Abstract
Around 20–30% of ovarian cancer patients exhibit chemoresistance, but there are currently no methods to predict whether a patient will respond to chemotherapy. Here, we discovered that chemoresistant ovarian cancer cells exhibit enhanced survival in a quiescent state upon experiencing the stress of physical confinement. When immobilized in stiff silica gels, most ovarian cancer cells die within days, but surviving cells exhibit hallmarks of single cell dormancy. Upon extraction from gels, the cells resume proliferation but demonstrate enhanced viability upon re-immobilization, indicating that initial immobilization selects for cells with a higher propensity to enter dormancy. RNA-seq analysis of the extracted cells shows they have signaling responses similar to cells surviving cisplatin treatment, and in comparison to chemoresistant patient cohorts, they share differentially expressed genes that are associated with platinum-resistance pathways. Furthermore, these extracted cells demonstrate greater resistance to cisplatin and paclitaxel, despite being proliferative. In contrast, serum starvation and hypoxia could not effectively select for chemoresistant cells upon removal of the environmental stress. These findings demonstrate that ovarian cancer chemoresistance and the ability to enter dormancy are linked, and immobilization rapidly distinguishes chemoresistant cells. This platform could be suitable for mechanistic studies, drug development, or as a clinical diagnostic tool.
Keywords: silica gel, ovarian cancer, chemoresistance, quiescence, immobilization
INTRODUCTION
Ovarian cancer is the fifth most common cancer in women, with 69% of patients ultimately succumbing to the disease (Cristea, Han, Salmon, & Morgan, 2010; Lengyel, 2010). Current standard of care involves surgery followed by combination chemotherapy with platinum and taxane agents (Cristea et al., 2010). However, this conventional course of treatment has limited long-term efficacy as the majority of patients will eventually relapse, and 20 – 30% will have chemoresistant disease, characterized by recurrence within six months of completing treatment (Agarwal & Kaye, 2003; Mantia-Smaldone, Edwards, & Vlad, 2011; Pfisterer & Ledermann, 2006). There is currently no rapid way to predict how responsive ovarian cancer patients will be to chemotherapy prior to treatment, as patient-derived xenograft (PDX) avatar models take months to establish (Scott, Mackay, & Haluska Jr., 2014). Recently, 3D in vitro models using patient tumor cells have shown promise as platforms for predicting patient drug response and require less time than PDX models (Raghavan et al., 2017; Rashidi et al., 2019); however, even these methods can take weeks. The ability to rapidly identify patients (within days) who would not respond to chemotherapy would enable clinicians to adjust monitoring strategies or suggest alternative therapies that are often only used once conventional treatment no longer works.
With many women initially responding to frontline treatments and some exhibiting complete remission, it is posited that recurrence and chemoresistance may be due to the presence of quiescent tumor cells that survive treatment (Chien, Kuang, Landen, & Shridhar, 2013; Kondoh et al., 2010), as it is traditionally thought that these cells are better able to evade chemotherapy. Quiescent cancer cells, existing in a growth-arrested state and consequently less susceptible to many anticancer chemotherapies, may be left behind as minimal residual disease after what appears to be successful treatment (Páez et al., 2012). Another potential source of quiescent cancer cells are disseminated tumor cells shed from the primary tumor, which can become growth-arrested after arriving at sites not permissive for proliferation (Aguirre-Ghiso, 2007). Moreover, not all cancer cells will enter quiescence under stress, with some exhibiting cell death instead, and it is unclear why only a unique subset of cells undergoes growth arrest to survive unfavorable conditions. More recently, a cancer stem cell hypothesis has been proposed suggesting that chemoresistant cancer cells with stem-like characteristics have this ability to enter quiescence and may reside within the primary tumor, persist through chemotherapy, and repopulate chemoresistant disease after months or years upon completing treatment (Chien et al., 2013). Although cancer cells which enter quiescence are known to evade chemotherapy and cause recurrence (Kondoh et al., 2010), the mechanisms by which they survive drug treatment remain unclear. Proposed mechanisms of chemoresistance in ovarian cancer involve enhanced DNA repair mechanisms, modified drug uptake and efflux by transmembrane proteins, and inhibition of pro-apoptosis pathways (Kalir & Kohtz, 2014), but further study is required to determine which mechanisms contribute to resistance in cells which readily undergo growth arrest in stressful conditions.
However, study of these cells and their drug-resistance mechanisms has been limited by the lack of suitable in vitro methods available to easily identify and isolate them. As entering dormancy is a mechanism for cells to survive in a microenvironment that is not conducive to growth, most in vitro platforms utilize environmental stress to induce quiescence. Oxygen-deficient or low-serum/low-glucose treatment methods are often used to mimic hypoxic and nutrient-deficient conditions in the body, and previous studies have demonstrated accumulation of growth-arrested cells in various tumor cell lines grown under these conditions (Carcereri de Prati et al., 2017; Oya, Zölzer, Werner, & Streffer, 2003). Recent work has shown that the stress of physical confinement upon immobilization in stiff yet porous alkoxide-based silica gels results in the ability to distinguish between cells that are readily able to enter dormancy and those that cannot, as breast cancer cell lines known to readily form tumors in vivo quickly die upon physical confinement while another cell line known to enter quiescence in vivo demonstrates enhanced survival in a growth-arrested state (Preciado, Reátegui, Azarin, Lou, & Aksan, 2017). As the quiescence-inducing stress in this model is matrix-based and interactions with the extracellular matrix (ECM) are also implicated in the responsiveness of ovarian cancer cells to chemotherapy (Chien et al., 2013), we sought to investigate whether the stress of physical confinement and the silica gel environment potentiate chemoresistance, in addition to quiescence, in ovarian cancer.
Here, we report the use of physical confinement in silica gels to identify ovarian cancer cells which exhibit quiescence when immobilized and resume proliferation upon extraction from the gel. This process selects for a population with a higher propensity to enter dormancy, as the extracted cells exhibit enhanced survival in the quiescent state upon re-immobilization. Interestingly, these extracted cells also demonstrate signaling responses similar to cells that have persisted following chemotherapy treatment, and accordingly the extracted cells are shown to be less sensitive to cisplatin and paclitaxel treatment than the original cell population. Furthermore, chemoresistant cells exhibit enhanced survival upon immobilization in the gels, indicating that they are more readily able to enter the dormant state. These findings demonstrate a linkage between chemoresistant and dormancy-capable phenotypes in ovarian cancer. As immobilization in silica gels can isolate populations with these phenotypes within days, this platform could be useful in future mechanistic or drug studies and could potentially be used as a tool to identify patients that may have chemoresistant disease and are at risk of recurrence.
MATERIALS AND METHODS
Cell Culture.
OVCAR-3 and SKOV-3 cells were purchased from the ATCC (HTB-161 and HTB-77). OVCAR-3 and SKOV-3 cells were cultured in RPMI 1640 media (Thermo Fisher Scientific) and McCoy’s 5A media (Thermo Fisher Scientific), respectively, supplemented with 10% fetal bovine serum (FBS) (Thermo Fisher Scientific). Cells were passaged when 80 – 90% confluence was reached. OVCAR-3 was chosen as the primary cell line since it was established from a patient who maintained progressive disease after completing combination chemotherapy and is resistant to clinically-relevant doses of cisplatin and other anticancer drugs (Hamilton et al., 1983). SKOV-3 is also known to be resistant to cisplatin and other drugs.
Immobilization of Cells by Silica Gel Encapsulation.
Silica gel matrices were formed from tetrakis(2-hydroxyethyl) orthosilicate (THEOS) and silica nanoparticles (SNPs) as described previously (Fig. S1A) (Preciado et al., 2017). Primary gelling solution consisted of FBS-supplemented cell culture media with 1% v/v penicillin-streptomycin (Thermo Fisher Scientific), 4-arm PEG (2 kDa, hydroxyl terminated, Creative PEGWorks), and NS125 silica nanoparticles (Nyacol) in a 0.9:0.1:0.02 volume ratio. Cells for encapsulation were trypsinized and cell solution at 2 × 106 cells per mL was added 1:1 with gelling solution and aliquoted to 50 μL per gel. Tetrakis (2-hydroxyethyl) orthosilicate (Gelest) was added to each aliquot at a 1:10 ratio and thoroughly mixed. Silica gel solution was quickly pipetted and spread onto a non-treated 35 mm polystyrene petri dish. Dishes were left untouched for a few minutes to allow complete gelling, after which media was added on top to prevent dehydration before incubating at 37 °C. OVCAR-3 cells were easily visualized within gels and observed to be intact and distributed primarily as single cells without forming large clusters (Fig. S1B).
Quantification of Immobilized Viable Cells.
Immobilized cells were stained with Calcein AM (Thermo Fisher Scientific) diluted 1:500 in phosphate buffered saline (PBS) for 30 minutes on a shaker platform at room temperature. Live cells were imaged using an EVOS FL Auto fluorescence microscope (Thermo Fisher Scientific) at 10x magnification. Ten images were taken for each timepoint (five images each from two separate gels), and the number of live cells was manually counted from each image. Fold change in viable cell number for each day was quantified as total number of live cells counted in each gel normalized to average total number of live cells counted in gels on Day 0.
Ki67 Immunofluorescence Analysis.
Washing was completed with PBS, and all incubation steps were at room temperature on a shaker platform. 2-D or silica gel samples were washed and then fixed with a 4% paraformaldehyde (PFA) solution for 12 or 20 minutes, respectively. Samples were washed again and incubated for 1 hour with blocking solution containing PBS with 5% normal goat serum (Thermo Fisher Scientific) and 0.3% Triton X-100 (Millipore Sigma). Ki67 primary antibody in antibody dilution buffer (PBS with 0.3% Triton x-100 and 1% bovine serum albumin (BSA) (Millipore Sigma)) was added to samples for overnight incubation at 4 °C. Samples were washed followed by addition of secondary antibody in antibody dilution buffer and incubated for 1 or 2 hours for 2-D or silica gel samples, respectively. All samples were washed before addition of 300 nM DAPI nuclear staining solution (Thermo Fisher Scientific) for 5 or 30 minutes for 2-D samples or silica gels, respectively. Samples were washed once more then imaged using an EVOS FL Auto fluorescence microscope with the same intensity and exposure time for all samples. See Table S1 in the supplemental information for antibody details.
Senescence-Associated β-galactosidase Staining.
Staining was completed using the Senescence β-Galactosidase Staining Kit #9860 from Cell Signaling Technologies according to the manufacturer’s instructions. Briefly, 2-D or silica gel samples were washed then fixed with 1x fixative solution for 12 or 20 minutes, respectively. Samples were washed again, and β-galactosidase staining solution was prepared, adjusted to a final pH between 5.9 and 6.1, and added. Samples were covered with parafilm to prevent evaporation, and 2-D or silica gel samples were placed in a dry 37 °C incubator overnight or for 24 hours, respectively. Brightfield images at 20x magnification were taken using an EVOS FL Auto fluorescence microscope. High passage human foreskin fibroblasts (ATCC; SCRC-1041) were used as a positive control.
Cell Extraction.
Gels were rinsed with PBS followed by incubation with 0.25% trypsin-EDTA for 7 minutes at 37 °C. Cells were then extracted from silica gels by pipetting with cell culture media to break apart the gels. The gel fragment and media solution was transferred to a conical tube and further pipetted to break up large fragments. This solution was transferred to a T75 flask containing cell culture media with 1% v/v penicillin-streptomycin and incubated overnight. Flasks were then washed two times with PBS to remove any remaining silica gel fragments, and fresh cell culture media with 1% v/v penicillin-streptomycin was added. For studies involving regrowth and treatment of extracted cells, extracted cells were imaged at the same locations (n ≥ 9 for each condition) for up to one week. Viable cell number was determined by manually counting cells from these images.
Western Blotting.
Protein was extracted from cells immobilized in silica gels for three days and cells maintained in standard 2-D culture using Halt Phosphatase Inhibitor Cocktail (Thermo Fisher Scientific) and Halt Protease Inhibitor Cocktail (Thermo Fisher Scientific), each diluted 1:100 in RIPA lysis buffer (Thermo Fisher Scientific). For protein separation, 10 μg of protein was loaded per well of a 4–15% Mini-Protean SDS-PAGE gel (Bio-Rad) for β-actin detection, and 30 μg of protein was loaded per well for detection of mitogen-activated protein kinase (MAPK) proteins. Proteins were then transferred to a polyvinylidene difluoride membrane (Bio-Rad), and membranes for MAPK proteins and β-actin were blocked in Tris-buffered saline containing Tween-20 (TBS-T, Millipore Sigma) with 5% BSA or 5% non-fat dry milk (Bio-Rad), respectively, for 1 hour at room temperature. Membranes were then incubated overnight at 4 °C with primary antibodies in TBS-T with 5% BSA or 5% milk for MAPKs and β-actin, respectively. MAPK membranes were then rinsed and incubated with secondary antibody diluted in TBS-T with 5% BSA for 1 hour at room temperature. SuperSignal West Pico Chemiluminescent substrate (Thermo Fisher Scientific) was used to detect protein bands, which were then imaged with a ChemiDoc Touch Imaging System (Bio-Rad). Western blot data was quantified using Image Lab software (Bio-Rad). The ratio of activated p38 is defined as the phosphorylated p38 MAPK “pp38” band intensity divided by the p38 MAPK “p38” band intensity. Similarly, the ratio of activated ERK is defined as the phosphorylated ERK1/2 MAPKs “pERK1/2” band intensity divided by the ERK1/2 MAPKs “ERK1/2” band intensity. The final p38:ERK activity ratio was then found by dividing these two ratios (ratio of activated p38/ratio of activated ERK). See supplemental information for antibody details (Table S1).
RNA-Sequencing Sample Preparation and Analysis.
Total RNA was collected from cells using the RNeasy Mini Kit (Qiagen) with two replicate samples collected for each condition (5 conditions, 10 samples total). 5 μg of RNA for each sample was submitted to the University of Minnesota Genomics Core for library creation and sequencing. Ten dual-indexed Illumina TruSeq stranded mRNA libraries were created for paired-end sequencing, and sequencing was completed in one lane of a HiSeq 2500 2×125-bp run using v4 chemistry, providing a total of ~22 million reads per sample. Trimming of low-quality ends and removal of adapter sequences was completed using Trimmomatic. Spliced Transcripts Alignment to a Reference (STAR) was used for mapping of reads to the human reference genome (GRCh38.91). The HTseq package in Python was used to obtain raw counts per million for each gene, then the EdgeR package in R was used to determine the fold change and significance of gene expression in pair-wise comparisons of the various experimental conditions relative to the control. Further analyses of these genes included Gene Ontology enrichment analysis using Database for Annotation, Visualization, and Integrated Discovery (DAVID v6.8, https://david.ncifcrf.gov/) for genes meeting criteria of fold change ≥ 2 and P-value < 0.05 and Ingenuity Pathway Analysis (IPA) (Qiagen) for genes with fold change ≥ 2, P-value < 0.05, and false discovery rate < 0.05. For comparison to patient data, genes identified as differentially expressed between chemoresistant and chemosensitive patients in the discovery group by Koti et al. and in their in silico analysis of patient datasets from The Cancer Genome Atlas were used (Koti et al., 2013). Differentially expressed genes from these analyses were compared to the differentially expressed genes in our “Extracted” group relative to control, and genes which were commonly up- or downregulated in both datasets were used in IPA to identify which canonical signaling pathways were most relevant to the common differential gene expression observed in the patient cohorts and our in vitro groups.
Hypoxia, Serum Starvation, and Drug Treatment.
Cells were seeded with 10% FBS-supplemented cell culture media and after 24 hours, plates were then placed into a hypoxia chamber (STEMCELL Technologies) maintaining 1% oxygen conditions for hypoxia treatment or media was aspirated and serum-free cell culture media was added to wells for serum starvation. For drug treatment studies, cells were seeded with 10% FBS-supplemented cell culture media and incubated at 37 °C overnight, and drug-containing media was freshly prepared prior to addition to cells for each treatment. Cells were grown in drug-containing media for 24 hours after each addition, then drug-containing media was aspirated and cells were rinsed with PBS followed by addition of fresh 10% FBS-supplemented cell culture media. Cisplatin (Millipore Sigma) was solubilized in ultrapure water by gentle stirring at 25 °C for 10–15 minutes then filtered and stored in the dark at room temperature as a 10 mM stock solution. Paclitaxel (Tocris) was reconstituted to a 100 mM stock solution with dimethyl sulfoxide, aliquoted, and stored at −20 °C.
Hypoxia or Serum Starvation Studies with Cisplatin Treatment.
Cells for standard culture, hypoxic, and serum starvation conditions were seeded at 100,000, 250,000, or 500,000 cells per well, respectively, on 6 well-plates in 10% FBS-supplemented cell culture media and incubated at 37 °C. Different seeding densities were chosen to ensure that the cell density for each condition was roughly the same at the start of cisplatin treatment. After 24 hours, cells were placed into a 1% oxygen hypoxia chamber or serum-free cell culture media was added to wells. After three days, media containing 0.5 μM cisplatin was added to wells, while control wells received drug-free media. After 24 hours of drug treatment, all wells were washed with PBS and fresh drug-free cell culture media was added. After three days, cells were stained with Trypan Blue (Thermo Fisher Scientific) and counted (Countess II Automated Cell Counter, Thermo Fisher Scientific).
Statistical Analysis.
Data are represented as mean ± S.D. of two or more biological replicates from one of three representative independent experiments or as mean ± S.D. from three combined independent experiments. P-values were determined from Student’s unpaired t-test using GraphPad Prism software, with P-value < 0.05 considered to be statistically significant.
RESULTS & DISCUSSION
Immobilized OVCAR-3 cells within silica gels exhibit hallmarks of single cell dormancy.
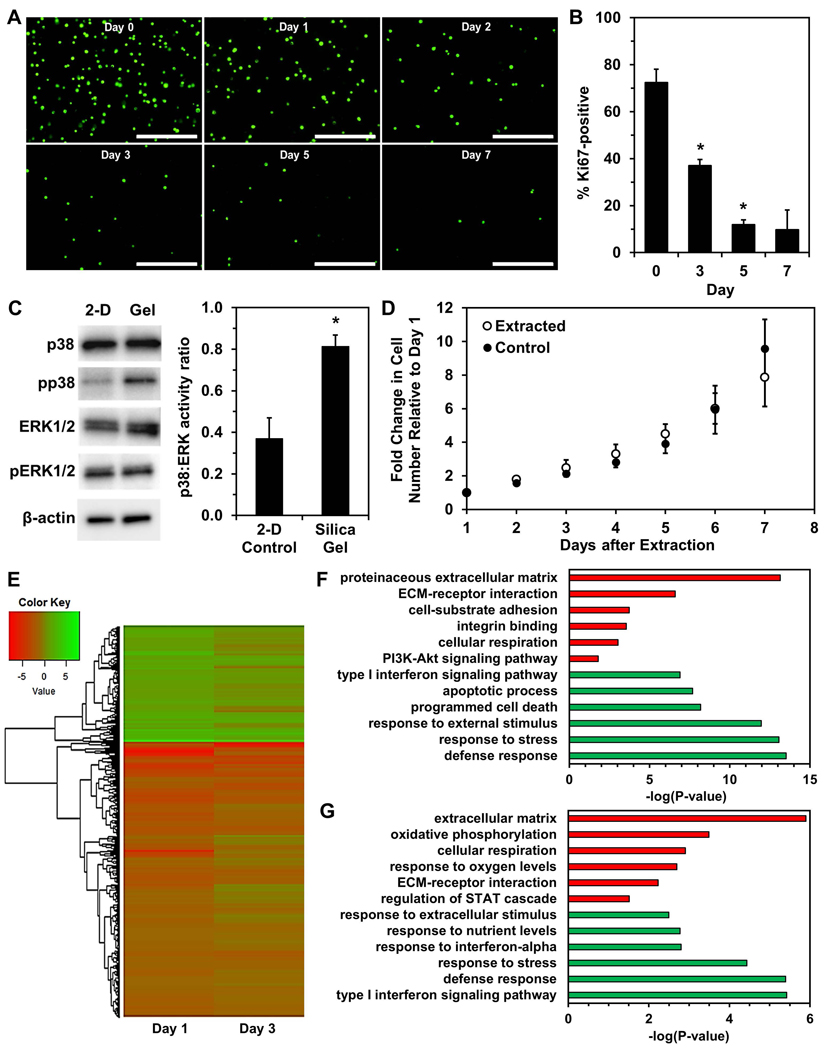
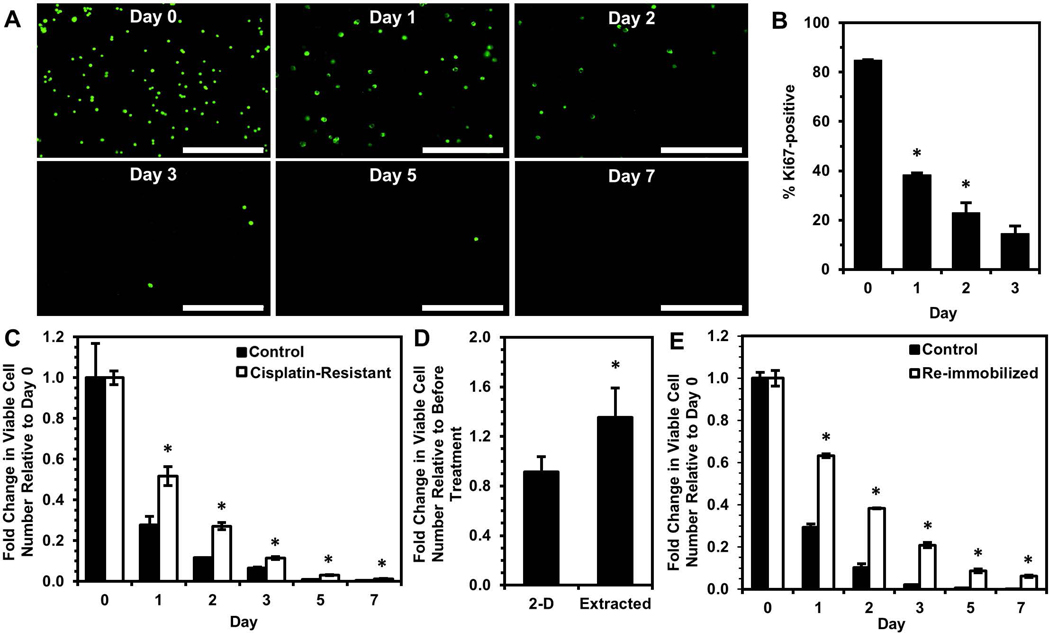
After immobilization in stiff yet porous silica gels that have previously been characterized and shown to inhibit cell motility, growth, and proliferation (Reátegui, Kasinkas, Kniesz, Lefebvre, & Aksan, 2014), a substantial decrease in viable OVCAR-3 cells was observed over time, with surviving cells remaining in a single-cell state and having no evident increase in size (Fig. 1A). The percentage of Ki67-positive cells significantly decreased by Day 3 of immobilization and continued to decrease at later timepoints (Fig. 1B & Fig. S2), which was consistent with our previous observations with the MCF-7 breast cancer cell line (Preciado et al., 2017). Ki67 is expressed in actively dividing cells and has been extensively characterized as a proliferation-specific marker, with loss of Ki67 expression indicating cell cycle exit into a resting (G0) phase (Gerdes et al., 1984; Scholzen & Gerdes, 2000). Immobilized live cells also demonstrated an increased p38:ERK activity ratio and higher expression of phosphorylated p38 MAPK (Fig. 1C). A high p38:ERK activity ratio is an indicator for cancer cell transition from metastatic growth to quiescence (Aguirre-Ghiso, 2007; Maria Soledad Sosa, Avivar-Valderas, Bragado, Wen, & Aguirre-Ghiso, 2011). Increased activation of p38 signaling is associated with response to environmental stress (Raingeaud et al., 1995) and tumor suppression (Brancho et al., 2003; V.Bulavin & Fornace Jr., 2004). Decreased ERK activity has been associated with small tumor nodules, while rapidly growing tumors and metastases exhibited high ERK activity in vivo (Aguirre-ghiso, Ossowski, & Rosenbaum, 2004). Further, ERK and p38 levels have previously been shown to predict dormancy in vivo in epidermoid carcinoma, fibrosarcoma, and other tumor types, and inhibition of p38 activity has even reversed dormant behavior (Aguirre-ghiso, Estrada, Liu, & Ossowski, 2003; Aguirre-ghiso, Liu, Mignatti, Kovalski, & Ossowski, 2001). Importantly, growth-arrested cells in silica gels were not senescent, as senescence-associated β-galactosidase expression levels did not change during immobilization (Fig. S3). When extracted from the silica gels after three days of immobilization, the cells resumed proliferation, exhibiting no significant differences in growth or morphology immediately after extraction compared to control cells (Fig. 1D & Fig. S4).
Figure 1. Immobilization of OVCAR-3 cells in silica gels results in survival of cells exhibiting quiescence.
(A) Fluorescence images of viable cells stained with calcein AM (green) at Days 0, 1, 2, 3, 5, and 7 of immobilization. Scale bar indicates 400 μm. (B) Percentage of Ki67-positive cells (number of Ki67-expressing cells normalized to number of DAPI-labeled cells) within silica gels over a one-week period quantified from fluorescence images (*P < 0.05 compared to previous timepoint). (C) Western blotting of p38 MAPK protein (“p38”), ERK1/2 MAPK proteins (“ERK1/2”), and their phosphorylated forms (“pp38” and “pERK1/2”) in cells immobilized for three days and control cells grown in standard 2-D culture conditions (*P < 0.05 compared to “2-D Control”). (D) Growth of cells extracted from silica gels after immobilization for 3 days and control cells cultured in 2-D. (E) Heatmap of log2FC of gene expression in cells immobilized for one or three days relative to control cells. (F,G) Gene ontology enrichment analysis using genes significantly upregulated (green) or downregulated (red) in “Day 1” (F) or “Day 3” (G) of immobilization relative to 2-D controls.
RNA sequencing analysis revealed 1,426 and 1,031 differentially expressed genes after 24 hours (“Day 1”) or 3 days (“Day 3”) of immobilization, respectively, relative to 2-D control cells, with 649 common differentially expressed genes between these groups (Fig. 1E). Gene Ontology (GO) enrichment analysis was next completed using differentially expressed genes (Fig. 1F&G). For significantly upregulated genes, terms related to stress and stimuli responses, along with interferon signaling, were enriched in both “Day 1” and “Day 3”, but apoptosis-related terms appeared only in “Day 1”. This result is unsurprising since the most significant reduction in viable cells occurs within the first two days of immobilization (Fig. 1A), and activation of apoptotic pathways would be present in many of these cells. Previous studies have also shown addition of exogenous interferon (IFN) induces cell cycle arrest in gastric (Gao et al., 2014) and renal (Tate, Patterson, Finkel-Jimenez, & Zea, 2013) carcinoma cells. Wall et al. showed IFN-gamma had an anti-proliferative effect on ovarian cancer cell lines in vitro, and they observed IFN treatment of platinum-refractory ovarian cancer patients caused significant cancer cell reduction although 2% residual disease remained, eventually leading to recurrence (Wall, Burke, Barton, Smyth, & Balkwill, 2003). GO analysis of downregulated genes resulted in terms related to cell adhesion, cell-protein interactions, and metabolism in both groups. Terms such as ECM-receptor interaction, integrin binding, and cell-substrate adhesion largely indicate a mechanotransduction signaling response of immobilized cells within silica gels, as cellular mechanotransduction is mediated by integrins and focal adhesions following cell receptor binding to extracellular matrix proteins (Katsumi, Orr, Tzima, & Schwartz, 2004; N. Wang, 2017). Reduced metabolism (Carcereri de Prati et al., 2017) and downregulation of PI3K-Akt signaling (Correa, Peart, Valdes, Dimattia, & Shepherd, 2012; María Soledad Sosa, Bragado, & Aguirre-Ghiso, 2014; Touil et al., 2013) are associated with quiescence. Previous work has shown that malignant cells resisting anchorage-dependent apoptosis also have inactivated PI3K-Akt signaling and increased entrance into the G0 phase (Daubriac et al., 2009). Additionally, patient ovarian cancer cells and OVCAR-3 cells entered quiescence and had decreased AKT phosphorylation in non-adherent culture conditions (Correa et al., 2012). These findings demonstrate the ability of silica gels to select for viable cells exhibiting dormant behavior under immobilization stress, while most of the starting population undergo cell death.
This demonstration of encapsulation in a stiff biomaterial prompting cell cycle exit recapitulates previous studies where gels derived from organic materials, such as collagen (Fang et al., 2016) or fibrin (Liu et al., 2018), showed increasing stiffness results in slowed cell growth and metabolism. Here, the silica gels are advantageous as they do not allow for subsequent growth or division once cells are immobilized, providing a harsh environment that causes most cells to quickly die. Another advantage of these silica gels is that immobilized OVCAR-3 cells are unable to degrade the silica matrix, while gels derived from natural materials may weaken from enzymatic degradation. Minimal protein amounts in the starting silica gel matrix also provides less sites for cell adhesion and an increasingly difficult environment for these cells, which may result in survival of those more likely to undergo growth arrest and resist apoptosis. Previously, 3-D bladder cancer spheroids cultured on low-adhesion aminoglycoside-derived hydrogels showed greater cell cycle arrest with increasing storage modulus (Pavan Grandhi, Potta, Nitiyanandan, Deshpande, & Rege, 2017). Another study has investigated ovarian cancer cell migration, invasion, and growth in cross-linked hyaluronan gels to determine whether the material could inhibit cancer cell adhesion and implantation in the pelvic cavity in vivo (Pang et al., 2018). Here, we showed the immobilization of cells in stiff silica gels – which also limits their growth and division – results in survival of cells which have entered a dormant state, potentially to tolerate the stress of growth/mitotic inhibition upon confinement.
Immobilization selects for a unique subpopulation with enhanced ability to enter dormancy.
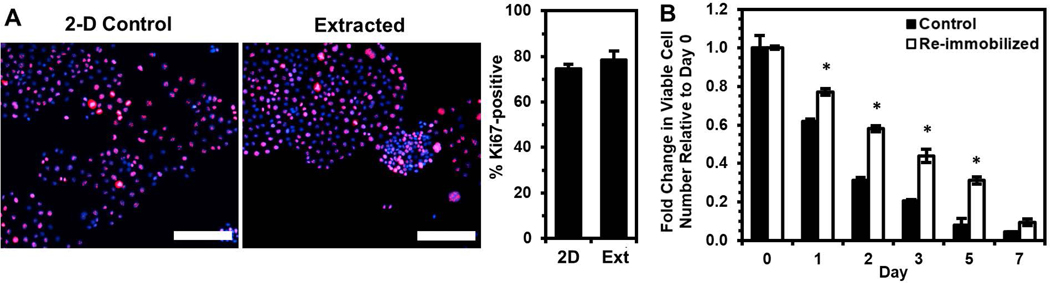
To test the hypothesis that immobilization selects for cells with a greater propensity to enter dormancy in order to survive the stress of physical confinement, we re-immobilized cells extracted from the silica gels. Immediately prior to re-immobilization, we confirmed via Ki67 expression that the extracted cells were as proliferative as control cells maintained in 2-D culture (Fig. 2A). Despite being proliferative just before returning to physical confinement, the extracted cells were found to have enhanced viability upon re-immobilization (Fig. 2B). These results indicate that immobilization of OVCAR-3 cells selects for a population that may have enhanced pro-survival mechanisms and a greater propensity to enter dormancy within gels.
Figure 2. Cells surviving silica gel immobilization demonstrate enhanced survival upon re-immobilization relative to the original control population.
Cells surviving immobilization for three days were extracted and cultured in 2-D for 1 week prior to immunostaining or re-immobilization. (A) Percentage of Ki67-positive cells (number of Ki67-expressing cells (red) normalized to number of DAPI-labeled cells (blue)) from immunofluorescence images (merged images shown) compared to 2-D control cells. Scale bar indicates 200 μm. (B) Fold change in viable cell number relative to Day 0 after re-immobilizing extracted cells in silica gels relative to cells immobilized for the first time (Control) (*P < 0.05 compared to “Control” for each timepoint).
Silica gel immobilization distinguishes OVCAR-3 cells with enhanced chemoresistance.
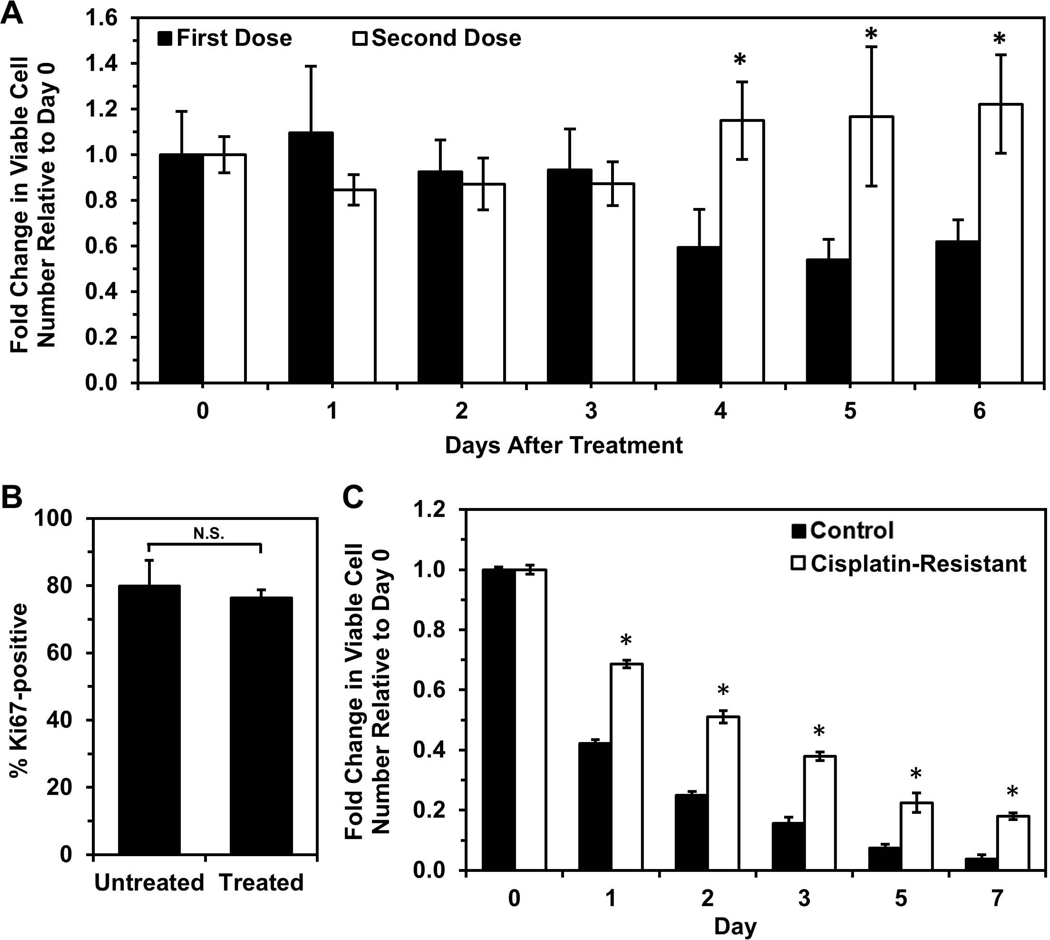
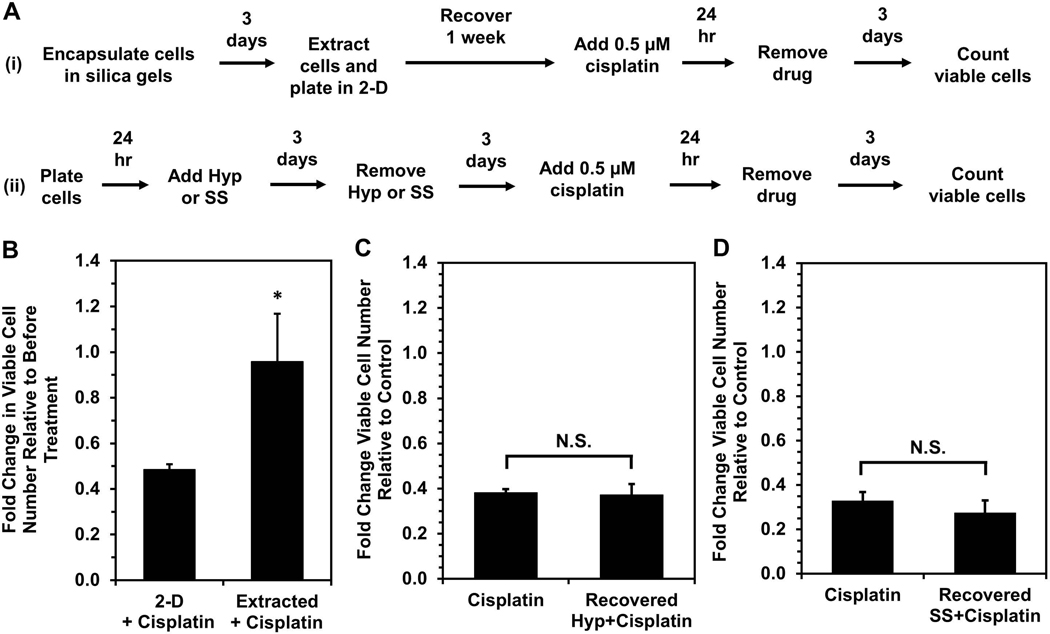
Because cells immobilized in silica gels exhibited differential gene expression associated with dysregulated cell-substrate interactions, which are also thought to play a role in chemoresistance (Chien et al., 2013; Yeh & Ramaswamy, 2015), we investigated whether cells with enhanced chemoresistance would exhibit a higher propensity to enter dormancy and survive upon immobilization. To enrich for cells more resistant to platinum chemotherapy, OVCAR-3 cells were treated with 0.5 μM cisplatin for 24 hours, and the surviving cells recovered for two weeks (Fig. S5). To verify that this procedure selected for chemoresistant cells, recovered cells were retreated with the same dose of cisplatin, and cell viability after re-treatment (“Second Dose”) was compared to viability following initial treatment (“First Dose”, Fig. 3A). Initial treatment caused a significant decrease in viability over a 6-day period, while cells undergoing re-treatment did not exhibit extensive cell death and continued to grow after Day 3, confirming their decreased sensitivity to the chemotherapy. Despite being in a proliferative state prior to immobilization (Fig. 3B), after immobilization the cisplatin-resistant cells had markedly enhanced survival relative to untreated cells immobilized in gels (Fig. 3C), suggesting cells with enhanced chemoresistance may also have greater capacity to enter dormancy when stressed by immobilization and the silica gel environment. Ultimately, these results indicate that silica gel technology could be used as a method to distinguish ovarian cancer cells with enhanced chemoresistance.
Figure 3. Cisplatin-resistant OVCAR-3 cells have higher tolerance for immobilization and silica gel environment.
(A) Cells retreated with a second dose of cisplatin are less susceptible to treatment than cells undergoing primary treatment, indicating the first dose selected for cisplatin-resistant cells. Each treatment dose was 0.5 μM cisplatin for 24 hours, and cells receiving a second dose were allowed a 2-week recovery period before treatment with the second dose. Viable cell number at Day 0 is the number of live cells counted immediately after drug removal. (*P < 0.05 compared to “First Dose”). (B) Cisplatin-resistant cells (“Treated”, i.e. treated with cisplatin and allowed to recover for 2 weeks) have similar Ki67 expression level as untreated cells. (C) Fold change in viable cell number relative to Day 0 of immobilized cisplatin-resistant cells and untreated control cells (*P < 0.05 compared to “Control” at each timepoint).
RNA sequencing analysis shows cells surviving silica gel immobilization demonstrate signaling responses associated with chemoresistance.
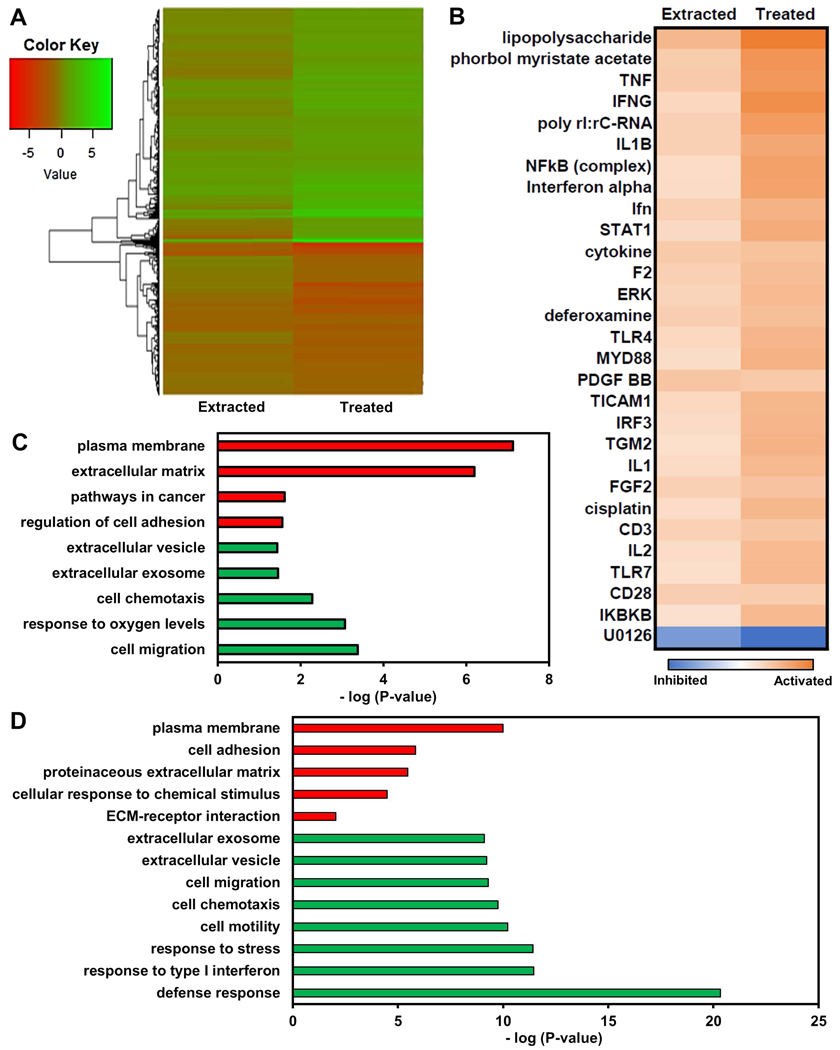
RNA sequencing analysis of cells which survived immobilization in silica gels and were extracted (“Extracted”) and cells which survived cisplatin treatment (“Treated”) was performed to identify potential mechanisms that allow cells to survive within gels or evade cisplatin treatment and which of these are shared between the two groups relative to 2-D untreated control cells (Fig. 4A). Many genes (1,424) were found to be differentially expressed in “Treated” cells relative to the 2-D untreated control, whereas only 328 genes were found to be differentially expressed in “Extracted” cells compared to the 2-D control. More than half of the genes differentially expressed in “Extracted” cells (63.7%, 209 genes) were also differentially expressed in “Treated” cells. Using Ingenuity Pathway Analysis (IPA), the Upstream Regulator function was used to discover transcriptional regulators which best explain the differential gene expression observed within the experimental data. The top 30 upstream regulators predicted to regulate the differential gene expression observed in both “Extracted” and “Treated” samples were primarily drugs or regulators associated with the inflammatory response (Fig. 4B). Interestingly, cisplatin was predicted as a regulator for “Treated” cells, as expected, but also for “Extracted” cells although they were never previously exposed to cisplatin, indicating that cells extracted from silica gels exhibit signaling responses as if they were treated by platinum chemotherapy. Fibroblast growth factor-2 (FGF-2) was additionally predicted to contribute to gene expression changes in surviving “Extracted” and “Treated” cells, and FGF-2 has previously been implicated as a mediator of chemoresistance by activating downstream survival pathways (Patel et al., 2015; Ware et al., 2013) and linked to cellular dormancy in the context of breast cancer (Fenig et al., 1997; H. Wang et al., 1997).
Figure 4. OVCAR-3 cells selected by immobilization regulate genes similarly to cells surviving cisplatin treatment.
(A) Heatmap of log2FC of gene expression in cells immobilized for three days, extracted, and cultured in standard 2-D conditions for two weeks (“Extracted”) and cells surviving treatment with 0.5 μM cisplatin for 24 hours and allowed to recover for 2 weeks in standard 2-D culture conditions (“Treated”) relative to control cells. (B) List of top 30 common upstream regulators (determined by average Z-score with P < 0.05) in “Extracted” and “Treated” cells based upon differentially expressed genes relative to control cells using Ingenuity Pathway Analysis. (C,D) Gene ontology enrichment analysis using genes significantly upregulated (green) or downregulated (red) in “Extracted” (C) or “Treated” (D) samples relative to 2-D controls.
GO enrichment analysis of significantly upregulated genes in “Extracted” and “Treated” cells (Fig. 4C&D) gave terms typically associated with more invasive (e.g. cell motility, migration, chemotaxis) and chemoresistant (e.g. extracellular exosomes/vesicles) phenotypes (Roussos, Condeelis, & Patsialou, 2014; Son & Moon, 2010; Zhang, Ji, Yang, Li, & Wang, 2018). Exosome and vesicle secretion are important for cell-cell communication in the tumor microenvironment and are known to regulate metastasis and chemoresistance in cancer (Zhang et al., 2018). Further, previous work has shown that extracellular vesicles secreted by cisplatin-treated ovarian cancer cells can induce invasion and protection from chemotherapy in other cells pre-treated with these vesicles (Samuel et al., 2018). In “Treated” cells, defense/stress responses and interferon signaling terms were enriched, which was also observed in “Day 1” and “Day 3” immobilization groups, indicating potential similarities in downstream signaling responses to silica gel immobilization and cisplatin treatment. Genes previously associated with tumor initiating cells or the cancer stem cell theory (e.g. CD133, CD44, CD117, EPCAM, ALDH1A1) (Kalir & Kohtz, 2014) were not observed to be consistently and significantly enriched for in experimental groups relative to control cells. Significantly downregulated genes enriched for terms related to the cell membrane, adhesion, and ECM in both “Extracted” and “Treated” cells. Previous studies have shown that cancer cells resisting apoptosis in non-adherent conditions are also more chemoresistant (Eguchi et al., 2015; Foley et al., 2015; Wheeler et al., 2018). Additionally in ovarian cancer, non-adherent cancer spheroids either created in vitro or isolated from in vivo ascites fluid also demonstrate enhanced chemoresistance (Hirst et al., 2018; Liao et al., 2014; Wheeler et al., 2018). Moreover, ovarian cancer spheroids are thought to be a unique subpopulation of tumor cells with self-renewal capability and enhanced chemoresistance (Liao et al., 2014).
Lastly, we compared the differential gene expression we observed in the “Extracted” population, which has been selected by immobilization to have a higher propensity to enter dormancy, with cancer cells from ovarian cancer patients who responded poorly to chemotherapy. Genes which were found to be significantly up- or downregulated in both the “Extracted” group relative to controls and chemoresistant patients relative to chemosensitive patients by Koti et al. and their fold change expression values were used in IPA to identify which pathways were most associated with these genes (Koti et al., 2013). Canonical signaling pathways previously associated with poor clinical outcome and response to chemotherapy in ovarian cancer were identified (e.g. STAT3 Pathway, NF-κB Signaling) (Abubaker et al., 2015; Annunziata et al., 2010; Barlin et al., 2013; Godwin et al., 2013; Permuth-Wey et al., 2016; Yoshikawa et al., 2018), and most notably, pathways responsible for regulation and repair of DNA, which are proposed as key mechanisms of chemoresistance (e.g. NER Pathway, BER Pathway, etc.), were also observed (Tables S2 & S3). Our findings demonstrate that cells extracted from silica gels experienced stresses inducing downstream signaling as if they were treated by cisplatin and display altered regulation of genes associated with pathways known to be important in chemoresistance.
OVCAR-3 cells selected by silica gel immobilization exhibit enhanced chemoresistance even when proliferative.
Because RNA sequencing analysis showed a significant overlap between dormancy-capable cells and chemoresistant phenotypes, we investigated whether dormancy-capable cells would be more resistant to cisplatin chemotherapy. Surviving OVCAR-3 cells were extracted from silica gels following three days of immobilization, and one week post-extraction they were treated with 0.5 μM cisplatin for 24 hours (Fig 5A, Scheme i). These extracted (dormancy-capable) cells demonstrated enhanced resistance to cisplatin treatment – despite being proliferative (Fig. 2A) – relative to control cells which were never previously encapsulated (Fig. 5B). Interestingly, when OVCAR-3 cells induced into quiescence via hypoxia (Carcereri de Prati et al., 2017; Lee, Leslie, & Azarin, 2018) or serum starvation (Oya et al., 2003) were treated with cisplatin, these cells exhibited enhanced chemoresistance (Fig. S6), consistent with earlier reports (Kondoh et al., 2010; Onozuka, Tsuchihara, & Esumi, 2011; Wosikowski, Silverman, Bishop, Mendelsohn, & Bates, 2000). However, after returning to a proliferative state upon removal of the stress (Fig. 5A, Scheme ii), these cells no longer had a survival advantage upon cisplatin treatment (Fig. 5C & D). These results demonstrate the uniqueness of physical confinement stress in silica gels, as it selects for cells which are chemoresistant even after the stress is lifted. This is further supported by our RNA sequencing analysis of “Extracted” cells, which demonstrated they regulate genes as if they had been treated with cisplatin and exhibit downstream signaling similar to “Treated” cells which survived cisplatin chemotherapy. It is possible that silica gels are a significantly harsher environment than hypoxia or serum starvation, and the stresses of physical confinement cause the majority of encapsulated cells to die, allowing for more efficient selection. Further, extracted cells may have enhanced chemoresistance due to mechanisms induced by growth inhibition and dysregulated cell-ECM substrate interactions experienced within gels. These cells may be able to mitigate pro-apoptotic signals that would be initiated under these stressful conditions and may instead sustain pro-survival pathways. The ability to resist programmed cell death would also be useful in resisting DNA damage-induced apoptosis via chemotherapy and could be a potential means for cells surviving confinement to also survive cisplatin treatment. Whether cells selected by immobilization are a unique subpopulation with innate capabilities or whether survival within silica gels is a learned adaptation by cells remains an important question for future study.
Figure 5. Silica gel immobilization selects for cells that are more chemoresistant even while proliferative.
(A) Timeline for cisplatin treatment studies of cells either (i) surviving silica gel immobilization or (ii) recovered from hypoxia treatment or serum starvation. Recovery periods of three days after hypoxia or serum starvation and one week post-extraction allowed for similar cell densities and return to a proliferative state at the start of cisplatin treatment. (B) Response to cisplatin treatment of cells surviving silica gel immobilization (“Extracted+Cisplatin”) relative to cells maintained in 2-D culture conditions (“2-D+Cisplatin”). Fold change in viable cell number indicates cell number at Day 3 post-cisplatin treatment relative to cell number before drug treatment (*P < 0.05 relative to 2-D). (C,D) Fold change in viable cell number for cells recovered from (C) hypoxia (“Recovered Hyp+Cisplatin”) or (D) serum starvation (“Recovered SS+Cisplatin”) and treated with 0.5 μM cisplatin for 24 hours relative to proliferating cells grown in standard culture conditions (“Cisplatin”) after treatment. Fold change in viable cell number indicates cell number at Day 3 post-cisplatin treatment relative to untreated controls for each respective condition (*P < 0.05 compared to “Cisplatin”).
Silica gel platform distinguishes cells with enhanced chemoresistance in other cell lines and towards taxane agents.
SKOV-3, another commonly used ovarian cancer model cell line for drug treatment study, ovarian adenocarcinoma cells were immobilized in silica gels to ensure that selection of cells with quiescent behavior and enhanced chemoresistance was not specific to OVCAR-3 cells. The number of viable SKOV-3 cells significantly decreased over time once immobilized in silica gels (Fig. 6A). The percentage of Ki67-positive SKOV-3 cells also decreased over time in gels (Fig. 6B), and surviving cells could be extracted and cultured in 2-D. SKOV-3 cells are known to be more resistant to cisplatin than OVCAR-3 cells (Kim et al., 2012) and required a higher dose (5 μM) of cisplatin for significant cell death with recovery of a small chemoresistant subpopulation. Upon immobilization in silica gels, this cisplatin-resistant SKOV-3 cell population exhibited enhanced survival relative to control cells with no previous drug treatment (Fig. 6C), consistent with the OVCAR-3 results. In separate studies, SKOV-3 cells surviving immobilization (dormancy-capable) were extracted from gels and found to have decreased sensitivity to cisplatin (Fig. 6D) along with enhanced viability upon re-immobilization (Fig. 6E), similar to OVCAR-3 cells. These results demonstrate that silica gel technology may be used to select for cells with enhanced chemoresistance and propensity to enter dormancy across ovarian cancer cell lines and may even show promise for application towards other cancer types.
Figure 6. Selection of unique subpopulation via silica gel immobilization is also observed with SKOV-3 ovarian cancer cell line.
(A) Calcein AM staining of viable SKOV-3 cells (green) immobilized for 0, 1, 2, 3, 5, and 7 days within silica gels. Scale bar indicates 400 μm. (B) Percentage of Ki67-positive SKOV-3 cells within silica gels over a three-day period (*P < 0.05 compared to previous timepoint). (C) Fold change in viable cell number relative to Day 0 of immobilized cisplatin-resistant SKOV-3 cells and untreated control cells (*P < 0.05 compared to “Control” at each timepoint). (D,E) SKOV-3 cells surviving immobilization for 24 hours were extracted and cultured in 2-D for 1 week and used to find (D) the fold change in viable cell number three days after 5 μM cisplatin treatment for 24 hours relative to before treatment (*P < 0.05 compared to “2-D”) and (E) the fold change in viable cell number relative to Day 0 after re-immobilizing extracted SKOV-3 cells in silica gels relative to proliferating cells immobilized for the first time (Control) (*P < 0.05 compared to “Control” at each timepoint).
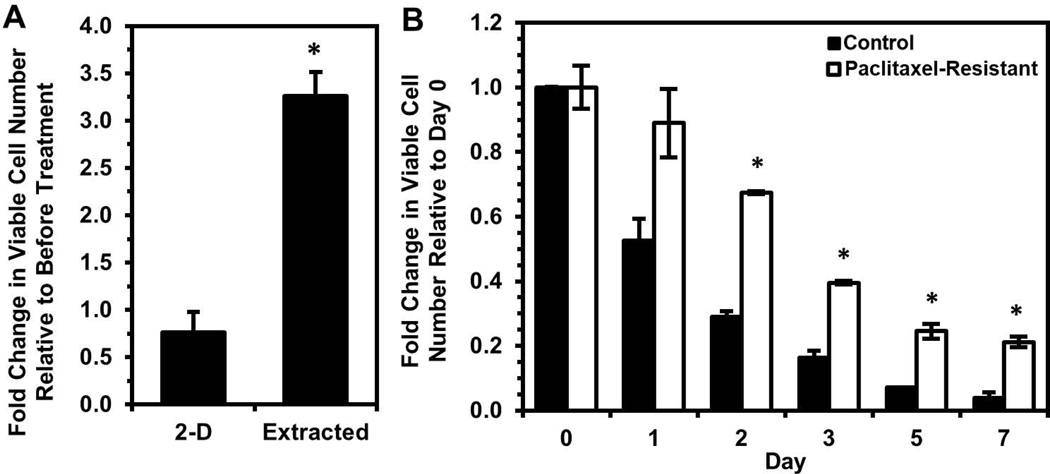
The taxane-based chemotherapeutic paclitaxel was also investigated in combination with silica gel immobilization, as addition of taxane agents in combination with platinum chemotherapy has been the standard regimen for the last decade (Cristea et al., 2010). OVCAR-3 cells extracted from silica gels were found to have decreased susceptibility to paclitaxel treatment relative to 2-D control cells, even though these extracted cells were proliferating at time of paclitaxel addition (Fig. 7A). Again, this result demonstrates that the subpopulation surviving immobilization has enhanced chemoresistance but not only towards platinum-based agents. Further, treatment of OVCAR-3 cells with paclitaxel followed by a two-week recovery period allowed for selection of a more paclitaxel-resistant population (Fig. S7). This population demonstrated significantly enhanced survival relative to untreated cells upon immobilization in silica gels (Fig. 7B), further demonstrating the ability of silica gels to identify ovarian cancer cells less susceptible to multiple types of chemotherapy.
Figure 7. Extracted cells are less sensitive to paclitaxel, and paclitaxel-resistant cells exhibit enhanced survival upon immobilization.
(A) Fold change in viable OVCAR-3 cell number three days after 30 nM paclitaxel for 24 hours relative to before treatment (*P < 0.05 compared to “2-D”). (B) Fold change in viable cell number relative to Day 0 of immobilized paclitaxel-resistant OVCAR-3 cells and untreated control cells (*P < 0.05 compared to “Control” at each timepoint).
These last findings demonstrate the broad applicability of our physical confinement approach using stiff silica gels to investigate a unique subset of cells exhibiting dormant behavior upon immobilization and enhanced chemoresistance once removed from gels. We also showed cells surviving immobilization and extracted from silica gels were found to have enhanced viability upon re-immobilization, indicating these cells are distinct from the original population. Further, RNA sequencing analysis revealed that extracted cells also exhibit downstream signaling activity as if previously exposed to cisplatin, and differentially expressed genes resulted in terms associated with a more chemoresistant phenotype using GO analysis. While cells released from quiescence using other in vitro platforms did not exhibit enhanced chemoresistance, cells selected by silica gels were less susceptible to platinum and taxane agents despite being in a proliferative state. This finding provides evidence that cells which have an enhanced ability to enter dormancy, not just cells that are quiescent at the time of treatment, may be a primary cause of chemoresistance. Future studies investigating pathways initiated by cells surviving immobilization which are also common to cells surviving cisplatin treatment could further elucidate the interplay between ability to enter dormancy and chemoresistance and ultimately identify new therapeutic targets for ovarian cancer. Moreover, as our results showed distinct behavior of cisplatin- and paclitaxel-resistant cells upon immobilization, future work immobilizing ovarian cancer cells isolated from patient biopsies would determine whether silica gels could be a facile method for identifying patients with increased risk of recurrence.
Supplementary Material
Acknowledgements
This work was supported by grant #IRG-16–189-58 from the American Cancer Society and Biotechnology Training Grant: NIHT32GM008347 (T.L.). The authors would like to acknowledge the University of Minnesota Genomics Center and the Minnesota Supercomputing Institute (MSI) for carrying out work and providing resources that contributed to the research results reported within this paper. The authors would also like to thank Hak Rae Lee for assistance with Western blotting experiments and Jennifer One for assistance with RNA sequencing analysis.
Footnotes
Conflict of Interest Statement
The authors declare that there are no conflicts of interests.
Data Availability
The sequencing data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (GEO) and are accessible through GEO Series accession number GSE144232.
References
- Abubaker K, Latifi A, Chan E, Luwor RB, Burns CJ, Thompson EW, … Ahmed N (2015). Enhanced activation of STAT3 in ascites-derived recurrent ovarian tumors: inhibition of cisplatin-induced STAT3 activation reduced tumorigenicity of ovarian cancer by a loss of cancer stem cell-like characteristics. Journal of Cancer Stem Cell Research. [Google Scholar]
- Agarwal R, & Kaye SB (2003). Ovarian cancer: strategies for overcoming resistance to chemotherapy. Nature Reviews Cancer, 3(7), 502–516. 10.1038/nrc1123 [DOI] [PubMed] [Google Scholar]
- Aguirre-Ghiso JA (2007). Models, mechanisms and clinical evidence for cancer dormancy. Nature Reviews Cancer, 7(11), 834–846. 10.1038/nrc2256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguirre-ghiso JA, Estrada Y, Liu D, & Ossowski L (2003). ERK MAPK Activity as a Determinant of Tumor Growth and Dormancy ; Regulation by p38 SAPK. Cancer Research, 63(7), 1684–1695. [PubMed] [Google Scholar]
- Aguirre-ghiso JA, Liu D, Mignatti A, Kovalski K, & Ossowski L (2001). Urokinase Receptor and Fibronectin Regulate the ERK MAPK to p38 MAPK Activity Ratios That Determine Carcinoma Cell Proliferation or Dormancy In Vivo. Molecular Biology of the Cell, 12, 863–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguirre-ghiso JA, Ossowski L, & Rosenbaum SK (2004). Green Fluorescent Protein Tagging of Extracellular Signal-Regulated Kinase and p38 Pathways Reveals Novel Dynamics of Pathway Activation during Primary and Metastatic Growth. Cancer Research, 418, 7336–7345. [DOI] [PubMed] [Google Scholar]
- Annunziata CM, Stavnes HT, Kleinberg L, Berner A, Hernandez LF, Birrer MJ, … Kohn EC (2010). NF-κB transcription factors are co-expressed and convey poor outcome in ovarian cancer. Cancer, 116(13). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlin JN, Jelinic P, Olvera N, Bogomolniy F, Bisogna M, Dao F, … Levine DA (2013). Validated gene targets associated with curatively treated advanced serous ovarian carcinoma. Gynecologic Oncology, 128(3), 512–517. [DOI] [PubMed] [Google Scholar]
- Brancho D, Tanaka N, Jaeschke A, Ventura J, Kelkar N, Tanaka Y, … Davis RJ (2003). Mechanism of p38 MAP kinase activation in vivo. Genes and Development, 17, 1969–1978. 10.1101/gad.1107303.on [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carcereri de Prati A, Butturini E, Rigo A, Oppici E, Rossin M, Boriero D, & Mariotto S (2017). Metastatic Breast Cancer Cells Enter Into Dormant State and Express Cancer Stem Cells Phenotype Under Chronic Hypoxia. Journal of Cellular Biochemistry, 118(10), 3237–3248. 10.1002/jcb.25972 [DOI] [PubMed] [Google Scholar]
- Chien J, Kuang R, Landen C, & Shridhar V (2013). Platinum-Sensitive Recurrence in Ovarian Cancer: The Role of Tumor Microenvironment. Frontiers in Oncology, 3(September), 1–6. 10.3389/fonc.2013.00251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa RJM, Peart T, Valdes YR, Dimattia GE, & Shepherd TG (2012). Modulation of AKT activity is associated with reversible dormancy in ascites-derived epithelial ovarian cancer spheroids. Carcinogenesis, 33(1), 49–58. 10.1093/carcin/bgr241 [DOI] [PubMed] [Google Scholar]
- Cristea M, Han E, Salmon L, & Morgan RJ (2010). Review: Practical considerations in ovarian cancer chemotherapy. Therapeutic Advances in Medical Oncology, 2(3), 175–187. 10.1177/1758834010361333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daubriac J, Fleury-Feith J, Kheuang L, Galipon J, Saint-Albin A, Renier A, … Jaurand M-C (2009). Malignant pleural mesothelioma cells resist anoikis as quiescent pluricellular aggregates. Cell Death and Differentiation, 16, 1146–1155. [DOI] [PubMed] [Google Scholar]
- Eguchi R, Fujita Y, Tabata C, Ogawa H, Wakabayashi I, Nakano T, & Fujimori Y (2015). Inhibition of Src family kinases overcomes anoikis resistance induced by spheroid formation and facilitates cisplatin-induced apoptosis in human mesothelioma cells. Oncology Reports, 34(5), 2305–2310. [DOI] [PubMed] [Google Scholar]
- Fang JY, Tan SJ, Wu YC, Yang Z, Hoang BX, & Han B (2016). From competency to dormancy : a 3D model to study cancer cells and drug responsiveness. Journal of Translational Medicine, 14(38), 1–13. 10.1186/s12967-016-0798-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenig E, Wieder R, Paglin S, Wang H, Persaud R, Haimovitz-Friedman A, … Yahalom J (1997). Basic fibroblast growth factor confers growth inhibition and mitogen-activated protein kinase activation in human breast cancer cells. Clinical Cancer Research, 3(1). [PubMed] [Google Scholar]
- Foley JM II, Monks DJS, Cherba NR, Monsma D, Davidson, P. DJ, … Steensma MR (2015). Anoikis-resistant subpopulations of human osteosarcoma display significant chemoresistance and are sensitive to targeted epigenetic therapies predicted by expression profiling. Journal of Translational Medicine, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Zhu M, Wu Y, Gao P, Qin Z, & Wang H (2014). Interferon-λ1 induces G1 phase cell cycle arrest and apoptosis in gastric carcinoma cells in vitro. Oncology Reports, 32(1), 199–204. [DOI] [PubMed] [Google Scholar]
- Gerdes J, Lemke H, Baisch H, Wacker HH, Schwab U, & Stein H (1984). Cell cycle analysis of a cell proliferation-associated human nuclear antigen defined by the monoclonal antibody Ki-67. Journal of Immunology, 133(4), 1710–1715. [PubMed] [Google Scholar]
- Godwin P, Baird AM, Heavey S, Barr MP, O’Byrne KJ, & Gately K (2013). Targeting Nuclear Factor-Kappa B to Overcome Resistance to Chemotherapy. Frontiers in Oncology, 3(120). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton TC, Young RC, Mckoy WM, Grotzinger KR, Green JA, Chu EW, … Ozols RF (1983). Characterization of a Human Ovarian Carcinoma Cell Line ( NIH : OVCAR-3 ) 1 with Androgen and Estrogen Receptors. Cancer Research, 43, 5379–5389. [PubMed] [Google Scholar]
- Hirst J, Pathak HB, Hyter S, Pessetto ZY, Ly T, Graw S, … Godwin AK (2018). Licofelone Enhances the Efficacy of Paclitaxel in Ovarian Cancer by Reversing Drug Resistance and Tumor Stem-like Properties. Cancer Research, 78(15). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalir TA, & Kohtz DS (2014). Chemoresistance, Dormancy and Recurrence in Platinum Drug Therapy of Ovarian Cancers. In Tumor Dormancy, Quiescence, and Senescence, Vol. 3 (Vol. 3, pp. 79–97). 10.1007/978-94-017-9325-4 [DOI] [Google Scholar]
- Katsumi A, Orr AW, Tzima E, & Schwartz MA (2004). Integrins in Mechanotransduction. Journal of Biological Chemistry, 279, 12001–12004. [DOI] [PubMed] [Google Scholar]
- Kim M, Pak JH, Choi WH, Park J-Y, Nam J-H, & Kim J-H (2012). The relationship between cisplatin resistance and histone deacetylase isoform overexpression in epithelial ovarian cancer cell lines. Journal of Gynecologic Oncology, 23(3), 182–189. 10.3802/jgo.2012.23.3.182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondoh E, Mori S, Yamaguchi K, Baba T, Matsumura N, Barnett JC, … Murphy SK (2010). Targeting slow-proliferating ovarian cancer cells. International Journal of Cancer, 126(10), 2448–2456. 10.1002/ijc.24919 [DOI] [PubMed] [Google Scholar]
- Koti M, Gooding RJ, Nuin P, Haslehurst A, Crane C, Weberpals J, … Squire JA (2013). Identification of the IGF1/PI3K/NF κB/ERK gene signalling networks associated with chemotherapy resistance and treatment response in high-grade serous epithelial ovarian cancer. BMC Cancer, 13, 1–11. 10.1186/1471-2407-13-549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HR, Leslie F, & Azarin SM (2018). A facile in vitro platform to study cancer cell dormancy under hypoxic microenvironments using CoCl 2. 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengyel E (2010). Ovarian Cancer Development and Metastasis. The American Journal of Pathology, 177(3), 1053–1064. 10.2353/ajpath.2010.100105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao J, Qian F, Tchabo N, Mhawech-Fauceglia P, Beck A, Qian Z, … Odunsi K (2014). Ovarian Cancer Spheroid Cells with Stem Cell-Like Properties Contribute to Tumor Generation, Metastasis and Chemotherapy Resistance through Hypoxia-Resistant Metabolism. PLoS ONE, 9(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Lv J, Liang X, Yin X, Zhang L, Chen D, … Huang B (2018). Fibrin Stiffness Mediates Dormancy of Tumor-Repopulating Cells via a Cdc42-Driven Tet2 Epigenetic Program. Cancer Research, 78(14), 3926–3938. 10.1158/0008-5472.CAN-17-3719 [DOI] [PubMed] [Google Scholar]
- Mantia-Smaldone GM, Edwards RP, & Vlad AM (2011). Targeted treatment of recurrent platinum-resistant ovarian cancer: Current and emerging therapies. Cancer Management and Research, 3(1), 25–38. 10.2147/CMR.S8759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onozuka H, Tsuchihara K, & Esumi H (2011). Hypoglycemic/hypoxic condition in vitro mimicking the tumor microenvironment markedly reduced the efficacy of anticancer drugs. Cancer Science, 102(5), 975–982. [DOI] [PubMed] [Google Scholar]
- Oya N, Zölzer F, Werner F, & Streffer C (2003). Effects of serum starvation on radiosensitivity, proliferation and apoptosis in four human tumor cell lines with different p53 status. Strahlentherapie Und Onkologie, 179(2), 99–106. 10.1007/s00066-003-0973-8 [DOI] [PubMed] [Google Scholar]
- Páez D, Labonte MJ, Bohanes P, Zhang W, Benhanim L, Ning Y, … Lenz HJ (2012). Cancer dormancy: A model of early dissemination and late cancer recurrence. Clinical Cancer Research, 18(3), 645–653. 10.1158/1078-0432.CCR-11-2186 [DOI] [PubMed] [Google Scholar]
- Pang J, Jiang P, Wang Y, Jiang L, Qian H, Tao Y, … Wu Y (2018). Cross-linked hyaluronan gel inhibits the growth and metastasis of ovarian carcinoma. Journal of Ovarian Research, 11(22). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel D, Gao Y, Son K, Siltanen C, Neve RM, Ferraraa K, & Revzin A (2015). Microfluidic co-cultures with hydrogel-based ligand trap to study paracrine signals giving rise to cancer drug resistance. Lab on a Chip, (24), 4614–4624. [DOI] [PubMed] [Google Scholar]
- Pavan Grandhi TS, Potta T, Nitiyanandan R, Deshpande I, & Rege K (2017). Chemomechanically engineered 3D organotypic platforms of bladder cancer dormancy and reactivation. Biomaterials, 142, 171–185. 10.1016/j.biomaterials.2017.07.008 [DOI] [PubMed] [Google Scholar]
- Permuth-Wey J, Fulp WJ, Reid BM, Chen Z, Georgeades C, Cheng JQ, … Lancaster JM (2016). STAT3 polymorphisms may predict an unfavorable response to first-line platinum-based therapy for women with advanced serous epithelial ovarian cancer. International Journal of Cancer, 138(3), 612–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfisterer J, & Ledermann JA (2006). Management of Platinum-Sensitive Recurrent Ovarian Cancer. Seminars in Oncology, 33(SUPPL. 6), 12–16. 10.1053/j.seminoncol.2006.03.012 [DOI] [PubMed] [Google Scholar]
- Preciado JA, Reátegui E, Azarin SM, Lou E, & Aksan A (2017). Immobilization platform to induce quiescence in dormancy-capable cancer cells. Technology, 05(03), 129–138. 10.1142/S2339547817500078 [DOI] [Google Scholar]
- Raghavan S, Mehta P, Ward MR, Bregenzer ME, Fleck EMA, Tan L, … Mehta G (2017). Personalized Medicine Based Approach to Model Patterns of Chemoresistance, and Tumor Recurrence Using Ovarian Cancer Stem Cell Spheroids. Clinical Cancer Research, 23(22), 6934–6945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raingeaud J, Gupta S, Rogers JS, Dickens M, Han J, Ulevitch RJ, & Davis RJ (1995). Pro-inflammatory Cytokines and Environmental Stress Cause p38 Mitogen-activated Protein Kinase Activation by Dual Phosphorylation on Tyrosine and Threonine. Journal of Biological Chemistry1, 270, 7420–7426. [DOI] [PubMed] [Google Scholar]
- Rashidi MRW, Mehta P, Bregenzer M, Raghavan S, Fleck EM, Horst EN, … Mehta G (2019). Engineered 3D Model of Cancer Stem Cell Enrichment and Chemoresistance. Neoplasia, 21(8), 822–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reátegui E, Kasinkas L, Kniesz K, Lefebvre MA, & Aksan A (2014). Silica–PEG gel immobilization of mammalian cells. J. Mater. Chem. B, 2(42), 7440–7448. 10.1039/C4TB00812J [DOI] [PubMed] [Google Scholar]
- Roussos ET, Condeelis JS, & Patsialou A (2014). Chemotaxis in cancer. Nature Reviews Cancer, 11(8), 573–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel P, Mulcahy LA, Furlong F, McCarthy HO, Brooks SA, Fabbri M, … Carter DRF (2018). Cisplatin induces the release of extracellular vesicles from ovarian cancer cells that can induce invasiveness and drug resistance in bystander cells. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences, 373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholzen T, & Gerdes J (2000). The Ki-67 protein: From the known and the unknown. Journal of Cellular Physiology, 182(3), 311–322. [DOI] [PubMed] [Google Scholar]
- Scott CL, Mackay HJ, & Haluska P Jr. (2014). Patient-derived xenograft models in gynecologic malignancies. In American Society of Clinical Oncology educational book (pp. 258–266). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son H, & Moon A (2010). Epithelial-mesenchymal Transition and Cell Invasion. Toxicological Research, 26(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosa Maria Soledad, Avivar-Valderas A, Bragado P, Wen HC, & Aguirre-Ghiso JA (2011). ERK1/2 and p38alpha/beta signaling in tumor cell quiescence: Opportunities to control dormant residual disease. Clinical Cancer Research, 17(18), 5850–5857. 10.1158/1078-0432.CCR-10-2574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosa María Soledad, Bragado P, & Aguirre-Ghiso JA (2014). Mechanisms of disseminated cancer cell dormancy: an awakening field. Nature Reviews Cancer, 14(9), 611–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate DJ, Patterson JR, Finkel-Jimenez B, & Zea AH (2013). Interferon (IFNγ) induces cell cycle arrest in RCC cell lines. Journal for ImmunoTherapy of Cancer, 1(1), 125. [Google Scholar]
- Touil Y, Zuliani T, Wolowczuk I, Kuranda K, Prochazkova J Andrieux J, Le Roy H, … Polakowska R (2013). The PI3K/AKT signaling pathway controls the quiescence of the low-Rhodamine123-retention cell compartment enriched for melanoma stem cell activity. Stem Cells, 31(4), 641–651. [DOI] [PubMed] [Google Scholar]
- V.Bulavin D, & Fornace AJ Jr. (2004). p38 MAP Kinase’s Emerging Role as a Tumor Suppressor. In Advances in Cancer Research (pp. 95–118). [DOI] [PubMed] [Google Scholar]
- Wall L, Burke F, Barton C, Smyth J, & Balkwill F (2003). IFN-γ Induces Apoptosis in Ovarian Cancer Cells in Vivo and in Vitro. Clinical Cancer Research, 9(3). [PubMed] [Google Scholar]
- Wang H, Rubin M, Fenig E, DeBlasio A, Mendelsohn J, Yahalom J, & Wieder R (1997). Basic Fibroblast Growth Factor Causes Growth Arrest in MCF-7 Human Breast Cancer Cells while Inducing both Mitogenic and Inhibitory G1 Events. Cancer Research, 57(9). [PubMed] [Google Scholar]
- Wang N (2017). Review of Cellular Mechanotransduction. Journal of Physics D: Applied Physics, 50(23). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware KE, Hinz TK, Kleczko E, Singleton KR, Marek LA, Helfrich BA, … Heasley LE (2013). A mechanism of resistance to gefitinib mediated by cellular reprogramming and the acquisition of an FGF2-FGFR1 autocrine growth loop. Oncogenesis, 2(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler LJ, Watson ZL, Qamar L, Yamamoto TM, Post MD, Berning AA, … Bitler BG (2018). CBX2 identified as driver of anoikis escape and dissemination in high grade serous ovarian cancer. Oncogenesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wosikowski K, Silverman JA, Bishop P, Mendelsohn J, & Bates SE (2000). Reduced growth rate accompanied by aberrant epidermal growth factor signaling in drug resistant human breast cancer cells. Biochim Biophys Acta, 1497(2), 215–226. 10.1016/S0167-4889(00)00062-8 [DOI] [PubMed] [Google Scholar]
- Yeh AC, & Ramaswamy S (2015). Mechanisms of Cancer Cell Dormancy—Another Hallmark of Cancer? Cancer Research, 75(23). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa T, Miyamoto M, Aoyama T, Soyama H, Goto T, Hirata J, … Takano M (2018). JAK2/STAT3 pathway as a therapeutic target in ovarian cancers. Oncology Letters, 15(4), 5772–5780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Ji Q, Yang Y, Li Q, & Wang Z (2018). Exosome: Function and Role in Cancer Metastasis and Drug Resistance. Technology in Cancer Research and Treatment, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.