each cell line. The phosphoproteomics analysis is the limiting factor for the sample size in the data integration analysis due to the multiplexing of tandem mass tag (TMT) 10-plex batching effects. Phosphorylated peptides are low abundant molecules and must be enriched for detection by mass spectrometry. Currently, the best approach for quantitative analysis of PTMs is to use TMT multiplexing, which is limited to a total of 10 samples within one batch. If the design includes more than 10 samples and two TMT-10plex batches are required, this introduces a strong batch effect due to the fact if a protein is missing in one batch it is now missing in all ten samples within the batch. Triplicate biological replicates for the DNA methylation, RNA-sequencing, protein, and phosphoproteomics analyses were utilized for data integration to satisfy the requirement of MixOmics DIABLO method, which requires the same samples for each method to be utilized.13 Table S1 (ESI†) indicates the biological replicates used for each method and the data for each sample and method can be found in the Supplemental files 1–4 (ESI†).
DNA methylation profiling
Genome-wide DNA methylation was assessed in bisulfite-converted genomic DNA using the Illumina Infinium® Methylation EPIC Bead Chip array (Illumina, San Diego, CA, USA).14 This technology interrogates over 850 000 methylation sites covering 99% of the RefSeq (NCBI Reference Sequence Database) genes, 96% of CpG islands (CGI) with a coverage across promoters, 5′ and 3′-UTRs, first exons and gene bodies.14 Genomic DNA (500 ng) was bisulfite treated and purified using the EZ DNA Methylation-Gold kit (Zymo Research, Irvine, CA) according to the manufacturer's protocol.14 The resultant bisulfite-converted DNA was processed and hybridized to the Illumina Infinium Methylation EPIC Bead Chip. Subsequently, it was fluorescently stained and scanned on an Illumina iScan according to the Infinium HD Assay Methylation Protocol Guide provided by Illumina.
Following an initial quality control step, the DNA methylation data was preprocessed and normalized using Bioconductor packages minfi and watermelon.15,16 Briefly, the function “preprocessNoob” (minfi) was used to correct for background fluorescence and dye biases within an array. Next, probes and samples with poor quality were identified and removed. Therefore, samples with more than 10% of probes that had detection p-values >1 × 10−5 or samples whose intensity distributions demonstrated irregularities were excluded.17 Furthermore, Illumina removed 1031 CpG probes when transitioning to their B1 version of the MethylationEPIC v1.0 manifest due to poor performance and additional probes in the transition from their B2 to their B3 version.18 In addition to probes flagged by Illumina, we also excluded probes with a median detection p-value > 0.05 from the subsequent statistical analysis. Next, we corrected the type II probe bias using the function “BMIQ” (wateRmelon) to achieve a comparable methylation distributions of type I and II probes.15 All single nucleotide polymorphism (SNP)-CpG interaction or cross-reactive probes were flagged and excluded from the downstream analyses in order to reduce the bias of the results. The DNA-methylation data were analyzed using the R statistical programming language (version 3.6).
The statistical analysis for probe-wise differential DNA methylation was performed following the limma workflow.19M-values were calculated by transforming β-values using the logit-transformation. Average M-values across promoter regions and associated genomic features were calculated for each “Promoter Associated” region annotated by the IlluminaHumanMethylationEPICanno.ilm10b4.hg19 R Package.20 The change in methylation at a promoter region was indicated by the difference between mean CpG methylation between treatments. A linear model was fitted with the “lmFit” and “eBayes” functions from the limma package. We individually adjusted the p-values for multiple testing across all CpG sites/promoter regions using the Benjamini–Hochberg method21 to control the False Discovery Rate (FDR). A CpG was considered differentially methylated if the FDR was <0.05.
RNA sequencing, gene expression analysis
Total RNA (500 ng) was extracted using the Qiagen RNAeasy kit (cat # 74104) and sequencing libraries were prepared at The Arkansas Children's Research Institute (ACRI) Genomics core by using the Illumina TruSeq Stranded mRNA Sample Preparation Kit v2. Libraries were validated on the advanced analytical fragment analyzer (AATI) for fragment size and quantified by use of a Qubit (Life Technologies) fluorometer. Equal amounts of each library was pooled for sequencing on the NextSeq 500 platform using a high output flow cell to generate approximately 25 million 75 base reads per sample.
RNA reads were checked for quality of sequencing using FastQC. The adaptors and low quality bases (Q < 20) were trimmed to a minimum of 36 base pairs using Trimmomatic. Reads that pass quality control were aligned to the GRCh38 Ensemble release 99 Homo sapiens reference genome using STAR sequence aligner. Raw counts were obtained from bam files using Subread's “featuresCount” and transformed to log2 counts per million (CPM).22 Low expressed genes were filtered out and libraries normalized by trimmed mean of M-values.23 Differential expression was performed using limma's “voom with quality weights” and p-values were corrected for multiple testing using the Benjamini–Hochberg procedure. Genes with a false discovery rate (FDR) p-value < 0.05 and fold change > 2 were considered significant.
Phosphoproteomics analysis
A protein and phosphopeptide analysis was performed as previously described in Storey et al.24 Briefly, 100 μg of protein from each cell line was reduced, alkylated, and purified by chloroform/methanol extraction prior to digestion with sequencing grade trypsin and LysC (Promega). The resulting peptides were labeled using a tandem mass tag 10-plex isobaric label reagent set (Thermo) and enriched using High-Select TiO2 and Fe-NTA phosphopeptide enrichment kits (Thermo) following the manufacturer's instructions. Eluted peptides were analyzed by mass spectrometry on an Orbitrap Eclipse Tribrid mass spectrometer (Thermo) using multi-notch MS3 parameters.
Proteins and phosphosites were identified and reporter ions quantified by searching the UniprotKB Homo sapiens database (November 2018) using MaxQuant (version 1.6.10.43; Max Planck Institute) with a parent ion tolerance of 3 ppm, a fragment ion tolerance of 0.5 Da, a reporter ion tolerance of 0.001 Da, trypsin enzyme with 2 missed cleavages, variable modifications including oxidation on M, acetyl on protein N-term, and phosphorylation on STY, and fixed modification of Carbamidomethyl on C. Protein and peptide identifications are accepted if they could be established with less than 1.0% false discovery. TMT MS3 reporter ion intensity values were analyzed for changes in total protein using the unenriched lysate sample. Phospho (STY) modifications were identified using the samples enriched for phosphorylated peptides. The enriched and un-enriched samples are multiplexed using two TMT10-plex batches, one for the enriched and one for the un-enriched samples.
Following data acquisition and database search, the results were normalized using cyclic loess normalization for both the protein and the phosphopeptide data sets.25
The normalized protein and phosphorylated peptide data was analyzed for differential abundance using the limma package by applying “lmFit” and “eBayes” functions. Protein kinases and their substrates were identified using PHOXTRACK.26 A similar approach was used for differential analysis of the phosphopeptides, with the addition of a few steps. The phosphosites were filtered to retain only peptides with a localization probability > 75%, filter peptides with zero values, and cyclic loess transformed. Limma was also used for differential analysis of single phosphosite peptides.
Histone post-translational modification analysis
The cell lines were grown to approximately 5 million cells, washed with 1× PBS, and the pellet was resuspended in 5 times the volume of the pellet with Radioimmunoprecipitation assay buffer (RIPA; 10 mM Tris-Cl, pH 8.0, 1 mM Ethylenediaminetetraacetic acid (EDTA), 0.5 mM ethylene glycol tetraacetic acid (EGTA), 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% sodium dodecyl sulfate, and 140 mM NaCl). The samples were incubated on ice for 30 min, spun at 4 °C for 10 min at max speed, and the supernatant was transferred to a new tube for the whole lysate total protein analysis. The pellet was washed with RIPA and incubated with 0.4 N H2SO4 at 4 °C overnight. The samples were spun at 4 °C for 10 min at max speed, the supernatant transferred to a new tube, added 66 μL of 100% TCA, incubated on ice for 30 min, spun at 4 °C for 10 min at max speed, washed the pellet in ice cold acetone, air dried the pellet, and resuspended the histones in 50 μL of H2O.
Histones were resolved (20 μg of histones per lane) by SDS-PAGE using 4–20% Novex Tris-glycine gradient gels (Life Technologies, Inc.) and stained with Thermo Fisher Scientific Pierce GelCode Blue stain reagent. The region of each gel lane containing the core histones was excised as one piece, diced into small pieces, destained, treated with d6-acetic anhydride to chemically block unmodified lysines and monomethylated lysines with an isotopically heavy acetyl, and digested in-gel with trypsin. Tryptic peptides were separated by reverse phase Jupiter Proteo resin (Phenomenex) on a 100 0.075 mm column using a nanoAcquity UPLC system (Waters). Peptides were eluted using a 40 min gradient from 97 : 3 to 35 : 65 buffer A/B ratio. (Buffer A consists of 0.1% formic acid, 0.5% acetonitrile; buffer B consists of 0.1% formic acid, 75% acetonitrile). Eluted peptides were ionized by electrospray (1.9 kV) followed by MS/MS analysis using collision induced dissociation on an Orbitrap Fusion Lumos mass spectrometer (Thermo Fisher Scientific). MS data were acquired using the Fourier Transform Mass Spectrometry analyzer in profile mode at a resolution of 60 000 over a range of 375 to 1500 m/z.
Proteins were identified by searching the UniProtKB Homo sapiens database (October 2019 database) using an in-house Mascot server (version 2.5.1; Matrix Science). Mascot search parameters were specified as follows: trypsin digestion with up to two missed cleavages; fixed carbamidomethyl modification of cysteine; variable modifications including methyl, dimethyl, trimethyl, acetyl, acetyl/2 H3, and methyl + acetyl/2 H3 modification of lysine; 2.0 ppm precursor ion tolerance; 0.50 Da fragment ion tolerance. A reverse sequence decoy search was also performed. Peptide and protein identifications were validated using Scaffold (version 4.8.7; Proteome Software). Peptide and protein identifications were accepted with a 1% FDR which was assigned by the Protein Prophet algorithm.27 The spectrum report was then exported for further analysis by PTMViz (https://github.com/ByrumLab/PTMViz). PTMViz was utilized to identify writers, erasers, and readers for methylation and acetylation (WERAM) modifying proteins for significant histone post-translational modifications. Briefly, PTMViz analyzes histone post-translational modification peptides (PTMs) using the relative abundances of PTMs and performs a limma moderated t-test to identify significant modifications.
Data integration
Each omics data set (DNA methylation of promoter regions, genes, proteins, and phosphopeptides) was normalized to account for technique specific variability and limitations. The methods of normalization are described above for each specific omics technique. The normalized data sets with consistent annotation were integrated using techniques such as Pearson correlation analysis and matrix factorization using the tools MixOmics and MOGSA.
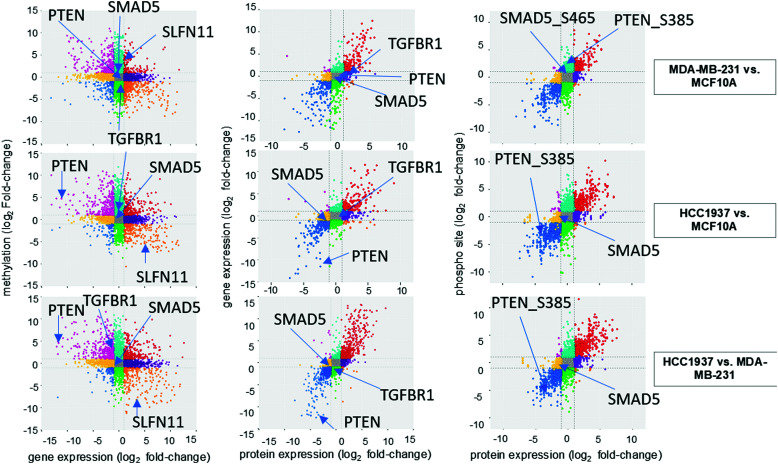
Multi-omics data annotation was curated by mapping the transcript version of the Illumina EPIC manifest Regulatory Features, the “gencode.v12.annotation.gtf.gz”, and the “genecode.v12.metadata.SwissProt.gz” files in order to retrieve consistent annotation for the CpGs grouped by promoter regions, gene expression, protein expression, and phosphopeptide data sets. The annotation data was mapped based on the transcript ID from each source to provide a final table consisting of the GenecodeCompV12 accession, Illumina Regulatory Feature name, transcript ID, gene ID, gene name, UniProtKB AC, and UniProtKB ID. The DNA methylation and gene expression data as well as the gene and protein data sets were matched based on Ensembl IDs. Proteins and phosphorylated peptides data sets were matched based on UniProtKB IDs. Pearson correlation analysis was performed using the significant features from each data set, which corresponds to the four outer quadrants on Fig. 3, to identify the relationships between DNA methylated promoters and the genes they regulate, between gene and protein expression, and between protein expression and phosphorylated proteins.
Fig. 3. Correlation of multi-omics data sets. The log2 fold change values for each multi-omics data set is displayed. Features with positive correlation (expression is significant and in the same direction) are shown in the top right and bottom left quadrants for RNAseq vs. protein and the protein vs phosphorylated peptides. Hypermethylated gene promoters show a positive fold change value but indicate gene repression and are correlated with negative gene expression fold change values (top left quadrant). Hypomethylated gene promoter are correlated with positive gene expression fold change values (bottom right quadrant). Key features discussed in the results are highlighted.
The DNA methylation promoter regions, gene, protein, and phosphopeptide normalized expression data was analyzed using MixOmics, which integrates multi-omics data to identify correlated variables measured on heterogeneous data sets and explain the categorical outcome of interest (supervised analysis). MixOmics applies matrix factorization using the supervised projection to latent structures (PLS) models for data integration to reduce the dimension, capture and explain the variation in the data that discriminate between MCF10A, MDA-MB-231, and the HCC1937 cell lines. We used the Data Integration Analysis for Biomarker discovery (DIABLO) with the sparse partial least squares regression (sPLS) method with tuning parameter set to keep the top 15 and 10 features for each data set corresponding to principal component 1 and 2, respectively.
Multi-omics gene-set analysis (MOGSA) was applied to the same data set as MixOmics, except only features with gene symbols were included, in order to investigate data integration at the pathway/gene set level. MOGSA is a multivariate single sample gene-set analysis method that integrates multiple experimental and molecular data types measured over the same set of samples.28 The method learns a low dimensional representation of most variant correlated features (genes, proteins, etc.) across multiple omics data sets, transforms the features onto the same scale and calculates an integrated gene-set score from the most informative features in each data type. A gene-set with a high GSS are driven by features that explain a large proportion of the global correlated information among data matrices and can be from one or all data matrices.
Results
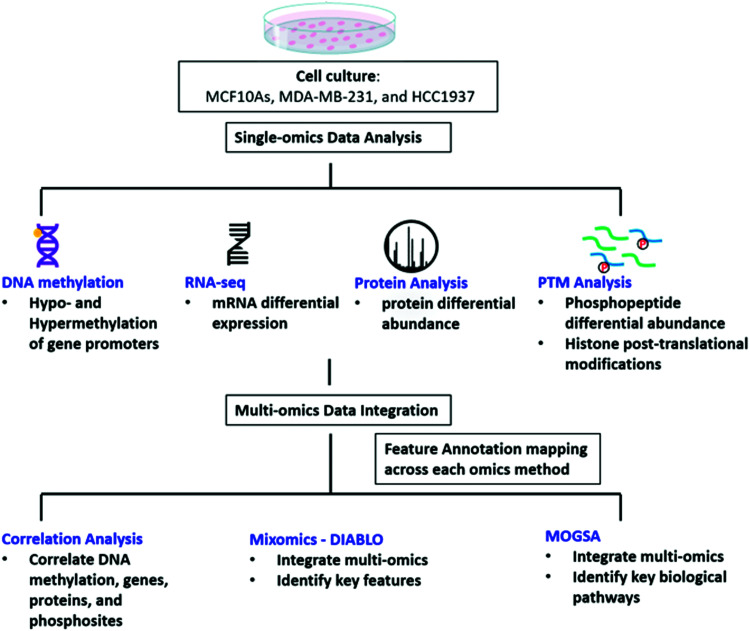
We investigated the regulatory mechanisms of non-tumorigenic human breast epithelial (MCF10A), triple negative breast cancer with a wild-type BRCA1 expression (MDA-MB-231) and triple negative breast cancer with a common homozygous BRCA1 5382insC mutation (HCC1937) using a multi-omics approach including DNA methylation, RNAseq, protein, phosphopeptides analysis, and histone post-translational modifications (Fig. 1). We first explored each omics data set individually and the data is provided in the ESI† (Supplemental files 1–5). We then curated the feature annotations for each data set so the Ensembl, Entrez Gene ID, Transcript ID, and UniProtKB IDs were the same among each of the data sets to allow for data integration using correlation analysis, MixOmics DIABLO, and MOGSA matrix factorization methods in order to identify key significant features and pathways distinguishing each of the biological conditions at various levels of regulation (Fig. 1 and Supplemental files 6–8, ESI†).
Fig. 1. Workflow of data analysis. MCF10A, MDA-MB-231, and HCC1937 cells were cultured and DNA, RNA, and protein were extracted for DNA methylation, RNAseq, protein expression, and analysis of phosphorylated peptides and histone post-translational modifications. Each single omics data set was individually analyzed. The feature annotations were then curated to match between each omics type and were integrated using correlation analysis, MixOmics, and MOGSA to find significant features of TNBC.
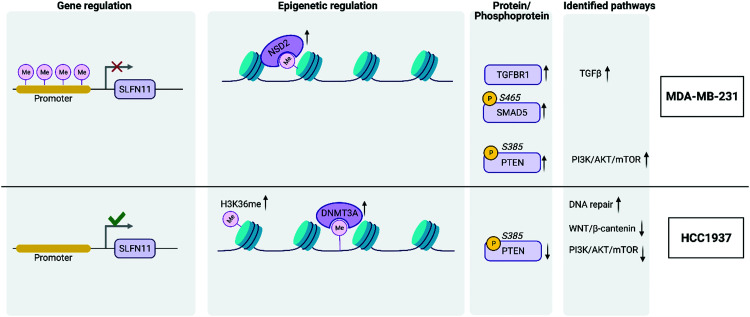
There were large differences in expression among each of the three cell lines providing for valuable data for integration and pathway analyses. First, the DNA methylation status of the promoters that regulate gene expression from each cell line was evaluated using Illumina EPIC Beadchip array technology. A total of 865 077 CpG probes were analyzed from the Beadchip array. We performed pair-wise comparisons of the three cell lines and found 96% of the significant CpGs were common among the pair-wise comparisons of the three cell lines. From these probes, 616 053 CpGs were associated with a gene and after averaging the M-values of the CpGs by promoter regions, a total of 86 168 regulatory features were analyzed for each group comparison. A total of 4175, 2992, 4050 significantly hypomethylated and 6393, 8528, 7663 significantly hypermethylated promoter regions were identified from the MDA-MB-231 vs. MCF10A, HCC1937 vs. MCF10A, and HCC1937 vs. MDA-MB-231 cell line comparisons, respectively. The M-values for the top significantly hypermethylated/hypomethylated promoter regions are shown as a heatmap that reveals methylation patterns across the three phenotypes (Fig. S1, ESI†). Certain genes are activated or repressed in TNBC compared to the control as well as some genes that are regulated differently in the HCC1937 BRCA1mu compared to both MDA-MB-231 BRCA1wt and the MCF10A control, which may be indicative of the more aggressive phenotype. One such gene is the SLFN11, in which the promoter is hypomethylated in HCC1937 compared with the other two cell lines, which are hypermethylated (Fig. S1, ESI†). SLFN11 serves as an S-phase checkpoint preventing the survival of cells after accumulation of DNA damage and replication stress.29 SLFN11 has shown a wide range of gene expression in TNBC; however, when studied together with BRCAness phenotype, it was shown that tumors with high SLFN11 expression and BRCA1 and BRCA2 germline mutations had increased response to irinotecan treatment, which is a Topoisomerase I inhibitor.29
RNA-sequencing and phosphoproteomic analysis was performed on the same cell lines and growth conditions as the DNA methylation data to investigate differential gene expression and protein abundance between the three cell lines. Out of a total of 16 718 genes analyzed, 7226, 7506, and 7724 genes were differentially regulated with an FDR p-value < 0.05 and an absolute fold change > 2 between the MDA-MB-231 vs. MCF10A, HCC1937 vs. MCF10A, and HCC1937 vs. MDA-MB-231 cell line comparisons, respectively (Fig. S2, ESI†). A total of 7634 proteins were quantified with 1350, 1997, and 1968 identified as differentially abundant in the same pair-wise comparisons above with a FDR adjusted p-value < 0.05 and an absolute fold change > 2 (Fig. S2, ESI†). Interestingly, SLFN11 was slightly elevated in the RNAseq gene expression data in MDA-MB-231 compared to MCF10As but had significantly higher expression in the HCC1937 cells compared to both MCF10As and MDA-MB-231 cells (Supplemental file 2, ESI†).
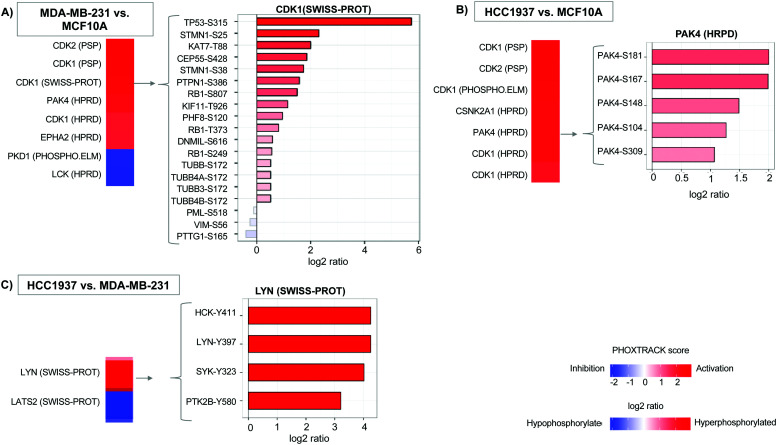
Additionally, we quantified 11 242 single phosphopeptide sites with 3243, 4490, and 4249 significantly differentiated between the MDA-MB-231 vs. MCF10A, HCC1937 vs. MCF10A, and HCC1937 vs. MDA-MB-231 cell line comparisons. Phosphorylated peptides were analyzed using PHOXTRACK (PHOsphosite-X-TRacing Analysis of Causal Kinases) to identify key regulating proteins by mapping phosphopeptides to putative kinases.26 Significant phosphopeptides from both MDA-MB-231 and HCC1937 TNBC cells compared to the control identified kinases such as CDK1, CDK2, and PAK4 (Fig. 2A and Supplemental file 4, ESI†). CDK1 kinase substrates include TP53-S315, KAT7-T88, STMN1-S25 and STMN1-S38, which were hyperphosphorylated in TNBC. PAK4 substrates PAK4-S181, S167, S104, S148, and S309 were also all hyperphosphorylated in both TNBC cell lines (Fig. 2B). PAK4 is an upstream regulator of JNK and is an important regulator of cytoskeleton remodeling. PAK4 is commonly overexpressed in human cancer30–32 and affects motility, invasion, metastasis and growth. Kinases regulating differentiating phosphopeptides between the subtypes of TNBC include LYN and LATS2. LYN is a non-receptor tyrosine kinases (nRTK) member of the SRC family and functions as an oncogene in breast cancer.4,6,33 The Lyn protein was found to be hyperphosphorylated at position Y397 in the BRCA1mu compared to BRCA1wt. In addition, a second SRC family member, HCK was also hyperphosphorylated at position Y411 by LYN kinase (Fig. 2C).
Fig. 2. Significant phosphorylated peptides and kinases. PHOXTRACK enrichment of known kinase targets from significantly differentiating phosphopeptides from (A) MDA-MB-231 compared to MCF10A, (B) HCC1937 compared to MCF10A, and (C) HCC1937 compared to MDA-MB-231. The PHOXTRACK score displays the predicted activation (red) or inactivation (blue) of a particular kinase in the vertical bar. A selected kinase and its substrates are shown in the horizontal bar plots. The plot displays the log2 ratio for hyper-phosphorylated (red) and hypo-phosphorylated (blue) peptides identified in the phosphoproteomics data set.
Other proteins of interest include those that write, read, and erase histone post-translational modifications. These proteins were identified from the Writers, Erasers, and Readers of Acetylation and Methylation (WERAM, version 1.0) database34 and are highlighted in Fig. S2 (ESI†). HDAC4 was significantly altered in HCC1937 compared to control, where it was reduced at both the transcription and protein level. This is not surprising since missense mutations were detected in HDAC4 specifically in HCC1937.35 In MDA-MB-231, only HDAC1 was significantly altered compared to control, where its transcription levels were elevated. Expression of HDACs is diverse and redundant across different TNBC subtypes. This redundancy is crucial especially for the BRCA1 mutant subtypes. Inhibition of HDACs has been shown to be lethal because this causes extensive DNA damage which cells deficient in BRCA1 cannot tolerate since their homologous recombination DNA repair mechanisms are impaired.36 HDAC4 has been shown to be involved in MTA1-mediated epigenetic regulation of ESR1 expression in breast cancer.35 This protein deacetylates HSPA1A and HSPA1B at Lys-77.
DNMT1 is overexpressed at the transcriptional level in HCC1937 compared to control, which is in line with the reported association between higher levels of DNMT1 and increased transformation.37,38 Interestingly, DNMT3A protein levels are increased in HCC1937 but decreased in MDA-MB-231 compared to control. DNMT3A is a histone H3K36me2 and H3K36me3 reader and acts as a transcriptional corepressor for ZBTB18.39 DNMTs function to methylate DNA and recruit NSD1 and NSD2 histone methyltransferases to specific genomic locations. NSD2 was found to be up-regulated in MDA-MB-231 compared to both HCC1937 and control cell lines and may be an indication of changes about to occur in the methylation patterns on H3K36 into H3K36me1/2. It is unclear the role of H3K36me and this modification was found to be increased greater than 2-fold in the HCC1937 cells compared to MCF10As.40 However, histone H3K36me2 has a clear role in double-strand break repair.41 Taken together, DNMTs work in conjunction with NSD2 to methylate H3K36 to H3K36me1/2, which plays a central role in transforming chromatin from heterochromatin into euchromatin; allowing for the upregulation of genes associated with cancer phenotypes.40 Histone H3K36me2 recruits DNMT3A and shapes the methylation landscape of intergenic regions.39 A previous study showed that DNMT protein levels, and not transcription levels, predict sensitivity to the demethylating agent, decitabine.42 Therefore, this discrepancy in DNMT levels among the TNBC cell lines may be of clinical significance, which warrants further research.
We also identified 2-fold enrichments of H3K9me2/3, H3K36me, H3K79me, and H4K20me modifications among the three cell lines (Table 1 and Supplemental file 5, ESI†). The log2 fold change values for each sample group comparison and each histone modification is listed in Table 1. We also included proteins identified as writers, readers, and erasers of the significant histone PTMs.
Significant histone PTMs.
| Histone PTM | log2 fold change (MDA-MB-231 vs. MCF10A) | log2 fold change (HCC1937 vs. MCF10A) | log2 fold change (HCC1937 vs. MDA-MB-231) | PTM modification proteins (WERAM) |
|---|---|---|---|---|
| H3k9me2 | 0.22 | −1.737 | −1.956 | MECOM, PRDM2,PRDM16, PRDM8, PRDM2, EHMT1, EHMT2, JHDM2A, JMJD2C, JMJD2B, TRIM28 |
| H3k9me3 | −3.319 | −4.735 | −1.416 | MECOM, PRDM2, PRDM16, PRDM8, PRDM2, EHMT1, EHMT2, JHDM2A, JMJD2C, JMJD2B, TRIM28 |
| H3k36me | 0.883 | 1.579 | 0.696 | WHSC1, NSD1, NSD2, SMYD2, SETMAR, SETD2, ASH1L, METNASE, SETD3 |
| H3k79me | 1.601 | 1.905 | 0.304 | DOT1 |
| H4k20me | −0.817 | −1.994 | −1.177 | WHSC1, SUV420H1, SUV420H2, SETD8, L3MBBTL1 |
Data integration
We investigated the correlation of features amongst DNA methylation status of promoter regions, gene expression, protein abundance, and phosphorylated peptides for each sample. A total of 10 393 features were in common based on Ensembl IDs between DNA methylation regulator features and gene expression data sets (Fig. 3 and Supplemental file 6, ESI†). The significant features from both single-omics analyses had a −0.39, −0.38, and −0.39 correlation between methylated/unmethylated and upregulated/downregulated genes for the MDA-MB-231 vs. MCF10A, HCC1937 vs. MCF10A, and HCC1937 vs. MDA-MB-231 group comparisons, respectively. These negative correlation values seems to indicate the two omics data sets are not correlated. However, it is actually significant since a positive beta value in DNA methylation indicates hypermethylation (positive delta beta) leading to decreased gene expression (negative fold change). We next investigated how much correlation was among gene and protein expression within the samples. There was a total of 7433 common features based on the Ensembl ID for genes and proteins with a positive correlation value of 0.82, 0.82, and 0.86 for the MDA-MB-231 vs. MCF10A, HCC1937 vs. MCF10A, and HCC1937 vs. MDA-MB-231 group comparisons, respectively (Fig. 3 and Supplemental file 7, ESI†). The majority of the expression of differentially significant genes and proteins had similar expression patterns. The positive correlation among proteins and phosphorylated peptides had even higher positive correlation values of 0.89, 0.9, and 0.9 among the same sample comparisons when analyzing a total of 9406 features common between the two data sets (based on UniProtKB IDs). The log2 fold change for the DNA methylation regulatory features (promoter regions) are shown on the y-axis, while the gene expression log2 fold change is on the x-axis. The significant features from each data set are plotted on the four outside quadrants of the correlation plot and the middle quadrants show the non-significant features (Fig. 3). Similar values are plotted for the gene and protein correlation and protein and phosphopeptides correlation for each pair-wise sample comparison (Fig. 3).
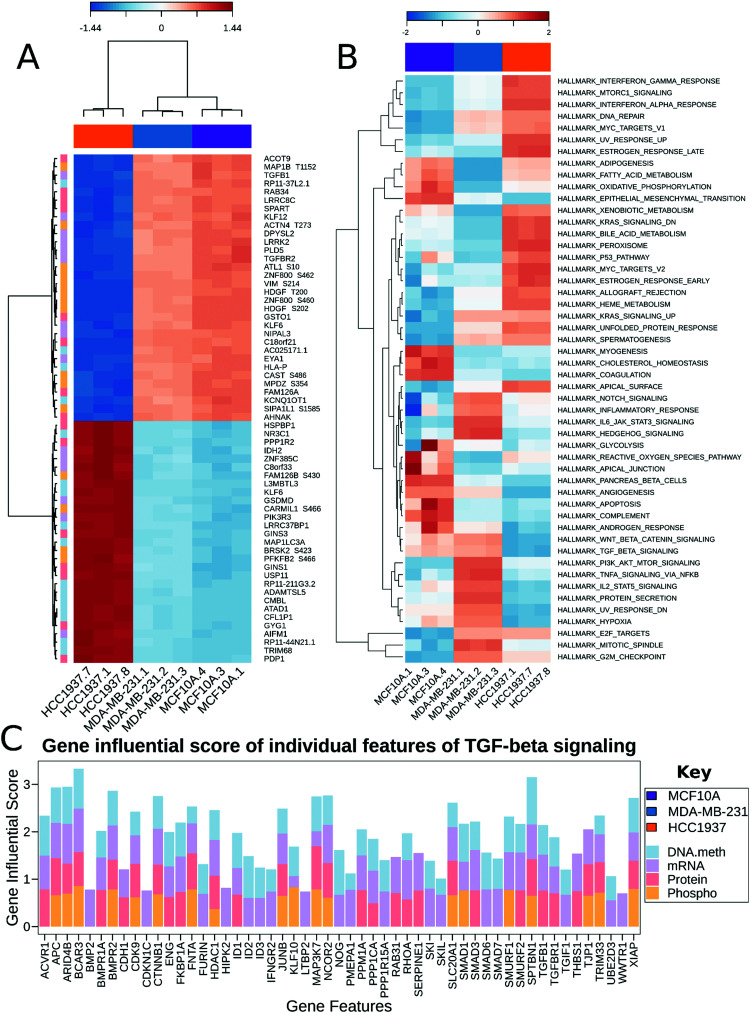
MixOmics supervised analysis was used to identify key features from multi-omics data that can clearly separate normal epithelial cells from triple negative breast cancer with and without a BRCA1 mutation. We used the established DIABLO method to integrate the same biological samples from each data set. The first principal component clearly separated the HCC1937 cell line from both MDA-MB-231 and MCF10As while the second principal component distinguishes between MDA-MB-231 and the HCC1937 and MCF10As. Important features in component 1 include TGFB1, TGFBR2, KLF6, KLF12, PIK3R3 and VIM, while features important from component 2 include NES, RASL11B, HOXC9, LAMB3, PRKCD, PRKCE, and MELK to name a few (Fig. 4A and Fig. S3, ESI†). TGFB1 expression and its receptors, TGFBR1 and TGFBR2, lead to the induction of canonical and noncanonical TGFβ signaling pathways and is correlated with oncogenic activity.43 In early stage breast cancer, TGFβ signaling shows tumor suppressive effects; however, in late stages, TGFB1 is linked with increased tumor progression, metastasis, and epithelial to mesenchymal transition (EMT).43 TGFB1 and TGFBR2 had decreased gene expression in HCC1937 compared to the other cell lines. KLF6 and KLF12 (Kruppel-like factors) are DNA-binding transcriptional regulators which play essential roles in proliferation, differentiation, apoptosis, and migration.44 In breast cancer, KLFs function in the process of EMT, invasion, and angiopoiesis.44 These genes were up-regulated in MCF10As and MDA-MB-231 cell lines. VIM is a marker for mesenchymal cell types and was found to be hyperphosphorylated at S214 in the MCF10A and MDA-MB-231 cell lines.45 NES was hyperphosphorylated at S768, S1496, and S1498 in the MDA-MB-231 cell line. NES promotes the disassembly of phosphorylated vimentin intermediate filaments during mitosis and is associated with reduced survival rates in basal-like subtypes of breast cancer.46 PRKCD, PRKCE, and MELK are non-receptor serine/threonine protein kinases and part of the VEGF and Wnt signaling pathways.47–50 Maternal embryonic leucine zipper kinase (MELK) shows increased gene expression in MDA-MB-231 cells. Silencing of this regulator leads to programmed cell death in MDA-MB-231 cells as it functions as a modulator of intercellular signaling through the apoptosis signal-regulating kinase/Jun N-terminal kinase (JNK) pathway, p38 signaling, and NF-kB pathway.51
Fig. 4. Multi-omics data integration. (A) Clustered Image Map for component 1. Represents the multi-omics signature in relation with the samples. The most important features in component one distinguishing between the three cell lines is shown as a clustered heatmap using the cimDiablo() function provided by MixOmics. The red and blue colors represent positive and negative Pearson correlations respectively, whereas grey represents small correlation values. (B) MOGSA heatmap showing the Gene Set Score (GSS) for significantly regulated gene-sets in the cell lines. The white colored blocks indicate the change of gene-sets are non-significant (FDR corrected p-value > 0.01). (C) The Gene Influential Score of individual features for the TGFβ signaling pathway.
Multi-omics gene-set analysis (MOGSA) was used to integrate DNA methylation at regulatory elements, gene expression, protein abundance, and phosphorylated peptides from the same set of samples. The method uses a low dimensional representation of most variant correlated features across the different data types and transforms the features to the same scale. The integrated gene-set score is then calculated from the most informative features in each data type.28 Interestingly, significant hallmark pathways differentiating the three cell lines include Notch signaling, PI3K/AKT/MTOR signaling, TNFα signaling via NFκB as up-regulated in TNBC BRCA1wt; KRAS signaling, MYC targets, and DNA repair as up-regulated in TNBC BRCA1mu (Fig. 4B). Epithelia Mesenchymal transition (EMT) was found to be increased in MCF10A and MDA-MB-231, which is consistent with the increased features VIM and TGFB1 from the MixOmics feature extraction analysis. TGFβ signaling was found to be down-regulated in the TNBC BRCA1mu cell line compared to both normal and BRCA1wt cell lines (Fig. 4B).
We investigated the features from each multi-omics level of regulation to see which data had the most influence in identifying TGFβ signaling as an important pathway. Fig. 4C displays the gene influential score for the TGFβ signaling gene features from DNA methylation, mRNA, protein, and the phosphorylation data sets. The data-wise decompose gene set score for a subset of these pathways is shown in Fig. S4 (ESI†). The majority of the features included data from at least two levels of regulation and all included data from RNAseq. By using a multi-omics approach, we are able to identify the majority of features in the pathway analysis but also identify the activated SMAD5 phosphorylation. It is worth noting that TGFβ-stimulated TGF-Beta Receptor Type I phosphorylates of SMAD1/5 to promote cancer cell migration but not in normal cells.52 Interestingly, we also observed that TGFBR1 protein expression and SMAD5 phosphorylation are up-regulated in the TNBC MDA-MB-231 cells compared to MCF10A, a normal mammary epithelial cell (Fig. 3 and Supplemental files 3 and 4, ESI†). This interesting finding raises the possibility to target TGFβ–TGFBR1–SMAD1/5 axis to inhibit the pro-tumorigenic functions of TGF-β signal transduction pathway. The DNA methylation and RNAseq data is freely available via GEO GSE171958 and the proteomics data is available via the ProteomeXchange PXD025238.
Discussion
Triple negative breast cancer is an aggressive cancer affecting younger women and has little therapeutic options. We sought to understand the underlying regulatory mechanisms of aggressiveness applying a multi-omics approach using well established cell lines: MCF10A (human breast epithelial), MDA-MB-231 (TNBC-BRCA1wt), and HCC1937 (TNBC-BRCA1 5382insC). By utilizing these cell lines, we limited the technical and biological variability associated with highly heterogeneous breast cancer patient samples deposited into large databases such as TCGA. This allows for the validation of the data integration methods with clear outcomes that can be applied to more complex patient samples in future work.
The emergence of concepts such as BRCAness further emphasize the need for understanding the systems-level biology of breast cancer. BRCAness refers to the phenomenon where tumors with functional BRCA1/2 display phenotypes similar to those of BRCA1/2-deficient tumors.53 This has been attributed to deficiency in homologous repair pathways in the BRCA-functional tumors, making them potentially susceptible to treatments usually reserved to BRCA1/2-mutant tumors such as PARP inhibitors (PARPi).53–55 Thus, strictly relying on the genomic information of the functional status of BRCA1/2, as opposed to considering the molecular mechanisms of the cells as a whole, may limit potentially impactful treatments for a subset of patients.
Our multi-omics integrations emphasize a stark discrepancy in expression of key signaling pathways among the TNBC subtypes. Specifically, the PI3K/AKT/mTOR pathway is differentially important for the two TNBC cell lines studied here, a conclusion that can be clearly drawn upon examining the MOGSA heatmap (Fig. 4B).56 Given that this pathway is among those most frequently mutated in TNBC, one may conclude that it is targetable. However, this treatment strategy may be ineffective in a subset of TNBCs (i.e., BRCA1 mutants) since this pathway is downregulated in the HCC1937 cell line (Fig. 4B). Our data highlight the inadequacy of this treatment strategy as this diversity among the cell lines is the underlying reason behind failure of treatments which targeted this pathway.54,56
The PI3K/AKT/mTOR pathway is frequently aberrant in cancer. Specifically, PIK3CA is enriched in 10% of TNBC cases,57–59 and loss-of-function PTEN occurs in one-third of TNBCs.60 Despite the high occurrence of mutations in PIK3CA among different TNBC subtypes, we did not observe a significant difference in its gene or protein expression levels.61 This may be explained by the observed upregulation of MYC target genes (Fig. 4B), as overexpression of MYC has been shown to downregulate the PI3K/AKT pathway.62 Additionally, we find that PTEN is of reduced transcription, protein and phosphorylation levels, and its promoter is hyper-methylated, in HCC1937 compared to the other two cell lines. Whereas PTEN transcription and protein levels were unchanged and its phosphorylation levels were increased when comparing MDA-MB-231 to control. This is in alignment with reports of the BL1 TNBC subtype, a subtype of MDA-MB-231 cells, expressing a low level of PTEN.61 The PI3K/AKT/mTOR pathway is activated by EGFR. This pathway is constitutively activated in some TNBC subtypes. Further analysis of the molecular mechanisms leading to this phenotype revealed that loss of PTEN and activating mutations in PI3KCA lead to activation of mTOR. The HCC1937 cell line is a known PTEN-null mutant63 – this is corroborated by our studies as mentioned above (Fig. 3). The loss of PTEN would suggest that the PI3K/AKT/mTOR pathway is hyper-activated since PTEN is a negative regulator of mTOR. However, our multi-omic data integration analyses indicate that this pathway is unexpectedly downregulated in HCC1937. Indeed, this apparent paradox may be explained by a report that demonstrated the lack of therapeutic benefits when combing EGFR and mTOR inhibitors (gefitinib and everolimus, respectively) to treat the HCC1937 cell line, whereas these drugs showed a favorable synergistic effect when treating TNBC cell lines with an activating PI3KCA mutation.64 Our data indicate that the HCC1937 cell line did not respond to the mTOR inhibitor, despite the absence of PTEN, possibly because the PI3K/AKT/mTOR pathway as a whole is downregulated (Fig. 4B). Additionally, PIK3CA mutant breast cancer cells are selectively sensitive to mTOR inhibitors but not the cells with PTEN loss of function. Suggesting two quite distinct functional consequences of the PI3K/AKT/mTOR pathway in breast cancer.65 Conversely, our integrated analysis demonstrate that PI3K/AKT/mTOR pathway is upregulated in the MDA-MB-231 cell line (Fig. 4B). This is in agreement with previous findings that this cell line responds to co-inhibition of EGFR and mTOR with lapatinib and rapamycin, respectively.66,67
Further, we offer a potential molecular basis for the lack of therapeutic advantage to combining EGFR and mTOR inhibitors to treat HCC1937.64 It would have been reasonable to target this TNBC subtype with such inhibitors since EGFR protein levels are elevated and PTEN is deleted in the HCC1937 cell line (Fig. 3B). However, our multi-omic analysis of this cell line clearly predicts the failure of co-inhibiting EGFR and mTOR as the PI3K/AKT/mTOR pathway is downregulated. Interestingly, the DNA methylation and gene expression data point to an increase in SLFN11 in the HCC1937 cells, suggesting a role for TOP1 inhibitor therapy for a subset of TNBC tumors.29
In contrast to reports which showed a concomitant increase in Wnt signaling along with reduced PTEN levels, we find that the Wnt/β-catenin pathway is downregulated in HCC1937 compared to MDA-MB-231.68 This may be explained by a previous report that established an interaction between BRCA1 and nuclear β-catenin.69 This report demonstrates that functional BRCA1 enhances the levels of the nuclear active form of β-catenin, whereas mutant BRCA1, such as that in HCC1937, limits its expression. Thus, the Wnt pathway genes, whose transcription is activated via β-catenin, may not be expressed in HCC1937 leading to the observed downregulation of the pathway in these cells. This further accentuates the heterogeneity even within TNBC subtypes as we demonstrate diversity in Wnt/β-catenin expression within the BL1 subtype. We do find however that MYC targets are increased in our integrated dataset, in correlation with the aforementioned report, which shows that high MYC pathway activity correlates with increased risk in the BL1 subtype (Fig. 4B).70
Overall, downregulation of signaling pathways downstream to EGFR is evident in HCC1937 compared to MDA-MB-231. This is reflected in their divergent sensitivities to inhibitors of key signaling pathways. For instance, MDA-MB-231 is sensitive to the MAPK pathway MEK1/2 inhibitor, selumetinib, whereas HCC1937 is not.71 In addition, it is not surprising that our integrated analyses show downregulation of key signaling pathways in HCC1937 cells. It is of note that the EGFR-activated pathway, PI3K/AKT/mTOR, and the TGFβ pathway interact through the MAPK pathway.72
Protein tyrosine kinases (PTK) are important regulators of several signal transduction pathways, shaping biological processes like cell proliferation, differentiation, migration, and apoptosis in response to internal and external stimuli.73 In breast cancer, amplification and activation of ERBB2 results in ERBB2 (HER2)-positive tumors. Additionally kinase-based gene expression signatures (GESs) is also reported in luminal BCs74 and in basal breast cancers.75 Several PTKs have been implicated and explored as potential therapeutic targets in cancer, nevertheless, the aberrant PTK signaling, and functional redundancy make them a hard target for therapeutic regiment. Recently, we reported that breast tumor kinase (BRK), promotes metastatic potential by phosphorylating SMAD4, the major component of TGFβ/SMAD signaling in mammary epithelial cells.76 However, the inhibition of the kinase activity of BRK with small molecules did not inhibit cancer cell growth77 – suggesting functional redundancy and kinase-independent roles of BRK in breast cancer. In this study, we also observed that besides ptk6/BRK (Supplemental files 2 and 4, ESI†), non-receptor protein tyrosine kinase LYN is hyperphosphorylated (Fig. 2C). Unfortunately, we were not able to detect SMAD4 phosphorylation in this data set. Of note, we will further interrogate whether LYN kinase beside BRK also regulates TGFβ/SMAD signaling in addition to BRK in TNBC cells (Fig. 5).
Fig. 5. Graphical summary. DNA methylation results reveal hypomethylation of SLFN11 promoter region in HCC1937 BRCA1 mutant cells. This combination has been shown to impact clinical treatment options. DNMT3A is a DNA methyltransferase and was elevated in HCC1937 cells along with Histone H3K36me. NSD2 is a histone methyltransferase elevated in MDA-MB-231 cells and is known to convert H3K36 to H3K36me1/2; transforming heterochromatin into euchromatin (active state). H3K36me2 recruits DNMT3A to shape the intergenic DNA methylation landscape. MDA-MB-231 cells showed increase TGFBR1, SMAD5 S465ph, and PTEN S385ph elevation leading to increased expression of TGFβ signaling and PI3K/AKT/mTOR pathways. HCC1937 cells had decreased expression of PTEN S385ph and decreased PI3K/AKT/mTOR and WNT/β-catenin pathways.
Conclusion
A multi-omics study design allows for the identification of information that may otherwise be missed due to technical limitations in the various technologies. For instance, we are able to detect function at the protein level but due to sequencing depth limitations, lower abundance proteins may not be detected and may miss crucial information. Therefore, information can be gained by analyzing the gene expression levels. In addition analyzing phosphorylated peptides provides information about changes in the activity of the proteins that may be functionally significant but not identified by protein abundance changes. We then add epigenetic modifications at the DNA methylation and histone post-translational levels that also regulates the expression of features. Together each level of regulation tells a story of the impact of signaling pathways in the context of disease.
For example, the value of conducting multi-omics analysis is demonstrated in Carbonic anhydrase 2 (CA2), where we observe a significant reduction in mRNA levels despite a significant increase in protein levels – this is true for both cancer cell lines compared to control (Fig. S2, ESI†). CA2 upregulation is associated with poor prognosis, enhanced tumor progression and metastasis of breast cancer.78 The reliance on mRNA levels alone may have missed the identification of this protein. The necessity of integrating the multi-omics profile of tumors is exemplified by the LOTUS trial. Patients with aberrant PI3K/AKT/mTOR pathways benefitted most and some with loss of PTEN did not benefit.66 These results solidify our observations that for some TNBC subtypes, aberrations of the PI3K/AKT/mTOR pathway do not necessarily arise with the loss of PTEN. This emphasizes the importance of carefully tailoring therapies to pathways activated in the specific tumor. This is best achieved by considering the genomic, transcriptomic and proteomic profile of TNBCs.
Author contributions
Kevin Chappell and Kanishka Manna analyzed the protein, histone PTM data, MixOmics, and MOGSA data, and helped write the manuscript. Charity L. Washam analyzed the correlation of each of the omics data sets. Stefan Graw and Jordan T. Bird analyzed the DNA methylation data set. Jordan T. Bird performed integrated data analysis using MixOmics and MOGSA. Allen Gies performed the RNAseq data analysis. Duah Alkam provided the summary figures and helped write the manuscript. Stephanie D. Byrum, Matthew Thompson, Maroof Khan Zafar, Lindsey Hazeslip, Christopher Randolph, and Alicia K Byrd helped to grow the cells, extract the DNA/RNA/protein, and performed the sequencing for each data set. Sayem Miah provided the biological expertise for triple negative breast cancer interpretation of the results and helped write the manuscript. Stephanie D. Byrum helped with the conceptual design, the editing of the R scripts, data analysis, writing of the manuscript, and overall guidance of the project. All authors read and edited the manuscript.
Conflicts of interest
No potential conflict of interest was reported by the authors.
Supplementary Material
Acknowledgments
This study was supported by the Arkansas Children's Research Institute, the Arkansas Biosciences Institute, and the Center for Translational Pediatric Research funded under the National Institutes of Health National Institute of General Medical Sciences (NIH/NIGMS) grant P20GM121293, National Science Foundation Award No. OIA-1946391, the UAMS Vice-Chancellor's for Research award, and the Little Rock Envoys Seeds of Science award. Sayem Miah is funded through UAMS Winthrop P. Rockefeller Cancer Institute.
Electronic supplementary information (ESI) available: Supplemental file 1: Illumina EPIC beadChip array DNA methylation normalized values and statistical results. Supplemental file 2: RNA-seq normalized counts and statistical results. Supplemental file 3: Protein normalized MS3 intensities and statistical results. Supplemental file 4: Phosphopeptide normalized MS3 intensities and statistical results. Supplemental file 5: Histone post-translational modifications normalized MS1 intensities. Supplemental file 6: DNA methylation of promoter regions and RNAseq gene expression common features and correlation results from Fig. 3. Supplemental file 7: RNAseq and protein common features and correlation results. Supplemental file 8: Protein and phosphorylated peptide features and correlation results. See DOI: 10.1039/d1mo00117e
References
- Turanli B. Karagoz K. Bidkhori G. Sinha R. Gatza M. L. Uhlen M. et al., Multi-Omic Data Interpretation to Repurpose Subtype Specific Drug Candidates for Breast Cancer. Front. Genet. 2019:10. doi: 10.3389/fgene.2019.00420. [DOI] [PMC free article] [PubMed] [Google Scholar]; , Available from: https://www.frontiersin.org/articles/10.3389/fgene.2019.00420/full
- Siegel R. L. Miller K. D. Jemal A. Cancer statistics. CA - Cancer J. Clin. 2019;69(1):7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- Reis-Filho J. S. Tutt A. N. J. Triple negative tumours: a critical review. Histopathology. 2008;52(1):108–118. doi: 10.1111/j.1365-2559.2007.02889.x. [DOI] [PubMed] [Google Scholar]
- Bianchini G. Balko J. M. Mayer I. A. Sanders M. E. Gianni L. Triple-negative breast cancer: challenges and opportunities of a heterogeneous disease. Nat. Rev. Clin. Oncol. 2016;13(11):674–690. doi: 10.1038/nrclinonc.2016.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogoda K. Niwińska A. Sarnowska E. Nowakowska D. Jagiełło-Gruszfeld A. Siedlecki J. et al., Effects of BRCA Germline Mutations on Triple-Negative Breast Cancer Prognosis. J. Oncol. 2020;2020:8545643. doi: 10.1155/2020/8545643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiorean R. Braicu C. Berindan-Neagoe I. Another review on triple negative breast cancer. Are we on the right way towards the exit from the labyrinth? Breast Edinb Scotl. 2013;22(6):1026–1033. doi: 10.1016/j.breast.2013.08.007. [DOI] [PubMed] [Google Scholar]
- Marra A. Trapani D. Viale G. Criscitiello C. Curigliano G. Practical classification of triple-negative breast cancer: intratumoral heterogeneity, mechanisms of drug resistance, and novel therapies. npj Breast Cancer. 2020;6:54. doi: 10.1038/s41523-020-00197-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y. Helenius M. Väänänen K. Bulanova D. Saarela J. Sokolenko A. et al., BRCA1-deficient breast cancer cell lines are resistant to MEK inhibitors and show distinct sensitivities to 6-thioguanine. Sci. Rep. 2016;6(1):28217. doi: 10.1038/srep28217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Y. Han B. Yu Y. Yao W. Bose S. Karlan B. Y. et al., Evaluation of MCF10A as a Reliable Model for Normal Human Mammary Epithelial Cells. PLoS One. 2015;10(7):e0131285. doi: 10.1371/journal.pone.0131285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MDA-MB-231 Cell Line – an overview|ScienceDirect Topics. [cited 2021 Apr 2]. Available from: https://www.sciencedirect.com/topics/medicine-and-dentistry/mda-mb-231-cell-line
- Huang Z. Yu P. Tang J. Characterization of Triple-Negative Breast Cancer MDA-MB-231 Cell Spheroid Model. OncoTargets Ther. 2020;13:5395–5405. doi: 10.2147/OTT.S249756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson G. E. Chen T. T.-L. Stastny V. A. Virmani A. K. Spillman M. A. Tonk V. Blum J. L. Schneider N. R. Wistuba I. I. Shay J. W. Minna J. D. Gazdar A. F. Characterization of a Breast Cancer Cell Line Derived from a Germ-Line BRCA1 Mutation Carrier. Cancer Res. 1998;58(15):3237–3242. [PubMed] [Google Scholar]
- Singh A. Shannon C. P. Gautier B. Rohart F. Vacher M. Tebbutt S. J. et al., DIABLO: an integrative approach for identifying key molecular drivers from multi-omics assays. Bioinformatics. 2019;35(17):3055–3062. doi: 10.1093/bioinformatics/bty1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M. Barnes B. Tsan C. Ho V. Klotzle B. Le J. M. et al., High density DNA methylation array with single CpG site resolution. Genomics. 2011;98(4):288–295. doi: 10.1016/j.ygeno.2011.07.007. [DOI] [PubMed] [Google Scholar]
- Pidsley R. Y. Wong C. C. Volta M. Lunnon K. Mill J. Schalkwyk L. C. A data-driven approach to preprocessing Illumina 450K methylation array data. BMC Genomics. 2013;14(1):293. doi: 10.1186/1471-2164-14-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aryee M. J. Jaffe A. E. Corrada-Bravo H. Ladd-Acosta C. Feinberg A. P. Hansen K. D. et al., Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinf. Oxf. Engl. 2014;30(10):1363–1369. doi: 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm-Benartzi C. S. Koestler D. C. Karagas M. R. Flanagan J. M. Christensen B. C. Kelsey K. T. et al., Review of processing and analysis methods for DNA methylation array data. Br. J. Cancer. 2013;109(6):1394–1402. doi: 10.1038/bjc.2013.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Infinium MethylationEPIC Product Files. [cited 2021 Apr 7]. Available from: https://support.illumina.com/downloads/infinium-methylationepic-v1-0-product-files.html
- Ritchie M. E. Phipson B. Wu D. Hu Y. Law C. W. Shi W. et al., limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortin J.-P. Triche Jr T. J. Hansen K. D. Preprocessing, normalization and integration of the Illumina HumanMethylationEPIC array with minfi. Bioinformatics. 2017;33(4):558–560. doi: 10.1093/bioinformatics/btw691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y. Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Methodol. 1995;57(1):289–300. [Google Scholar]
- Liao Y. Smyth G. K. Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinf. Oxf. Engl. 2014;30(7):923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Robinson M. D. Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11(3):R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey A. J. Naceanceno K. S. Lan R. S. Washam C. L. Orr L. M. Mackintosh S. G. et al., ProteoViz: a tool for the analysis and interactive visualization of phosphoproteomics data. Mol. Omics. 2020;16(4):316–326. doi: 10.1039/c9mo00149b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graw S. Tang J. Zafar M. K. Byrd A. K. Bolden C. Peterson E. C. et al., proteiNorm − A User-Friendly Tool for Normalization and Analysis of TMT and Label-Free Protein Quantification. ACS Omega. 2020:9. doi: 10.1021/acsomega.0c02564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidner C. Fischer C. Sauer S. PHOXTRACK–a tool for interpreting comprehensive datasets of post-translational modifications of proteins. Bioinformatics. 2014;30(23):3410–3411. doi: 10.1093/bioinformatics/btu572. [DOI] [PubMed] [Google Scholar]
- Nesvizhskii A. I. Keller A. Kolker E. Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 2003;75(17):4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- Meng C. Basunia A. Peters B. Gholami A. M. Kuster B. Culhane A. C. MOGSA: Integrative Single Sample Gene-set Analysis of Multiple Omics Data. Mol. Cell. Proteomics. 2019;18(8 suppl 1):S153–S168. doi: 10.1074/mcp.TIR118.001251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coussy F. El-Botty R. Château-Joubert S. Dahmani A. Montaudon E. Leboucher S. et al., BRCAness, SLFN11, and RB1 loss predict response to topoisomerase I inhibitors in triple-negative breast cancers. Sci. Transl. Med. 2020;12(531):eaax2625. doi: 10.1126/scitranslmed.aax2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y.-G. Ning K. Li F. Group II p21-activated kinases as therapeutic targets in gastrointestinal cancer. World J. Gastroenterol. 2016;22(3):1224–1235. doi: 10.3748/wjg.v22.i3.1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. Liu F. Li F. PAK as a therapeutic target in gastric cancer. Expert Opin. Ther. Targets. 2010;14(4):419–433. doi: 10.1517/14728221003642019. [DOI] [PubMed] [Google Scholar]
- Kumar R. and Li D.-Q., Chapter Four – PAKs in Human Cancer Progression: From Inception to Cancer Therapeutic to Future Oncobiology in Advances in Cancer Research. ed. K. D. Tew and P. B. Fisher, Academic Press, 2016. pp. 137–209. Available from: https://www.sciencedirect.com/science/article/pii/S0065230X16300021 [DOI] [PubMed] [Google Scholar]
- Foulkes W. D. Smith I. E. Reis-Filho J. S. Triple-negative breast cancer. N. Engl. J. Med. 2010;363(20):1938–1948. doi: 10.1056/NEJMra1001389. [DOI] [PubMed] [Google Scholar]
- Xu Y. Zhang S. Lin S. Guo Y. Deng W. Zhang Y. et al., WERAM: a database of writers, erasers and readers of histone acetylation and methylation in eukaryotes. Nucleic Acids Res. 2017;45(Database issue):D264–D270. doi: 10.1093/nar/gkw1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clocchiatti A. Di Giorgio E. Ingrao S. Meyer-Almes F.-J. Tripodo C. Brancolini C. Class IIa HDACs repressive activities on MEF2-depedent transcription are associated with poor prognosis of ER+ breast tumors. FASEB J. 2013;27(3):942–954. doi: 10.1096/fj.12-209346. [DOI] [PubMed] [Google Scholar]
- Zhang B. Lyu J. Yang E. J. Liu Y. Wu C. Pardeshi L. et al., Class I histone deacetylase inhibition is synthetic lethal with BRCA1 deficiency in breast cancer cells. Acta Pharm. Sin. B. 2020;10(4):615–627. doi: 10.1016/j.apsb.2019.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belinsky S. A. Nikula K. J. Baylin S. B. Issa J. P. Increased cytosine DNA-methyltransferase activity is target-cell-specific and an early event in lung cancer. Proc. Natl. Acad. Sci. U. S. A. 1996;93(9):4045–4050. doi: 10.1073/pnas.93.9.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J. Issa J. P. Herman J. Bassett D. E. Nelkin B. D. Baylin S. B. Expression of an exogenous eukaryotic DNA methyltransferase gene induces transformation of NIH 3T3 cells. Proc. Natl. Acad. Sci. U. S. A. 1993;90(19):8891–8895. doi: 10.1073/pnas.90.19.8891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg D. N. Papillon-Cavanagh S. Chen H. Yue Y. Chen X. Rajagopalan K. N. et al., The histone mark H3K36me2 recruits DNMT3A and shapes the intergenic DNA methylation landscape. Nature. 2019;573(7773):281–286. doi: 10.1038/s41586-019-1534-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaghi M. Broccoli V. Sessa A. H3K36 Methylation in Neural Development and Associated Diseases. Front. Genet. 2020;10 doi: 10.3389/fgene.2019.01291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fnu S. Williamson E. A. De Haro L. P. Brenneman M. Wray J. Shaheen M. et al., Methylation of histone H3 lysine 36 enhances DNA repair by nonhomologous end-joining. Proc. Natl. Acad. Sci. U. S. A. 2011;108(2):540–545. doi: 10.1073/pnas.1013571108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J. Qin B. Moyer A. M. Nowsheen S. Liu T. Qin S. et al., DNA methyltransferase expression in triple-negative breast cancer predicts sensitivity to decitabine. J. Clin. Invest. 2018;128(6):2376–2388. doi: 10.1172/JCI97924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarzynska J. M. Two Faces of TGF-Beta1 in Breast Cancer. Mediators Inflamm. 2014 doi: 10.1155/2014/141747. [DOI] [PMC free article] [PubMed] [Google Scholar]; , available from
- Zhang J. Li G. Feng L. Lu H. Wang X. Krüppel-like factors in breast cancer: Function, regulation and clinical relevance. Biomed. Pharmacother. 2020;123:109778. doi: 10.1016/j.biopha.2019.109778. [DOI] [PubMed] [Google Scholar]
- Mendez M. G. Kojima S.-I. Goldman R. D. Vimentin induces changes in cell shape, motility, and adhesion during the epithelial to mesenchymal transition. FASEB J. 2010;24(6):1838–1851. doi: 10.1096/fj.09-151639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asleh K. Won J. R. Gao D. Voduc K. D. Nielsen T. O. Nestin expression in breast cancer: association with prognosis and subtype on 3641 cases with long-term follow-up. Breast Cancer Res. Treat. 2018;168(1):107–115. doi: 10.1007/s10549-017-4583-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi K. Banasavadi-Siddegowda Y. Mo X. Kim S.-H. Mao P. Kig C. et al., MELK-dependent FOXM1 phosphorylation is essential for proliferation of glioma stem cells. Stem Cells Dayt. Ohio. 2013;31(6):1051–1063. doi: 10.1002/stem.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. Lee J.-K. Ahn S. H. Lee J. Nam D.-H. WNT signaling in glioblastoma and therapeutic opportunities. Lab. Investig. J. Technol. Methods Pathol. 2016;96(2):137–150. doi: 10.1038/labinvest.2015.140. [DOI] [PubMed] [Google Scholar]
- Duong M. Yu X. Teng B. Schroder P. Haller H. Eschenburg S. et al., Protein kinase C ∈ stabilizes β-catenin and regulates its subcellular localization in podocytes. J. Biol. Chem. 2017;292(29):12100–12110. doi: 10.1074/jbc.M117.775700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dissanayake S. K. Weeraratna A. T. Detecting PKC phosphorylation as part of the Wnt/calcium pathway in cutaneous melanoma. Methods Mol. Biol. 2008;468:157. doi: 10.1007/978-1-59745-249-6_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G. Yang M. Zuo L. Wang M.-X. MELK as a potential target to control cell proliferation in triple-negative breast cancer MDA-MB-231 cells. Oncol. Lett. 2018;15(6):9934–9940. doi: 10.3892/ol.2018.8543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu I. M. Schilling S. H. Knouse K. A. Choy L. Derynck R. Wang X.-F. TGFβ-stimulated Smad1/5 phosphorylation requires the ALK5 L45 loop and mediates the pro-migratory TGFβ switch. EMBO J. 2009;28(2):88–98. doi: 10.1038/emboj.2008.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord C. J. Ashworth A. BRCAness revisited. Nat. Rev. Cancer. 2016;16(2):110–120. doi: 10.1038/nrc.2015.21. [DOI] [PubMed] [Google Scholar]
- Marra A. Trapani D. Viale G. Criscitiello C. Curigliano G. Practical classification of triple-negative breast cancer: intratumoral heterogeneity, mechanisms of drug resistance, and novel therapies. npj Breast Cancer. 2020;6:54. doi: 10.1038/s41523-020-00197-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer H. McCabe N. Lord C. J. Tutt A. N. J. Johnson D. A. Richardson T. B. et al., Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434(7035):917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- Bianchini G. Balko J. M. Mayer I. A. Sanders M. E. Gianni L. Triple-negative breast cancer: challenges and opportunities of a heterogeneous disease. Nat. Rev. Clin. Oncol. 2016;13(11):674–690. doi: 10.1038/nrclinonc.2016.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis C. Shah S. P. Chin S.-F. Turashvili G. Rueda O. M. Dunning M. J. et al., The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486(7403):346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y.-Z. Ma D. Suo C. Shi J. Xue M. Hu X. et al., Genomic and Transcriptomic Landscape of Triple-Negative Breast Cancers: Subtypes and Treatment Strategies. Cancer Cell. 2019;35(3):428–440. doi: 10.1016/j.ccell.2019.02.001. [DOI] [PubMed] [Google Scholar]
- Nik-Zainal S. Davies H. Staaf J. Ramakrishna M. Glodzik D. Zou X. et al., Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016;534(7605):47–54. doi: 10.1038/nature17676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D.-Y. Jiang Z. Ben-David Y. Woodgett J. R. Zacksenhaus E. Molecular stratification within triple-negative breast cancer subtypes. Sci. Rep. 2019;9(1):19107. doi: 10.1038/s41598-019-55710-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellmann K. Martel J. Poirier D. J. P. Labrie M. M. Landry J. Downregulation of the PI3K/Akt survival pathway in cells with deregulated expression of c-Myc. Apoptosis. 2006;11(8):1311–1319. doi: 10.1007/s10495-006-8205-1. [DOI] [PubMed] [Google Scholar]
- Neve R. M. Chin K. Fridlyand J. Yeh J. Baehner F. L. Fevr T. et al., A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10(6):515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Guerrab A. Bamdad M. Bignon Y.-J. Penault-Llorca F. Aubel C. Co-targeting EGFR and mTOR with gefitinib and everolimus in triple-negative breast cancer cells. Sci. Rep. 2020;10(1):6367. doi: 10.1038/s41598-020-63310-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigelt B. Warne P. H. Downward J. PIK3CA mutation, but not PTEN loss of function, determines the sensitivity of breast cancer cells to mTOR inhibitory drugs. Oncogene. 2011;30(29):3222–3233. doi: 10.1038/onc.2011.42. [DOI] [PubMed] [Google Scholar]
- Costa R. L. B. Han H. S. Gradishar W. J. Targeting the PI3K/AKT/mTOR pathway in triple-negative breast cancer: a review. Breast Cancer Res. Treat. 2018;169(3):397–406. doi: 10.1007/s10549-018-4697-y. [DOI] [PubMed] [Google Scholar]
- Liu T. Yacoub R. Taliaferro-Smith L. D. Sun S.-Y. Graham T. R. Dolan R. et al., Combinatorial effects of lapatinib and rapamycin in triple-negative breast cancer cells. Mol. Cancer Ther. 2011;10(8):1460–1469. doi: 10.1158/1535-7163.MCT-10-0925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persad A. Venkateswaran G. Hao L. Garcia M. E. Yoon J. Sidhu J. et al., Active β-catenin is regulated by the PTEN/PI3 kinase pathway: a role for protein phosphatase PP2A. Genes Cancer. 2016;7(11–12):368–382. doi: 10.18632/genesandcancer.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. Sekine M. Tung N. Avraham H. K. Wild-type BRCA1, but not mutated BRCA1, regulates the expression of the nuclear form of beta-catenin. Mol. Cancer Res. 2010;8(3):407–420. doi: 10.1158/1541-7786.MCR-09-0403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilir B. Kucuk O. Moreno C. S. Wnt signaling blockage inhibits cell proliferation and migration, and induces apoptosis in triple-negative breast cancer cells. J. Transl. Med. 2013;11:280. doi: 10.1186/1479-5876-11-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y. Helenius M. Väänänen K. Bulanova D. Saarela J. Sokolenko A. et al., BRCA1-deficient breast cancer cell lines are resistant to MEK inhibitors and show distinct sensitivities to 6-thioguanine. Sci. Rep. 2016;6:28217. doi: 10.1038/srep28217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundqvist A. Vasilaki E. Voytyuk O. Bai Y. Morikawa M. Moustakas A. et al., TGFβ and EGF signaling orchestrates the AP-1- and p63 transcriptional regulation of breast cancer invasiveness. Oncogene. 2020;39(22):4436–4449. doi: 10.1038/s41388-020-1299-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul M. K. Mukhopadhyay A. K. Tyrosine kinase – Role and significance in Cancer. Int. J. Med. Sci. 2004;1(2):101–115. doi: 10.7150/ijms.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertucci F. Finetti P. Rougemont J. Charafe-Jauffret E. Cervera N. Tarpin C. et al., Gene expression profiling identifies molecular subtypes of inflammatory breast cancer. Cancer Res. 2005;65(6):2170–2178. doi: 10.1158/0008-5472.CAN-04-4115. [DOI] [PubMed] [Google Scholar]
- Sabatier R. Finetti P. Mamessier E. Raynaud S. Cervera N. Lambaudie E. et al., Kinome expression profiling and prognosis of basal breast cancers. Mol. Cancer. 2011;10:86. doi: 10.1186/1476-4598-10-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miah S. Banks C. A. S. Ogunbolude Y. Bagu E. T. Berg J. M. Saraf A. et al., BRK Phosphorylates SMAD4 for proteasomal degradation and inhibits tumor suppressor FRK to control SNAIL, SLUG and metastatic potential. Sci. Adv. 2019;5(10):eaaw3113. doi: 10.1126/sciadv.aaw3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu L. Levine K. Gajiwala K. S. Cronin C. N. Nagata A. Johnson E. et al., Small molecule inhibitors reveal PTK6 kinase is not an oncogenic driver in breast cancers. PLoS ONE. 2018;13(6):e0198374. doi: 10.1371/journal.pone.0198374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mboge M. Y. Mahon B. P. McKenna R. Frost S. C. Carbonic Anhydrases: Role in pH Control and Cancer. Metabolites. 2018;8(1):19. doi: 10.3390/metabo8010019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.