Objective:
Some HIV+ patients, virally suppressed on ART, show occasional ‘blips’ of detectable HIV-1 plasma RNA. We used a new highly sensitive assay of cell-associated HIV-1 RNA to measure transcriptional activity in PBMCs and production of infectious virus from the viral reservoir, in patients with and without ‘blips’.
Design/methods:
RNA and DNA extracted from cells in 6 ml of peripheral blood, from suppressed patients with one to two ‘blip’ episodes over the past 2 years of ART (n = 55), or no ‘blips’ (n = 52), were assayed for HIV-1 RNA transcripts and proviral DNA targeting the highly conserved ‘R’ region of the LTR. Follow-up samples were also collected. Purified CD4+ T cells were cultured with anti-CD3/CD28/CD2 T-cell activator to amplify transcription and measure replication competent virus.
Results:
HIV-1 RNA transcripts ranged from 1.3 to 5415 copies/106 white blood cells. ‘Blip’ patients had significantly higher levels vs. without blips (median 192 vs. 49; P = 0.0007), which correlated with: higher levels of inducible transcripts after activation in vitro, sustained higher HIV-1 transcription levels in follow-up samples along with increasing HIV-1 DNA in some, and production of replication-competent HIV-1.
Conclusion:
Viral ‘blips’ are significant reflecting higher transcriptional activity from the reservoir and contribute to the reservoir over time. This sensitive assay can be used in monitoring the size and activity of the HIV-1 reservoir and will be useful in HIV-1 cure strategies.
Keywords: activation of CD4+ T cells, blip episode, HIV reservoir, HIV-1 transcriptional activity
Introduction
Most HIV-1-infected individuals on antiretroviral therapy (ART) successfully reduce HIV-1 RNA plasma viral load (pVL) to less than 20 copies/ml, the lower limit of detection for standard diagnostic RT-PCR assays [1,2]. Nonetheless, occasional viral blips (elevated pVL between 20 and 200 copies/ml) occur [3–6]. These blips could be related to assay variation, given the extremely sensitive nature of the pVL diagnostic test [7–11] or a brief cytokine/antigen-driven increase in HIV-1 replication from the HIV-1 reservoir in the context of an unrelated illness [12], or residual activation from the reservoir [13–16]. Importantly, no new resistance mutations have been seen before, during, or shortly after blips [17].
The clinical significance of viral blips is unclear. Some studies have shown an association with progression of disease [3,13–16,18], whereas most do not [4–6,19]. Recent evidence suggests that they impede the ART-related decline in the HIV-1 latent reservoir [20]. A better understanding of the precise nature of blips would help address these uncertainties.
The research-based highly sensitive single copy RNA assay of HIV-1 has quantified very low levels of residual pVL on ART [18,21–26]. However, this methodology is technically challenging, because of the very low concentration of HIV virions in suppressed patients in plasma, on ART. Other assays have been developed to quantify the intact proviral HIV-1 DNA reservoir [27–31], including the Intact Proviral DNA Assay (IPDA) [27] and the quadruplex qPCR (Q4PCR) assay [28] but are impractical for routine clinical use. The latter two have been designed to increase the probability of quantifying only intact HIV-1 DNA using two or four highly conserved regions (reviewed in [29]). However, infected cells containing replication-competent provirus are very rare, usually containing only one copy of HIV proviral DNA per infected cell.
Instead, cell-associated RNA assays can detect many copies of spliced and unspliced RNA [30,31]. Furthermore, measurements of cell-associated HIV-1 RNA in PBMC cultures activated in vitro with either anti-CD3/CD28 [32,33] or PMA/ionomycin [34], have found HIV-1 is able to newly infect reporter CD4+ T-cell lines, thereby corresponding to replication-competent HIV-1 in circulating cells. Indeed, levels of cell-associated HIV RNA may be a surrogate for reservoir activity: higher levels prior to ART discontinuation correlate with quicker pVL rebound during treatment interruptions [35,36].
We used a recently described, extremely sensitive assay to detect intracellular HIV-1 RNA and total HIV-1 DNA [37], targeting the highly conserved ‘R’ region of the LTR. This ‘Double-R assay’ is able to detect all HIV-1 mRNA transcription, including spliced mRNA and unspliced mRNA as well as total integrated HIV-1 DNA. Herein we used this sensitive assay to better understand HIV-1 transcriptional activity and DNA in PBMCs from blip-experienced and blip-negative patients. We addressed the significance of the ex-vivo HIV-1 transcript activity in peripheral blood and compared it with transcript activity of in-vitro activated purified CD4+ T cells.
Methods
Study design, DNA and RNA extraction
This study was approved by the St Vincent's Hospital Human Research Ethics Committee (HREC LNR/16/SVH/327) and used standard-of-care samples sent to the NSW State Reference laboratory for HIV, St Vincent's Hospital between 2017 and 2020 for monitoring pVL and CD4+ T-cell counts. Demographics and HIV-1 disease characteristics available from St Vincent's Hospital patient database, are shown in Table S1. Also, an elite controller study was approved by South Western Sydney Local Health District Human Research Ethics Committee (HREC 2020/ETH00235). We defined viral blips as elevated routine patient monitoring pVL between 20 and 200 copies/ml. The median interval between analysis time points for this observational study was 6 months [interquartile range (IQR) = 5–8]. White blood cells (WBCs) were prepared from 6 ml of fresh anticoagulated whole blood in ACD (acid citrate dextrose) tubes, as previously described [37]. DNA and RNA were extracted using the Maxwell RSC automated extraction platform (Promega, Madison, Wisconsin, USA), with the Maxwell RSC Buffy Coat DNA kit (Promega) and Maxwell RSC Simply RNA Tissue kit (Promega), respectively, according to the manufacturer's protocol. WBCs were counted after red blood cell lysis using TC20 Automated Cell Counter (Bio-Rad, Hercules, California, USA), and these counts were used to normalize the HIV-1 DNA and RNA results as copy numbers/106 cells.
The double-R assay based on πCode End-Point PCR assay
We used a recently reported assay to detect intracellular HIV-1 transcription and total HIV-1 DNA [37], targeting the highly conserved ‘R’ region in both the 5′-‘LTR’ and 3′-‘LTR’ regions (Fig. S1). The Double-R assay is based on the precision image pi-code (πCode) MicroDiscs detection platform [37–39], and is at least 27 times more sensitive than the current gold standard Real-Time PCR assay [37]. HIV-1 copy number per patient sample, in duplicate, was determined from a standard curve generated with HIV-1 plasmid controls, with 8 points of 0.73--1600 HIV-1 copies/μl. The HIV-1 copy number was normalized per one million of WBCs and used as the standardized unit.
Analysis of intact sequences in proteinase/reverse transcriptase and integrase region
To confirm the presence of intact HIV-1 sequences, protease/reverse transcriptase (PR/RT) and integrase regions were amplified with the in-house Drug Resistance assay, at the NSW State Reference laboratory for HIV, St Vincent's Hospital [37,40]. HIV-1 subtype was identified using the HIV Drug Resistance Database, Stanford University [41]. Identification of insertions, deletions, stop codons and APOBEC3G-related mutations in PR/RT and integrase regions was conducted using the same Stanford HIV Drug Resistance Database.
Isolation of CD4+ T cells by negative selection and culture with ex-vivo T-cell activation
CD4+ T cells were isolated from 3 to 4 ml of fresh ACD-anticoagulated whole blood using the RosetteSep CD4 Enrichment Cocktail (StemCell Technologies, Vancouver BC, Canada), following the manufacturer's instructions. Briefly, 50 μl of RosetteSep reagent was mixed per milliliter of whole blood, incubated at room temperature (RT) for 20 min, and then layered on Ficoll-Hypaque for density centrifugation. The layer of CD4+-enriched cells was washed twice with PBS and resuspended in 2 ml of RPMI containing 20% FCS and 20 IU/ml of IL-2 (Roche, Basel, Switzerland; 10799068001). Cells were counted using TruCount tubes (BD Biosciences, San Jose, California, USA) and simultaneously analyzed for purity, by flow analysis on a 5-laser Fortessa as previously described [42]. Typically, 1.5–2 × 106 cells at more than 96% purity of CD45+CD4+ cells were obtained and incubated in two wells of a 48-well plate (Corning, Corning, New York, USA). One well was left with IL-2 only, and to the second well was added 5 μl of anti-CD3/CD28/CD2 (StemCell Technologies). Cells were cultured for 3 days in 5% CO2 at 37 °C, before RNA extraction, using the Maxwell RSC SimplyRNA Tissue kit. HIV-1 RNA transcript numbers in these cultures were normalized to copy numbers/input 106 CD4+ T cells.
Reverse transcriptase assay of supernatants from activated CD4+ T and SupT1 cells
Reverse transcriptase activity in culture supernatants was determined as previously described [43,44]. Reverse transcriptase activity was used to measure the amount of HIV-1 in 10 μl of culture supernatant from activated CD4+ T cells, collected after days 6 and 9 of culture (see Fig. S7 for further details).
Statistical analysis
Standard curves from known concentrations of HIV-1 plasmid copy numbers were generated with GraphPad Prism v7 (GraphPad Software, San Diego, California, USA). HIV-1 transcription levels in blip versus without blip groups were analyzed by nonparametric Mann--Whitney test. Pearson correlation was used for analysis of HIV-1 transcription and HIV-1 DNA levels. HIV-1 transcription levels in IL-2-only cultures versus IL-2 with the anti-CD3/CD28/CD2 activator cultures were analyzed by the parametric paired t test.
Results
Intracellular HIV-1 transcript activity and total HIV-1 DNA levels in peripheral blood
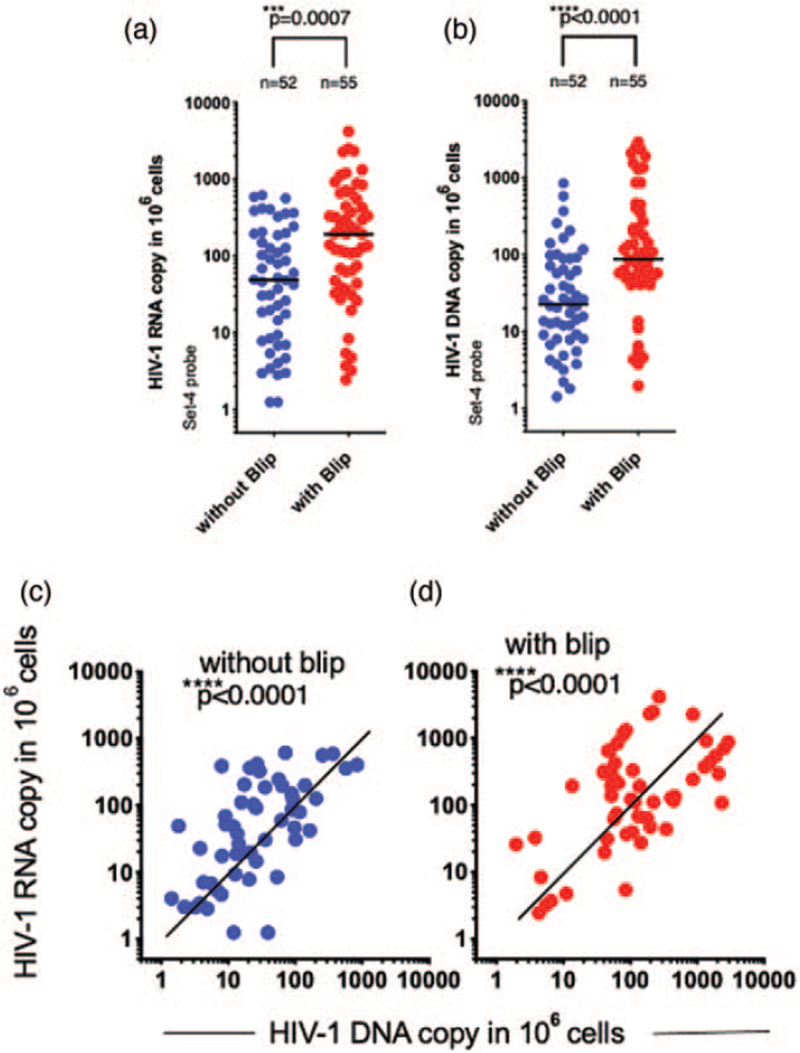
The Double-R assay (Fig. S1a and b) detected cell-associated HIV-1 transcriptional activity from both patient groups (see Table S1 for patient group details). Individual patient HIV-1 transcriptional activity, expressed as copies/106 WBCs, had a range greater than 3 log10, from 1.3 to 5415 copies/106 WBCs (Fig. 1a and b and Fig. S2a and b). The median HIV-1 RNA transcription levels in the ‘blip’ group was significantly higher than the ‘without blip’ group (Fig. 1a: 192 vs. 49 copies/106 cells; P = 0.0007 and Fig. S2a). The median total HIV-1 DNA levels in the blip group was also significantly higher than the without blip group (Fig. 1b: 88 vs. 23 copies/106 cells; P < 0.0001 and Fig. S2b).
Fig. 1.
Detection of HIV-1 RNA transcription and total HIV-1.
(a) HIV-1 RNA transcriptional levels and (b) HIV-1 DNA levels, detected by the Double-R assay for patients with (red symbols) and without blips (blue symbols). Values were normalized to 106 WBC cells. Medians are shown with a bar. (c) Correlation analysis of HIV-1 RNA transcription and HIV-1 DNA in without blip-experienced patients and (d) with blip-experienced patients. These results show values obtained using the set-4 probe. The set-6 probe analyses confirming the set-4 probe results, are shown in Fig. S2a--d.
We then assessed the correlation between HIV-1 RNA transcription levels and HIV-1 DNA levels in each of the two groups, and identified a strong correlation between intracellular HIV-1 RNA transcripts and HIV-1 DNA for each patient group (Fig. 1c and d; P < 0.0001 and Fig. S2c and d).
Importantly, Fig. 1c also shows that, at any given HIV-1 DNA level, the number of transcripts varied by up to 3 log10 between patients.
Follow-up sample analysis in HIV-1 transcript activity and total HIV-1 DNA levels
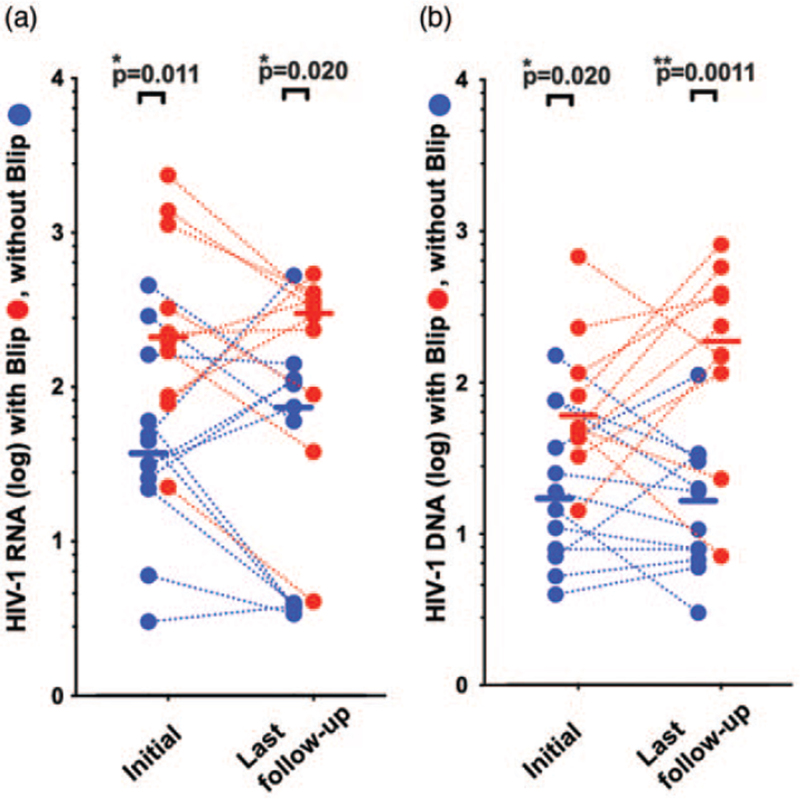
We further investigated whether the initial high levels of HIV-1 transcription and DNA were also seen in follow-up samples over a median of 17 months (IQR = 11–20). Ten blip and twelve without blip patients had multiple samples from different time points assessed (Fig. 2a and b and Fig. S3, 4).
Fig. 2.
Follow-up sample analyses of blip and without blip-experienced patients. WBC, white blood cells.
(a) Difference in HIV-1 RNA transcriptional levels and (b) HIV-1 DNA levels for blip (red symbols; n = 10) and without blip patients (blue symbols; n = 12). HIV-1 RNA transcripts and DNA levels are shown for initial analyses (left, M = 0) and for last follow-up time point analyses (right). Values were normalized to 106 WBC cells. Median values are shown as bars for with blip patients and without blip experienced patients, respectively. WBC, white blood cells.
Overall, the significant elevation of HIV-1 transcripts in blip patients compared to without blip patients was maintained (Fig. 2); initially with median 209 vs. 38 respectively; P = 0.011) and at last follow-up with median 305 vs. 67, respectively; P = 0.020. HIV-1 total DNA was significantly higher in blip patients initially (median 50 vs. 17; P = 0.020) and at last follow-up (median 118 vs. 15; P = 0.0011) (see Fig. S3 and Fig. S4 for details)
Isolation and in-vitro activation of purified CD4+ T cells
As HIV-1 infection is known to persist in peripheral blood memory CD4+ T cells [32,45–48], we investigated the effect on HIV transcripts of using strong polyclonal stimulation of purified CD4+ T cells. In 15 consecutive patients, constituting nine blip patients and six without blip patients, CD4+ T cells were isolated from 3 to 4 ml of whole blood samples, using negative selection, to achieve greater than 96% purity (Fig. S5a). The purified CD4+ T cells were set up as 3-day cultures containing IL-2 in the presence of a highly effective T-cell activator combination of anti-CD3, anti-CD28 and anti-CD2 monoclonal antibodies (anti-CD3/CD28/CD2 activator). A parallel culture containing only IL-2 was also studied for each patient sample.
The anti-CD3/CD28/CD2 activator combination induced large clusters of blasts in cultures from all patients (Fig. S5b and c and Table S2) and up-regulated cell surface expression of the activation markers CD25 and CD134 in a very high proportion of CD4+ T cells by day 2 (Fig. S6a and b), consistent with a very high level of polyclonal TCR stimulation [49]. In the presence of the anti-CD3/CD28/CD2 T-cell activator with IL-2, there were increased numbers of cells, at day 3 vs. the inactivated IL-2-only culture (Table S2). The increased cell number was associated with proliferation of the majority of CD4+ T cells, up to three to four cell divisions, by day 3 (Fig. S6c-e).
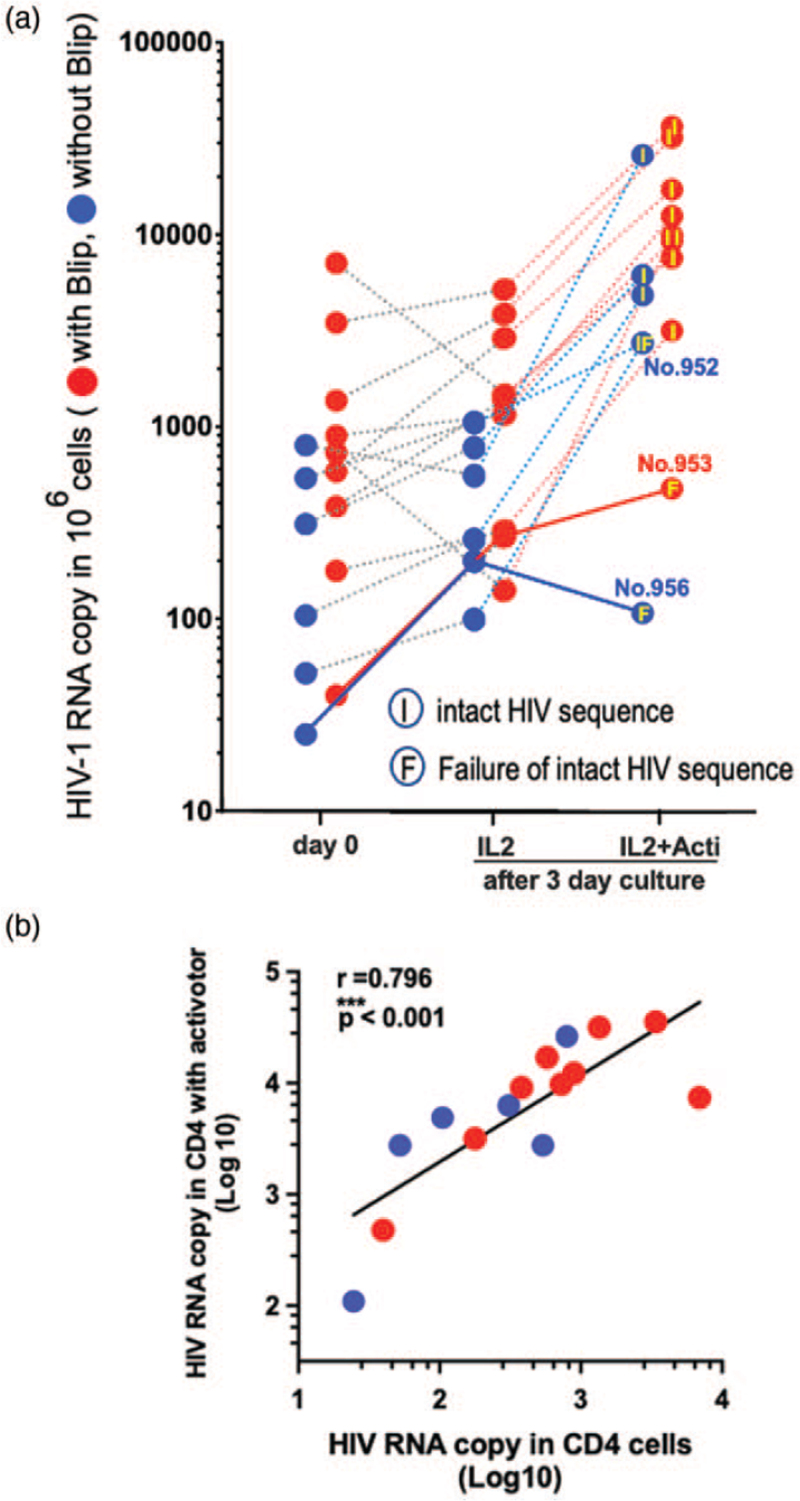
HIV-1 transcripts were detected in CD4+ T cells ex vivo, prior to culture, as described above, for these 15 patients (day 0, left in Fig. 3a). The purified CD4+ T cells cultured with IL-2 had approximately similar transcript levels to the ex-vivo results from the same patients (middle column versus first column in Fig. 3a). With the anti-CD3/CD28/CD2 T-cell activator and IL-2, the transcription levels were greatly increased in all but two patients (right column in Fig. 3a).
Fig. 3.
Presence of replication-competent HIV-1 in latent reservoir after in-vitro culture of activated CD4+ T cells.
(a) Day-0 data was HIV-1 RNA transcription levels, ex vivo, normalized to 106 CD4+ T cells, in original peripheral blood, prior to culture (left). HIV-1 RNA transcription levels were also measured after 3-day ex-vivo culture of CD4+ T cells in the presence of IL2 only (middle) and in the presence of IL2 with anti-CD3/CD28/CD2 T-cell activator (right), normalized to 106 CD4+ T cells. Intact HIV-1 RNA sequences were identified in intracellular mRNA extracted from the activated CD4+ T cells. HIV-1 RNA sequence in Protease and Reverse Transcriptase (PR/RT) and integrase (IN) regions were analyzed using intracellular mRNA extracted from the activated CD4+ T cells. After Sanger Sequence analysis, we identified presence of intact HIV-1 RNA sequences in three PR, RT, and IN regions, indicated with ‘I’ in the activated dots (Table S3). Two patients, ID953, ID956, showing the lowest transcriptional activity at day 0, also showed the lowest transcriptional activity after in-vitro culture. After Sanger Sequence analysis, we failed to identify the presence of intact HIV-1 RNA sequences from these samples, indicated with ‘F’ within the dot. The patient ID952 was labelled as ‘F’ because of failure to identify intact HIV-1 sequences in IN region; however, we identified the presence of intact HIV-1 sequences in both PR and RT regions (Table S2). (b) Correlation of HIV-1 RNA transcription in CD4+ T cells after in vitro 3-day culture in the presence of IL2 and T-cell activator, versus HIV-1 RNA transcription per 106 CD4+ T cells in peripheral blood, ex vivo, prior to culture. Blip patients are shown by red symbols and without blip-experienced patients are shown by blue symbols.
Overall, transcriptional activity in the CD4+ T cells ex vivo, prior to culture, highly correlated with transcriptional activity in the purified CD4+ T cells cultured in vitro in the presence of IL2 and anti-CD3/CD28/CD2 activator (∗∗∗P < 0.001, Fig. 3b).
Presence of replication-competent HIV-1 in latent reservoir
We extended the analysis of the intracellular mRNA obtained from the activated CD4+ T cells, using HIV-1 Proteinase/Reverse Transcriptase and integrase region sequences (Fig. 3a, Table S2). The sequence data revealed that the HIV-1 promoter was able to transcribe long sequences of ‘intact HIV-1 mRNA’ in most of the activated cultures: 13 out of 15 samples in HIV-1 PR/RT region and 12 out of 15 samples in HIV-1 IN region. We could not identify any insertions, deletions, stop codons, or APOBEC3G-related mutations from a total ≈3500 bp long sequence analyses (Table S2 and Table S3). The anti-CD3/CD28/CD2 activator combination was able to induce intact, nonmutated HIV-1 RNA sequences from its promoter in all but three patients (Fig. 3a).
Two samples (ID953, 956) out of 15 for the HIV-1 PR/RT region, and three (ID952, 953, 956) out of 15 samples for the HIV-1 integrase region, failed to amplify by PCR for HIV-1 sequencing (Table S3). Out of these three samples, two showed a less than two-fold increase of HIV-1 transcriptional activity after anti-CD3/CD28/CD2 activation, compared with the IL-2-only culture (Fig. 3a and Table S2). Importantly, the two patients with the lowest levels of intracellular HIV-1 transcriptional activity in CD4+ T cells at day 0 showed the lowest levels of in-vitro reactivation from latent infection of HIV-1 within CD4+ T cells, and failed to show intact HIV-1 RNA sequences in the PR/RT and integrase regions (patients ID953 and ID956 in Fig. 3a).
Therefore, transcriptional activity detected by the double-R assay in CD4+ T cells ex vivo, prior to culture, correlated with the production of replication-competent HIV-1 in cultures with maximal activation in vitro.
Consistent with intact sequences, viral outgrowth occurred in the anti-CD3/CD28/CD2-activated cultures. HIV-1 was detected in the culture supernatants of the activated CD4+ T cells on days 6–10 (Fig. S7a), by reverse transcriptase assay. Furthermore, inoculation from the activated CD4+ T-cell cultures into secondary cultures of the CD4+ SupT1 cell line demonstrated productive infection, by reverse transcriptase assay (Fig. S7b). These results were also confirmed in analyses done on an elite controller showing extremely low HIV-1 transcripts by the double R assay and validated by purified CD4+ T-cell culture activity (Table S4; Table S5; Table S6 and Fig. S8 for further details).
Discussion
In this study, patients with past viral blips had significantly increased cell-associated HIV-1 transcriptional activity temporally remote from the blip episodes, compared to without blip patients as assessed by the Double-R assay. This significant increase was maintained in follow-up samples, and in some, there was an increase in HIV-1 DNA in PBMCs reflecting an increase in that reservoir. Further, the transcriptional activity in ex-vivo CD4+ T cells had significant correlation with transcriptional activity in in-vitro activated purified CD4+ T cells, harbouring HIV-1 intact HIV-1 sequences and production of replication-competent HIV-1.
Our results are in accord with recent studies and also extend these studies. Several groups have shown that blips may be associated with high-baseline pVL or progression of disease [3,13–16]. A large Swiss longitudinal HIV Cohort Study with 1057 individuals with analysis of total HIV-1 DNA in PBMCs showed clear evidence that blips impede the decline in the HIV-1 latent reservoir [20]. The current study extends these data by showing that regardless of the timing of blip episodes, high-level HIV transcripts are sustained in blip patients. Even some without blip patients at follow-up after prolonged suppression of pVL (>31months) had borderline levels of HIV-1 transcripts (between 49 and 192 RNA copies/WBCs: definition of borderline in Fig. S3).
The single copy RNA assay has shown that most clinically suppressed patients have persistent low-level viremia but it has only been used in research and clinical trial contexts [18,21,23–26] and has the disadvantage of a limited range from 0 to 20 copies. The Double-R assay has an improved dynamic range of at least 3 log10 copies making it more suitable for routine clinical monitoring.
Indeed, the Double-R assay has significant advantages over other methodologies in terms of detecting any residual HIV-1 transcripts to reflect HIV-1 promotor activity. The Double-R assay detects HIV-1 promoter activity at both ends of 5′-LTR and 3′-LTR of all transcripts including un-spliced and spliced intracellular HIV-1 RNA. It requires only 4–6 ml of whole blood and is not labour-intensive or resource-intensive, in contrast to the previous research-based viral outgrowth or limiting dilution assays. The Double-R assay targets the ‘R’ region, just downstream of the HIV-1 promoter ‘U3’ region in the LTR; it is a highly conserved and essential region for HIV-1 integration [50,51]. The U3 region is also essential for HIV-1 transcription but can be modulated by promoter-targeted short hairpin RNA to induce HIV-1 transcriptional gene silencing akin to a latent HIV-1 infection [52–56]. It is likely that when the promoter region in the HIV-1 LTR DNA mutates, HIV-1 would not be able to efficiently produce HIV-1 transcripts.
Our target ‘R’ region sequence to detect HIV-1 transcripts is quite different from previously described targets in un-spliced and spliced HIV-1 RNA assays [32,57–59]. Previous studies, that clearly showed a correlation between relatively higher cell-associated HIV RNA levels in PBMC and more rapid rebound during treatment interruptions, were mainly based on unspliced transcripts but in close to half of patients during ART, these were at or below their assay's limit of detection [35,36].
Several HIV-1 DNA-based analyses of the memory CD4+ T-cell reservoir [60] using next-generation sequencing (NGS) have reported that the majority (>90%) of the integrated HIV-1 genome is replication-incompetent [27,48,61], with newer approaches now used to identify intact proviral genomes [62–70], particularly in Effector Memory CD4+ T cells [62]. However, the disadvantage of these approaches is that intact virus is present close to, or below, the lower limit of detection [62]. In contrast, our HIV-1 RNA transcript analysis following very efficient activation of CD4+ T cells, with greatly increased intracellular mRNA because of the additional CD2 co-stimulation that increases signalling proximal to the T-cell receptor, separate from CD28 co-stimulation via PI3K/Akt [71,72], had the distinct advantage of being able to detect activatable and intact HIV-1 sequences at very low levels.
Analysis of transcriptional activity by the Double-R assay has some limitations and the current study was not designed to quantify exactly the proportion of replication-competent virus relative to the HIV-1 transcriptional activity in CD4+ T cells. The amount of transcriptional activity detected by the Double-R assay would almost certainly be an overestimate compared with the amount of intact HIV-1 transcripts in the CD4+ T cells in peripheral blood, which will be the subject of future studies.
Our results have several significant consequences, which are hypothesis-generating. First, the variability in transcriptional activity may relate to particular HIV-1 characteristics or possibly differential effects of antiretroviral drugs. Further research addressing these possibilities is required. Second, measurement of transcriptional activity in peripheral blood CD4+ T cells can act as a surrogate marker for the HIV-1 reservoir. Whilst we did not measure the reservoir in other tissues or cell types, a recent study examining many separate tissue reservoirs in autopsy samples suggests that CD4+ T cells in the peripheral blood are nevertheless responsible for dissemination of HIV-1 from reservoir sites of tissue replication [73]. Third, the reservoir is dynamic rather than stable, with a bi-directional relationship between blips and reservoir size. Blips may both contribute to, and reflect, activity and size of the reservoir. This residual transcription may eventually represent a new target of antiretroviral therapy.
In conclusion, viral blips are significant. High-sensitivity detection of HIV-1 transcripts within the circulating reservoir of memory CD4+ T cells will enable new scalable opportunities to more closely monitor treated patients experiencing blips, and also study the efficacy of treatments aimed at achieving HIV eradication.
Acknowledgements
Author contributions: K.S. conceived the project. K.S. and J.Z. designed the experiments and analysis. A.L., J.Z., B.J.B. and K.S. conducted experiments. A.L., J.Z., B.J.B. and K.S wrote the manuscript. J.Y. and J.C. analyzed the Sanger Sequence Analysis. J.Y., M.S., J.C., R.W., N.R., T.S., D.D., S.P., C.F., I.H., L.P.M. and P.C. coordinated and provided clinical samples and patient data. Z.L., J.Z. and K.S. analyzed stats. T.I. and C.-S.H. provided the precision image pi-code (πCode) MicroDiscs detection platform. V.O., L.E., G.S. and B.J.B. provided clinical support for the study and valuable critical analysis and discussion. All authors reviewed and approved the manuscript.
This research was funded by the St Vincent's Clinic Foundation Research Grant and AMR Translational Research Grant with partial support from NHMRC grant #1105808. K.S. receives research funds from Denka Co. Ltd.
Conflicts of interest
K.S. receives research funds from Denka Co. Ltd. K.S. is the original inventor under WO2018/045425 (PTC/AU2017/050974) patent, titled ‘Methods of detecting Lentivirus’ of HIV-1 detection targeting ‘R’ region.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Footnotes
Supplemental digital content is available for this article.
References
- 1.Palella FJ, Jr, Delaney KM, Moorman AC, Loveless MO, Fuhrer J, Satten GA, et al. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. HIV Outpatient Study Investigators. N Engl J Med 1998; 338:853–860. [DOI] [PubMed] [Google Scholar]
- 2.Maldarelli F, Palmer S, King MS, Wiegand A, Polis MA, Mican J, et al. ART suppresses plasma HIV-1 RNA to a stable set point predicted by pretherapy viremia. PLoS Pathog 2007; 3:e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sorstedt E, Nilsson S, Blaxhult A, Gisslen M, Flamholc L, Sonnerborg A, Yilmaz A. Viral blips during suppressive antiretroviral treatment are associated with high baseline HIV-1 RNA levels. BMC Infect Dis 2016; 16:305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Teira R, Vidal F, Munoz-Sanchez P, Geijo P, Viciana P, Ribera E, et al. VACH Study Group. Very low level viraemia and risk of virological failure in treated HIV-1-infected patients. HIV medicine 2017; 18:196–203. [DOI] [PubMed] [Google Scholar]
- 5.den Oudsten M, van Kampen J, Rijnders B, van de Vijver D, van der Ende M. Is HIV-1 viraemia below 20 copies/mL in antiretroviral-treated patients associated with virologic outcome?. Infect Dis (Lond) 2019; 51:259–267. [DOI] [PubMed] [Google Scholar]
- 6.Widdrington J, Payne B, Medhi M, Valappil M, Schmid ML. The significance of very low-level viraemia detected by sensitive viral load assays in HIV infected patients on HAART. J Infect 2011; 62:87–92. [DOI] [PubMed] [Google Scholar]
- 7.Ruelle J, Debaisieux L, Vancutsem E, De Bel A, Delforge ML, Pierard D, Goubau P. HIV-1 low-level viraemia assessed with 3 commercial real-time PCR assays show high variability. BMC Infect Dis 2012; 12:100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murray JM, Zaunders JJ, Koelsch KK, Natarajan V, Badralmaa Y, McBride K, et al. Short communication: HIV blips while on antiretroviral therapy can indicate consistently detectable viral levels due to assay underreporting. AIDS Res Hum Retroviruses 2013; 29:1621–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wojewoda CM, Spahlinger T, Harmon ML, Schnellinger B, Li Q, Dejelo C, et al. Comparison of Roche Cobas AmpliPrep/Cobas TaqMan HIV-1 test version 2.0 (CAP/CTM v2.0) with other real-time PCR assays in HIV-1 monitoring and follow-up of low-level viral loads. J Virol Methods 2013; 187:1–5. [DOI] [PubMed] [Google Scholar]
- 10.Wiesmann F, Ehret R, Naeth G, Daumer M, Fuhrmann J, Kaiser R, et al. Multicenter evaluation of two next-generation HIV-1 quantitation assays, Aptima Quant Dx and Cobas 6800, in comparison to the RealTime HIV-1 Reference Assay. J Clin Microbiol 2018; 56:e00292-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.White K, Garner W, Wei L, Eron JJ, Zhong L, Miller MD, et al. Repeat testing of low-level HIV-1 RNA: assay performance and implementation in clinical trials. AIDS 2018; 32:1053–1057. [DOI] [PubMed] [Google Scholar]
- 12.Zoufaly A, Kiepe JG, Hertling S, Hufner A, Degen O, Feldt T, et al. Immune activation despite suppressive highly active antiretroviral therapy is associated with higher risk of viral blips in HIV-1-infected individuals. HIV Med 2014; 15:449–457. [DOI] [PubMed] [Google Scholar]
- 13.Taiwo B, Hunt PW, Gandhi RT, Ellingson A, McKenna M, Jacobson JM, et al. CD8+ T-cell activation in HIV-1-infected patients experiencing transient low-level viremia during antiretroviral therapy. J Acquir Immune Defic Syndr 2013; 63:101–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pernas B, Grandal M, Pertega S, Canizares A, Castro-Iglesias A, Mena A, et al. Any impact of blips and low-level viraemia episodes among HIV-infected patients with sustained virological suppression on ART?. J Antimicrob Chemother 2016; 71:1051–1055. [DOI] [PubMed] [Google Scholar]
- 15.Hermans LE, Moorhouse M, Carmona S, Grobbee DE, Hofstra LM, Richman DD, et al. Effect of HIV-1 low-level viraemia during antiretroviral therapy on treatment outcomes in WHO-guided South African treatment programmes: a multicentre cohort study. Lancet Infect Dis 2018; 18:188–197. [DOI] [PubMed] [Google Scholar]
- 16.Kehoe K, Boulle A, Tsondai PR, Euvrard J, Davies MA, Cornell M. Long-term virologic responses to antiretroviral therapy among HIV-positive patients entering adherence clubs in Khayelitsha, Cape Town, South Africa: a longitudinal analysis. J Int AIDS Soc 2020; 23:e25476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nettles RE, Kieffer TL, Kwon P, Monie D, Han Y, Parsons T, et al. Intermittent HIV-1 viremia (Blips) and drug resistance in patients receiving HAART. JAMA 2005; 293:817–829. [DOI] [PubMed] [Google Scholar]
- 18.Hey-Cunningham WJ, Murray JM, Natarajan V, Amin J, Moore CL, Emery S, et al. PINT study team. Early antiretroviral therapy with raltegravir generates sustained reductions in HIV reservoirs but not lower T-cell activation levels. AIDS 2015; 29:911–919. [DOI] [PubMed] [Google Scholar]
- 19.Farmer A, Wang X, Ganesan A, Deiss RG, Agan BK, O’Bryan TA, et al. Factors associated with HIV viral load ‘blips’ and the relationship between self-reported adherence and efavirenz blood levels on blip occurrence: a case-control study. AIDS Res Ther 2016; 13:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bachmann N, von Siebenthal C, Vongrad V, Turk T, Neumann K, Beerenwinkel N, et al. Swiss HIV Cohort Study. Determinants of HIV-1 reservoir size and long-term dynamics during suppressive ART. Nature communications 2019; 10:3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dahl V, Peterson J, Spudich S, Lee E, Shacklett BL, Price RW, Palmer S. Single-copy assay quantification of HIV-1 RNA in paired cerebrospinal fluid and plasma samples from elite controllers. AIDS 2013; 27:1145–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palmer S, Wiegand AP, Maldarelli F, Bazmi H, Mican JM, Polis M, et al. New real-time reverse transcriptase-initiated PCR assay with single-copy sensitivity for human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol 2003; 41:4531–4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tosiano MA, Jacobs JL, Shutt KA, Cyktor JC, Mellors JW, Simpler A. More sensitive single-copy HIV-1 RNA assay for quantification of persistent HIV-1 viremia in individuals on suppressive antiretroviral therapy. J Clin Microbiol 2019; 57:e01714–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Margot N, Koontz D, McCallister S, Mellors JW, Callebaut C. Measurement of plasma HIV-1 RNA below the limit of quantification (<20 copies/mL) of commercial assays with the integrase HIV RNA single-copy assay. J Clin Virol 2018; 108:50–52. [DOI] [PubMed] [Google Scholar]
- 25.Wang XQ, Palmer S. Single-molecule techniques to quantify and genetically characterise persistent HIV. Retrovirology 2018; 15:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jacobs JL, Tosiano MA, Koontz DL, Staines B, Worlock A, Harrington K, et al. Automated multireplicate quantification of persistent HIV-1 viremia in individuals on antiretroviral therapy. J Clin Microbiol 2020; 58:e01442–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bruner KM, Wang Z, Simonetti FR, Bender AM, Kwon KJ, Sengupta S, et al. A quantitative approach for measuring the reservoir of latent HIV-1 proviruses. Nature 2019; 566:120–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gaebler C, Lorenzi JCC, Oliveira TY, Nogueira L, Ramos V, Lu CL, et al. Combination of quadruplex qPCR and next-generation sequencing for qualitative and quantitative analysis of the HIV-1 latent reservoir. J Exp Med 2019; 216:2253–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Falcinelli SD, Ceriani C, Margolis DM, Archin NM. New frontiers in measuring and characterizing the HIV reservoir. Front Microbiol 2019; 10:2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pasternak AO, Berkhout B. What do we measure when we measure cell-associated HIV RNA. Retrovirology 2018; 15:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abdel-Mohsen M, Richman D, Siliciano RF, Nussenzweig MC, Howell BJ, Martinez-Picado J, et al. BEAT-HIV Delaney Collaboratory to Cure HIV-1 infection. Recommendations for measuring HIV reservoir size in cure-directed clinical trials. Nat Med 2020; 26:1339–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Massanella M, Yek C, Lada SM, Nakazawa M, Shefa N, Huang K, Richman DD. Improved assays to measure and characterize the inducible HIV reservoir. EBioMedicine 2018; 36:113–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Z, Gurule EE, Brennan TP, Gerold JM, Kwon KJ, Hosmane NN, et al. Expanded cellular clones carrying replication-competent HIV-1 persist, wax, and wane. Proc Natl Acad Sci U S A 2018; 115:E2575–E2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Procopio FA, Fromentin R, Kulpa DA, Brehm JH, Bebin AG, Strain MC, et al. A novel assay to measure the magnitude of the inducible viral reservoir in HIV-infected individuals. EBioMedicine 2015; 2:874–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li JZ, Etemad B, Ahmed H, Aga E, Bosch RJ, Mellors JW, et al. The size of the expressed HIV reservoir predicts timing of viral rebound after treatment interruption. AIDS 2016; 30:343–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pasternak AO, Grijsen ML, Wit FW, Bakker M, Jurriaans S, Prins JM, et al. Cell-associated HIV-1 RNA predicts viral rebound and disease progression after discontinuation of temporary early ART. JCI Insight 2020; 5:e134196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suzuki K, Hu JY, Ishida T, Lin Y, Minote S, Liu Z, et al. Development of an ultrasensitive HIV-1 DNA detection assay based on an automated (Code End-Point PCR system). J AIDS HIV Treat 2019; 1:68. [Google Scholar]
- 38.Chen CL, Chen CK, Ho CL, Chi WM, Yeh CH, Hu SP, et al. Clinical evaluation of IntelliPlex KRAS G12/13 mutation kit for detection of KRAS mutations in codon 12 and 13: a novel multiplex approach. Mol Diagn Ther 2019; 23:645–656. [DOI] [PubMed] [Google Scholar]
- 39.Kao JH, Lin CY, Chuang WL, Cheng YY, Hu JY, Liang WK, et al. Clinical evaluation of IntelliPlex(TM) HCV genotyping kit for hepatitis C virus genotyping. Diagn Microbiol Infect Dis 2019; 94:344–348. [DOI] [PubMed] [Google Scholar]
- 40.Koelsch KK, Rasmussen TA, Hey-Nguyen WJ, Pearson C, Xu Y, Bailey M, et al. Impact of allogeneic hematopoietic stem cell transplantation on the HIV reservoir and immune response in 3 HIV-infected individuals. J Acquir Immune Defic Syndr 2017; 75:328–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shafer RW. Rationale and uses of a public HIV drug-resistance database. J Infect Dis 2006; 194 Suppl 1:S51–S58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zaunders J, Munier CML, McGuire HM, Law H, Howe A, Xu Y, et al. Mapping the extent of heterogeneity of human CCR5+ CD4+ T cells in peripheral blood and lymph nodes. AIDS 2020; 34:833–848. [DOI] [PubMed] [Google Scholar]
- 43.Suzuki K, Craddock BP, Okamoto N, Kano T, Steigbigel RT. Detection of human immunodeficiency virus (HIV) by colorimetric assay for reverse transcriptase activity on magnetic beads. Biotechnol Appl Biochem 1993; 18 (Part 1):37–44. [PubMed] [Google Scholar]
- 44.Suzuki K, Craddock BP, Okamoto N, Kano T, Steigbigel RT. Poly A-linked colorimetric microtiter plate assay for HIV reverse transcriptase. J Virol Methods 1993; 44 (2–3):189–198. [DOI] [PubMed] [Google Scholar]
- 45.Murray JM, Zaunders JJ, McBride KL, Xu Y, Bailey M, Suzuki K, et al. PINT Study Team. HIV DNA subspecies persist in both activated and resting memory CD4+ T cells during antiretroviral therapy. J Virol 2014; 88:3516–3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murray JM. Latent HIV dynamics and implications for sustained viral suppression in the absence of antiretroviral therapy. J Virus Erad 2018; 4:91–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maria CV, Kathleen LC. The impact of cellular proliferation on the HIV-1 reservoir. Viruses 2019; 12:1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pinzone MR, VanBelzen DJ, Weissman S, Bertuccio MP, Cannon L, Venanzi-Rullo E, et al. Longitudinal HIV sequencing reveals reservoir expression leading to decay which is obscured by clonal expansion. Nat Commun 2019; 10:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zaunders JJ, Munier ML, Seddiki N, Pett S, Ip S, Bailey M, et al. High levels of human antigen-specific CD4+ T cells in peripheral blood revealed by stimulated coexpression of CD25 and CD134 (OX40). J Immunol 2009; 183:2827–2836. [DOI] [PubMed] [Google Scholar]
- 50.Lesbats P, Engelman AN, Cherepanov P. Retroviral DNA integration. Chem Rev 2016; 116:12730–12757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Richetta C, Thierry S, Thierry E, Lesbats P, Lapaillerie D, Munir S, et al. Two-long terminal repeat (LTR) DNA circles are a substrate for HIV-1 integrase. J Biol Chem 2019; 294:8286–8295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Suzuki K, Shijuuku T, Fukamachi T, Zaunders J, Guillemin G, Cooper D, et al. Prolonged transcriptional silencing and CpG methylation induced by siRNAs targeted to the HIV-1 promoter region. J RNAi Gene Silencing 2005; 1:66–78. [PMC free article] [PubMed] [Google Scholar]
- 53.Suzuki K, Juelich T, Lim H, Ishida T, Watanebe T, Cooper D, et al. Closed chromatin architecture is induced by an RNA duplex targeting the HIV-1 promoter region. J Biol Chem 2008; 283:23353–23363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Suzuki K, Ishida T, Yamagishi M, Ahlenstiel C, Swaminathan S, Marks K, et al. Transcriptional gene silencing of HIV-1 through promoter targeted RNA is highly specific. RNA Biol 2011; 8:1035–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Suzuki K, Hattori S, Marks K, Ahlenstiel C, Maeda Y, Ishida T, et al. Promoter targeting shRNA suppresses HIV-1 infection in vivo through transcriptional gene silencing. Mol Ther Nucleic Acids 2013; 2:e137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Higaki K, Hirao M, Kawana-Tachikawa A, Iriguchi S, Kumagai A, Ueda N, et al. Generation of HIV-resistant macrophages from IPSCs by using transcriptional gene silencing and promoter-targeted RNA. Mol Ther Nucleic Acids 2018; 12:793–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pasternak AO, Lukashov VV, Berkhout B. Cell-associated HIV RNA: a dynamic biomarker of viral persistence. Retrovirology 2013; 10:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Eriksson S, Graf EH, Dahl V, Strain MC, Yukl SA, Lysenko ES, et al. Comparative analysis of measures of viral reservoirs in HIV-1 eradication studies. PLoS Pathog 2013; 9:e1003174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kiselinova M, Pasternak AO, De Spiegelaere W, Vogelaers D, Berkhout B, Vandekerckhove L. Comparison of droplet digital PCR and seminested real-time PCR for quantification of cell-associated HIV-1 RNA. PLoS One 2014; 9:e85999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Siliciano RF, Greene WC. HIV latency. Cold Spring Harb Perspect Med 2011; 1:a007096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lorenzi JC, Cohen YZ, Cohn LB, Kreider EF, Barton JP, Learn GH, et al. Paired quantitative and qualitative assessment of the replication-competent HIV-1 reservoir and comparison with integrated proviral DNA. Proc Natl Acad Sci U S A 2016; 113:E7908–E7916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hiener B, Horsburgh BA, Eden JS, Barton K, Schlub TE, Lee E, et al. Identification of genetically intact HIV-1 proviruses in specific CD4(+) T cells from effectively treated participants. Cell Rep 2017; 21:813–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Horsburgh BA, Palmer S. For viral reservoir studies, timing matters. Trends Microbiol 2019; 27:809–810. [DOI] [PubMed] [Google Scholar]
- 64.Lee E, Bacchetti P, Milush J, Shao W, Boritz E, Douek D, et al. Memory CD4 + T cells expressing HLA-DR contribute to HIV persistence during prolonged antiretroviral therapy. Front Microbiol 2019; 10:2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.De Scheerder MA, Van Hecke C, Zetterberg H, Fuchs D, De Langhe N, Rutsaert S, et al. Evaluating predictive markers for viral rebound and safety assessment in blood and lumbar fluid during HIV-1 treatment interruption. J Antimicrob Chemother 2020; 75:1311–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Garcia M, Morcilla V, Navarrete-Munoz MA, Fisher K, Cabello A, Lopez-Bernaldo JC, et al. HIV-DNA content in pTfh cells is associated with residual viremia in elite controllers. AIDS 2020; 35:393–398. [DOI] [PubMed] [Google Scholar]
- 67.Horsburgh BA, Lee E, Hiener B, Eden JS, Schlub TE, von Stockenstrom S, et al. High levels of genetically intact HIV in HLA-DR+ memory T cells indicates their value for reservoir studies. AIDS 2020; 34:659–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lee E, von Stockenstrom S, Morcilla V, Odevall L, Hiener B, Shao W, et al. Impact of antiretroviral therapy duration on HIV-1 infection of T cells within anatomic sites. J Virol 2020; 94: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Neidleman J, Luo X, Frouard J, Xie G, Hsiao F, Ma T, et al. Phenotypic analysis of the unstimulated in vivo HIV CD4 T cell reservoir. Elife 2020; 9:e60933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Palmer S, Dijkstra M, Ket JC, Wahome EW, Walimbwa J, Gichuru E, et al. Acute and early HIV infection screening among men who have sex with men, a systematic review and meta-analysis. J Int AIDS Soc 2020; 23 Suppl 6:e25590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Skanland SS, Moltu K, Berge T, Aandahl EM, Tasken K. T-cell co-stimulation through the CD2 and CD28 co-receptors induces distinct signalling responses. Biochem J 2014; 460:399–410. [DOI] [PubMed] [Google Scholar]
- 72.Green L. Development of an anti-CD2/CD3/CD28 bead-based T-cell proliferation assay. Biosci Horizons 2014; 7:hzu012. [Google Scholar]
- 73.Chaillon A, Gianella S, Dellicour S, Rawlings SA, Schlub TE, De Oliveira MF, et al. HIV persists throughout deep tissues with repopulation from multiple anatomical sources. J Clin Invest 2020; 130:1699–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.