Abstract
Introduction
The early diagnosis and detection could greatly improve the clinical outcome of gastric cancer (GC) patients. However, the non-invasive biomarkers for GC detection remain to be identified.
Method
We used online databases (GEPIA, UALCAN, Kaplan-Meier plotter, TIMER, and MEXPRESS) to explore the association between H19 or metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) expression in tissues and the occurrence, development, prognosis, the levels of immune cell infiltration, and methylation of GC; the correlation between mRNA expression and DNA methylation levels of genes were also examined. Methylation levels of H19 or MALAT1 in peripheral blood were compared between 150 GC patients and 100 healthy controls (HCs). Predictive nomograms were constructed among female and male groups for GC diagnosis. The calibration curves, Hosmer–Lemeshow test, and decision curve analysis were also used to examine the nomograms’ predictive ability and clinical values.
Results
Using multiple online databases, we found that the mRNA expressions of H19 and MALAT1 in tissues were related to the occurrence of GC, and such expressions were associated with immune cell infiltration of GC and negatively correlated with DNA methylation levels of H19 and MALAT1. H19 gene, H19C island, and MALAT1B island, as well as 20 CpG sites were hypermethylated in peripheral blood of GC patients compared with HCs; similar results were also found in female and male groups (P < .05 for all). The combination of H19c3, H19c4, MALAT1b12, and age, as well as the combination of H19b7, H19c1, H19c5, and age in the nomograms could distinguish GC patients from HCs in the female group and male group, respectively.
Conclusion
We found statistically significant hypermethylation of H19 and MALAT1 promoters in GC patients, and meaningful sensitivity and specificity of MALAT1 and H19 methylation in discriminating GC and HCs were observed in both female and male groups, which indicates that the peripheral blood-based DNA methylation of H19 and MALAT1 could act as potential non-invasive biomarkers for the diagnosis of GC.
Keywords: DNA methylation, peripheral blood, H19, metastasis-associated lung adenocarcinoma transcript 1, gastric cancer
Introduction
Following behind the malignancies of lung, breast, colorectum, and prostate, gastric cancer (GC) remains the fifth most common carcinoma and the third leading disease of tumor-related deaths globally.1,2 The estimated new cases and deaths of GC were 1000 000 and 783 000 in 2018 worldwide. 3 It is well accepted that GC patients with early-stage have a great long-term outcome, with the reported 5-year overall survival (OS) between 84% and 97%.4,5 Endoscopy and biopsy are known as the primary diagnostic method for GC; however, such procedures are invasive and expensive and are challenging to clinicians; few people take these examinations. 6 Therefore, nearly 3 quarters of GC patients are diagnosed at an advanced stage, missing the chance to get the gastrectomy and endoscopic resection treatment, which is limited to patients with early GC. 7 Although there is significant progress in understanding the molecular mechanisms of GC, the precise diagnostic biomarkers for early GC detection remain limited. In the last few decades, epigenetic and genetic alterations had been identified as several significant factors that could induce GC, including DNA methylation, 8 non-coding RNAs (lncRNAs), 9 histone modifications, 10 and microRNAs. 9 Aberrant DNA methylation, up to now, is the most well-studied deregulated molecular mechanism in the carcinogenesis of GC. Nevertheless, most of the studies on methylation related to GC’s molecular markers were based on tissues; few of them focused on samples that can be collected non-invasively, such as blood and urine.10-13
Long lncRNAs are regulatory RNAs that exceed 200 nucleotides (nt) in length and cannot encode proteins. 14 Increasing evidence suggests that the expression of lncRNAs contribute to many types of human cancers,15-17 and the regulation of DNA methylation by lncRNAs plays an important role in controlling gene expression in the process of tumor progression and prognosis. 18 As a highly conserved representative of lncRNA in mammals, metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) is an oncogene that is regulated by DNA methylation and is highly expressed in many carcinomas such as breast, prostate, and GC.19,20 Growing evidence has shown that MALAT1 has the potential to be not only a diagnostic but also a prognostic biomarker in GC.21,22 H19 is a transcript subjected to genomic imprinting and located on chromosome 11p15.5. 23 As an imprinted gene, lncRNAH19 has been widely studied and was found to participate in nearly all stages of tumorigenesis. 24 In GC, upregulated expression of H19 was observed, and higher expression was positively correlated with a worse prognosis of GC. 23 Although the role of MALAT1 and H19 expression in the occurrence and prognosis of GC had been studied, bioinformatic evidence that supported these findings is limited, and the correlation between the expression and methylation of these genes had not been explored. It is also unclear whether the methylation levels of these 2 genes in peripheral blood can be used as non-invasive markers for the early diagnosis of GC. Therefore, we first applied a bioinformatic analysis to explore the role of H19 and MALAT1 expression in the tumorigenesis, progression, and prognosis of GC. As we hypothesize that methylation levels of H19 and MALAT1 in peripheral blood have the potential to be biomarkers for GC detection, we also evaluated the correlation between the methylation and expression levels of these 2 genes in the online database. We performed a case-control study to examine the relationship between carcinogenesis of GC and methylation levels of MALAT1 and H19 in peripheral blood, which are helpful to identify precise biomarkers for early diagnosis of GC.
Methods
Bioinformatic Analysis of H19 and MALAT1
In the present study, bioinformatic analysis was utilized to verify the reported association between MALAT1 and H19 expression and the occurrence, development, and prognosis of GC. Database of GEPIA 25 (Student’s t-test, cutoff P-value: .05) and UALCAN 26 (Student’s t-test, cutoff P-value: .05) was first used to test the expression of H19 and MALAT1, comparing GC tissues and the relevant normal tissues. The associations between MALAT1 or H19 expression and the pathological stages were explored by GEPIA and UALCAN, while the relationship of these 2 genes and the survival status of GC patients was explored by Kaplan–Meier plotter and UALCAN, respectively (since the data of H19 expression and the survival of GC patients were not available on Kaplan–Meier plotter, 27 we thus performed the survival analysis on UALCAN database). As both MALAT1 28 and H19 29 were reported to be involved in immune cell infiltration, the correlations between gene expression and the levels of immune infiltration were also evaluated within the TIMER database. 30 Finally, we utilized the MEXPRESS database 31 to investigate the potential association between the methylation and expression levels of MALAT1 and H19. The methylation level of each probe was represented with a beta value, and the Pearson correlation coefficient value (R) and the adjusted P-value (Benjamini–Hochberg) were also obtained.
Patients and Samples
Among the Chinese Han population, we carried out a case-control study that enrolled 150 GC patients from the First Affiliated Hospital of Anhui Medical University. Patients who were diagnosed by histopathology and had no previous history of malignant tumors in any organs and healthy controls (HC) who were age- and gender-matched with GC patients were recruited to test the DNA methylation levels. Five milliliters of each participant’s peripheral blood samples were obtained, and face-to-face interviews were performed by investigators with a comprehensive questionnaire among GC patients only. Clinical and demographic information such as age, gender, tumor site, TNM stage, tumor differentiation, smoking, drinking, family history, and the dynamic changes of the psychological situation during the treatment are contained in this questionnaire and were described previously. All procedures conducted by this study were approved by Anhui Medical University’s ethical community and abide by the 1964 Helsinki Declaration and the later amendments. All patients had signed written informed consent.
DNA Methylation Detection
The methods of DNA extraction and bisulfite conversion are described at length.32,33 Briefly, we used the QIAGEN kit to extract the genomic DNA from peripheral blood leukocytes of GC and HCs following the manufacturer’s protocols. According to previously reported criteria for CpG islands (CGIs) and CpG island shores (CGI shores) selection, 34 we finally selected for sequencing 60 CpG methylation sites on 3 CGIs (H19A, H19B, H19C) and 1 CGI shore (H19D) from H19 promoter, as well as 31 CpG methylation sites on 2 CGIs (MALAT1A, MALAT1B) and 1 CGI shore (MALAT1C) of MALAT1 promoter (the position and the primer sequences of the CGIs, CGI shores, and CpG sites are shown in Supplementary Tables S1 and S2).
After DNA extraction and CpG site selection, 400 ng genomic DNA was bisulfite-converted using the EZ DNA Methylation™-GOLD kit (Zymo Research) for methylation analysis. Utilizing indexed primers, we amplified the bisulfite-modified DNA sequence with multiplex PCR. We then utilized agarose electrophoresis to separate these PCR amplicons and purified them with a QIAquick Gel Extraction kit (QIAGEN). The methylation levels of H19 and MALAT1 were analyzed using an NGS-based multiple-targeted CpG methylation analysis method manufactured by MethylTarget™ (Genesky Biotechnologies Inc, Shanghai, China), and the methylation detection was conducted on an Illumina Hiseq/Miseq 2000 following the manufacturer’s instructions.
Statistical Analysis
The skewed data in the current study were presented as median with interquartile range (IQR), while the normally distributed data were depicted as mean ± standard deviation (SD). Gender and age differences between GC and HCs were compared by Student’s t-test, and the Mann–Whitney U test was used to explore the differences of methylation levels between the 2 categories. Binary and Multinomial regression analyses were carried out to identify the associations of methylation levels between GC and HCs and between various clinicopathological variables of GC. The OR (odds ratios) and 95% CI (confidence intervals) were then calculated. All statistical analysis was conducted using SPSS 24.0 and R software, and Graphpad Prism 8.0 was also used to generate the figures.
Diagnostic Nomogram for GC
Among GC and HCs in female and male groups, a receiver operating curve (ROC) was implemented to evaluate the predictive value of CpG sites of H19 and MALAT1 promoters as a biomarker for the diagnosis of GC. The area under curve (AUC) was calculated. The first 3 CpG sites with the highest AUC values among female and male groups were combined with age to devise GC’s diagnostic nomogram. We used the calibration curves and the Hosmer–Lemeshow test (P > .05) to examine the calibration ability of the nomograms. The discriminative ability of the nomograms was evaluated by ROC. 35 We also carried out a decision curve analysis (DCA) to evaluate the nomograms’ clinical values using the R library rmda package; the method for calculating the net benefit of the nomograms was described in detail elsewhere. 36
Results
Bioinformatic Analysis of MALAT1 and H19 in GC
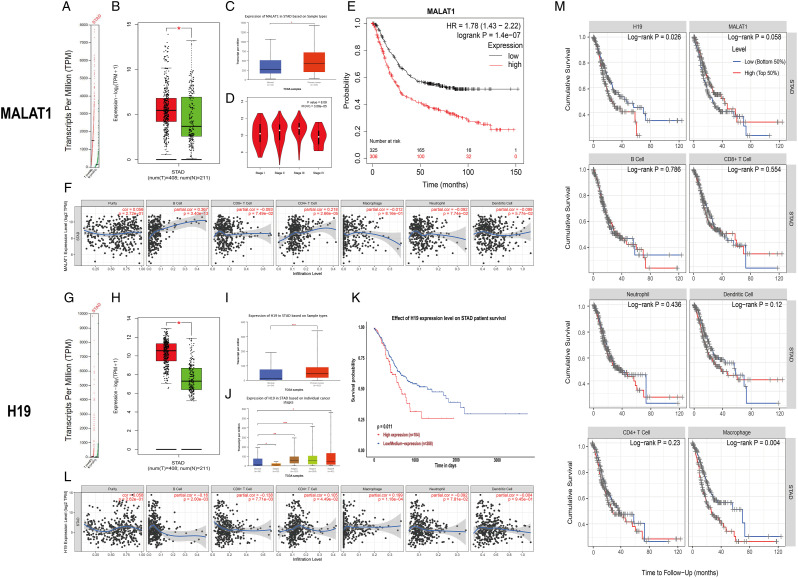
To evaluate the potential diagnostic and prognostic value of MALAT1 and H19 in GC, we assessed the expression levels of these 2 genes in GC tissues and normal tissues in GEPIA and UALCAN. As expected, the transcriptional levels of MALAT1 and H19 in GC tissues were significantly elevated (Figures 1A-1C and 1G-1IA-CG-IFigures 1A-1C and 1G-1IFigures 1A-1C and 1G-1IFigures 1A-1C and 1G-1I). Using GEPIA and UALCAN, we then explored the associations between the expression of variously expressed MALAT1 or H19 and the pathological stages of GC; significant correlations were observed in the databases. In general, GC tumors of more advanced stages expressed higher levels of MALAT1 and H19 (Figures 1D and 1J). The prognostic values of the expression of MALAT1 and H19 in GC patients were also explored. As shown in Figures 1E and 1K, higher expressions of MALAT1 and H19 were significantly correlated with a shorter survival probability of GC (Kaplan–Meier plotter, P = 1.4 × 10−7; UALCAN database, P = .011).
Figure 1.
Bioinformatic analysis of metastasis-associated lung adenocarcinoma transcript 1 and H19 in GC. (A-C, G-I) The expression of metastasis-associated lung adenocarcinoma transcript 1 and H19 in GC tissues and adjacent normal tissues: A and G, scatter diagram (GEPIA); B and H, box plot (GEPIA); C and I, box plot (UALCN). (D, J) Correlation between metastasis-associated lung adenocarcinoma transcript 1 and H19 expression and tumor stages in GC patients (D, GEPIA; J, UALCA). (E, K) Prognostic value of metastasis-associated lung adenocarcinoma transcript 1 and H19 expression in GC (E, Kaplan-Meier plotter; K, UALCA). (F, L) Correlation between metastasis-associated lung adenocarcinoma transcript 1 and H19 expression and immune cell infiltration of GC (TIMER). (M) Survival differences for immune infiltrates and the expression of H19 and MALAT1 (TIMER). GC: gastric cancer.
As for the correlation between the immune cell infiltration and the expressions of MALAT1 and H19, we found the expression of MALAT1 was positively correlated with the infiltration of B cells (Cor = .367, P = 3.40 × 10−13) and CD4+ T cells (Cor = .218, P = 2.66 × 10−5) (Figure 1F), while the expression of H19 and the infiltration of B cells (Cor = −.16, P = 2.00 × 10−3) and CD8+ T cells (Cor = −.138, P = 7.71 × 10−3) (Figure 1F) were negatively correlated (Figure 1L). Interestingly, positively correlations between the expression of H19 and the infiltration of CD4+ T cells (Cor = .105, P = 4.49 × 10−2) and macrophages (Cor = .199, P = 1.18 × 10−4) were also observed (Figure 1L). In addition, using the TIMER database, we also drew Kaplan–Meier plots for immune infiltrates and the expression of H19 and MALAT1 to visualize the survival differences, which are shown in Figure 1M. We observed that H19 (P = .026) expression and macrophage (P = .04) were significantly correlated with the clinical outcome of GC patients.
With respect to the correlation between methylation and expression levels of H19 and MALAT1, we observed negative statistical correlations between MALAT1 or H19 gene expression and DNA methylation at numerous probes, such as cg26489875 (R = − .407, P < .001) in MALAT1 and cg011716026 in H19 (R = −.427, P < .001) (Figure 2).
Figure 2.
Correlation between metastasis-associated lung adenocarcinoma transcript 1 and H19 DNA methylation and gene expression in MEXPRESS.
Characteristics of Participants in the Study
The demographic information (gender, age) of 150 GC patients and 100 controls and the clinical and pathological characteristics of GC patients were presented in our previous study. 37 No significant difference was detected in gender and age between GC patients and HCs (P = .592, P = .413, respectively). We redivided the GC patients according to the various metastasis status. Among 150 patients, lymph node or visceral metastasis was found in 68 patients (45.33%), 53 patients did not metastasize (35.33%), and 29 patients (19.33%) had both lymph node and visceral metastasis. Body mass index (BMI) and HER-2 status were also included in the current study. Underweight to normal patients (BMI < 25) accounted for 74% of all patients, and only 39 patients had tested their HER-2 status (positive: 9 cases, negative: 30 cases).
Association Between Methylation Levels of H19, MALAT1 Promoters in Peripheral Blood, and GC Risk Among all Participants
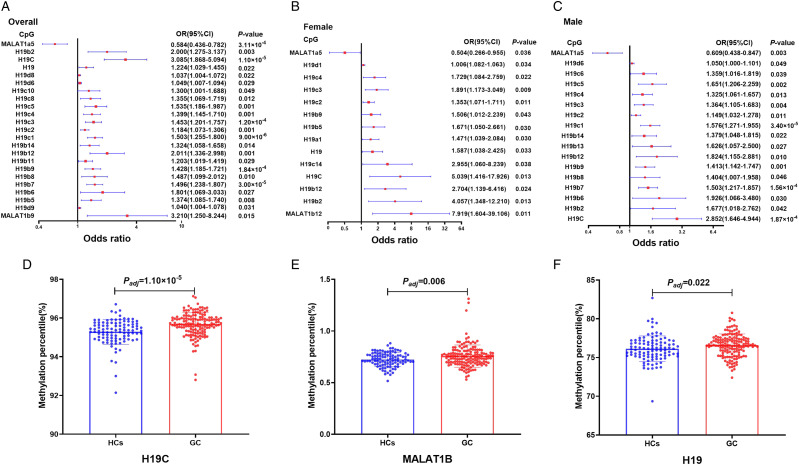
A total of 91 CpG sites of H19 and MALAT1 promoters were detected for the differential methylation analysis. Differences in the methylation levels of H19 and MALAT1 promoters between GC and HCs are shown in Supplementary Table S3. The gene/CpG regions/CpG sites with statistical significance (all P < .05) are shown in Table 1. Among all the CpG sites, 20 CpG sites in the H19B island, H19C island, H19D island shore, and MALAT1B island were hypermethylated in GC patients compared with HCs (OR > 1, P < .05 for all), while 1 CpG site (MALAT1a5) in MALAT1A island was hypermethylated in HCs conversely (OR = .584, P = 3.11 × 10−4) (Figure 3A, Table 1). To further explore the methylation situations of H19 and MALAT1 in GC patients, the methylation levels of the CpG region (CGI or CGI shore) were calculated by averaging the methylation levels of all nearby CpG sites of H19 and MALAT1 promoters (since the range of CI of MALAT1B island was too broad, we did not include this site in Figure 3A. The CI values of MALAT1B island can be found in Table 1). Similarly, the hypermethylation of H19 C island and MALAT1B island was observed in the GC group (Mann–Whitney U test: P = 1.10 × 10−5, P = .006, respectively) (Figures 3D and 3E, Table 1). Using the same way, we calculated the overall methylation level of H19 and MALAT1 genes by averaging the methylation levels of all nearby CpG regions of H19 and MALAT1, and only hypermethylation of the H19 gene in the GC group was observed (Mann–Whitney U test: P = .022) (Figure 3F, Table 1).
Table 1.
Association between methylation levels of H19, MALAT1 promoters in peripheral blood and GC risk* (P < .05).
| Name | Methylation level a | Logistic regression analysis | ||||
|---|---|---|---|---|---|---|
| GC | HCs | Odds ratio | 95%CI | P-value | ||
| Genes | H19 | 76.61 (75.71,77.44) | 76.10 (75.22,76.98) | 1.224 | 1.029–1.455 | .022 |
| Regions | H19C | 95.72 (95.38,96.03) | 95.36 (94.95,95.72) | 3.085 | 1.868–5.094 | 1.10 × 10 −5 |
| MALAT1B | .75 (.69,0.80) | .72 (.68,0.77) | 94.912 | 3.629–2482.615 | .006 | |
| H19b2 | 97.41 (96.99,97.85) | 97.20 (96.85,97.58) | 2.000 | 1.275–3.137 | .003 | |
| H19b5 | 93.68 (92.74,94.30) | 93.36 (92.42,94.03) | 1.374 | 1.085–1.740 | .008 | |
| H19b6 | 97.97 (97.62,98.32) | 97.87 (97.49,98.09) | 1.801 | 1.069–3.033 | .027 | |
| H19b7 | 92.45 (91.29,93.30) | 91.77 (90.48,92.49) | 1.496 | 1.238–1.807 | 3.00 × 10 −5 | |
| H19b8 | 96.22 (95.75,96.67) | 95.86 (95.35,96.42) | 1.487 | 1.099–2.012 | .010 | |
| H19b9 | 94.60 (93.77,95.54) | 94.18 (92.82,95.04) | 1.428 | 1.185–1.721 | 1.84 × 10 −4 | |
| H19b11 | 90.66 (89.71,91.52) | 90.14 (89.28,91.24) | 1.203 | 1.019–1.419 | .029 | |
| Sites | H19b12 | 97.13 (96.61,97.48) | 96.74 (96.36,97.15) | 2.001 | 1.336–2.998 | .001 |
| H19b14 | 95.30 (94.67,95.91) | 94.93 (94.35,95.46) | 1.324 | 1.058–1.658 | .014 | |
| H19c1 | 93.74 (92.81,94.67) | 92.68 (91.9,93.68) | 1.503 | 1.255–1.800 | 9.00 × 10 −6 | |
| H19c2 | 86.69 (84.89,88.78) | 85.27 (83.62,87.49) | 1.184 | 1.073–1.306 | .001 | |
| H19c3 | 95.85 (94.94,96.54) | 94.96 (93.97,95.9) | 1.453 | 1.201–1.757 | 1.20 × 10 −4 | |
| H19c4 | 95.63 (94.78,96.43) | 94.95 (94.02,95.69) | 1.399 | 1.145–1.710 | .001 | |
| H19c5 | 96.34 (95.55,96.94) | 95.81 (95.18,96.36) | 1.535 | 1.186–1.987 | .001 | |
| H19c8 | 96.99 (96.50,97.67) | 96.57 (96.14,97.05) | 1.355 | 1.069–1.719 | .012 | |
| H19c10 | 96.80 (96.22,97.44) | 96.61 (96.04,97.09) | 1.300 | 1.001–1.688 | .049 | |
| H19d6 | 82.48 (78.51,87.10) | 80.37 (76.76,84.83) | 1.049 | 1.007–1.094 | .022 | |
| H19d8 | 70.66 (65.50,76.87) | 68.63 (63.94,73.20) | 1.037 | 1.004–1.072 | .029 | |
| H19d9 | 76.82 (71.17,82.47) | 74.51 (70.21,79.43) | 1.040 | 1.004–1.078 | .031 | |
| MALAT1a5 | 4.94 (4.46,5.51) | 5.45 (4.82,6.06) | .584 | .436–.782 | 3.11 × 10 −4 | |
| MALAT1b9 | .76 (.54,0.92) | .66 (.52,0.82) | 3.210 | 1.250–8.244 | .015 | |
*Adjusted for age and gender.
aMethylation level is expressed as a percentage. Data was expressed as median (P25, P75). GC: gastric cancer, HCs: health controls, MALAT1: metastasis-associated lung adenocarcinoma transcript 1.
Figure 3.
Correlation analysis of H19 and metastasis-associated lung adenocarcinoma transcript 1 methylation of peripheral blood between GC patients and HCs. (A-C) Methylation of H19 and metastasis-associated lung adenocarcinoma transcript 1 (including gene, CpG regions, and CpG sites) in the whole group of participants, in the female group, and in the male group. The squares and horizontal lines correspond to the odds ratio (OR) and 95% confidence interval (CI), respectively. (D-F) Comparisons of the methylation levels of H19 gene, H19C island, and MALAT1B island between GC patients and HCs. GC: gastric cancer, HCs: healthy controls.
Association Between Methylation Levels of H19, MALAT1 Promoters in Peripheral Blood, and GC Risk in Female and Male Groups
Gender variations in the occurrence and mortality of GC had been reported, with larger, higher stage, and worse OS of GC occurring in the male group. 38 We thus performed subgroup analysis to evaluate the differences of H19 or MALAT1 methylation levels between GC and HCs in female and male groups. In the female group, 1 CpG site in the H19A island, 4 CpG sites in the H19B island, 4 CpG sites in the H19C island, 1 CpG site in the H19D island shore, and 1 CpG site in the MALAT1B island were hypermethylated in GC patients compared with that in HCs (OR > 1, P < .05 for all) (Figure 3B, Supplementary Table S4). Interestingly, hypermethylation of MALAT1a5 was also observed in the HCs group (OR = .504, P = .036). In the male group, similar results were observed compared with those in the female group. Hypermethylation of 8 CpG sites in the H19B island, 6 CpG sites in the H19C island, 1 CpG site in the H19D island shore, and 1 CpG site in the MALAT1B island were found in GC (OR > 1, P < .05 for all), while MALAT1a5 in the MALAT1A island was hypomethylated (OR = .609, P = .003) (Figure 3C, Supplementary Table S5). Identically, overall methylation levels of H19 and MALAT1 CpG regions and genes were calculated. We found hypermethylation of H19C island, MALAT1B island, and H19 gene in GC patients in the female group. While in the male group, such hypermethylation was observed in the H19C island (OR > 1, P < .05 for all) (since the range of CI of MALAT1B island and MALAT1b5 cite was too wide, we did not include them in Figure 3. The CI values of MALAT1B island and MALAT1b5 site can be found in Supplementary Tables S4 and S5) (Figures 3B and 3C, Supplementary Tables S4 and S5).
Differences of Methylation Levels of H19, MALAT1 Promoters in Peripheral Blood Between GC Patients With Different Clinical Characteristics
To explore the association between H19 or MALAT1 methylation in GC with different clinical characteristics, GC patients were divided into the following groups: poorly differentiated group and well/moderately differentiated group; stage I–III group and stage IV group; patients without metastasis and patients with metastasis (including patients with lymph node or visceral metastases and patients with both lymph node and visceral metastases); underweight to normal group (BMI < 25) and overweight/obese group (BMI ≥ 25); upper third (tumor site) group, middle third group, lower third group, and diffused type group; patients with age ≤ mean group and patients with age > mean group; patients with HER-2 negative group and positive group (patients who had not tested their HER-2 status were not included). Significant differences were observed for the methylation levels of H19 and MALAT1 between patients with various differentiations, TNM stages, with and without metastasis, BMI levels, tumor sites, and age groups (Supplementary Tables S6-11), while no significant difference was observed for the methylation levels of H19 and MALAT1 between patients with HER-2 negative and positive (Supplementary Table S12).
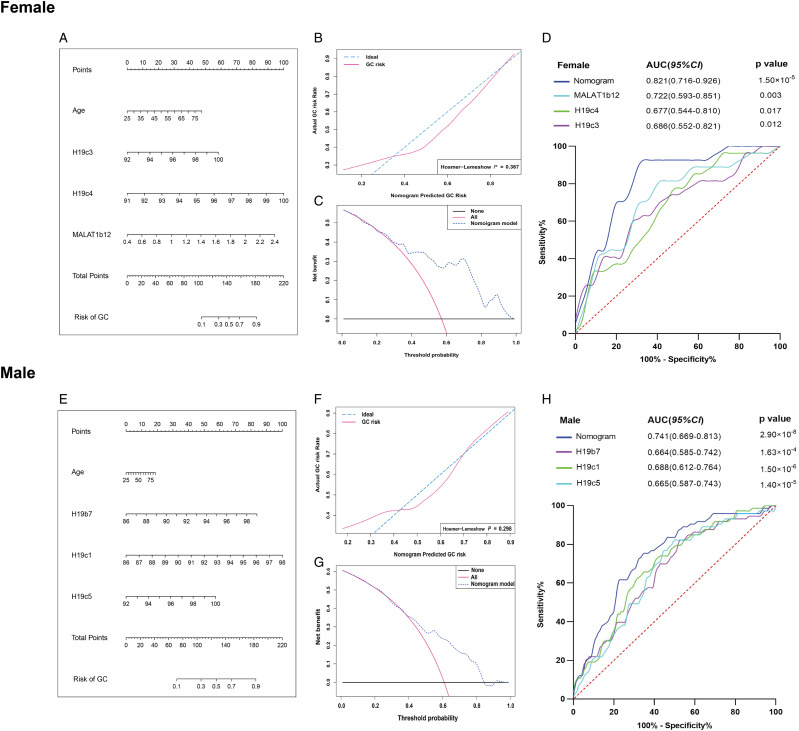
Diagnostic Nomograms and Model Performance of GC in Female and Male Groups
To examine the potential diagnostic value of H19 and MALAT1 methylation as non-invasive biomarkers for GC, ROC curve analysis was performed. Taking age and the first 3 CpG sites with the highest AUC values among the female (H19c3, H19c4, MALAT1b12) and male groups (H19b1, H19c1, H19c5) into consideration, we generated 2 diagnostic nomograms (Figures 4A and 4E). Based on the sum of the nomograms, the higher the total points, the higher the GC risk. The calibration curve of the nomograms displayed accurate predictive ability for the diagnosis of GC both in the female and male groups, and no significant differences exist between the predictive GC risk calibration curve and the ideal curve for GC from the Hosmer–Lemeshow test (P = .367, P = .298, respectively) (Figures 4B and 4F). The clinical values of the nomograms were evaluated by DCA; the results demonstrated wide ranges of the threshold and probabilities of the nomograms in the diagnosis of GC in both female (Figure 4C) and male (Figure 4G) groups. The AUC of the nomogram was .821 (95% CI: .716 - .926, P = 1.50 × 10−5), with the sensitivity and specificity as 69.4% and 92.6% in the female group and .741 (95% CI: .669 - .813, P = 2.90 × 10−8), with the sensitivity and specificity as 65.8% and 75.3% in the male group, respectively (Figures 4D and 4H, Supplementary Table S13). We found the nomograms generated in the female and male groups had higher AUC, indicating more potential in GC diagnosis than that of each CpG site alone. The AUC of MALAT1b12, H19c4, H19c3 in the female group were .722, .677, .686, respectively, while AUC of H19b7, H19c1, H19c5 in the male group were .664, .688, .665, respectively (Figures 4D and 4H).
Figure 4.
The logistic regression-based nomogram model for predicting the risk of GC in the female and male groups. The methylation levels of H19c3, H19c4, MALAT1b12, and age were used to construct the nomogram for predicting the risk of GC in female group (A-D), while the methylation levels of H19b7, H19c1, and H19c5 were used to construct the nomogram for predicting the risk of GC in male group (E-H). (A, D) The construction of the nomogram. The value of each variable was given a score on the point scale axis. A total score could be calculated by adding each single score, and by projecting the total score to the lower total point scale, we were able to estimate the probability of GC in female and male groups. (B, E) The calibration curve and the Hosmer–Lemeshow (H-L) test of the nomogram. The calibration curve with 1000 bootstrap resampling was made for an assessment of the nomogram-predicted probabilities of GC in female and male groups. The H-L test was used to examine how well the percentage of the actual probability of the GC matched the percentage of the nomogram-predicted probability of GC over deciles of predicted risk in the female and male groups. (C, F) Decision curve analysis of the nomogram. The decision curve analysis was performed to compare the standardized net benefit of the nomogram at the different threshold probabilities. The ranges of the standardized net benefit of the nomogram were very wide and practical. (D, H) Receiver operating characteristic curve. The area under receiver (AUC) operating characteristic curve values for the constructed nomogram of GC in female and male groups was .821 and .741, respectively. GC: gastric cancer.
Discussion
Previous studies have shown that aberrant DNA methylation plays an important role in the occurrence and development of GC; however, most of these studies conducted their research on tumor tissues.10-13 Non-invasive diagnostic biomarkers for GC are urgently needed. In the current study, we found that the expressions of MALAT1 and H19 were associated with the tumorigenic and prognostic of GC and may also regulate the immune cell infiltration in GC. Moreover, H19 and MALAT1 expression were found significantly negatively correlated with their methylation levels, which indicates a significant role of the DNA methylation of H19 and MALAT1 in GC’s occurrence and development. Our case-control study results show significant differences in the peripheral blood-based DNA methylation levels of MALAT1 and H19 between GC patients and HCs, and such methylation is also associated with the characteristics of GC. Meaningful sensitivity and specificity of MALAT1 and H19 methylation in discriminating GC and HCs were observed in both female and male groups.
The alteration of DNA methylation may activate oncogenes, suppress suppressor genes, and induce chromosome instabilities, which initiates tumorigenesis and stimulates cancer development. 39 During cancer progression, an important mechanism for controlling gene expression is regulating DNA methylation by lncRNAs. 18 Accumulating evidence has shown that DNA methylation in peripheral blood can be used as biomarkers for early diagnosis of several tumor types, such as malignant pleural mesothelioma, breast cancer, and GC.40-42
Abnormal expression of the MALAT1 oncogene was first reported in metastatic non-small cell lung carcinoma. 19 Higher MALAT1 expression was also observed in liver, breast, pancreatic, colorectal, and prostate cancer.43-45 Consistent with our results, a similar higher expression of MALAT1 in gastric carcinoma tissues and cell lines was previously reported. 18 Downregulated expression of UPF1, negatively correlated with MALAT1, was found in GC, and the downregulation may be due to the hypermethylation in the promoter region. 46 Like MALAT1, aberrant expression of H19 was detected in various mammalian tumors,47,48 and the upregulated expression of H19 in GC is positively correlated with the worse clinical outcome of GC patients. 23 As an imprinted gene, H19 exhibits a dynamic expression pattern during human embryonic development, and the methylation levels of the imprinted control regions (ICRs) of the H19 promoter are considered the main regulator of its expression during mammalian development.49,50 Therefore, it is reasonable to speculate that the higher methylation of MALAT1 and H19 promoters in GC and the associations of their methylation with GC’s clinical characteristics (eg, tumor differentiation, TNM stages) may associate with their aberrant expressions. In the current study, we found the H19 gene, H19 C island, and MALAT1B island, as well as 20 CpG sites in the H19B island, H19C island, H19D island shore, and MALAT1B island were hypermethylated in GC patients compared with HCs; similar results were also found in the female and the male groups. Moreover, evidence from nomogram analysis suggests that combined detection of multiple CpG sites of MALAT1 and H19 genes in peripheral blood are helpful for diagnosing GC. Particularly in the female group, the combination of H19c3, H19c4, MALAT1b12, and age could be used to predict GC’s occurrence, with high sensitivity and specificity, which suggests the peripheral blood-based DNA methylation of H19 and MALAT1 have the potential to serve as non-invasive biomarkers for GC diagnosis.
Previous studies reported that one of the crucial determinant factors that influence immunotherapy and clinical prognosis of cancer is immune cell infiltration, which is associated with tumor recurrence and progression. 51 In the current study, we observed significant correlations between H19 or MALAT1 expression and macrophages, B cells, CD8+ T cells, and CD4+ T cells infiltration, which implies that the expression of H19 and MALAT1 may also affect the immune status in GC. As one of the crucial forms of epigenetic modifications, DNA methylation is tightly involved in regulating the development, function, and differentiation of the immune system. 52 Recently, a divergent application of DNA methylation between immune cells was detected by investigators. 53 Hence, we hypothesize that the methylation of H19 and MALAT1 promoter in peripheral blood may be associated not only with H19 and MALAT1 expression but also with immune cell infiltration of GC. However, further experiments in vivo and in vitro are needed to clarify the correlation between the methylation and expression of H19 and MALAT1 and GC’s immune cell infiltration.
Strengths and Limitations
This is the first study utilizing the quantitative method to detect the methylation levels of H19 and MALAT1 in peripheral blood, which covered all CpG sites of all CpG regions, and the diagnostic value of each CpG site and regions could thereby be evaluated. However, there are several potential limitations needed to be recognized. First, we did not detect the expressions of H19 and MALAT1 in the involved participants, and the immune status of the GC patients was not examined. Second, we did not perform sample size estimations, despite the great potential of H19 and MALAT1 DNA methylation in peripheral blood to be non-invasive biomarkers for GC diagnosis; further larger-multicenter systematic, unbiased prospective studies are required to validate our observed results and develop a novel feasible peripheral blood-based assay for clinical application.
Conclusion
In the current study, we found a hyperexpression of H19 and MALAT1 in GC tissues, and such expression is correlated with DNA methylation and immune cell infiltration of GC. Moreover, we discovered that H19 and MALAT1 are significantly hypermethylated in the peripheral blood of GC patients, and the combination of multiple CpG sites of H19 and MALAT1 promoters could be used to distinguish GC patients from HCs, especially in the female group. This suggests that the peripheral blood-based DNA methylation of H19 and MALAT1 could act as potential non-invasive biomarkers for GC diagnosis.
Supplemental Material
Supplemental Material, sj-pdf-1-ccx-10.1177_10732748211043667 for Peripheral Blood-Based DNA Methylation of Long Non-Coding RNA H19 and Metastasis-Associated Lung Adenocarcinoma Transcript 1 Promoters are Potential Non-Invasive Biomarkers for Gastric Cancer Detection by Dingtao Hu, Xiaoqi Lou, Nana Meng, Zhen Li, Ying Teng, Yanfeng Zou and Fang Wang in Cancer Control
Acknowledgments
We thank all the people who offer help for this study.
Author Contributions: Fang Wang conceived the study idea. Dingtao Hu collected the data. Xiaoqi Lou, Nana Meng, Zhen Li, Ying Teng, Yanfeng Zou, and Fang Wang contributed to the analysis of the data. Dingtao Hu, and Xiaoqi Lou wrote the initial draft with all authors providing critical feedback and edits to subsequent revisions. All authors approved the final draft of the article. All authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Fang Wang is the guarantor. The corresponding author attests that all listed authors meet authorship criteria and that no others meeting the criteria have been omitted.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the grant from the National Natural Science Foundation of China (grant numbers 81602115), the Foundation of Supporting Program for the Excellent Young Faculties in University of Anhui Province in China (gxyq2019012), grants for Scientific Research of BSKY from the First Affiliated Hospital of Anhui Medical University and grants for Outstanding Youth from the First Affiliated Hospital of Anhui Medical University.
Ethical Statement: Our study was approved by The Ethics Committee of Anhui Medical University (approval no. PJ2019-01-12). All patients provided written informed consent prior to enrollment in the study.
Supplemental Material: Supplemental material for this article is available online.
ORCID iD
Fang Wang https://orcid.org/0000-0003-0871-780X
References
- 1.Yang L, Zheng R, Zheng R, et al. Incidence and mortality of stomach cancer in China, 2014, Chin J Canc Res. 2018;30(3):291-298. doi: 10.21147/j.issn.1000-9604.2018.03.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thrift AP, El-Serag HB. Burden of gastric cancer. Clin Gastroenterol Hepatol. 2020;18(3):534-542. Epub 2019 Jul 27; doi: 10.1016/j.cgh.2019.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: Cancer J for Clin. 2018;68(6):394-424. Epub 2018 Sep 12. Erratum in: CA Cancer. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 4.Isomoto H, Shikuwa S, Yamaguchi N, et al. Endoscopic submucosal dissection for early gastric cancer: a large-scale feasibility study. Gut. 2009;58(3):331-336. doi: 10.1136/gut.2008.165381. Epub 2008 Nov 10. [DOI] [PubMed] [Google Scholar]
- 5.Gotoda T, Iwasaki M, Kusano C, Seewald S, Oda I. Endoscopic resection of early gastric cancer treated by guideline and expanded National Cancer Centre criteria. Br J Surg. 2010;97(6):868-871. doi: 10.1002/bjs.7033. [DOI] [PubMed] [Google Scholar]
- 6.Wu D, Zhang P, Ma J, et al. Serum biomarker panels for the diagnosis of gastric cancer. Cancer Med. 2019;8(4):1576-1583. doi: 10.1002/cam4.2055. Epub 2019 Mar 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Digklia A, Wagner AD. Advanced gastric cancer: current treatment landscape and future perspectives. World J Gastroenterol. 2016;22(8):2403-2414. doi: 10.3748/wjg.v22.i8.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ebrahimi V, Soleimanian A, Ebrahimi T, et al. Epigenetic modifications in gastric cancer: focus on DNA methylation. Gene. 2020;742:144577. Epub 2020 Mar 18. doi: 10.1016/j.gene.2020.144577. [DOI] [PubMed] [Google Scholar]
- 9.Li D, Wang J, Zhang M, et al. LncRNA MAGI2-AS3 is regulated by BRD4 and promotes gastric cancer progression via maintaining ZEB1 overexpression by sponging miR-141/200a. Mol Ther Nucleic Acids. 2020;19:109-123. 10.1016/j.omtn.2019.11.003. Epub 2019 Nov 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calcagno DQ, Gigek CO, Chen ES, Burbano RR, Smith Mde A, DNA and histone methylation in gastric carcinogenesis. World J Gastroenterol. 2013;19(8):1182-1192. doi: 10.3748/wjg.v19.i8.1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deng P, Chang X-J, Gao Z-M, et al. Downregulation and DNA methylation of ECRG4 in gastric cancer. OncoTargets Ther. 2018;11:4019-4028. doi: 10.2147/OTT.S161200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suzuki H, Tokino T, Shinomura Y, Imai K, Toyota M. DNA methylation and cancer pathways in gastrointestinal tumors. Pharmacogenomics. 2008;9(12):1917-1928. doi: 10.2217/14622416.9.12.1917. [DOI] [PubMed] [Google Scholar]
- 13.Enomoto S, Maekita T, Tsukamoto T, et al. Lack of association between CpG island methylator phenotype in human gastric cancers and methylation in their background non-cancerous gastric mucosae. Canc Sci. 2007;98(12):1853-1861. Epub 2007 Sep 26. doi: 10.1111/j.1349-7006.2007.00625.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu X, Tian X, Yu C, et al. A long non-coding RNA signature to improve prognosis prediction of gastric cancer. Mol Canc. 2016;15(3):60. doi: 10.1186/s12943-016-0544-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yuan J-h., Yang F, Wang F, et al. A long noncoding RNA activated by TGF-β promotes the invasion-metastasis cascade in hepatocellular carcinoma. Canc Cell. 2014;25(5):666-681. doi: 10.1016/j.ccr.2014.03.010. Epub 2014 Apr 24. [DOI] [PubMed] [Google Scholar]
- 16.Zhang X, Rice K, Wang Y, et al. Maternally expressed gene 3 (MEG3) noncoding ribonucleic acid: Isoform structure, expression, and functions. Endocrinology. 2010;151(3):939-947. doi: 10.1210/en.2009-0657. Epub 2009 Dec 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Z, Jin Y, Ren H, Ma X, Wang B, Wang Y. Downregulation of the long non-coding RNA TUSC7 promotes NSCLC cell proliferation and correlates with poor prognosis. Am J Tourism Res. 2016;8(2):680-687. [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou Z, Lin Z, Pang X, et al. Epigenetic regulation of long non-coding RNAs in gastric cancer. Oncotarget. 2017:9(27):19443-19458. doi: 10.18632/oncotarget.23821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo F, Guo L, Li Y, Zhou Q, Li Z. MALAT1 is an oncogenic long non-coding RNA associated with tumor invasion in non-small cell lung cancer regulated by DNA methylation. Int J Clin Exp Pathol. 2015;8(12):15903-15910. [PMC free article] [PubMed] [Google Scholar]
- 20.Ji P, Diederichs S, Wang W, Böing S, et al. MALAT-1, a novel noncoding RNA, and thymosin β4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22(39):8031-8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 21.Wang J, Su L, Chen X, et al. MALAT1 promotes cell proliferation in gastric cancer by recruiting SF2/ASF. Biomed Pharmacother. 2014;68(5):557-564. doi: 10.1016/j.biopha.2014.04.007. Epub 2014 Apr 28. [DOI] [PubMed] [Google Scholar]
- 22.Li L, Geng Y, Feng R, et al. The human RNA surveillance factor UPF1 modulates gastric cancer progression by targeting long non-coding RNA MALAT1. Cell Physiol Biochem. 2017;42(6):2194-2206. doi: 10.1159/000479994. Epub 2017 Aug 15. [DOI] [PubMed] [Google Scholar]
- 23.Zhang L, Zhou Y, Huang T, et al. The interplay of LncRNA-H19 and its binding partners in physiological process and gastric carcinogenesis. Int J Mol Sci. 2017;18(2):450. doi: 10.3390/ijms18020450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raveh E, Matouk IJ, Gilon M, Hochberg A. The H19 long non-coding RNA in cancer initiation, progression and metastasis - a proposed unifying theory. Mol Canc. 2015;14:184. doi: 10.1186/s12943-015-0458-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98-W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19(8):649-658. doi: 10.1016/j.neo.2017.05.002. Epub 2017 Jul 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Szász AM, Lánczky A, Nagy Á, et al. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget. 2016;7(31):49322-49333. doi: 10.18632/oncotarget.10337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Q-M, Lian G-Y, Song Y, Huang Y-F, Gong Y. LncRNA MALAT1 promotes tumorigenesis and immune escape of diffuse large B cell lymphoma by sponging miR-195. Life Sci. 2019;231:116335. doi: 10.1016/j.lfs.2019.03.040. Epub 2019 Mar 18. [DOI] [PubMed] [Google Scholar]
- 29.Li X, Wang H, Zhang Y, et al. Overexpression of lncRNA H19 changes basic characteristics and affects immune response of bovine mammary epithelial cells. PeerJ. 2019;7:e6715. doi: 10.7717/peerj.6715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li T, Fan J, Wang B, et al. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Canc Res. 2017;77(21):e108-e110. doi: 10.1158/0008-5472.CAN-17-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koch A, Jeschke J, Van Criekinge W, van Engeland M, De Meyer T. MEXPRESS update 2019. Nucleic Acids Res. 2019;47(W1):W561-W565. doi: 10.1093/nar/gkz445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo S, Yan F, Xu J, et al. Identification and validation of the methylation biomarkers of non-small cell lung cancer (NSCLC). Clin Epigenet. 2015;7(1):3. doi: 10.1186/s13148-014-0035-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao Y, Guo S, Sun J, et al. Methylcap-seq reveals novel DNA methylation markers for the diagnosis and recurrence prediction of bladder cancer in a Chinese population. PloS One. 2012;7(4):e35175. doi: 10.1371/journal.pone.0035175. Epub 2012 Apr 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang X, Lu J, Pan Z, et al. DNA methylation and transcriptome signature of the IL12B gene in ankylosing spondylitis. Int Immunopharm. 2019;71:109-114. doi: 10.1016/j.intimp.2019.03.026. Epub 2019 Mar 16. [DOI] [PubMed] [Google Scholar]
- 35.Iasonos A, Schrag D, Raj GV, Panageas KS. How to build and interpret a nomogram for cancer prognosis. J Clin Oncol. 2008;26(8):1364-1370. doi: 10.1200/JCO.2007.12.9791. [DOI] [PubMed] [Google Scholar]
- 36.Vickers AJ, Cronin AM, Elkin EB, Gonen M. Extensions to decision curve analysis, a novel method for evaluating diagnostic tests, prediction models and molecular markers. BMC Med Inf Decis Making. 2008;8:53. doi: 10.1186/1472-6947-8-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lou X, Hu D, Li Z, et al. Associations of BNIP3 and DAPK1 gene polymorphisms with disease susceptibility, clinicopathologic features, anxiety, and depression in gastric cancer patients. Int J Clin Exp Pathol. 2021;14(5):633-645. In Press. [PMC free article] [PubMed] [Google Scholar]
- 38.Li H, Wei Z, Wang C, Chen W, He Y, Zhang C. Gender differences in gastric cancer survival: 99,922 cases based on the SEER database. J Gastrointest Surg. 2020;24(8):1747-1757. doi: 10.1007/s11605-019-04304-y. Epub 2019 Jul 25. [DOI] [PubMed] [Google Scholar]
- 39.Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 2007;8(4):286-298. doi: 10.1038/nrg2005. Epub 2007 Mar 6. [DOI] [PubMed] [Google Scholar]
- 40.Guarrera S, Viberti C, Cugliari G, et al. Peripheral blood DNA methylation as potential biomarker of malignant pleural mesothelioma in asbestos-exposed subjects. J Thorac Oncol. 2019;14(3):527-539. doi: 10.1016/j.jtho.2018.10.163. Epub 2018 Nov 5. [DOI] [PubMed] [Google Scholar]
- 41.Xu Z, Sandler DP, Taylor JA. Blood DNA methylation and breast cancer: a prospective case-cohort analysis in the sister study. J Natl Cancer Inst. 2020;112(1):87-94. doi: 10.1093/jnci/djz065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun H, Zhou H, Zhang Y, et al. Aberrant methylation of FAT4 and SOX11 in peripheral blood leukocytes and their association with gastric cancer risk. J Canc. 2018;9(13):2275-2283. doi: 10.7150/jca.24797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Malakar P, Shilo A, Mogilevsky A, et al. Long Noncoding RNA MALAT1 promotes hepatocellular carcinoma development by SRSF1 upregulation and mTOR activation. Canc Res. 2017;77(5):1155-1167. doi: 10.1158/0008-5472.CAN-16-1508. Epub 2016 Dec 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin R, Maeda S, Liu C, Karin M, Edgington TS. A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene. 2007;26(6):851-858. doi: 10.1038/sj.onc.1209846. Epub 2006 Jul 31. [DOI] [PubMed] [Google Scholar]
- 45.Meseure D, Vacher S, Lallemand F, et al. Prognostic value of a newly identified MALAT1 alternatively spliced transcript in breast cancer. Br J Canc. 2016;114(12):1395-1404. doi: 10.1038/bjc.2016.123. Epub 2016 May 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li L, Geng Y, Feng R, et al. The human RNA surveillance factor UPF1 modulates gastric cancer progression by targeting long non-coding RNA MALAT1. Cell Physiol Biochem. 2017;42(6):2194-2206. doi: 10.1159/000479994. Epub 2017 Aug 15. [DOI] [PubMed] [Google Scholar]
- 47.Hao Y, Crenshaw T, Moulton T, Newcomb E, Tycko B. Tumour-suppressor activity of H19 RNA. Nature. 1993;365(6448):764-767. doi: 10.1038/365764a0. [DOI] [PubMed] [Google Scholar]
- 48.Luo M, Li Z, Wang W, Zeng Y, Liu Z, Qiu J. Long non-coding RNA H19 increases bladder cancer metastasis by associating with EZH2 and inhibiting E-cadherin expression. Canc Lett. 2013;333(2):213-221. doi: 10.1016/j.canlet.2013.01.033. Epub 2013 Jan 24. [DOI] [PubMed] [Google Scholar]
- 49.Lustig O, Ariel I, Ilan J, Lev-Lehman E, De-Groot N, Hochberg A. Expression of the imprinted gene H19 in the human fetus. Mol Reprod Dev. 1994;38(3):239-246. doi: 10.1002/mrd.1080380302. [DOI] [PubMed] [Google Scholar]
- 50.Sasaki H, Ishihara K, Kato R. Mechanisms of Igf2/H19 imprinting: DNA methylation, chromatin and long-distance gene regulation. J Biochem. 2000;127(5):711-715. doi: 10.1093/oxfordjournals.jbchem.a022661. [DOI] [PubMed] [Google Scholar]
- 51.Liu X, Wu S, Yang Y, Zhao M, Zhu G, Hou Z. The prognostic landscape of tumor-infiltrating immune cell and immunomodulators in lung cancer. Biomed Pharmacother. 2017;95:55-61. doi: 10.1016/j.biopha.2017.08.003. Epub 2017 Aug 18. [DOI] [PubMed] [Google Scholar]
- 52.Morales-Nebreda L, McLafferty FS, Singer BD. DNA methylation as a transcriptional regulator of the immune system. Transl Res. 2019;204:1-18. doi: 10.1016/j.trsl.2018.08.001. Epub 2018 Aug 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schuyler RP, Merkel A, Raineri E, et al. Distinct trends of DNA methylation patterning in the innate and adaptive immune systems. Cell Rep. 2016;17(8):2101-2111. doi: 10.1016/j.celrep.2016.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material, sj-pdf-1-ccx-10.1177_10732748211043667 for Peripheral Blood-Based DNA Methylation of Long Non-Coding RNA H19 and Metastasis-Associated Lung Adenocarcinoma Transcript 1 Promoters are Potential Non-Invasive Biomarkers for Gastric Cancer Detection by Dingtao Hu, Xiaoqi Lou, Nana Meng, Zhen Li, Ying Teng, Yanfeng Zou and Fang Wang in Cancer Control