Abstract
Background
Childhood diarrhoea, a major cause of morbidity and mortality in low-income regions, remains scarcely studied in many countries, such as Guinea-Bissau. Stool sample drying enables later qPCR analyses of pathogens without concern about electricity shortages.
Methods
Dried stool samples of children under five years treated at the Bandim Health Centre in Bissau, Guinea-Bissau were screened by qPCR for nine enteric bacteria, five viruses, and four parasites. The findings of children having and not having diarrhoea were compared in age groups 0–11 and 12–59 months.
Results
Of the 429 children– 228 with and 201 without diarrhoea– 96.9% and 93.5% had bacterial, 62.7% and 44.3% viral, and 52.6% and 48.3% parasitic pathogen findings, respectively. Enteroaggregarive Escherichia coli (EAEC; 60.5% versus 66.7%), enteropathogenic E. coli (EPEC; 61.4% versus 62.7%), Campylobacter (53.2% versus 51.8%), and enterotoxigenic E. coli (ETEC; 54.4% versus 44.3%) were the most common bacterial pathogens. Diarrhoea was associated with enteroinvasive E. coli (EIEC)/Shigella (63.3%), ETEC (54.4%), astrovirus (75.0%), norovirus GII (72.6%) and Cryptosporidium (71.2%). The only pathogen associated with severe diarrhoea was EIEC/Shigella (p<0.001). EAEC was found more frequent among the infants, and EIEC/Shigella, Giardia duodenalis and Dientamoeba fragilis among the older children.
Conclusions
Stool pathogens proved common among all the children regardless of them having diarrhoea or not.
Author summary
Diarrhoeal diseases rank second as cause of childhood mortality and morbidity in low-income countries, yet prospective cohort studies in children with and without diarrhea covering the large variety of diarrhoeal pathogens are limited. While some studies have been conducted among Guinea-Bissauan children, many of them were from the 1990s, when the coverage of the various pathogens was less extensive and the diagnostic methods less sensitive than the modern qPCR methods.
We conducted an observational study with a large cohort and covered concomitantly the various bacterial, viral and parasitic agents, and analyzed their associations with the presence/absence of diarrhoeal symptoms and age groups. Importantly, the assay performed well with dried stool samples and, therefore, appears applicable for epidemiological studies in resource-poor regions.
A pathogen finding was recorded for almost all (98%) children: bacteria in 97%, viruses in 59% and parasites in 51%. Ongoing diarrhoea was associated with findings of enteroninvasive Escherichia coli/Shigella, enterotoxigenic E. coli, astrovirus, norovirus GII and Cryptosporidium. Differences were seen between age groups, infants and young children. The only pathogen associated with severe diarrhoea was enteroninvasive E. coli/Shigella.
Introduction
Diarrhoeal diseases rank second as cause of childhood mortality and morbidity in low-income countries (LICs) worldwide. In the African region, diarrhoea accounts for more than 10% of deaths among children under five years of age [1–3]. The aetiological agents for childhood diarrhoea comprise a variety of bacteria, viruses and parasites [4] which mostly spread through unclean water and food, a consequence of poor overall hygiene [4,5]. Information on the aetiology is needed for epidemiological surveillance, devising preventive measures such as vaccinations, and empiric treatments [6], yet national surveillance programmes are either lacking or non-functional. Thus, the current understanding is based on limited research data merely covering a handful of countries [7,8]. In only a few instances are bacterial, viral, and parasitic pathogens scrutinized using the same study setting [4,9,10].
Guinea-Bissau with an under-five mortality rate of 78.5/1000 live births [11] is among the 27 countries categorized by the World Bank [12] as low-income economies. Prospective cohort studies of Guinea-Bissauan children investigating infections with diarrhoeal pathogens report findings of rotavirus (3–35%) [13–16], Campylobacter (1.8%), Shigella (0.6–2%), and diarrhoeagenic Escherichia coli strains (4–58%) [15–17], Vibrio cholerae (1.2.8%, 1168–14 228 cases) [15,18,19], Giardia duodenalis [15,16,20], Cryptosporidium (0.1–12.5%) [15,16,21,22], and Entamoeba histolytica/dispar (2–26.6%) [15,16]. Many of these studies conducted in the 1990s applied contemporary, less sensitive analysis methods not covering as many pathogens as the modern qPCR assays: for example, research into diarrhoeal viruses only looked at rotaviruses [13–16,23]. Now that co-infections have proved common, methods with broad coverage are needed [23].
This research was undertaken to determine the frequency of potentially pathogenic viruses, bacteria and parasites and their association with diarrhoea among Guinea-Bissauan children.
Materials and methods
Ethics statement
The study protocol was approved by the Ethics Committee of Helsinki University Hospital, Finland and Comité Nacional de Ética na Saúde, Instituto de Pública, Guinea-Bissau (No: 031/CNES/2010). The children’s parents or carers were informed about the aim of study and they signed a written informed consent.
We conducted an observational cohort study among children under five years of age having and not having diarrhoea who were treated at the Bandim Health Centre in the suburb Bandim in Bissau, the capital of Guinea-Bissau.
Bandim belongs to a health project study area comparable to most parts of Bissau in terms of socio-economic status, including income and standard of living. The country has a tropical climate with the rainy season lasting from June to October.
Study population and sample material
The study was conducted between November 2010 and October 2012. All children less than five years seeking medical care at the Bandim Health Centre were screened and invited to participate (Fig 1). Those with ongoing diarrhoea were included in the diarrhoea group. Those with no diarrhoeal symptoms over the past 7 days were eligible in the control group. Children whose condition required transfer to hospital were not eligible. The parents/carers of the children recruited filled in a questionnaire gathering information on symptoms (those with diarrhoea) as well as socio-economic parameters. One fresh stool specimen was collected from each child. Applied on filter paper (Hemoccult, Becman Coulter, Co. Clare, Ireland), the samples were dried at room temperature and stored in separate plastic bags with desiccants.
Fig 1. Study protocol.
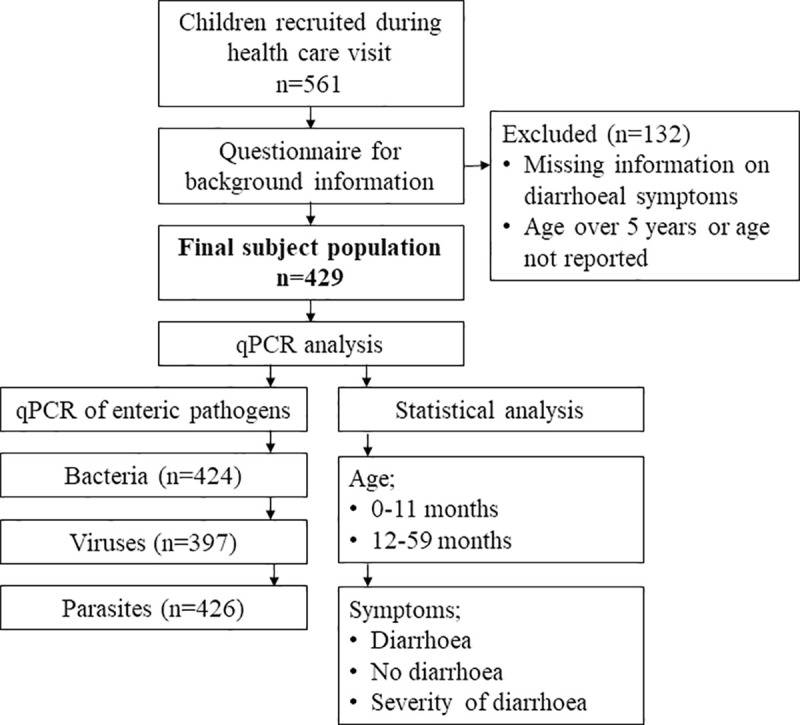
A total of 561 children were recruited into the study. Samples of children over five years of age or those with no information about age or diarrhoea were not included in the analyses.
A total of 561 children were recruited. The inclusion criteria were age under 5 years, information of presence/absence of diarrhoea at the time of sampling, and a stool sample available for analyses (Fig 1). Diarrhoea as defined by World Health Organization (WHO) [3]: passage of three or more loose or liquid stools per day or more frequently than is normal for the individual. Children meeting this criterion were included in the diarrhoeal cohort, the others in that not having diarrhoea. Severe diarrhoea was defined as six or more unformed diarrhoeal stools per day with fever, or haemorrhagic stools; other kinds of diarrhoea were categorized as non-severe.
Total nucleic acid Isolation
Total nucleic acid was extracted from the stool specimens with the NucliSENS kit (Durham, NC) using the Tecan Freedom EVO 200 liquid handling system (Tecan, Männedorf, Switzerland). Extraction script was scheduled using Tecan Evoware 2 software based on easyMAG (bioMérieux, Marcy l´Etoile, France) automatic nucleic acid purification platform following the manufacturer´s instructions. Briefly, the filter papers were cut into pieces measuring 1x1 cm and moistened with 2 ml lysis buffer overnight at room temperature. After vortexing, the filter papers were removed from the buffer and the remaining liquid was used for nucleic acid isolation [24]. The extraction was performed using 900 μl of the lysed sample and eluted to a volume of 100 μl.
PCR Amplification for detection of diarrhoeal pathogens
The samples were analyzed by three different qPCR assays: (1) a test screening for nine enteric bacteria [25], covering Campylobacter sp., Salmonella sp., Y. enterocolitica, Vibrio cholerae, enteroinvasive E. coli (EIEC)/ Shigella, enterohaemorrhagic E. coli (EHEC), enteroaggregative E. coli (EAEC), enteropathogenic E. coli (EPEC) and enterotoxigenic E. coli (ETEC), utilizing the Stratagene MxPro 3005P instrument (Agilent Technologies Inc., CA) according to manufacturer´s instructions; (2) a test screening for four enteric parasites (Amplidiag Stool Parasites; Mobidiag Ltd, Helsinki, Finland), covering G. duodenalis, Cryptosporidium sp., E. histolytica and Dientamoeba fragilis; and (3) a test screening for five viruses (Amplidiag Viral GE; Mobidiag, Finland), covering norovirus GI and GII, rotavirus A, sapovirus, astrovirus, and adenovirus 40 and 41. The latter two assays were carried out on the Bio-Rad CFX96 detection system with Bio-Rad CFX Manager software (Bio-Rad Laboratories, CA) according to the manufacturer´s instructions, and run files were analyzed automatically with Amplidiag Analyzer (Mobidiag, Finland) software.
All the tests are CE marked under IVDD, including testing for limit-of-detection, cross-reactivity, inclusivity, repeatability, reproducibility, stability, and external clinical performance evaluation study. All details are readily available in the kit information for users, and they are several independent publications for the kits. For further information, see reference [26]. An average LoD for target is approx. 103–105 CFU/ ml, which is line with other commercial tests on the market. The tests do not cross-react with any organism in commercial flora or closely related, non-pathogenic organisms. Based on original clinical performance evaluation study, the Viral GE kit has an overall sensitivity of 98.1% and specificity of 98.8%, the respective figures for the Bacterial GE kit are 98.7% and 99.6%, and for the Stool Parasite kit 96,9% and 99,4%, respectively.
Statistical analysis
Pearson’s chi-square test, Fisher’s exact test, or binary logistic regression analysis was used to compare categorical variables, when applicable. Statistical significance was defined as p < 0.05 or 95% CIs ranging only either above or below 1. The statistical analyses were carried out using SPSS 22 software (IBM Corp, Armonk, NY).
Results
Demographics
A total of 561 individuals were recruited for the study, 429 of whom met the inclusion criteria. The final study population comprised 228 children having and 201 not having diarrhoea. There were 192 infants (0–11 months) and 253 young children (12–59 months) (Table 1).
Table 1. Demographic data of the children recruited.
| Total n (%) |
No diarrhoea n (%) |
Diarrhoea n (%) |
|
|---|---|---|---|
| Total | 429 (100) | 201 (46.9) | 228 (53.1) |
| Sex a | |||
| Male | 204 (49.2) | 85 (41.7) | 119 (58.3) |
| Female | 211 (50.8) | 105 (49.8) | 106 (50.2) |
| Age (years) | |||
| mean (range) | 1.6 (0.0–5.0) | 1.7 (0.1–5.0) | 1.4 (0.0–4.4) |
| Infants (0–11 months) | 192 (45.0) | 93 (48.4) | 99 (51.6) |
| Young children (12–59 months) | 235 (55.0) | 106 (45.1) | 129 (54.9) |
| Diarrhoea during preceding year | |||
| any | 269 (65.9) | 105 (39.0) | 164 (61.0) |
| 0–2 times | 157 (38.8) | 60 (38.2) | 97 (61.8) |
| ≥3 times | 109 (26.9) | 45 (41.3) | 64 (58.7) |
Data is missing, n (%)
a) 14 (3.3)
Symptoms of diarrhoea
The average duration of diarrhoea before presentation was 2.7 days (range 1–14) and mean maximum number of diarrhoeal stools per day 4.9 (range 1–10). Diarrhoea was reported for 65.9% at least once during the preceding year, and 26.9% had experienced three episodes or more (Table 1). The stools were watery in 43.4%, mucous in 37.5% and bloody in 5.6% of the cases. Of the other symptoms, the most commonly recorded was fever (40.5%) followed by vomiting (19.0%), and stomach pain (14.3%). Prior medical consultations for the same illness before contacting the Bandim Health Center were rare (5.1%).
Diarrhoeal pathogens in stool samples
The microbial findings from all participants are shown in Table 2. For 1.6% of the children no pathogens were identified. Only one pathogen was found in 2.2% and 6.0% of children with or without diarrhoea, respectively. No differences were seen in the number of pathogen types between those having and not having diarrhoea (Table 2).
Table 2. Microbial findings in Guinea-Bissauan children with and without diarrhoea.
| Total n (%) |
No diarrhoea n (%) |
Diarrhoea n (%) |
Diarrhoea versus No diarrhoea OR (95% CI) p-value |
||
|---|---|---|---|---|---|
| Total | 429 (100) | 201 (46.9) | 228 (53.1) | ||
| No pathogen | 7 (1.6) | 5 (2.5) | 2 (0.9) | ref | 0.063 |
| Any pathogen | 422 (98.4) | 194 (96.5) | 226 (99.1) | 2.9 (0.6–15.2) | 0.176 |
| One pathogen | 17 (4.0) | 12 (6.0) | 5 (2.2) | 1.0 (0.1–7.3) | 0.967 |
| Multiple pathogens | 405 (94.4) | 184 (91.5) | 221 (96.9) | 3.0 (0.6–15.8) | 0.188 |
| Any bacteria a | 409 (96.5) | 188 (93.5) | 221 (96.9) | 1.5 (0.5–4.6) | 0.404 |
| Campylobacter | 225 (53.1) | 107 (53.2) | 118 (51.8) | 0.9 (0.6–1.3) | 0.553 |
| EAEC | 272 (64.2) | 134 (66.7) | 138 (60.5) | 0.7 (0.5–1.0) | 0.113 |
| EHEC | 6 (1.4) | 5 (2.4) | 1 (0.4) | 0.2 (0.0–1.5) | 0.076 |
| EIEC/Shigella | 98 (23.1) | 36 (17.9) | 62 (27.2) | 1.7 (1.0–2.6) | 0.032 |
| EPEC | 266 (62.7) | 126 (62.7) | 140 (61.4) | 0.9 (0.6–1.3) | 0.607 |
| ETEC | 213 (50.2) | 89 (44.3) | 124 (54.4) | 1.4 (1.0–2.1) | 0.066 |
| Salmonella | 11 (2.6) | 6 (3.0) | 5 (2.2) | 0.7 (0.2–2.4) | 0.574 |
| V. cholerae | 2 (0.5) | 0 (0.0) | 2 (0.9) | NA | 0.502 |
| Yersinia | 3 (0.7) | 1 (0.5) | 2 (0.9) | 1.7 (0.2–19.2) | 0.557 |
| Any virus b | 233 (58.7) | 89 (44.3) | 143 (62.7) | 2.2 (1.5–3.3) | 0.001 |
| Adenovirus 40, 41 | 73 (18.4) | 32 (15.9) | 40 (17.5) | 1.1 (0.7–1.8) | 0.723 |
| Astrovirus | 40 (10.1) | 10 (5.0) | 30 (13.2) | 2.9 (1.4–6.0) | 0.004 |
| Norovirus GI/GII | 92 (23.2) | 30 (14.9) | 62 (27.2) | 2.1 (1.3–3.4) | 0.003 |
| Norovirus GI | 21 (5.3) | 10 (5.0) | 11 (4.8) | 1.0 (0.4–2.3) | 0.903 |
| Norovirus GII | 73 (18.4) | 20 (10.0) | 53 (23.2) | 2.7 (1.6–4.8) | 0.001 |
| Rotavirus A | 93 (23.4) | 40 (19.9) | 53 (23.2) | 1.2 (0.7–1.9) | 0.463 |
| Sapovirus | 29 (7.3) | 10 (10.0) | 19 (8.3) | 1.7 (0.8–3.8) | 0.184 |
| Any parasite c | 217 (50.9) | 97 (48.3) | 120 (52.6) | 1.2 (0.8–1.8) | 0.341 |
| Cryptosporidium sp. | 59 (13.8) | 17 (8.5) | 42 (18.4) | 2.4 (1.3–4.4) | 0.003 |
| D. fragilis | 43 (10.7) | 20 (10.0) | 23 (10.1) | 1.0 (0.5–1.9) | 0.999 |
| E. histolytica | 3 (0.7) | 3 (1.5) | 0 (0.0) | NA | 0.099 |
| G. duodenalis | 159 (37.3) | 74 (36.8) | 85 (37.3) | 1.0 (0.7–1.5) | 0.849 |
Data is missing, n (%)
a) 5 (1.2)
b) 32 (7.5)
c) 3 (0.7)
OR = odds ratio (logistic regression); CI = confidence interval; bolding indicates statistically significant at p<0.05 (Pearson χ2 test or Fisher´s exact test); ref = reference group; NA = not applicable
Bacterial pathogens
Bacterial pathogens were detected for 96.5% of children. EAEC and EPEC were the most common findings (64.2% and 62.7%, respectively), followed by Campylobacter (53.1%), ETEC (50.2%), and EIEC/Shigella (23.1%) (Table 2).
When comparing the subgroups with and without diarrhoea, EIEC/Shigella was found more frequently among those with than without (27.2% versus 17.9%, OR 1.7 [1.1–2.6], p = 0.03); with ETEC the rates were 50.2% versus 44.3% (OR 1.4 [1.0–2.1], p = 0.066).
Comparisons between the two age groups showed higher rates of EAEC among infants than young children (72.4% versus 56.6%, p<0.001), and higher rates of EIEC/Shigella among young children than infants (29.9% versus 14.7%, p<0.001). As for the other bacteria, no differences were recorded between the various age groups (S1 Table).
Table 3 depicts the greater prevalence of EAEC detected for infants without diarrhoea than those with it (73/93 versus 66/99, p = 0.035). When comparing young children having and not having diarrhoea, those having it showed higher rates of EIEC/Shigella (48/129 versus 22/106, p = 0.007) and ETEC (68/129 versus 41/106, p = 0.038).
Table 3. Diarrhoeal pathogens in 429 children with and without diarrhoea divided into two age groups: infants (0–11 months) and young children (12–59 months).
| 0–11 months | 12–59 months | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Total n (%) |
No diarrhoea n (%) |
Diarrhoea n (%) |
Diarrhoea versus No diarrhoea OR (95% CI) p-value |
No diarrhoea n (%) |
Diarrhoea n (%) |
Diarrhoea versus No diarrhoea OR (95% CI) p-value |
|||
| Total | 429 (100) | 94 (48.4) | 99 (51.6) | 107 (45.1) | 129 (54.9) | ||||
| Any pathogen | 422 (98.8) | 91 (97.8) | 98 (99.0) | 2.2 (0.2–24.2) | 0.612 | 103 (97.2) | 128 (99.2) | 3.7 (0.4–36.4) | 0.330 |
| Any bacteria a | 409 (95.8) | 89 (95.7) | 95 (96.0) | 0.5 (0.1–3.0) | 0.684 | 98 (92.5) | 126 (97.7) | 3.9 (0.8–19.5) | 0.144 |
| Campylobacter | 225 (53.1) | 49 (52.7) | 47 (47.5) | 0.8 (0.4–1.4) | 0.380 | 58 (54.7) | 71 (55.0) | 1.0 (0.6–1.7) | 0.963 |
| EAEC | 272 (64.2) | 73 (78.5) | 66 (66.7) | 0.5 (0.3–1.0) | 0.035 | 60 (56.6) | 72 (55.8) | 0.9 (0.6–1.6) | 0.825 |
| EHEC | 6 (1.4) | 1 (1.1) | 0 (0.0) | NA | 0.479 | 4 (3.8) | 1 (0.8) | 2.0 (0.2–1.8) | 0.176 |
| EIEC/Shigella | 98 (23.1) | 14 (15.1) | 14 (14.1) | 0.9 (0.4–2.0) | 0.809 | 22 (20.8) | 48 (37.2) | 2.2 (1.2–4.0) | 0.007 |
| EPEC | 266 (62.7) | 58 (62.4) | 65 (65.7) | 1.1 (0.6–2.0) | 0.782 | 67 (63.2) | 75 (58.1) | 0.8 (0.5–1.3) | 0.365 |
| ETEC | 213 (50.2) | 48 (51.6) | 56 (56.6) | 1.2 (0.7–2.1) | 0.597 | 41 (38.7) | 68 (52.7) | 1.7 (1.0–2.9) | 0.038 |
| Salmonella | 11 (2.6) | 5 (5.4) | 3 (3.0) | 0.5 (0.1–2.3) | 0.483 | 1 (0.9) | 2 (1.6) | 1.6 (1.4–18.3) | 1.000 |
| V. cholerae | 2 (0.5) | 0 (0.0) | 1 (1.0) | NA | 1.000 | 0 (0.0) | 1 (0.8) | NA | 1.000 |
| Yersinia | 3 (0.7) | 1 (1.1) | 1 (1.0) | 0.9 (0.1–14.9) | 1.000 | 0 (0.0) | 1 (0.8) | NA | 1.000 |
| Any viruses b | 233 (54.6) | 51 (54.8) | 65 (65.7) | 1.7 (0.9–3.2) | 0.092 | 38 (35.8) | 78 (60.5) | 2.8 (1.6–4.9) | <0.001 |
| Adenovirus 40, 41 | 73 (18.4) | 20 (21.5) | 17 (17.2) | 0.8 (0.4–1.6) | 0.456 | 12 (11.3) | 23 (17.8) | 1.7 (0.8–3.5) | 0.186 |
| Astrovirus | 40 (10.1 | 4 (4.3) | 16 (16.2) | 4.4 (1.4–13.6) | 0.007 | 6 (5.7) | 14 (10.9) | 2.0 (0.7–5.4) | 0.172 |
| Norovirus GI | 21 (5.3) | 8 (8.6) | 5 (5.1) | 0.5 (0.2–1.8) | 0.333 | 2 (1.9) | 6 (4.7) | 2.5 (0.5–12.5) | 0.305 |
| Norovirus GII | 73 (18.4) | 12 (12.9) | 28 (28.3) | 2.7 (1.3–5.8) | 0.008 | 8 (7.5) | 25 (19.4) | 2.9 (1.2–6.8) | 0.011 |
| Rotavirus A | 93 (23.4) | 22 (23.7) | 24 (24.2) | 1.0 (0.5–2.0) | 0.903 | 18 (17.0) | 29 (22.5) | 1.4 (0.7–2.7) | 0.338 |
| Sapovirus | 29 (7.3) | 6 (6.5) | 8 (8.1) | 1.3 (0.4–3.9) | 0.654 | 4 (3.8) | 11 (8.5) | 2.3 (0.7–7.5) | 0.151 |
| Any parasites c | 217 (50.8) | 35 (37.6) | 40 (40.4) | 1.4 (0.6–2.0) | 0.653 | 60 (56.6) | 80 (62.0) | 1.2 (0.7–2.0) | 0.503 |
| Cryptosporidium sp. | 59 (13.8) | 10 (10.8) | 21 (21.2) | 2.3 (1.0–5.1) | 0.045 | 7 (6.6) | 21 (16.3) | 2.7 (1.0–6.6) | 0.026 |
| D. fragilis | 159 (37.3) | 4 (4.3) | 5 (5.1) | 1.2 (0.3–4.7) | 1.000 | 16 (15.1) | 18 (14.0) | 0.9 (0.4–1.8) | 0.724 |
| E. histolytica | 43 (10.7) | 1 (1.1) | 0 (0.0) | NA | 0.487 | 2 (1.9) | 0 (0.0) | NA | 0.198 |
| G. duodenalis | 3 (0.7) | 24 (25.8) | 21 (21.2) | 0.8 (0.4–1.5) | 0.476 | 48 (45.3) | 64 (49.6) | 1.1 (0.7–1.9) | 0.599 |
Data is missing, n (%)
a) 5 (1.2)
b) 32 (7.5)
c) 3 (0.7)
OR = odds ratio (logistic regression); CI = confidence interval; bolding indicates statistically significant at p<0.05 (Pearson χ2 test or Fisher´s exact test); NA = not applicable
Viral pathogens
Viral pathogens were detected in 58.7% of all the children, more commonly among those having diarrhoea than those not having the disease (62.7% versus 44.3%, OR 2.2 [1.5–3.3], p = 0.001). The most frequently found viruses were rotavirus A (23.4%), norovirus GII (18.4%), and adenovirus (18.4%) (Table 2).
Table 2 presents a comparison between those with and without the disease. In the diarrhoea group, a higher frequency was recorded for astrovirus (13.2% versus 5.0%, OR 2.9 [1.4–6.0], p = 0.004) and norovirus GII (23.2% versus 10.0%, OR 2.7 [1.6–4.8], p = 0.001) (Table 3), particularly among infants (astrovirus 4.3% versus 16.2%; OR 4.4 [1.4–13.6], p = 0.007] and norovirus GII (12.9% versus 28.3%; OR 2.7 [1.3–5.8], p = 0.008).
Parasitic pathogens
In 50.9% of all the children parasites were detected, most frequently Giardia (37.3%) followed by Cryptosporidium (13.8%), D. fragilis (10.7%), and E. histolytica (0.7%). When comparing differences between the subgroups with and without diarrhoea, the only significant difference was seen for Cryptosporidium which proved more prevalent among children having (18.4%) than those not having the disease (8.5%) (Table 2). The finding applied to both age groups (Table 3) (21.2% versus 10.8% in infants OR 2.3 [1.0–5.1], p = 0.045 and 16.3% versus 6.6% in young children OR 2.7 [1.1–6.6] p = 0.026). When analyzed by age-groups, young children had higher rates of Giardia and D. fragilis than infants (S1 Table) (23.4% versus 48.5%; OR 3.0 [2.0–4.7], p<0.001 and 4.7% versus 14.5%; OR 3.5 [1.6–7.5], p = 0.001). For other parasites no differences were detected.
Association with severe diarrhoea
The severity analyses (Table 4) covered the six most common bacterial pathogens (Campylobacter, EAEC, EIEC/Shigella, EPEC, ETEC, Salmonella), the two most common viruses (norovirus GII, rotavirus A), and the two most common parasites (Cryptosporidium, Giardia).
Table 4. Severity of diarrhoea.
For severity analyses, the findings were selected that involved the six most common bacterial pathogens (Campylobacter, EAEC, EIEC/Shigella, EPEC, ETEC, Salmonella), the two most common viruses (norovirus GII, rotavirus A) and the two most common parasites (Cryptosporidium, G. duodenalis).
| Total n (%) |
Nonsevere n (%) |
Severe* n (%) |
Severe versus Nonsevere OR (95% CI) p-value |
||
|---|---|---|---|---|---|
| Total | 223 (100) | 152 (68.2) | 71 (31.8) | ||
| Any pathogen | 221 (99.1) | 150 (98.7) | 71 (100) | NA | 1.000 |
| Any bacteria | 216 (96.9) | 146 (96.1) | 70 (98.6) | 2.4 (0.3–20.9) | 0.667 |
| Campylobacter | 116 (52.0) | 81 (53.3) | 35 (49.3) | 0.8 (0.5–1.5) | 0.545 |
| EAEC | 136 (61.0) | 87 (57.2) | 49 (69.0) | 1.6 (0.9–3.0) | 0.104 |
| EIEC/Shigella | 61 (27.4) | 26 (17.1) | 32 (45.1) | 3.5 (1.9–6.4) | <0.001 |
| EPEC | 137 (61.4) | 95 (62.5) | 42 (59.2) | 0.9 (0.5–1.5) | 0.591 |
| ETEC | 121 (54.3) | 77 (50.7) | 44 (62.0) | 1.6 (0.9–2.8) | 0.126 |
| Salmonella | 5 (2.2) | 2 (1.3) | 3 (4.2) | 3.3 (0.5–20.1) | 0.330 |
| Any virus | 141 (63.2) | 100 (65.8) | 41 (57.7) | 0.9 (0.5–1.7) | 0.688 |
| Norovirus GII | 53 (26.0) | 40 (26.3) | 13 (18.3) | 0.7 (0.3–1.4) | 0.318 |
| Rotavirus A | 53 (26.0) | 35 (23.0) | 18 (25.3) | 1.3 (0.7–2.5) | 0.460 |
| Any parasite | 118 (52.9) | 84 (55.3) | 34 (47.9) | 0.7 (0.4–1.3) | 0.281 |
| Cryptosporidium | 41 (18.4) | 29 (19.1) | 12 (16.9) | 0.9 (0.4–1.8) | 0.680 |
| Giardia | 85 (38.1) | 65 (42.8) | 20 (28.2) | 0.5 (0.3–1.0) | 0.033 |
*Severe diarrhoea was defined as six or more unformed diarrhoeal stools per day with fever, or haemorrhagic stools.
OR = odds ratio (logistic regression); CI = confidence interval; bolding indicates statistically significant at p<0.05 (Pearson χ2 test or Fisher´s exact test); NA = not applicable
Only EIEC/Shigella was associated with severe diarrhoea, (45.1% versus 17.1%; OR 3.5 [1.9–6.4], p<001), also when analyzed by age groups (infants: 30.0% versus 7.5%, p = 0.009, young children: 56.1% versus 28.2%, p = 0.003) (S2 Table), while Giardia was less frequently found in severe than non-severe diarrhoea (28.2% versus 42.8%; OR 0.5 [0.3–1.0], p = 0.033); by age-group, the difference was only statistically significant among infants (3.3% versus 29.9%; OR 0.1 [0.0–0.6], p = 0.003), not among young children (46.3% versus 52.9%, OR 0.8 [0.4–1.6], p = 0.488).
Other species showed no statistical significance, neither when analyzed for all participants nor both of the age groups (S2 Table).
Discussion
General characteristics
Hardly any of the participants had stool samples free of pathogens: qPCR detected enteropathogens in almost all the children regardless of whether they had diarrhoea (99.1%) or not (96.5%). These data accord with other studies carried out in low-income countries [27]: the pathogen rates for Angolan children with and without diarrhoea were 97% and 84%, and for Rwandan children 94% and 79%, respectively [28]. These investigations agree with others which, besides demonstrating the high sensitivity of PCR methods, show that children in LICs are throughout their early years constantly exposed to a multitude of pathogens [4,29]. Bacterial pathogens were identified more commonly than viruses and parasites which were not rare either. The findings are discussed in detail below.
Bacterial pathogens
Among all the children, regardless of them having diarrhoea or not, the pathogens detected most frequently were EAEC, EPEC, Campylobacter, and ETEC. These findings agree with data for Burkina Faso [30] and earlier results from 1996–98 for Guinea-Bissau reporting EAEC as the most prevalent pathogen in those with and without diarrhoea alike, followed by diffusely adherent E. coli (DAEC), EPEC, Giardia, ETEC, rotavirus, and Salmonella [23]. This investigation undertaken in Guinea-Bissau ten years before ours only associated rotavirus, ETEC, EIEC/Shigella, and Cryptosporidium with diarrhoeal symptoms, however. Compared to those older data, our Guinea-Bissauan children had a higher prevalence of Campylobacter and less Salmonella. While diarrhoeagenic E. coli are also frequent findings in travellers to Sub-Saharan Africa [31,32], it is noteworthy that they relatively seldom have EIEC/Shigella and Campylobacter [31,33]. The differences between visitors and children living in LICs may simply be explained by differing exposure: travellers mostly stay in places with a higher standard of hygiene. In Southeast Asia Campylobacter is common among locals and travellers alike [31,34].
Campylobacter was found in almost equal proportions among children with (51.8%) and without (53.2%) diarrhoea. Campylobacter have been reported to be shed for up to four weeks after resolution of symptoms [35]. Long-term asymptomatic carriage may lead to underestimates of its aetiologic role in childhood diarrhoea [36]. V. cholerae was only detected in two children with diarrhoea. The previous large cholera epidemic in Guinea-Bissau occurred in 2008, lasting from May to January 2009 with a total of 14 228 suspected cases [18]. Our study was conducted nearly two years after it, from November 2010 to October 2012, and the next smaller outbreak only took place between 2012 and 2014 [37].
Viral pathogens
Our high rates of rotavirus (23.4%) presumably relate to the timing of our samples collection before Guinea-Bissau introduced rotavirus vaccine into its national immunization programme in December 2015 [38]; after that the virus’s prevalence has probably declined. In our data norovirus GII and adenovirus were also frequent findings (18.4%), while the rates of sapovirus and norovirus GI were fairly low. Noroviruses altogether (23.2%) were almost as common as rotavirus, but only the GII genotype was associated with diarrhoea. These results accord with previous studies conducted in Gambia [39] and Burkina Faso [40]. Noroviruses are considered a major cause of viral gastroenteritis across all age groups worldwide [41]. Among African children, the impact and epidemiology of norovirus infections remain scarcely investigated [42]. Our results highlight the role of viruses in childhood diarrhoea affecting inhabitants of a low-income country.
Parasitic pathogens
In Sub-Saharan Africa, up to half of the children are afflicted with soil-transmitted protozoan parasites and helminthes infections [43]. In our study, parasitic pathogens were identified in 51% of the children. However, the total burden of parasites may have been even higher since helminths were not covered by our qPCR.
Giardia (37%) and Cryptosporidium (14%) were the most common, and Giardia proved prevalent also among those asymptomatic. Asymptomatic carriage of Giardia and Cryptosporidium found common in Sub-Saharan Africa [4,44] has been associated with growth retardation [45].
As regards children, the prevalence of D. fragilis in Africa has scarcely been researched. In our young study population it proved equally common in the symptomatic and asymptomatic groups. It should be noted that the present investigation focused on acute diarrhoea, not on milder prolonged gastrointestinal symptoms typical to dientamoebiasis [46].
Pathogen findings in two age groups
In the multisite birth cohort study MAL-ED [29], norovirus GII and rotavirus had the strongest association with diarrhoea over the first year of life. Our investigation associated norovirus GII and astrovirus with diarrhoea in infants, and EIEC/Shigella, ETEC, norovirus GII, and Cryptosporidium with diarrhoea in young children. These findings agree with the results of the GEMS research [4] and previous Guinea-Bissauan studies [17,23]. Giardia and D. fragilis were most common among young children, but not associated with acute diarrhoea in either of the two age groups. According with our findings, Valentiner-Branth et al. have shown in a study among Guinea-Bissauan children that already during the first 30 days of their life, 50% of them become infected with EAEC, followed by DAEC, EPEC, and ETEC [23]. Human breast milk contains an array of anti-infectious and anti-inflammatogenic factors. Thus, breastfed infants may become colonized by diarrhoeal pathogens and still stay healthy [47]. Indeed, WHO recommends breastfeeding for the first six months exclusively and, complemented with solid foods at six months, at least till the age of two years [48]. Unfortunately, our questionnaire did not cover the duration of breastfeeding. However, by Guinea-Bissauan custom, mothers breastfeed their infants for two years (P-E. Kofoed, personal communication).
Severity of diarrhoea
Severe diarrhoea was associated with EIEC/Shigella but not other pathogen findings among both infants and young children. Other studies have detected rotavirus, Cryptosporidium, Shigella, and ETEC in cases of moderate-to-severe diarrhoea. For countries whose national programme does not cover rotavirus vaccination, association with rotavirus has also been reported [4,29]. In our study, the association between moderate or severe diarrhoea and Cryptosporidium did not reach statistical significance (p = 0.7). In LICs, Giardia infections are generally considered self-limiting [4,29], which accords with our results: Giardia proved less common in severe than nonsevere diarrhoea.
Limitations
We did not relate our findings to Cycle threshold (Ct)-values of qPCR. Such analysis could possibly show differences between children having/not having diarrhoea [34]. Indeed, because the highly sensitive qPCR analyses may detect minuscule amounts of pathogens without clinical significance, further research into the cut-off Ct-values in relation to symptomatic infections may be warranted. However, many pathogens such as noroviruses, EAEC, Salmonella, or Campylobacter may colonize the intestine and be found in stools for weeks after the resolution of clinical symptoms [49]. Furthermore, as qPCR measures nucleic acids, the assay does not distinguish between live and dead pathogens. Since the stools were not cultured in our study, we could not distinguish Shigella from EIEC. Dried samples may yield lower discovery rates than fresh ones, but they have been shown to be higher than those for frozen samples [50]. In regions with an unreliable electric supply, dried samples remain the only option to investigate the aetiological agents of diarrhoea comprehensively. For our assessment of the severity of diarrhoea, information on the degree of dehydration was not available.
Conclusions
EAEC and EPEC were the pathogens detected most frequently among Guinea-Bissauan children regardless of whether they had diarrhoea or not. Campylobacter were found in symptomatic and asymptomatic children alike. As this pathogen can be shed for prolonged periods after symptoms have resolved, its role in the aetiology of diarrhoea in LICs may have been underestimated. The introduction of rotavirus vaccine into Guinea-Bissau’s national immunization programme after our study is likely to have brought down the high rates of rotavirus in a few years. The problem with the other stool pathogens persists, however.
Supporting information
(DOCX)
(DOCX)
(XLSX)
Abbreviations
- Ct
Cycle threshold
- DAEC
diffusely adherent Escherichia coli
- EAEC
enteroaggregative E. coli
- EIEC
enteroinvasive E. coli
- EHEC
enterohaemorrhagic E. coli
- EPEC
enteropathogenic E. coli
- ETEC
enterotoxigenic E. coli
- GEMS
Global Enteric Multicenter Study
- LICs
low-income countries
- qPCR
quantitative polymerase chain reaction
- WHO
World Health Organization
Data Availability
All relevant data are in the manuscript and its Supporting Information files. The individuals' personal data cannot be shared publicly because of research regulations. Contact details for further information: Meilahti Vaccine Research Center, MeVac Biomedicum 1, Helsinki University Hospital and University of Helsinki, POB 700, 00029 HUS, email vaccine@hus.fi.
Funding Statement
This work was supported by the Finnish Governmental Subsidy for Health Science Research (AK), the SSAC Foundation (AK), the Sigrid Jusélius Foundation (AK) and the University of Helsinki Doctoral Programme in Population Health Foundation (SM). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Liu L, Johnson HL, Cousens S, Perin J, Scott S, Lawn JE, et al. Global, regional, and national causes of child mortality: an updated systematic analysis for 2010 with time trends since 2000. Lancet. 2012; 379(9832):2151–61. doi: 10.1016/S0140-6736(12)60560-1 [DOI] [PubMed] [Google Scholar]
- 2.Walker CLF, Rudan I, Liu L, Nair H, Theodoratou E, Bhutta ZA, et al. Global burden of childhood pneumonia and diarrhoea. Lancet. 2013; 381(9875):1405–16. doi: 10.1016/S0140-6736(13)60222-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Diarrhoeal disease. [Internet]. [accessed 7 July, 2021]. Available from: https://www.who.int/news-room/fact-sheets/detail/diarrhoeal-disease.
- 4.Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S, et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet. 2013; 382(9888):209–22. doi: 10.1016/S0140-6736(13)60844-2 [DOI] [PubMed] [Google Scholar]
- 5.Oloruntoba EO, Folarin TB, Ayede AI. Hygiene and sanitation risk factors of diarrhoeal disease among under-five children in Ibadan, Nigeria. Afr Health Sci. 2014; 14(4):1001–11. doi: 10.4314/ahs.v14i4.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colston JM, Ahmed AMS, Soofi SB, Svensen E, Haque R, Shrestha J, et al. Seasonality and within-subject clustering of rotavirus infections in an eight-site birth cohort study. Epidemiol Infect. 2018; 146(6):688–97. doi: 10.1017/S0950268818000304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reddy C, Kuonza L, Ngobeni H, Mayet NT, Doyle TJ, Williams S. South Africa field epidemiology training program: developing and building applied epidemiology capacity, 2007–2016. BMC Public Health. 2019. -05-October;19(Suppl 3):469. doi: 10.1186/s12889-019-6788-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wurapa F, Afari E, Ohuabunwo C, Sackey S, Clerk C, Kwadje S, et al. One Health concept for strengthening public health surveillance and response through Field Epidemiology and Laboratory Training in Ghana. Pan Afr Med J. 2011; 10 Supp 1:6. [PMC free article] [PubMed] [Google Scholar]
- 9.Fletcher SM, Stark D, Ellis J. Prevalence of gastrointestinal pathogens in Sub-Saharan Africa: systematic review and meta-analysis. J Public Health Africa. 2011; 2(2). doi: 10.4081/jphia.2011.e30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oppong TB, Yang H, Amponsem-Boateng C, Kyere EKD, Abdulai T, Duan G, et al. Enteric pathogens associated with gastroenteritis among children under 5 years in Sub-Saharan Africa: a systematic review and meta-analysis.Epidemiol Infect. 2020. 2;148. doi: 10.1017/S0950268820000618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Country profiles Guinea-Bissau [Internet]. [accessed 7 July, 2021]. Available from: https://data.unicef.org/country/gnb/.
- 12.World Bank Country and Lending Groups [Internet]. [accessed 7 July, 2021]. Available from: https://datahelpdesk.worldbank.org/knowledgebase/articles/906519-world-bank-country-and-lending-groups.
- 13.Rodrigues A, de Carvalho M, Monteiro S, Mikkelsen CS, Aaby P, Molbak K et al. Hospital surveillance of rotavirus infection and nosocomial transmission of rotavirus disease among children in Guinea-Bissau. Pediatr Infect Dis J. 2007; 26(3):233–7. doi: 10.1097/01.inf.0000254389.65667.8b [DOI] [PubMed] [Google Scholar]
- 14.Fischer TK, Aaby P, Mølbak K, Rodrigues A. Rotavirus disease in Guinea-Bissau, West Africa: a review of longitudinal community and hospital studies. J Infect Dis. 2010; 202 Suppl:239. doi: 10.1086/653568 [DOI] [PubMed] [Google Scholar]
- 15.Mølbak K, Wested N, Højlyng N, Scheutz F, Gottschau A, Aaby P, et al. The etiology of early childhood diarrhea: a community study from Guinea-Bissau. J Infect Dis. 1994; 169(3):581–7. doi: 10.1093/infdis/169.3.581 [DOI] [PubMed] [Google Scholar]
- 16.Steenhard NR, Ørnbjerg N, Mølbak K. Concurrent infections and socioeconomic determinants of geohelminth infection: a community study of schoolchildren in periurban Guinea-Bissau. Trans R Soc Trop Med Hyg. 2009; 103(8):839–45. doi: 10.1016/j.trstmh.2009.05.005 [DOI] [PubMed] [Google Scholar]
- 17.Steinsland H, Valentiner-Branth P, Perch M, Dias F, Fischer TK, Aaby P, et al. Enterotoxigenic Escherichia coli infections and diarrhea in a cohort of young children in Guinea-Bissau. J Infect Dis. 2002; 186(12):1740–7. doi: 10.1086/345817 [DOI] [PubMed] [Google Scholar]
- 18.Luquero FJ, Banga CN, Remartínez D, Palma PP, Baron E, Grais RF. Cholera epidemic in Guinea-Bissau (2008): the importance of "place". PLoS ONE. 2011; 6(5):e19005. doi: 10.1371/journal.pone.0019005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gunnlaugsson G, Angulo FJ, Einarsdóttir J, Passa A, Tauxe RV. Epidemic cholera in Guinea-Bissau: the challenge of preventing deaths in rural West Africa. Int J Infect Dis. 2000; 4(1):8–13. doi: 10.1016/s1201-9712(00)90059-6 [DOI] [PubMed] [Google Scholar]
- 20.Ferreira FS, Centeno-Lima S, Gomes J, Rosa F, Rosado V, Parreira R, et al. Molecular characterization of Giardia duodenalis in children from the Cufada Lagoon Natural Park, Guinea-Bissau. Parasitol Res. 2012; 111(5):2173–7. doi: 10.1007/s00436-012-3068-6 [DOI] [PubMed] [Google Scholar]
- 21.Perch M, Sodemann M, Jakobsen MS, Valentiner-Branth P, Steinsland H, Fischer TK, et al. Seven years’ experience with Cryptosporidium parvum in Guinea-Bissau, West Africa. Ann Trop Paediatr. 2001; 21(4):313–8. doi: 10.1080/07430170120093490 [DOI] [PubMed] [Google Scholar]
- 22.Carstensen H, Hansen HL, Kristiansen HO, Gomme G. The epidemiology of cryptosporidiosis and other intestinal parasitoses in children in southern Guinea-Bissau. Trans R Soc Trop Med Hyg. 1987;81(5):860–4. doi: 10.1016/0035-9203(87)90054-x [DOI] [PubMed] [Google Scholar]
- 23.Valentiner-Branth P, Steinsland H, Fischer TK, Perch M, Scheutz F, Dias F, et al. Cohort study of Guinean children: incidence, pathogenicity, conferred protection, and attributable risk for enteropathogens during the first 2 years of life. J Clin Microbiol. 2003; 41(9):4238–45. doi: 10.1128/JCM.41.9.4238-4245.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mero S, Kirveskari J, Antikainen J, Ursing J, Rombo L, Kofoed PE, et al. Multiplex PCR detection of Cryptosporidium sp, Giardia lamblia and Entamoeba histolytica directly from dried stool samples from Guinea-Bissauan children with diarrhoea. Infect Dis (Lond). 2017; 49(9):655–63. doi: 10.1080/23744235.2017.1320728 [DOI] [PubMed] [Google Scholar]
- 25.Antikainen J, Kantele A, Pakkanen SH, Lääveri T, Riutta J, Vaara M, et al. A quantitative polymerase chain reaction assay for rapid detection of 9 pathogens directly from stools of travelers with diarrhea. Clin Gastroenterol Hepatol. 2013; 11(10):1300,1307.e3. doi: 10.1016/j.cgh.2013.03.037 [DOI] [PubMed] [Google Scholar]
- 26.Mobidiag. [Internet], (accessed 7 July, 2021). Available from: https://mobidiag.com/products/amplidiag/
- 27.Pelkonen T, Dos Santos MD, Roine I, Dos Anjos E, Freitas C, Peltola H, et al. Potential Diarrheal Pathogens Common Also in Healthy Children in Angola. Pediatr Infect Dis J. 2018; 37(5):424–8. doi: 10.1097/INF.0000000000001781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kabayiza J, Andersson ME, Nilsson S, Bergström T, Muhirwa G, Lindh M. Real-time PCR identification of agents causing diarrhea in Rwandan children less than 5 years of age. Pediatr Infect Dis J. 2014; 33(10):1037–42. doi: 10.1097/INF.0000000000000448 [DOI] [PubMed] [Google Scholar]
- 29.Platts-Mills JA, Babji S, Bodhidatta L, Gratz J, Haque R, Havt A, et al. Pathogen-specific burdens of community diarrhoea in developing countries: a multisite birth cohort study (MAL-ED). Lancet Glob Health. 2015; 3(9):564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bonkoungou IJO, Lienemann T, Martikainen O, Dembelé R, Sanou I, Traoré AS, et al. Diarrhoeagenic Escherichia coli detected by 16-plex PCR in children with and without diarrhoea in Burkina Faso. Clin Microbiol Infect. 2012; 18(9):901–6. doi: 10.1111/j.1469-0691.2011.03675.x [DOI] [PubMed] [Google Scholar]
- 31.Lääveri T, Vilkman K, Pakkanen SH, Kirveskari J, Kantele A. A prospective study of travellers’ diarrhoea: analysis of pathogen findings by destination in various (sub)tropical regions. Clin Microbiol Infect. 2018; 24(8):908.e9,908.e16. doi: 10.1016/j.cmi.2017.10.034 [DOI] [PubMed] [Google Scholar]
- 32.Leung AKC, Leung AAM, Wong AHC, Hon KL. Travelers’ Diarrhea: A Clinical Review. Recent Pat Inflamm Allergy Drug Discov. 2019; 13(1):38–48. doi: 10.2174/1872213X13666190514105054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kotloff KL, Riddle MS, Platts-Mills JA, Pavlinac P, Zaidi AKM. Shigellosis. Lancet. 2018; 391(10122):801–12. doi: 10.1016/S0140-6736(17)33296-8 [DOI] [PubMed] [Google Scholar]
- 34.Lertsethtakarn P, Silapong S, Sakpaisal P, Serichantalergs O, Ruamsap N, Lurchachaiwong W, et al. Travelers’ Diarrhea in Thailand: A Quantitative Analysis Using TaqMan Array Card. Clin Infect Dis. 2018; 67(1):120–7. doi: 10.1093/cid/ciy040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bloomfield SJ, Midwinter AC, Biggs PJ, French NP, Marshall JC, Hayman DTS, et al. Long-term Colonization by Campylobacter jejuni Within a Human Host: Evolution, Antimicrobial Resistance, and Adaptation. J Infect Dis. 2017; 217(1):103–11. doi: 10.1093/infdis/jix561 [DOI] [PubMed] [Google Scholar]
- 36.Liu J, Platts-Mills JA, Juma J, Kabir F, Nkeze J, Okoi C, et al. Use of quantitative molecular diagnostic methods to identify causes of diarrhoea in children: a reanalysis of the GEMS case-control study. Lancet. 2016; 388(10051):1291–301. doi: 10.1016/S0140-6736(16)31529-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Regional Cholera Platforms in Guinea-Bissau [Internet]. [accessed 7 July, 2021]. Available from: https://plateformecholera.info/index.php/bonus-page-2/national-strategies-plans/guinea-bissau.
- 38.Mwenda JM, Burke RM, Shaba K, Mihigo R, Tevi-Benissan MC, Mumba M, et al. Implementation of Rotavirus Surveillance and Vaccine Introduction—World Health Organization African Region, 2007–2016. MMWR Morb Mortal Wkly Rep. 2017; 66(43):1192–6. doi: 10.15585/mmwr.mm6643a7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lekana-Douki SE, Kombila-Koumavor C, Nkoghe D, Drosten C, Drexler JF, Leroy EM. Molecular epidemiology of enteric viruses and genotyping of rotavirus A, adenovirus and astrovirus among children under 5 years old in Gabon. Int J Infect Dis. 2015; 34:90–5. doi: 10.1016/j.ijid.2015.03.009 [DOI] [PubMed] [Google Scholar]
- 40.Ouédraogo N, Kaplon J, Bonkoungou IJO, Traoré AS, Pothier P, Barro N, et al. Prevalence and Genetic Diversity of Enteric Viruses in Children with Diarrhea in Ouagadougou, Burkina Faso. PLoS ONE. 2016; 11(4):e0153652. doi: 10.1371/journal.pone.0153652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mans J, Armah GE, Steele AD, Taylor MB. Norovirus Epidemiology in Africa: A Review. PLoS ONE. 2016; 11(4):e0146280. doi: 10.1371/journal.pone.0146280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Munjita SM. Current Status of Norovirus Infections in Children in Sub-Saharan Africa. J Trop Med. 2015; 2015:309648. doi: 10.1155/2015/309648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hotez PJ, Kamath A. Neglected tropical diseases in sub-saharan Africa: review of their prevalence, distribution, and disease burden. PLoS Negl Trop Dis. 2009; 3(8):e412. doi: 10.1371/journal.pntd.0000412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aldeyarbi HM, Abu El-Ezz, Nadia M. T., Karanis P. Cryptosporidium and cryptosporidiosis: the African perspective. Environ Sci Pollut Res Int. 2016; 23(14):13811–21. doi: 10.1007/s11356-016-6746-6 [DOI] [PubMed] [Google Scholar]
- 45.Garzón M, Pereira-da-Silva L, Seixas J, Papoila AL, Alves M. Subclinical Enteric Parasitic Infections and Growth Faltering in Infants in São Tomé, Africa: A Birth Cohort Study. Int J Environ Res Public Health. 2018; 4;15(4). doi: 10.3390/ijerph15040688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pietilä J, Meri T, Siikamäki H, Tyyni E, Kerttula AM, Pakarinen L, et al. Dientamoeba fragilis—the most common intestinal protozoan in the Helsinki Metropolitan Area, Finland, 2007 to 2017. Euro Surveill. 2019; 24(29). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wold AE, Adlerberth I. Breast feeding and the intestinal microflora of the infant—implications for protection against infectious diseases. Adv Exp Med Biol. 2000; 478:77–93. doi: 10.1007/0-306-46830-1_7 [DOI] [PubMed] [Google Scholar]
- 48.Infant and young child feeding [Internet]. [accessed 7 July, 2021]. Available from: https://www.who.int/en/news-room/fact-sheets/detail/infant-and-young-child-feeding.
- 49.Levine MM, Robinson-Browne RM. Factors that explain excretion of enteric pathogens by persons without diarrhea. Clin Infect Dis. 2012; 55 Suppl 4:S303–11. doi: 10.1093/cid/cis789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.von Huth S, Thingholm LB, Bang C, Rühlemann MC, Franke A, Holmskov U. Minor compositional alterations in faecal microbiota after five weeks and five months storage at room temperature on filter papers. Sci Rep. 2019; 9(1):19008. doi: 10.1038/s41598-019-55469-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(XLSX)
Data Availability Statement
All relevant data are in the manuscript and its Supporting Information files. The individuals' personal data cannot be shared publicly because of research regulations. Contact details for further information: Meilahti Vaccine Research Center, MeVac Biomedicum 1, Helsinki University Hospital and University of Helsinki, POB 700, 00029 HUS, email vaccine@hus.fi.


