Abstract
Objective
To explore the hypermethylated long non-coding (lnc)RNAs involved in bladder carcinogenesis and prognosis.
Methods
Reduced representation bisulfite sequencing and RNA sequencing were performed on five paired tumor and adjacent normal tissue samples from bladder cancer patients. The differentially methylated regions around transcription start sites and differentially expressed genes, including lncRNAs, were analyzed. Correlations between DNA methylation modifications and the expression of lncRNAs were examined. Survival analysis was surveyed on the GEPIA web server.
Results
We identified 19,560 hypomethylated and 68,781 hypermethylated differentially methylated regions around transcription start sites in bladder cancer tissues. In total, 2321 differentially expressed genes were found in bladder tumors, among which, 367 were upregulated and 1954 were downregulated. There were 141 downregulated genes involving eight lncRNAs that were consistently hypermethylated, while 24 upregulated genes were consistently hypomethylated. Survival analysis demonstrated that hypermethylation of lncRNAs LINC00683 and MSC-AS1 were associated with poor overall survival in bladder cancer patients.
Conclusion
Some lncRNAs are controlled by DNA methylation in bladder cancer and they might be important factors in bladder carcinogenesis. Hypermethylated lncRNAs including LINC00683 and MSC-AS1 have the potential to be prognostic biomarkers for bladder cancer.
Keywords: DNA methylation, bladder cancer, long non-coding RNA, reduced representation bisulfite sequencing, RNA sequencing, biomarker
Background
Bladder cancer is one of the most frequent urological malignancies worldwide, with approximately 430,000 new cases diagnosed in 2012 and 165,000 bladder cancer-related deaths. 1 In China, bladder cancer rates increased rapidly between 2005 and 2015, reaching a morbidity of 5.8 per 100,000 in 2015. 2 Despite the recent emergence of multiple new diagnostic and treatment methodologies including trans urethral resection of bladder tumor, radical cystectomy, chemotherapy, radiotherapy, and immunotherapy, bladder cancer, especially muscle-invasive bladder cancer, remains associated with a poor 5-year survival rate (<60%) and a high recurrence rate (50%–70%). Due to the unclear cause of bladder cancer, a deeper understanding of the potential molecular mechanisms that drive malignancy would benefit current and future therapies.
Epigenetics has been revealed to be an important factor in bladder cancer carcinogenesis. 3 There are three common types of epigenetics: DNA methylation, non-coding RNA-mediated gene silencing, and histone modifications.4,5 Among these, DNA methylation is the most common form of epigenetics in mammals that alters gene expression without changing DNA sequence.6,7
Some oncogenes and tumor suppressor genes have been shown to play important roles in bladder tumorigenesis. 8 Altered methylation of 5′ CpG islands is a mechanism through which tumor suppressors are inhibited or oncogenes are activated in bladder cancer. 3 It has been suggested that hypermethylation of tumor suppressors, which inhibits gene expression, plays a vital role in human tumor progression. 3
Long non-coding RNAs (lncRNAs) are RNAs greater than 200 nucleotides in length that do not encode proteins and whose transcripts are located in non-protein coding regions. 9 In human cells, more than 3000 lncRNAs have been shown to be involved in chromatin modifications, alternative splicing, translation, and miRNA sponges.9,10 LncRNAs regulate the expression of downstream target genes by binding to proteins, and thus play oncogenic or tumor suppressive roles. 11 Previous studies have reported that lncRNAs are atypically expressed in various tumors, such as breast cancer, 12 lung cancer, 13 bladder cancer, 14 and prostate cancer. 15 For instance, lncRNA HOTAIR was reported to be highly expressed in metastatic breast cancer, where it acts as an oncogene by inhibiting HOXD when combined with PRC2. 16 LncRNA urothelial cancer-associated 1 was reported to be highly expressed in bladder cancer cells and was proven to enhance bladder cancer cell migration and invasion in part through the hsa-miR-145/ZEB1/2/FSCN1 pathway. 17 Seitz et al. showed that the novel lncRNAs LINC00958 and LINC01296 were candidate oncogenes in bladder cancer. 18 We have previously reported that lncRNA UBC1 plays an oncogenic role in bladder cancer by recruiting the chromatin modification complex PRC2 to regulate histone methylation of downstream target genes. 19 Our previous study also showed that lncRNA UCA1 was upregulated in bladder cancer specimens, which may promote invasion and EMT in bladder cancer cells via regulating the miR-143/HMGB1 pathway. 14
Increasing evidence has demonstrated that the expression of lncRNAs is correlated with aberrant DNA methylation in human cancers.20–22 It has also been confirmed that methylation of lncRNA ESRP2 is related to breast cancer prognosis. 23 However, it remains unclear which lncRNAs are involved in human bladder cancer and the association between lncRNAs and DNA methylation.
In this study, reduced representation bisulfite sequencing (RRBS) was conducted on five paired bladder cancer and normal adjacent tissues to screen for methylated-RNAs (including lncRNAs). The association between differentially methylated regions (DMRs), the expression of lncRNAs, and bladder cancer prognosis were then further analyzed.
Methods and materials
Tissue specimens
All tissue samples were collected from bladder cancer patients, immediately transferred to liquid nitrogen, and then stored at −80°C until DNA and RNA extraction. The diagnosis of each bladder cancer patient was confirmed by pathologists. This study was approved by the Institute Research Ethics Committee of Peking University Shenzhen Hospital (No. 20090017). Written informed consent was given by all patients before surgery.
RRBS protocol
Genomic DNA was extracted using the DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany) and subjected to RRBS library preparation using the Rapid RRBS Library Prep Kit (Cat. No. AG0422, Shenzhen Ace Gene Technology Co., Ltd., Shenzhen, China) following the manufacturer’s instructions. Briefly, 100 ng of genomic DNA was digested using MspI, end-repaired, 3′-dA-tailed, and ligated to 5-methylcytosine-modified adapters. After bisulfite treatment, the DNA was amplified with 12 cycles of polymerase chain reaction using 8-bp dual index primers (Illumina, San Diego, CA, USA). Size selection was performed using a dual-SPRI protocol to obtain DNA fractions containing MspI-digested products ranging from 100 to 350 bp. The constructed RRBS libraries were then analyzed using a Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA) and sequenced on Illumina platforms using a 150 × 2 paired-end sequencing protocol.
DMR analysis
Raw RRBS data were processed according to the standard procedures of Shenzhen Ace Gene Technology Co., Ltd. Metilene software was applied to scan for DMRs between the bladder cancer and normal tissue groups (MWU-test and 2D KS-test). Differential hyper- and hypo-methylated regions were counted for the gene body, and 2 kb upstream and downstream. A violin boxplot and heatmap were created to identify hyper- and hypo-methylated regions between the two groups.
RNA sequencing
Total RNA was isolated from five paired bladder cancer and normal adjacent tissues using TRIzol and following the manufacturer's instructions. The concentration and quality of RNA was determined using a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and a Bioanalyzer 2100 (Agilent), respectively. The TruSeq RNA Sample Preparation Kit V2 (Illumina) was used for next generation sequencing library construction following manufacturer's protocols. Briefly, mRNA was purified from 1000 ng total RNA using oligo-dT magnetic beads, and then fragmented. First-strand cDNA synthesis was performed using random hexamer priming, followed by second-strand cDNA synthesis; 3′ adenylation was then performed on the double stranded cDNA for end repair. Illumina adaptors were ligated to both ends of the cDNA, purified by gel electrophoresis, and then amplified with adaptor sequence-specific polymerase chain reaction primers to generate amplicons of 200 to 500 bp. Finally, the amplified libraries were hybridized to the Illumina paired-end flow cell and amplified using cBot (Illumina) at 10 pM per lane. Paired-end reads of 150 nt were generated for each sample, and the alignment was performed using the reference genome.
Analysis of differentially expressed genes (DEGs)
Clean RNA-sequencing data were processed according to the standard procedures of Shenzhen Ace Gene Technology Co., Ltd. DEG-seq software was used to scan the DEGs between the bladder cancer and normal groups. Volcano plots were used to show differences in gene expression profiles between tissues. The correlation between promoter methylation level and gene expression were analyzed.
Association between DMR-related genes (anchored promoter) and DEGs
Data from RRBS and RNA sequencing were collected to analyze the correlation between DMRs and DEGs of promoter regions (from 2 kb upstream to 500 bp downstream of the transcriptional start site [TSS]). Genes were classified as follows: up-regulated consistently and hypomethylated or down-regulated consistently and hypermethylated.
Analyses of Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways
To discover the biological processes, cellular components, molecular functions, metabolic and signal transduction pathways, analyses of GO and KEGG were used to characterize the DMRs, DEGs, and DMR-related DEGs between groups (fold change>2 and p < 0.05).
Survival analysis
Disease-free survival and overall survival time of bladder cancer patients with different expression levels of hypermethylated-lncRNAs were analyzed using the GEPIA web server (http://gepia.cancer- pku.cn/). The log-rank test was used to assess survival times between groups. 24
Results
Genome-wide distribution of TSS methylation in bladder cancer tissues
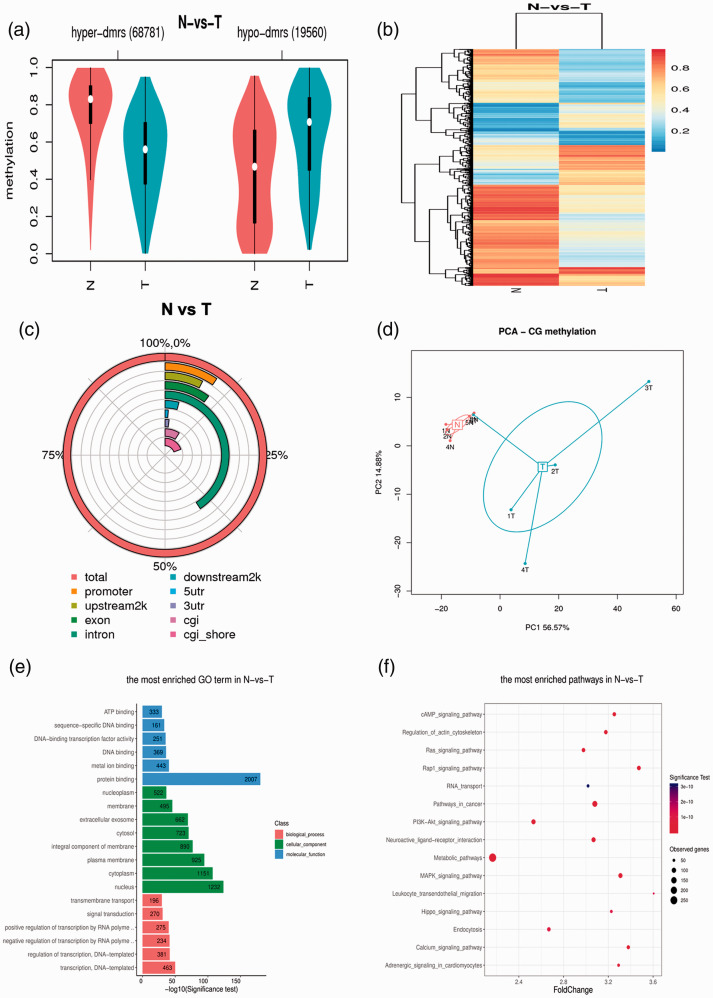
Five paired bladder cancer and normal adjacent tissues were obtained from Peking University Shenzhen Hospital between 2018 and 2019. In total, 19,560 hypomethylated and 68,781 hypermethylated DMRs were detected around TSS regions in bladder cancer tissues compared with normal tissues (Figure 1a). DMR methylation levels in bladder cancer tissues were lower than in normal tissues (Figure 1b). Methylation was enriched in the promoter and intron regions rather than the untranslated regions or gene body flanking regions (Figure 1c). Furthermore, principal component analysis (PCA) of genome-wide methylation levels showed that methylation status was different between cancerous and normal tissues (Figure 1d).
Figure 1.
Genome-wide distribution of transcriptional start site methylation in bladder cancer tissues. (a) Violin boxplot showing the average methylation levels of differentially methylated regions (DMRs) around transcriptional start sites. (b) Heatmap showing the methylation levels of DMRs between groups. (c) Distribution scale of DMRs. (d) Principal component analysis (PCA) of genome-wide methylation levels. Gene Ontology (GO) (e) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway (f) analyses of DMR-related genes.
GO and KEGG pathway analysis were conducted to examine the potential role of DMRs around promoter regions. GO analysis revealed that the top five enriched terms were protein binding (2007 genes), nucleus (1232 genes), cytoplasm (1151 genes), plasma membrane (925 genes) and integral component of membrane (890 genes) (Figure 1e). KEGG pathway analysis revealed that metabolic pathways (264 genes), pathways in cancer (124 genes), MAPK signaling pathways (86 genes), neuroactive ligand-receptor interaction (86 genes), and PI3K-Akt signaling pathways (89 genes) were the top five pathways (Figure 1f).
Global changes in RNA expression in bladder cancer tissues
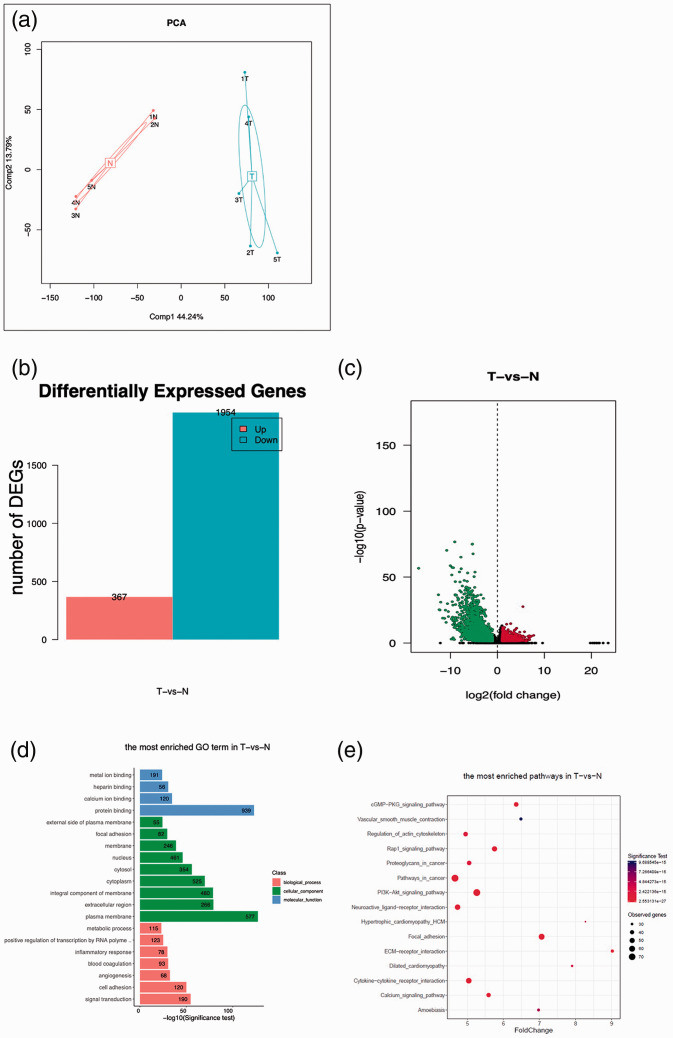
RNA sequencing was used to examine RNA expression profiles of bladder cancer and normal bladder tissues. PCA of all RNA transcripts showed that gene expression differed between cancerous and normal tissues (Figure 2a). In total, 2321 DEGs were found in tumor tissues compared with normal tissues (Figure 2b). Among these genes, 367 were upregulated and 1954 were downregulated. Volcano plots showed that the expression profiles of bladder cancers differed from that of normal bladder tissues (Figure 2c).
Figure 2.
Global changes in RNA expression in bladder cancer tissues. (a) Principal component analysis (PCA) of whole RNA data. Histogram (b) and volcano plot (c) showing the differentially expressed genes (DEGs), with red representing upregulated genes, and green signifying downregulated genes. Fold change >2, p < 0.00001. Gene Ontology (GO) (d) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway (E) analyses of the DEGs.
GO and KEGG enrichment were used to investigate the possible roles of the DEGs. These results showed that protein binding (939 genes), plasma membrane (577 genes), cytoplasm (525 genes), integral component of membrane (480 genes), nucleus (461 genes), and cytosol (354 genes) were the top five GO terms (Figure 2d). KEGG enrichment analysis revealed that PI3K/Akt signaling pathways, focal adhesion, cytokine-cytokine receptor interaction, and Rap1 signaling pathway were correlated with cancer tissues (Figure 2e).
The expression of lncRNAs is regulated by methylation of gene promoters
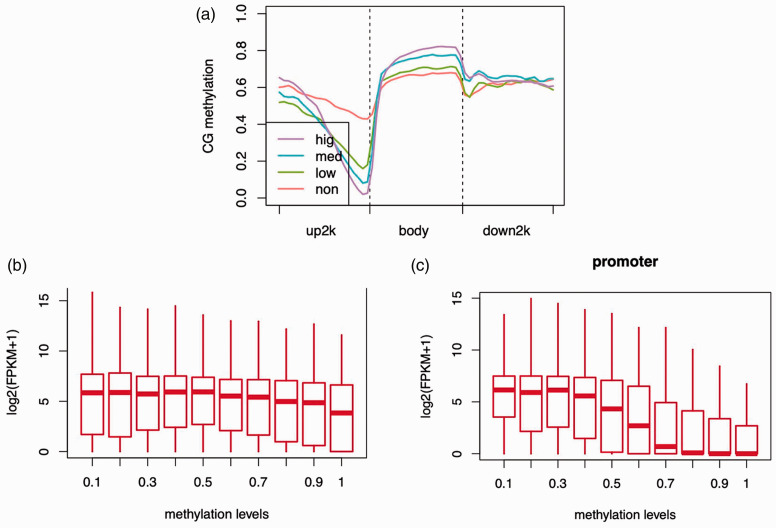
To explore the potential link between gene expression and methylation levels, the gene expression profiles and methylation status of bladder cancer samples were analyzed. Methylation levels were negatively correlated with gene expression levels, especially near TSS regions (Figure 3a). Gene expression levels were decreased with increased methylation level. The association between promoter methylation level and gene expression was stronger than that of gene body methylation levels (Figure 3b and 3c).
Figure 3.
Relationship between methylation and gene expression levels. (a) Methylation levels were negatively correlated with gene expression levels, especially near transcriptional start sites (TSS). Expression levels were graded as follows: <0.001, non; 0.001 to 5, low; 5 to 50, medium; >50, high. (b) and (c) Correlation between promoter methylation level and gene expression were stronger than that of gene body methylation.
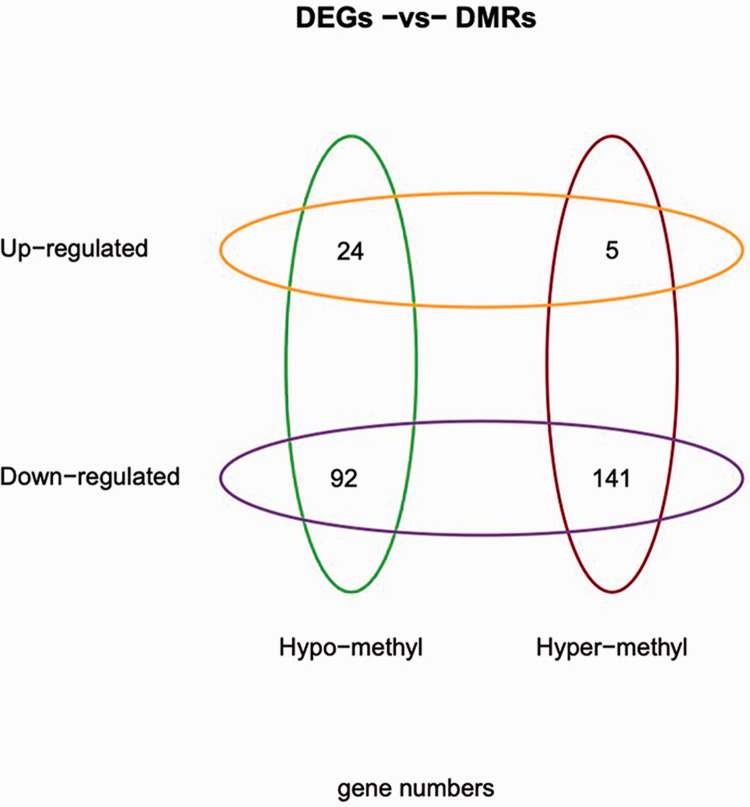
The combination of RRBS and RNA sequencing data showed that 141 genes were consistently downregulated and hypermethylated, while 24 genes were consistently upregulated and hypomethylated (Figure 4). Among these genes with consistent changes in expression and methylation, eight lncRNAs were discovered to be involved that were consistently downregulated and hypermethylated (Table 1).
Figure 4.
Correlation between the genes that showed differential methylation and those that showed differential expression.
Table 1.
Methylated lncRNAs that showed differential expression in bladder cancer tissues.
| lncRNA | Methylation status | Expression status |
|---|---|---|
| HAND2-AS1 | Hyper | Down |
| LINC00092 | Hyper | Down |
| CPEB1-AS1 | Hyper | Down |
| LINC00683 | Hyper | Down |
| VIM-AS1 | Hyper | Down |
| FOXD3-AS1 | Hyper | Down |
| LINC00403 | Hyper | Down |
| MSC-AS1 | Hyper | Down |
lncRNA, long non-coding RNA.
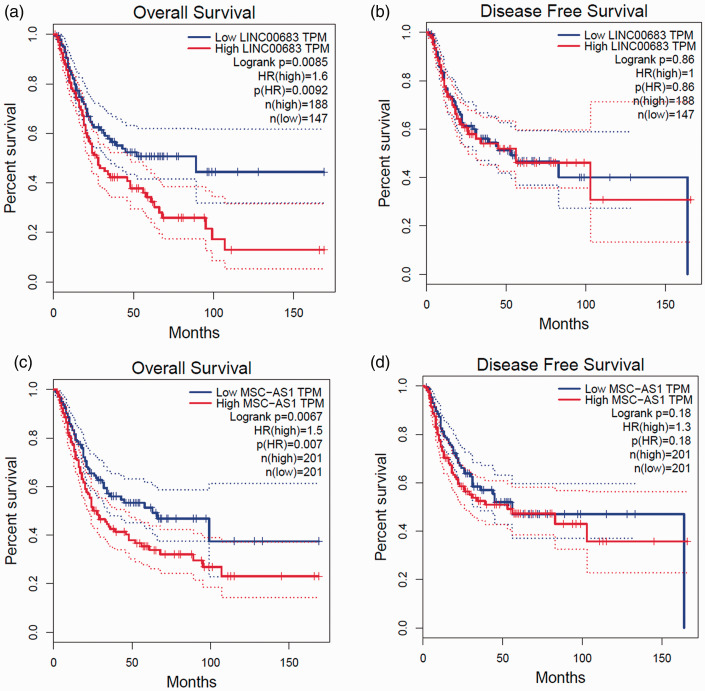
Furthermore, GEPIA analysis was used to examine the relationship between lncRNAs and bladder cancer survival. High expression of LINC00683 and MSC-AS1 (hypomethylated) were both associated with poor overall survival (LINC00683: hazard ratio=1.6, p = 0.0092; MSC-AS1: hazard ratio=1.5, p = 0.007) (Figure 5a and 5c). This suggested shorter overall survival time in patients with higher expression of LINC00683 or MSC-AS1. No significant association was identified between LINC00683 or MSC-AS1 with disease-free survival (Figure 5b and 5d).
Figure 5.
Two differentially expressed lncRNAs, LINC00683 and MSC-AS1 were associated with poor overall survival. (a) and (b) Overall and disease-free survival in bladder cancer patients according to LINC00683 expression. (c) and (d) Overall and disease-free survival in bladder cancer patients according to MSC-AS1 expression. Survival analyses were conducted using the online GEPIA tool (http://gepia.cancer-pku.cn/).
Discussion
DNA methylation is a crucial epigenetic regulatory mechanism that is involved in human diseases, especially cancers. 3 RRBS is a method of sequencing methylation within a gene promoter region and is more conducive to finding changes in promoter methylation in tumors compared with whole genome bisulfite sequencing. Therefore, RRBS is the preferred method to examine methylation status in genes. In this study, we used RRBS to examine the methylation status of bladder cancer patients.
Studies have shown that in human cancers, the tumor genome is generally hypomethylated. 25 Our RRBS experiments confirmed that the overall bladder cancer genome was hypomethylated, which led to increased genome instability, making this an important mechanism of bladder carcinogenesis. Studies have also shown that the increased gene methylation that results from malignant transformation, especially CpG island methylation in TSS regions, can inhibit the expression of tumor suppressors. 3 DMRs have been identified in the reprogramming and developmental stages of embryos; therefore, DMRs have also been used to study methylation levels between bladder tumors and normal bladder tissues.3,26 This study demonstrated that a differential methylation clustering heatmap of DMRs could be generated between cancerous and normal tissues.
Studies have shown that there are many DEGs between bladder tumors and normal bladder tissues, including upregulated oncogenes and downregulated tumor suppressors, which may be related to bladder tumorigenesis. 8 We collected five paired bladder cancer and adjacent normal tissues and used RNA sequencing to find 2321 DEGs in bladder cancer, of which 367 were upregulated, and 1954 were downregulated. KEGG analysis showed that the downregulated genes were primarily involved in cancer pathways, including PI3K/Akt signaling, focal adhesion, cytokine receptor interaction, and Rap1 signaling. On the basis of these results, we suggest that the downregulated genes may be involved in bladder carcinogenesis as tumor suppressors.
To further explore the relationship between gene expression and promoter methylation in bladder cancer, RRBS and RNA-sequencing data were analyzed. These results showed that 141 genes were consistently downregulated and hypermethylated in bladder cancer. KEGG analysis showed that the downregulated genes were mostly related to proteoglycans in cancer, including those involved in PI3K/Akt signaling and other pathways. This indicated increased methylation of tumor suppressor gene promoters in bladder cancer, resulting in their downregulation, which may be an important mechanism of bladder cancer tumorigenesis.
Among the hypermethylated genes that were downregulated in their promoter regions, there were eight lncRNAs with significant differences: HAND2-AS1, LINC00092, CPEB1-AS1, LINC00683, VIM-AS1, FOXD3-AS1, LINC00403, and MSC-AS1.
The function of the identified hypermethylated-lncRNAs have not been well studied in bladder cancer. One lncRNA, HAND2-AS1, is downregulated in rectal cancer and may play a tumor-suppressive role by recruiting mir-1275 to inhibit the progression of rectal cancer. 27 HAND2-AS1 has also been reported to be downregulated in cervical cancer, 28 melanoma, 29 chronic myeloid leukemia, 30 non-small cell lung cancer, 31 and other malignant tumors, in which it is thought to inhibit tumor development by binding to microRNAs or mRNAs. Research regarding the role of HAND2-AS1 in bladder cancer has not been reported; therefore, subsequent studies will examine the role of HAND2-AS1 in bladder cancer.
Two other identified hypermethylated-lncRNAs, MSC-AS1 and LINC00683, do not have reported roles in bladder cancer. It has been reported that MSC-AS1 is upregulated in renal clear cell carcinoma, where it is correlated with a poor prognosis. 32 Sun et al. demonstrated that the MSC-AS1/miR-29b-3p axis modulates cell proliferation and gemcitabine-induced apoptosis in pancreatic cancer. 33 Additionally, lncRNA MSC-AS1 may be a biomarker of laryngeal cancer. 34 LINC00683 was remarkably downregulated in prostate cancer samples and strongly correlated with patient survival, which suggests LINC00683 might be a new diagnostic biomarker and therapeutic target for prostate cancer treatments. 35
In this study, the GEPIA database was used to survey the relationship between these differentially expressed lncRNAs and bladder cancer prognosis. We found that a gain of promoter region methylation and loss of LINC00683 and MSC-AS1 expression were associated with reduced survival in bladder cancer patients. These findings suggested that LINC00683 and MSC-AS1 might be biomarkers involved in bladder cancer prognosis. Thus, in the future, we might predict the prognosis of bladder cancer patients using LINC00683 and MSC-AS1 expression. It would be better if more samples were involved in this study. More research is needed to more fully determine the association between methylated-lncRNAs and bladder cancer prognosis.
Conclusions
Taken together, our findings revealed that some lncRNAs are under control of DNA methylation. Hypermethylated-lncRNAs might be important factors in bladder carcinogenesis. Specifically, LINC00683 and MSC-AS1 may act as biomarkers for bladder cancer prognosis.
Supplemental Material
Supplemental material, sj-pdf-1-imr-10.1177_03000605211049946 for Role of hypermethylated-lncRNAs in the prognosis of bladder cancer patients by Junhua Luo, Jinming Xu, Longhua Ou, Yingchen Zhou, Haichao Yun, Yu Yang, Xionghui Wu and Yan Wang in Journal of International Medical Research
Acknowledgements
We thank the Guangdong and Shenzhen Key Laboratory of Male Reproductive Medicine and Genetics, Institute of Urology, Peking University Shenzhen Hospital.
Footnotes
Declaration of conflicting interest: The authors declare that there is no conflict of interest.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by a grant from the Medical Scientific Research Foundation of Guangdong Province (A2021126), the Shenzhen High-level Hospital Construction Fund, and ‘San-ming’ Project of Medicine in Shenzhen (SZSM201612066).
ORCID iD: Junhua Luo https://orcid.org/0000-0002-1885-4595
References
- 1.Antoni S, Ferlay J, Soerjomataram I, et al . Bladder cancer incidence and mortality: A global overview and recent trends. Eur Urol 2017; 71: 96–108. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, et al . Cancer statistics in China, 2015. CA Cancer J Clin 2016; 66: 115–132. [DOI] [PubMed] [Google Scholar]
- 3.Sardi I, Dal Canto M, Bartoletti R, et al. Abnormal C-Myc oncogene DNA methylation in human bladder cancer: Possible role in tumor progression. Eur Urol 1997; 31: 224–230. [DOI] [PubMed] [Google Scholar]
- 4.Gui Y, Guo G, Huang Y, et al . Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat Genet 2011; 43: 875–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baylin SB, Jones PA. A decade of exploring the cancer epigenome — biological and translational implications. Nat Rev Cancer 2011; 11: 726–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taby R, Issa JP. Cancer epigenetics. CA Cancer J Clinic 2010; 60: 376–392. [DOI] [PubMed] [Google Scholar]
- 7.Tsai HC, Baylin SB. Cancer epigenetics: Linking basic biology to clinical medicine. Cell Res 2011; 21: 502–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brandau S, Böhle A. Bladder cancer. I. Molecular and genetic basis of carcinogenesis. Eur Urol 2001; 39: 491–497. [DOI] [PubMed] [Google Scholar]
- 9.Guttman M, Amit I, Garber M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009; 458: 223–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khalil AM, Guttman M, Huarte M, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U.S.A 2009; 106: 11667–11672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nagano T, Fraser P. No-nonsense functions for long noncoding RNAs. Cell 2011; 145: 178–181. [DOI] [PubMed] [Google Scholar]
- 12.Liang Y, Song X, Li Y, et al. LncRNA BCRT1 promotes breast cancer progression by targeting miR-1303/PTBP3 axis. Mol Cancer 2020; 19: 85. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Zhen Q, Gao LN, Wang RF, et al . LncRNA DANCR Promotes Lung Cancer by Sequestering miR-216a. Cancer Control 2018; 25: 1073274818769849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luo J, Chen J, Li H, et al . LncRNA UCA1 promotes the invasion and EMT of bladder cancer cells by regulating the miR‐143/HMGB1 pathway. Oncol Lett 2017; 14: 5556–5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shang Z, Yu J, Sun L, et al. LncRNA PCAT1 activates AKT and NF-κB signaling in castration-resistant prostate cancer by regulating the PHLPP/FKBP51/IKKα complex. Nucleic Acids Res 2019; 47: 4211–4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010; 464: 1071–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xue M, Pang H, Li X, et al. Long non-coding RNA urothelial cancer-associated 1 promotes bladder cancer cell migration and invasion by way of the hsa-miR-145-ZEB1/2-FSCN1 pathway. Cancer Sci 2016; 107: 18–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seitz AK, Christensen LL, Christensen E, et al. Profiling of long non-coding RNAs identifies LINC00958 and LINC01296 as candidate oncogenes in bladder cancer. Sci Rep 2017; 7: 395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He W, Cai Q, Sun F, et al. Linc-UBC1 physically associates with polycomb repressive complex 2 (PRC2) and acts as a negative prognostic factor for lymph node metastasis and survival in bladder cancer. Biochim Biophys Acta 2013; 1832: 1528–1537. [DOI] [PubMed] [Google Scholar]
- 20.Guo W, Liu S, Dong Z, et al . Aberrant methylation-mediated silencing of lncRNA CTC-276P9.1 is associated with malignant progression of esophageal squamous cell carcinoma. Clin Exp Metastasis 2018; 35: 53–68. [DOI] [PubMed] [Google Scholar]
- 21.Dong Z, Zhang A, Liu S, et al . Aberrant methylation-mediated silencing of lncRNA MEG3 functions as a ceRNA in esophageal cancer. Mol Cancer Res 2017; 15: 800–810. [DOI] [PubMed] [Google Scholar]
- 22.Pan W, Zhang N, Liu W, et al. The long noncoding RNA GAS8-AS1 suppresses hepatocarcinogenesis by epigenetically activating the tumor suppressor GAS8. J Biol Chem 2018; 293: 17154–17165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heilmann K, Toth R, Bossmann C, et al. Genome-wide screen for differentially methylated long noncoding RNAs identifies Esrp2 and lncRNA Esrp2-as regulated by enhancer DNA methylation with prognostic relevance for human breast cancer. Oncogene 2017; 36: 6446–6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang Z, Li C, Kang B, et al. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017; 45: W98–W102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Irizarry RA, Ladd-Acosta C, Wen B, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet 2009; 41: 178–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meissner A, Mikkelsen TS, Gu H, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 2008; 454: 766–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou J, Lin J, Zhang H, et al. LncRNA HAND2-AS1 sponging miR-1275 suppresses colorectal cancer progression by upregulating KLF14. Biochem Biophys Res Commun 2018; 503: 1848–1853. [DOI] [PubMed] [Google Scholar]
- 28.Chen S, Wang J. HAND2-AS1 inhibits invasion and metastasis of cervical cancer cells via microRNA-330-5p-mediated LDOC1. Cancer Cell Int 2019; 19: 353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu P, Du R, Yu X. LncRNA HAND2-AS1 overexpression inhibits cancer cell proliferation in melanoma by downregulating ROCK1. Oncol Lett 2019; 18: 1005–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang JR, Shi MX, Zeng Y. LncRNA HAND2-AS1 inhibits proliferation and promotes apoptosis of chronic myeloid leukemia cells by sponging with micRNA-1275. Eur Rev Med Pharmacol Sci 2019; 23: 2103–2111. [DOI] [PubMed] [Google Scholar]
- 31.Miao F, Che J, Shi M, et al. LncRNA HAND2-AS1 inhibits non-small cell lung cancer migration, invasion and maintains cell stemness through the interactions with TGF-β1. Biosci Rep 2019; 39: BSR20181525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu Z, Li L, Cheng P, et al. lncRNA MSC-AS1 activates Wnt/β-catenin signaling pathway to modulate cell proliferation and migration in kidney renal clear cell carcinoma via miR-3924/WNT5A. J Cell Biochem 2020; 121: 4085–4093. [DOI] [PubMed] [Google Scholar]
- 33.Sun Y, Wang P, Yang W, et al. The role of lncRNA MSC-AS1/miR-29b-3p axis-mediated CDK14 modulation in pancreatic cancer proliferation and gemcitabine-induced apoptosis. Cancer Biol Ther 2019; 20: 729–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Y, Meng W, Cao H, et al . Identification of MSC-AS1, a novel lncRNA for the diagnosis of laryngeal cancer. Eur Arch Otorhinolaryngol 2021; 278: 1107–1118. [DOI] [PubMed] [Google Scholar]
- 35.Liu Y, Yang B, Su Y, et al . Downregulation of long noncoding RNA LINC00683 associated with unfavorable prognosis in prostate cancer based on TCGA. J Cell Biochem 2019; 120: 14165–14174. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-imr-10.1177_03000605211049946 for Role of hypermethylated-lncRNAs in the prognosis of bladder cancer patients by Junhua Luo, Jinming Xu, Longhua Ou, Yingchen Zhou, Haichao Yun, Yu Yang, Xionghui Wu and Yan Wang in Journal of International Medical Research