Abstract
The mission of translational science is to bring predictivity and efficiency to the development and dissemination of interventions that improve human health. Ten years ago this year, the National Center for Advancing Translational Sciences was founded to embody, conduct, and support this new discipline. The Center’s first decade has brought substantial progress across a broad range of translational areas, from diagnostic and drug development to clinical trials to implementation science to education. The origins of the translational science and advances to this point are reviewed here and allow the establishment of an ambitious future research agenda for the field.
INTRODUCTION
A foundational premise of modern biomedical research is that fundamental discovery provides the basis for the development of interventions to ameliorate and cure disease. 1 It is largely this premise that has led to the growth of the biomedical scientific enterprise over the last 60 years, and its abiding support from the public. Although undoubtedly true, this construct is also manifestly incomplete. Some of the most commonly used and still most effective medications—morphine for pain, aspirin for inflammation, and lithium for bipolar disorder among them—were developed without knowledge of these diseases’ underlying biology. In addition, more recently, the spectacular achievements of fundamental science have not been accompanied by commensurate improvements in our ability to diagnose and treat most diseases. These observations do not serve to undermine the value of fundamental biomedical science or the importance of maintaining it as a robust and growing enterprise. Rather, it suggests that our common conception of how treatments and cures are developed and deployed is incomplete; in other words, as happens so often in science, the prevailing model has turned out to be overly simplistic.
“Translation” is the term commonly used to describe the process by which a biomedical observation is turned into an intervention that improves health. “Translational science” is the field which studies the translational process in order to establish its governing scientific principles, thereby transforming translation, as has occurred in other sciences before it, from empiricism to predictivity. 2 Translational science begins, therefore, as all sciences do, with observations that cannot be explained by current theory—in this case, the observation that the avalanche of successful fundamental discovery has not led to the expected therapeutic windfall. Understanding and ameliorating the causes of this observed divergence is essential to the promise of science reaching all patients in need, and in no small measure will determine how the public views the success of the medical science enterprise and their investment in it.
The origins of translation
Although the term translation is relatively new in the biomedical lexicon, becoming widely used only in the 1970s, the activity is ancient, tracing its lineage to efforts by physicians and traditional healers in antiquity to identify and codify substances and procedures that could heal the sick. Importantly, therefore, translation is rooted in the immediate medical needs of patients, not in science. It has always esteemed practical if anecdotal successes over theory‐driven systematic learning. The practical focus of translation was reinforced in the pharmaceutical industry, which grew out of apothecaries and dye and chemical companies where commercial demands required prioritization of saleable successes; understanding of molecular mechanisms and success/failure factors were considered secondarily or not at all. Until just the last century, translational efforts began with public health or clinical observations, which led to identification of behavioral interventions or isolation of active biological substances of varying purity; the oft‐cited examples of John Snow’s water pump handle intervention in the 1854 London cholera epidemic, and liver extract to treat pernicious anemia in the 1920s, are archetypes of the clinicoempirical nature of most translational history. Only with the development of increasingly reductionistic scientific techniques during the 20th century—from physiology to pharmacology to microbiology to protein chemistry to molecular cloning—did it become possible to begin a translational effort from a nonclinical starting point. In addition, only in the last several decades has it become routine to start translational efforts with molecular targets having only circumstantial connection to a disease.
Thus, whereas progress in both fundamental discovery and translation remained tightly coupled to clinical observations for most of their histories, breathtaking progress in basic biomedical science has far outpaced translational or clinical science in the last 40 years. The striking divergence in the pace of progress in these previously coupled fields is evident in everyday experience: whereas the capacities and activities of the basic research laboratory are virtually unrecognizable compared to 40 years ago, the capacities and activities of most medical practice remain largely unchanged, and translation has become progressively less productive over that time. 3
THE BEST AND WORST OF TIMES
Biomedical research in the third decade of the 21st century is thus beset by paradoxes. On the one hand, we are fantastically knowledgeable about genetic, cellular, and organismal physiology in health and disease, thanks to the epochal achievements of fundamental science because the current model for support of research was established at the end of World War II. Forty years ago, the number of human diseases the molecular basis of which was understood was less than 20; today it is almost 7000. This avalanche of discovery has fundamentally transformed our knowledge of ourselves in health and disease. However, with a few notable exceptions—mainly in cancer and infectious disease—our ability to effectively treat the diseases we now understand remains limited: of the ~ 8000 diseases that affect humans, less than 600 have any regulatorily approved treatment, and most of these are symptomatic (i.e., not disease‐modifying). The health of the American population remains frustratingly poor overall. 4 This failure of translation is not for lack of effort; over $50 billion are spent on translational efforts in the public and private sectors every year. Opportunities for improvement abound: the cost of developing a new drug is now over $2 billion, continues to increase faster than inflation, and has increased relentlessly since 1950 despite enormous advances in science 5 ; the clinical trials process is widely acknowledged to be inefficient 6 , 7 ; after a drug or intervention is shown to be useful, its dissemination to all patients who could benefit is slow and variable, 8 , 9 and patient adherence to those interventions remains suboptimal and limits the health benefits of the interventions developed. 10 In all, the time required for the end‐to‐end translational process, from an idea in the laboratory to a drug or other intervention based on that idea reaching all patients who could benefit, is currently over 20 years, and its success rate is below 1%. Inspiring individual examples of remarkably effective therapies—including enzyme replacement therapies for particularly rare diseases, small molecule correctors for cystic fibrosis, PCSK9 inhibitors for elevated cholesterol, and gene therapies for forms of inherited blindness and spinal muscular atrophy—demonstrate what a future therapeutic world might hold. But those successes remain the exceptions, with development times still measured in decades and resulting products having extremely high costs that are arguably unsustainable, and certainly undesirable. In addition, while there have recently been some encouraging signs of improved overall timelines and success rates, productivity (success per unit of effort) and timelines have worsened over the last 40 years, in stark contrast to the remarkable increase in productivity in fundamental research during this time.
These divergences have been recognized for some time, 11 , 12 and a variety of efforts have been undertaken to address it. In the private sector, multiple mergers and acquisitions were undertaken in an attempt to achieve economies of scale, and the multicompany collaborative TransCelerate Biopharma was formed in 2012 to create platforms for sharing among companies. Public‐private partnerships, including the Innovative Medicines Initiative (IMI) in Europe, and the Accelerating Medicines Partnerships (AMP) in the United States, have produced substantial datasets that are benefitting the entire research ecosystem. In the academic sector, the National Institutes of Health (NIH) Roadmap aimed to “redefine the ways that medical research is conducted and, ultimately, how research leads to improvements in health.” 13 Among the Roadmap programs, were several aimed at the translational divide. The Molecular Libraries Initiative 14 created a robust small molecule assay development, screening, and chemistry probe development capacity in the public sector for the first time, and was remarkably productive during its 10‐year lifetime. 15 The Clinical and Translational Science Awards (CTSA) program 16 has been similarly transformative, increasing the academic standing of the clinical and translational sciences by creating intellectual “homes” for these disciplines in academia, and providing critical clinical trial, biostatistics, informatics, and regulatory support, and innovative education programs.
For the most part, these efforts were focused on structural issues that had prevented efficient translational and clinical research, and collaborative sharing of capacities and insights. They changed how translational and clinical research is done for the better, allowing more projects to be carried out. But with rare exceptions they did not change what was done in the translational process; and so, despite notable individual successes from these and other efforts, overall success rates in translation did not improve. A new approach was needed, 17 and this led to the formation of a new NIH center, the National Center for Advancing Translational Sciences (Box 1).
BOX 1. Creation of the National Center for Advancing Translational Sciences.
In 2010, an advisory group of the National Institutes of Health proposed a new component of NIH be created, focused on the science of translation (https://smrb.od.nih.gov//documents/reports/TMAT_122010.pdf). That proposal led to the formation of the National Center for Advancing Translational Sciences (NCATS) in 2011. Created in a time of fiscal restraint, NCATS was not appropriated new funding when it was authorized by Congress; rather, it was formed via an NIH internal reorganization, bringing together four programs that previously were resident in other parts of NIH. These included the CTSA program from the former National Center for Research Resources, the NIH Center for Translational Therapeutics from the National Human Genome Research Institute, the Office of Rare Diseases Research from the NIH Office of the Director, and the Cures Acceleration Network, also from the NIH Office of the Director.
The mission stated for the new NCATS was to “catalyze the generation of innovative methods and technologies” to enhance the development, testing, and implementation of diagnostics and therapeutics across human diseases and conditions. It was to transform the translational process and eliminate the basic‐health divide, delivering on the promise of science for medicine. It was to focus not on incremental science, but rather on “disruptive translational innovation.” 17
The first priority of the new Center was to define and promulgate its mission, which was initially poorly understood. 18 The term that defined NCATS’ mission—“translation”—had other common scientific and lay meanings, leading to misunderstandings that the NCATS’ mission was linguistics, or how proteins are produced from mRNA. Therefore, the Center undertook a deliberate process to define “translation” and its congeners “translational research” and “translational science,” 2 and emphasized that while it studies the first (translation) as a process, and performs the second (translational research), what distinguishes the Center from any other organization in the United States or internationally is its focus on the third—translational science—as a discipline.
The initial focus areas for the new center were formulated via a series of meetings and consultations with experts on particular topics (https://ncats.nih.gov/about/center). These led to the creation of the first NCATS Strategic Plan (https://ncats.nih.gov/strategicplan), and as well as a series of long‐term goals to guide its activities.
THE NEED FOR A SCIENCE OF TRANSLATION
The contrasting productivity of basic and translational research have been mirrored by opposing conceptual approaches in the 2 fields over the last 75 years. Since the publication of Science: The Endless Frontier 1 in 1945, the dominant—and successful—ethos of basic biomedical science has been the need to systematically understand the processes which underlie physiological function and dysfunction. In contrast, translation has its historical roots in medical practice, so has traditionally been empirical and practical. Identification of new pharmaceutical, behavioral, or surgical interventions was driven by individual, often initially serendipitous, observations that became accepted often more because of common use than solid data or science‐driven understanding. The legacy of this history is that translation is, even today, an almost relentlessly empirical process. Efforts to increase productivity over the last 3 decades have repeatedly been to front‐load the translational system, with more genes/targets (the Human Genome Project), more chemical starting points (combinatorial chemistry), more biologically active compounds (high throughput screening), more “technology transfer” of basic discoveries to companies, or more projects (“shots on goal”). Given the enormous complexity of living systems, the multistep nature of translation, and the increase in degrees of freedom from protein to cell to organ to animal to human to community, a trial‐and‐error approach was always unlikely to succeed, and indeed failure is the most common outcome, leading to the “valley of death” characterization of translation. But when faced with this reality, the translational community has tended to double down on empiricism, with increased the numbers of projects, or a new structure of the people doing them, remaining the dominant solutions. As a result, understanding of underlying phenomena sufficient to enable increasing predictability has largely not occurred in translation.
Inherent in the empirical approach is a tacit acceptance of the historical reality that translational process has been immutably unpredictable, a “zone of chaos” or irreducible complexity in the words of one experienced and chastened pharmaceutical scientist. 19 In contrast, translational science posits that although highly complex, the translational process is governed by general laws that have to date resisted characterization, and that translational efficiency will increase only when those laws are defined. Many fields of research, from physics to engineering to genetics, have transitioned from observation to understanding to prediction by elucidating the fundamental scientific and operational principles that govern behavior of systems in their fields. This is the transition that is required for translation, and is the goal of the discipline of translational science.
This concept of “translational science” is distinct from how the term has been used previously (e.g., Zerhouni, 2005 20 ). Whereas previous writers have used the term to describe the practice of a variety of scientific disciplines (chemistry, biostatistics, regulatory, clinical trials, etc.) to do translation for a particular target or disease, NCATS uses the term to describe the study of translation itself, as a means to accelerate the progress in the performance of translation. In this way, NCATS uses the term “science” in its usual connotation as an “intellectual and practical activity encompassing the systematic study of the structure and behaviour of the physical and natural world through observation and experiment” (Oxford).
Although widespread translational research exists in the public and private sectors, translational science currently does not. Virtually every component of the NIH, and every biopharmaceutical company, performs translational research in order to meet their missions to diagnose and treat the diseases under their purview. NCATS’ “disease” is the translational process itself, which has a high project “mortality rate” and is exorbitantly costly. NCATS aims to identify, understand, and treat these causes of “translational disease,” just as other organizations do particular disorders. Although this approach is new to translation, it is the repeated history of science that elucidation of general principles has led to dramatic increases in predictivity, efficiency, and capability. The transformation of genetics from an observational endeavor to powerful and efficient driver of science and medicine provides an instructive example driven by discovery of the rules of DNA base pairing, replication, and the genetic code, which provided the basis for the field of genome science. More recently, the field of data science has begun to revolutionize the use of data. In just such a way, understanding of general scientific principles will transform translation from an empirical, failure‐prone, costly process to a predictive and productive science. The potential of such a translational science approach has been well‐demonstrated in the success of the Lipinski “Rule of 5,” 21 the AstraZeneca “5Rs,” 22 and the recent description of Biological Activity‐Based Modeling (BABM) from NCATS. 23 In each case, these general principles were deduced from the results of many individual translational research projects across targets and diseases, by researchers whose main goals were to produce individual drugs or probes, not general principles. In order for these kinds of principles to be defined more commonly and broadly, and ongoingly tested and refined as all models must be, prospectively designed projects to elucidate general predictive principles, performed by translational scientists whose primary goal is such principles, will be essential. Additionally, because the data that would allow such deduction exists across organizations, more widespread data sharing of both successful and unsuccessful translational research experiments, and the development of a robust field of translational data science, will be required.
CREATION OF A TRANSLATIONAL MAP
Bringing science to translation is needed not only within individual steps of the process, but across this currently highly complex endeavor. This aspect of translational science is a systems’ engineering problem. Translation is frequently diagrammed as a linear progression of a small number of steps, and referred to as a “pipeline,” connoting a vessel through which starting material progresses at a rate proportional to the diameter of the conduit, and exits unchanged with 100% fidelity. In fact, translation is a multistep, multidomain, multidiscipline recursive web that has more in common with a metabolic pathway map, but in which the product bears no resemblance to the starting material, and is rarely successfully produced. Realizing that the traditional oversimplified ways of representing and referring to the translational process had become limiting to understanding and re‐engineering, a group of academic, industry, government, and patient advocate investigators under the auspices of the Institute of Medicine (now National Academy of Medicine) created comprehensive translational maps (Figure 1) 24 for both small molecule and biologics development, that have now been greatly expanded and converted from static paper to dynamic electronic form, complete with major problem areas identified and resources available to investigators to collaboratively overcome them (https://ncats.nih.gov/translation/maps). Illustrating the enormous complexity of the translational process, the current “4D” (Dynamic Drug Discovery and Development) map has over 10,000 nodes in over a dozen domains of activity. This map has provided individual translational researchers a guide to planning, provided translational science educators an important educational tool, and provided the field of translational science a research agenda that focuses on developing solutions to the most difficult problematic steps to translational progress.
FIGURE 1.
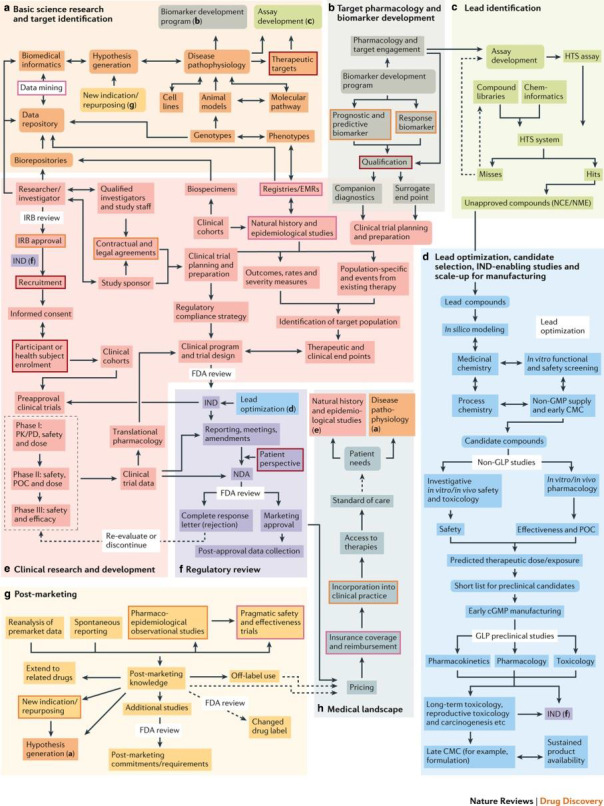
“The Drug Discovery, Development and Deployment Map (small molecule version)” by Wagner et al. 24 The map is licensed to the public under the Creative Commons Attribution‐Share Alike 4.0. cGMP, current good manufacturing practice; EMR, electronic medical record; FDA, US Food and Drug Administration; GLP, good laboratory practice; GMP, good manufacturing practice; HTS, high‐throughput screening; IND, investigational new drug; IRB, institutional review board; NCE/NME, new chemical entity/new molecular entity; PK/PD, pharmacokinetic/pharmacodynamic; POC, point of care
The map illustrates the complexity that has to date frustrated efforts to increase overall yield. The translational process is seen to be a greater than 20‐step process, with much work having documented yields of 0.1 to 0.7 at each step (e.g., Paul et al. 12 ), the overall success rate observed of less than 1% is exactly what would be expected. However, the solution to any low‐yield systems engineering problem is not to add more starting material (“shots on goal”) to an inefficient process, but rather to identify the rate‐limiting steps of the process and develop catalysts or new methods that increase the yield of those steps by a log or more, which when combined increase overall yield substantially. The latter is the translational science strategy: identifying and developing understanding, technologies, and paradigms that aim to increase by 10‐fold or more the efficiency of the rate‐limiting steps in the translational map.
A TRANSLATIONAL SCIENCE RESEARCH AGENDA
The 4D Map process and input from the translational community allowed creation of an initial translational “problem list” of prevalent causes of translational research failure, the rate‐limiting steps to translational efficiency (Figure 2). It was evident that translational projects fail for both “hard” science (e.g., biology, chemistry, pharmacology, and toxicology) and social science (e.g., incentive structures, credit allocation, economics, and intellectual property) reasons, so translational science needed to innovate in both areas. Additionally, it was noted that these issues all fell into the “tragedy of the commons” 25 —problems common to all disease areas and sectors, so not perceived by any organization, public or private, as their responsibility to solve. This latter insight helped to explain why these fundamental problems had not been tackled before—and reinforced the importance and potential of this new field of translational science.
FIGURE 2.

Major rate‐limiting translational problems that are the focus of Translational Science. *Identified in the Drug Development Map (18, Supplemental Figure SB) as being particularly prone to failure, delay in progression, and/or high cost and therefore high priority for innovation. IP, intellectual property; IRB, institutional review board
This process also made it clear that not only did the function of translational science need to be different from other fields of science, but that its operational characteristics and structure were necessarily different as well. NCATS created Strategic Principles (Figure 3; https://ncats.nih.gov/strategicplan/principles) to articulate these distinctions and establish core characteristics of exemplary translational science. In structure, translation absolutely requires a team as its unit of operation, because over 20 distinct scientific disciplines are needed to traverse the translational journey from target to intervention to patient to practice. This creates challenges for the current individual‐focused incentive structure of most academic organizations and many nonprofits. It necessitates the incorporation of scientifically expert and skilled project managers to act as “coaches” for the translational team. Because the team has as its deliverable an intervention that improves the health of patients, inclusion of patients and communities intended to benefit on the research team is a fundamental feature of translation. Because neither optimal scientific team operation or best practices for inclusion of patients and community members on it are so far defined, they are key components of the translational science agenda.
FIGURE 3.

NCATS Strategic Principles. NCATS, National Center for Advancing Translational Sciences
Operationally, translational science must advance by the definition of principles and technologies that allow translation to be driven by prediction rather than empiricism. Such paradigms and technologies must be generally useful (i.e., target/disease agnostic), but improved effectiveness must not simply be prophetic, but demonstrated in use cases chosen for their generalizability. In addition, if improvement is shown, they must be broadly and intentionally disseminated to enable the entire research community to apply them in their own translational research. This operating schema NCATS has formalized as the “3Ds”: Develop, Demonstrate, and Disseminate. All translational science projects must develop a technology or insight or paradigm to improve the efficiency or effectiveness of a rate‐limiting translational roadblock, demonstrate its utility in achieving that improvement in one or more use cases, and then actively disseminate these improvements. Given the nature of translation, which focuses on tangibly useful products—drugs, devices, behavioral interventions, and medical procedures—that directly improve human health, this dissemination includes not only the publications in the scientific literature and patents, but also widespread data sharing, education and training, and implementation of health improvements at the individual and community levels.
An additional rationale for the 3Ds paradigm is a practical one, bearing on how translational scientists choose disease applications with which to demonstrate the utility of their technologies. Translational science is often referred to as “disease agnostic,”, which is true but limited. It is more properly described as “disease universal” because it addresses the scientific and operational bottlenecks that are common to translational research for most or all diseases. Because the majority of human diseases currently have limited or no specific treatment, translational science encompasses all human diseases. This points up key features of ideal translational science demonstration projects: they are chosen not only because there is an unmet medical need—this will apply to many potential projects—but also because they are well‐suited to test the utility of the new technology or paradigm, and because that utility would be applicable to many other if not all human diseases. This is what is meant by translational science projects needing to be “catalytic” and demonstrating “generalizable principles.”
All of these features have been codified in the NCATS Translational Spectrum (Figure 4), which illustrates that (a) translation is a circular, rather than linear, process, which can start at any stage of the translational process and proceed clockwise or counterclockwise; (b) each of the five stages must typically be traversed in order but in particular situations stages may be skipped, accelerating progress; (c) patients are at the center of every activity in translation, both as members of the research team and as necessary beneficiaries; and (d) the “3Ds” govern translational science efforts to decrease the time and effort required to circumnavigate the spectrum.
FIGURE 4.
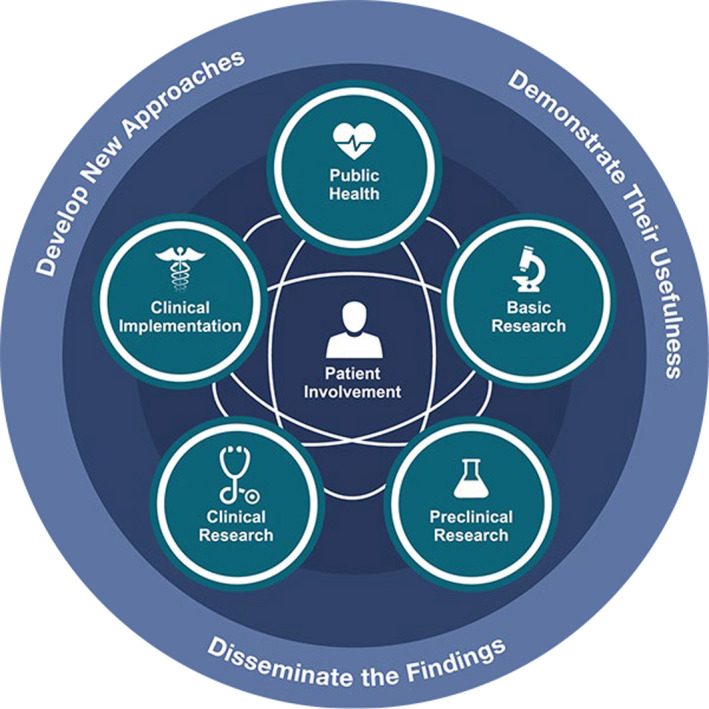
NCATS Translational Science Spectrum. NCATS, National Center for Advancing Translational Sciences
A RESEARCH AGENDA FOR THE NEXT DECADE OF TRANSLATIONAL SCIENCE
Translational science has made remarkable progress in the last 10 years, making possible the enumeration of a much more ambitious agenda for the next decade. The accomplishment of this agenda will advance the transformation of translation into a predictive science, thereby increasing its efficiency and effectiveness and getting more treatments to more people more quickly, efficiently, and effectively.
Scientific priorities for translational science
Predictive efficacy
Up to 90% of investigational drugs that enter human testing are never approved for any indication 26 ; the most common reason for this failure is lack of efficacy in humans despite promising efficacy in animal or cellular models of the indication. Although animal models continue to be invaluable for understanding fundamental biology, their ability to accurately predict responses to therapeutic interventions—whether those be pharmacological or behavioral—is quite limited. Because this preclinical (nonhuman) to clinical (human) transition is where much translational failure occurs, it must be a major focus of translational science. But the need for preclinical models that are more predictive of human responses is not the only predictive efficacy research priority. Rates of attrition are high at each step of the ladder of increasing complexity in the preclinical stage (e.g., biochemical to cell‐based activity of a compound; high‐throughput cell line testing to primary or induced pluripotent stem cell (iPSC)‐derived cell activity; single cell to multicellular in vitro system; and in vitro system of any complexity to in vivo animal model). At each stage, empirical testing of dozens to millions of molecules is required because our understanding of how to predict efficacy at the next more complex stage is rudimentary and unreliable. Human cell‐based models based on iPSCs, 3D multicellular aggregate or printed models, and microphysiological systems have all shown promise but our understanding of their predictivity of human responses remains limited. 27 Challenges in cell production and differentiation are considerable, 28 and capacity to model complex in vivo interactions, or long‐term disease states or pharmacological responses, remain areas of needed development.
Predictive toxicology
Toxicity of drugs is a frequent cause of failure in drug development and withdrawal after market approval. Many compounds fail for toxicity in preclinical development, and of the 90% of investigational drugs that fail in human testing, 26 about 30% of that failure is due to unanticipated toxicity. Although much progress has been made in identifying the structural features of various interventional molecule types (small molecules, peptides/proteins, oligonucleotides, and gene therapy vectors) that predispose to in vitro or in vivo toxicity, our ability to predict adverse events, particularly less common idiosyncratic events, remains limited. Much research in fundamental in vivo and in vitro mechanistic toxicology is needed to develop a systematic understanding of the relationships between molecular structure and adverse events in living systems. Such a systematic understanding would allow candidate interventional molecules of all types to be made with low adverse event liability from the beginning, the current practice of making many molecules with unpredictable liabilities, most of which fail in later stages of testing. Several industry‐based and public‐private partnerships have made early progress in predictive toxicology via massive data generation and sharing efforts, including the International Consortium for Innovation and Quality in Pharmaceutical Development (IQ) biopharmaceutical collaborative drug safety program (e.g., Treem et al. 29 ); the BioCelerate subsidiary of the pharmaceutical company collaborative TransCelerate BioPharma Inc.; the Critical Path Institute's Predictive Safety Testing Consortium (e.g., Schomaker et al. 30 ), and the Innovative Medicines Initiative eTox 31 and TransQST programs. The Toxicology in the 21st Century program, a collaborative effort among NCATS, the National Toxicology Program, the Environmental Protection Agency, and the US Food and Drug Administration (FDA) has also made promising advances in this area, 32 but much more needs to be achieved before accurate and reliable prediction of common and idiosyncratic toxicities is possible.
A complementary approach to the predictive toxicology problem is the development of new assay types based on human cells that can identify potential toxicities more accurately and efficiently than current animal testing methods. These include human primary cells and those differentiated from iPSCs; 3D models including spheroids, organoids, and printed tissues; and microphysiological systems (MPS; or “tissue chips”). Multiple components of the NIH as well as dozens of provider companies and a newly organized IQ MPS Affiliate are conducting and supporting research in all these areas, and early results are promising (e.g.., Wei al. 33 ).
De‐risking undrugged targets/currently untreatable diseases
The third major reason for discontinuation of intervention development projects is “business reasons,” 26 which refers to the judgment that the risk of failure is too high or the anticipated return is insufficient, so resources would be better applied to other projects. These reasons apply to both private and public sector organizations, because in both cases reasonably anticipated financial return must be sufficient, whether via revenue in the private sector, or via grant funding in the public sector. For this reason, risk aversion is widespread in both settings, and investigators, funding agencies, and companies prefer to work on a small subset of diseases and targets rather than assume the risk involved with addressing new target or therapeutic areas.
This cause of translational failure requires the development of technologies, knowledge, and paradigms that reduce the risk of new targets and diseases, so that funding agencies and companies will fund them. Examples of such efforts are the NIH Illuminating the Druggable Genome (IDG) program 34 ; the Structural Genomics Consortium Structure and Chemical Probes programs, 35 and Target2035. 36
The “Chemical Space” problem
Currently, less than 10% of human and pathogen targets have been “drugged,” and modulating new classes of targets is likely to require new chemical structures. However, fewer than 108 of the potential 1060 drug‐like compounds have ever been synthesized, and technologies do not exist to efficiently explore the vast chemical space which remains unmined for drug development. Although progress has been made in such prediction for specific target classes (“privileged structures” for kinases and GPCRs, for example), and ligand‐ and structure‐based drug design is increasingly useful, most potential targets are not currently addressable via these methods. The ultimate goal must be understanding of the general rules that govern interaction of small molecules (and other modalities) and their targets, which would allow a priori prediction of chemical structures likely to modulate any desired biological target. On a practical level, chemical space exploration is limited by the efficiency of synthesis of quality small molecules, and isolation of active compounds from natural product extracts, has not increased appreciably in the last 100 years, remaining roughly one compound/week/chemist. Medicinal chemistry optimization remains a largely empirical process guided by experience and intuition of the individual chemist rather than data‐driven, generally applicable, predictive syntheses based on universal and fundamental understanding structure‐activity relationships. In an era in which chemistry’s partner disciplines of biology and informatics have increased their throughput by many orders of magnitude, chemistry is the rate‐limiting factor in many translational projects, and is thus an important focus for translational science innovation. Early efforts in this area include increasingly sophisticated efforts in automated chemistry, 37 DNA‐encoded libraries, 38 and the NCATS ASPIRE project 39 and similar projects in industry including the Eli Lilly Automated Synthesis Laboratory. 40
New therapeutic modalities to reach currently inaccessible disease space
Over the last several decades, therapeutic modalities have broadened from small molecules, medical/surgical procedures, and behavior modification to include a variety of therapeutic peptides, proteins, antibodies, oligonucleotides, viral vector‐delivered gene replacement, cell therapies including engineered (e.g., CAR‐T), embryonic, adult, and iPSCs and their derivatives, exosomes, devices, and science‐driven behavioral change technologies. Despite these new technologies, a large number of disease‐causing abnormalities remain inaccessible, including those caused by aberrant protein‐protein interactions, missense mutations causing misfolding, and nonsense and splice‐site mutations. New types of chemistries (e.g., peptoids 41 and protacs 42 ) and new genetic therapy modalities including modified anti‐sense oligonucleotides (ASOs) and morpholinos, CRISPR‐Cas9, and other somatic cell gene editing technologies, 43 and more targeted and immune‐evading gene therapy viral and nonviral vectors are all needed areas of translational science innovation. In addition, for the new gene therapy, gene editing and cell therapies to reach their potential as widespread and accessible treatments, scale‐up, manufacturing, reproducibility, impurity, qualification, and other limitations typically referred to as “Chemistry, Manufacturing, and Controls” (CMC) will need to be overcome. Given that these new therapeutic modalities have the potential to be transformative for many patients with currently untreatable or poorly treatable diseases, these CMC issues must be a major focus for translational science.
New methodologies to increase efficiency in preclinical development
New methods to increase the efficiency of the preclinical therapeutic development, from early assay development and high‐throughput screening to later investigational new drug (IND)‐enabling studies, are needed. With the attenuation of many of these activities in large pharmaceutical companies and the increasing “virtualization” of preclinical development in pharma and biotech, innovation in these areas is particularly important for academic translational science. NCATS has fostered a number of new such approaches, including quantitative high throughput screening, 44 matrix library screening, 45 the Predictor project, 46 biological activity‐based modeling (Huang et al., 2021), and novel organizational/management models that have demonstrated substantial improvements in success rates and costs in small molecule drug development. 47 , 48
Harnessing pleiotropy and promiscuity for therapeutic development
A fundamental feature of living systems is their use of a relatively limited number of biochemical and signaling pathways to perform diverse functions in different cellular or organ system contexts. A fundamental feature of therapeutic molecules is that they tend to act on multiple targets in vivo. These two factors—pleiotropy and promiscuity—led to the frequent occurrence of a therapeutic molecule developed for one indication being useful in treating others. Such “repurposing” has historically most often been the result of serendipitous clinical observations, and, if applied broadly, may have the potential to provide treatments for many diseases with the current pharmacopeia, obviating the difficulties of new chemical entity development. Translational science seeks to develop technologies to make a priori repurposing more predictive—“systematizing serendipity”—based on fundamental chemical and biological knowledge. But because obstacles to repurposing of drugs are not only scientific, but include funding, legal, regulatory, policy, and reimbursement hurdles, translational science must innovate on these issues as well, particularly for the majority of drugs that no longer have patent protection. The NCATS Pharmaceutical Collection project 49 and Cures Within Reach (https://www.cureswithinreach.org/) have made initial strides in this area, and NCATS and the FDA recently held a joint workshop on off‐patent repurposing to devise a research agenda for this area. 50 A related research area is the prospective targeting of multiple diseases from the start based on shared molecular etiology, a concept that would bring multiplexing—a concept widely applied in other areas of biology—to therapeutic development. 51
Patient/Community engagement
In virtually every area of consumer product development and dissemination outside biomedical research, the people who are the intended consumers of a new product are included in the design and development of the product, to ensure its relevance and subsequent use. One of the most effective “disruptive technologies” in translational science is the involvement of patients and communities in the development and implementation of new health interventions, which can bring increased relevance, urgency, applicability, and ultimately adoption of interventions successfully developed. For this reason, NCATS established as one of its founding principles that patients, patient advocacy groups, and communities should be involved as members of the research team from the start of every project NCATS undertakes or supports (e.g., Merkel et al. 52 and Walkley et al. 53 ). But like many aspects of translational science, this aspiration is more easily stated than achieved, because the field currently lacks robust generalizable strategies for ongoing patient and community collaboration that are demonstrated to shorten the time and/or improve the efficiency of the translational process. This patient/community engagement science agenda has already produced important advances via the community engagement research programs of the CTSA hubs, the required patient advocacy group partnerships in the RDCRN, the NCATS Toolkit for Patient‐Focused Therapy Development (https://ncats.nih.gov/toolkit), among other initiatives.
Biomarkers for human clinical response
When an exogenous molecule, device, or procedure is initially administered to humans with the target disease, reliable, robust, and reproducible predictors of ultimate durable clinical response are critical to avoiding downstream clinical efficacy failure in larger trials. However, such reliable indicators of reversal of the pathologic process are currently difficult to identify and qualify for either nonregulatory or regulatory applications, and their absence is a frequent cause of translational failure. 54 The Biomarkers Consortium, 55 IQ Consortium, Critical Path Institute Biomarker Qualification Program (https://c‐path.org/biomarker‐qualification‐program‐tables/), and the FDA CDER Biomarker Qualification Program (https://www.fda.gov/drugs/drug‐development‐tool‐ddt‐qualification‐programs/biomarker‐qualification‐program) have made strides in the development of particular biomarkers, but new more efficient and generalizable technologies for biomarker identification, utilization, qualification, and prediction are urgently needed and are a major area for translational science innovation.
Disease natural history, registries, and clinical outcome criteria
For many disorders, especially rare/orphan diseases (defined as having <200,000 US prevalence), the clinical phenotype spectrum, disease course, and causes of morbidity and mortality are poorly understood, and are often based on small numbers of the most severely affected patients. Without accurate and representative clinical characteristics and natural history, intervention development is difficult, particularly in trials for rare diseases that are increasingly utilizing historical rather than placebo controls. Natural history studies and their patient registries also enable biomarker identification and efficient clinical trial recruitment. Technologies are needed for cost‐effective and interoperable patient registries and natural history studies, which will both advance translation for individual diseases but also allow identification of commonalities among them that would catalyze application of therapeutics developed for one disease to be rapidly repurposed for others. The use of common ontologies and common data elements will be key to this effort. 56
Clinical trial designs
The randomized, controlled, double‐blind clinical trial became the standard for demonstration of clinical efficacy of therapeutics in the late 1940s and has served medical science well. However, as the number of therapeutics to be tested and the number of types of diseases being addressed have increased and diversified, the need for additional trial designs that are able to generate reliable results with smaller numbers of patients, multiple therapeutics, or indications simultaneously, and more cheaply and efficiently have become increasingly urgent. Adaptive, Bayesian, master protocol, umbrella, basket, historical control, and pragmatic trial designs among others have been developed. But as with other areas of translational limitation, the need for both continued innovation and more widespread implementation of such designs is needed. In addition to scientific limitations, such designs are perceived to carry increased risk of failure for regulatory or medical adoption reasons, so many trial sponsors are reluctant to adopt them. So translational science must also focus on de‐risking and implementation science strategies for these new technologies. The recent coronavirus disease 2019 (COVID‐19) pandemic led to a quantal increase in the frequency of such trial designs due to increased risk tolerance brought on by the pandemic, with multiple adaptive master protocol trials being rapidly instituted around the world, including as part of the Accelerating COVID‐19 Therapeutic Interventions and Vaccines (ACTIV) initiative. 57
Clinical trial operational efficiency
In addition to scientific and statistical innovations, translational science must also address operational limitations to clinical trial efficiency. Per‐patient cost now averages over $40,000, with those costs doubling every decade (https://aspe.hhs.gov/system/files/pdf/77166/rpt_erg.pdf). Many clinical trials are not completed on time or on budget, or fail for futility due to inability to recruit the number of participants required 58 ; cycle time for large clinical trials, from funding to reporting of results, can be 10 years or more. Further, many trials, whether sponsored by industry or public sector funders, may not answer clinical questions that are needed for improved care of patients, but instead answer questions of interest to the academic investigator or to a company seeking marketing advantage. 7 Clinical trial efficiency and relevance are absolutely required for interventions to reach patients who need them, and thus are major areas for translational science innovation. Particular focus is needed in the primary causes of delay or cost: multisite institutional review board (IRB) review and contracting, site and investigator qualification, recruitment, surge capacity, and adequacy and timeliness of results reporting.
Single/Harmonized IRBs
It has not been unusual for multisite trials to experience start delays of 12–24 months due to the need for the IRB and legal teams at each site—which may number 50 or more—to agree on and independently certify the protocol. Such multiple review has been shown not to improve safety, is a large drain on institutional resources, and during the time required patients’ disease may progress rendering them ineligible for the trial by the time it is approved. To address these issues, in 2016, NCATS launched SMART IRB, 59 which now has over 800 institutional signatories who have agreed to single IRB (sIRB) review and reliance. Reported improvement in approval times has been stunning, with one large center (Harvard Catalyst) reporting times to reliance and approval routinely in 2 weeks, with some trials approved in days—an over 10‐fold improvement in efficiency. Beginning in January 2018, the NIH required all new human subjects research it funds to use an sIRB, and beginning in January 2020, the Common Rule began requiring that US institutions engaged in multisite human subjects’ research to rely on an sIRB. Although these developments have begun to solve the IRB problem and are a major accomplishment for translational science, much remaining scientific, legal, and operational innovation is needed to make the new system universally efficient and thus realize its promise for translational effectiveness and the health of patients.
Clinical trial participant recruitment, retention, and diversity
It has been appreciated for many years that up to 50% of clinical trials end in futility after failing to recruit and retain the required number of participants to yield a statistically valid result. 58 In addition, racial and ethnic minorities are very frequently under‐represented in clinical trials compared to their population and disease prevalence. 60 These numbers have not improved appreciably over the last 25 years despite reporting mandates from the NIH and others. Clearly, recruitment and retention strategies, particularly for under‐represented minorities, are a major area of scientific and operational innovation in translational science. The NCATS‐funded Recruitment Innovation Center (https://trialinnovationnetwork.org/recruitment‐innovation‐center/) and Accrual to Clinical Trials network (https://www.actnetwork.us/; https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6241502/) are two programs in this area.
Clinical trial networks
The increase in preclinical translation activity in private and public sectors has created an increased need for human subjects’ research, both observational to define the natural history of diseases, and interventional to test drugs, devices, medical procedures, and behavioral therapies, which aim to slow or reverse that natural history. At the current time, most publicly funded multicenter trials are performed in disease‐specific networks (e.g., the AIDS Clinical Trials Group (https://actgnetwork.org/), NCI Experimental Therapeutics Clinical Trials Network (https://ctep.cancer.gov/initiativesPrograms/etctn.htm, NHLBI PETAL https://petalnet.org/) and NIDA CTN (https://www.drugabuse.gov/about‐nida/organization/cctn/clinical‐trials‐network‐ctn), or in site groups created specifically for each trial and disbanded upon completion. Industry funded trials are generally run through contract research organizations that have relationships with hundreds of hospitals and clinics, most of which are not utilized for publicly funded trials. The segmented and often ephemeral nature of these trial networks has prevented efficiencies from cross‐trial and cross‐disease insights and data harmonization, operational efficiencies from an integrated learning clinical trials system, and sharing of capacities so important for system flexibility and surge capacity needed to adapt to public health needs. The recent COVID‐19 pandemic has brought these issues into relief, given the unprecedented need for rapid creation, execution, and coordination of multisite clinical trials, and the difficulty of the current atomized system to respond effectively. The European Clinical Research Infrastructure Network (ECRIN) was established in 2004 to address the need for ongoing disease‐agnostic clinical research network capacity in Europe. 61 The Patient‐Centered Clinical Research Network (PCORnet) was established in 2014 with an emphasis on capacity for real‐world evidence studies, pragmatic clinical trials, population health research, and health systems research. 62 More recently, the NCATS CTSA Trial Innovation Network (TIN; https://trialinnovationnetwork.org/) was started in 2017 as a national network for translational medicine, able to carry out clinical studies of any stage on any disease, with a focus on scientific and operational innovation and efficiency. 63 The TIN has assisted with dozens of clinical trials across virtually every therapeutic area in the last 4 years, and played a major role in clinical trials of immunomodulators and convalescent plasma in the recent COVID‐19 pandemic. The NCATS also coordinates a separate clinical network for rare diseases, the Rare Diseases Clinical Research Network (RDCRN; https://www.rarediseasesnetwork.org/), that is supported by 10 NIH Institutes and Centers. These networks provide the start to the capacity, efficiency, and flexibility needed in the clinical trials enterprise.
Data interoperability
The burgeoning of data production in biomedical research has been accompanied by proliferation of data types, data models, standards, and usage conditions that suit the needs of particular data producers but are not compatible or interoperable with the data needs or standards of others. The resulting thicket of data types and standards prevents the field from benefitting from insights that could be derived from integration of these data. Although this issue affects all fields of science, it is particularly problematic in translation given that translational science is a fundamentally integrative discipline, aiming to deduce general principles from diverse disease examples and connect insights across the translational spectrum from pathophysiology to interventions to health of communities. Thus, data interoperability and integration are critical areas of innovation in translational science. The NCATS has such initiatives in all of its activities, but two large‐scale flagship projects illustrate the approach: The Biomedical Data Translator 64 and the National COVID‐19 Cohort Collaborative (N3C). 65 The Translator project is connecting and integrating data across data types, from genomics to cell biology to pathology to clinical to environment in a purpose‐built, open‐source platform to enhance human reasoning via sophisticated open‐ended and iterative computational investigation, enable identification of currently unappreciated connections among diseases and their pathophysiology, and advancing the shift from the current symptom‐based disease nosology to one based on molecular and cellular abnormalities that can be targeted by specific preventive and therapeutic interventions. 66 The N3C, in contrast, is connecting, harmonizing, and integrating large amounts of data of a single type—clinical electronic health record (EHR) data—that are currently represented in multiple different data models in many separate institutions, preventing system‐wide insights that could inform better patient care. The N3C currently contains over 4 billion rows of data on over 3.5 million patients from 42 academic medical institutions across the United States, of whom over 800,000 have been diagnosed with COVID‐19; its eventual scope will be over twice this large. Hundreds of qualified researchers across the world are now using the N3C data to answer important questions on acute COVID‐19 symptomatology, patient comorbidities, ethnic and racial disparities, therapeutic responses, and the characteristics and prevalence of persistent symptoms following COVID‐19 infection (i.e., the Post‐Acute Sequelae of COVID‐19, or “Long COVID”). The scope and scale of the N3C are unprecedented, as was the speed of its creation over just a few months in the summer of 2020. The reasons this was possible illustrate important principles of translational science: collaboration among very diverse research teams, focus on transformational capacities to solve rate‐limiting steps to translational efficiency, and patient‐relevant deliverables. The N3C was enabled by a deliberate strategy to create secure scientific collaborative platforms starting in 2016 at NCATS, and an equally deliberate building of interoperable nationwide clinical informatics capacities in the CTSA program hubs and National Center for Data to Health (CD2H, https://cd2h.org/). N3C is another prominent example of the NCATS “3Ds” paradigm: although the current use case is COVID‐19, the platform is generally applicable to any disease.
Shortening time of intervention adoption
It has been estimated that an average of 17 years currently elapses between the time an intervention is shown to be effective and the time it reaches all patients who could benefit, 9 and that medical care continues to frequently not follow evidence‐based recommendations. 67 This “last mile” of the translational process is particularly complex, because it involves not only scientists and physicians, but also sectors not primarily focused on medical research (payors, public health departments, and social determinants of health including housing and food). This is a critical area for translational science innovation, because translation is only successful when it improves health of individuals and communities in tangible and measurable ways.
Fostering the adoption of individual interventions for particular diseases is performed by public health campaigns by governments and nonprofits, disease‐specific professional societies, and NIH institutes, and is the focus of pharmaceutical and device company marketing. However, the area of scientific investigation that seeks to understand the general principles and practices of successful intervention adoption in clinical and public health settings is the field of dissemination and implementation (D&I) science, 68 and is a crucial component of translational science that is only beginning to have the emphasis and support it needs from the research community. The NIH Office of Disease Prevention supports a wide spectrum of D&I research (https://prevention.nih.gov/research‐priorities/dissemination‐implementation), as does the Agency for Healthcare Research and Quality (AHRQ). The NCATS is supporting a wide spectrum of D&I research (e.g., Brownson et al. 69 ) and projects that have, for example, improved dissemination of health interventions in high‐need communities 70 and improved medication adherence. 71
Organizational innovation priorities for translational science
Translation is by nature a cross‐stage and cross‐discipline, and often cross‐culture and cross‐value, activity. The word translation itself—meaning “to carry across”—implies a gulf between the participants that must be bridged. The many different scientific, medical, and patient disciplines that comprise the translational team can lead to miscommunication and misalignment of priorities that regularly impede the progress of translational projects, or cause outright failure. Such disjoints can occur in any realm of science, but in translational science they are a feature of every project. Progressing a translational intervention from the laboratory to clinical testing to medical practice to public health requires no less than 20 distinct scientific disciplines, each with its own vocabulary, incentive structures, and valued outcomes. Many translational projects require transition from public sector to private sector, sometimes multiple times, and involve patients and researchers, all of whom ultimately desire the same result—an intervention that successfully improves health—but whose priorities and short‐term reward structures are often starkly different. These dynamics mitigate against the sustained team effort that is obligatory for translational success.
Although reliable figures are lacking, it is common experience that a substantial number of translational projects fail not for the “hard” science reasons enumerated in the previous section, but for social science and organizational reasons. This has substantial implications for the field of translational science: because the field seeks to understand, and develop solutions to, all the causes of translational inefficiency, ineffectiveness, and failure, it must include innovation in the social and organizational science etiologies of that failure just as much it does the bioscience causes. NCATS therefore stresses innovation in both. Here, I enumerate some of these social and organizational science priorities for translational science.
Understanding of translation
Historically, most biomedical translation occurred in the pharmaceutical industry, which for proprietary reasons did not publish or otherwise promulgate most of its science. Drugs and devices are presented as finished products, with very little description of how these interventions were developed. As a result of this “black box” tradition of therapeutic development, most of the scientific and lay public have little or no knowledge of the activities and distinct scientific disciplines of translation, and the field lacks a robust scholarly literature that could drive improvements in understanding and productivity as has happened with other fields of science. Together, these dynamics have led to widespread mistrust of biopharmaceutical sector, and persistent misperceptions about translation which hamper solutions that would benefit health and the public. 72 Therefore, the first priority in the social/organizational domain is in education and communication at all levels, to instantiate understanding of translation by scientists, physicians, policymakers, and the public. This understanding is a prerequisite for making other priorities possible.
Understanding of translational science
The concept of a “science of translation,” distinct from the process of translation for a particular intervention for a specific target or disease (often referred to as “translational research”), is relatively new and still unfamiliar to many. Translational science is a field of investigation which seeks to understand the fundamental scientific principles underlying each step of the translational process, in order to make it more predictable and efficient; it is as different from translation as, for example, data science is from data. 2 Understanding of this difference has been hampered by the unfamiliarity of many outside the biopharmaceutical industry of what translation entails, its currently empirical nature, and a resulting belief that translation is a rote process that involves little science as conventionally defined—when in fact the opposite is true. 72 Understanding that a science of translation is needed to solve the well‐appreciated inefficiencies and high failure rate of new intervention development, and academic and training programs to support and foster it, must be a major priority for the coming years of translational science.
Academic discipline of translational science
Many of the needs and opportunities described above will only be addressable in the context of a new academic discipline of translational science. Departments of Translational Science will need to have subdisciplines not commonly represented in academic settings (e.g., toxicology, process chemistry, regulatory science, clinical trial design innovation, and implementation science) and have distinct organization and reward structure, incentivizing the team science and multidiscipline collaboration required for successful translation. Demonstrated improvements in the understanding, efficiency, effectiveness, and health impacts of translation would be the primary outcome. Measures of success would include publications and patents but would explicitly recognize the contributions and expertise of all team members, including consideration of an alphabetical author lists as is the practice in other fields. Additional measures would be equally important in considerations of promotion, including deliverables that are demonstrably useful to researchers in other fields, other organizations in the translational ecosystem, including regulators, companies, and payors, and most importantly patients. Systems for assigning credit for contributions to long‐term, multistage, multidisciplinary projects would need to be instituted in departments of Translational Science, utilizing the lessons from such systems in genome science, physical science, and biopharmaceutical organizations. New funding streams for translational science, and mechanisms to support the careers of academic translational scientists after their training, are critically needed for academic departments of translational science to develop and prosper. NCATS is the principal US funder of translational science and although it supports robust training in the discipline, it currently cannot support the research careers of translational scientists. Resolving this absence of academic translational science career funding must be an urgent strategic priority for the field (Box 2).
BOX 2. Why translational science, and much translational research, must be an academic discipline.
Some have questioned whether academic translational research projects, and an academic discipline of translational science, are necessary or appropriate given that the private‐sector biopharmaceutical industry has as its mission the development of drugs and devices to prevent and treat disease. 18 Conversely, some have suggested that biopharma must be unaware of, or uncaring about, the medical needs and translational inefficiencies that academic translational research and science address. Both misapprehend the incentives and imperatives of the private sector, and the complementary nature of public and private in driving translational advances. Because private sector organizations have a legal fiduciary responsibility to their investors (i.e., funders) to provide their initial investment back and a financial gain to compensate them for the use of their investment dollars, companies must choose projects that have a reasonable likelihood of delivering that financial return in the relatively short term (typically 3–5 years). Diseases with low prevalence, and targets that are poorly understood (“unvalidated”) are generally viewed as being too risky to support private sector investment due to the low expected financial return and/or excess likelihood of project failure. The majority of human diseases and novel targets have risk profiles that make them unattractive to private sector investment.
The Orphan Drug Act of 1983 and a variety of regulatory voucher programs have had some success in altering the financial risk calculus and making more diseases attractive to private sector investment. The scientific risk of unvalidated targets can only be mitigated by work not reliant on financial return on investment (i.e., public sector funding). Because 95% of human diseases currently have no specific FDA‐approved treatment, and 90% of disease targets remain unvalidated, public sector research to “de‐risk”—that is, develop to the point of proof of principle—is required to allow adoption by the private sector. These are the necessary and proper province of public sector translational research, which historically has been remarkably productive. 73 Translational science, which produces fundamental knowledge to improve the understanding, efficiency, and effectiveness of the translation for all in the research ecosystem, also does not produce the financial return required for private sector investment. Academic translational science will increase knowledge and efficiencies for all, and academic translational research will de‐risk projects sufficiently for private sector investment, and create interventions for diseases that cannot support a return on investment model. By working in complementary roles, public and private sector biomedical research together advance the health and well‐being of the public.
Academic incentives/credit for team science
Translation is obligatorily collaborative, given the multiplicity and diversity of disciplines needed to successfully develop and implement an intervention, and the science to make translation more effective and efficient. The organization, operation, credit allocation, and incentives in academic institutions often mitigate against those required for translation, and as academic institutions become more active in translation, this form/function mismatch has become limiting to productivity at many institutions. Most academic promotion and tenure committees, even those at institutions with policies that encourage academic credit for team science, adhere to the traditional metric of “independent” productivity as measured by first‐ or last‐author publications in high impact factor journals, and grant funding that supplies not only salary and supplies but the indirect costs that fund much of academic infrastructure. While this metric remains appropriate for many areas of science, particularly in the basic sciences, it is an impediment to academic translational research and translational science. In addition, although academic institutions can do much to adapt their incentive and reward structures to the needs of translational science, public funding agencies have also increasingly recognized the incentives inherent in the dominant “principal investigator” model, which have perpetuated the traditional model of promotion, recognition, and tenure in funded institutions. Multiple institutes and centers at the NIH, including NCATS, have instituted novel funding mechanisms to support academic team science (e.g., https://www.nigms.nih.gov/grants/RM1/Pages/Collaborative‐Program‐Grant‐for‐Multidisciplinary‐Teams‐(RM1).aspx).
Integration of project management
One of NCATS’ mantras is the “Translation is a Team Sport.” One of the implications of this reality is the need for scientifically expert project managers who are also skilled in the people and deliverable management required for a translational team to be successful. Whereas long a feature of biopharmaceutical teams, albeit not always with the scientific skills or stature needed, project management has been virtually nonexistent in the academic and nonprofit sectors until relatively recently. In academia particularly, the invaluable role that effective project managers play on the translational team has not been generally recognized, and they rarely are accorded academic stature. NCATS has instituted a robust project management structure and has supported our grantees to do the same.
Education/Training
Like any new field, translational science requires the building of an educational and training framework, heuristics, and professoriate that will train investigators new to translation and translational science. The fundamental characteristics of a translational scientist have defined by an international consortium and are now widely adopted. 74 Robust and successful training programs in translational research developed by the CTSA program provide innovative curricula, competencies, and shared educational platforms across the consortium. 75 Complementing these established efforts with translational science training is just beginning. 76 Because translational science has not traditionally been an academic discipline, much of the scholarly literature and training materials, particularly in preclinical translation, will need to be expanded and generalized, adapted from those that many biopharmaceutical companies created to educate and train their scientists, or created de novo. Traditionally, training in preclinical translation and regulatorily directed clinical trials occurred in pharmaceutical companies, with those companies assuming that their new hires would have no knowledge or training in translation in their academic institutions from which they came. This training practice has waned over the last 20 years, putting increased importance on robust and innovative translational training programs in academic institutions.
An example of the training materials needed is the NCATS Assay Guidance Manual. 77 The NIH Chemical Genomics Center, now part of NCATS, partnered with Eli Lilly and Co. beginning in 2005 to adapt and externalize Lilly’s internal training materials. This remarkable public‐private educational partnership has since evolved into a freely available and regularly updated electronic book with an international editorial board and over 40 chapters with detailed rationales and protocols for virtually every stage of the preclinical drug development process, used by over 30,000 researchers worldwide ever year. This Assay Guidance Manual (AGM) serves much the same function as the “Maniatis” Molecular Cloning manual 78 did for the those in the then‐new field of molecular biology. The AGM has been adapted into a course that is now widely attended by both academic and pharmaceutical scientists. A new course on translational science has also been designed and initiated by the NCATS Education Branch, using the case study method. Every CTSA hub also has robust training programs that have evolved over the last decade to include nontraditional fields, including team science, regulatory science, and entrepreneurship. For many trainees, translational science is an attractive field because it offers a host of new scientific opportunities, and embodies the teamwork and medical and social impact that trainees increasingly seek in their chosen field.
Data transparency/sharing
In addition to the technical challenges to data sharing discussed above, there are myriad policy and organizational issues that limit data sharing, and that are in need of study and innovative solutions. Although progress has recently been made on policies for sharing of genomic and other types of data from NIH‐funded research (https://grants.nih.gov/grants/policy/data_sharing/), sharing of translationally relevant data remains problematic, including, for example, data on drugs or other interventions intended for or currently being commercialized, data on drugs and devices that were approved by regulatory agencies, particularly those that are now off‐patent, and data of various types from EHRs. In the academic domain, the NIH National Library of Medicine ClinicalTrials.gov database, Drugs@FDA and NCATS Inxight Drugs resources (https://drugs.ncats.io/), Open Data Portal of drug screening and animal model data (https://opendata.ncats.nih.gov/), and N3C enclave for COVID‐19 EHR data (https://covid.cd2h.org/) are examples of the kind of data transparency needed. Likewise, industry data sharing platforms and registries such as the Yale Open Data Access (YODA) Project (https://yoda.yale.edu) and the TransCelerate platform for clinical trial data (https://www.transceleratebiopharmainc.com/initiatives/historical‐trial‐data‐sharing/) are exemplars of such translational data sharing.
Intellectual property management
Given that translation often involves commercialization, intellectual property (IP) and potential liability are frequent source of disagreement, delays, and outright project failure. This may be due to differing valuation of IP by the partners (e.g., IP considered late‐stage and therefore valuable by an academic partner but early stage and therefore less valuable by a company partner), unfamiliarity with incentives, policies, or laws governing the partner, or lack of template agreements that would facilitate negotiations, among other causes. NCATS has instituted bidirectional education, innovative IP management policies based on data‐driven milestones, and templating of agreements, and has achieved substantial improvements in productivity (https://ncats.nih.gov/files/NCATS_Innovation_Patents_2018_2020.pdf).
Collaborative structures
Related to, but independent of, IP management is the need for innovation in collaborative structures to support the broad and frequent partnerships that characterize translation. Because every project at NCATS is a collaboration with at least one partner across the public, nonprofit, and for‐profit sectors, it has prioritized creation of innovative partnership models that can deliver effective agreements and products more efficiently. The most public expression of the NCATS approach to collaborative agreements is in the name of the office responsible for them: the Office of Strategic Alliances (OSA), rather than the commonly used “technology transfer.” Like other translational terms (e.g., “pipeline” 24 ), “technology transfer” may perpetuate a misconception, in this case that academic inventions are generally immediately suitable for commercialization with minimal additional development by the partner. In practice, this is rarely the case, and what is needed is an alliance to jointly investigate whether the invention can be developed successfully. Using this guiding ethos, the NCATS OSA has produced a quantity and complexity of collaborative agreements equal to those of organizations many times its size, demonstrating the kind of increased efficiency and innovation that is the hallmark of successful translational science programs. Among the breakthrough agreements OSA has put in place are a widely used template agreement for company‐academic partnerships to investigate new disease indications for existing pharmaceutical assets (https://ncats.nih.gov/alliances/forms), and an umbrella agreement covering all of NIH with the Pfizer Centers for Therapeutic Innovation (https://ncats.nih.gov/cti/about).
CHALLENGES AND PROSPECTS
We are fortunate to be living in a time of seemingly endless possibilities in biomedical science. It is a time in which what research questions we ask is no longer limited principally by our technical capabilities. This puts an increased onus on the research community to not only ask the all‐important basic questions that will drive our understanding of ourselves and our world and produce the “seed corn” for future translation, but also ask how we deliver on the promise of this amazing science to our fellow human beings, millions of whom suffer from poorly treatable diseases, the causes of which are now in many cases well‐understood. Translational science aims to enable the answering of this latter question though the same strategy that has succeeded in transforming our capacities in so many other ways: science.
The oft‐used moniker for translation is the “Valley of Death,” invoking the often inexplicable failure of the vast majority of projects in their attempts to traverse the translational journey from scientific potential into medical reality. This daunting situation will not be solved by doubling down on current approaches, sending more projects into the Valley of Death. It will be solved by scientific understanding of the reasons for failure and new technologies to avoid or overcome them. The ultimate goal of translational science is not to traverse the Valley of Death. It is to eliminate the Valley of Death entirely, through scientific understanding and innovation. This is the scientific challenge of our time, much as fundamental physics, communications and computing, and molecular biology and genomics were in times past. Each of these seemingly intractable challenges yielded to bold vision and sustained innovative execution, and in so doing immeasurably transformed our world for better. Translational science is in many ways an even greater challenge, and its realization will require the same sustained bold vision and execution. A world in which a patient never again has to hear from their doctor, “there is nothing we can do for you,” is the ultimate promise of translational science. There is no more important, more urgent, or more noble mission.
CONFLICT OF INTEREST
The author declares no competing interests for this work.
DISCLAIMER
As Chief Science Advisor of Clinical and Translational Science, Christopher P. Austin was not involved in the review or decision process for this paper.
Funding information
No funding was received for this work.
REFERENCES
- 1. Bush V. Science: The Endless Frontier. Washington, DC: United States Government Printing Office; 1945. [Google Scholar]
- 2. Austin CP. Translating translation. Nat Rev Drug Discov. 2018;17:455‐456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Scannell JW, Blanckley A, Boldon H, Warrington B. Diagnosing the decline in pharmaceutical R&D efficiency. Nat Rev Drug Discov. 2012;11:191‐200. [DOI] [PubMed] [Google Scholar]
- 4. National Research Council (US); Institute of Medicine (US) . U.S. Health in International Perspective: Shorter Lives, Poorer Health. In Woolf SH, Aron L, editors. Washington, DC: National Academies Press (US); 2013. [PubMed] [Google Scholar]
- 5. DiMasi JA, Grabowski HG, Hansen RW. Innovation in the pharmaceutical industry: New estimates of R&D costs. J Health Econ. 2016;47:20‐33. [DOI] [PubMed] [Google Scholar]
- 6. Sung NS, Crowley WF Jr, Genel M, et al. Central challenges facing the national clinical research enterprise. JAMA. 2003;289:1278‐1287. [DOI] [PubMed] [Google Scholar]
- 7. Zarin DA, Goodman SN, Kimmelman J. Harms from Uninformative Clinical Trials. JAMA. 2019;322:813‐814. [DOI] [PubMed] [Google Scholar]
- 8. Berwick DM. Disseminating innovations in health care. JAMA. 2003;289:1969‐1975. [DOI] [PubMed] [Google Scholar]
- 9. Morris ZS, Wooding S, Grant J. The answer is 17 years, what is the question: understanding time lags in translational research. J R Soc Med. 2011;104:510‐520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Osterberg L, Blaschke T. Adherence to medication. N Engl J Med. 2005;353:487‐497. [DOI] [PubMed] [Google Scholar]
- 11. Rosenberg RN. Translating biomedical research to the bedside: A national crisis and a call to action. JAMA. 2003;289:1305‐1306. [DOI] [PubMed] [Google Scholar]
- 12. Paul SM, Mytelka DS, Dunwiddie CT, et al. How to improve R&D productivity: the pharmaceutical industry's grand challenge. Nat Rev Drug Discov. 2010;9:203‐214. [DOI] [PubMed] [Google Scholar]
- 13. Zerhouni E, The NIH. Roadmap. Science. 2003;302:63‐72. [DOI] [PubMed] [Google Scholar]
- 14. Austin CP, Brady LS, Insel TR, Collins FS. NIH molecular libraries initiative. Science. 2004;306:1138‐1139. [DOI] [PubMed] [Google Scholar]
- 15. Schreiber SL, Kotz JD, Li M, et al. Advancing biological understanding and therapeutics discovery with small‐molecule probes. Cell. 2015;161:1252‐1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zerhouni EA. Translational and clinical science–time for a new vision. N Engl J Med. 2005;353:1621‐1623. [DOI] [PubMed] [Google Scholar]
- 17. Collins FS. Reengineering translational science: the time is right. Sci Transl Med. 2011;3(90):90cm17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wadman M. NIH director grilled over translational research centre. Nature News Blog. 2012; Mar 20. http://blogs.nature.com/news/2012/03/nih‐director‐grilled‐over‐translational‐research‐center.html [Google Scholar]
- 19. Sigman S. BioPharm Summit 2003: Why Drugs Fail; San Francisco. Bio‐IT World, October 29, 2003.
- 20. Zerhouni EA. US biomedical research: basic, translational, and clinical sciences. JAMA. 2005;294:1352‐1358. [DOI] [PubMed] [Google Scholar]
- 21. Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 2001;46:3‐26. [DOI] [PubMed] [Google Scholar]
- 22. Morgan P, Brown DG, Lennard S, et al. Impact of a five‐dimensional framework on R&D productivity at AstraZeneca. Nat Rev Drug Discov. 2018;17:167‐181. [DOI] [PubMed] [Google Scholar]
- 23. Huang R, Xu M, Zhu H, et al. Massive‐scale biological activity‐based modeling identifies novel antiviral leads against SARS‐CoV‐2. Nat Biotechnol. https://doi.org/ 10.1038/s41587-021-00839-1. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wagner J, Dahlem AM, Hudson LD, et al. A dynamic map for learning, communicating, navigating and improving therapeutic development. Nat Rev Drug Discov. 2018;17:150. [DOI] [PubMed] [Google Scholar]
- 25. Hardin G. The tragedy of the commons. Science. 1968;162:1243‐1248. [PubMed] [Google Scholar]
- 26. Harrison R. Phase II and phase III failures: 2013–2015. Nat Rev Drug Discov. 2016;15:817‐818. [DOI] [PubMed] [Google Scholar]
- 27. Low LA, Mummery C, Berridge BR, Austin CP, Tagle DA. Organs‐on‐chips: into the next decade. Nat. Rev. Drug Discovery. https://doi.org/ 10.1038/s41573-020-0079-3. [DOI] [PubMed] [Google Scholar]
- 28. Tristan CA, Ormanoglu P, Slamecka J, et al. Robotic high‐throughput biomanufacturing and functional differentiation of human pluripotent stem cells. bioRxiv [Preprint]. 2020:2020.08.03.235242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Treem WR, Palmer M, Lonjon‐Domanec I, et al. Consensus Guidelines: Best practices for detection, assessment and management of suspected acute drug‐induced liver injury during clinical trials in adults with chronic viral hepatitis and adults with cirrhosis secondary to hepatitis B. C and nonalcoholic steatohepatitis. Drug Saf. 2021;44:133‐165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schomaker S, Potter D, Warner R, et al. Serum glutamate dehydrogenase activity enables early detection of liver injury in subjects with underlying muscle impairments. PLoS One. 2020;15:e0229753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sanz F, Pognan F, Steger‐Hartmann T, et al. Legacy data sharing to improve drug safety assessment: the eTOX project. Nat Rev Drug Discov. 2017;16:811‐812. [DOI] [PubMed] [Google Scholar]
- 32. Tice RR, Austin CP, Kavlock RJ, Bucher JR. Improving the human hazard characterization of chemicals: a Tox21 update. Environ Health Perspect. 2013;121:756‐765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wei Z, Liu X, Ooka M, et al. Two‐dimensional cellular and three‐dimensional bio‐printed skin models to screen topical‐use compounds for irritation potential. Front Bioeng Biotechnol. 2020;8:109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rodgers G, Austin C, Anderson J, et al. Glimmers in illuminating the druggable genome. Nat Rev Drug Discov. 2018;17:301‐302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Arrowsmith CH, Audia JE, Austin C, et al. The promise and peril of chemical probes. Nat Chem Biol. 2015;11:536‐541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Carter AJ, Kraemer O, Zwick M, Mueller‐Fahrnow A, Arrowsmith CH, Edwards AM. Target 2035: probing the human proteome. Drug Discov Today. 2019;24:2111‐2115. [DOI] [PubMed] [Google Scholar]
- 37. Trobe M, Burke MD. The molecular industrial revolution: Automated synthesis of small molecules. Angew Chem Int Ed Engl. 2018;57:4192‐4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Franzini RM, Randolph C. Chemical Space of DNA‐Encoded Libraries. J Med Chem. 2016;59(14):6629‐6644. [DOI] [PubMed] [Google Scholar]
- 39. Sittampalam GS, Rudnicki DD, Tagle DA, Simeonov A, Austin CP. Mapping biologically active chemical space to accelerate drug discovery. Nat Rev Drug Discov. 2019;18:83‐84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Godfrey AG, Masquelin T, Hemmerle H. A remote‐controlled adaptive Medchem lab: an innovative approach to enable drug discovery in the 21st Century. Drug Discov Today. 2013;18:795‐802. [DOI] [PubMed] [Google Scholar]
- 41. Parthasarathy A, Anandamma SK, Kalesh KA. The medicinal chemistry of therapeutic peptides: Recent developments in synthesis and design optimizations. Curr Med Chem. 2019;26:2330‐2355. [DOI] [PubMed] [Google Scholar]
- 42. Coll‐Martínez B, Delgado A, Crosas B. The potential of proteolytic chimeras as pharmacological tools and therapeutic agents. Molecules. 2020;25(24):5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Perry ME, Valdes KM, Wilder E, Austin CP, Brooks PJ. Genome editing to ‘re‐write’ wrongs. Nat Rev Drug Discov. 2018;17:689‐690. [DOI] [PubMed] [Google Scholar]
- 44. Inglese J, Auld DS, Jadhav A, et al. Quantitative high‐throughput screening: A titration‐based approach that efficiently identifies biological activities in large chemical libraries. Proc Natl Acad Sci USA. 2006;103:11473‐11478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mathews Griner LA, Guha R, et al. High‐throughput combinatorial screening identifies drugs that cooperate with ibrutinib to kill activated B‐cell‐like diffuse large B‐cell lymphoma cells. Proc Natl Acad Sci USA. 2014;111:2349‐2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zakharov AV, Zhao T, Nguyen DT, et al. Novel consensus architecture to improve performance of large‐scale multitask deep learning QSAR models. J Chem Inf Model. 2019;59:4613‐4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fagnan DE, Yang NN, McKew JC, Lo AW. Financing translation: analysis of the NCATS rare‐diseases portfolio. Sci Transl Med. 2015;7(276):276ps3. [DOI] [PubMed] [Google Scholar]
- 48. Das S, Huang S, Lo AW. Acceleration of rare disease therapeutic development: a case study of AGIL‐AADC. Drug Discov Today. 2019;24:678‐684. [DOI] [PubMed] [Google Scholar]
- 49. Huang R, Zhu H, Shinn P, et al. The NCATS Pharmaceutical Collection: a 10‐year update. Drug Discov Today. 2019;24:2341‐2349. [DOI] [PubMed] [Google Scholar]
- 50. Austin CP. Repurposing challenges: Conceptualization of a research agenda. DIA Global Forum, June 2020.
- 51. Brooks PJ, Tagle DA, Groft S. Expanding rare disease drug trials based on shared molecular etiology. Nat Biotechnol. 2014;32:515‐518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Merkel PA, Manion M, Gopal‐Srivastava R, et al. Rare Diseases Clinical Research Network. The partnership of patient advocacy groups and clinical investigators in the rare diseases clinical research network. Orphanet J Rare Dis. 2016;11(1):66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Walkley SU, Davidson CD, Jacoby J, et al. Fostering collaborative research for rare genetic disease: the example of Niemann‐Pick type‐C disease. Orphanet J Rare Dis. 2016;11:161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Galson S, Austin CP, Khandekar E, et al. The failure to fail smartly. Nat Rev Drug Discov. 2021;20:259‐260. [DOI] [PubMed] [Google Scholar]
- 55. Menetski JP, Austin CP, Brady LS, et al. The FNIH Biomarkers Consortium embraces the BEST. Nat Rev Drug Discov. 2019;18:567‐568. [DOI] [PubMed] [Google Scholar]
- 56. Haendel MA, Chute CG, Robinson PN. Classification, ontology, and precision medicine. N Engl J Med. 2018;379:1452‐1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Collins FS, Stoffels P. Accelerating COVID‐19 Therapeutic Interventions and Vaccines (ACTIV): An Unprecedented Partnership for Unprecedented Times. JAMA. 2020;323:2455‐2457. [DOI] [PubMed] [Google Scholar]
- 58. Kitterman DR, Cheng SK, Dilts DM, Orwoll ES. The prevalence and economic impact of low‐enrolling clinical studies at an academic medical center. Acad Med. 2011;86:1360‐1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Cobb N, Witte E, Cervone M, et al. The SMART IRB platform: A national resource for IRB review for multisite studies. J Clin Transl Sci. 2019;3:129‐139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Gilmore‐Bykovskyi A, Jackson JD, Wilkins CH. The urgency of justice in research: Beyond COVID‐19. Trends Mol Med. 2021;27(2):97‐100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Djurisic S, Rath A, Gaber S, et al. Barriers to the conduct of randomised clinical trials within all disease areas. Trials. 2017;18:360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Fleurence RL, Curtis LH, Califf RM, Platt R, Selby JV, Brown JS. Launching PCORnet, a national patient‐centered clinical research network. J Am Med Inform Assoc. 2014;21:578‐582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bernard GR, Harris PA, Pulley JM, et al. A collaborative, academic approach to optimizing the national clinical research infrastructure: The first year of the Trial Innovation Network. J Clin Transl Sci. 2018;2:187‐192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Austin CP, Colvis CM, Southall NT. Deconstructing the translational Tower of Babel. Clin Transl Sci. 2019;12:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Bennett TD, Moffitt RA, Hajagos JG, et al. The National COVID Cohort Collaborative: Clinical characterization and early severity prediction. medRxiv [Preprint]. https://doi.org/ 10.1101/2021.01.12.21249511. [DOI] [Google Scholar]
- 66. National Research Council (US) Committee on A Framework for Developing a New Taxonomy of Disease . Toward Precision Medicine: Building a Knowledge Network for Biomedical Research and a New Taxonomy of Disease. Washington, DC: National Academies Press (US); 2011. [PubMed] [Google Scholar]
- 67. Levine DM, Linder JA, Landon BE. The quality of outpatient care delivered to adults in the United States, 2002 to 2013. JAMA Intern Med. 2016;176:1778‐1790. [DOI] [PubMed] [Google Scholar]
- 68. Estabrooks PA, Brownson RC, Pronk NP. Dissemination and implementation science for public health professionals: An overview and call to action. Prev Chronic Dis. 2018;15:E162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Brownson RC, Proctor EK, Luke DA, et al. Building capacity for dissemination and implementation research: one university’s experience. Implementation Sci. 2017;12:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Victor RG, Lynch K, Li N, et al. A cluster‐randomized trial of blood‐pressure reduction in Black barbershops. N Engl J Med. 2018;378:1291‐1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Labovitz DL, Shafner L, Reyes Gil M, Virmani D, Hanina A. Using artificial intelligence to reduce the risk of nonadherence in patients on anticoagulation therapy. Stroke. 2017;48:1416‐1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Austin CP. Translational misconceptions. Nat Rev Drug Discov. https://doi.org/ 10.1038/d41573-021-00008-8. [DOI] [PubMed] [Google Scholar]
- 73. Stevens AJ, Jensen JJ, Wyller K, Kilgore PC, Chatterjee S, Rohrbaugh ML. The role of public‐sector research in the discovery of drugs and vaccines. N Engl J Med. 2011;364:535‐541. [DOI] [PubMed] [Google Scholar]
- 74. Gilliland CT, White J, Gee B, et al. The fundamental characteristics of a translational scientist. ACS Pharmacol Transl Sci. 2019;2:213‐216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Pusek S, Knudson B, Tsevat J, et al. Personalized training pathways for translational science trainees: building on a framework of knowledge, skills, and abilities across the translational science spectrum. J Clin Transl Sci. 2020;4:102‐107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Tsevat J, Smyth SS. Training the translational workforce: Expanding beyond translational research to include translational science. J Clin Transl Sci. 2020;4:360‐362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Coussens NP, Sittampalam GS, Guha R, et al. The Assay Guidance Manual: Quantitative biology and pharmacology in preclinical drug discovery. Clin Transl Sci. 2018;11:461‐470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]


