Abstract
Neurodegenerative disorders emerge from the failure of intricate cellular mechanisms, which ultimately lead to the loss of vulnerable neuronal populations. Research conducted across several laboratories has now provided compelling evidence that pathogenic proteins can also contribute to non-cell autonomous toxicity in several neurodegenerative contexts, including Alzheimer’s, Parkinson’s, and Huntington’s diseases as well as Amyotrophic Lateral Sclerosis. Given the nearly ubiquitous nature of abnormal protein accumulation in such disorders, elucidating the mechanisms and routes underlying these processes is essential to the development of effective treatments. To this end, physiologically relevant human in vitro models are critical to understand the processes surrounding uptake, release and nucleation under physiological or pathological conditions. This review explores the use of human-induced pluripotent stem cells (iPSCs) to study prion-like protein propagation in neurodegenerative diseases, discusses advantages and limitations of this model, and presents emerging technologies that, combined with the use of iPSC-based models, will provide powerful model systems to propel fundamental research forward.
Subject terms: Neuroscience, Stem cells
The origin of the prion hypothesis for neurodegenerative diseases
The prion disorder scrapie was first described in animals as early as the 18th century, but the disease-causing agent, the prion protein (PrP), was only identified in the 1980s [1, 2]. In the intervening time, a number of disorders involving the same protein were reported in mammals, including chronic wasting disease in cervids, bovine spongiform encephalopathy in cows and Creutzfeldt–Jakob’s disease (CJD) in humans [1]. These disorders, known as transmissible spongiform encephalopathies, first came to the public eye when the bovine spongiform encephalopathy epidemic made front page news in the early 2000s. Research into PrP gained momentum when evidence of a novel variant of CJD was linked to ingestion of infected meat [3]. Unlike other disease-causing agents, such as viruses and bacteria, the key information leading to pathogenicity of prion and prion-like proteins is not encoded in the genetic material but instead, in the structure and biophysical properties of the misfolded protein. In general, the formation of dimers and oligomers is the rate-limiting-step in the protein aggregation process [4]. The presence of the misfolded protein can alter this by acting as a catalyst for the conformational change of the endogenous protein [4]. While these mechanisms of protein infectivity were first described for the PrP protein, the majority, if not all proteins associated with neurodegenerative diseases such as Alzheimer’s disease (AD), Parkinson’s disease (PD), Huntington’s disease (HD) and Amyotrophic Lateral Sclerosis (ALS), are now recognized to have prion-like properties (Fig. 1a).
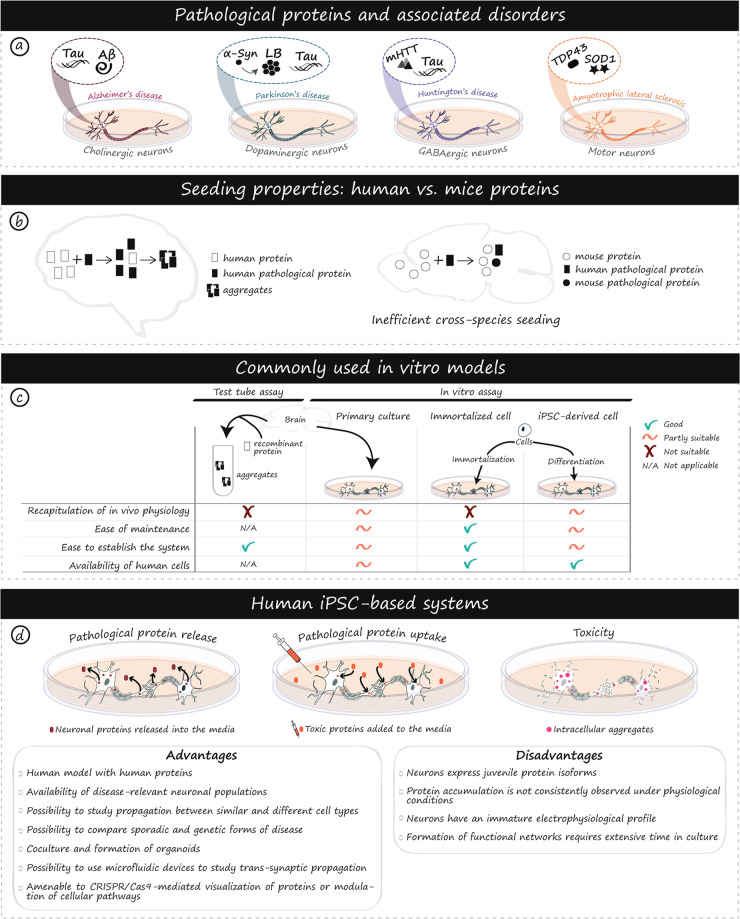
Fig. 1. Prion-like proteins in human diseases and iPSC models.
Protein aggregation is associated with multiple neurodegenerative disorders, involves disease-specific proteins, and affects unique populations of neurons (a). Variability in the propensity to aggregate and inefficient cross-species seeding between human and mouse protein homologs have been observed (b). Ex vivo/test tube and in vitro assays including primary cultures, immortalized cells and iPSC-derived cells are commonly used to study protein propagation. Advantages and disadvantages of each model are listed (c). iPSC-based models enable the production of human neuronal populations of interest that better recapitulate physiological features of human diseases compared to rodent-based models. This unprecedented access to human cells opens new avenues for the study of mechanisms associated with propagation, seeding, and toxicity of pathological proteins (d). As with any disease model, a number of advantages and disadvantages need to be carefully considered to inform and guide experimental choices (d). α-Syn alpha-synuclein, Aβ amyloid beta, AD Alzheimer’s disease, ALS Amyotrophic lateral sclerosis, Cas9 CRISPR associated protein 9, CRISPR Clustered Regularly Interspaced Short Palindromic Repeats, GABA gamma aminobutyric acid, HD Huntington’s disease, LB Lewy body, mHTT mutant huntingtin, PD Parkinson’s disease, SOD1 superoxide dismutase 1, TDP-43 TAR DNA-binding 43 protein.
Evidence of prion-like properties for proteins such as α-Synuclein (α-Syn), tau, amyloid-β (Aβ), huntingtin (HTT), superoxide dismutase (SOD1) and TAR DNA-binding 43 protein (TDP-43) have slowly been accruing over decades of research through the use of diverse techniques such as in vitro protein conversion assays and immunohistochemical assessment of post-mortem tissue. Some of the earliest indications of prion-like properties came from the study of brains of PD patients who had received fetal tissue transplants. Detailed analysis of the grafts unveiled the presence of aggregated α-Syn within this previously healthy fetal tissue [5–8]. Seminal work conducted by Braak and colleagues on the characterization of the spread of α-Syn across disease stages provided an additional indication that proteins may indeed propagate between cells. From a systematic study of post-mortem tissue, Braak et al. hypothesized that α-Syn pathology follows a specific pattern of propagation, originating in the medulla oblangata and migrating through synaptically connected regions, including into the substantia nigra pars compacta [9]. These findings offered a strong conceptual framework for subsequent studies of the prion-like behavior of α-Syn. For example, it was thereafter reported that injections of either PD brain homogenates or α-Syn fibrils into the brains of mice and non-human primates were sufficient to induce the loss of dopaminergic neurons and the development of motor impairments [10–12]. More recent experimental paradigms have expanded on this notion by demonstrating that injection of α-Syn into the peripheral nervous system is sufficient to cause central disease in rodents [13, 14]. Meanwhile, detection of pathogenic α-Syn seeds in the brains or cerebral spinal fluid of PD patients has been shown to be a selective and sensitive diagnostic tool [15, 16], further supporting the relationship between misfolded species and disease progression.
The tracking of tau spreading throughout the progression of AD was also performed by Braak et al. several years prior to the description of α-Syn pathology in PD [17]. More recently, other proteins associated with common neurodegenerative disorders have been administered via injection of brain homogenates derived from post-mortem tissue of AD and frontotemporal lobe dementia (FTD) patients and were reported to lead to Aβ, tau, and TDP-43 pathology [18–21]. Furthermore, tau [22], Aβ [23], α-Syn [10, 24], mutant HTT (mHTT) [25], and SOD1 [26] oligomers/fibrils have been used in a wide variety of in vitro and in vivo experiments to demonstrate their neurotoxicity. There is now ample evidence that prion-like proteotoxicity is sufficient to provoke the manifestation of common neurodegenerative diseases in a non-cell autonomous manner. However, despite extensive research, much remains to be learned about the mechanisms of prion-like properties as well as the degree to which they contribute to disease manifestation and/or progression. Disease-relevant models are needed to unravel the molecular mechanisms of prion pathogenesis and thereby design novel protein-targeted therapies. One powerful model system with the potential to greatly enhance our understanding of the contributions of pathological proteins to neurodegenerative disorders involves the use of human-induced pluripotent stem cells (iPSCs). In this review, we discuss molecular and species-specific advantages of iPSCs, studies that have previously been performed in iPSCs to model prion-like protein propagation, current limitations that need to be considered, as well as recently developed technologies that are likely to advance the field.
Considerations for modeling properties of pathological proteins
Species-specific cellular differences
Mouse models have been developed and widely used to study environmental and genetic determinants of neurodegenerative diseases [27, 28]. However, these genetic models fail to reproduce all pathological features of human neurodegenerative disorders. For example, rodent models of AD, PD and HD rarely display overt neuronal cell death or the progressive neurodegeneration which characterizes these human conditions [27, 29, 30]. In addition, evidence demonstrating intrinsic functional differences between human and mouse cells has begun to emerge, including differences in developmental stages, the rate of protein degradation during development, RNA processing, glial cell biology and global transcriptomes [28, 31–34]. These species-specific differences likely account for some of the challenges faced by mouse-based systems in modeling human diseases.
Species-specific protein differences
While all of these differences have important consequences for the understanding and interpretation of studies using mice, one particularly salient difference, in the context of prion-like disorders, is variations in the peptide sequences, isoform expression, or post-translational modification of proteins associated with common neurodegenerative diseases (Fig. 1b). The details of the most frequent modifiers of protein toxicity and/or aggregation, as well as differences between mice and humans, are summarized in Table 1. While such difficulties can be overcome using humanized mice, purified fibrils, or by adding high doses of seed-competent proteins into a culture system, the inherent structural differences in the proteins can be efficaciously circumvented through the use of human cell models (Fig. 1c). For these reasons, neurons and glia derived from iPSCs are attractive alternatives to rodent primary cultures as they produce human-specific cell types.
Table 1.
 |
α-Syn alpha-synuclein, Aβ amyloid beta, AD Alzheimer’s disease, ALS Amyotrophic lateral sclerosis, AP amyloid plaques, APP amyloid precursor protein gene, CAA cerebral amyloid angiopathy, CBD corticobasal degeneration, CHR chromosome, CJD Creutzfeldt–Jakob’s disease, CP cored plaques, COP cored-only plaques, CTE chronic traumatic encephalopathy, DLB dementia with Lewy bodies, DN dystrophic neurites, DNP/CNP dense neuritic plaques/cored neuritic plaques, DP diffuse plaques, DS Down’s syndrome, FI fatal insomnia, FTLD frontotemporal lobar degeneration, FTDP-17 frontotemporal dementia with parkinsonism linked to chromosome 17, GFT glial fibrillary tangles, GGI globular glial inclusions, GI glial inclusions, GSS Gerstmann–Sträussler–Scheinker syndrome, HD Huntington’s disease, HpScl hippocampal sclerosis, HTT huntingtin protein, Htt huntingtin gene, MAPT microtubule-associated protein tau gene, mHTT mutant huntingtin, MSA multi-system atrophy, Namino-terminal inserts, NCI neural cytoplasmic inclusions, NFL neurofibrillary lesions, NFT neurofibrillary tangles, NII neuronal intranuclear inclusions, NP neuritic plaques, NT neuropil threads, PiD Pick’s disease, PD Parkinson’s disease, PDD Parkinson’s disease dementia, PHF paired helical filaments, PMA progressive muscular atrophy, PolyQ polyglutamine, PSP supranuclear palsy, PRNP gene encoding for PrP, PrP prion protein, PrP-CAA pure prion protein cerebral amyloid angiopathy, PrP-SA PrP‐systemic amyloidosis, SF straight filaments, SNCA gene encoding for α-Syn, SOD1 superoxide dismutase protein, SOD1 superoxide dismutase gene, S serine, SORT1 transmembrane receptor sortilin, T threonine, TARDP TDP-43 gene, TDP-43 TAR-binding domain protein-43, TsAs thorn-shaped astrocytes, VPSPr variably protease-sensitive prionopathy, Y tyrosine, 3R 3 repeats, 4R 4 repeats.
PrP
Prion diseases are rare amongst neurodegenerative proteinopathies in that they are not human-specific conditions, as other mammalian species also spontaneously develop transmissible spongiform encephalopathies (e.g. cervids) [35]. Most human neurodegenerative disorders do not have a naturally occurring mammalian equivalent, thus in vivo studies generally involve investigation of the human pathological protein expressed in the model organism of interest. However, the presence of bona fide transmissible spongiform encephalopathies (e.g. CJD, kuru and scrapie) in multiple species has diversified the sources of misfolded proteins available in the prion field. PrP scrapie (PrPSc), the infectious form of PrP, contained in homogenates from sheep, cows and humans are all routinely tested in multiple mammalian species to better understand the factors essential to interspecies disease transmission. Such studies have identified the amino acid sequence, folding conformation and glycosylation of the prion protein as factors that contribute to the presence or absence of species barriers [36–38]. One recent study has even suggested that single amino acid residues in horse and dog PrP contribute to the relative resistance of these animals to experimentally-induced disease [39]. While the gene encoding for PrP (PRNP) is highly conserved in mammals, substitutions have occurred across evolution. Mice and humans differ by only 20 amino acids [40] (Table 1), but this is sufficient to limit transmission of disease from humans to mice. To overcome this, transgenic animals have been generated to express a chimeric form of PrP which facilitates human to mouse transmission [41].
Tau
While the tau protein shares similarities between mice and humans, not all splice variants are equally represented in each species and the murine N-terminal peptide sequence lacks 11 residues present in the canonical human homolog [42, 43] (Table 1). In the human adult central nervous system (CNS), tau has 6 common isoforms that derive from alternative splicing of Exons 2, 3 and 10. The tau variant that includes Exon 10 has four microtubule-binding repeats and is referred to as 4R tau, while exclusion of Exon 10 results in 3R tau which contains only three microtubule-binding domains [42]. Inclusion or exclusion of Exon 10 is developmentally regulated in humans, with 3R tau demonstrating dominant expression in the juvenile nervous system, while 3R and 4R tau are found in similar proportions in the adult CNS. In mice, this change is more dramatic with 3R tau being entirely absent from the adult mouse brain [42] (Table 1). In humans, the balance between 3R and 4R is very important. In fact, mutations in MAPT affecting alternative splicing of Exon 10 lead to the development of tauopathies with preferential accumulation of either 3R or 4R, depending on the mutation [44–47]. The presence or absence of the extra microtubule binding domain also impacts spreading and seeding of tau, with analysis of cross-seeding between 4R and 3R tau, suggesting that 3R tau can recruit 4R tau, but that the reverse does not occur [48]. Furthermore, both 3R and 4R tau have the capacity to trigger aggregation, but injected 4R tau can spread to more remote areas in vivo [49]. Given the complex relationship between 3R and 4R tau in human disease, the absence of 3R tau in the adult mouse CNS may have important ramifications. Recent studies have additionally described discrepancies in the N-terminal portion of mouse and human tau. These changes are suspected to alter interactions between tau and other proteins, including end-binding proteins, which may impact the release of tau as well as the formation of aggregates [43, 50].
Aβ
Unlike tau, no differences in Aβ isoform expression have been reported between humans and mice. Structurally, mouse and human Aβ are very similar, differing by only 3 amino acid substitutions in the N-terminal domain [51, 52] (Table 1). Despite the similarity in sequence, cell culture analyses with human and mouse Aβ show both differential seeding capacity and toxicity [51]. After 48 h of exposure to human Aβ, congo red positive amyloid fibrils are visible in cultured SH-SY5Y cells, while no aggregates are detected 72 h following treatment with rodent Aβ [51]. Animal studies show that overexpression of mouse Aβ is insufficient to induce pathological changes or to exacerbate plaque formation when mouse and human Aβ are simultaneously overexpressed [52]. A transgenic background is also required for Aβ-containing homogenates from AD patients to induce behavioral and neuropathological phenotypes [53].
α-Syn
Comparison of α-Syn between species has revealed that only 7 amino acids differ between canonical human and mouse homologs (Table 1). However, these changes impact the electrophoretic mobility, conformation of fibrils, cleavage by proteases and cross-seeding [54, 55]. Both human and mouse α-Syn can efficiently seed proteins from the same species, suggesting no inherent differences in the kinetics of aggregation. Instead, the alterations in the amino acid sequence seem to only influence cross-species seeding [55]. Importantly, the rate of cross-seeding can be modified by adjusting the amino acid sequence [54, 55]. More recent work has shown that mouse and human α-Syn produce distinct fibrillar structures with human protein forming twisted fibrils and the mouse protein generating straight fibrils [54]. These structures could explain why mouse α-Syn can inhibit aggregation of the human protein (Table 1) [56]. This inhibitory effect was clearly demonstrated in one series of experiments where human α-Syn was expressed in mice in the presence or absence of mouse α-Syn within the genetic background. Primary neurons expressing only human α-Syn formed Lewy-body like inclusions, which were absent when mouse α-Syn was also expressed [56]. Similar results were observed in the brains of transgenic animals, where the lack of mouse α-Syn increased the tendency of human α-Syn to aggregate [56].
HTT
Multiple changes in the N-terminal structure have been detected between canonical mouse and human homologs of HTT, including differences in the average number of polyglutamine repeats and in the organization of the polyproline region [57] (Table 1). The potential consequences of the fluctuations in the polyglutamine number are inferred from clinical data indicating that the age of onset is inversely correlated with the number of CAG repeats [58]. The consequences of variations in the polyproline region are less apparent, but they result in an altered structure of the mouse protein [57] which may in turn change HTT’s capacity to misfold. Further evidence of the relevance of this polyproline stretch comes from the heightened phenotype of chimeric knock-in mouse models expressing the pathogenic human HTT exon fragment 1 (HTTExon 1) in the normal mouse gene [59], as opposed to the milder phenotype detected in a non-chimeric knock-in mouse model with equivalent CAG expansions [60].
SOD1
Between mice and humans, 25 amino acids differ in the canonical SOD1 protein [61]. Similar to what has been reported for Aβ, these differences result in a mouse protein which is less prone to aggregate than its human homolog [62]. Furthermore, when human SOD1 is added into the brains of SOD1 transgenic mice, the mouse form of the protein is absent from the aggregates, while the endogenous human protein is present [61].
TDP-43
TDP-43 has very high homology between mice and humans, with only 14 of 414 amino acids differing between the canonical sequences of the two species [63]. Despite this overlap, there are numerous physiological differences. For example, human and mouse TDP-43 are transcribed differently such that human TDP-43 has nine less mRNA transcript variants and one fewer protein isoform than mouse TDP-43 [64]. These features have important functional consequences, which are highlighted by differences in progranulin release in mouse and human cells. Progranulin release is largely dependent on the receptor protein sortilin 1, whose alternative splicing is influenced by TDP-43. In mouse cells, elimination of TDP-43 results in increased expression of Sort1ex17b, while in human cells the SORT1Δex17b isoform is increased [65]. This altered isoform ratio in the two species leads to a difference in the number of receptors detectable within the membrane, with murine cells displaying constant expression levels, while human cells show an increase in the number of receptors [65].
Current in vitro systems modeling the behavior of prion-like proteins
Non-iPSC-based models
Studies evaluating the capacity of different proteins to seed pathology have frequently used test tube-based measures such as the amyloid seeding assay (ASA) [66], real-time quaking-induced conversion (RT-QuiC) [67] and protein misfolding cyclic amplification (PMCA) [68]. Test-tube assays are valuable methods for determining whether seed-competent forms of proteins are present within biological samples, as well as measuring the kinetics of the seeding reaction [69, 70] (Fig. 1c). However, such assays are specialized towards assessment of seeding under controlled conditions and do not represent the rate of aggregate formation within cellular systems. As this review is focused on cellular models, test-tube assays will not be discussed further.
Assessment of spreading, toxicity and seeding, under physiological conditions, has traditionally been performed in primary and immortalized cells (Fig. 1c). Primary cells are isolated directly from humans or model organisms, and generally retain characteristics of their tissue of origin. Challenges with these cells include the presence of multiple cell types and low yields. Cell number is further limited by the lack of cell division of some primary cells in culture [71]. While this is representative of physiological conditions, it does prevent expansion of cells after isolation. Alternatively, several groups use immortalized cell lines [72]. These cells are homogenous and easy to grow, as they have the capacity to expand over extended periods of time, although the presence of genetic mutations may not represent normal physiology (Fig. 1c).
Despite these drawbacks, work with cell culture models has identified multiple routes by which pathogenic proteins can spread, including through the release and uptake of extracellular vesicles or soluble proteins, synapses, glial phagocytosis and tunneling nanotubes [73–75]. While the basic uptake and release of specific proteins seem to be well conserved, the rate of uptake and response differs between models depending on the tissue source and the metabolic state of the cells, with more active non-terminally differentiated precursor cells demonstrating greater uptake than mature neurons [76]. Some toxicity experiments have further shown differential susceptibility of cell lines and primary cells to toxicity induced by aggregates [77]. In genomic studies, primary cells have been compared to their immortalized equivalents and, when cell type-specific genes were considered, the R2 value approached 0.5, indicating a poor concordance between the two cell types [78]. Genes related to cell division and apoptosis were particularly affected, with a strong enrichment in immortalized cells. A similar phenomenon has been described for the human neuroblastoma cell line SH-SY5Y. SH-SY5Y cells have elevated N-Myc levels, which has been suggested to modify their response to apoptotic stimuli [79]. Such changes render toxicity studies in immortalized cells challenging. Primary cells are further problematic as availability of human primary brain cells is limited and cells from animal models can differ from their human counterparts (Fig. 1c).
Human iPSC-based systems
Pluripotent stem cells were successfully isolated and cultured from mice embryos for the first time in the early 1980s [80]. Their capacity to maintain pluripotency in cell culture, almost indefinitely, has made them an important research tool. The potential of these cells to contribute to both basic and clinical research was enhanced when a landmark study identified c-Myc, Klf4, Sox2 and Oct3/4 as the four essential transcription factors for inducing pluripotency in adult mouse somatic cells [81]. Subsequent work confirmed that these same factors induced pluripotency in adult human somatic cells [82, 83]. This discovery has led to the development of numerous protocols for the differentiation of various cell types, including cells of the CNS such as astrocytes [84], neurons [85], oligodendrocytes [86] and microglia [87].
The feasibility and utility of iPSC models to study prion-like proteins have been demonstrated in studies assessing the specific toxicity profiles of different pools of Aβ derived from control or AD patient brains [88], as well as the spontaneous aggregation of tau harboring P301L and V337M disease-associated mutations [89]. In addition to the advantages of having a model with human proteins, iPSCs can mimic multiple pathological features observed in patients, including selective vulnerability of neuronal subpopulations [90, 91], mitochondrial dysfunction [92, 93] or impairment of protein degradation pathways [94, 95]. Modeling neurodegenerative diseases with iPSC-derived cells is a powerful approach that offers a human-based system to elucidate mechanisms of disease onset and progression, including pathogenic prion-like seed formation and trans-cellular protein propagation (Fig. 1c).
Protein release, uptake and propagation in iPSC-based systems
A significant number of studies related to protein spreading have focused on characterizing the release, subsequent uptake and response of cells to pathogenic proteins; major events that contribute to protein propagation. After uptake by naive cells, pathological proteins act as seeds by interacting with homologous proteins to increase the likelihood of a shift from a normal to a pathological conformation [96]. To date, iPSCs have been used to study how disease-associated proteins such as tau [74, 97, 98], Aβ [99], α-Syn [100], mHTT [101] and SOD1 [26] are released and taken up by neighboring cells (Table 2). Unless otherwise stated, the studies reviewed in the following sections refer to human iPSCs.
Table 2.
Summary of iPSC and prion-like mechanistic studies.
 |
α-SYN alpha-synuclein, Aβ amyloid beta, AD Alzheimer’s disease, CJD Creutzfeldt-Jakob’s disease, E glutamic acid, GSS Gerstmann–Sträussler–Scheinker syndrome, GABAergic gamma aminobutyric acid expressing neurons, iPSC induced pluripotent stem cells, K lysine, mHTT mutant huntingtin, PRNP gene encoding for PrP, PrPC cellular non-pathogenic prion protein, PrPSC pathogenic prion protein, ROS reactive oxygen species, S serine, SNCA gene encoding for α-SYN, SOD1 superoxide dismutase 1, WT HTT wild-type huntingtin.
Release of pathogenic proteins
Studies assessing protein release from iPSCs have largely focused on tau with only one study addressing Aβ. Studies related to tau have demonstrated that healthy iPSC-derived neurons release both soluble and aggregated forms of the protein. The secretion of soluble forms of tau occurs independently of the classic endoplasmic reticulum and Golgi apparatus secretory pathway, and appears to be temperature-dependent [97, 98, 102]. AD and control iPSC lines have been used for these studies and both models demonstrated that release occurred independently of cell death. However, AD-related lines indicate that different forms of tau may be released under pathological conditions. Expression of an AD-relevant mutation in the Presenilin 1 (PSEN1) gene was associated with a change in the lengths of the tau fragments released into the media [98]. Similarly, iPSC lines with genetic mutations in the AD-related genes Aβ precursor protein (APP) or PSEN1 have been found to release more Aβ42 than Aβ40 [99]. While these two studies involved PSEN1, mutations in the tau repeat domain (tau-P301L-V337M) have also been shown to alter release of pathological proteins. Specifically, such cells were described to spontaneously form and secrete aggregated tau proteins [89], which is in contrast to healthy cells where the majority of released tau was non-aggregated and free-floating [98, 102, 103]. Collectively, these studies indicate that protein release is a physiological mechanism that is not specific to pathogenic processes, but that the form of the protein released can be influenced by disease-causing mutations.
Uptake of pathogenic proteins
The biology of tau uptake has also been a focus of research related to protein propagation. In particular, it has been reported that misfolded and native forms of tau are internalized to similar levels in control iPSC-derived cortical neurons, although there are some differences in the cellular machinery involved in uptake [74]. Indeed, healthy iPSC-derived excitatory cortical neurons internalize P301S tau monomers via dynamin-dependent endocytosis or macropinocytosis, while P301S tau aggregates are primarily taken up by micropinocytosis [74]. Additional studies on the mechanisms of prion-like propagation of tau have shown that exogenous tau oligomers or preformed fibrils can increase the level of pathological phosphorylated tau within healthy or MAPT P301L-expressing iPSC-derived cortical neurons, trigger a conformational change, and recruit endogenous tau to promote aggregation [104, 105] (Table 2).
Disease models using iPSCs have also contributed to the elucidation of mechanisms of uptake of SOD1 aggregates. For example, iPSC-derived motor neurons exposed to SOD1-preformed aggregates undergo Rac1-mediated membrane ruffling and blebbing characteristic of macropinocytosis, and subsequent internalization of these protein aggregates [26]. Finally, the propagation of mHTT aggregates was highlighted using an ex vivo chimeric model of HD, where organotypic brain slices were prepared from a severe transgenic mouse model of HD (R6/2) and injected with control iPSC-derived human neurons. mHTT aggregates could be transferred from mouse tissue to healthy human neurons and affect their morphology, suggesting that trans-cellular spread of mHTT from mouse to human neurons mediated neuronal impairment [101].
Propagation and associated toxicity of pathogenic proteins
While a significant amount of work has been conducted to understand the factors that influence prion-like protein release, uptake and the mechanisms mediating these processes, few studies have addressed the functional consequences of toxic proteins on iPSC survival and function. Two different methods have been utilized for these experiments; genetic mutation/overexpression and addition of exogenous pathological proteins. Studies using genetic manipulation demonstrated that SNCA duplication, E46K and E57K α-Syn, as well as mutated forms of tau, tau-A152T and tau-P301L-V337M, result in spontaneous α-Syn oligomerization or tau aggregation [89, 106, 107]. Impairments in the anterograde axonal transport of iPSC-derived cortical neurons along with synaptic degeneration accompanied α-Syn oligomerization, recapitulating elements of early neurodegeneration in synucleinopathies [106]. Similarly, iPSC-derived cortical neurons expressing tau-A152T to model FTD showed that accumulation of intracellular insoluble tau and phospho-tau (S396) correlated with neuronal vulnerability to cellular stressors, including exogenous Aβ (1–42) [107]. While the mutation of tau facilitated accumulation of pathological proteins in the aforementioned publication, not all genetic modifications studied have shown similar results. For example, iPSC-derived neurons harboring the rare Y218N PRNP mutation associated with Gerstmann-Sträussler-Scheinker syndrome internalized, but failed to replicate and propagate PrPSC [108].
Exogenous administration of tau, Aβ, α-Syn and mHTT have all been tested experimentally. Specifically, addition of tau increased aggregation/phosphorylation of the endogenous tau protein and triggered conformational changes within neurons [104, 105]. Addition of Aβ oligomers promoted the production of harmful reactive oxygen species in control cortical neurons [109]. Addition of mHTT fibrils to GABAergic cells induced neuronal atrophy and neurite shortening [25] while addition of α-Syn ribbons and fibrils caused changes in spontaneous calcium oscillations and mitochondrial morphology [110]. One very recent study further evaluated possible interactions between PrP and other prion-like proteins. iPSC-derived cortical neurons genetically edited to suppress PrP expression and their isogenic controls were exposed to α-Syn, Aβ or tau soluble aggregates, as well as pathogenic human brain lysates from AD, Pick’s disease or dementia with Lewy bodies. Cells lacking PrP had no neurodegenerative phenotypes, while isogenic controls exhibited functional and morphological alterations [111]. These intriguing results suggest that PrP mediates, at least in part, the toxicity triggered by other prion-like proteins and could serve as a therapeutic target for a number of neurodegenerative diseases. Together, these studies provide strong support for the use of iPSC models to study the effects of pathological proteins on human neurons by mirroring phenotypes observed in disease settings.
Glia and pathological proteins
Neuron-to-neuron spread of prion-like proteins is an essential aspect of pathology, but evidence suggests that microglia and astrocytes may also play a role, either through attempted degradation of proteins or via glia-to-neuron or glia-to-glia transfer.
Glia-mediated protein clearance
Phagocytic abilities of microglia and astrocytes are essential to maintain tissue homeostasis and several studies using non-iPSC models, as well as in vivo models, have demonstrated their ability to recognize and uptake α-Syn [112–115] tau [116–118], Aβ [119, 120] or mHTT [121] aggregates. Recently, studies using iPSC-based systems have also contributed to this body of work by demonstrating that expression of APOE4 [122], the PSEN1ΔE9 mutation [123], a homozygous KM670/671NL mutation in APP [124] or knockout of APP [124] is sufficient to disrupt uptake of Aβ1-42 by astrocytes. Furthermore, astrocytes expressing the PSEN1ΔE9 mutation not only lost their ability to uptake the Aβ1–42 fragment, but also increased the number of Aβ fragments they released into the media [123].
Few studies on the functional consequences of disease-related mutations on iPSC-derived microglia are available. However, two recent reports have shown that microglia differentiated from iPSCs expressing either the APOE4 variant or containing a mutated C9orf72 gene, a known risk factor for ALS and FTD, have a reduced ability to degrade internalized Aβ1-40 fragments, leading to pathological Aβ1-40 accumulation [122, 125]. When internalized by a recipient glial cell, pathological proteins can accumulate, form inclusion bodies and subsequently trigger a series of cellular dysfunctions that further drive the propagation process.
Protein spread
In addition to their role in protein clearance, microglia and astrocytes may also exacerbate pathological protein spread by participating in cell-to-cell transfer. Embryonic stem cell-derived astrocytes exposed to aggregated α-Syn produce tunneling nanotubes to communicate with neighboring healthy astrocytes and recover healthy mitochondria from these cells. This process results in the passage of accumulated α-Syn from the damaged cell to the healthy neighbor [126]. Exposure of iPSC-derived astrocytes with different PRNP variants to prions originating from the brain of CJD patients expressing the same PRNP variant also results in the internalization, self-propagating replication and accumulation of PrPsc [127]. Lysates prepared from PrPsc-positive astrocytes infect and propagate the PrPsc pathology to naive astrocytes. In addition to passing pathological proteins to other astrocytes, iPSC-derived astrocytes with the PD-related mutation LRRK2 G2019S convey endogenous α-Syn to iPSC-derived dopaminergic neurons [91]. While studies on transfer between glia and other cells is limited, the evidence to date suggests that they may be important contributors and more models incorporating multiple cell types could provide valuable information.
Idiopathic vs. familial etiology
A unique application of iPSC-based technologies is to compare the role and behavior of pathological proteins in idiopathic vs. familial forms of neurodegenerative diseases. For example, studies of sporadic and familial AD have shown differences in the levels of phospho tau (Thr 231), secreted Aβ40 and Aβ42/Aβ40 ratios [128, 129]. While few changes were reported in familial vs. sporadic neuronal cultures, a greater Aβ42/Aβ40 ratio in familial AD was observed. For example, motor neurons differentiated from 32 sporadic ALS-derived iPSCs (e.g., SOD1 and TDP-43 mutants) displayed shorter neurites, higher levels of lactate dehydrogenase and abnormal production of protein aggregates as compared to control neurons [130]. However, the severity of these phenotypes varied between different sporadic patients, and overall the cells did not recapitulate the SOD1 protein aggregation observed in SOD1 familial ALS, but instead demonstrated formation of cytosolic TDP-43 aggregates. This study therefore indicates that sporadic ALS can be modeled using iPSC-based technologies and that these systems do not necessarily match phenotypes from familial forms of disease. Such methods have the capacity to not only increase our understanding of disease mechanisms, but also to screen for potential drug candidates that mitigate neurodegeneration across all forms of ALS.
While such investigations are still nascent, they show great promise towards improving our understanding of the genetic determinants of protein spreading and seeding and offer unique models to design personalized medicine. Genetic-based models may not be representative of the more common idiopathic forms of neurodegenerative diseases and iPSC systems could provide a platform to model idiopathic disease as well. However, further validation of iPSC models generated from idiopathic cases will be required to confirm that they accurately recapitulate pathology across the spectrum of etiologies and neurodegenerative diseases.
Challenges and perspectives of iPSC-derived models
Biochemical maturation and aging of iPSC-derived neurons
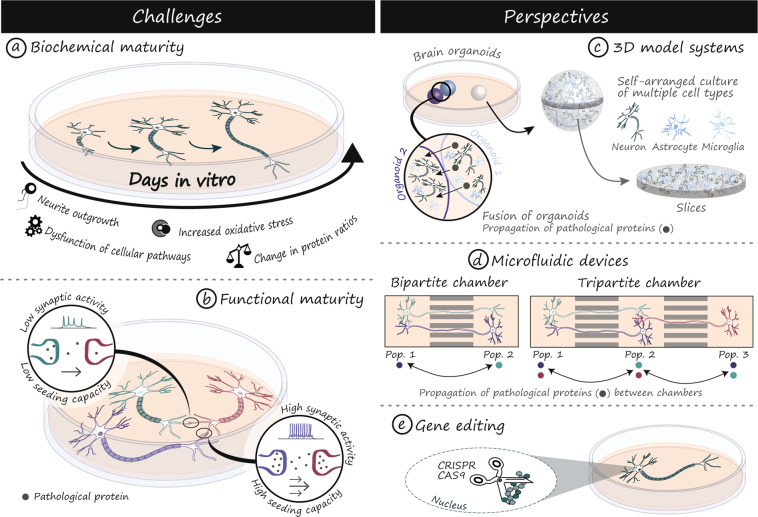
Functional maturity and age-related cellular changes are paramount considerations when studying prion-like proteins (Fig. 2), as some proteins, such as tau (Table 1), are characterized by changes in splicing or expression throughout development [42]. Since iPSC-derived neurons show an immature phenotype, they tend to express more juvenile forms of proteins including predominant expression of 3R tau [131]. Although iPSCs are generally derived from adult cells, the process of reprogramming resets markers of cellular age such that the cells effectively return to an embryonic state [132]. In addition to altering the pattern of expression of proteins such as tau, age is the greatest risk factor for developing neurodegenerative disorders. While the mechanisms are still unknown, age has been associated with increased protein spreading [133]. To some extent, co-culture with glia, 3D-culturing and increased time in culture can overcome these limitations but these may not be sufficient to fully replicate processes that take decades [131, 134]. An alternative emerging technology involves the expression of progerin, a mutated form of lamin A responsible for the premature aging phenotype observed in patients suffering from the Hutchinson-Gilford progeria syndrome. When progerin-related aging is induced in dopaminergic neurons derived from PD patients, the cells acquire characteristic disease phenotypes, including altered morphology and protein inclusions [135]. Another interesting approach involves the direct conversion of adult cells into neurons, thereby avoiding a pluripotent intermediate stage [136–139]. This method has been shown, in the case of HD, to generate induced neurons which retain markers of cellular age and display stronger disease phenotypes, including overt neuronal degeneration and protein aggregation [140]. While this technology is still quite novel, there are indications that it may more faithfully mimic age-dependent molecular mechanisms of degeneration and could therefore provide new insights into the processes of protein release, uptake and seeding.
Fig. 2. Challenges and perspectives of iPSCs for modeling proteinopathies.
Neurons differentiated from iPSCs tend to have an immature phenotype, which can impact cellular functions critical to the successful modeling of prion-like protein propagation and seeding (a, b). New techniques such as brain organoids (c), microfluidic devices (d) and CRISPR/Cas9 gene editing (e) are important tools to overcome these limitations and further establish iPSCs as a model of choice to study the properties and role of prion-like proteins in neurodegenerative diseases. Cas9 CRISPR associated protein 9, CRISPR Clustered Regularly Interspaced Short Palindromic Repeats, Pop. 1 population 1, Pop. 2 population 2, Pop. 3 population 3, 3D three-dimensional.
As the field moves forward, models that consistently display spontaneous formation of protein aggregates will become invaluable resources to mimic disease phenotypes and elucidate early events responsible for protein misfolding and formation of pathogenic seeds. Spontaneous formation of intracellular misfolded/aggregated proteins has been observed in both sporadic and familial forms of disease [89, 106, 107, 141], however, these results have been variable. In one study, sixteen lines of iPSC with five clones each were generated from sporadic ALS and only three of them displayed TDP-43 protein aggregation, while none of the eight lines with SOD1 mutations generated detectable aggregates [141]. Even in the lines that displayed aggregation, this was not detected in every clonal line from the same patient [141]. In AD, a review citing fourteen different iPSC studies, the majority of which used familial forms of the disease, reported oligomers or aggregates in only four of the studies [142]. Together, the available literature suggests that detection of protein aggregation does not readily occur in early iPSC-derived neurons and currently requires large numbers of patients and clones, which is beyond the capabilities of many groups. These challenges could be at least partially overcome by implementing the maturation strategies delineated above.
Functional maturation of iPSC-derived cell types
Neuronal activity is an important parameter that regulates the propagation of pathogenic prion-like proteins such as tau [143, 144], Aβ [145] and α-Syn [146, 147]. For example, activation of AMPA receptors, release of pre-synaptic glutamate, active synapses and synaptic contacts are important mechanisms regulating the trans-synaptic and trans-cellular propagation of tau [144, 148, 149]. To model prion-like protein propagation using iPSC-derived neurons, it is critical to ensure that the cells produced display appropriate physiological electrical properties (Fig. 2). Methods commonly used to determine the functional maturity of neurons differentiated from iPSCs include whole-cell patch-clamp recordings, electrode arrays and assessment of functional receptors (NMDA, AMPA, GABAA) by calcium imaging [150, 151]. At early stages of differentiation, iPSC-derived neurons have an immature electrophysiological phenotype, but in vitro maturation and formation of functional neural networks can be achieved by extending the time in culture [150, 151]. The method used to produce neurons has important consequences on electrophysiological maturity. An alternative approach using NGN2-mediated neuronal induction of iPSCs coupled with glia co-culture demonstrated significant electrical maturity as early as 14 days post-induction [152, 153]. In addition, direct conversion of fibroblasts to neurons results in populations that tend to lack spontaneous and evoked neurotransmission and have passive membrane properties similar to immature neurons after 3–4 weeks in culture [154]. However, electrically active induced neurons can be obtained by increasing the time in culture to 85–100 days and co-culturing them with glia [140, 155, 156]. Therefore, ensuring the functional maturity of neurons is critical to accurately recapitulate disease progression and protein propagation.
Astrocyte maturity is an equally important aspect of disease modeling, as an increasing number of studies implicate astrocytes in the progression of neurodegenerative disorders [91, 126, 157, 158]. Several astrocyte differentiation protocols have been established, but they differ in the timing (4-weeks to several months) and nature of the differentiation cocktail [159]. Overall, these protocols produce cells that express known markers of astrocytes (e.g. GFAP, S100β), respond to inflammatory stimuli, support neuronal growth and enhance neuron survival [159–163]. However, they fail to recapitulate the complex stellate morphology composed of branches and fine processes typical of astrocytes in vivo. In contrast to neurons, the molecular and functional characteristics of mature astrocyte and the impact of aging on their neuroprotective or neurotoxic properties have not been extensively studied. Given the essential role of astrocytes in the maintenance of appropriate neuronal functions and immune processes of the brain, maturity may have important ramifications in regards to the spreading of pathological proteins.
Genomic variability
Disease modeling using iPSCs is often intended to leverage a patient’s genetic background to induce a disease phenotype. However, while highly penetrant disease-associated mutations may drive strong in vitro phenotypic alterations, genetic variants with a low penetrance or small allelic fold change could result in smaller effects that may be masked by the inter-individual or reprogramming-induced variability intrinsic to iPSC-based models [164]. For example, it has been estimated that the comparison of iPSC-derived cells prepared from 20 to 80 unrelated individuals would be necessary to accurately recapitulate phenotypes induced by variants with small allelic fold changes [165]. However, implementing studies with large cohorts of patient-derived cells is costly and technically challenging, which has consequently limited most reports to a small number of iPSC lines.
Microenvironment and connectivity
In their native brain tissue, neurons and non-neuronal cells evolve in a complex 3D microenvironment where cell-extracellular matrix interactions, direct cell-cell contacts and paracrine signals mediate tissue homeostasis, cellular functions, and survival [166–168]. While organoids provide a technological advance in 3D modeling, they more accurately represent a developing rather than a mature CNS tissue [169] and lack functional and structural brain connectivity, two features associated with clinical phenotypes in neurodegenerative diseases [170]. Furthermore, organoids do not contain vasculature that could promote long-term survival and maturation via oxygen and nutrient distribution, as well as through direct contributions of endothelial cells to organogenesis [171]. A number of groups have recently reported the successful integration of vascular-like networks in cultured organoids [172–175], which hold great promises for the future development of increasingly complex 3D models of the CNS. In the following section, we will discuss in more details the potential of organoids as a new tool to model prion-like protein propagation.
Modeling the behavior of prion-like proteins in complex systems
iPSC-derived organoids
iPSCs self-organize and form 3D human-like cerebral tissue in vitro (Fig. 2). These spheroid structures, or organoids, can be patterned to produce regionalized in vitro human brain tissue including the midbrain [176], neocortex [177] and even a complex cerebral-like circuit composed of interconnected, but distinct brain regions [178]. The unique organization of the human brain has made the modeling of human diseases in mice difficult, but these new in vitro culture systems have the potential to overcome such limitations. For example, 3D cerebral-like organoids can recapitulate features of human brain development with the presence of cell layers of the subventricular zone unique to the human brain and absent in mice [178]. Microcephaly has traditionally been difficult to study in mice, possibly because of the different behaviors of human and mice neural progenitor cells. However, cerebral-like organoids successfully modeled this human-specific disorder [178], highlighting the powerful potential of 3D tissue models to study neurological disorders. Human cerebral-like organoids are multicellular structures comprising progenitors, neurons, astrocytes and oligodendrocytes [179–181]. Microglia originate from a different developmental lineage than neuronal cells and are consequently, often absent in organoids. Interestingly, recent studies reported the presence of microglia that developed within organoids, suggesting that adaptations of existing protocols can be implemented to obtain organoids comprising various cell populations [180, 182]. Furthermore, microglia can be differentiated separately and added exogenously to the cultured organoids. These microglia penetrate organoids and acquire in vivo-like functions, such as the ability to clear Aβ aggregates [122]. While organoids are a powerful new tool, it should be noted that the complexity of the model tends to lead to significant variability in the response to experimental conditions, which generally requires the analysis of an increased number of samples to ensure the reproducibility and validity of the results.
The potential of organoids can be harnessed to study prion-like proteins and mechanisms of propagation. For example, control cerebral organoids can be inoculated with brain homogenates prepared from CJD patients and display features indicative of PrP infectivity, self-seeding and propagation [183]. Future studies could also fuse different brain organoids to produce fully connected structures that support cell migration and material exchange from one organoid to another [184, 185]. Such systems could be used to monitor the propagation of prion-like proteins from a source to a naive organoid to determine the cell types involved, the resulting modifications in 3D morphology and any potential tissue reorganization. Changes occurring within an organoid can be monitored by live-cell imaging either in a full organoid [186] or in organoid slices [187] (Fig. 2). Live-cell imaging in whole organoids requires the use of bright fluorescent reporter proteins to compensate from the loss of signal intensity due to the considerable gap between the objective lens and the structure of interest and the presence of a gel-like extracellular matrix surrounding the organoid [186]. Several non-invasive optical sectioning microscopes such as spinning disk confocal [186], two-photon [188] and light-sheet microscopes [189, 190] have been tested and ensured low phototoxicity, rapid image acquisition and sufficient sensitivity to enable appropriate spatial and temporal resolution. End-point 3D imaging of fixed whole organoids is also possible and can be improved by optical clearing techniques [191]. Whole organoid 3D imaging is a powerful approach to visualize tissue architecture and cell-cell interactions that are lost in 2D cultures and sectioned tissues. A combination of organoid-based models and advanced microscopy techniques are powerful, yet unexplored tools, to study prion-like protein propagation and associated cellular dysfunction in a unique, human-based multicellular 3D model.
Microfluidic technologies
Axon isolation chambers are ideal devices to study mechanisms of prion-like protein propagation and uptake by isolating neuronal somas, axons and synaptic compartments in separated, but interconnected chambers (Fig. 2). These devices can be designed to accommodate several populations of neurons in bi- and tripartite chambers that are independent but synaptically interconnected through microgroove arrays [143, 149] (Fig. 2). Pathogenic seeds of prion-like proteins can be added to a fluidically isolated chamber without diffusing to the other compartments. Microfluidic devices can be used to address a unique range of biological questions such as elucidating the effects of prion-like proteins in distinct compartments of the neurons, the mechanisms and direction of their axonal transport [192], as well as to discover antibodies or screen for inhibitors of trans-neuronal protein propagation [193–195]. These devices have been extensively used to demonstrate in vitro trans-cellular propagation of proteins including α-Syn [192, 196] and tau [143, 149, 197] in primary mouse neuronal cultures. For example, it was found that α-Syn (1–120) preformed fibrils propagate to distal parts of the neuronal somas and compartmentalized neurites. When somas are treated with preformed fibrils, phosphorylated α-Syn (p-α-Syn) aggregates are found in the soma followed by propagation to the neurites. When neurites are treated with the preformed fibrils, p-α-Syn pathology is identified in the neurites followed by propagation back to the soma where aggregates are formed [192]. Other studies showed that tau seeds can be transferred from a donor to a recipient neuron via anterograde transport from the soma [198], and in a tripartite chamber device, tau pathology propagates from the first population of neurons to a second and third population in a trans-synaptic fashion [143]. Overall, the use of microfluidic axon isolation devices has revealed important information on the propagation potential of different species of prion-like proteins and their modes of transport. Future experiments with human iPSC-derived neurons will hopefully confirm these findings in a human model as well as provide an opportunity to study how cells derived from patients with neurodegenerative diseases impact the findings. Further progress can also be made via the coupling of microfluidics devices with advanced microscopy techniques to resolve spatial and temporal events leading to pathology. One example of this is a recent study that cultured motor neurons derived from ALS patient iPSCs in a microfluidic device to understand how organelle trafficking rate and direction was changed in these cells [199]. The coupling of this system with live-cell microscopy permitted isolation of distal and proximal regions of the axons [199]. To further determine the mechanisms of protein propagation, axon isolation chambers partnered with microelectrode arrays and iPSC-derived neurons will be powerful systems that could be used to study the propagation of prion-like proteins in response to neuronal stimulation [200].
Combining gene editing with iPSC modeling
Gene editing technologies are powerful methods to manipulate a gene of interest (e.g. gene knockdown) or to visualize a protein (e.g. tagged endogenous protein). Among the multiple gene editing technologies available, such as Zinc Finger Nuclease (ZFN), Transcription Activator-Like Effector Nuclease (TALEN) and Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR), CRISPR offers distinct advantages such as broad applicability, low cost, relative ease of use and rapid implementation [201–203]. The CRISPR system is well suited to the study of prion proteins and has been adopted as the gene-editing method of choice in studies that address a diversity of biological questions [91, 204, 205] (Fig. 2). It enables the insertion of fluorescent protein tags to endogenous proteins of interest and alleviates the need to overexpress tagged proteins that could result in protein mislocalization or misfolding, protein imbalance and possibly, dysregulation of cellular processes [201, 206]. Fluorescent labeling of prion-like proteins is a widely used approach to study their behavior, functions and modes of propagation [207]. However, methods using viral vectors or plasmid transfection to transiently or stably express the exogenously tagged protein of interest lead to protein overexpression. To overcome limitations associated with overexpression, CRISPR/Cas9 was used to introduce a FLAG tag into the endogenous SNCA locus, leading to the production of cells that expresses FLAG-tagged α-Syn at endogenous levels [91]. The CRISPR system can also be used to identify proteins involved in a pathway of interest by modulating their endogenous expression levels using CRISPR interference (CRISPRi) and CRISPR activation (CRISPRa) of single genes, or as a screening strategy [208]. Greater silencing efficiencies can be obtained by CRISPRi compared to RNA interference (RNAi), as demonstrated by an incomplete 40% knockdown of mHTT by RNAi [209] and a complete knockdown with CRISPRi [210]. Furthermore, CRISPRi can be used as a screening strategy to uncover pathways essential to the prion-like propagation of proteins. For example, a CRISPRi screen in an immortalized cell line successfully identified key proteins and cellular pathways that mediate tau propagation [205]. Other prion-like proteins, such as α-Syn, have been successfully deleted [211] or tagged [91] in iPSC lines using CRISPR-based technologies to characterize their function and identify drug candidates.
Another important advantage of gene-editing technologies in the study of prion-like proteins in iPSC-based model is the generation of isogenic lines, i.e. the correction of disease-causing mutations to produce mutant and control lines that differ only at the specific mutation of interest. The implementation of isogenic controls ensures genetically defined conditions, particularly relevant to the study of genetic forms of neurodegenerative diseases. Gene correction has been applied successfully to repair mutations in prion-like proteins, including the N276K mutation in tau [212], abnormal CAG repeats in mHTT [210] and SNCA triplication [213]. Alternatively, an iPSC line originating from a healthy donor can be mutated at a specific locus to produce a disease iPSC line on a control background [214, 215]. However, it should be noted that gene editing of iPSCs has also proven somewhat challenging, with limited uptake, low editing efficiency and significant cell death associated with transfection of the plasmid DNA elements required for CRISPR/Cas9-mediated gene correction. Furthermore, a majority of the corrections tend to be monoallelic and require a second round of manipulations to achieve biallelic gene editing [216, 217]. In the past few years, new strategies have emerged to improve iPSC survival, reduce off-target mutations and increase the frequency of biallelic gene corrections. Recent examples using CRISPR/Cas9 gRNA ribonucleoprotein complexes showed that directly delivering precomplexed Cas9-gRNA, instead of plasmid DNA, has the potential to solve specific technical challenges associated with the editing of iPSCs [216, 218].
Conclusions
To date, iPSCs have frequently been used to validate the physiological relevance of findings from cell lines and to investigate the different mechanisms involved in various aspects of prion-like protein spreading and seeding. While the resulting findings have been informative, challenges related to the immaturity of neurons and the absence of supportive non-neuronal cells remain. New technological advances such as 3D modeling and induced neurons will likely contribute to overcome these challenges and advance our understanding of prion-like mechanisms of neurodegeneration. Furthermore, human iPSC-based models provide unlimited access to vulnerable neuronal populations that were traditionally unavailable and offer a human-specific background to study the properties of proteins of interest. In light of the current literature and despite limitations such as neuronal immaturity and the absence of a native microenvironment, iPSC-based models provide exciting perspectives and unique advantages to elucidate the role of pathological prion-like proteins in neurodegenerative diseases and the molecular mechanisms regulating propagation and neurotoxicity.
Acknowledgements
FC is a recipient of a Researcher Chair from the Fonds de Recherche du Québec en Santé (FRQS) providing salary support and operating funds and receives funding from the Canadian Institutes of Health Research (CIHR) to conduct her HD-related research. MA is supported by post-doctoral fellowships from both CIHR and FRQS and HLD is supported by an FRQS doctoral research award.
Author contributions
ARJ and MA researched data for the article and wrote the manuscript. HLD designed the figures and tables. FC contributed to discussions and helped edit the manuscript. All authors reviewed the content before publication.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
The original online version of this article was revised due to a retrospective Open Access order.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
3/9/2021
A Correction to this paper has been published: 10.1038/s41380-021-01055-8
Contributor Information
Francesca Cicchetti, Email: francesca.cicchetti@crchudequebec.ulaval.ca.
Melanie Alpaugh, Email: melanie.alpaugh@crchudequebec.ulaval.ca.
References
- 1.Das AS, Zou WQ. Prions: beyond a single protein. Clin Microbiol Rev. 2016;29:633–58. doi: 10.1128/CMR.00046-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prusiner SB, Groth DF, Cochran SP, Masiarz FR, McKinley MP, Martinez HM. Molecular properties, partial purification, and assay by incubation period measurements of the hamster scrapie agent. Biochem. 1980;19:4883–91. doi: 10.1021/bi00562a028. [DOI] [PubMed] [Google Scholar]
- 3.Ironside JW. Neuropathological findings in new variant CJD and experimental transmission of BSE. FEMS Immunol Med Mic. 1998;21:91–95. doi: 10.1016/S0928-8244(98)00020-0. [DOI] [PubMed] [Google Scholar]
- 4.Lee CC, Nayak A, Sethuraman A, Belfort G, McRae GJ. A three-stage kinetic model of amyloid fibrillation. Biophys J. 2007;92:3448–58. doi: 10.1529/biophysj.106.098608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ahn TB, Langston JW, Aachi VR, Dickson DW. Relationship of neighboring tissue and gliosis to alpha-synuclein pathology in a fetal transplant for Parkinson’s disease. Am J Neurodegen Dis. 2012;1:49–59. [PMC free article] [PubMed] [Google Scholar]
- 6.Chu Y, Kordower JH. Lewy body pathology in fetal grafts. Ann NY Acad Sci. 2010;1184:55–67. doi: 10.1111/j.1749-6632.2009.05229.x. [DOI] [PubMed] [Google Scholar]
- 7.Kordower JH, Chu Y, Hauser RA, Freeman TB, Olanow CW. Lewy body-like pathology in long-term embryonic nigral transplants in Parkinson’s disease. Nat Med. 2008;14:504–6. doi: 10.1038/nm1747. [DOI] [PubMed] [Google Scholar]
- 8.Li JY, Englund E, Holton JL, Soulet D, Hagell P, Lees AJ, et al. Lewy bodies in grafted neurons in subjects with Parkinson’s disease suggest host-to-graft disease propagation. Nat Med. 2008;14:501–3. doi: 10.1038/nm1746. [DOI] [PubMed] [Google Scholar]
- 9.Braak H, Del Tredici K, Rub U, de Vos RA, Jansen Steur EN, Braak E. Staging of brain pathology related to sporadic Parkinson’s disease. Neurobiol Aging. 2003;24:197–211. doi: 10.1016/S0197-4580(02)00065-9. [DOI] [PubMed] [Google Scholar]
- 10.Luk KC, Kehm VM, Zhang B, O’Brien P, Trojanowski JQ, Lee VM. Intracerebral inoculation of pathological alpha-synuclein initiates a rapidly progressive neurodegenerative alpha-synucleinopathy in mice. J Exp Med. 2012;209:975–86. doi: 10.1084/jem.20112457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Recasens A, Dehay B, Bove J, Carballo-Carbajal I, Dovero S, Perez-Villalba A, et al. Lewy body extracts from Parkinson disease brains trigger alpha-synuclein pathology and neurodegeneration in mice and monkeys. Ann Neurol. 2014;75:351–62. doi: 10.1002/ana.24066. [DOI] [PubMed] [Google Scholar]
- 12.Dehay B, Bezard E. Intrastriatal injection of alpha-synuclein fibrils induces Parkinson-like pathology in macaques. Brain. 2019;142:3321–2. doi: 10.1093/brain/awz329. [DOI] [PubMed] [Google Scholar]
- 13.Kim S, Kwon SH, Kam TI, Panicker N, Karuppagounder SS, Lee S, et al. Transneuronal propagation of pathologic alpha-synuclein from the gut to the brain models Parkinson’s disease. Neuron. 2019;103:627–41 e627. doi: 10.1016/j.neuron.2019.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Challis C, Hori A, Sampson TR, Yoo BB, Challis RC, Hamilton AM, et al. Gut-seeded alpha-synuclein fibrils promote gut dysfunction and brain pathology specifically in aged mice. Nat Neurosci. 2020;23:327–36. doi: 10.1038/s41593-020-0589-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Groveman BR, Orru CD, Hughson AG, Raymond LD, Zanusso G, Ghetti B, et al. Rapid and ultra-sensitive quantitation of disease-associated alpha-synuclein seeds in brain and cerebrospinal fluid by alphaSyn RT-QuIC. Acta Neuropathol Commun. 2018;6:7. doi: 10.1186/s40478-018-0508-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singh S, DeMarco ML. In vitro conversion assays diagnostic for neurodegenerative proteinopathies. JALM. 2020;5:142–157. doi: 10.1373/jalm.2019.029801. [DOI] [PubMed] [Google Scholar]
- 17.Braak H, Braak E, Bohl J. Staging of Alzheimer-related cortical destruction. Eur Neurol. 1993;33:403–8. doi: 10.1159/000116984. [DOI] [PubMed] [Google Scholar]
- 18.Meyer-Luehmann M, Coomaraswamy J, Bolmont T, Kaeser S, Schaefer C, Kilger E, et al. Exogenous induction of cerebral beta-amyloidogenesis is governed by agent and host. Science. 2006;313:1781–4. doi: 10.1126/science.1131864. [DOI] [PubMed] [Google Scholar]
- 19.Gary C, Lam S, Herard AS, Koch JE, Petit F, Gipchtein P, et al. Encephalopathy induced by Alzheimer brain inoculation in a non-human primate. Acta Neuropathol Commun. 2019;7:126. doi: 10.1186/s40478-019-0771-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clavaguera F, Akatsu H, Fraser G, Crowther RA, Frank S, Hench J, et al. Brain homogenates from human tauopathies induce tau inclusions in mouse brain. PNAS. 2013;110:9535–40. doi: 10.1073/pnas.1301175110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Porta S, Xu Y, Restrepo CR, Kwong LK, Zhang B, Brown HJ, et al. Patient-derived frontotemporal lobar degeneration brain extracts induce formation and spreading of TDP-43 pathology in vivo. Nat Commun. 2018;9:4220. doi: 10.1038/s41467-018-06548-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lasagna-Reeves CA, Castillo-Carranza DL, Sengupta U, Clos AL, Jackson GR, Kayed R. Tau oligomers impair memory and induce synaptic and mitochondrial dysfunction in wild-type mice. Mol Neurodegen. 2011;6:39. doi: 10.1186/1750-1326-6-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mulder CK, Dong Y, Brugghe HF, Timmermans HA, Tilstra W, Westdijk J, et al. Immunization with small amyloid-beta-derived cyclopeptide conjugates diminishes amyloid-beta-induced neurodegeneration in mice. J Alzheimer’s Dis. J Alzheimer’s Dis. 2016;52:1111–23. doi: 10.3233/JAD-151136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Polinski NK, Volpicelli-Daley LA, Sortwell CE, Luk KC, Cremades N, Gottler LM, et al. Best practices for generating and using Alpha-synuclein pre-formed fibrils to model Parkinson’s disease in rodents. J Parkinson’s Dis. 2018;8:303–22. doi: 10.3233/JPD-171248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Masnata M, Sciacca G, Maxan A, Bousset L, Denis HL, Lauruol F, et al. Demonstration of prion-like properties of mutant huntingtin fibrils in both in vitro and in vivo paradigms. Acta Neuropathol. 2019;137:981–1001. doi: 10.1007/s00401-019-01973-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zeineddine R, Pundavela JF, Corcoran L, Stewart EM, Do-Ha D, Bax M, et al. SOD1 protein aggregates stimulate macropinocytosis in neurons to facilitate their propagation. Mol Neurodegen. 2015;10:57. doi: 10.1186/s13024-015-0053-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blesa J, Przedborski S. Parkinson’s disease: animal models and dopaminergic cell vulnerability. Front Neuroanat. 2014;8:155. doi: 10.3389/fnana.2014.00155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dawson TM, Golde TE, Lagier-Tourenne C. Animal models of neurodegenerative diseases. Nat Neurosci. 2018;21:1370–9. doi: 10.1038/s41593-018-0236-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holley SM, Kamdjou T, Reidling JC, Fury B, Coleal-Bergum D, Bauer G, et al. Therapeutic effects of stem cells in rodent models of Huntington’s disease: Review and electrophysiological findings. CNS Neurosci Ther. 2018;24:329–42. doi: 10.1111/cns.12839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jankowsky JL, Zheng H. Practical considerations for choosing a mouse model of Alzheimer’s disease. Mol Neurodegen. 2017;12:89. doi: 10.1186/s13024-017-0231-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oberheim NA, Takano T, Han X, He W, Lin JH, Wang F, et al. Uniquely hominid features of adult human astrocytes. J Neurosci. 2009;29:3276–87. doi: 10.1523/JNEUROSCI.4707-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han X, Chen M, Wang F, Windrem M, Wang S, Shanz S, et al. Forebrain engraftment by human glial progenitor cells enhances synaptic plasticity and learning in adult mice. Cell Stem Cell. 2013;12:342–53. doi: 10.1016/j.stem.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y, Sloan SA, Clarke LE, Caneda C, Plaza CA, Blumenthal PD, et al. Purification and characterization of progenitor and mature human astrocytes reveals transcriptional and functional differences with mouse. Neuron. 2016;89:37–53. doi: 10.1016/j.neuron.2015.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rayon T, Stamataki D, Perez-Carrasco R, Garcia-Perez L, Barrington C, Melchionda M, et al. Species-specific pace of development is associated with differences in protein stability. Science. 2020;369:eaba7667. doi: 10.1126/science.aba7667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prusiner SB. Prions. PNAS. 1998;95:13363–83. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barria MA, Balachandran A, Morita M, Kitamoto T, Barron R, Manson J, et al. Molecular barriers to zoonotic transmission of prions. Emerg Infect Dis. 2014;20:88–97. doi: 10.3201/eid2001.130858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wiseman FK, Cancellotti E, Piccardo P, Iremonger K, Boyle A, Brown D, et al. The glycosylation status of PrPC is a key factor in determining transmissible spongiform encephalopathy transmission between species. J Virol. 2015;89:4738–47. doi: 10.1128/JVI.02296-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hasegawa K, Mohri S, Yokoyama T. Comparison of the local structural stabilities of mammalian prion protein (PrP) by fragment molecular orbital calculations. Prion. 2013;7:185–91. doi: 10.4161/pri.23122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanchez-Garcia J, Fernandez-Funez P. D159 and S167 are protective residues in the prion protein from dog and horse, two prion-resistant animals. Neurobiol Dis. 2018;119:1–12. doi: 10.1016/j.nbd.2018.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barron RM, Thomson V, Jamieson E, Melton DW, Ironside J, Will R, et al. Changing a single amino acid in the N-terminus of murine PrP alters TSE incubation time across three species barriers. EMBO J. 2001;20:5070–8. doi: 10.1093/emboj/20.18.5070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Korth C, Kaneko K, Groth D, Heye N, Telling G, Mastrianni J, et al. Abbreviated incubation times for human prions in mice expressing a chimeric mouse-human prion protein transgene. PNAS. 2003;100:4784–9. doi: 10.1073/pnas.2627989100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hernandez F, Merchan-Rubira J, Valles-Saiz L, Rodriguez-Matellan A, Avila J. Differences between human and murine tau at the N-terminal end. Front Aging Neurosci. 2020;12:11. doi: 10.3389/fnagi.2020.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hernandez F, Cuadros R, Olla I, Garcia C, Ferrer I, Perry G, et al. Differences in structure and function between human and murine tau. Biochim Biophys Acta Mol Basis Dis. 2019;1865:2024–30. doi: 10.1016/j.bbadis.2018.08.010. [DOI] [PubMed] [Google Scholar]
- 44.Stanford PM, Shepherd CE, Halliday GM, Brooks WS, Schofield PW, Brodaty H, et al. Mutations in the tau gene that cause an increase in three repeat tau and frontotemporal dementia. Brain. 2003;126(Pt 4):814–26. doi: 10.1093/brain/awg090. [DOI] [PubMed] [Google Scholar]
- 45.Hutton M, Lendon CL, Rizzu P, Baker M, Froelich S, Houlden H, et al. Association of missense and 5′-splice-site mutations in tau with the inherited dementia FTDP-17. Nature. 1998;393:702–5. doi: 10.1038/31508. [DOI] [PubMed] [Google Scholar]
- 46.Metrick MA, II, Ferreira NDC, Saijo E, Kraus A, Newell K, Zanusso G, et al. A single ultrasensitive assay for detection and discrimination of tau aggregates of Alzheimer and Pick diseases. Acta Neuropathol Commun. 2020;8:22. doi: 10.1186/s40478-020-0887-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hu WT, Parisi JE, Knopman DS, Boeve BF, Dickson DW, Ahlskog JE, et al. Clinical features and survival of 3R and 4R tauopathies presenting as behavioral variant frontotemporal dementia. Alzheimer Dis Assoc Disord. 2007;21:S39–43. doi: 10.1097/WAD.0b013e31815bf5e5. [DOI] [PubMed] [Google Scholar]
- 48.Dinkel PD, Siddiqua A, Huynh H, Shah M, Margittai M. Variations in filament conformation dictate seeding barrier between three- and four-repeat tau. Biochemistry. 2011;50:4330–6. doi: 10.1021/bi2004685. [DOI] [PubMed] [Google Scholar]
- 49.Levarska L, Zilka N, Jadhav S, Neradil P, Novak M. Of rodents and men: the mysterious interneuronal pilgrimage of misfolded protein tau in Alzheimer’s disease. J Alzheimer’s Dis. 2013;37:569–77. doi: 10.3233/JAD-131106. [DOI] [PubMed] [Google Scholar]
- 50.Sayas CL, Medina M, Cuadros R, Olla I, Garcia E, Perez M, et al. Role of tau N-terminal motif in the secretion of human tau by End Binding proteins. PLoS ONE. 2019;14:e0210864. doi: 10.1371/journal.pone.0210864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ueno H, Yamaguchi T, Fukunaga S, Okada Y, Yano Y, Hoshino M, et al. Comparison between the aggregation of human and rodent amyloid beta-proteins in GM1 ganglioside clusters. Biochemistry. 2014;53:7523–30. doi: 10.1021/bi501239q. [DOI] [PubMed] [Google Scholar]
- 52.Jankowsky JL, Younkin LH, Gonzales V, Fadale DJ, Slunt HH, Lester HA, et al. Rodent A beta modulates the solubility and distribution of amyloid deposits in transgenic mice. J Biol Chem. 2007;282:22707–20. doi: 10.1074/jbc.M611050200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kane MD, Lipinski WJ, Callahan MJ, Bian F, Durham RA, Schwarz RD, et al. Evidence for seeding of beta -amyloid by intracerebral infusion of Alzheimer brain extracts in beta -amyloid precursor protein-transgenic mice. J Neurosci. 2000;20:3606–11. doi: 10.1523/JNEUROSCI.20-10-03606.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tanaka G, Yamanaka T, Furukawa Y, Kajimura N, Mitsuoka K, Nukina N. Sequence- and seed-structure-dependent polymorphic fibrils of alpha-synuclein. Biochim Biophys Acta Mol Basis Dis. 2019;1865:1410–20. doi: 10.1016/j.bbadis.2019.02.013. [DOI] [PubMed] [Google Scholar]
- 55.Luk KC, Covell DJ, Kehm VM, Zhang B, Song IY, Byrne MD, et al. Molecular and biological compatibility with host alpha-synuclein influences fibril pathogenicity. Cell Rep. 2016;16:3373–87. doi: 10.1016/j.celrep.2016.08.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fares MB, Maco B, Oueslati A, Rockenstein E, Ninkina N, Buchman VL, et al. Induction of de novo alpha-synuclein fibrillization in a neuronal model for Parkinson’s disease. PNAS. 2016;113:E912–921. doi: 10.1073/pnas.1512876113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tartari M, Gissi C, Lo Sardo V, Zuccato C, Picardi E, Pesole G, et al. Phylogenetic comparison of huntingtin homologues reveals the appearance of a primitive polyQ in sea urchin. Mol Biol Evol. 2008;25:330–8. doi: 10.1093/molbev/msm258. [DOI] [PubMed] [Google Scholar]
- 58.Walker FO. Huntington’s disease. Lancet. 2007;369:218–28. doi: 10.1016/S0140-6736(07)60111-1. [DOI] [PubMed] [Google Scholar]
- 59.Menalled LB, Sison JD, Dragatsis I, Zeitlin S, Chesselet MF. Time course of early motor and neuropathological anomalies in a knock-in mouse model of Huntington’s disease with 140 CAG repeats. J Comp Neurol. 2003;465:11–26. doi: 10.1002/cne.10776. [DOI] [PubMed] [Google Scholar]
- 60.Heng MY, Detloff PJ, Albin RL. Rodent genetic models of Huntington disease. Neurobiol Dis. 2008;32:1–9. doi: 10.1016/j.nbd.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 61.Seetharaman SV, Taylor AB, Holloway S, Hart PJ. Structures of mouse SOD1 and human/mouse SOD1 chimeras. Arch Biochem Biophys. 2010;503:183–90. doi: 10.1016/j.abb.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Karch CM, Borchelt DR. Aggregation modulating elements in mutant human superoxide dismutase 1. Arch Biochem Biophys. 2010;503:175–82. doi: 10.1016/j.abb.2010.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang HY, Wang IF, Bose J, Shen CK. Structural diversity and functional implications of the eukaryotic TDP gene family. Genomics. 2004;83:130–9. doi: 10.1016/S0888-7543(03)00214-3. [DOI] [PubMed] [Google Scholar]
- 64.D’Alton S, Altshuler M, Lewis J. Studies of alternative isoforms provide insight into TDP-43 autoregulation and pathogenesis. RNA. 2015;21:1419–32. doi: 10.1261/rna.047647.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gumina V, Onesto E, Colombrita C, Maraschi A, Silani V, Ratti A. Inter-species differences in regulation of the progranulin-Sortilin axis in TDP-43 Cell models of neurodegeneration. Int J Mol Sci. 2019;20:5866. doi: 10.3390/ijms20235866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Colby DW, Zhang Q, Wang S, Groth D, Legname G, Riesner D, et al. Prion detection by an amyloid seeding assay. PNAS. 2007;104:20914–9. doi: 10.1073/pnas.0710152105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wilham JM, Orru CD, Bessen RA, Atarashi R, Sano K, Race B, et al. Rapid end-point quantitation of prion seeding activity with sensitivity comparable to bioassays. PLoS Pathog. 2010;6:e1001217. doi: 10.1371/journal.ppat.1001217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Saborio GP, Permanne B, Soto C. Sensitive detection of pathological prion protein by cyclic amplification of protein misfolding. Nature. 2001;411:810–3. doi: 10.1038/35081095. [DOI] [PubMed] [Google Scholar]
- 69.Raymond GJ, Hope J, Kocisko DA, Priola SA, Raymond LD, Bossers A, et al. Molecular assessment of the potential transmissibilities of BSE and scrapie to humans. Nature. 1997;388:285–8. doi: 10.1038/40876. [DOI] [PubMed] [Google Scholar]
- 70.Caughey B, Orru CD, Groveman BR, Hughson AG, Manca M, Raymond LD, et al. Amplified detection of prions and other amyloids by RT-QuIC in diagnostics and the evaluation of therapeutics and disinfectants. Prog Mol Biol Transl Sci. 2017;150:375–88. doi: 10.1016/bs.pmbts.2017.06.003. [DOI] [PubMed] [Google Scholar]
- 71.Soto-Gamez A, Quax WJ, Demaria M. Regulation of survival networks in senescent cells: from mechanisms to interventions. J Mol Biol. 2019;431:2629–43. doi: 10.1016/j.jmb.2019.05.036. [DOI] [PubMed] [Google Scholar]
- 72.Kaur G, Dufour JM. Cell lines: valuable tools or useless artifacts. Spermatogenesis. 2012;2:1–5. doi: 10.4161/spmg.19885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Holmes BB, DeVos SL, Kfoury N, Li M, Jacks R, Yanamandra K, et al. Heparan sulfate proteoglycans mediate internalization and propagation of specific proteopathic seeds. PNAS. 2013;110:E3138–3147. doi: 10.1073/pnas.1301440110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Evans LD, Wassmer T, Fraser G, Smith J, Perkinton M, Billinton A, et al. Extracellular monomeric and aggregated tau efficiently enter human neurons through overlapping but distinct pathways. Cell Rep. 2018;22:3612–24. doi: 10.1016/j.celrep.2018.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jansen AH, Batenburg KL, Pecho-Vrieseling E, Reits EA. Visualization of prion-like transfer in Huntington’s disease models. Biochim Biophys Acta Mol Basis Dis. 2017;1863:793–800. doi: 10.1016/j.bbadis.2016.12.015. [DOI] [PubMed] [Google Scholar]
- 76.Jurgielewicz BJ, Yao Y, Stice SL. Kinetics and specificity of HEK293T extracellular vesicle uptake using imaging flow cytometry. Nanoscale Res Lett. 2020;15:170. doi: 10.1186/s11671-020-03399-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Heravi M, Dargahi L, Parsafar S, Tayaranian Marvian A, Aliakbari F, Morshedi D. The primary neuronal cells are more resistant than PC12 cells to alpha-synuclein toxic aggregates. Neurosci Lett. 2019;701:38–47. doi: 10.1016/j.neulet.2019.01.055. [DOI] [PubMed] [Google Scholar]
- 78.Deng L, Pollmeier L, Zhou Q, Bergemann S, Bode C, Hein L, et al. Gene expression in immortalized versus primary isolated cardiac endothelial cells. Sci Rep. 2020;10:2241. doi: 10.1038/s41598-020-59213-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kang JH, Rychahou PG, Ishola TA, Qiao J, Evers BM, Chung DH. MYCN silencing induces differentiation and apoptosis in human neuroblastoma cells. Biochem Biophys Res Commun. 2006;351:192–7. doi: 10.1016/j.bbrc.2006.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Evans M. Discovering pluripotency: 30 years of mouse embryonic stem cells. Nat Rev Mol Cell Biol. 2011;12:680–6. doi: 10.1038/nrm3190. [DOI] [PubMed] [Google Scholar]
- 81.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 82.Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–6. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 83.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–20. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 84.Suga M, Kondo T, Inoue H. Modeling neurological disorders with human pluripotent stem cell-derived astrocytes. Int J Mol Sci. 2019;20:3862. doi: 10.3390/ijms20163862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chang CY, Ting HC, Liu CA, Su HL, Chiou TW, Lin SZ, et al. Induced pluripotent stem cell (iPSC)-based neurodegenerative disease models for phenotype recapitulation and drug screening. Molecules. 2020;25:2000. doi: 10.3390/molecules25082000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chanoumidou K, Mozafari S, Baron-Van Evercooren A, Kuhlmann T. Stem cell derived oligodendrocytes to study myelin diseases. Glia. 2020;68:705–20. doi: 10.1002/glia.23733. [DOI] [PubMed] [Google Scholar]
- 87.Hasselmann J, Blurton-Jones M. Human iPSC-derived microglia: a growing toolset to study the brain’s innate immune cells. Glia. 2020;68:721–39. doi: 10.1002/glia.23781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hong W, Wang Z, Liu W, O’Malley TT, Jin M, Willem M, et al. Diffusible, highly bioactive oligomers represent a critical minority of soluble Abeta in Alzheimer’s disease brain. Acta Neuropathol. 2018;136:19–40. doi: 10.1007/s00401-018-1846-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Reilly P, Winston CN, Baron KR, Trejo M, Rockenstein EM, Akers JC, et al. Novel human neuronal tau model exhibiting neurofibrillary tangles and transcellular propagation. Neurobiol Dis. 2017;106:222–34. doi: 10.1016/j.nbd.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nagai M, Re DB, Nagata T, Chalazonitis A, Jessell TM, Wichterle H, et al. Astrocytes expressing ALS-linked mutated SOD1 release factors selectively toxic to motor neurons. Nat Neurosci. 2007;10:615–22. doi: 10.1038/nn1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.di Domenico A, Carola G, Calatayud C, Pons-Espinal M, Munoz JP, Richaud-Patin Y, et al. Patient-specific iPSC-derived astrocytes contribute to non-cell-autonomous neurodegeneration in parkinson’s disease. Stem Cell Rep. 2019;12:213–29. doi: 10.1016/j.stemcr.2018.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cooper O, Seo H, Andrabi S, Guardia-Laguarta C, Graziotto J, Sundberg M, et al. Pharmacological rescue of mitochondrial deficits in iPSC-derived neural cells from patients with familial Parkinson’s disease. Sci Transl Med. 2012;4:141ra190. doi: 10.1126/scitranslmed.3003985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li L, Roh JH, Chang EH, Lee Y, Lee S, Kim M, et al. iPSC modeling of presenilin1 mutation in Alzheimer’s disease with cerebellar ataxia. Exp Neurobiol. 2018;27:350–64. doi: 10.5607/en.2018.27.5.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lee JK, Jin HK, Park MH, Kim BR, Lee PH, Nakauchi H, et al. Acid sphingomyelinase modulates the autophagic process by controlling lysosomal biogenesis in Alzheimer’s disease. J Exp Med. 2014;211:1551–70. doi: 10.1084/jem.20132451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Webster CP, Smith EF, Bauer CS, Moller A, Hautbergue GM, Ferraiuolo L, et al. The C9orf72 protein interacts with Rab1a and the ULK1 complex to regulate initiation of autophagy. EMBO. 2016;35:1656–76. doi: 10.15252/embj.201694401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Scialo C, De Cecco E, Manganotti P, Legname G. Prion and prion-like protein strains: deciphering the molecular basis of heterogeneity in neurodegeneration. Viruses. 2019;11:261. doi: 10.3390/v11030261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chai X, Dage JL, Citron M. Constitutive secretion of tau protein by an unconventional mechanism. Neurobiol Dis. 2012;48:356–66. doi: 10.1016/j.nbd.2012.05.021. [DOI] [PubMed] [Google Scholar]
- 98.Bright J, Hussain S, Dang V, Wright S, Cooper B, Byun T, et al. Human secreted tau increases amyloid-beta production. Neurobiol Aging. 2015;36:693–709. doi: 10.1016/j.neurobiolaging.2014.09.007. [DOI] [PubMed] [Google Scholar]
- 99.Moore S, Evans LD, Andersson T, Portelius E, Smith J, Dias TB, et al. APP metabolism regulates tau proteostasis in human cerebral cortex neurons. Cell Rep. 2015;11:689–96. doi: 10.1016/j.celrep.2015.03.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Reyes JF, Olsson TT, Lamberts JT, Devine MJ, Kunath T, Brundin P. A cell culture model for monitoring alpha-synuclein cell-to-cell transfer. Neurobiol Dis. 2015;77:266–75. doi: 10.1016/j.nbd.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 101.Pecho-Vrieseling E, Rieker C, Fuchs S, Bleckmann D, Esposito MS, Botta P, et al. Transneuronal propagation of mutant huntingtin contributes to non-cell autonomous pathology in neurons. Nat Neurosci. 2014;17:1064–72. doi: 10.1038/nn.3761. [DOI] [PubMed] [Google Scholar]
- 102.Kanmert D, Cantlon A, Muratore CR, Jin M, O’Malley TT, Lee G, et al. C-terminally truncated forms of Tau, but not full-length Tau or its C-terminal fragments, are released from neurons independently of cell death. J Neurosci. 2015;35:10851–65. doi: 10.1523/JNEUROSCI.0387-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Guix FX, Corbett GT, Cha DJ, Mustapic M, Liu W, Mengel D, et al. Detection of aggregation-competent tau in neuron-derived extracellular vesicles. Int J Mol Sci. 2018;19:663. doi: 10.3390/ijms19030663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Usenovic M, Niroomand S, Drolet RE, Yao L, Gaspar RC, Hatcher NG, et al. Internalized Tau oligomers cause neurodegeneration by inducing accumulation of pathogenic tau in human neurons derived from induced pluripotent stem cells. J Neurosci. 2015;35:14234–50. doi: 10.1523/JNEUROSCI.1523-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Verheyen A, Diels A, Dijkmans J, Oyelami T, Meneghello G, Mertens L, et al. Using human iPSC-derived neurons to model TAU aggregation. PLoS ONE. 2015;10:e0146127. doi: 10.1371/journal.pone.0146127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Prots I, Grosch J, Brazdis RM, Simmnacher K, Veber V, Havlicek S, et al. Alpha-Synuclein oligomers induce early axonal dysfunction in human iPSC-based models of synucleinopathies. PNAS. 2018;115:7813–8. doi: 10.1073/pnas.1713129115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Silva MC, Cheng C, Mair W, Almeida S, Fong H, Biswas MHU, et al. Human iPSC-derived neuronal model of Tau-A152T frontotemporal dementia reveals tau-mediated mechanisms of neuronal vulnerability. Stem Cell Rep. 2016;7:325–40. doi: 10.1016/j.stemcr.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Matamoros-Angles A, Gayosso LM, Richaud-Patin Y, di Domenico A, Vergara C, Hervera A, et al. iPS cell cultures from a Gerstmann-Straussler-Scheinker patient with the Y218N PRNP mutation recapitulate tau pathology. Mol Neurobiol. 2018;55:3033–48. doi: 10.1007/s12035-017-0506-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Jarosz-Griffiths HH, Corbett NJ, Rowland HA, Fisher K, Jones AC, Baron J, et al. Proteolytic shedding of the prion protein via activation of metallopeptidase ADAM10 reduces cellular binding and toxicity of amyloid-beta oligomers. J Biol Chem. 2019;294:7085–97. doi: 10.1074/jbc.RA118.005364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gribaudo S, Tixador P, Bousset L, Fenyi A, Lino P, Melki R, et al. Propagation of alpha-Synuclein Strains within human reconstructed neuronal network. Stem Cell Rep. 2019;12:230–44. doi: 10.1016/j.stemcr.2018.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Corbett GT, Wang Z, Hong W, Colom-Cadena M, Rose J, Liao M, et al. PrP is a central player in toxicity mediated by soluble aggregates of neurodegeneration-causing proteins. Acta Neuropathol. 2020;139:503–26. doi: 10.1007/s00401-019-02114-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lee HJ, Suk JE, Patrick C, Bae EJ, Cho JH, Rho S, et al. Direct transfer of alpha-synuclein from neuron to astroglia causes inflammatory responses in synucleinopathies. J Biol Chem. 2010;285:9262–72. doi: 10.1074/jbc.M109.081125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lindstrom V, Gustafsson G, Sanders LH, Howlett EH, Sigvardson J, Kasrayan A, et al. Extensive uptake of alpha-synuclein oligomers in astrocytes results in sustained intracellular deposits and mitochondrial damage. Mol Cell Neuro. 2017;82:143–56. doi: 10.1016/j.mcn.2017.04.009. [DOI] [PubMed] [Google Scholar]
- 114.Braidy N, Gai WP, Xu YH, Sachdev P, Guillemin GJ, Jiang XM, et al. Uptake and mitochondrial dysfunction of alpha-synuclein in human astrocytes, cortical neurons and fibroblasts. Transl Neuro. 2013;2:20. doi: 10.1186/2047-9158-2-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Cavaliere F, Cerf L, Dehay B, Ramos-Gonzalez P, De Giorgi F, Bourdenx M, et al. In vitro alpha-synuclein neurotoxicity and spreading among neurons and astrocytes using Lewy body extracts from Parkinson disease brains. Neurobiol Dis. 2017;103:101–12. doi: 10.1016/j.nbd.2017.04.011. [DOI] [PubMed] [Google Scholar]
- 116.Hopp SC, Lin Y, Oakley D, Roe AD, DeVos SL, Hanlon D, et al. The role of microglia in processing and spreading of bioactive tau seeds in Alzheimer’s disease. J Neuroinflam. 2018;15:269. doi: 10.1186/s12974-018-1309-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Luo W, Liu W, Hu X, Hanna M, Caravaca A, Paul SM. Microglial internalization and degradation of pathological tau is enhanced by an anti-tau monoclonal antibody. Sci Rep. 2015;5:11161. doi: 10.1038/srep11161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Martini-Stoica H, Cole AL, Swartzlander DB, Chen F, Wan YW, Bajaj L, et al. TFEB enhances astroglial uptake of extracellular tau species and reduces tau spreading. J Exp Med. 2018;215:2355–77. doi: 10.1084/jem.20172158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wyss-Coray T, Loike JD, Brionne TC, Lu E, Anankov R, Yan F, et al. Adult mouse astrocytes degrade amyloid-beta in vitro and in situ. Nat Med. 2003;9:453–7. doi: 10.1038/nm838. [DOI] [PubMed] [Google Scholar]
- 120.Paresce DM, Ghosh RN, Maxfield FR. Microglial cells internalize aggregates of the Alzheimer’s disease amyloid beta-protein via a scavenger receptor. Neuron. 1996;17:553–65. doi: 10.1016/S0896-6273(00)80187-7. [DOI] [PubMed] [Google Scholar]
- 121.Pearce MMP, Spartz EJ, Hong W, Luo L, Kopito RR. Prion-like transmission of neuronal huntingtin aggregates to phagocytic glia in the Drosophila brain. Nat Commun. 2015;6:6768. doi: 10.1038/ncomms7768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lin YT, Seo J, Gao F, Feldman HM, Wen HL, Penney J, et al. APOE4 causes widespread molecular and cellular alterations associated with Alzheimer’s disease phenotypes in human iPSC-derived brain cell types. Neuron. 2018;98:1141–54 e1147. doi: 10.1016/j.neuron.2018.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Oksanen M, Petersen AJ, Naumenko N, Puttonen K, Lehtonen S, Gubert Olive M, et al. PSEN1 mutant iPSC-derived model reveals severe astrocyte pathology in Alzheimer’s disease. Stem Cell Rep. 2017;9:1885–97. doi: 10.1016/j.stemcr.2017.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Fong LK, Yang MM, Dos Santos Chaves R, Reyna SM, Langness VF, Woodruff G, et al. Full-length amyloid precursor protein regulates lipoprotein metabolism and amyloid-beta clearance in human astrocytes. J Biol Chem. 2018;293:11341–57. doi: 10.1074/jbc.RA117.000441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lorenzini I, Alsop E, Levy J, Gittings LM, Rabichow BE, lall d, et al. Activated iPSC-microglia from C9orf72 ALS/FTD patients exhibit endosomal-lysosomal dysfunction. bioRxiv. 2020:277459.
- 126.Rostami J, Holmqvist S, Lindstrom V, Sigvardson J, Westermark GT, Ingelsson M, et al. Human astrocytes transfer aggregated Alpha-Synuclein via tunneling nanotubes. J Neurosci. 2017;37:11835–53. doi: 10.1523/JNEUROSCI.0983-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Krejciova Z, Alibhai J, Zhao C, Krencik R, Rzechorzek NM, Ullian EM, et al. Human stem cell-derived astrocytes replicate human prions in a PRNP genotype-dependent manner. J Exp Med. 2017;214:3481–95. doi: 10.1084/jem.20161547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Israel MA, Yuan SH, Bardy C, Reyna SM, Mu Y, Herrera C, et al. Probing sporadic and familial Alzheimer’s disease using induced pluripotent stem cells. Nature. 2012;482:216–20. doi: 10.1038/nature10821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ochalek A, Mihalik B, Avci HX, Chandrasekaran A, Teglasi A, Bock I, et al. Neurons derived from sporadic Alzheimer’s disease iPSCs reveal elevated TAU hyperphosphorylation, increased amyloid levels, and GSK3B activation. Alzheimer’s Res Ther. 2017;9:90. doi: 10.1186/s13195-017-0317-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Fujimori K, Ishikawa M, Otomo A, Atsuta N, Nakamura R, Akiyama T, et al. Modeling sporadic ALS in iPSC-derived motor neurons identifies a potential therapeutic agent. Nat Med. 2018;24:1579–89. doi: 10.1038/s41591-018-0140-5. [DOI] [PubMed] [Google Scholar]
- 131.Miguel L, Rovelet-Lecrux A, Feyeux M, Frebourg T, Nassoy P, Campion D, et al. Detection of all adult Tau isoforms in a 3D culture model of iPSC-derived neurons. Stem Cell Res. 2019;40:101541. doi: 10.1016/j.scr.2019.101541. [DOI] [PubMed] [Google Scholar]
- 132.Studer L, Vera E, Cornacchia D. Programming and reprogramming cellular age in the era of induced pluripotency. Cell Stem Cell. 2015;16:591–600. doi: 10.1016/j.stem.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wegmann S, Bennett RE, Delorme L, Robbins AB, Hu M, McKenzie D, et al. Experimental evidence for the age dependence of tau protein spread in the brain. Sci Adv. 2019;5:eaaw6404. doi: 10.1126/sciadv.aaw6404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Nekrasov ED, Vigont VA, Klyushnikov SA, Lebedeva OS, Vassina EM, Bogomazova AN, et al. Manifestation of Huntington’s disease pathology in human induced pluripotent stem cell-derived neurons. Mol Neurodegen. 2016;11:27. doi: 10.1186/s13024-016-0092-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Miller JD, Ganat YM, Kishinevsky S, Bowman RL, Liu B, Tu EY, et al. Human iPSC-based modeling of late-onset disease via progerin-induced aging. Cell Stem Cell. 2013;13:691–705. doi: 10.1016/j.stem.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Sudhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–41. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Pfisterer U, Kirkeby A, Torper O, Wood J, Nelander J, Dufour A, et al. Direct conversion of human fibroblasts to dopaminergic neurons. PNAS. 2011;108:10343–8. doi: 10.1073/pnas.1105135108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Caiazzo M, Dell’Anno MT, Dvoretskova E, Lazarevic D, Taverna S, Leo D, et al. Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature. 2011;476:224–7. doi: 10.1038/nature10284. [DOI] [PubMed] [Google Scholar]
- 139.Abernathy DG, Kim WK, McCoy MJ, Lake AM, Ouwenga R, Lee SW, et al. MicroRNAs induce a permissive chromatin environment that enables neuronal subtype-specific reprogramming of adult human fibroblasts. Cell Stem Cell. 2017;21:332–48 e339. doi: 10.1016/j.stem.2017.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Victor MB, Richner M, Olsen HE, Lee SW, Monteys AM, Ma C, et al. Striatal neurons directly converted from Huntington’s disease patient fibroblasts recapitulate age-associated disease phenotypes. Nat Neurosci. 2018;21:341–52. doi: 10.1038/s41593-018-0075-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Burkhardt MF, Martinez FJ, Wright S, Ramos C, Volfson D, Mason M, et al. A cellular model for sporadic ALS using patient-derived induced pluripotent stem cells. Mol Cell Neurosci. 2013;56:355–64. doi: 10.1016/j.mcn.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Arber C, Lovejoy C, Wray S. Stem cell models of Alzheimer’s disease: progress and challenges. Alzheimer’s Res Ther. 2017;9:42. doi: 10.1186/s13195-017-0268-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wu JW, Hussaini SA, Bastille IM, Rodriguez GA, Mrejeru A, Rilett K, et al. Neuronal activity enhances tau propagation and tau pathology in vivo. Nat Neurosci. 2016;19:1085–92. doi: 10.1038/nn.4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Yamada K, Holth JK, Liao F, Stewart FR, Mahan TE, Jiang H, et al. Neuronal activity regulates extracellular tau in vivo. J Exp Med. 2014;211:387–93. doi: 10.1084/jem.20131685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Pignataro A, Middei S. Trans-synaptic spread of amyloid-beta in Alzheimer’s disease: paths to beta-amyloidosis. Neural Plast. 2017;2017:5281829. doi: 10.1155/2017/5281829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Fortin DL, Nemani VM, Voglmaier SM, Anthony MD, Ryan TA, Edwards RH. Neural activity controls the synaptic accumulation of alpha-synuclein. J Neurosci. 2005;25:10913–21. doi: 10.1523/JNEUROSCI.2922-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Paillusson S, Clairembault T, Biraud M, Neunlist M, Derkinderen P. Activity-dependent secretion of alpha-synuclein by enteric neurons. J Neurochem. 2013;125:512–7. doi: 10.1111/jnc.12131. [DOI] [PubMed] [Google Scholar]
- 148.Pooler AM, Phillips EC, Lau DH, Noble W, Hanger DP. Physiological release of endogenous tau is stimulated by neuronal activity. EMBO Rep. 2013;14:389–94. doi: 10.1038/embor.2013.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Calafate S, Buist A, Miskiewicz K, Vijayan V, Daneels G, de Strooper B, et al. Synaptic contacts enhance cell-to-cell Tau pathology propagation. Cell Rep. 2015;11:1176–83. doi: 10.1016/j.celrep.2015.04.043. [DOI] [PubMed] [Google Scholar]
- 150.Pruunsild P, Bengtson CP, Bading H. Networks of cultured iPSC-derived neurons reveal the human synaptic activity-regulated adaptive gene program. Cell Rep. 2017;18:122–35. doi: 10.1016/j.celrep.2016.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Amin H, Maccione A, Marinaro F, Zordan S, Nieus T, Berdondini L. Electrical responses and spontaneous activity of human iPS-derived neuronal networks characterized for 3-month culture with 4096-electrode arrays. Front Neurosci. 2016;10:121. doi: 10.3389/fnins.2016.00121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Zhang Y, Pak C, Han Y, Ahlenius H, Zhang Z, Chanda S, et al. Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron. 2013;78:785–98. doi: 10.1016/j.neuron.2013.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Nehme R, Zuccaro E, Ghosh SD, Li C, Sherwood JL, Pietilainen O, et al. Combining NGN2 programming with developmental patterning generates human excitatory neurons with NMDAR-mediated synaptic transmission. Cell Rep. 2018;23:2509–23. doi: 10.1016/j.celrep.2018.04.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Koppensteiner P, Boehm S, Arancio O. Electrophysiological profiles of induced neurons converted directly from adult human fibroblasts indicate incomplete neuronal conversion. Cell Reprogram. 2014;16:439–46. doi: 10.1089/cell.2014.0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Drouin-Ouellet J, Lau S, Brattas PL, Rylander Ottosson D, Pircs K, Grassi DA, et al. REST suppression mediates neural conversion of adult human fibroblasts via microRNA-dependent and -independent pathways. EMBO Mol Med. 2017;9:1117–31. doi: 10.15252/emmm.201607471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Xue Y, Qian H, Hu J, Zhou B, Zhou Y, Hu X, et al. Sequential regulatory loops as key gatekeepers for neuronal reprogramming in human cells. Nat Neurosci. 2016;19:807–15. doi: 10.1038/nn.4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.de Rus Jacquet A, Tancredi JL, Lemire AL, DeSantis MC, Li W-P, O’Shea EK. The LRRK2 G2019S mutation alters astrocyte-to-neuron communication via extracellular vesicles and induces neuron atrophy in a human iPSC-derived model of Parkinson’s disease. bioRxiv. 2020:178574. [DOI] [PMC free article] [PubMed]
- 158.Birger A, Ben-Dor I, Ottolenghi M, Turetsky T, Gil Y, Sweetat S, et al. Human iPSC-derived astrocytes from ALS patients with mutated C9ORF72 show increased oxidative stress and neurotoxicity. EBioMedicine. 2019;50:274–89. doi: 10.1016/j.ebiom.2019.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Chandrasekaran A, Avci HX, Leist M, Kobolak J, Dinnyes A. Astrocyte differentiation of human pluripotent stem cells: new tools for neurological disorder research. Front Cell Neurosci. 2016;10:215. doi: 10.3389/fncel.2016.00215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.de Rus Jacquet A. Preparation and co-culture of ipsc-derived dopaminergic neurons and astrocytes. Curr Protoc Cell Biol. 2019;85:e98. doi: 10.1002/cpcb.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Tcw J, Wang M, Pimenova AA, Bowles KR, Hartley BJ, Lacin E, et al. An efficient platform for astrocyte differentiation from human induced pluripotent stem cells. Stem Cell Rep. 2017;9:600–14. doi: 10.1016/j.stemcr.2017.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Li X, Tao Y, Bradley R, Du Z, Tao Y, Kong L, et al. Fast generation of functional subtype astrocytes from human pluripotent stem cells. Stem Cell Rep. 2018;11:998–1008. doi: 10.1016/j.stemcr.2018.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Hedegaard A, Monzon-Sandoval J, Newey SE, Whiteley ES, Webber C, Akerman CJ. Pro-maturational effects of Human iPSC-derived cortical astrocytes upon iPSC-derived cortical neurons. Stem Cell Rep. 2020;15:38–51. doi: 10.1016/j.stemcr.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Volpato V, Webber C. Addressing variability in iPSC-derived models of human disease: guidelines to promote reproducibility. Dis Model Mech. 2020;13:dmm042317. doi: 10.1242/dmm.042317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Schwartzentruber J, Foskolou S, Kilpinen H, Rodrigues J, Alasoo K, Knights AJ, et al. Molecular and functional variation in iPSC-derived sensory neurons. Nat Genet. 2018;50:54–61. doi: 10.1038/s41588-017-0005-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Barros CS, Franco SJ, Muller U. Extracellular matrix: functions in the nervous system. Cold Spring Harb Perspect Biol. 2011;3:a005108. doi: 10.1101/cshperspect.a005108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Spampinato SF, Bortolotto V, Canonico PL, Sortino MA, Grilli M. Astrocyte-derived paracrine signals: relevance for neurogenic niche regulation and blood-brain barrier integrity. Front Pharm. 2019;10:1346. doi: 10.3389/fphar.2019.01346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Kornberg TB, Roy S. Communicating by touch–neurons are not alone. Trends Cell Biol. 2014;24:370–6. doi: 10.1016/j.tcb.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Luo C, Lancaster MA, Castanon R, Nery JR, Knoblich JA, Ecker JR. Cerebral organoids recapitulate epigenomic signatures of the human fetal brain. Cell Rep. 2016;17:3369–84. doi: 10.1016/j.celrep.2016.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Pievani M, Filippini N, van den Heuvel MP, Cappa SF, Frisoni GB. Brain connectivity in neurodegenerative diseases–from phenotype to proteinopathy. Nat Rev Neurol. 2014;10:620–33. doi: 10.1038/nrneurol.2014.178. [DOI] [PubMed] [Google Scholar]
- 171.Vargas-Valderrama A, Messina A, Mitjavila-Garcia MT, Guenou H. The endothelium, a key actor in organ development and hPSC-derived organoid vascularization. J Biomed Sci. 2020;27:67. doi: 10.1186/s12929-020-00661-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Pham MT, Pollock KM, Rose MD, Cary WA, Stewart HR, Zhou P, et al. Generation of human vascularized brain organoids. Neuroreport. 2018;29:588–93. doi: 10.1097/WNR.0000000000001014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Cakir B, Xiang Y, Tanaka Y, Kural MH, Parent M, Kang YJ, et al. Engineering of human brain organoids with a functional vascular-like system. Nat Methods. 2019;16:1169–75. doi: 10.1038/s41592-019-0586-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Worsdorfer P, Dalda N, Kern A, Kruger S, Wagner N, Kwok CK, et al. Generation of complex human organoid models including vascular networks by incorporation of mesodermal progenitor cells. Sci Rep. 2019;9:15663. doi: 10.1038/s41598-019-52204-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Grebenyuk S, Ranga A. Engineering organoid vascularization. Front Bioeng Biotechnol. 2019;7:39. doi: 10.3389/fbioe.2019.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Monzel AS, Smits LM, Hemmer K, Hachi S, Moreno EL, van Wuellen T, et al. Derivation of human midbrain-specific organoids from neuroepithelial stem cells. Stem Cell Rep. 2017;8:1144–54. doi: 10.1016/j.stemcr.2017.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Camp JG, Badsha F, Florio M, Kanton S, Gerber T, Wilsch-Brauninger M, et al. Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. PNAS. 2015;112:15672–7. doi: 10.1073/pnas.1520760112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME, et al. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–9. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Pasca AM, Sloan SA, Clarke LE, Tian Y, Makinson CD, Huber N, et al. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat Methods. 2015;12:671–8. doi: 10.1038/nmeth.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Marton RM, Miura Y, Sloan SA, Li Q, Revah O, Levy RJ, et al. Differentiation and maturation of oligodendrocytes in human three-dimensional neural cultures. Nat Neurosci. 2019;22:484–91. doi: 10.1038/s41593-018-0316-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Qian X, Song H, Ming GL. Brain organoids: advances, applications and challenges. Development. 2019;146:dev166074. doi: 10.1242/dev.166074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Ormel PR, Vieira de Sa R, van Bodegraven EJ, Karst H, Harschnitz O, Sneeboer MAM, et al. Microglia innately develop within cerebral organoids. Nat Commun. 2018;9:4167. doi: 10.1038/s41467-018-06684-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Groveman BR, Foliaki ST, Orru CD, Zanusso G, Carroll JA, Race B, et al. Sporadic Creutzfeldt-Jakob disease prion infection of human cerebral organoids. Acta Neuropathol Commun. 2019;7:90. doi: 10.1186/s40478-019-0742-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Xiang Y, Tanaka Y, Patterson B, Kang YJ, Govindaiah G, Roselaar N, et al. Fusion of regionally specified hPSC-derived organoids models human brain development and interneuron migration. Cell Stem Cell. 2017;21:383–98 e387. doi: 10.1016/j.stem.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Bagley JA, Reumann D, Bian S, Levi-Strauss J, Knoblich JA. Fused cerebral organoids model interactions between brain regions. Nat Methods. 2017;14:743–51. doi: 10.1038/nmeth.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Bolhaqueiro ACF, van Jaarsveld RH, Ponsioen B, Overmeer RM, Snippert HJ, Kops G. Live imaging of cell division in 3D stem-cell organoid cultures. Methods Cell Biol. 2018;145:91–106. doi: 10.1016/bs.mcb.2018.03.016. [DOI] [PubMed] [Google Scholar]
- 187.Bershteyn M, Nowakowski TJ, Pollen AA, Di Lullo E, Nene A, Wynshaw-Boris A, et al. Human iPSC-derived cerebral organoids model cellular features of lissencephaly and reveal prolonged mitosis of outer radial glia. Cell Stem Cell. 2017;20:435–49 e434. doi: 10.1016/j.stem.2016.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Browne AW, Arnesano C, Harutyunyan N, Khuu T, Martinez JC, Pollack HA, et al. Structural and functional characterization of human stem-cell-derived retinal organoids by live imaging. Invest Ophthalmol Vis Sci. 2017;58:3311–8. doi: 10.1167/iovs.16-20796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Held M, Santeramo I, Wilm B, Murray P, Levy R. Ex vivo live cell tracking in kidney organoids using light sheet fluorescence microscopy. PLoS ONE. 2018;13:e0199918. doi: 10.1371/journal.pone.0199918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Andilla J, Jorand R, Olarte OE, Dufour AC, Cazales M, Montagner YL, et al. Imaging tissue-mimic with light sheet microscopy: a comparative guideline. Sci Rep. 2017;7:44939. doi: 10.1038/srep44939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Dekkers JF, Alieva M, Wellens LM, Ariese HCR, Jamieson PR, Vonk AM, et al. High-resolution 3D imaging of fixed and cleared organoids. Nat Protoc. 2019;14:1756–71. doi: 10.1038/s41596-019-0160-8. [DOI] [PubMed] [Google Scholar]
- 192.Volpicelli-Daley LA, Luk KC, Patel TP, Tanik SA, Riddle DM, Stieber A, et al. Exogenous alpha-synuclein fibrils induce Lewy body pathology leading to synaptic dysfunction and neuron death. Neuron. 2011;72:57–71. doi: 10.1016/j.neuron.2011.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Nobuhara CK, DeVos SL, Commins C, Wegmann S, Moore BD, Roe AD, et al. Tau antibody targeting pathological species blocks neuronal uptake and interneuron propagation of Tau in vitro. Am J Pathol. 2017;187:1399–412. doi: 10.1016/j.ajpath.2017.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Tran HT, Chung CH, Iba M, Zhang B, Trojanowski JQ, Luk KC, et al. Alpha-synuclein immunotherapy blocks uptake and templated propagation of misfolded alpha-synuclein and neurodegeneration. Cell Rep. 2014;7:2054–65. doi: 10.1016/j.celrep.2014.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Cheong R, Paliwal S, Levchenko A. High-content screening in microfluidic devices. Expert Opin Drug Discov. 2010;5:715–20. doi: 10.1517/17460441.2010.495116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Freundt EC, Maynard N, Clancy EK, Roy S, Bousset L, Sourigues Y, et al. Neuron-to-neuron transmission of alpha-synuclein fibrils through axonal transport. Ann Neurol. 2012;72:517–24. doi: 10.1002/ana.23747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Takeda S, Wegmann S, Cho H, DeVos SL, Commins C, Roe AD, et al. Neuronal uptake and propagation of a rare phosphorylated high-molecular-weight tau derived from Alzheimer’s disease brain. Nat Commun. 2015;6:8490. doi: 10.1038/ncomms9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Dujardin S, Lecolle K, Caillierez R, Begard S, Zommer N, Lachaud C, et al. Neuron-to-neuron wild-type Tau protein transfer through a trans-synaptic mechanism: relevance to sporadic tauopathies. Acta Neuropathol Commun. 2014;2:14. doi: 10.1186/2051-5960-2-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Pal A, Glass H, Naumann M, Kreiter N, Japtok J, Sczech R, et al. High content organelle trafficking enables disease state profiling as powerful tool for disease modelling. Sci Data. 2018;5:180241. doi: 10.1038/sdata.2018.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Lewandowska MK, Radivojevic M, Jackel D, Muller J, Hierlemann AR. Cortical axons, isolated in channels, display activity-dependent signal modulation as a result of targeted stimulation. Front Neurosci. 2016;10:83. doi: 10.3389/fnins.2016.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Bukhari H, Muller T. Endogenous fluorescence tagging by CRISPR. Trends Cell Biol. 2019;29:912–28. doi: 10.1016/j.tcb.2019.08.004. [DOI] [PubMed] [Google Scholar]
- 202.Pickar-Oliver A, Gersbach CA. The next generation of CRISPR-Cas technologies and applications. Nat Rev Mol Cell Biol. 2019;20:490–507. doi: 10.1038/s41580-019-0131-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Adli M. The CRISPR tool kit for genome editing and beyond. Nat Commun. 2018;9:1911. doi: 10.1038/s41467-018-04252-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Kaczmarczyk L, Mende Y, Zevnik B, Jackson WS. Manipulating the prion protein gene sequence and expression levels with CRISPR/Cas9. PLoS ONE. 2016;11:e0154604. doi: 10.1371/journal.pone.0154604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Chen JJ, Nathaniel DL, Raghavan P, Nelson M, Tian R, Tse E, et al. Compromised function of the ESCRT pathway promotes endolysosomal escape of tau seeds and propagation of tau aggregation. J Biol Chem. 2019;294:18952–66. doi: 10.1074/jbc.RA119.009432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Rice AM, McLysaght A. Dosage-sensitive genes in evolution and disease. BMC Biol. 2017;15:78. doi: 10.1186/s12915-017-0418-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Bae EJ, Yang NY, Song M, Lee CS, Lee JS, Jung BC, et al. Glucocerebrosidase depletion enhances cell-to-cell transmission of alpha-synuclein. Nat Commun. 2014;5:4755. doi: 10.1038/ncomms5755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Kampmann M. CRISPRi and CRISPRa screens in mammalian cells for precision biology and medicine. ACS Chem Biol. 2018;13:406–16. doi: 10.1021/acschembio.7b00657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Stanek LM, Sardi SP, Mastis B, Richards AR, Treleaven CM, Taksir T, et al. Silencing mutant huntingtin by adeno-associated virus-mediated RNA interference ameliorates disease manifestations in the YAC128 mouse model of Huntington’s disease. Hum Gene Ther. 2014;25:461–74. doi: 10.1089/hum.2013.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Xu X, Tay Y, Sim B, Yoon SI, Huang Y, Ooi J, et al. Reversal of phenotypic abnormalities by CRISPR/Cas9-mediated gene correction in huntington disease patient-derived induced pluripotent stem cells. Stem Cell Rep. 2017;8:619–33. doi: 10.1016/j.stemcr.2017.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Chen Y, Dolt KS, Kriek M, Baker T, Downey P, Drummond NJ, et al. Engineering synucleinopathy-resistant human dopaminergic neurons by CRISPR-mediated deletion of the SNCA gene. Eur J Neurosci. 2019;49:510–24. doi: 10.1111/ejn.14286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Hallmann AL, Arauzo-Bravo MJ, Mavrommatis L, Ehrlich M, Ropke A, Brockhaus J, et al. Astrocyte pathology in a human neural stem cell model of frontotemporal dementia caused by mutant TAU protein. Sci Rep. 2017;7:42991. doi: 10.1038/srep42991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Heman-Ackah SM, Manzano R, Hoozemans JJM, Scheper W, Flynn R, Haerty W, et al. Alpha-synuclein induces the unfolded protein response in Parkinson’s disease SNCA triplication iPSC-derived neurons. Hum Mol Genet. 2017;26:4441–50. doi: 10.1093/hmg/ddx331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Soldner F, Laganiere J, Cheng AW, Hockemeyer D, Gao Q, Alagappan R, et al. Generation of isogenic pluripotent stem cells differing exclusively at two early onset Parkinson point mutations. Cell. 2011;146:318–31. doi: 10.1016/j.cell.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Reinhardt P, Schmid B, Burbulla LF, Schondorf DC, Wagner L, Glatza M, et al. Genetic correction of a LRRK2 mutation in human iPSCs links parkinsonian neurodegeneration to ERK-dependent changes in gene expression. Cell Stem Cell. 2013;12:354–67. doi: 10.1016/j.stem.2013.01.008. [DOI] [PubMed] [Google Scholar]
- 216.Jackow J, Guo Z, Hansen C, Abaci HE, Doucet YS, Shin JU, et al. CRISPR/Cas9-based targeted genome editing for correction of recessive dystrophic epidermolysis bullosa using iPS cells. PNAS. 2019;116:26846–52. doi: 10.1073/pnas.1907081116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Li XL, Li GH, Fu J, Fu YW, Zhang L, Chen W, et al. Highly efficient genome editing via CRISPR-Cas9 in human pluripotent stem cells is achieved by transient BCL-XL overexpression. Nucleic Acids Res. 2018;46:10195–215. doi: 10.1093/nar/gky804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Vakulskas CA, Dever DP, Rettig GR, Turk R, Jacobi AM, Collingwood MA, et al. A high-fidelity Cas9 mutant delivered as a ribonucleoprotein complex enables efficient gene editing in human hematopoietic stem and progenitor cells. Nat Med. 2018;24:1216–24. doi: 10.1038/s41591-018-0137-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Prusiner SB. Molecular biology of prion diseases. Science. 1991;252:1515–22. doi: 10.1126/science.1675487. [DOI] [PubMed] [Google Scholar]
- 220.Makrinou E, Collinge J, Antoniou M. Genomic characterization of the human prion protein (PrP) gene locus. Mamm Genome. 2002;13:696–703. doi: 10.1007/s00335-002-3043-0. [DOI] [PubMed] [Google Scholar]
- 221.Schmitz M, Lullmann K, Zafar S, Ebert E, Wohlhage M, Oikonomou P, et al. Association of prion protein genotype and scrapie prion protein type with cellular prion protein charge isoform profiles in cerebrospinal fluid of humans with sporadic or familial prion diseases. Neurobiol Aging. 2014;35:1177–88. doi: 10.1016/j.neurobiolaging.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 222.Giannopoulos PN, Robertson C, Jodoin J, Paudel H, Booth SA, LeBlanc AC. Phosphorylation of prion protein at serine 43 induces prion protein conformational change. J Neurosci. 2009;29:8743–51. doi: 10.1523/JNEUROSCI.2294-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Parchi P, Chen SG, Brown P, Zou W, Capellari S, Budka H, et al. Different patterns of truncated prion protein fragments correlate with distinct phenotypes in P102L Gerstmann-Straussler-Scheinker disease. PNAS. 1998;95:8322–7. doi: 10.1073/pnas.95.14.8322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Baiardi S, Rossi M, Capellari S, Parchi P. Recent advances in the histo-molecular pathology of human prion disease. Brain Pathol. 2019;29:278–300. doi: 10.1111/bpa.12695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Zhong Q, Congdon EE, Nagaraja HN, Kuret J. Tau isoform composition influences rate and extent of filament formation. J Biol Chem. 2012;287:20711–9. doi: 10.1074/jbc.M112.364067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Hefti MM, Farrell K, Kim S, Bowles KR, Fowkes ME, Raj T, et al. High-resolution temporal and regional mapping of MAPT expression and splicing in human brain development. PLoS ONE. 2018;13:e0195771. doi: 10.1371/journal.pone.0195771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Lacovich V, Espindola SL, Alloatti M, Pozo Devoto V, Cromberg LE, Carna ME, et al. Tau Isoforms Imbalance Impairs the Axonal Transport of the Amyloid Precursor Protein in Human Neurons. J Neurosci. 2017;37:58–69. doi: 10.1523/JNEUROSCI.2305-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Liu C, Gotz J. Profiling murine tau with 0N, 1N and 2N isoform-specific antibodies in brain and peripheral organs reveals distinct subcellular localization, with the 1N isoform being enriched in the nucleus. PLoS ONE. 2013;8:e84849. doi: 10.1371/journal.pone.0084849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Neddens J, Temmel M, Flunkert S, Kerschbaumer B, Hoeller C, Loeffler T, et al. Phosphorylation of different tau sites during progression of Alzheimer’s disease. Acta Neuropathol Commun. 2018;6:52. doi: 10.1186/s40478-018-0557-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Ercan-Herbst E, Ehrig J, Schondorf DC, Behrendt A, Klaus B, Gomez Ramos B, et al. A post-translational modification signature defines changes in soluble tau correlating with oligomerization in early stage Alzheimer’s disease brain. Acta Neuropathol Commun. 2019;7:192. doi: 10.1186/s40478-019-0823-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Katsumoto A, Takeuchi H, Tanaka F. Tau pathology in chronic traumatic encephalopathy and Alzheimer’s disease: similarities and differences. Front Neurol. 2019;10:980. doi: 10.3389/fneur.2019.00980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Barthelemy NR, Bateman RJ, Hirtz C, Marin P, Becher F, Sato C, et al. Cerebrospinal fluid phospho-tau T217 outperforms T181 as a biomarker for the differential diagnosis of Alzheimer’s disease and PET amyloid-positive patient identification. Alzheimer’s Res Ther. 2020;12:26. doi: 10.1186/s13195-020-00596-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Barthelemy NR, Mallipeddi N, Moiseyev P, Sato C, Bateman RJ. Tau phosphorylation rates measured by mass spectrometry differ in the intracellular brain vs. extracellular cerebrospinal fluid compartments and are differentially affected by Alzheimer’s disease. Front Aging Neurosci. 2019;11:121. doi: 10.3389/fnagi.2019.00121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Vuono R, Winder-Rhodes S, de Silva R, Cisbani G, Drouin-Ouellet J, Network RIotEHsD. et al. The role of tau in the pathological process and clinical expression of Huntington’s disease. Brain. 2015;138(Pt 7):1907–18. doi: 10.1093/brain/awv107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Bamburg JR, Bernstein BW, Davis RC, Flynn KC, Goldsbury C, Jensen JR, et al. ADF/Cofilin-actin rods in neurodegenerative diseases. Curr Alzheimer Res. 2010;7:241–50. doi: 10.2174/156720510791050902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Spillantini MG, Goedert M, Crowther RA, Murrell JR, Farlow MR, Ghetti B. Familial multiple system tauopathy with presenile dementia: a disease with abundant neuronal and glial tau filaments. PNAS. 1997;94:4113–8. doi: 10.1073/pnas.94.8.4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Ferrer I, Lopez-Gonzalez I, Carmona M, Arregui L, Dalfo E, Torrejon-Escribano B, et al. Glial and neuronal tau pathology in tauopathies: characterization of disease-specific phenotypes and tau pathology progression. J Neuropathol Exp Neurol. 2014;73:81–97. doi: 10.1097/NEN.0000000000000030. [DOI] [PubMed] [Google Scholar]
- 238.Strang KH, Golde TE, Giasson BI. MAPT mutations, tauopathy, and mechanisms of neurodegeneration. Lab Invest. 2019;99:912–28. doi: 10.1038/s41374-019-0197-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Leong YQ, Ng KY, Chye SM, Ling APK, Koh RY. Mechanisms of action of amyloid-beta and its precursor protein in neuronal cell death. Metab Brain Dis. 2020;35:11–30. doi: 10.1007/s11011-019-00516-y. [DOI] [PubMed] [Google Scholar]
- 240.Rezaei-Ghaleh N, Amininasab M, Kumar S, Walter J, Zweckstetter M. Phosphorylation modifies the molecular stability of beta-amyloid deposits. Nat Commun. 2016;7:11359. doi: 10.1038/ncomms11359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Kumar S, Wirths O, Stuber K, Wunderlich P, Koch P, Theil S, et al. Phosphorylation of the amyloid beta-peptide at Ser26 stabilizes oligomeric assembly and increases neurotoxicity. Acta Neuropathol. 2016;131:525–37. doi: 10.1007/s00401-016-1546-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Thal DR, Walter J, Saido TC, Fandrich M. Neuropathology and biochemistry of Abeta and its aggregates in Alzheimer’s disease. Acta Neuropathol. 2015;129:167–82. doi: 10.1007/s00401-014-1375-y. [DOI] [PubMed] [Google Scholar]
- 243.Takahashi RH, Nagao T, Gouras GK. Plaque formation and the intraneuronal accumulation of beta-amyloid in Alzheimer’s disease. Pathol Int. 2017;67:185–93. doi: 10.1111/pin.12520. [DOI] [PubMed] [Google Scholar]
- 244.Bassil F, Brown HJ, Pattabhiraman S, Iwasyk JE, Maghames CM, Meymand ES, et al. Amyloid-Beta (Abeta) plaques promote seeding and spreading of alpha-synuclein and Tau in a mouse model of lewy body disorders with abeta pathology. Neuron. 2020;105:260–75 e266. doi: 10.1016/j.neuron.2019.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Batarseh YS, Duong QV, Mousa YM, Al Rihani SB, Elfakhri K, Kaddoumi A. Amyloid-beta and astrocytes interplay in amyloid-beta related disorders. Int J Mol Sci. 2016;17:338. doi: 10.3390/ijms17030338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Irwin DJ, Hurtig HI. The Contribution of Tau, Amyloid-Beta and Alpha-Synuclein Pathology to Dementia in Lewy Body Disorders. JADP. 2018;8:444. doi: 10.4172/2161-0460.1000444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Polymeropoulos MH, Lavedan C, Leroy E, Ide SE, Dehejia A, Dutra A, et al. Mutation in the alpha-synuclein gene identified in families with Parkinson’s disease. Science. 1997;276:2045–7. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- 248.Simon-Sanchez J, Schulte C, Bras JM, Sharma M, Gibbs JR, Berg D, et al. Genome-wide association study reveals genetic risk underlying Parkinson’s disease. Nat Genet. 2009;41:1308–12. doi: 10.1038/ng.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Beyer K, Ariza A. Alpha-Synuclein posttranslational modification and alternative splicing as a trigger for neurodegeneration. Mol Neurobiol. 2013;47:509–24. doi: 10.1007/s12035-012-8330-5. [DOI] [PubMed] [Google Scholar]
- 250.Burre J, Sharma M, Sudhof TC. Cell biology and pathophysiology of alpha-Synuclein. Cold Spring Harb Perspect Med. 2018;8:a024091. doi: 10.1101/cshperspect.a024091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Villar-Pique A, Lopes da Fonseca T, Outeiro TF. Structure, function and toxicity of alpha-synuclein: the Bermuda triangle in synucleinopathies. J Neurochem. 2016;139(Suppl 1):240–55. doi: 10.1111/jnc.13249. [DOI] [PubMed] [Google Scholar]
- 252.Calo L, Wegrzynowicz M, Santivanez-Perez J, Grazia, Spillantini M. Synaptic failure and alpha-synuclein. Mov Disord. 2016;31:169–77. doi: 10.1002/mds.26479. [DOI] [PubMed] [Google Scholar]
- 253.Peng C, Gathagan RJ, Lee VM. Distinct alpha-Synuclein strains and implications for heterogeneity among alpha-Synucleinopathies. Neurobiol Dis. 2018;109(Pt B):209–18. doi: 10.1016/j.nbd.2017.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. The Huntington’s Disease Collaborative Research Group. Cell. 1993;72:971–83. [DOI] [PubMed]
- 255.Ruzo A, Ismailoglu I, Popowski M, Haremaki T, Croft GF, Deglincerti A, et al. Discovery of novel isoforms of huntingtin reveals a new hominid-specific exon. PLoS ONE. 2015;10:e0127687. doi: 10.1371/journal.pone.0127687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Mort M, Carlisle FA, Waite AJ, Elliston L, Allen ND, Jones L, et al. Huntingtin exists as multiple splice forms in human brain. J Huntington’s Dis. 2015;4:161–71. doi: 10.3233/JHD-150151. [DOI] [PubMed] [Google Scholar]
- 257.Neueder A, Landles C, Ghosh R, Howland D, Myers RH, Faull RLM, et al. The pathogenic exon 1 HTT protein is produced by incomplete splicing in Huntington’s disease patients. Sci Rep. 2017;7:1307. doi: 10.1038/s41598-017-01510-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Romo L, Mohn ES, Aronin N. A fresh look at huntingtin mRNA processing in Huntington’s disease. J Huntington’s Dis. 2018;7:101–8. doi: 10.3233/JHD-180292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Ehrnhoefer DE, Sutton L, Hayden MR. Small changes, big impact: posttranslational modifications and function of huntingtin in Huntington disease. Neuroscientist. 2011;17:475–92. doi: 10.1177/1073858410390378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Cicchetti F, Lacroix S, Cisbani G, Vallieres N, Saint-Pierre M, St-Amour I, et al. Mutant huntingtin is present in neuronal grafts in Huntington disease patients. Ann Neurol. 2014;76:31–42. doi: 10.1002/ana.24174. [DOI] [PubMed] [Google Scholar]
- 261.Hartlage-Rubsamen M, Ratz V, Zeitschel U, Finzel L, Machner L, Koppen J, et al. Endogenous mouse huntingtin is highly abundant in cranial nerve nuclei, co-aggregates to Abeta plaques and is induced in reactive astrocytes in a transgenic mouse model of Alzheimer’s disease. Acta Neuropathol Commun. 2019;7:79. doi: 10.1186/s40478-019-0726-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, Hentati A, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- 263.Groner Y, Lieman-Hurwitz J, Dafni N, Sherman L, Levanon D, Bernstein Y, et al. Molecular structure and expression of the gene locus on chromosome 21 encoding the Cu/Zn superoxide dismutase and its relevance to Down syndrome. Ann NY Acad Sci. 1985;450:133–56. doi: 10.1111/j.1749-6632.1985.tb21489.x. [DOI] [PubMed] [Google Scholar]
- 264.Zinman L, Liu HN, Sato C, Wakutani Y, Marvelle AF, Moreno D, et al. A mechanism for low penetrance in an ALS family with a novel SOD1 deletion. Neurology. 2009;72:1153–9. doi: 10.1212/01.wnl.0000345363.65799.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Fay JM, Zhu C, Proctor EA, Tao Y, Cui W, Ke H, et al. A phosphomimetic mutation stabilizes SOD1 and rescues cell viability in the context of an ALS-associated mutation. Structure. 2016;24:1898–906. doi: 10.1016/j.str.2016.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Jiang H, Shimizu H, Shiga A, Tanaka M, Onodera O, Kakita A, et al. Familial amyotrophic lateral sclerosis with an I104F mutation in the SOD1 gene: Multisystem degeneration with neurofilamentous aggregates and SOD1 inclusions. Neuropathology. 2017;37:69–77. doi: 10.1111/neup.12324. [DOI] [PubMed] [Google Scholar]
- 267.Milani P, Gagliardi S, Cova E, Cereda C. SOD1 transcriptional and posttranscriptional regulation and its potential implications in ALS. Neurol Res Int. 2011;2011:458427. doi: 10.1155/2011/458427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Prasad A, Bharathi V, Sivalingam V, Girdhar A, Patel BK. Molecular mechanisms of TDP-43 misfolding and pathology in amyotrophic lateral sclerosis. Front Mol Neurosci. 2019;12:25. doi: 10.3389/fnmol.2019.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Xiao S, Sanelli T, Chiang H, Sun Y, Chakrabartty A, Keith J, et al. Low molecular weight species of TDP-43 generated by abnormal splicing form inclusions in amyotrophic lateral sclerosis and result in motor neuron death. Acta Neuropathol. 2015;130:49–61. doi: 10.1007/s00401-015-1412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Hasegawa M, Arai T, Nonaka T, Kametani F, Yoshida M, Hashizume Y, et al. Phosphorylated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Ann Neurol. 2008;64:60–70. doi: 10.1002/ana.21425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Robinson JL, Geser F, Stieber A, Umoh M, Kwong LK, Van Deerlin VM, et al. TDP-43 skeins show properties of amyloid in a subset of ALS cases. Acta Neuropathol. 2013;125:121–31. doi: 10.1007/s00401-012-1055-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Lin WL, Dickson DW. Ultrastructural localization of TDP-43 in filamentous neuronal inclusions in various neurodegenerative diseases. Acta Neuropathol. 2008;116:205–13. doi: 10.1007/s00401-008-0408-9. [DOI] [PMC free article] [PubMed] [Google Scholar]