Abstract
Objective
We investigated the impact of human immunodeficiency virus (HIV) infection and anti-retroviral therapy (ART) on the gut microbiota of children.
Design
This cross-sectional study investigated the gut microbiota of children with and without HIV.
Methods
We collected fecal samples from 59 children with HIV (29 treated with ART [ART(+)] and 30 without ART [HIV(+)]) and 20 children without HIV [HIV(–)] in Vietnam. We performed quantitative RT-PCR to detect 14 representative intestinal bacteria targeting 16S/23S rRNA molecules. We also collected the blood samples for immunological analyses.
Results
In spearman’s correlation analyses, no significant correlation between the number of dominant bacteria and age was found among children in the HIV(−) group. However, the number of sub-dominant bacteria, including Streptococcus, Enterococcus, and Enterobacteriaceae, positively correlated with age in the HIV(−) group, but not in the HIV(+) group. In the HIV(+) group, Clostridium coccoides group positively associated with the CD4+ cell count and its subsets. In the ART(+) group, Staphylococcus and C. perfringens positively correlated with CD4+ cells and their subsets and negatively with activated CD8+ cells. C. coccoides group and Bacteroides fragilis group were associated with regulatory T-cell counts. In multiple linear regression analyses, ART duration was independently associated with the number of C. perfringens, and Th17 cell count with the number of Staphylococcus in the ART(+) group.
Conclusions
HIV infection and ART may influence sub-dominant gut bacteria, directly or indirectly, in association with immune status in children with HIV.
Introduction
The gut microbiota comprises approximately 100 trillion microbes from more than 1000 bacterial species [1, 2]. The gut microbiota plays a major role in nutrient absorption, food metabolism, intestinal barrier protection from pathogens, and the modulation of gut immune function [3–5]. Although the composition of the gut microbiota may be influenced by age, diet, genetics, and geography, four phyla (i.e., Firmicutes, Bacteroidetes, Actinobacteria, and Proteobacteria) are dominant and stable in healthy individuals [4, 6, 7]. CD4+ T cells and their subsets, such as type 1 helper T cells (Th1), Th2, Th17, and regulatory T (Treg) cells, have been associated with the gut microbiota, and their interactions are associated with various diseases, such as inflammatory bowel disease, rheumatoid arthritis, and cancer [8, 9].
Gut-associated lymphoid tissue (GALT) is the largest replication site, and it serves as a reservoir of human immunodeficiency virus type 1 (HIV) [10–12]. Progressive HIV infections are characterized by a depletion of CD4+ T cells in the GALT, followed by microbial translocation, gut microbiota dysbiosis, and chronic immune activation [11, 13–17]. Despite the sustained viral suppression and immune recovery provided by anti-retroviral therapy (ART), the imbalance in gut microbiota is, at best, only partially restored for a long time after initiating ART in adults [15, 16, 18].
In the gut microbiota of healthy children, the dominant phyla are Bacteroidetes and Actinobacteria, particularly the Bifidobacterium genus of Actinobacteria. These bacteria have a functional composition similar to that of healthy adults [7, 19–22]. A few studies from Africa and India have shown reduced bacterial diversity in the gut microbiota of children with HIV and children treated with ART compared to the microbiota of children without HIV [23–25]. However, no consensus exists on whether ART in children with HIV may restore the gut microbiota to the state observed in children without HIV [23–25]. The impacts of HIV and ART on the gut microbiota in children remain poorly understood.
In Vietnam, no study has focused on understanding the gut microbiota in children with HIV. Therefore, the current study aimed to investigate the impact of HIV infection and ART on the gut microbiota among children in Vietnam.
Methods
Study population
This non-randomized, cross-sectional study was conducted at the Vietnam National Children’s Hospital (VNCH) in Hanoi, Vietnam, in 2012 [26, 27]. Children with HIV who did not start ART [HIV(+) group], children with HIV who received ART [ART(+) group], and children without HIV infection [HIV(−) group, control] were recruited.
The inclusion criteria for children with HIV were followed at the VNCH and ≥2 years old. Exclusion criteria were progression of HIV to acquired immunodeficiency syndrome (AIDS), treatment with anything that may influence the immune system, any antibiotics except cotrimoxazole, hospitalization in the prior 8 weeks, or symptoms of gastrointestinal infection, such as diarrhea, nausea, and vomiting, with fever, at the time of recruitment. The children in the ART(+) group resided at an orphanage near Hanoi. The children in the HIV(+) group resided at their own homes. The children in the HIV(−) group resided at a different orphanage in Hanoi [26, 27].
Collection and preparation of fecal samples
Immediately after defecation, fecal samples were collected in a plastic container (Sarstedt AG & Co., Nümbrecht, Germany), kept at 4°C, and transferred to the laboratory using containers maintained at 4°C. At the laboratory, the fecal samples were weighed, suspended in RNA-stabilizing solution (RNAlater; Ambion, Inc., Austin, TX, USA), and homogenized (20 mg of feces/mL). The fecal homogenate (200 μL) was added to 1 mL of Dulbecco’s Phospahte Buffered Saline (Nissui Pharmaceutical Co., Ltd., Tokyo, Japan). After centrifuging the mixture at 12,000 × g for 5 min, the pellet was stored at −80°C until used for RNA extraction. The whole process was completed within 24 hours after defecation [28].
Quantification of bacteria in human feces by RT-qPCR
Total RNA extraction and subsequent reverse-transcription and quantitative polymerase chain reaction (RT-qPCR) assays were performed using the methods described by Matsuda et al. [29, 30]. Briefly, 4 mg of feces were subjected to total RNA extraction, and each extracted RNA sample was serially diluted 10-fold. Three serial dilutions of the extracted RNA samples (corresponding to 1/400, 1/4,000, and 1/40,000 of the extracted RNA) were subjected to RT-qPCR with specific primer sets that targeted the 16S or 23S rRNA of the 14 representative intestinal bacteria in four main phyla, including: Firmicutes, such as Clostridium coccoides group, C. leptum subgroup, C. difficile, C. perfringens, Lactobacillus spp., Streptococcus, Enterococcus, and Staphylococcus; Actinobacteria, such as Bifidobacterium and Atopobium cluster; Bacteroidetes, such as Bacteroides fragilis group and Prevotella; and Proteobacteria, such as Enterobacteriaceae and Pseudomonas [29–32]. The counts of Lactobacillus spp. obtained with RT-qPCR were expressed as the sum of six Lactobacillus subgroups and two species. In the same experiment, a standard curve was generated with the RT-qPCR data (by threshold cycle: CT value) and the cell counts (by DAPI staining) of the dilution series of total RNA from the standard strain for each bacterial target. The CT values from fecal samples were normalized to the standard curve to obtain the bacterial count per gram wet weight of feces.
In addition, the individual bacteria in the fecal microbiota are present at different microbial cell counts. Previous reports revealed that the average total bacterial count is approximately 1011 cells/g of feces [29, 31]. We regarded the threshold for dominance in abundance at 1.0% of the total bacterial count, and the threshold in counts was set at 109 cells/g [6, 33–35].
Laboratory methods
White blood cell (WBC) counts, WBC differentiation, hemoglobin level (Beckman Coulter, Lh 780, USA), total cholesterol level, and fasting blood sugar (Olympus AU640, Japan) were measured at the clinical laboratory of VNCH. Plasma HIV viral loads were measured as described previously [26].
Immunological analysis
Immunological investigations were performed with blood samples as described previously [26]. Briefly, whole blood samples were stained with a combination of monoclonal antibodies to detect cell surface molecules within 6 hours after collection and analyzed using a JSAN flow cytometer (Bay Bioscience, Kobe, Japan). The obtained data were analyzed by Flowjo V.7.5.5 (FLOWJO, OR, USA). We defined CD38+HLA-DR+CD8+ cells as activated CD8+ cells, CXCR3+CCR6−CD25lowCD4+ cells as Th1, CXCR3−CCR6−CD25lowCD4+ cells as Th2, CXCR3−CCR6+CD25lowCD4+ cells as Th17 cells, and CD25highCD4+ cells as Treg cells [36–38]. The gating strategy for cell staining analysis by flow cytometry is shown as S1 Fig. The absolute cell count was calculated as WBC count × percentage of lymphocytes × percentage of target cells obtained by flow cytometry.
Statistical analysis
Statistical analyses were performed using SPSS version 25 (IBM SPSS Statistics, USA) and R version 3.6.2 [39]. The chi-squared test or Fisher’s exact test was performed to assess the differences in bacterial detection rates. The gut microbiota patterns were presented as biplots with the principal component analysis (PCA) using the prcomp function from the ggbiplot package in R. The number of bacteria was compared between the groups using the Mann-Whitney U test. Spearman’s rank test was used to analyze the pairwise correlations between bacteria and possibly related factors, such as age, ART duration, CD4+ cells and their subsets, CD8+ cells, the proportion of activated CD8+ cells, and the use of cotrimoxazole. The correlations were visualized as a heatmap using the corrplot function in R. Simple linear regression was used to assess the linear relationship of the significantly correlated pairs. The significant relationship was confirmed in multiple linear stepwise regression analysis. CD4+ cells were not included in the multiple linear regression analysis due to the multicollinearity with their subsets. P < 0.05 was considered significant.
Study approval
This study was carried out according to the World Medical Association’s Declaration of Helsinki, the Japanese Ethics Guidelines for Human Genome/Gene Analysis Research, and the Vietnamese Ethics Guidelines. The protocol was approved by the Ethics Committee of Kanazawa University [2011–080 (5775)] and the Ethics Committee of the VNCH (09/2012/BVNTWW-HD3), Hanoi, Vietnam. Each child’s parents or guardians were informed, and written consent was obtained for all participants. This study is registered at UMIN-CTR: UMIN000015044.
Results
Recruitment and characteristics of the study population
Approximately 500 children with HIV were followed at the VNCH in Hanoi, Vietnam, in 2012, and 40 of them did not start ART according to the Guidelines of the Ministry of Health in Vietnam (No. 3003/QĐ-BYT dated 19/08/2009) [40]. We invited all 40 of the children who did not receive ART, 30 of whom consented to participate in this study [13 females and 17 males; median age 5.9 years, range 2.0–8.8 years; HIV(+) group]. We tried to recruit age- and gender-matched children with HIV who were on ART [n = 29: 12 females and 17 males; median age 6.1 years, range 3.6–8.6 years; median duration of ART: 3.5 years, range 0.8–5.8 years; ART(+) group] and children without HIV as a control [n = 20, 8 females and 12 males; median age 4.1, range 2.0–8.3 years; HIV(–) group], though we could only recruit a smaller number of children without HIV who were 2 years younger than the HIV(+) and ART(+) groups (P = 0.048 and P = 0.009, respectively). Their detailed demographic characteristics and immune status are provided in Table 1 and elsewhere [26, 27].
Table 1. Characteristics and immune status of each study group [25].
| Characteristic | HIV(−) (n = 20) | HIV(+) (n = 30) | ART(+) (n = 29) | P-values | ||
|---|---|---|---|---|---|---|
| HIV(+) vs. HIV(−) | ART(+) vs. HIV(−) | HIV(+) vs. ART(+) | ||||
| Age (years) | 4.1 (2.0–8.3) | 5.9 (2.0–8.8) | 6.1 (3.6–8.6) | 0.048 | 0.009 | 0.44 |
| Gender, F/M | 8/12 | 13/17 | 12/17 | 0.82 | 0.92 | 0.89 |
| Height (cm) | 110 (80–130) | 107.5 (77–129.5) | 110 (90–130) | 0.88 | 0.42 | 0.41 |
| Body weight (kg) | 16 (9–35) | 17.3 (10–27) | 19.8 (12.0–32.8) | 0.47 | 0.14 | 0.14 |
| Hemoglobin (g/L) | 130.5 (114–141) | 121.5 (83–139) | 129 (104–157) | 0.001 | 0.74 | 0.001 |
| Total cholesterol (mmol/L) | 3.9 (3.2–5.0) | 2.8 (1.8–4.3) | 3.8 (2.8–5.3) | <0.001 | 0.57 | <0.001 |
| Fasting blood sugar (mmol/L) | 4.9 (4.2–5.3) | 3.8 (2.6–8.3) | 5.1 (3.5–6.0) | < 0.001 | 0.03 | <0.001 |
| WHO clinical stage, 2/1 | 1/29 | 5/24 | 0.10 | |||
| ART duration (years) | 3.5 (0.8–5.8) | |||||
| Age of ART initiation (years) | 2.7 (0.4–6.9) | |||||
| Viral load (log10 copies/μL) | 5.0 (3.2–6.5) | 3.6 (2.4–4.4)* | <0.001 | |||
| CD4+ cell count (cells/μL) | 1050 (693–2688) | 691 (97–1784) | 894 (244–1711) | 0.003 | 0.018 | 0.43 |
| Th1 count (cells/μL) | 136 (74–220) | 78 (25–227) | 147 (49–211) | 0.004 | 0.61 | 0.003 |
| Th2 count (cells/μL) | 822 (413–2196) | 532 (63–1375) | 553 (119–1369) | 0.017 | 0.009 | 0.98 |
| Th17 count (cells/μL) | 109 (51–192) | 45 (6–116) | 58 (23–144) | <0.001 | <0.001 | 0.02 |
| Treg count (cells/μL) | 48 (16–94) | 14 (0–133) | 30 (9–71) | <0.001 | 0.004 | <0.001 |
| CD8+ cell count (cells/μL) | 1101 (634–2874) | 1417 (470–3127) | 1212 (769–2064) | 0.24 | 0.29 | 0.64 |
| Activated CD8+ cells (%) | 12.9 (5.8–38.6) | 28.3 (12.2–53.3) | 10.2 (5.0–27.7) | <0.001 | 0.33 | <0.001 |
| CD4+/CD8+ ratio | 1.03 (0.45–2.34) | 0.49 (0.06–1.18) | 0.66 (0.20–1.42) | <0.001 | 0.001 | 0.19 |
Values are given as the median (range) or the number of patients. F: female: M: male; HIV(+): children with HIV and not treated with ART; ART(+): children with HIV and treated with ART; HIV(−): children not infected with HIV. P-values are based on the Mann-Whitney U test, except for the gender and WHO clinical stage comparison, which is based on the chi-squared test or Fisher’s exact test.
*Viral load was undetectable in 22 children in the ART(+) group.
The 29 children in the ART(+) group were all treated with two nucleoside reverse transcriptase inhibitors (NRTIs); 25 were also treated with one non-nucleoside reverse transcriptase inhibitor and the remaining 4 with one protease inhibitor (PI): 8 received zidovudine/lamivudine/nevirapine; 7 received stavudine/lamivudine/nevirapine; 6 received zidovudine/lamivudine/efavirenz; 4 received stavudine/lamivudine/efavirenz; 2 received zidovudine/lamivudine/lopinavir boosted with ritonavir; 1 received abacavir/lamivudine/lopinavir boosted with ritonavir; and 1 received abacavir/didanosine/lopinavir boosted with ritonavir. Fifteen children in the HIV(+) group and nine children in the ART(+) group received cotrimoxazole according to the Guidelines of the Ministry of Health in Vietnam (No. 3003/QĐ-BYT dated 19/08/2009) [40].
Gut microbiota profiles
The dominant bacteria in the gut microbiota (≥109 cells/g of feces) included C. coccoides group, C. leptum subgroup, Bifidobacterium, Atopobium cluster, B. fragilis group, and Prevotella. The sub-dominant gut microbiota (<109 cells/g) included C. difficile, C. perfringens, Streptococcus, Enterobacteriaceae, Lactobacillus spp., Enterococcus, Staphylococcus, and Pseudomonas (S1 Table). Due to the low detection frequencies of C. difficile and Pseudomonas (3.4% to 20% in all groups), these two bacteria were not included in further analyses (S2 Table).
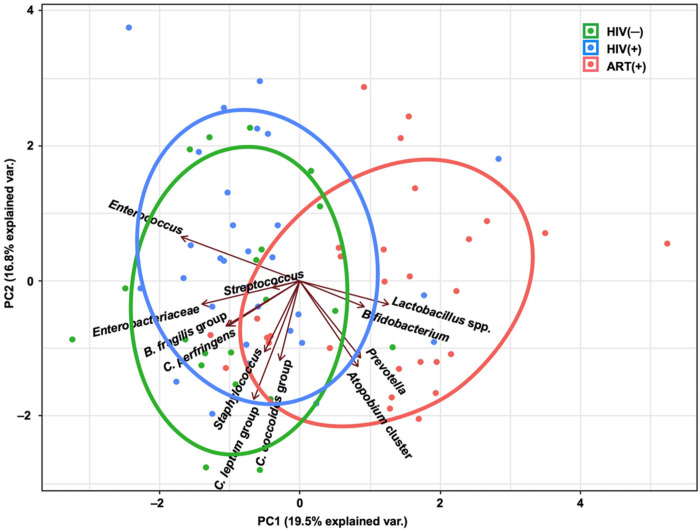
PCA revealed that the HIV(−) and HIV(+) groups had similar gut microbiota structures. The gut microbiota structure of the ART(+) group was different from the other groups and characterized by the abundance of Atopobium cluster, Bifidobacterium, Prevotella, and Lactobacillus spp. (Fig 1).
Fig 1. Principal component analysis based on the overall structure of the gut microbiota in three groups of children.
Each data point represents an individual sample. Ellipses represent the 95% confidence level. Treatment groups are color-coded: green, HIV(−); blue, HIV(+); and red, ART(+). Arrows indicate characteristic vectors of the 12 bacterial factors.
The number of bacteria in each group is shown in Fig 2. In the HIV(+) group, the numbers of C. perfringens and Atopobium cluster were significantly lower (P = 0.02 and P = 0.048, respectively) and the number of Lactobacillus spp. significantly higher (P = 0.02) than in the HIV(–) group. In the ART(+) group, the numbers of Enterococcus, B. fragilis group, and Enterobacteriaceae were significantly lower (P < 0.001, P = 0.04, P = 0.002, respectively) and the numbers of Bifidobacterium and Atopobium cluster significantly higher (both P < 0.001) than in the HIV(+) group (Fig 2 and S1 Table).
Fig 2. Box plots showing the abundance of bacteria in the gut microbiota of the three study groups.

Phylum Firmicutes: Clostridium coccoides group, C. leptum subgroup, C. perfringens, Lactobacillus spp., Streptococcus, Enterococcus, and Staphylococcus; phylum Actinobacteria: Bifidobacterium and Atopobium cluster; phylum Bacteroidetes: Bacteroides fragilis group and Prevotella; and phylum Proteobacteria: Enterobacteriaceae. The lines and error bars correspond to the medians ± 95% confidence intervals. White box, HIV(−) group; oblique line box, HIV(+) group; gray box, ART(+) group. *P < 0.05, Mann-Whitney U test. C. difficile and Pseudomonas were not included due to the low detection frequencies (3.4% to 20% in all three groups; C. difficile, median = 1.15 log10 cells/g feces, and Pseudomonas, median = 1.45 log10 cells/g feces, S2 Table).
Association between age and gut microbiota
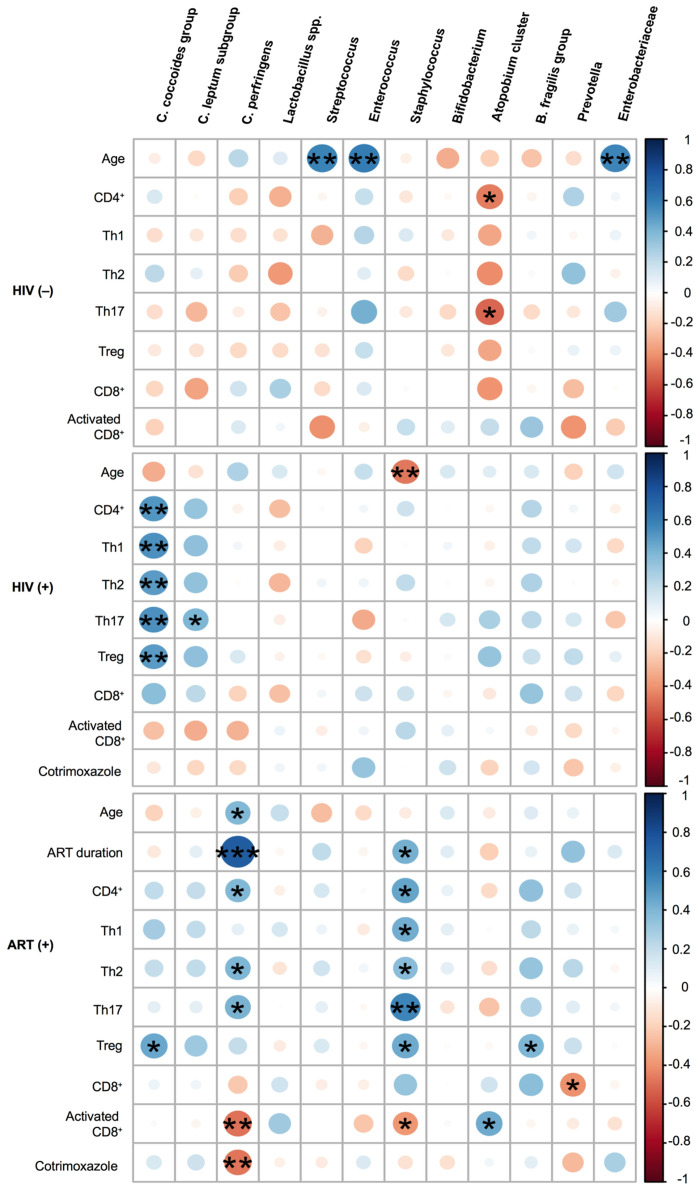
In the HIV(−) group, but not in the HIV(+) group, the numbers of Streptococcus, Enterococcus, and Enterobacteriaceae positively correlated with age (Rho = 0.59, P = 0.006; Rho = 0.61, P = 0.005; and Rho = 0.57, P = 0.008, respectively; Fig 3 and S3 Table). The number of Staphylococcus inversely correlated with age in the HIV(+) group (Rho = −0.47, P = 0.009).
Fig 3. Heatmap representing the correlation of gut microbiota with age, anti-retroviral therapy (ART) duration, immune status, and use of cotrimoxazole.
Blue shading indicates a positive association and red shading a negative association. The scale indicates the strengths of associations. C. difficile and Pseudomonas were not included due to the low detection frequencies (3.4% to 20% in all three groups: C. difficile, median = log10 1.15 cells/g feces, and Pseudomonas, median = log10 1.45 cells/g feces, S2 Table). The color intensity and size of the circles are proportional to the correlation coefficients. *P < 0.05, **P < 0.01, ***P < 0.001, based on Spearman’s rank-test.
Association between gut microbiota and immune status
In the HIV(−) group, the number of Atopobium cluster inversely correlated with CD4+ cell and Th17 counts (Rho = −0.46, P = 0.04 and Rho = −0.51, P = 0.02, respectively). In the HIV(+) group, the number of C. coccoides group significantly correlated with the CD4+ cell count and its subsets (CD4+ cells: Rho = 0.51, P = 0.004; Th1: Rho = 0.54, P = 0.002; Th2: Rho = 0.50, P = 0.005; Th17: Rho = 0.54, P = 0.002; and Treg: Rho = 0.49, P = 0.006).
In the ART(+) group, the number of Staphylococcus significantly correlated with the CD4+ cell count and its subsets (CD4+ cells: Rho = 0.46, P = 0.01; Th1: Rho = 0.44, P = 0.02; Th2: Rho = 0.37, P = 0.047; Th17: Rho = 0.58, P = 0.001; Treg: Rho = 0.45, P = 0.02), the percentage of activated CD8+ cells (Rho = −0.39, P = 0.04), and the ART duration (Rho = 0.42, P = 0.02). The number of C. perfringens significantly correlated with age (Rho = 0.39, P = 0.03), CD4+ cell count (Rho = 0.39, P = 0.04), Th2 count (Rho = 0.40, P = 0.03), Th17 count (Rho = 0.42, P = 0.03), and percentage of activated CD8+ cells (Rho = −0.49, P = 0.01), and most strongly with the ART duration (Rho = 0.75, P < 0.001). The C. coccoides group and B. fragilis group were associated with Treg cell count (Rho = 0.45, P = 0.01 and Rho = 0.40, P = 0.03, respectively). Prevotella was negatively associated with the CD8+ cell count (Rho = −0.41, P = 0.03; Fig 3 and S3 Table).
Impact of cotrimoxazole on the gut microbiota of children with HIV
Fifteen (50.0%) of the 30 children in the HIV(+) group and 9 (31%) of the 29 children in the ART(+) group received cotrimoxazole. Cotrimoxazole treatment did not significantly affect the gut microbiota profile in the HIV(+) group. However, in the ART(+) group, the number of C. perfringens was significantly lower among children treated with cotrimoxazole than those not treated with cotrimoxazole [with cotrimoxazole: 3.4 (2.2−5.4) vs. without cotrimoxazole: 6.2 (4.8−7.5), P = 0.01; S4 Table].
Independent predictors of the gut microbiota in children with HIV
The multiple linear regression analyses including age, ART duration, immune status, and use of cotrimoxazole showed that the ART duration was independently associated with the number of C. perfringens (Beta coefficient = 0.726, P < 0.001), and Th17 count with the number of Staphylococcus (Beta coefficient = 0.428, P = 0.02) in the ART(+) group (Tables 2 and 3). The linear regression analysis for C. coccoides group in the HIV(+) group showed no significant association (S5 Table).
Table 2. Linear regression analysis of Clostridium perfringens with age, ART duration, immune status, and use of cotrimoxazole in the ART(+) group.
| Unadjusted linear regression | Adjusted linear regression | |||||
|---|---|---|---|---|---|---|
| Variable | Beta | SE | P-value | Beta | SE | P-value |
| Age (years) | 0.323 | 0.261 | 0.09 | |||
| ART duration (years) | 0.726 | 0.166 | <0.001 | 0.726 | 0.166 | <0.001 |
| Th2 count | 0.418 | 0.001 | 0.024 | |||
| Th17 count | 0.371 | 0.015 | 0.048 | |||
| Activated CD8+ cells (%) | -0.484 | 0.068 | 0.008 | |||
| Cotrimoxazole use (yes vs. no) | -0.446 | 0.809 | 0.015 | |||
ART: anti-retroviral therapy; Beta: regression coefficient; SE: standard error.
The factors with P < 0.05 in the simple linear regression analysis were included in the stepwise multiple linear regression analysis. P-values in bold are significant.
Table 3. Linear regression analysis of Staphylococcus with ART duration and immune status in the ART(+) group.
| Unadjusted linear regression | Adjusted linear regression | |||||
|---|---|---|---|---|---|---|
| Variable | Beta | SE | P-value | Beta | SE | P-value |
| ART duration (years) | 0.370 | 0.109 | 0.048 | |||
| Th1 count | 0.309 | 0.005 | 0.103 | |||
| Th2 count | 0.279 | 0.001 | 0.142 | |||
| Th17 count | 0.428 | 0.007 | 0.020 | 0.428 | 0.007 | 0.020 |
| Treg count | 0.244 | 0.012 | 0.202 | |||
| Activated CD8+ cells (%) | -0.305 | 0.036 | 0.108 | |||
ART: anti-retroviral therapy; Treg: regulatory T cells; Beta: regression coefficient; SE: standard error. The factors with P < 0.05 in the simple linear regression analysis were included in the stepwise multiple linear regression analysis. P-values in bold are significant.
Discussion
In the present study, we investigated the impact of HIV infection and ART on the gut microbiota of Vietnamese children using RT-qPCR. We found that several sub-dominant gut bacteria were positively associated with age in children without HIV, but this was not observed in the children with HIV. In addition, Staphylococcus negatively correlated with age, i.e. the duration of HIV infection, in the children vertically infected with HIV, and ART duration had an independent positive association with C. perfringens and Th17 count with Staphylococcus in the HIV-infected children on ART. These findings indicate an impact of HIV infection and ART on the sub-dominant gut microbiota, such as C. perfringens and Staphylococcus, in children. Our findings highlight the importance of investigating the role of the sub-dominant gut microbiota in the pathogenesis of HIV infection.
To the best of our knowledge, this study is the first to apply RT-qPCR techniques to evaluate the gut microbiota, particularly sub-dominant bacteria, in children with and without HIV. The sum of the six dominant bacterial groups in fecal samples detected by RT-qPCR was previously reported to cover 71.3% of the total intestinal bacterial count estimated by hybridization with a universal probe [29, 41]. This finding ensures the validity of using RT-qPCR methods to identify the main gut microbiota profile in this study, though our results may not be comparable directly to the results of the other studies using NGS, since the RT-qPCR method is not appropriate to calculate the diversity indices and the relative abundance of the selected bacteria. In addition, the RT-qPCR approach can detect and enumerate the gut bacteria at the population level between 102 and 1011 cells/g of feces, whereas the lower detection limit of next generation sequencing (NGS) methods is 107 to 108 cells/g [33]. The counts of the sub-dominant bacteria, including Lactobacilli and potential pathogens, such as C. perfringens, were near the detection limit of NGS or lower [33]; thus, we took advantage of RT-qPCR to estimate the counts of these less abundant, but clinically significant, targets.
In this study, the numbers of dominant gut bacteria, including C. coccoides group, C. leptum subgroup, Bifidobacterium, and Atopobium cluster, did not correlate with age in the HIV(−) group, i.e., children aged 2 to 8 years. This finding is consistent with previous findings that the gut microbiota of healthy children stabilizes and becomes similar to that of adults at around 2 or 3 years of age [7, 21, 22, 33]. In contrast, several sub-dominant gut bacteria, including Streptococcus, Enterococcus, and Enterobacteriaceae, positively correlated with age in the HIV(−) group. This finding is also consistent with previous findings in Japanese children evaluated using the same RT-qPCR methods [33]. These results suggest that the dominant gut microbiota may reach stable levels by 2 or 3 years of age, whereas the sub-dominant bacteria may still be in transition in children aged 2 to 8 years.
We found that C. coccoides group positively correlated with the CD4+ cell count and its subsets in the HIV(+) group, as well as the Treg cell count in the ART(+) group. The C. coccoides spp. are known to promote the expansion and differentiation of Treg cells, which play a central role in regulating gut inflammation through the production of butyrate, a short-chain fatty acid (SCFA), in mice [42, 43]. In the ART(+) group, the B. fragilis group positively correlated with the Treg cell count. This finding is consistent with a previous study that found that B. fragilis promotes Treg cell function by producing polysaccharide A [44]. Thus, HIV infection and ART may also influence the immune status by changing the levels of gut bacterial metabolites, such as SCFAs [45, 46]. Further studies on bacterial metabolites and their networks in the gut microbiota of children with HIV treated with and without ART may elucidate the underlying mechanisms of immune modulation in HIV infection and ART interventions.
Multiple regression analysis showed a positive association between gut Staphylococcus and Th17 count in the ART(+) group, which was shown for the first time. Th17 cells produce interleukin-17, which is important for promoting neutrophil recruitment to clear bacteria and has a specific role in the host defense against Staphylococcus aureus skin infection [47]. Thus, it would be interesting to investigate the interaction between Th17 and gut Staphylococcus in order to understand the pathophysiology of HIV infection in children who are on ART.
The use of cotrimoxazole reportedly influences some gut bacteria and reduces gut inflammation in children with HIV [48–50]. In the current study, the use of cotrimoxazole was associated only with C. perfringens in the ART(+) group. However, in multiple regression analysis, we found that ART duration, but not the use of cotrimoxazole, was independently associated with C. perfringens, which is a potentially harmful bacterium [51]. These findings suggest that a novel therapeutic approach, such as ingesting probiotics and/or prebiotics, may be necessary to restore gut microbiota homeostasis in children with HIV who are on ART [52].
In this study, all of the children in the ART(+) group were treated with NRTIs as backbone drugs and only four also received a PI. Therefore, the positive correlation between the number of C. perfringens and ART duration may be due to the use of NRTIs, but not PIs, even though PIs are known to lower the diversity of the gut microbiota [53, 54]. Our study is consistent with the previous study that ART, especially NRTI-including regimen, had more suppressive impacts on the composition and the variability of the gut microbiota [55]. Further study is needed to investigate whether NRTIs influence the gut microbiota, directly or indirectly, through the restoration of immune status in children with HIV who are on ART.
This study has some limitations. First, the children in the HIV(−) group were 2 years younger than the other groups. The diets were not controlled among the groups, though the children in the HIV(−) and ART(+) groups who resided at orphanages were provided the same diets. The children in the HIV(+) group who resided in their own homes appeared to have poorer nutritional status than the children in other groups, which could be due to the uncontrolled diet and/or HIV infection [26]. Considering the influence of age and diet factors on the gut microbiota [7, 21, 22, 33, 56–58], we did not focus on comparing the gut microbiota between the groups, but highlighted the factors associated with the gut microbiota in each group. Second, the number of patients recruited in the present study was relatively small, which may limit the significance of our findings. Third, we have not mentioned the HIV-exposure history of the children in the HIV(−) group because we could not confirm the history via documents, though they were reportedly born to mothers with HIV. Machiavelli et al. reported that the gut microbiota of HIV-exposed but uninfected children is similar to that of HIV-unexposed and uninfected children at the age of 18 months except in several taxa [59]. Therefore, the impact of HIV-exposure history on the gut microbiota in the children without HIV over 2 years of age would be limited. These findings require confirmation in longitudinal studies that compare the gut microbiome between age-matched children with and without HIV, with and without HIV-exposure history, and/or before and after initiating ART to assess the effect of ART on the composition of the gut microbiota in children with HIV.
This study provided new insights into the alterations in the gut microbiota, particularly the sub-dominant groups of bacteria, among children with HIV. Our results suggest that HIV infection and ART influence the sub-dominant gut microbiota, directly or indirectly, in association with the immune status of children with HIV.
Supporting information
CD8+ cell activations was defined as the CD38+HLA-DR+ population. Regulatory T (Treg) cells were defined as CD25highCD4+ cells, Th1 as CXCR3+CCR6−CD25lowCD4+ cells, Th2 as CXCR3−CCR6−CD25lowCD4+ cells, and Th17 as CXCR3−CCR6+CD25lowCD4+ cells. Unstained cells were used for gating controls.
(TIF)
Values are the median counts (IQR), based on RT-qPCR, expressed in units of log10 cells/g feces. The Lactobacillus spp. counts were obtained with RT-qPCR and are expressed as the sum of the six subgroups and two species; ART: anti-retroviral therapy; C.: Clostridium; L.: Lactobacillus; B.: Bacteroides. P-values in bold are statistically significant, based on the Man-Whitney U test.
(DOCX)
Values are the detection frequency, defined as the % of samples that harbored detectable microbiota, among all samples in a given group. The Lactobacillus spp. counts were obtained with RT-qPCR and are expressed as the sum of the six subgroups and two species; ART: anti-retroviral therapy; C.: Clostridium; L.: Lactobacillus; B.: Bacteroides; P-values in bold are statistically significant, based on the Chi-square test or Fisher’s exact probability test.
(DOCX)
ART: anti-retroviral therapy; Treg: regulatory T cell; C.: Clostridium; B.: Bacteroides; P-values in bold are statistically significant, based on Spearman’s rank correlation analysis.
(DOCX)
Values are the median counts (IQR), based on RT-qPCR, expressed in units of log10 cells/g feces; ART: anti-retroviral therapy; C.: Clostridium; B.: Bacteroides; P-values in bold are statistically significant, based on the Mann−Whitney U test.
(DOCX)
ART: anti-retroviral therapy; Treg: regulatory T cells; Beta: regression coefficient; SE: standard error. Stepwise multiple linear regression analysis was not done, since no factor with P < 0.05 was found in the simple linear regression analysis.
(DOCX)
Acknowledgments
We would like to thank the children who participated in this study for their invaluable support through the use of their samples. We would also like to thank Ms. Thuy Thi Thanh Giang and other staff at the Department of Infectious Diseases and Molecular Laboratory of VNCH, who contributed enormously to this work.
Data Availability
All relevant data are within the manuscript and its Supporting information files.
Funding Statement
This study was supported, in part, by the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) in Japan (the Program of Founding Research Centers for Emerging and Reemerging Infectious Disease; grant number 23406013, https://www.mext.go.jp/en/publication/whitepaper/title03/detail03/sdetail03/sdetail03/1372928.htm) and the Kanazawa University President Strategic Research Fund (https://www.kanazawa-u.ac.jp/research_bulletin/contact.html). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464(7285):59–65. doi: 10.1038/nature08821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124(4):837–48. doi: 10.1016/j.cell.2006.02.017 [DOI] [PubMed] [Google Scholar]
- 3.Kamada N, Nunez G. Regulation of the immune system by the resident intestinal bacteria. Gastroenterology. 2014;146(6):1477–88. doi: 10.1053/j.gastro.2014.01.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flint HJ, Scott KP, Louis P, Duncan SH. The role of the gut microbiota in nutrition and health. Nat Rev Gastroenterol Hepatol. 2012;9(10):577–89. doi: 10.1038/nrgastro.2012.156 [DOI] [PubMed] [Google Scholar]
- 5.Maslowski KM, Mackay CR. Diet, gut microbiota and immune responses. Nat Immunol. 2011;12(1):5–9. doi: 10.1038/ni0111-5 [DOI] [PubMed] [Google Scholar]
- 6.Mariat D, Firmesse O, Levenez F, Guimaraes V, Sokol H, Dore J, et al. The Firmicutes/Bacteroidetes ratio of the human microbiota changes with age. BMC Microbiol. 2009;9:123. doi: 10.1186/1471-2180-9-123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486(7402):222–7 doi: 10.1038/nature11053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng D, Liwinski T, Elinav E. Interaction between microbiota and immunity in health and disease. Cell Res. 2020;30(6):492–506. doi: 10.1038/s41422-020-0332-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kamada N, Seo SU, Chen GY, Núñez G. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol. 2013;13(5):321–35. doi: 10.1038/nri3430 [DOI] [PubMed] [Google Scholar]
- 10.Sengupta S, Siliciano RF. Targeting the Latent Reservoir for HIV-1. Immunity. 2018;48(5):872–95. doi: 10.1016/j.immuni.2018.04.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mehandru S, Poles MA, Tenner-Racz K, Horowitz A, Hurley A, Hogan C, et al. Primary HIV-1 infection is associated with preferential depletion of CD4+ T lymphocytes from effector sites in the gastrointestinal tract. J Exp Med. 2004;200(6):761–70. doi: 10.1084/jem.20041196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Veazey RS, DeMaria M, Chalifoux LV, Shvetz DE, Pauley DR, Knight HL, et al. Gastrointestinal tract as a major site of CD4+ T cell depletion and viral replication in SIV infection. Science. 1998;280(5362):427–31. doi: 10.1126/science.280.5362.427 [DOI] [PubMed] [Google Scholar]
- 13.Brenchley JM, Paiardini M, Knox KS, Asher AI, Cervasi B, Asher TE, et al. Differential Th17 CD4 T-cell depletion in pathogenic and nonpathogenic lentiviral infections. Blood. 2008;112(7):2826–35. doi: 10.1182/blood-2008-05-159301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hunt PW. Th17, gut, and HIV: therapeutic implications. Curr Opin HIV AIDS. 2010;5(2):189–93. doi: 10.1097/COH.0b013e32833647d9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou Y, Ou Z, Tang X, Zhou Y, Xu H, Wang X, et al. Alterations in the gut microbiota of patients with acquired immune deficiency syndrome. J Cell Mol Med. 2018;22(4):2263–71. doi: 10.1111/jcmm.13508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lozupone CA, Li M, Campbell TB, Flores SC, Linderman D, Gebert MJ, et al. Alterations in the gut microbiota associated with HIV-1 infection. Cell Host Microbe. 2013;14(3):329–39. doi: 10.1016/j.chom.2013.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dillon SM, Lee EJ, Kotter CV, Austin GL, Dong Z, Hecht DK, et al. An altered intestinal mucosal microbiome in HIV-1 infection is associated with mucosal and systemic immune activation and endotoxemia. Mucosal Immunol. 2014;7(4):983–94. doi: 10.1038/mi.2013.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McHardy IH, Li X, Tong M, Ruegger P, Jacobs J, Borneman J, et al. HIV Infection is associated with compositional and functional shifts in the rectal mucosal microbiota. Microbiome. 2013;1(1):26. doi: 10.1186/2049-2618-1-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ringel-Kulka T, Cheng J, Ringel Y, Salojarvi J, Carroll I, Palva A, et al. Intestinal microbiota in healthy U.S. young children and adults—a high throughput microarray analysis. PLoS One. 2013;8(5):e64315. doi: 10.1371/journal.pone.0064315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Derrien M, Alvarez AS, de Vos WM. The Gut Microbiota in the First Decade of Life. Trends Microbiol. 2019;27(12):997–1010. doi: 10.1016/j.tim.2019.08.001 [DOI] [PubMed] [Google Scholar]
- 21.Stewart CJ, Ajami NJ, O’Brien JL, Hutchinson DS, Smith DP, Wong MC, et al. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature. 2018;562(7728):583–8. doi: 10.1038/s41586-018-0617-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, et al. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A. 2011;108 Suppl 1:4578–85. doi: 10.1073/pnas.1000081107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaur US, Shet A, Rajnala N, Gopalan BP, Moar P, D H, et al. High Abundance of genus Prevotella in the gut of perinatally HIV-infected children is associated with IP-10 levels despite therapy. Sci Rep. 2018;8(1):17679. doi: 10.1038/s41598-018-35877-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flygel TT, Sovershaeva E, Classen-Weitz S, Hjerde E, Mwaikono KS, Odland JO, et al. Composition of gut microbiota of children and adolescents with perinatal HIV infection taking antiretroviral therapy in Zimbabwe. J Infect Dis. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abange WB, Martin C, Nanfack AJ, Yatchou LG, Nusbacher N, Nguedia CA, et al. Alteration of the gut fecal microbiome in children living with HIV on antiretroviral therapy in Yaounde, Cameroon. Sci Rep. 2021;11(1):7666 doi: 10.1038/s41598-021-87368-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bi X, Ishizaki A, Nguyen LV, Matsuda K, Pham HV, Phan CT, et al. Impact of HIV Infection and Anti-Retroviral Therapy on the Immune Profile of and Microbial Translocation in HIV-Infected Children in Vietnam. Int J Mol Sci. 2016;17(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ishizaki A, Bi X, Nguyen LV, Matsuda K, Pham HV, Phan CTT, et al. Effects of Short-Term Probiotic Ingestion on Immune Profiles and Microbial Translocation among HIV-1-Infected Vietnamese Children. Int J Mol Sci. 2017;18(10). doi: 10.3390/ijms18102185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kurakawa T, Kubota H, Tsuji H, Matsuda K, Asahara T, Takahashi T, et al. Development of a sensitive rRNA-targeted reverse transcription-quantitative polymerase chain reaction for detection of Vibrio cholerae/mimicus, V. parahaemolyticus/alginolyticus and Campylobacter jejuni/coli. Microbiol Immunol. 2012;56(1):10–20. doi: 10.1111/j.1348-0421.2011.00405.x [DOI] [PubMed] [Google Scholar]
- 29.Matsuda K, Tsuji H, Asahara T, Matsumoto K, Takada T, Nomoto K. Establishment of an analytical system for the human fecal microbiota, based on reverse transcription-quantitative PCR targeting of multicopy rRNA molecules. Appl Environ Microbiol. 2009;75(7):1961–9. doi: 10.1128/AEM.01843-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsuda K, Tsuji H, Asahara T, Takahashi T, Kubota H, Nagata S, et al. Sensitive quantification of Clostridium difficile cells by reverse transcription-quantitative PCR targeting rRNA molecules. Appl Environ Microbiol. 2012;78(15):5111–8. doi: 10.1128/AEM.07990-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matsuki T, Watanabe K, Fujimoto J, Takada T, Tanaka R. Use of 16S rRNA gene-targeted group-specific primers for real-time PCR analysis of predominant bacteria in human feces. Appl Environ Microbiol. 2004;70(12):7220–8. doi: 10.1128/AEM.70.12.7220-7228.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sakaguchi S, Saito M, Tsuji H, Asahara T, Takata O, Fujimura J, et al. Bacterial rRNA-targeted reverse transcription-PCR used to identify pathogens responsible for fever with neutropenia. J Clin Microbiol. 2010;48(5):1624–8. doi: 10.1128/JCM.01724-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsuji H, Matsuda K, Nomoto K. Counting the Countless: Bacterial Quantification by Targeting rRNA Molecules to Explore the Human Gut Microbiota in Health and Disease. Front Microbiol. 2018;9:1417. doi: 10.3389/fmicb.2018.01417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harmsen HJ, Raangs GC, He T, Degener JE, Welling GW. Extensive set of 16S rRNA-based probes for detection of bacteria in human feces. Appl Environ Microbiol. 2002;68(6):2982–90. doi: 10.1128/AEM.68.6.2982-2990.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.González-Hernández LA, Ruiz-Briseño MDR, Sánchez-Reyes K, Alvarez-Zavala M, Vega-Magaña N, López-Iñiguez A, et al. Alterations in bacterial communities, SCFA and biomarkers in an elderly HIV-positive and HIV-negative population in western Mexico. BMC Infect Dis. 2019;19(1):234. doi: 10.1186/s12879-019-3867-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hunt PW, Martin JN, Sinclair E, Bredt B, Hagos E, Lampiris H, et al. T cell activation is associated with lower CD4+ T cell gains in human immunodeficiency virus-infected patients with sustained viral suppression during antiretroviral therapy. J Infect Dis. 2003;187(10):1534–43. doi: 10.1086/374786 [DOI] [PubMed] [Google Scholar]
- 37.Maecker HT, McCoy JP, Nussenblatt R. Standardizing immunophenotyping for the Human Immunology Project. Nat Rev Immunol. 2012;12(3):191–200. doi: 10.1038/nri3158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kinter A, McNally J, Riggin L, Jackson R, Roby G, Fauci AS. Suppression of HIV-specific T cell activity by lymph node CD25+ regulatory T cells from HIV-infected individuals. Proc Natl Acad Sci U S A. 2007;104(9):3390–5. doi: 10.1073/pnas.0611423104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Team R. R:A language and environment for statistical computing. R Foundation for Statistical Computing Vienna, Austria; 2018. https://www.r-project.org.
- 40.Ministry of Health, Vietnam. Guidelines for HIV/AIDS Diagnosis and Treatment; 2009. https://www.who.int/hiv/pub/guidelines/vietnam_art.pdf?ua=1.
- 41.Hasegawa S, Goto S, Tsuji H, Okuno T, Asahara T, Nomoto K, et al. Intestinal Dysbiosis and Lowered Serum Lipopolysaccharide-Binding Protein in Parkinson’s Disease. PLoS One. 2015;10(11):e0142164. doi: 10.1371/journal.pone.0142164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504(7480):446–50. doi: 10.1038/nature12721 [DOI] [PubMed] [Google Scholar]
- 43.Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500(7461):232–6. doi: 10.1038/nature12331 [DOI] [PubMed] [Google Scholar]
- 44.Telesford KM, Yan W, Ochoa-Reparaz J, Pant A, Kircher C, Christy MA, et al. A commensal symbiotic factor derived from Bacteroides fragilis promotes human CD39(+)Foxp3(+) T cells and Treg function. Gut Microbes. 2015;6(4):234–42. doi: 10.1080/19490976.2015.1056973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Serrano-Villar S, Rojo D, Martínez-Martínez M, Deusch S, Vázquez-Castellanos JF, Bargiela R, et al. Gut Bacteria Metabolism Impacts Immune Recovery in HIV-infected Individuals. EBioMedicine. 2016;8:203–16. doi: 10.1016/j.ebiom.2016.04.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vázquez-Castellanos JF, Serrano-Villar S, Latorre A, Artacho A, Ferrús ML, Madrid N, et al. Altered metabolism of gut microbiota contributes to chronic immune activation in HIV-infected individuals. Mucosal Immunol. 2015;8(4):760–72. doi: 10.1038/mi.2014.107 [DOI] [PubMed] [Google Scholar]
- 47.Miller LS, Cho JS. Immunity against Staphylococcus aureus cutaneous infections. Nature Reviews Immunology. 2011;11(8):505–518. doi: 10.1038/nri3010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bourke CD, Gough EK, Pimundu G, Shonhai A, Berejena C, Terry L, et al. Cotrimoxazole reduces systemic inflammation in HIV infection by altering the gut microbiome and immune activation. Sci Transl Med. 2019;11(486). doi: 10.1126/scitranslmed.aav0537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gough EK, Bourke CD, Berejena C, Shonhai A, Bwakura-Dangarembizi M, Prendergast AJ, et al. Strain-level analysis of gut-resident pro-inflammatory viridans group Streptococci suppressed by long-term cotrimoxazole prophylaxis among HIV-positive children in Zimbabwe. Gut Microbes. 2020;11(4):1104–15. doi: 10.1080/19490976.2020.1717299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kyosiimire-Lugemwa J, Anywaine Z, Abaasa A, Levin J, Gombe B, Musinguzi K, et al. Effect of Stopping Cotrimoxazole Preventive Therapy on Microbial Translocation and Inflammatory Markers Among Human Immunodeficiency Virus-Infected Ugandan Adults on Antiretroviral Therapy: The COSTOP Trial Immunology Substudy. J Infect Dis. 2020;222(3):381–90. doi: 10.1093/infdis/jiz494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kiu R, Hall LJ. An update on the human and animal enteric pathogen Clostridium perfringens. Emerg Microbes Infect. 2018;7(1):141. doi: 10.1038/s41426-018-0144-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Monteiro CRAV, do Carmo MS, Melo BO, Alves MS, Dos Santos CI, Monteiro SG, et al. In Vitro Antimicrobial Activity and Probiotic Potential of. Nutrients. 2019;11(2). doi: 10.3390/nu11020448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pinto-Cardoso S, Klatt NR, Reyes-Terán G. Impact of antiretroviral drugs on the microbiome: unknown answers to important questions. Curr Opin HIV AIDS. 2018;13(1):53–60. doi: 10.1097/COH.0000000000000428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Villanueva-Millan MJ, Perez-Matute P, Recio-Fernandez E, Lezana Rosales JM, Oteo JA. Differential effects of antiretrovirals on microbial translocation and gut microbiota composition of HIV-infected patients. J Int AIDS Soc. 2017;20(1):21526. doi: 10.7448/IAS.20.1.21526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Imahashi M, Ode H, Kobayashi A, Nemoto M, Matsuda M, Hashiba C, et al. Impact of long-term antiretroviral therapy on gut and oral microbiotas in HIV-1-infected patients. Sci Rep. 2021;11(1):960. doi: 10.1038/s41598-020-80247-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Singh RK, Chang HW, Yan D, Lee KM, Ucmak D, Wong K, et al. Influence of diet on the gut microbiome and implications for human health. J Transl Med. 2017;15(1):73. doi: 10.1186/s12967-017-1175-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nguyen DT, Van Hoorde K, Cnockaert M, De Brandt E, Aerts M, Binh Thanh L, et al. A description of the lactic acid bacteria microbiota associated with the production of traditional fermented vegetables in Vietnam. Int J Food Microbiol. 2013;163(1):19–27. doi: 10.1016/j.ijfoodmicro.2013.01.024 [DOI] [PubMed] [Google Scholar]
- 58.Surono IS, Widiyanti D, Kusumo PD, Venema K. Gut microbiota profile of Indonesian stunted children and children with normal nutritional status. PLoS One. 2021;16(1):e0245399. doi: 10.1371/journal.pone.0245399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Machiavelli A, Duarte RTD, Pires MMS, Zárate-Bladés CR, Pinto AR. The impact of in utero HIV exposure on gut microbiota, inflammation, and microbial translocation. Gut Microbes. 2019;10(5):599–614. doi: 10.1080/19490976.2018.1560768 [DOI] [PMC free article] [PubMed] [Google Scholar]