Abstract
Developmental dyslexia (DD) is a learning disorder affecting the ability to read, with a heritability of 40–60%. A notable part of this heritability remains unexplained, and large genetic studies are warranted to identify new susceptibility genes and clarify the genetic bases of dyslexia. We carried out a genome-wide association study (GWAS) on 2274 dyslexia cases and 6272 controls, testing associations at the single variant, gene, and pathway level, and estimating heritability using single-nucleotide polymorphism (SNP) data. We also calculated polygenic scores (PGSs) based on large-scale GWAS data for different neuropsychiatric disorders and cortical brain measures, educational attainment, and fluid intelligence, testing them for association with dyslexia status in our sample. We observed statistically significant (p < 2.8 × 10−6) enrichment of associations at the gene level, for LOC388780 (20p13; uncharacterized gene), and for VEPH1 (3q25), a gene implicated in brain development. We estimated an SNP-based heritability of 20–25% for DD, and observed significant associations of dyslexia risk with PGSs for attention deficit hyperactivity disorder (at pT = 0.05 in the training GWAS: OR = 1.23[1.16; 1.30] per standard deviation increase; p = 8 × 10−13), bipolar disorder (1.53[1.44; 1.63]; p = 1 × 10−43), schizophrenia (1.36[1.28; 1.45]; p = 4 × 10−22), psychiatric cross-disorder susceptibility (1.23[1.16; 1.30]; p = 3 × 10−12), cortical thickness of the transverse temporal gyrus (0.90[0.86; 0.96]; p = 5 × 10−4), educational attainment (0.86[0.82; 0.91]; p = 2 × 10−7), and intelligence (0.72[0.68; 0.76]; p = 9 × 10−29). This study suggests an important contribution of common genetic variants to dyslexia risk, and novel genomic overlaps with psychiatric conditions like bipolar disorder, schizophrenia, and cross-disorder susceptibility. Moreover, it revealed the presence of shared genetic foundations with a neural correlate previously implicated in dyslexia by neuroimaging evidence.
Subject terms: Genetics, Neuroscience, Psychiatric disorders
Introduction
Developmental dyslexia (DD) is a specific learning disorder affecting the ability to read that is not better accounted for by intellectual disabilities, uncorrected visual or auditory acuity, other mental or neurological disorders, or inadequate educational instruction [1]. People with dyslexia show difficulties in accurate and/or fluent word recognition, decoding, spelling, and/or reading comprehension [2]. The prevalence of DD is reported to be around 5–10% among school-aged children, depending on the criteria used for diagnosis [3]. DD tends to recur in families [4, 5] and most twin studies have reported a heritability (h2) between 40 and 60% [2, 6]. A similar range of heritability has been reported for several cognitive skills representing/underlying reading ability, such as word reading, spelling, and phoneme awareness (h2 ~40–70%) [7–9]. Of note, a large proportion of this heritability remains unexplained, and DD shows a complex architecture, with multiple genetic and environmental factors playing a role in its aetiology [10].
Linkage and candidate gene association studies have identified a small number of candidate susceptibility genes, most of which have been associated not only with dyslexia, but also with continuous interindividual variation in relevant cognitive skills like word reading, spelling, and others (as reviewed in [11–13]). The most robust candidate genes identified so far include DYX1C1 (15q21) [14], DCDC2 and KIAA0319 (6p22.3) [15–18], GCFC2 and MRPL19 (2p12) [19], and ROBO1 (3p12.3-p12.3) [20–22]. DD and reading-related cognitive traits have also been investigated via genome-wide association studies (GWAS), which involve analyses of many single-nucleotide polymorphisms (SNPs) spread across the genome. A few such studies have been reported, using either a case-control design [23–25] or a continuous trait analysis approach [26–30]. However, only two of these studies identified associations that met criteria for genome-wide significance [27, 28]. The first was a GWAS of multiple cognitive skills related to reading ability, which revealed a genome-wide significant association at rs17663182 (MIR924HG; 18q12.2) with rapid automatized naming (RAN), in nine cohorts of reading-impaired and typically developing subjects of European ancestry (maximum N = 3468) [28]. More recently, in a north-American cohort of non-European ancestry (N = 1331), Truong et al. [27] identified a genome-wide significant multivariate association of rs1555839 (10q23.31; upstream from the RPL7P34 gene) with RAN and rapid alternating stimulus, and replicated the association with RAN in an independent cohort of European ancestry [27].
Here, we carried out a case-control GWAS meta-analysis involving 2274 dyslexia cases and 6272 controls from nine different countries that partly overlap with those from the prior Gialluisi et al. study of continuous traits (≤2500 overlapping samples) [28]. We performed association testing at the single variant, gene, and pathway level, and estimated SNP-based heritability. Moreover, we analyzed associations of polygenic scores (PGS) derived from large-scale GWAS data from other related neuropsychiatric disorders, as well as intelligence, educational attainment, and cortical brain measures.
Subjects and methods
Datasets
The datasets involved in the present study were collected in nine different populations of European ancestry, with six different languages (see Table 1). Subsets have already been tested for association with continuous reading-related traits [28]. Ethical approval was obtained for each cohort at the local level, and written informed consent was obtained for all the participants or their parents.
Table 1.
General descriptive statistics and recruitment criteria of the datasets analyzed in the study.
| Dataset | Recruitment | Language | Age range, years (mean, SD)b | IQ inclusion criteria | N after QC (cases:controls) |
|---|---|---|---|---|---|
| AGSa | Unrelated cases and controls | German | 8–19 (10.7, 2.4) | Age-appropriate WISC Block Design score [99, 100] ≥7 Age-appropriate WISC Similarities score [99, 100] ≥6 | 1454 (1047:407) |
| Finland | Finnish | 324 (161:163) | |||
| France | French | 163 (106:57) | |||
| Hungary | Hungarian | 241 (90:151) | |||
| Netherlands | Dutch | 284 (136:148) | |||
| ENall1b | Related cases from Colorado + related cases from UK (Oxford) + unrelated unscreened controls from UK general population (WTCC2_1958) | English | 8–19 (11.5, 2.7) | Colorado: average score of age-adjusted WISC-R/WAIS-R verbal IQ and performance IQ, multiple subtests) [101] ≥80 [28] UK: average of age-adjusted standardized BAS/WAIS-R similarities subtest and BAS matrices subtest score [102, 103] ≥80 [28] | 3313 (554:2759) |
| ENall2b | Unrelated cases from UK (Cardiff) + unrelated unscreened controls from UK general population (WTCC2_NBS) | English | 5–31 (11.8, 3.6) | Average of age-adjusted standardized BAS/WAIS-R similarities subtest and BAS matrices subtest score [102, 103] ≥80 [28] | 2767 (180:2587) |
IQ intelligence quotient, DD developmental dyslexia, WISC Wechsler Intelligence Scale for Children, WAIS Wechsler Adult Intelligence Scale—Revised, BAS British Ability Scale.
aAustria–Germany–Switzerland.
bIn the English-speaking datasets (i.e., ENall1 and ENall2) age and IQ information refers only to cases, since no such data were available from the general population control cohorts.
Unrelated DD cases and controls with IQ in the normal range were recruited in Austria (N = 374), Finland (N = 336), France (N = 165), Germany (N = 1454), Hungary (N = 243), The Netherlands (N = 311), and Switzerland (N = 67) (see Table 1). DD cases were defined as participants showing low performance on tests of word reading (standardized score ≤−1.25), with the exception of 148 German cases, which were defined based on a ≥1.5 standard deviation discrepancy between the observed and expected spelling score based on their IQ (see [31, 32] and Supplementary methods). Controls were defined as individuals with standardized word reading scores >−0.85 [33, 34].
Samples from Austria, Germany, and Switzerland were merged together into a single dataset (hereafter called AGS), since they shared language and genetic ancestry [28]. Two additional datasets were included in the study, made up of native English speakers. One of them consisted of DD cases selected from two sibling-based cohorts, namely the Colorado Reading Disability Cohort [26, 35] and an independent cohort from Oxford, UK [28, 36]. These cases were merged to form a single case-control dataset with unscreened controls from the Wellcome Trust Case Control Consortium 2 (WTCCC2) 1958 British birth cohort (WTCCC2_1958), a sample of sequential live births in the UK during 1 week in 1958 [37]. The other English-speaking dataset consisted of unrelated DD cases recruited in Cardiff, UK and the WTCCC2 National Blood Service (WTCCC2_NBS) cohort, a collection of subjects who have donated blood to the UK blood service. These datasets, hereafter called ENall1 (N = 3531) and ENall2 (N = 2947), met the same word reading-based inclusion criteria as above for cases, while controls were unscreened, as in other prominent studies [38, 39].
Genotype quality control (QC) and imputation
Genotyping array platforms used for the different datasets are reported in Table S1a. These included Illumina HumanHap 300k, 550k, 660k, OmniExpress Human CoreExome and BeadChips, and Illumina 1.2 M chips. Genotype QC was carried out, as previously described [28], in PLINK v1.90b3s [40] and QCTOOL v1.4 (see URLs). Briefly, SNPs were excluded if they showed a variant call rate <98%, a minor allele frequency (MAF) <5%, or a Hardy–Weinberg equilibrium (HWE) exact test p value < 10−6. Samples showing a genotyping rate <98%, mismatches between genetic and pedigree-based sex, cryptic relatedness (in datasets of unrelated subjects), or identity-by-descent not corresponding to the available pedigree information (in datasets including related cases) were also discarded. Similarly, we discarded genetic ancestry outliers detected in a multidimensional scaling (MDS) analysis of pairwise genetic distance and samples with extreme genome-wide heterozygosity values (see Table S1b).
For imputation, genotypes of autosomal SNPs were aligned to the 1000 Genomes phase I v3 reference panel (ALL populations, June 2014 release) [41] and pre-phased using SHAPEIT v2 (r837) [42]. Imputation was then performed using IMPUTE2 v2.3.2 [43] in 5 Mb chunks with 500 kb buffers, filtering out variants that were monomorphic in the 1000 Genomes EUR (European) samples. Chunks with <51 genotyped variants or concordance rates <92% were fused with neighbouring chunks and re-imputed. Finally, imputed variants (genotype probabilities) were filtered for IMPUTE2 INFO metric ≥0.8, as well as MAF and HWE thresholds as above. We re-evaluated genetic ancestry and genome-wide heterozygosity outliers after imputation and observed substantial concordance with pre-imputation QC. After QC, 2274 dyslexia cases and 6272 controls were left for analysis (see Tables S1c, d for a power computation).
Genetic association test and meta-analysis
After genotype QC and imputation, we tested autosomal variant allelic dosages for association with case-control status within each dataset. In all the datasets except ENall1, we ran association tests through logistic regression in PLINK, using the first ten genetic ancestry (MDS) components as covariates. To account for the genetic relationship among related subjects in ENall1, we modelled a generalized linear mixed-effects model association test through FastLMM v2.07 [44], using a genetic relationship matrix as a random effect, while disabling normalization to unit variance for tested SNPs. Then we combined the results of the association tests in the different datasets through a fixed-effects sample size-based meta-analysis in METAL v25-03-2011 (“Stouffer” method) [45]. This was done in order to overcome the heterogeneity of scales of the association tests used in the different datasets. The genome-wide significance threshold was set to α = 5 × 10−8. To obtain an estimate of the odds ratio (OR) for the top association identified, we performed a Wald association test in the ENall1 dataset through a logistic mixed model approach in GMMAT [46], which was not possible to perform at the genome-wide level due to the high computational load implied. We then meta-analyzed the resulting association statistics across datasets, through a fixed-effects inverse variance-weighted method in METAL [45].
Gene- and pathway-based enrichment tests
We performed a gene-based association analysis on the results of the GWAS meta-analysis in MAGMA v1.06 [47]. First, we assigned genetic variants to protein-coding genes based on their position according to the NCBI 37.3 (hg19) build, extending the region of annotation to 10 kb from the 3′-/5′-UTR (untranslated region). In total, 18,013 genes (out of 19,427 genes available) included at least one variant that passed internal QC and were thus tested for enrichment of single-variant associations, using default settings. For this analysis, we set a genome-wide significance threshold α = 2.8 × 10−6, correcting for 18,013 genes tested.
We used the results of the gene-based association analysis to carry out a pathway-based enrichment test for associations with DD, through a competitive gene-set analysis in MAGMA v1.06. We tested for enrichment 1329 canonical pathways (i.e., classical representations of biological processes compiled by domain experts) from the Molecular Signatures Database website (MSigDB v5.2, collection C2, subcollection CP; see URLs). To correct enrichment statistics for testing of multiple pathways, we used an adaptive permutation procedure with default settings (up to a maximum of 10,000 permutations). Hence, in this analysis we set the significance threshold to α = 0.05.
Estimation of heritability
We used the summary statistics from the DD case-control GWAS to compute SNP-based heritability of the disorder, through LD score regression [48, 49]. For this analysis, we used only common SNPs tested in the GWAS and present in the HapMap 3 reference panel [50], excluding the MHC region, since these variants show a good imputation quality (r2 > 0.9) in most studies. All the analyses presented below were performed on these variants (1,025,494 SNPs), using LD information based on the 1000 G phase 1 v3 EUR panel (see URLs).
We first computed the proportion of genetic variance explained by all SNPs mentioned above on the observed scale, and then repeated the analysis using a liability threshold model, i.e., assuming that the binary trait that we use is determined by an unobserved normally distributed liability threshold [48, 49]. This analysis requires specification of the proportion of cases in the GWAS (27%), and the estimated prevalence of the disorder in the reference population, which has been reported to be 5–10% among school-aged children [3]. Hence, we carried out the analysis using the limits of this prevalence range, namely 0.05 and 0.10, respectively.
To extrapolate biological information from our GWAS summary statistics, we computed partitioned heritability for 53 overlapping functional annotation categories identified in the genome [51], irrespective of the cell types analyzed (baseline model). These annotations include DNase I hypersensitivity sites, coding regions, untranslated regions, enhancers, promoters and several histone marks as defined by different public resources (see “Results” section and [51] for a complete list). Similarly, we carried out a stratified LD score regression using only central nervous system (CNS) cell-specific annotations of four histone marks—H3K4me1, H3K4me3, H3K9ac, and H3K27ac—to identify a specific enrichment of functional elements associated with transcriptional activity in these cells. We performed this analysis both for all the CNS cells pooled together and singularly for each cell type available in brain tissues, while correcting for the contribution of all functional annotation categories previously tested in the baseline model, as suggested by the developer [51]. Thereby, we could identify the contribution of common variants annotated to histone marks which are specifically enriched in nervous cells. Finally, we computed partitioned heritability for diverse sets of genes whose expression is specifically enriched in 13 different brain regions, based on RNA-seq data from the Genotype-Tissue Expression portal (GTEx v6) [52, 53]. The brain regions available included amygdala, anterior cingulate cortex, caudate nucleus, cerebellar hemispheres, cerebellum, cortex, frontal cortex, hippocampus, hypothalamus, nucleus accumbens, putamen, spinal cord, and substantia nigra.
Polygenic score (PGS) analyses
Genetic liability to neuropsychiatric disorders, intelligence and education
We investigated potential genetic links between dyslexia and related and/or comorbid neuropsychiatric disorders, including attention deficit hyperactivity disorder (ADHD) [54–56], autism spectrum disorder (ASD) [57], major depressive disorder (MDD) [58], bipolar disorder (BD) [59], and schizophrenia (SCZ) [60], as well as with genetic liability shared across different neuropsychiatric disorders, including ADHD, ASD, BD, MDD, SCZ, anorexia nervosa, obsessive-compulsive disorder and Tourette syndrome [61]. Moreover, we tested association with fluid intelligence [62] and educational attainment (years of education completed, EduYears) [63], which are phenotypically correlated with reading ability [64, 65]. To this end, we performed a PGS analysis in our sample using summary statistics available from previous independent GWAS studies of the other traits of interest (hereafter called training GWAS) [61–63, 66–70]. PGSs were computed with PRSice-2 v2.2.11 [71], using only summary statistics based on samples of European ancestry in the training GWAS and quality controlled variants in a random extraction of one individual per family from our dyslexia (target) GWAS (MAF ≥ 5%; HWE p ≥ 10−6; variant call rate ≥95%; N = 8456). We further pruned SNPs through LD-clumping (pairwise r2 < 0.05 within sliding 300 kb windows) and removed those variants with discordant coordinates/alleles between the training and the target GWAS. We then computed average (default) PGS using only variants with association p value < 0.05 in the training GWAS (as in [28, 72, 73]), since this represents a reasonable trade-off between goodness-of-fit of the PGS and the risk of introducing noise in the model by including genetic variants meeting more lenient association thresholds. We then built generalized linear models (glm) of dyslexia vs PGS adjusted for sex and genetic ancestry (10 MDS components) in the same set of unrelated subjects used above (2184 cases and 6272 controls). To check for robustness of our findings, we repeated the analysis at different association significance thresholds in each training GWAS (with p < 5 × 10−8, 1 × 10−5, 0.001, 0.05, 0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, 0.9 and 1.0). A Bonferroni-corrected significance threshold was set to α = 4.5 × 10−4 for this analysis, conservatively correcting for eight (six binary neuropsychiatric and two continuous) training traits, and 14 significance thresholds tested.
Polygenic scores of brain cortical measures
We carried out an exploratory analysis to test associations with PGSs influencing the surface area (SA) and thickness (T) of 34 brain cortical regions (June 2020 release), recently analyzed in a GWAS involving 33,992 participants of European ancestry [74]. We tested PGSs (at p < 0.05 in the training GWAS) of both SA and T of all the cortical regions adjusted for global measures (total SA and average T, respectively), both separately and jointly in a multivariable setting. This choice was motivated by the fact that different structural alterations have been described in dyslexic subjects [75] and a complex brain network of different structures is thought to underlie dyslexia phenotypes and related skills [76]. To insure against potential overfitting bias in a conventional ordinary least squares regression with a high number of predictors, we applied two alternative multivariable models. First, a stepwise regression through the stepaic() function of the MASS package, which retains only variables associated with a decrease in the Akaike information criterion, representing a trade-off between goodness-of-fit and parsimony of the model. Then, an elastic net regression, using the glmnet and caret packages, as in [77]. To this end, we divided our dataset into a random training and test set (80:20 ratio), then trained the elastic net and carried out hyperparameter (α and λ) tuning in the training set, with tenfold cross-validation. Finally, we tested the performance of the optimized model, assessing classification accuracy in the independent test set (N = 1690). All the models involving cortical PGSs were adjusted for MDS components and sex, as explained above. For this analysis, we considered associations as statistically robust only if they showed significant and similar effect sizes across the different models tested (using a significance threshold α = 8.3 × 10−4, correcting for 60 independent cortical measures, as in [74]).
Results
Single-variant genome-wide associations
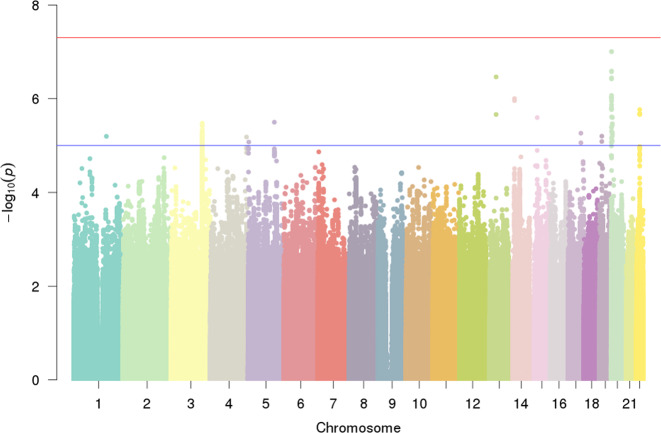
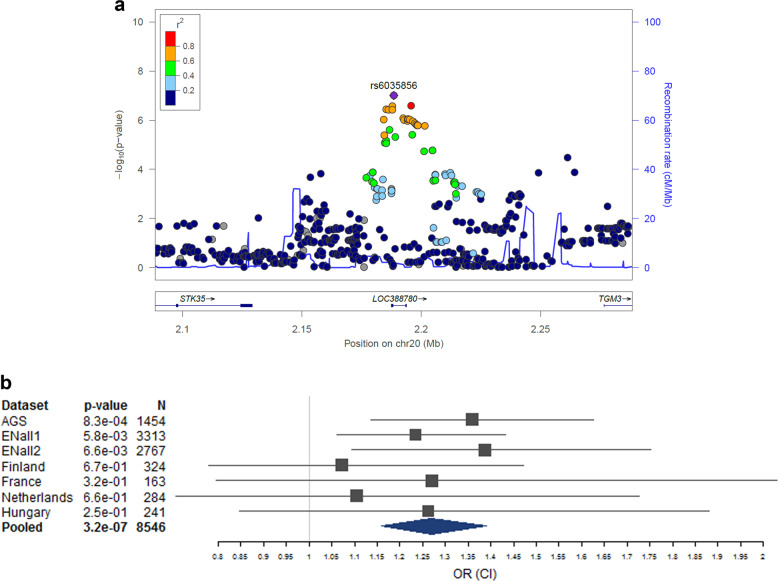
No single-variant association with DD reached genome-wide significance (Figs. 1 and S1). The strongest single-variant associations detected in the GWAS are reported in Table 2 (p < 5 × 10−7) and, more extensively, in Table S2a (p < 10−5). The top hit was detected at rs6035856 (G/T, MAF = 0.45; p = 9.9 × 10−8), an intronic variant located within the gene LOC388780 (chr20p13; Fig. 2a). Following logistic mixed modelling and inverse variance-based meta-analysis of the rs6035856 association, we computed an OR [confidence interval] of 1.27[1.16; 1.39] for the major allele G (p = 3.2 × 10−7). In all datasets, the major allele G was associated with increased DD risk (Fig. 2b and Table S2b). Although this SNP was not directly genotyped, it showed high quality imputation statistics across datasets (INFO metric in the range 0.89–0.95). Other SNPs in the vicinity of rs6035856 were also associated with DD (see Table 2) and were all in moderate/high LD with the top hit (r2 > 0.6; see Fig. 2a).
Fig. 1. Manhattan plot of the GWAS pooled analysis.
The blue and red line represent the genome-wide (α = 5 × 10−8) and suggestive significance (α = 1 × 10−5) threshold.
Table 2.
Most significant single-variant associations (p < 5 × 10−7) detected in the present GWAS.
| SNP | REF allele | ALT allele | REF allele frequencya,b | Z-scoreb | p | Directionb | I2 (p)c | Position (chr:bp) | Position within gene | Gene symbol | Distance from gene (bp) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs6035856 | G | T | 0.55 | 5.329 | 9.88 × 10−8 | +++++++ | 0 (0.91) | 20:2188542 | intron | LOC388780 | – |
| rs6035857 | A | C | 0.55 | 5.329 | 9.88 × 10−8 | +++++++ | 0 (0.91) | 20:2188544 | intron | LOC388780 | – |
| rs6082416 | G | C | 0.55 | 5.153 | 2.56 × 10−7 | +++++++ | 0 (0.85) | 20:2195832 | downstream | LOC388780 | 2035 |
| rs6137325 | G | T | 0.59 | 5.147 | 2.65 × 10−7 | +++++++ | 0 (0.91) | 20:2187943 | exon | LOC388780 | – |
| rs2094530 | C | T | 0.22 | 5.098 | 3.43 × 10−7 | ?--???- | 0 (0.82) | 13:51564457 | downstream | GUCY1B2 | 4190 |
| rs6047381 | C | T | 0.58 | 5.089 | 3.59 × 10−7 | +++++++ | 0 (0.84) | 20:2185357 | upstream | LOC388780 | 2217 |
| rs6137326 | C | T | 0.59 | 5.083 | 3.72 × 10−7 | +++++++ | 0 (0.91) | 20:2187944 | exon | LOC388780 | – |
| rs6132418 | A | T | 0.58 | 5.08 | 3.78 × 10−7 | +++++++ | 0 (0.85) | 20:2186281 | upstream | LOC388780 | 1293 |
aAverage allele frequency computed for reference (REF) alleles over all the datasets analyzed.
bAllele frequencies, meta-analysis Z-scores, and directions of effect refer to REF alleles. Directions of effect are reported for each single dataset analyzed, in the following order: AGS, Finland, France, Holland, Hungary, ENall1, ENall2. “+” means that the major allele is the risk allele, while “?” indicates that the variant was not tested in the corresponding dataset.
cI2 test for heterogeneity of genetic effect across datasets (the closer to “0”, the more homogenous is the genetic effect).
Fig. 2. Details of the genome-wide top hit rs6035856.
a Local association and b forest plot of the genome-wide top variant (rs6035856). The forest plot shows the odds ratio (OR) and 95% confidence intervals (CI) on the x-axis, by dataset and for the pooled analysis. Detailed OR statistics can be found in Table S2b. Note to forest plot: the sibling-based dataset ENall1 was analyzed genome-wide through linear mixed modelling (in FastLMM) for computational reasons, while its OR, as shown here, was computed via a Wald test in a logistic mixed model (GMMAT), to make it comparable to the other ORs produced through logistic regression (PLINK). Hence, the result of the pooled analysis—which here was performed through the inverse variance-based method—is slightly discrepant from the original genome-wide analysis (see Table 2).
Gene- and pathway-based enrichment analyses
Gene-level analysis of genome-wide single-variant association signals with DD revealed two significant enrichments, after correcting for the 18,013 genes tested across the genome (p < 2.8 × 10−6; Table S2c). These enrichments were observed for the gene VEPH1 (ventricular zone expressed PH domain-containing 1; 3q25; Z = 5.63; permutation-based p = 8 × 10−8) and for the gene LOC388780, where top GWAS variant mapped to (Z = 5.26; p = 1.7 × 10−7). However, the analysis of 1329 canonical pathways from the MSigDB website did not reveal any significant enrichment (Table S2d).
SNP-based heritability
We computed the SNP-based heritability (h2SNP) of DD through LD score regression, using the summary statistics of HapMap 3 SNPs analysed in the GWAS. This analysis yielded an estimate of h2SNP (SE) = 0.19(0.06) on the observed scale, while, on the liability scale, we observed h2SNP (SE) of 0.20(0.06) (assuming a dyslexia prevalence of 0.05) and of 0.25(0.08) (for prevalence 0.1; see Table S3a).
We next computed partitioned heritability for different functional categories in the genome, through stratified LD score regression. The analysis of 53 overlapping functional annotation categories in the baseline model (i.e., including functional annotations irrespective of the cell type) revealed no statistically significant enrichments of heritability for such general annotation classes (see Table S3b). Similarly, the stratified LD score regression applied to annotations specific to CNS cell types detected no significant contribution to SNP-based heritability of the four histone marks tested (H3K4me1, H3K4me3, H3K9ac, and H3K27ac; Table S3c). When we analysed partitioned heritability by sets of specifically overexpressed genes in 13 different brain regions available in the GTEx database (see “Methods” section), we observed no significant contributions to h2SNP surviving correction for multiple testing (Table S3d).
Polygenic scores and dyslexia risk
We report in Table 3 the results of the main PGS analysis on neuropsychiatric disorders, intelligence and educational attainment, including only variants with association p < 0.05 in the training GWAS (i.e. at pT = 0.05), while the results at the different association significance (pT) thresholds tested are reported in Table S4a–h. At pT = 0.05, glm logistic regressions revealed that standardized PGS of EDUyears and fluid intelligence were significantly associated with dyslexia risk in our sample, surviving correction for multiple testing, with OR = 0.86[0.82; 0.91] (R2 = 0.39%; p = 1.95 × 10−7) and 0.72[0.68; 0.76] (1.79%; 9.40 × 10−29), respectively. Also, we observed significant associations with dyslexia risk for three of the neuropsychiatric disorders analyzed: ADHD (1.23[1.16; 1.3]; 0.73%; 7.66 × 10−13), BD (1.53[1.44; 1.63]; 2.80%; 1.33 × 10−43) and SCZ (1.36[1.28; 1.45]; 1.35%; 3.65 × 10−22). Similarly, we identified a significant association with common genetic liability shared across different psychiatric disorders (1.23 [1.16; 1.30]; 0.69%; 3.12 × 10−12). These associations were concordant across all tested significance thresholds, and p values decreased when more inclusive criteria were used (i.e., for pT ranging between 0.1 and 1, see Table S4a–h).
Table 3.
Results of the polygenic score (PGS) analysis for the different training traits/disorders tested.
| Trait/disorder PGS | OR [95% CI] | R2 (%) | p | Training GWAS (Reference) | Training GWAS N (cases/controls) |
|---|---|---|---|---|---|
| ADHD | 1.23 [1.16; 1.3] | 0.73 | 7.66 × 10−13 | [67] | 53,293 (19,099/34,194) |
| ASD | 1.01 [0.96; 1.07] | <0.01 | 0.69 | [68] | 46,351 (18,382/27,969) |
| BD | 1.53 [1.44; 1.63] | 2.80 | 1.33 × 10−43 | [66] | 51,710 (20,352/31,358) |
| MDD | 1.01 [0.95; 1.06] | <0.01 | 0.83 | [70] | 500,199 (170,756/329,443) |
| SCZ | 1.36 [1.28; 1.45] | 1.35 | 3.65 × 10−22 | [69] | 77,096 (33,640/43,456) |
| Cross-Disorder | 1.23 [1.16; 1.30] | 0.69 | 3.12 × 10−12 | [61] | 438,997 (162,151/276,846) |
| EduYears | 0.86 [0.82; 0.91] | 0.39 | 1.95 × 10−7 | [63] | 766,345 |
| Intelligence | 0.72 [0.68; 0.76] | 1.79 | 9.40 × 10−29 | [62] | 269,867 |
We report odds ratios (OR) for dyslexia with 95% confidence intervals (95% CI) per standardized PGS in our dataset, along with relevant R2 and p values, at pT = 0.05 in the training GWAS. Full results for the different pT thresholds tested are reported in Table S4a–h. Statistically significant associations (p < 4.5 × 10−4) are highlighted in bold.
ADHD attention deficit hyperactivity disorder, ASD autism spectrum disorder, MDD major depressive disorder, BD bipolar disorder, SCZ schizophrenia, CROSS-DISORDER shared genetic basis of ADHD, ASD, BD, MDD, SCZ, anorexia nervosa, obsessive-compulsive disorder and Tourette syndrome based on the GWAS meta-analysis by the Cross-Disorder Group of the Psychiatric Genomics Consortium [61], EduYears years of education completed.
The analysis of PGS for SA and T of 34 brain cortical regions revealed an association of the transverse temporal gyrus T with prevalent DD risk, which remained significant after correction for multiple testing (OR = 0.90[0.86; 0.96]; p = 4.53 × 10−4; Table S4i). This association was confirmed in a multivariable setting, both in stepwise (0.90[0.85; 0.95]; p = 2.45 × 10−4; Table S4j) and in elastic net regression (OR = 0.92; Table S4k). However, the variance explained by this PGS was low (0.17% in univariate regression) and all the cortical PGS selected in elastic net regression jointly conferred only a modest gain in dyslexic classification accuracy, compared to the null model including only covariates (0.4%).
Discussion
To the best of our knowledge, the present work reports the largest case-control GWAS study conducted on dyslexia to date, involving 2274 DD cases and 6272 controls from nine different populations of European ancestry, speaking six different languages.
We identified a suggestive association at rs6035856 (p~10−8), an intronic variant located within the gene LOC388780 (20p13), ~400 bp downstream of exon 1. This small (~6 kb) gene encodes a non-coding RNA which has not been functionally characterized yet, but is expressed in different organs, including the CNS [52]. Gene-based association testing supported the implication of LOC388780 in DD genetic risk, showing a genome-wide significant enrichment of associations for this gene. Based on the Roadmap Epigenome 25-state model using 12 imputed marks, this region is classified as a Promoter Upstream Transcription Start Site (2_PromU chromatin state) in several brain cell types, including those from middle hippocampus, anterior caudate, cingulate gyrus, inferior frontal lobe, and dorsolateral prefrontal cortex [78], suggesting potential roles in transcriptional regulation.
Gene-based analysis also detected significant evidence of enrichment for the gene VEPH1 (ventricular zone expressed PH domain-containing 1; 3q25), coding for a partly characterized protein which promotes brain development [79], probably through regulation of the TGF-β signalling pathway [80]. However, we did not observe any significant enrichment of associations for TGF-β-related pathways.
The analysis of SNP-based heritability indicated that 20–25% of the total variance in DD could be explained by common variants in our dataset. This estimate is lower than typical heritability estimates for dyslexia provided by twin studies (40–60%) [2]. As with other complex traits, the discrepancy between twin- and SNP-based heritability suggests that part of dyslexia risk may be due to the genetic effects of variants other than SNPs, such as common copy number (CNVs) and rare variants. Although the relationship of CNVs and rare variants with DD and reading-related traits has not been extensively investigated to date, this hypothesis is partly supported by some recent findings. First, rare CNVs have often been implicated in familial forms of dyslexia [81] and the candidate gene DYX1C1 was first identified through a rare chromosomal rearrangement which co-segregated with dyslexia in a Finnish family [82]. Similarly, a targeted high-throughput sequencing study of 96 reading-impaired subjects reported an excess of putatively damaging rare variants in the candidate susceptibility loci DYX2 and CCDC136/FLNC [83]. Second, CNVs associated with neuropsychiatric disorders showed a significant influence on different cognitive traits in a large Icelandic population-based sample (N~102,000) [84]. In particular, a recurrent deletion of 15q11.2 was associated with a history of dyslexia and dyscalculia [84] and, in a later study, with cognitive, structural, and functional correlates of these impairments [85]. Third, a study reported >50% of the heritability of general cognition (IQ) and educational attainment (EduYears) to be explained by genetic variants in low LD with SNPs commonly genotyped on microarrays, especially rare variants. Indeed, SNP-based heritability of these traits approached the total heritability estimates from previous studies, when including also rare variants [86]. This suggests a substantial contribution of rare genetic variants to individual differences in intelligence and education, which may also extend to correlated cognitive traits such as reading ability.
PGS analyses revealed several significant associations between dyslexia risk and genetic liability to psychiatric disorders and other correlates.
First, we observed that PGSs for educational attainment and fluid intelligence were significantly associated with DD in our sample, in line with previous studies [30, 72, 87]. Luciano et al. [87] observed that PGSs of word reading, nonword repetition, and reading–spelling from GWAS studies of ~6600 children from UK and Australia showed significant positive associations with both verbal-numerical reasoning and educational attainment (college or university degree) in the UK Biobank cohort. Similarly, a PGS based on EduYears accounted for 2–5% of the variance in reading efficiency and comprehension in an independent UK sample (N = 5825) [72]. In the same study, Selzam et al. reported a PGSs of childhood general cognitive ability and adult verbal-numerical reasoning to explain a small but significant proportion (0.1–1.1%) of the variance in reading efficiency and comprehension at several developmental stages [72]. We later replicated these findings in a GWAS of reading-related cognitive skills partly overlapping with the present study (Nmax = 3468), extending the evidence of genetic overlap to cognitive predictors of dyslexia risk like phoneme awareness and digit span [28]. More recently, a GWAS of word reading in 4430 US children presenting in hospitals/clinics provided a further replication, reporting higher fractions of variance explained by EduYears (18%) and intelligence PGSs (7%) [30]. Together, the various PGS-based studies strongly support the existence of shared genetic factors influencing educational attainment, general cognition, and more specialized abilities like reading [88].
Second, genetic liability to ADHD was significantly associated with an increased DD risk, explaining 0.73% of its variance (at pT = 0.05). This finding is in line with the hypothesis of shared genetic bases between these disorders, initially suggested by twin studies [54, 56], and with evidence of genomic overlap reported for ADHD and the key cognitive features of dyslexia in our previous GWAS [28]. Recently, Price et al. [30] replicated the inverse association between ADHD-PGS and word reading in US children, as did Verhoef et al. [89] in a British longitudinal cohort (Nmax = 5919) for reading accuracy/comprehension at age 7, reading and spelling accuracy at age 9.
Third, we detected genetic links between two other neuropsychiatric disorders—BD and SCZ—and dyslexia. Standardized BD- and SCZ-PGS were associated with an increased DD risk, explaining 2.8% and 1.4% of its variance, respectively. Comorbidity of DD with a number of psychiatric disorders—including also BD and SCZ—has been previously reported [59, 60], and siblings of dyslexic subjects showed a high relative risk of being affected by ADHD, BD, SCZ, depression and autism, among others [59]. Although no significant associations between a SCZ-PGS and continuous reading-related traits were observed in a smaller independent dataset [87], the association between SCZ genetic risk and DD is in line with the reported genetic influence of SCZ risk variants on reading problems in the general population [84]. To the best of our knowledge, no evidence of a common genetic basis for BD and reading difficulties has been reported so far, although shared familial (and potentially genetic) risks have been previously suggested [59, 90]. Of note, we detected no significant associations between MDD-/ASD-PGS and dyslexia, but we did observe this for psychiatric cross-disorder genetic liability. These findings open up new scenarios in psychiatric genetics, suggesting a shared genetic and biological foundation across many different neurodevelopmental and neuropsychiatric conditions of phenotypically and clinically different nature.
Finally, the analysis of PGS influencing different brain cortical regions revealed a small, but robust and significant, protective effect against DD risk for a PGS increasing thickness of the transverse temporal gyrus. This region, also known as Heschl’s gyrus, is located within the primary auditory cortex—which is fundamental for auditory discrimination and speech perception [91]—and has been previously implicated in dyslexia by neuroimaging evidence, although not always consistently across studies [92–95]. Moreover, it overlaps with the left perisylvian regions where Galaburda et al. detected neuronal ectopias in four post-mortem dyslexic brains [96]. Here, we provide evidence of a genetic overlap between dyslexia risk and potential brain structural features proposed from non-genetic studies, although caution is suggested in the interpretation of these findings due to the inconsistencies across neuroimaging studies and to the potential role of regional brain asymmetries in the measures analyzed, which here were not taken into account due to the unavailability of GWAS summary statistics for separate hemispheres [75].
In spite of strengths like the wealth of cohorts and languages analyzed, and a relative homogeneity of recruitment, phenotypic assessment, and QC procedures, the present study also shows some limitations. In particular, there was a non-optimal case:control ratio in some datasets and a lack of properly screened controls in the English-speaking datasets. Although we acknowledge these would be preferred to improve power, the use of unscreened population controls is common where large numbers are needed, and has been exploited elsewhere [38, 39, 97]. Indeed, while for very common diseases the use of unscreened controls may notably affect power, for less common disorders/statuses (with prevalence <0.2) the loss of power is reduced and counterbalanced by the larger sample size which can be achieved through the use of unscreened populations [98]. Also, the PGS approach is based on the assumption that population structure and other possible confounds are well controlled in the training and target GWAS, which we implemented by adjusting all analyses for sex and genetic ancestry. However, independent replication of these results is warranted to substantiate the novel findings coming from the PGS analysis. Moreover, although the present study represents to our knowledge the largest GWAS on dyslexia to date [98], its sample size is relatively low compared to other studies in the neuropsychiatric field [66–70], which limited the power of analyses. Larger collaborative efforts are being implemented to improve these aspects to further enlighten the genetic epidemiology of dyslexia.
URLs
Wellcome Trust Case Control Consortium 2: https://www.wtccc.org.uk/ccc2/
PLINK: https://www.cog-genomics.org/plink2
QCTOOL: http://www.well.ox.ac.uk/~gav/qctool/
METAL: http://www.sph.umich.edu/csg/abecasis/Metal/index.html
MAGMA: http://ctg.cncr.nl/software/magma
MSigDB: http://software.broadinstitute.org/gsea/msigdb;
LD score regression: https://github.com/bulik/ldsc
Per-variant LD scores: https://data.broadinstitute.org/alkesgroup/LDSCORE/
Genotype-Tissue Expression portal (GTEx): http://www.gtexportal.org/home/
Brain eQTL Almanac (Braineac): http://www.braineac.org/
LocusZoom: http://www.locuszoom.org/
The R Project: https://www.r-project.org/
MASS package: https://cran.r-project.org/web/packages/MASS/index.html
Glmnet package: https://cran.r-project.org/web/packages/glmnet/index.html
Caret package: https://cran.r-project.org/web/packages/caret/index.html
Supplementary information
Acknowledgements
AG and TFMA were supported by the Munich Cluster for Systems Neurology (SyNergy). AG was supported by Fondazione Umberto Veronesi. SP is a Royal Society University Research fellow. BMM, CF, BSP and SEF are supported by the Max Planck Society. AW, BM and HK were funded by the Fraunhofer Society and the Max Planck Society within the “Pakt für Forschung und Innovation”. HK was also supported by LIFE—Leipzig Research Center for Civilization Diseases funded by means of the European Union; the European Regional Development Fund (ERDF); and the Free State of Saxony within the excellence initiative. FR is supported by Agence Nationale de la Recherche (ANR-06-NEURO-019-01, ANR-17-EURE-0017 IEC, ANR-10-IDEX-0001-02 PSL, ANR-11-BSV4-014-01), European Commission (LSHM-CT-2005-018696). TFMA was supported by the B.M.B.F. through the DIFUTURE consortium of the Medical Informatics Initiative Germany (grant 01ZZ1804A) and by the European Union’s Horizon 2020 Research and Innovation Programme (grant MultipleMS, EU RIA 733161). We would also like to acknowledge our project partners Catherine Billard, Caroline Bogliotti, Vanessa Bongiovanni, Laure Bricout, Camille Chabernaud, Isabelle Comte-Gervais, Florence Delteil-Pinton, Florence George, Christophe-Loïc Gérard, Marie Lageat, Marie-France Leheuzey, Marie-Thérèse Lenormand, Marion Liébert, Emilie Longeras, Emilie Racaud, Isabelle Soares-Boucaud, Sylviane Valdois, Nadège Villiermet, and Johannes Ziegler. This study makes use of data generated by the WTCCC. A full list of the investigators who contributed to the generation of the data is available at www.wtccc.org.uk. Funding for the WTCCC project was provided by the Wellcome Trust under awards 076113 and 085475. Open Access funding enabled and organized by Projekt DEAL.
Author contributions
AG, TFMA, NM-S, DC and KM contributed to genotype QC and imputation, and to phenotype QC. AG and TFMA carried out statistical analyses. AG, BMM and GSK wrote the paper. AG, TFMA, NM-S, KM, JB, PH, KUL, DC, BSP, FH, DT, VC, GH, YC, SI, JFD, APMor., JH, EGW, JCD, RKO, SDS, BP, AV, UM, HL, MPJ, PHTL, DB, MB, JFS, JBT, FF, AW, HK, BM, CF, TB, APMon., FR, KL, JK, TSS, SP, SEF, JS, MMN, BMM, GSK contributed to collection, phenotypic assessment and/or genotyping of the datasets included in the present study, and critically reviewed the manuscript. BMM and GSK supervised the present work.
Data availability
Summary statistics data supporting the findings of the present study are available upon request to the corresponding authors.
Code availability
Bioinformatic codes supporting the findings of the present study are available upon request to the corresponding authors.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Till F. M. Andlauer, Nazanin Mirza-Schreiber, Kristina Moll
These authors contributed equally: Johannes Schumacher, Markus M. Nöthen, Bertram Müller-Myhsok, Gerd Schulte-Körne
Contributor Information
Bertram Müller-Myhsok, Email: bmm@psych.mpg.de.
Gerd Schulte-Körne, Email: Gerd.Schulte-Koerne@med.uni-muenchen.de.
Supplementary information
The online version of this article (10.1038/s41380-020-00898-x) contains supplementary material, which is available to authorized users.
References
- 1.American Psychiatric Association. Diagnostic and statistical manual of mental disorders. American Psychiatric Association: Washington, DC; 2013.
- 2.Raskind WH, Peter B, Richards T, Eckert MM, Berninger VW. The genetics of reading disabilities: from phenotypes to candidate genes. Front Psychol. 2013;3:1–20. doi: 10.3389/fpsyg.2012.00601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pennington BF, Bishop DVM. Relations among speech, language, and reading disorders. Annu Rev Psychol. 2009;60:283–306. doi: 10.1146/annurev.psych.60.110707.163548. [DOI] [PubMed] [Google Scholar]
- 4.Schulte-Körne G, Deimel W, Müller K, Gutenbrunner C, Remschmidt H. Familial aggregation of spelling disability. J Child Psychol Psychiatry. 1996;37:817–22. doi: 10.1111/j.1469-7610.1996.tb01477.x. [DOI] [PubMed] [Google Scholar]
- 5.Gilger JW, Hanebuth E, Smith SD, Pennington BF. Differential risk for developmental reading disorders in the offspring of compensated versus noncompensated parents. Read Writ. 1996;8:407–17. [Google Scholar]
- 6.Fisher SE, DeFries JC. Developmental dyslexia: genetic dissection of a complex cognitive trait. Nat Rev Neurosci. 2002;3:767–80. doi: 10.1038/nrn936. [DOI] [PubMed] [Google Scholar]
- 7.Scerri TS, Schulte-Körne G. Genetics of developmental dyslexia. Eur Child Adolesc Psychiatry. 2010;19:179–97. doi: 10.1007/s00787-009-0081-0. [DOI] [PubMed] [Google Scholar]
- 8.Gayan J, Olson RK. Genetic and environmental influences on individual differences in printed word recognition. J Exp Child Psychol. 2003;84:97–123. doi: 10.1016/S0022-0965(02)00181-9. [DOI] [PubMed] [Google Scholar]
- 9.Francks C, MacPhie IL, Monaco AP. The genetic basis of dyslexia. Lancet Neurol. 2002;1:483–90. doi: 10.1016/S1474-4422(02)00221-1. [DOI] [PubMed] [Google Scholar]
- 10.Peterson RL, Pennington BF. Developmental dyslexia. Annu Rev Clin Psychol. 2015;11:283–307. doi: 10.1146/annurev-clinpsy-032814-112842. [DOI] [PubMed] [Google Scholar]
- 11.Carrion-Castillo A, Franke B, Fisher SE. Molecular genetics of dyslexia: an overview. Dyslexia. 2013;19:214–40. doi: 10.1002/dys.1464. [DOI] [PubMed] [Google Scholar]
- 12.Kere J. The molecular genetics and neurobiology of developmental dyslexia as model of a complex phenotype. Biochem Biophys Res Commun. 2014;452:236–43. doi: 10.1016/j.bbrc.2014.07.102. [DOI] [PubMed] [Google Scholar]
- 13.Paracchini S, Diaz R, Stein J. Advances in dyslexia genetics—new insights into the role of brain asymmetries. Adv Genet. 2016;96:53–97. doi: 10.1016/bs.adgen.2016.08.003. [DOI] [PubMed] [Google Scholar]
- 14.Taipale M, Kaminen N, Nopola-Hemmi J, Haltia T, Myllyluoma B, Lyytinen H, et al. A candidate gene for developmental dyslexia encodes a nuclear tetratricopeptide repeat domain protein dynamically regulated in brain. Proc Natl Acad Sci USA. 2003;100:11553–8. doi: 10.1073/pnas.1833911100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Francks C, Paracchini S, Smith SD, Richardson AJ, Scerri TS, Cardon LR, et al. A 77-kilobase region of chromosome 6p22.2 is associated with dyslexia in families from the United Kingdom and from the United States. Am J Hum Genet. 2004;75:1046–58. doi: 10.1086/426404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cope N, Harold D, Hill G, Moskvina V, Stevenson J, Holmans P, et al. Strong evidence that KIAA0319 on chromosome 6p is a susceptibility gene for developmental dyslexia. Am J Hum Genet. 2005;76:581–91. doi: 10.1086/429131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meng H, Smith SD, Hager K, Held M, Liu J, Olson RK, et al. DCDC2 is associated with reading disability and modulates neuronal development in the brain. Proc Natl Acad Sci USA. 2005;102:17053–8. doi: 10.1073/pnas.0508591102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schumacher J, Anthoni H, Dahdouh F, König IR, Hillmer AM, Kluck N, et al. Strong genetic evidence of DCDC2 as a susceptibility gene for dyslexia. Am J Hum Genet. 2005;78:52–62. doi: 10.1086/498992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anthoni H, Zucchelli M, Matsson H, Müller-Myhsok B, Fransson I, Schumacher J, et al. A locus on 2p12 containing the co-regulated MRPL19 and C2ORF3 genes is associated to dyslexia. Hum Mol Genet. 2007;16:667–77. doi: 10.1093/hmg/ddm009. [DOI] [PubMed] [Google Scholar]
- 20.Hannula-Jouppi K, Kaminen-Ahola N, Taipale M, Eklund R, Nopola-Hemmi J, Kääriäinen H, et al. The axon guidance receptor gene ROBO1 is a candidate gene for developmental dyslexia. PLoS Genet. 2005;1:0467–74. doi: 10.1371/journal.pgen.0010050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bates TC, Luciano M, Medland SE, Montgomery GW, Wright MJ, Martin NG. Genetic variance in a component of the language acquisition device: ROBO1 polymorphisms associated with phonological buffer deficits. Behav Genet. 2011;41:50–7. doi: 10.1007/s10519-010-9402-9. [DOI] [PubMed] [Google Scholar]
- 22.Tran C, Wigg KG, Zhang K, Cate-Carter TD, Kerr E, Field LL, et al. Association of the ROBO1 gene with reading disabilities in a family-based analysis. Genes, Brain Behav. 2014;13:430–8. doi: 10.1111/gbb.12126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meaburn EL, Harlaar N, Craig IW, Schalkwyk LC, Plomin R. Quantitative trait locus association scan of early reading disability and ability using pooled DNA and 100K SNP microarrays in a sample of 5760 children. Mol Psychiatry. 2008;13:729–40. doi: 10.1038/sj.mp.4002063. [DOI] [PubMed] [Google Scholar]
- 24.Field LL, Shumansky K, Ryan J, Truong D, Swiergala E, Kaplan BJ. Dense-map genome scan for dyslexia supports loci at 4q13, 16p12, 17q22; suggests novel locus at 7q36. Genes Brain Behav. 2013;12:56–69. doi: 10.1111/gbb.12003. [DOI] [PubMed] [Google Scholar]
- 25.Eicher JD, Powers NR, Miller LL, Akshoomoff N, Amaral DG, Bloss CS, et al. Genome-wide association study of shared components of reading disability and language impairment. Genes Brain Behav. 2013;12:792–801. doi: 10.1111/gbb.12085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gialluisi A, Newbury DF, Wilcutt EG, Olson RK, DeFries JC, Brandler WM, et al. Genome-wide screening for DNA variants associated with reading and language traits. Genes Brain Behav. 2014;13:686–701. doi: 10.1111/gbb.12158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Truong DT, Adams AK, Paniagua S, Frijters JC, Boada R, Hill DE, et al. Multivariate genome-wide association study of rapid automatised naming and rapid alternating stimulus in Hispanic American and African–American youth. J Med Genet. 2019. jmedgenet-2018-105874. [DOI] [PMC free article] [PubMed]
- 28.Gialluisi A, Andlauer TFM, Mirza-Schreiber N, Moll K, Becker J, Hoffmann P, et al. Genome-wide association scan identifies new variants associated with a cognitive predictor of dyslexia. Transl Psychiatry. 2019;9:77. doi: 10.1038/s41398-019-0402-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luciano M, Evans DM, Hansell NK, Medland SE, Montgomery GW, Martin NG, et al. A genome-wide association study for reading and language abilities in two population cohorts. Genes, Brain Behav. 2013;12:645–52. doi: 10.1111/gbb.12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Price KM, Wigg KG, Feng Y, Blokland K, Wilkinson M, He G, et al. Genome-wide association study of word reading: overlap with risk genes for neurodevelopmental disorders. Genes, Brain Behav. 2020;19:e12648. doi: 10.1111/gbb.12648. [DOI] [PubMed] [Google Scholar]
- 31.Roeske D, Ludwig KU, Neuhoff N, Becker J, Bartling J, Bruder J, et al. First genome-wide association scan on neurophysiological endophenotypes points to trans-regulation effects on SLC2A3 in dyslexic children. Mol Psychiatry. 2009;16:97. doi: 10.1038/mp.2009.102. [DOI] [PubMed] [Google Scholar]
- 32.Schulte-Körne G, Ziegler A, Deimel W, Schumacher J, Plume E, Bachmann C, et al. Interrelationship and familiality of dyslexia related quantitative measures. Ann Hum Genet. 2007;71:160–75. doi: 10.1111/j.1469-1809.2006.00312.x. [DOI] [PubMed] [Google Scholar]
- 33.Landerl K, Ramus F, Moll K, Lyytinen H, Leppänen PHT, Lohvansuu K, et al. Predictors of developmental dyslexia in European orthographies with varying complexity. J Child Psychol Psychiatry. 2013;54:686–94. doi: 10.1111/jcpp.12029. [DOI] [PubMed] [Google Scholar]
- 34.Moll K, Ramus F, Bartling J, Bruder J, Kunze S, Neuhoff N, et al. Cognitive mechanisms underlying reading and spelling development in five European orthographies. Learn Instr. 2014;29:65–77. doi: 10.1016/j.learninstruc.2013.09.003. [DOI] [Google Scholar]
- 35.Willcutt EG, Pennington BF, Olson RK, Chhabildas N, Hulslander J. Neuropsychological analyses of comorbidity between reading disability and attention deficit hyperactivity disorder: in search of the common deficit. Dev Neuropsychol. 2005;27:35–78. doi: 10.1207/s15326942dn2701_3. [DOI] [PubMed] [Google Scholar]
- 36.Brandler WM, Morris AP, Evans DM, Scerri TS, Kemp JP, Timpson NJ, et al. Common variants in left/right asymmetry genes and pathways are associated with relative hand skill. PLoS Genet. 2013;9:e1003751. doi: 10.1371/journal.pgen.1003751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rucker JJH, Breen G, Pinto D, Pedroso I, Lewis CM, Cohen-Woods S, et al. Genome-wide association analysis of copy number variation in recurrent depressive disorder. Mol Psychiatry. 2013;18:183–9. doi: 10.1038/mp.2011.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huckins LM, Hatzikotoulas K, Southam L, Thornton LM, Steinberg J, Aguilera-Mckay F, et al. Investigation of common, low-frequency and rare genome-wide variation in anorexia nervosa. Mol Psychiatry. 2018;23:1169–80. doi: 10.1038/mp.2017.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.International T, Against L, Consortium E, Epilepsies C. Genome-wide mega-analysis identifies 16 loci and highlights diverse biological mechanisms in the common epilepsies. Nat Commun. 2018;9:5269. doi: 10.1038/s41467-018-07524-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chang CC, Chow CC, Tellier LCAM, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Consortium T 1000 GP. Auton A, Abecasis GR, Altshuler DM, Durbin RM, Abecasis GR, et al. A global reference for human genetic variation. Nature. 2015;526:68. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Delaneau O, Zagury J-F, Marchini J. Improved whole-chromosome phasing for disease and population genetic studies. Nat Meth. 2013;10:5–6. doi: 10.1038/nmeth.2307. [DOI] [PubMed] [Google Scholar]
- 43.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lippert C, Listgarten J, Liu Y, Kadie CM, Davidson RI, Heckerman D. FaST linear mixed models for genome-wide association studies. Nat Meth. 2011;8:833–5. doi: 10.1038/nmeth.1681. [DOI] [PubMed] [Google Scholar]
- 45.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–1. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen H, Wang C, Conomos MP, Stilp AM, Li Z, Sofer T, et al. Control for population structure and relatedness for binary traits in genetic association studies via logistic mixed models. Am J Hum Genet. 2016;98:653–66. doi: 10.1016/j.ajhg.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol. 2015;11:e1004219. doi: 10.1371/journal.pcbi.1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bulik-Sullivan B, Loh PR, Finucane HK, Ripke S, Yang J, Patterson N, et al. LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47:291–5. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bulik-Sullivan B, Finucane HK, Anttila V, Gusev A, Day FR, Loh PR, et al. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47:1236–41. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.The International HapMap Consortium. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Finucane HK, Bulik-Sullivan B, Gusev A, Trynka G, Reshef Y, Loh PR, et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet. 2015;47:1228–35. doi: 10.1038/ng.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Consortium TGte. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–5. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Finucane HK, Reshef YA, Anttila V, Slowikowski K, Gusev A, Byrnes A, et al. Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types. Nat Genet. 2018;50:621–9. doi: 10.1038/s41588-018-0081-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Willcutt EG, Pennington BF, DeFries JC. Twin study of the etiology of comorbidity between reading disability and attention-deficit/hyperactivity disorder. Am J Med Genet. 2000;96:293–301. doi: 10.1002/1096-8628(20000612)96:3<293::AID-AJMG12>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 55.Willcutt EG, Betjemann RS, McGrath LM, Chhabildas NA, Olson RK, DeFries JC, et al. Etiology and neuropsychology of comorbidity between RD and ADHD: the case for multiple-deficit models. Cortex. 2010;46:1345–61. doi: 10.1016/j.cortex.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Willcutt EG, Pennington BF, Olson RK, DeFries JC. Understanding comorbidity: a twin study of reading disability and attention-deficit/hyperactivity disorder. Am J Med Genet Part B Neuropsychiatr Genet. 2007;144B:709–14. doi: 10.1002/ajmg.b.30310. [DOI] [PubMed] [Google Scholar]
- 57.Russell G, Pavelka Z. Co-occurrence of developmental disorders: children who share symptoms of autism, dyslexia and attention deficit hyperactivity disorder. In: Fitzgerald M, editor. Recent advances in autism spectrum disorders—vol. I. Rijeka: InTech; 2013. p. 17.
- 58.Visser L, Kalmar J, Linkersdörfer J, Görgen R, Rothe J, Hasselhorn M, et al. Comorbidities between specific learning disorders and psychopathology in elementary school children in Germany. Front Psychiatry. 2020;11:292. doi: 10.3389/fpsyt.2020.00292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cederlöf M, Maughan B, Larsson H, D’Onofrio BM, Plomin R. Reading problems and major mental disorders—co-occurrences and familial overlaps in a Swedish nationwide cohort. J Psychiatr Res. 2017;91:124–9. doi: 10.1016/j.jpsychires.2017.03.014. [DOI] [PubMed] [Google Scholar]
- 60.Whitford V, O’Driscoll GA, Titone D. Reading deficits in schizophrenia and their relationship to developmental dyslexia: a review. Schizophr Res. 2017. 10.1016/j.schres.2017.06.049. [DOI] [PubMed]
- 61.Lee PH, Anttila V, Won H, Feng Y-CA, Rosenthal J, Zhu Z, et al. Genomic relationships, novel loci, and pleiotropic mechanisms across eight psychiatric disorders. Cell. 2019;179:1469–82.e11. doi: 10.1016/j.cell.2019.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Savage JE, Jansen PR, Stringer S, Watanabe K, Bryois J, de Leeuw CA, et al. Genome-wide association meta-analysis in 269,867 individuals identifies new genetic and functional links to intelligence. Nat Genet. 2018;50:912–9. doi: 10.1038/s41588-018-0152-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee JJ, Wedow R, Okbay A, Kong E, Maghzian O, Zacher M, et al. Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nat Genet. 2018;50:1112–21. doi: 10.1038/s41588-018-0147-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Peng P, Wang T, Wang C, Lin X. A meta-analysis on the relation between fluid intelligence and reading/mathematics: effects of tasks, age, and social economics status. Psychol Bull. 2019;145:189–236. doi: 10.1037/bul0000182. [DOI] [PubMed] [Google Scholar]
- 65.Ritchie SJ, Bates TC, Plomin R. Does learning to read improve intelligence? A longitudinal multivariate analysis in identical twins from age 7 to 16. Child Dev. 2015;86:23–36. doi: 10.1111/cdev.12272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stahl EA, Breen G, Forstner AJ, McQuillin A, Ripke S, Trubetskoy V, et al. Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat Genet. 2019;51:793–803. doi: 10.1038/s41588-019-0397-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Demontis D, Walters RK, Martin J, Mattheisen M, Als TD, Agerbo E, et al. Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat Genet. 2019;51:63–75. doi: 10.1038/s41588-018-0269-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Grove J, Ripke S, Als TD, Mattheisen M, Walters RK, Won H, et al. Identification of common genetic risk variants for autism spectrum disorder. Nat Genet. 2019;51:431–44. doi: 10.1038/s41588-019-0344-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–7. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Howard DM, Adams MJ, Clarke T-K, Hafferty JD, Gibson J, Shirali M, et al. Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat Neurosci. 2019;22:343–52. doi: 10.1038/s41593-018-0326-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Choi SW, O’Reilly PF. PRSice-2: Polygenic Risk Score software for biobank-scale data. Gigascience. 2019;8:1–6. doi: 10.1093/gigascience/giy165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Selzam S, Dale PS, Wagner RK, DeFries JC, Cederlöf M, O’Reilly PF, et al. Genome-wide polygenic scores predict reading performance throughout the school years. Sci Stud Read. 2017;21:334–49. doi: 10.1080/10888438.2017.1299152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Halldorsdottir T, Piechaczek C, Soares de Matos AP, Czamara D, Pehl V, Wagenbuechler P, et al. Polygenic risk: predicting depression outcomes in clinical and epidemiological cohorts of youths. Am J Psychiatry. 2019;176:615–25. doi: 10.1176/appi.ajp.2019.18091014. [DOI] [PubMed] [Google Scholar]
- 74.Grasby KL, Jahanshad N, Painter JN, Colodro-Conde L, Bralten J, Hibar DP, et al. The genetic architecture of the human cerebral cortex. Science. 2020;367:eaay669. doi: 10.1126/science.aay6690. [DOI] [Google Scholar]
- 75.Ramus F, Altarelli I, Jednoróg K, Zhao J, Scotto, di Covella L. Neuroanatomy of developmental dyslexia: pitfalls and promise. Neurosci Biobehav Rev. 2018;84:434–52. doi: 10.1016/j.neubiorev.2017.08.001. [DOI] [PubMed] [Google Scholar]
- 76.Gialluisi A, Guadalupe T, Francks C, Fisher SE. Neuroimaging genetic analyses of novel candidate genes associated with reading and language. Brain Lang. 2017;172:9–15. doi: 10.1016/j.bandl.2016.07.002. [DOI] [PubMed] [Google Scholar]
- 77.Krapohl E, Patel H, Newhouse S, Curtis CJ, Von Stumm S, Dale PS, et al. Multi-polygenic score approach to trait prediction. Mol Psychiatry. 2018;23:1368–74. doi: 10.1038/mp.2017.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Roadmap Epigenomics C, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Muto E, Tabata Y, Taneda T, Aoki Y, Muto A, Arai K, et al. Identification and characterization of Veph, a novel gene encoding a PH domain-containing protein expressed in the developing central nervous system of vertebrates. Biochimie. 2004;86:523–31. doi: 10.1016/j.biochi.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 80.Yi JJ, Barnes AP, Hand R, Polleux F, Ehlers MD. TGF-β signaling specifies axons during brain development. Cell. 2010;142:144–57. doi: 10.1016/j.cell.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gialluisi A, Visconti A, Willcutt EG, Smith SD, Pennington BF, Falchi M, et al. Investigating the effects of copy number variants on reading and language performance. J Neurodev Disord. 2016. 10.1186/s11689-016-9147-8. [DOI] [PMC free article] [PubMed]
- 82.Nopola-Hemmi J, Taipale M, Haltia T, Lehesjoki A-E, Voutilainen A, Kere J. Two translocations of chromosome 15q associated with dyslexia. J Med Genet. 2000;37:771–5. doi: 10.1136/jmg.37.10.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Adams AK, Smith SD, Truong DT, Willcutt EG, Olson RK, DeFries JC, et al. Enrichment of putatively damaging rare variants in the DYX2 locus and the reading-related genes CCDC136 and FLNC. Hum Genet. 2017;136:1395–405. doi: 10.1007/s00439-017-1838-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Stefansson H, Meyer-Lindenberg A, Steinberg S, Magnusdottir B, Morgen K, Arnarsdottir S, et al. CNVs conferring risk of autism or schizophrenia affect cognition in controls. Nature. 2014;505:361–6. doi: 10.1038/nature12818. [DOI] [PubMed] [Google Scholar]
- 85.Ulfarsson MO, Walters GB, Gustafsson O, Steinberg S, Silva A, Doyle OM, et al. 15q11.2 CNV affects cognitive, structural and functional correlates of dyslexia and dyscalculia. Transl Psychiatry. 2017;7:e1109. doi: 10.1038/tp.2017.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hill WD, Arslan RC, Xia C, Luciano M, Amador C, Navarro P, et al. Genomic analysis of family data reveals additional genetic effects on intelligence and personality. Mol Psychiatry. 2018. 10.1038/s41380-017-0005-1. [DOI] [PMC free article] [PubMed]
- 87.Luciano M, Hagenaars SP, Cox SR, Hill WD, Davies G, Harris SE, et al. Single nucleotide polymorphisms associated with reading ability show connection to socio-economic outcomes. Behav Genet. 2017;47:469–79. doi: 10.1007/s10519-017-9859-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Davis OSP, Haworth CMA, Plomin R. Learning abilities and disabilities: generalist genes in early adolescence. Cogn Neuropsychiatry. 2009;14:312–31. doi: 10.1080/13546800902797106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Verhoef E, Demontis D, Burgess S, Shapland CY, Dale PS, Okbay A, et al. Disentangling polygenic associations between attention-deficit/hyperactivity disorder, educational attainment, literacy and language. Transl Psychiatry. 2019;9:35. doi: 10.1038/s41398-018-0324-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.McDonough-Ryan P, DelBello M, Shear PK, Ris MD, Soutullo C, Strakowski SM. Academic and cognitive abilities in children of parents with bipolar disorder: a test of the nonverbal learning disability model. J Clin Exp Neuropsychol. 2002;24:280–5. doi: 10.1076/jcen.24.3.280.980. [DOI] [PubMed] [Google Scholar]
- 91.Cai DC, Fonteijn H, Guadalupe T, Zwiers M, Wittfeld K, Teumer A, et al. A genome-wide search for quantitative trait loci affecting the cortical surface area and thickness of Heschl’s gyrus. Genes, Brain Behav. 2014;13:675–85. doi: 10.1111/gbb.12157. [DOI] [PubMed] [Google Scholar]
- 92.Clark KA, Helland T, Specht K, Narr KL, Manis FR, Toga AW, et al. Neuroanatomical precursors of dyslexia identified from pre-reading through to age 11. Brain. 2014;137:3136–41. doi: 10.1093/brain/awu229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Altarelli I, Leroy F, Monzalvo K, Fluss J, Billard C, Dehaene-Lambertz G, et al. Planum temporale asymmetry in developmental dyslexia: revisiting an old question. Hum Brain Mapp. 2014;35:5717–35. doi: 10.1002/hbm.22579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ma Y, Koyama MS, Milham MP, Castellanos FX, Quinn BT, Pardoe H, et al. Cortical thickness abnormalities associated with dyslexia, independent of remediation status. NeuroImage Clin. 2015;7:177–86. doi: 10.1016/j.nicl.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Leonard C, Eckert M, Given B, Virginia B, Eden G. Individual differences in anatomy predict reading and oral language impairments in children. Brain. 2006;129:3329–42. doi: 10.1093/brain/awl262. [DOI] [PubMed] [Google Scholar]
- 96.Galaburda AM, Sherman GF, Rosen GD, Aboitiz F, Geschwind N. Developmental dyslexia: four consecutive patients with cortical anomalies. Ann Neurol. 1985;18:222–33. doi: 10.1002/ana.410180210. [DOI] [PubMed] [Google Scholar]
- 97.Zabaneh D, Krapohl E, Gaspar HA, Curtis C, Lee SH, Patel H, et al. A genome-wide association study for extremely high intelligence. Mol Psychiatry. 2018;23:1226–32. doi: 10.1038/mp.2017.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Moskvina V, Holmans P, Schmidt KM, Craddock N. Design of case-controls studies with unscreened controls. Ann Hum Genet. 2005;69:566–76. doi: 10.1111/j.1529-8817.2005.00175.x. [DOI] [PubMed] [Google Scholar]
- 99.Wechsler D. The Wechsler Intelligence Scale for Children. 3rd ed. London: The Psychological Corporation; 1992.
- 100.Wechsler D. Wechsler Intelligence Scale for Children. 4th ed. San Antonio, TX: Psychological Corporation; 2003.
- 101.Wechsler D. Manual for the Wechsler Intelligence Scale for Children—Revised. New York, NY: The Psychological Corporation; 1974.
- 102.Wechsler D. Manual for the Wechsler Adult Intelligence Scale—Revised. New York, NY: Psychological Corporation; 1981.
- 103.Elliot Murray DJ, Pearson LSCD. The British Ability Scales. Slough, UK: NFER; 1979. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Summary statistics data supporting the findings of the present study are available upon request to the corresponding authors.
Bioinformatic codes supporting the findings of the present study are available upon request to the corresponding authors.