Abstract
Auto-inhibition is a common transcriptional control mechanism that is well characterized in the regulatory transcription factor Ets-1. Autoinhibition of Ets-1 DNA binding works through an inhibitory module that exists in two conformations. DNA binding requires a change in the inhibitory module from the packed to disrupted conformation. This structural switch provides a mechanism to tightly regulate Ets-1 DNA binding. We report that the Ets-1 partner protein core-binding factor α2 (CBFα2; also known as AML1 or PEBP2) stimulates Ets-1 DNA binding and counteracts auto-inhibition. Support for this conclusion came from three observations. First, the level of cooperative DNA binding (10-fold) was similar to the level of repression by auto-inhibition (10- to 20-fold). Next, a region necessary for cooperative DNA binding mapped to the inhibitory module. Third, an Ets-1 mutant with a constitutively disrupted inhibitory module did not bind DNA cooperatively with CBFα2. Furthermore, two additional lines of evidence indicated that CBFα2 affects the structural switch by direct interactions with Ets-1. First, the retention of cooperative DNA binding on nicked duplexes eliminated a potential role of through-DNA effects. Second, cooperative DNA binding was observed on composite sites with altered spacing or reversed orientation. We suggest that only protein interactions can accommodate this observed flexibility. These findings provide a mechanism by which CBF relieves the auto-inhibition of Ets-1 and illustrates one strategy for the synergistic activity of regulatory transcription factors.
Auto-inhibition modulates a variety of transcription factor activities (21). In this regulatory mechanism, inhibitory sequences act in cis to repress such functions as DNA binding, transcriptional activation, nuclear localization, and ligand interaction. The picture emerging from the study of several transcription factors suggests that protein partnerships can counteract auto-inhibition. For example, serum response factor counteracts the auto-inhibition of Elk-1 DNA binding (36). The DNA binding of Pip, which contains an auto-inhibitory domain, requires interactions with PU.1 (9), and Pbx activates the DNA binding of its partner protein Hoxb (10, 11). Similarly, DNA binding by the yeast repressor a1 requires formation of a ternary complex with α2 (63). Examples within basal transcriptional machinery include the auto-inhibition of the ς subunit of Escherichia coli RNA polymerase. Interaction with core RNA polymerase counters this inhibitory effect (13). Also, the inhibitory function of the amino terminus of TATA-binding protein is abrogated by interactions with SNAPc (40). In each of these DNA binding scenarios, auto-inhibition generates a tighter control switch from the off to on state. To decipher the molecular basis of the interplay between auto-inhibition and protein partnerships, we focus on Ets-1, for which auto-inhibition is well characterized at the mechanistic and structural levels (21).
Auto-inhibition, together with protein partnerships, can provide specificity within a family of highly related transcription factors (22). Ets-1 belongs to the ets gene family, which includes over 50 genes throughout the metazoa. The ets proteins display common DNA binding properties due to conservation in the ETS domain (22, 56). Nevertheless, each ets protein appears to have a specific function, presumably by regulating unique target genes. For example, Ets-1 is in the mesodermal compartment of several tissues in the mouse embryo (33) and in T and B lymphocytes, natural killer cells, endothelial cells, and the brain of the adult mouse (4, 7, 17, 18, 73). Targeted disruption of ets-1 in the mouse results in abnormal T- and B-cell function, as well as defective natural killer cell development (4, 8, 42). However, there are other ETS domain proteins found in these tissues. For example, T lymphocytes contain the ets proteins TEL, PU.1, Elf-1, Fli-1, Erg, Ets-2, and GABP, in addition to Ets-1 (1). Thus, some level of control beyond DNA binding must determine specificity. Auto-inhibition of DNA binding provides a regulatory framework with which to generate this specificity.
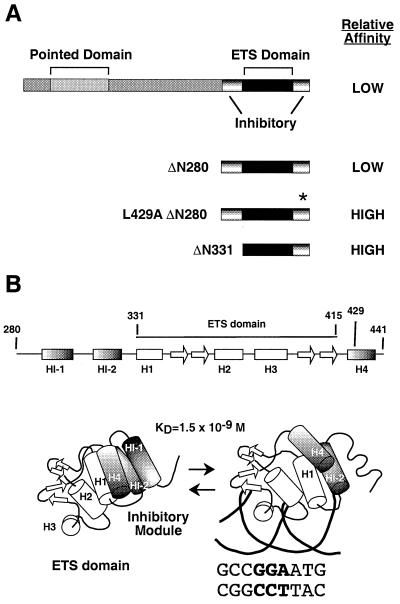
We have developed a structural and mechanistic model of Ets-1 auto-inhibition based on biophysical and biochemical studies (21) (Fig. 1). Ets-1 binds DNA through the ETS domain, which consists of three α helices and four β strands folded into a winged helix-turn-helix motif (5, 15, 32, 41, 72). Two regions that flank the ETS domain work together to repress the DNA binding activity of Ets-1. Deletion of either region leads to derepression, resulting in DNA binding affinity that is 10- to 20-fold higher than that measured for full-length Ets-1 (25, 27, 35, 51, 71). Nuclear magnetic resonance analyses indicate that the flanking inhibitory regions consist of three α helices, HI-1 and HI-2 amino terminal and H4 carboxyl terminal to the ETS domain (15, 59). These data support a model in which the three inhibitory helices and H1 of the ETS domain pack together in a four-helix bundle to form an inhibitory module. Upon DNA binding, helix HI-1 unfolds, causing a disruption of the inhibitory module (27, 51). We have proposed that this conformational change is responsible for the relatively low DNA binding affinity of the native Ets-1. In this study, we tested whether a partnership with a second DNA binding protein counteracts the auto-inhibition of Ets-1.
FIG. 1.
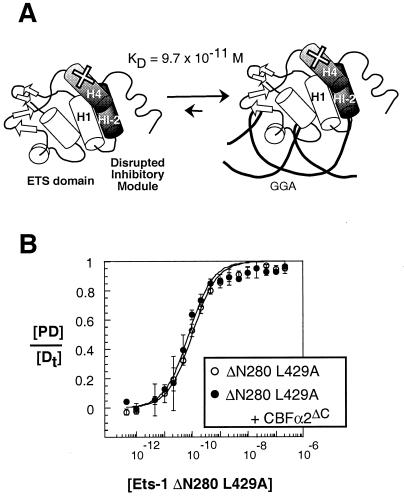
Ets-1 domains and conformational change. (A) Schematic of the structural and functional domains of full-length Ets-1, Ets-1ΔN280, and Ets-1ΔN331. The asterisk indicates the mutation in helix H4 of Ets-1ΔN280;L429A. (B) Schematic representation of the secondary structure of Ets-1ΔN280 as determined by nuclear magnetic resonance spectroscopy (14, 59). Shown are the tertiary structure of the ETS domain (15) and proposed structure of the inhibitory module in the absence (left) and presence (right) of DNA (15, 21, 59). The KD value is for the SC1/core composite site.
We have chosen the Ets-1–core-binding factor (CBF) partnership as a model system. CBF represents a small family of heterodimeric proteins (62). Three genes in vertebrate genomes (in humans, CBFA1, CBFA2 [AML1], and CBFA3) encode α subunits that contain a conserved DNA binding domain, termed the Runt domain (28). A single vertebrate gene, CBFB, encodes the non-DNA-binding subunit, CBFβ (2, 3, 28, 34, 48, 49, 70). CBFα2 is found in most hematopoietic lineages and is required for hematopoiesis (45, 46, 50, 53, 54, 68, 69). Thus, both Ets-1 and CBF are important proteins in mammalian immune system development and function.
Experimental approaches demonstrate the functionality of the Ets-1 and CBF partnership. Several cellular and viral enhancers contain adjacent Ets-1 and CBF binding sites, including the T-cell receptor (TCR) α and β enhancers, immunoglobulin μ heavy-chain enhancer, osteopontin promoter, and enhancers of the polyomavirus and Moloney murine leukemia virus (Mo-MLV) (16, 24, 29, 39, 55, 74). Mutation of the Ets-1 and CBF binding sites in the Mo-MLV enhancer reduces viral enhancer activity and alters viral disease specificity (60, 61). In vivo footprinting assays show occupancy of the Mo-MLV Ets-1 and CBF binding sites in T cells, thus confirming the in vivo function of these adjacent sites (20). Ets-1 and members of the CBF family synergistically activate the TCRα and TCRβ enhancers, the immunoglobulin μ heavy-chain enhancer, the osteopontin promoter, as well as the Mo-MLV enhancer in vivo (16, 19, 31, 39, 55, 65, 74). Cooperative DNA binding between Ets-1 and CBFα2 is detected in in vitro studies of the TCRα and TCRβ enhancers (19, 31, 39, 74). Furthermore, the accompanying report quantifies the level of cooperativity and demonstrates that enhancement of Ets-1 and CBFα2 DNA binding is reciprocal (23). These findings suggest that DNA binding cooperativity is a mechanism for transcriptional synergy.
This study links the auto-inhibition phenomenon of Ets-1 to the DNA binding cooperativity of the Ets-1 and CBFα2 partnership. The regions of Ets-1 necessary for cooperativity and auto-inhibition overlap. In addition, disruption of the inhibitory module abrogates the effect of CBFα2 on Ets-1 DNA binding. Finally, the binding sites can be topologically uncoupled and widely spaced, suggesting that through-DNA effects do not mediate cooperativity. These findings support a mechanistic model of the Ets-1 and CBF protein partnership in which protein interactions counteract the auto-inhibition mechanism.
MATERIALS AND METHODS
Protein synthesis and purification.
Full-length Ets-1 was synthesized in bacteria and purified from the insoluble fraction by conventional chromatography (27). Ets-1ΔN280 (wild type and L429A mutant) and Ets-1ΔN331 were produced in bacteria and purified from the soluble fraction by conventional chromatography (51). CBFα2 with amino acids 1 to 331 of the 451-amino-acid native CBF residues, termed CBFα2ΔC, was produced in Sf9 insect cells with a baculovirus expression system and purified by affinity chromatography with FLAG and His tag methodology (23). Purified Ets-1 and CBFα2 proteins were incubated first with 1/10 volume of fresh 0.1 M dithiothreitol at 4°C for 30 min before each use to reverse any artifactual oxidation. Activity of protein preparations was determined by DNA titration experiments (23, 27).
Synthetic oligonucleotides.
Oligonucleotides were synthesized (Applied Biosystems model 394 or 3948 apparatus) and purified either by gel purification as described previously (27) or by reverse-phase chromatography on an automated DNA synthesizer (Applied Biosystems model 3948) followed by gel filtration (BioSpin-6; Bio-Rad). Radiolabeling of 5′ termini was performed with T4 polynucleotide kinase and [γ32P]ATP (7,000 Ci/mol) prior to annealing of complementary oligonucleotides, as described previously (27).
Synthetic complementary oligonucleotides containing the composite Ets-1 and CBF binding sites were used for the DNA binding assays. The sequences of the Mo-MLV enhancer (ets/cbf) oligonucleotides were 5′-GATCCCAAACAGGATATCTGTGGTAAGCA-3′ (top strand) and 5′-GATCTGCTTACCACAGATATCCTGTTTGG-3′ (bottom strand). Sequences of the SC1/core oligonucleotides were 5′-GGCCAAGCCGGAAGTGTGTGGTAAACACTTT-3′ (top strand) and 5′-AAAGTGTTTACCACACACTTCCGGCTTGGCC-3′ (bottom strand). Mutant versions of these duplexes are listed in Tables 1 and 2 with only the top strand presented. In the case of the oligonucleotides in which the binding site orientation was reversed, 9 and 14 bp for the Ets-1 and CBF binding sites, respectively, were reversed to accommodate both requisite core sequences (underscored in Table 2) and the preferred flanking sequences. This engineering altered the native spacing. Nicked duplexes were generated by annealing two separate oligonucleotides that represented the top strand to an intact complementary bottom strand. This DNA duplex has a nick at the junction of the two upper-strand oligonucleotides. Furthermore, the 5′ phosphate in the nick (junction) of the two upper strands is missing.
TABLE 1.
Autoinhibition on the Mo-MLV and the SC1 Ets-1 binding sites
| Site | Composite site sequencea | CBF | Mean KD (nM) ± SEb
|
|||
|---|---|---|---|---|---|---|
| Ets-1 | ΔN280 | ΔN331 | ΔN280 L429A | |||
| MLV ets/cbf | 5′-GATCCCAAACAGGATATCTGTGGTAAGCA-3′ | − | 7.10 ± 0.58 | 0.25 ± 0.0090 | ||
| + | 0.34 ± 0.028 | |||||
| MLV ets−/cbf | 5′-GATCCCAAACATTATATCTGTGGTAAGCA-3′ | − | 430 ± 44 | |||
| + | 380 ± 55 | |||||
| SC1/core | 5′-GGCCAAGCCGGAAGTGTGTGGTAAACACTTT-3′ | − | 0.85c ± 0.13 | 1.50 ± 0.22 | 0.044 ± 0.18 | 0.097 ± 0.0090 |
| + | 0.086c ± 0.0060 | 0.15 ± 0.020 | 0.047 ± 0.0040 | 0.077 ± 0.0080 | ||
| SC1 | 5′-TCGACGGCCAAGCCGGAAGTGAGTGCC-3′ | − | 0.20d ± 0.020 | 0.44e | 0.0085d ± 0.00070 | |
Underscored sequences indicate Ets-1 and CBFα2 recognition sequences.
Values are the means of two to three independent experiments. SE expresses the accuracy of the curve fit rather than the error reflected in the error bars (as described in Materials and Methods).
From accompanying report (23).
Published data (27).
Published data (51).
TABLE 2.
Effects of altered spacing and orientation of Ets-1 and CBFα2 binding sites
| Site | Composite site sequencea | Mean KD (nM) ± SEb
|
|
|---|---|---|---|
| Ets-1 | Ets-1 + CBF | ||
| SC1/core | 5′-GGCCAAGCCGGAAGTGTGTGGTAAACACTTT-3′ | 0.85c ± 0.13 | 0.086c ± 0.0060 |
| Nicked SC1/core | 5′-GGCCAAGCCGGAAG TGTGTGGTAAACACTTT-3′ | 3.30 ± 0.61 | 0.21 ± 0.010 |
| invCBF | 5′-GGCCAAGCCGGAAGTCCTCGTTTACCACACCACTTT-3′ | 1.90 ± 0.29 | 0.14 ± 0.0010 |
| invEts | 5′-GGCCAACTTCCGGCTCCTCGGTGTGGTAAACACTTT-3′ | 0.67 ± 0.050 | 0.19 ± 0.0080 |
| Δ+5 | 5′-GGCCAAGCCGGAAGTGCATCGTGTGGTAAACACTTT-3′ | 1.90 ± 0.26 | 0.24 ± 0.0020 |
| Δ+10 | 5′-GGCCAAGCCGGAAGTGCATTCGCTCGTGTGGTAAACACTTT-3′ | 2.20 ± 0.23 | 0.25 ± 0.0030 |
| Δ−4 | 5′-GGCCAAGCCGGATGTGGTAAACACTTT-3′ | 2.30 ± 0.28 | 0.069 ± 0.0050 |
| Nicked Δ−4 | 5′-GGCCAAGCCGGATGTGGTAAACACTTT-3′ | 6.80 ± 0.41 | 0.027 ± 0.0050 |
Underscored sequences indicate the Ets-1 and CBFα2 recognition sequences.
Values are a result of two to three independent experiments. SE expresses the accuracy of the curve fit rather than the error reflected in the error bars (as described in Materials and Methods).
From accompanying report (23).
EMSA.
Equilibrium dissociation constants (KDs) of CBFα2ΔC and Ets-1 were determined by electrophoretic mobility shift assays (EMSA) using conditions described previously (47). In brief, the reaction mixtures were in TGEK60 buffer, which consisted of 25 mM Tris-Cl (pH 7.9), 10% glycerol, 6 mM MgCl2, 0.5 mM EDTA, and 60 mM KCl with 0.5 mM dithiothreitol and 200 μg of bovine serum albumin per ml. Following a 20-min incubation on ice (40 min for Ets-1ΔN280), reaction products were loaded onto a native polyacrylamide gel (acrylamide:bisacrylamide, 30:0.8, 45 mM Tris-borate [pH 8.3], 1 mM EDTA). The full-length Ets-1 protein was resolved on 6% polyacrylamide gels (16 cm), while the truncated Ets-1 proteins were resolved on 8% gels (20 cm). The gels were dried and exposed to a PhosphorImager screen (Molecular Dynamics), and relative radioactivity was quantified as the volume integration of individual bands. When protein titrations were used, the concentrations were in a range that resulted in approximately 0 to 100% binding. For experiments in which CBFα2ΔC was added in saturating amounts, the concentration was at least 10-fold higher than the KD of CBFα2ΔC for its specific site (≥2 × 10−8 M). This ensured >90% DNA occupancy. In all assays, the DNA concentrations were at least 10-fold below the estimated KD of the Ets-1 species (≤10−11 M), ensuring that the total Ets-1 species concentration [Pt] was an accurate estimate of free Ets-1 species concentration [P].
Quantitative analysis of DNA binding.
For assays containing only a single binding species, Ets-1, the KDs were measured as described previously (27). Specifically, the fraction of free DNA, [D]/[Dt], was determined by measuring the ratio of the free DNA signal analyzed at each protein concentration to the DNA signal in a control lane containing no protein. The fraction of DNA in complex with protein, [PD]/[Dt], was derived from the relationship [PD]/[Dt] = 1 − [D]/[Dt]. To derive the KD with standard error (SE), the data were fit to the rearranged mass action equation, [PD]/[Dt] = 1/(1 + KD/[P]), using nonlinear least squares analyses (Kaleidagraph; Synergy Software). Multiple experiments were performed with the same range of protein concentrations to provide mean and SE values for each data point. Mean values were used for curve fitting. SEs of means are displayed as error bars. SEs of the KD values, which were provided by the curve fitting, are presented in tabular form.
To measure cooperative DNA binding, the apparent DNA binding affinities of Ets-1 species were determined in the presence of a large molar excess of the CBFα2ΔC fragment. To perform curve fitting with the equation [PD]/[Dt] = 1/(1 + KD/[P]), several assumptions were made. (i) The disappearance of the binary complex (DNA + CBFα2ΔC) was the key parameter to be measured; therefore, [Dt] was defined as the binary complex signal in a control lane that contained DNA and only CBFα2ΔC. (ii) The binary complex signal (DNA + CBFα2ΔC) was used as [D] for reaction mixtures with DNA + CBFα2ΔC + Ets-1 species. (iii) The fraction of DNA bound in the ternary complex was defined as [PD]/[Dt], which was derived from 1 − [D]/[Dt].
RESULTS
Ets-1 binding to the Mo-MLV enhancer is repressed by auto-inhibition and enhanced by CBFα2.
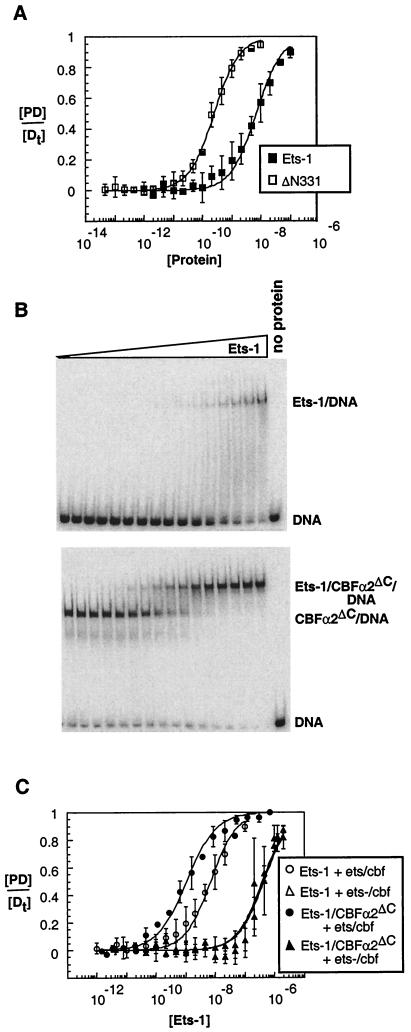
The initial goal was to provide a biological context for the hypothesis that CBF counteracts Ets-1 autoinhibition. Our approach was to document the phenomena of auto-inhibition and cooperativity on a biologically relevant site that displays synergistic transcriptional activation by Ets-1 and CBF. Our previous studies of Ets-1 DNA binding often used an artificial, high-affinity binding site, termed SC1 (47). For our initial experiments, we switched to the composite element in the Mo-MLV enhancer that binds both Ets-1 and CBFα2 (24, 65). To investigate the autoinhibition of Ets-1 on the Mo-MLV enhancer site, we measured the DNA binding affinity of full-length Ets-1 and the amino-terminal deletion mutant, Ets-1ΔN331, which lacks the amino-terminal inhibitory helices (Fig. 1). DNA binding by purified proteins was detected by EMSA. KDs were determined from full binding curves. The affinity of Ets-1 was 27-fold lower than that of Ets-1ΔN331 (Fig. 2A; Table 1). This result is similar to the 23-fold auto-inhibition observed previously on the SC1 site (Table 1) (27). Also, as expected from previous studies, Ets-1 affinity for the Mo-MLV site was 10-fold lower than that for the high-affinity SC1 site (24, 43, 47). The detection of autoinhibition on the Mo-MLV enhancer establishes that this phenomenon is not dependent on the sequence or affinity of the binding site.
FIG. 2.
Ets-1 autoinhibition and DNA cooperativity with CBFα2ΔC on the Mo-MLV enhancer. (A) Measurement of auto-inhibition. Equilibrium DNA binding curves for Ets-1 (filled squares) and Ets-1ΔN331 (open squares) were obtained from EMSA as described in Materials and Methods. [DP]/[Dt] is presented as the mean (±SE) of two or three independent experiments. The KD (molar) was derived by fitting the data to the equation [PD]/[Dt] = 1/(1 + (KD/[P])), using nonlinear least squares analysis. (B) EMSA of equilibrium DNA binding studies of Ets-1 titrated onto DNA alone (top) or in the presence of a constant (∼2 × 10−8 M) amount of CBFα2ΔC (bottom). The wedge indicates increasing amounts of Ets-1 in each binding assay as indicated in panel C. (C) Equilibrium DNA binding curves for Ets-1 (data from panel B), as well as from a repeat of these experiments performed on a mutant Mo-MLV site, ets−/cbf.
To characterize cooperative DNA binding of Ets-1 and CBF on the Mo-MLV enhancer site, Ets-1 DNA binding was measured in the presence and absence of saturating levels of CBFα2. We used a fragment of CBFα2 (spanning amino acids 1 to 331) designated CBFα2ΔC, which is the largest version of CBFα2 obtainable in a pure state from a baculovirus expression system (23). The presence of CBFα2ΔC enhanced Ets-1 DNA binding affinity 20-fold (Fig. 2B and C; Table 1). This result is similar to the 10-fold enhancement of Ets-1 affinity for the artificial composite site, SC1/core, which contains the high-affinity Ets-1 binding site SC1 juxtaposed with native spacing to the CBF site found in the Mo-MLV enhancer (Table 1) (23). DNA binding of Ets-1 and CBFα2ΔC also was measured on a Mo-MLV enhancer site, ets−/cbf, in which the Ets-1 recognition sequence was mutated (Fig. 2C; Table 1). Nonspecific Ets-1 binding was detected, but only at the highest concentrations of Ets-1. Dual occupancy, but no cooperative DNA binding, was observed with CBFα2ΔC. In conclusion, cooperative binding of Ets-1 and CBFα2ΔC is independent of the sequence and affinity of the binding site; however, sequence-specific DNA binding by Ets-1 is required. In summary, the Mo-MLV composite site displays both autoinhibition and DNA binding cooperativity. Thus, these initial findings link the auto-inhibition and DNA binding cooperativity phenomena to a biologically relevant enhancer element.
Regions required for cooperativity and auto-inhibition overlap.
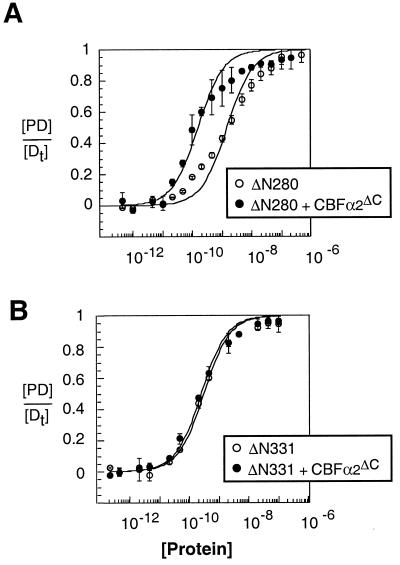
In studies of the Mo-MLV enhancer site, the level of auto-inhibition (10- to 20-fold) was in the same range as the level of cooperative binding. This was our first clue that CBFα2 might enhance Ets-1 DNA binding by affecting auto-inhibition. These results also suggested that both phenomena might involve the same region of Ets-1. To map the regions of Ets-1 that are necessary for cooperativity with CBFα2, the DNA binding affinities of amino-terminal deletion mutants Ets-1ΔN280 and Ets-1ΔN331 were measured with and without CBFα2ΔC (Fig. 1). The composite site, SC1/core, composed of a high-affinity Ets-1 site juxtaposed to the CBF site of the Mo-MLV enhancer, was used because the higher affinity leads to sharper bands on the EMSA gel and thus an increase in the precision of the assay. Ets-1ΔN280 displayed a 10-fold enhancement of DNA binding in the presence of CBFα2ΔC, demonstrating cooperativity similar to that of full-length Ets-1 (Fig. 3A; Table 1) (23). In contrast, Ets-1ΔN331 and CBFα2ΔC did not bind DNA cooperatively (Fig. 3B; Table 1). Our results indicate that sequences necessary for cooperativity lie between positions 280 and 331. Two amino-terminal inhibitory helices, HI-1 and HI-2, that play a key role in auto-inhibition are found in this region (Fig. 1B) (59). Because this region is part of a structural domain that includes these two helices as well as the carboxyl-terminal inhibitory helix, H4, and the ETS domain (Fig. 1B), we did not attempt to further delineate functional sequences. In conclusion, the structural domain required for auto-inhibition is also required for DNA binding cooperativity.
FIG. 3.
Mapping regions required for cooperativity between Ets-1 and CBFα2ΔC. (A) Equilibrium DNA binding curves for Ets-1ΔN280 obtained from EMSA in the absence (open circles) and presence (filled circles) of a constant (∼2 × 10−8 M) amount of CBFα2ΔC. (B) Equilibrium DNA binding curves for Ets-1ΔN331 obtained from EMSA in the absence (open circles) and presence (filled circles) of a constant (∼2 × 10−8 M) amount of CBFα2ΔC.
Analyses of DNA determinants for cooperativity implicate flexible protein-protein interactions.
Two potential mechanisms could mediate cooperativity. CBFα2 could enhance Ets-1 DNA binding indirectly through the topological connection between the two binding sites. This would represent a through-DNA effect, with CBFα2 causing a change in the DNA conformation that affects the binding of Ets-1. Alternatively, CBFα2 could directly contact Ets-1 and mediate its effect through protein interactions. In this case, DNA could play a relatively passive role by simply tethering the two proteins for optimal interaction.
To distinguish between protein-protein interactions and through-DNA effects, we investigated the DNA determinants for cooperativity. To test the proposed through-DNA mechanism, the two sites were topologically uncoupled by nicking one strand of the composite binding site, SC1/core. This break in the phosphodiester backbone did not lower the level of cooperativity (Table 2). This result suggests that DNA binding by one protein does not indirectly influence the binding of the second protein. It is worth noting that this experiment does not address the long-distance effects of DNA conformation that have been detected in ternary complexes formed on more widely spaced binding sites (66).
The DNA determinants for cooperative binding also were explored by manipulating the orientation of the binding sites (Table 2). Both the Ets-1–DNA and CBFα2-DNA binary complexes are expected to show polarity because the Runt domain and the ETS domain each bind DNA as a monomer with no apparent pseudosymmetry. In the case of Ets-1, structural data demonstrate the asymmetry of the ETS domain-DNA interaction (5, 32, 41, 72). Structural information for CBFα2 also predicts an asymmetric complex (6, 44). Thus, a change in the orientation of either binding site could alter the putative protein-protein interface. Surprisingly, changing the orientation of the CBF site (invCBF) did not change cooperative DNA binding (Table 2). These results indicate that considerable flexibility exists in the CBF elements that participate in the cooperativity and implicate sequences outside the highly structured Runt domain as being important for cooperativity. This proposal is consistent with mapping data in the accompanying report (23). We propose that these flanking elements display sufficient flexibility or perhaps are located centrally within the structure, such that reversal of site orientation does not affect the CBFα2 and Ets-1 interactions. In contrast, reversal of the Ets-1 binding site orientation (invEts) reduced cooperativity from 10- to 3.5-fold (Table 2). We propose that the asymmetric location of the inhibitory module within Ets-1ΔN280 causes this reduction (Fig. 1). Because sequences within this module are necessary for cooperativity, the optimal interface between Ets-1 and CBF may be compromised by the change in binding site orientation.
DNA requirements were investigated further by varying the spacing between the recognition sequences for Ets-1 and CBF. To disrupt or retain helical phasing, the spacing of 4 bp between the SC1 and core sites was expanded by 5 (Δ+5) or 10 (Δ+10) bp (Table 2). In addition, we tested a duplex in which the spacing was eliminated (Δ−4). The changes in spacing reflect the variability of spacing in native sites. For example, the polyomavirus enhancer and osteopontin promoters have the added 9 or 10 bp, whereas the TCRβ enhancer is identical to Δ−4 (19, 55, 65, 74). Both the Δ+5 and Δ+10 duplexes displayed cooperativity between Ets-1 and CBFα2 (Table 2). (These results provide an important control for the site orientation experiments above, in which spacing was altered.) Because locally altered DNA conformation does not easily lead to long-distance effects (75), this retention of cooperativity supports the conclusion that through-DNA effects are not involved in the 10-fold enhancement of Ets-1 binding. Again, these results imply that protein elements that mediate cooperativity must accommodate considerable structural flexibility.
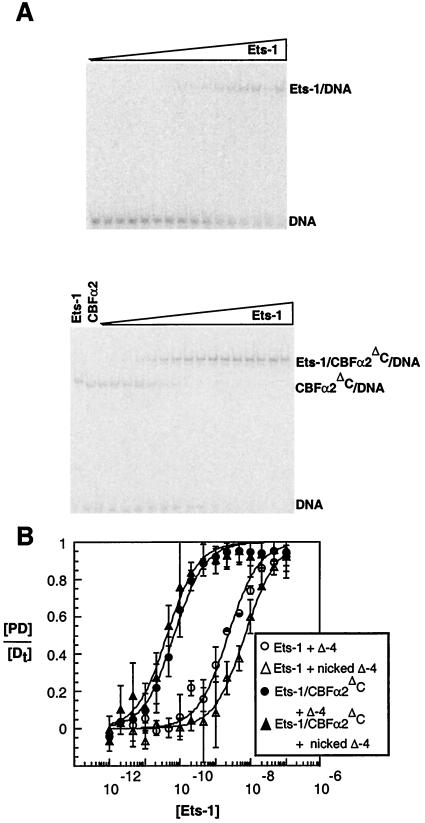
In contrast to results from widely spaced binding sites, the Δ−4 duplex displayed a higher level of cooperative DNA binding than the SC1/core composite site (Fig. 4; Table 2). The 33-fold cooperativity could be mediated by additional direct contacts that would be possible due to the closer proximity of the proteins. Alternatively, the proximity of the recognition sequences could facilitate through-DNA effects. To test this latter alternative, a nick was introduced into one strand of the Δ−4 duplex (Table 2). Nicking did not lower the level of cooperativity. The retention of cooperativity eliminates an indirect mechanism that uses the topological coupling of the binding sites. Instead, the nicked binding site displayed an increase in cooperative DNA binding, from the 33-fold effect observed with Δ−4 to a 250-fold effect (Fig. 4; Table 2). There are several possible reasons for this increase. A torsional strain in the DNA backbone could develop due to the proximity of the sites, and the nick relieves the strain by uncoupling the sites. Alternatively, the loss of phosphate at the site of the nick could affect the energetics of binding. These findings suggest that an additional mechanism contributes to cooperativity of closely apposed sites. Future investigations will explore this possibility.
FIG. 4.
Effects of topological uncoupling of DNA binding sites for Ets-1 and CBFα2ΔC. (A) EMSA of equilibrium DNA binding studies of Ets-1 titrated onto nicked Δ−4 DNA alone (top) or in the presence of a constant (∼2 × 10−8 M) amount of CBFα2ΔC (bottom). (B) Equilibrium DNA binding curves for Ets-1 obtained from EMSA in the absence (open symbols) and presence (filled symbols) of CBFα2ΔC. Cooperative DNA binding was measured on either the Δ−4 (circles) or nicked Δ−4 (triangles) DNA duplex (Table 2).
Derepressed Ets-1 mutant is resistant to CBF cooperative effects.
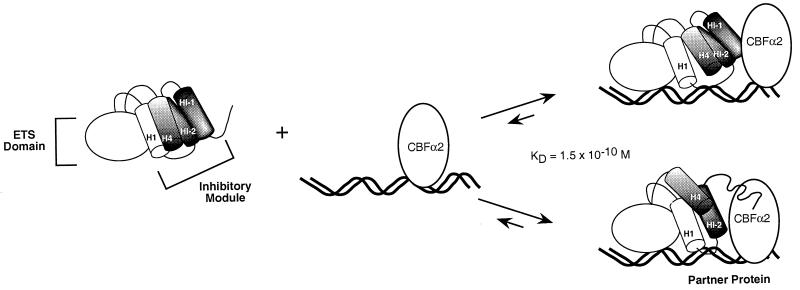
The results of quantitative binding assays and mapping experiments suggested that cooperativity could function by counteracting auto-inhibition of Ets-1. Furthermore, the analysis of DNA determinants suggested that the mechanism involves protein interactions rather than through-DNA effects. Finally, considerable flexibility is expected in these protein interactions. There are two possible models to explain how the Ets-1–CBFα2 partnership counteracts auto-inhibition through such flexible protein interactions. In one scenario, CBFα2 could cause a conformational change in the folded inhibitory module. We envision this as a repositioning of structural elements such that unfolding of helix HI-1 is no longer necessary. Alternatively, CBFα2 could favor a shift in the equilibrium toward the disrupted inhibitory module. Unfolding and/or refolding of helix HI-1 could be affected.
To gain additional insight into the mechanism of cooperativity, we tested a version of Ets-1 in which the inhibitory module is constitutively disrupted and that displays derepressed DNA binding. Ets-1ΔN280;L429A bears an amino acid substitution in the inhibitory helix H4 (Fig. 5A). Proteolysis experiments indicate that helix HI-1 is constitutively unfolded in this mutant version of Ets-1 (data not shown). In quantitative binding assays, Ets-1ΔN280;L429A bound DNA with a 15-fold-higher affinity than Ets-1ΔN280 (Fig. 5B; Table 1). More importantly, CBFα2ΔC did not enhance the affinity of this mutant version of Ets-1 (Fig. 5B; Table 1). The presence of a constitutively unfolded inhibitory module resulted in insensitivity to CBFα2 effects. This finding strongly suggests that the DNA binding cooperativity mechanism involves regulating the conformation of the inhibitory module. As detailed in Discussion, these results are consistent with either mechanistic model. In conclusion, the behavior of this mutant demonstrates that the inhibitory module is required for DNA binding cooperativity, thus supporting our proposal that DNA binding cooperativity counteracts auto-inhibition.
FIG. 5.
Derepressed Ets-1 variant displays no DNA binding cooperativity with CBFα2. (A) Schematic representation of the constitutive unfolding of Ets-1ΔN280;L429A due to a leucine-to-alanine substitution in helix H4, as shown (×). The KD value is for the SC1/core. (B) Equilibrium DNA binding curves for Ets-1ΔN280;L429A obtained from EMSA in the absence (open circles) and presence (filled circles) of a constant (∼2 × 10−8 M) amount of CBFα2ΔC.
DISCUSSION
Auto-inhibition provides a mechanism by which protein activity can be regulated tightly. In the case of Ets-1, this auto-regulation decreases the DNA binding affinity 10- to 20-fold. We predicted that a regulatory mechanism would counteract this auto-inhibition and enable the full DNA binding potential of the Ets-1 ETS domain to be used within a biological context. Ets-1 functions in association with other transcription factors, implicating cooperative DNA binding with other proteins as a mechanism for rescinding auto-inhibition. Both this study and the accompanying report (23) present quantitative analyses of the DNA binding cooperativity between Ets-1 and CBFα2. The quantitative approach of these experiments is distinctive, representing one of only a few such comprehensive studies of a DNA binding partnership in a eukaryotic system. These experiments also laid the groundwork for our mechanistic model of CBF rescinding Ets-1 auto-inhibition.
Mechanistic model of CBFα2 and Ets-1 cooperative DNA binding.
There are two general mechanisms by which CBFα2 could affect Ets-1 DNA binding. Due to the close apposition of the Ets-1 and CBF binding sites, we considered a through-DNA mechanism that would require no direct protein interactions. This model was discounted by the observations that topological uncoupling of the two sites did not disrupt cooperativity. In addition, there was considerable flexibility in the spacing and orientation of Ets-1 and CBFα2 binding sites. Consistent with these observations, selection of consensus binding sites for Ets-1 and CBFα2 also detected flexibility in site configuration (74). The alternative mechanism evokes direct protein interactions between the partners. Protein-protein interactions could accommodate the observed flexibility. This has been proposed previously for the ets protein Elk-1 and its partner serum response factor, which display DNA binding cooperativity on composite sites with a variety of spacing configurations and orientations (67). Our study provides a rigorous quantitative analysis of this type of flexibility.
Our proposed mechanism for DNA binding cooperativity is built on the model of Ets-1 auto-inhibition (Fig. 1) (21). DNA binding is accompanied by a conformational change that includes unfolding of one of the three inhibitory helices. We have proposed that this requisite conformational change reduces the DNA binding affinity of full-length Ets-1 and other repressed species such as Ets-1ΔN280. This report links auto-inhibition and DNA binding cooperativity. Quantitative binding studies showed that DNA binding cooperativity requires the presence of the amino-terminal inhibitory helices. Because this region is part of the inhibitory module, we envision that CBFα2 could affect the conformational state of this structural domain. Two possible models are envisioned (Fig. 6). First, CBFα2 could alter the position of the inhibitory module such that the unfolding of HI-1 is no longer necessary for DNA binding. This repositioning model predicts a protein interface between Ets-1 and CBFα2 that would stabilize the inhibitory helices in the absence of their usual contact to the ETS domain. Such direct contacts could be made within any of the inhibitory helices. Alternatively, CBFα2 could stimulate the rate of unfolding and/or prevent refolding of helix HI-1. In contrast to the first model, this disruption model need not evoke a specific interface between the disrupted inhibitory module and CBFα2 since no structural elements are being stabilized. Either of these scenarios accommodates the flexibility of the DNA determinants for cooperativity.
FIG. 6.
Model of Ets-1 auto-inhibition and CBFα2 cooperativity. The inhibitory module is composed of three inhibitory helices, HI-1, HI-2, and H4, that pack with helix H1 of the ETS domain. Ets-1 DNA binding in the absence of CBF is accompanied by unfolding of helix HI-1 (Fig. 1). We propose two alternative models by which CBFα2 can counteract auto-inhibition. First, CBFα2 repositions the inhibitory elements such that the unfolding of helix HI-1 is no longer necessary. Alternatively, CBFα2 enhances the unfolding or represses the refolding of helix HI-1. The KD value is for the ternary complex on the SC1/core composite site.
The behavior of the constitutively activated mutant, the L429A derivative of Ets-1ΔN280, can be explained by both models. The inhibitory module of this mutant protein is constitutively disrupted and thus predominantly in the unfolded state. If CBFα2 alters the position of the inhibitory module by making specific contacts with structural elements, the disrupted inhibitory helices in Ets-1ΔN280;L429A would not provide the required protein interface, consistent with the repositioning model. The behavior of Ets-1ΔN280;L429A also can be explained by the model in which CBFα2 shifts the conformational equilibrium of helix HI-1 toward the unfolded state. This equilibrium would not be influenced in the constitutively unfolded inhibitory module. Recall that this disruption model proposes no specific binding interface between the disrupted inhibitory module and CBF in the ternary complex. If such contacts were to exist, the L429A variant might be expected to retain the interface and, thus, cooperativity. Thus, the lack of cooperativity between CBFα2 and Ets-1ΔN280;L429A is also consistent with the disruption model. To distinguish between these alternative mechanisms, structural studies are under way to determine the conformational state of the inhibitory module within the ternary complex.
Our preliminary analyses suggest that any interaction between Ets-1 and CBFα2 at physiologically relevant concentrations occurs only on DNA. Specifically, surface plasmon resonance experiments that were performed at protein concentrations in the 10 nM range failed to detect interactions (data not shown). The interaction could have such a low affinity that it will be difficult to detect by this approach. Alternatively, DNA binding by one or the other partner may expose residues necessary for the interaction. In this regard, it is interesting that reciprocal cooperativity, which enhances CBFα2 DNA binding, was observed only under conditions in which CBFα2 was incubated with DNA before the addition of Ets-1 (23). Similarly, the enhancement of Ets-1 binding reported here and in the accompanying report (23) was detected under conditions in which CBFα2 is likely to bind before Ets-1. Although all components were added simultaneously to the reaction mixtures, the high concentration of CBFα2 (∼10−8 M) is expected to cause a rapid association of CBFα2, facilitating preassembly of a DNA-CBFα2 binary complex. The model in Fig. 6 reflects this proposed sequence of events.
There are two reports that describe direct interactions between Ets-1 and CBFα2 in the absence of DNA. In one study, the amino-terminal sequences of Ets-1 (amino acids 123 to 240), which lie outside the regions mapped here, interact directly with CBFα2 in a glutathione S-transferase pulldown assay (19). In a second study, glutathione S-transferase pull-down assays detected CBFα2 interactions with only the carboxyl-terminal half of Ets-1 (31). The concentration and folded state of the proteins were not measured in these assays. Thus, neither the affinity nor the specificity of these interactions has been rigorously determined. High-resolution structural and genetic analyses are necessary to describe the interface of Ets-1 and CBFα2 in the ternary complex.
In the accompanying report (23), which focuses on the DNA binding properties of CBF, we report that Ets-1 enhances the DNA binding activity of CBFα2ΔC at least sevenfold. Thus, as expected from thermodynamic principles, the DNA binding cooperativity between Ets-1 and CBFα2 is reciprocal. Several studies report that CBFα2 also displays auto-inhibition (23, 30, 31). We suggest that Ets-1 counteracts the auto-inhibition of the CBFα2 Runt domain, perhaps also by altering an inhibitory conformation. Interestingly, the cooperative binding of CBFα2 with Ets-1 is an alternative to the enhancement of CBFα2 DNA binding by its heterodimeric partner CBFβ. For additional discussion of the interplay between Ets-1 and CBFβ see the accompanying report (23).
By necessity our quantitative studies were performed with a truncated version of CBFα2 that could be obtained highly purified from a baculovirus expression system. We were initially concerned that this experimental design could compromise our study. However, recently a less quantitative analysis of DNA binding cooperativity between Ets-1 and CBFα2 that utilized full length CBFα2 prepared by in vitro transcription-translation was reported (31). Importantly, the two studies concur on the regions of Ets-1 that are important for DNA binding cooperativity. These findings thus lend support to our conclusions that are based on the behavior of truncated CBFα2.
Combinatorial control of transcription.
Partnerships between two transcription factors are the building blocks of combinatorial control of gene expression. These types of interactions are predicted to enhance the specificity of regulatory transcription factors. In the case of Ets-1 and CBFα2, each binds a sequence-specific region of only 9 to 10 bp as a monomer, whereas the ternary complex binds to a composite site that spans 18 to 20 bp of specific sequences. This extended region of preferred sequence provides a higher degree of sequence specificity and thus could direct Ets-1 to function preferentially on enhancer elements with CBFα2 binding sites, or vice versa. Interestingly, in the case of Ets-1, there are other putative DNA binding partners, including Jun/Fos, TFE3, USF, and NF-κB (12, 37, 58, 66). Preliminary quantitative studies indicate that NF-κB has effects on Ets-1 DNA binding comparable to those of CBFα2 (12) (T. Goetz, unpublished data). This promiscuity of Ets-1 could reflect multiple pathways to transcriptional synergy, including alternative mechanisms for DNA binding cooperativity. However, these other partnerships could work also by counteracting auto-inhibition. Indeed, a recent report implicates the basic helix-loop-helix protein TFE3 in this role on the immunoglobulin μ enhancer (66). The mechanistic models developed here could accommodate Ets-1 interactions with these other partner proteins.
The maximal activity of complex enhancers requires large assemblies of DNA binding proteins. This phenomenon is well illustrated by the TCRα enhancer, in which a basic helix-loop-helix protein (either USF or TFE3) and the high-mobility-group domain protein Lef-1 function with Ets-1 and CBFα2 (19, 26, 39). Therefore, the Ets-1 and CBFα2 partnership works in the context of a much larger multiprotein complex. Another such example is the function of Ets-1 on the human immunodeficiency virus type 1 viral enhancer in collaboration with Sp1, NF-κB, Lef-1, and USF (or TFE3) (57, 64). Such complexes provide additional opportunities to enhance the specificity and affinity of regulatory transcription factors.
An additional interesting feature of this network of interactions is the family membership of Ets-1 and CBFα2. As described earlier, there are at least 20 vertebrate ets genes and three CBFA genes. Other ets proteins also synergize with CBFα2 (e.g., Fli-1, PU.1, GABP, and MEF) (38, 52, 65). Similarly, another CBFA protein, CBFA1, is functionally linked to Ets-1 (55). Furthermore, multiple members of the ets family of transcription factors display auto-inhibition of DNA binding (22). It is possible that the mechanisms of auto-inhibition and DNA binding cooperativity described here are sufficiently conserved to provide a framework for understanding many of these combinations of transcription factors.
ACKNOWLEDGMENTS
We acknowledge research support from the National Institutes of Health to B.J.G. (GM38663) and training grant support for T.L.G. (CA090602). National Institutes of Health support to the Huntsman Cancer Institute (CA42014) is also acknowledged. N.A.S. was supported by National Institutes of Health grants CA58343 and CA75611.
We are grateful to Don Ayer, John Bushweller, and Lawrence McIntosh for critical comments on the manuscript.
REFERENCES
- 1.Anderson M K, Hernandez-Hoyos G, Diamond R A, Rothenberg E V. Precise developmental regulation of Ets family transcription factors during specification and commitment to the T cell lineage. Development. 1999;126:3131–3148. doi: 10.1242/dev.126.14.3131. [DOI] [PubMed] [Google Scholar]
- 2.Bae S-C, Takahashi E, Zhang Y, Ogawa E, Shigesada K, Namba Y, Satake M, Ito Y. Cloning, mapping and expression of PEBP2αC, a third gene encoding the mammalian Runt domain. Gene. 1995;159:245–248. doi: 10.1016/0378-1119(95)00060-j. [DOI] [PubMed] [Google Scholar]
- 3.Bae S-C, Yamaguchi-Iwai Y, Ogawa E, Maruyama M, Inuzuka N, Kagoshima H, Shigesada K, Satake M, Ito Y. Isolation of PEBP2αB cDNA representing the mouse homolog of human acute myeloid leukemia gene, AML1. Oncogene. 1993;8:809–814. [PubMed] [Google Scholar]
- 4.Barton K, Muthusamy N, Fischer C, Ting C-N, Walunas T, Lanier L, Leiden J. The Ets-1 transcription factor is required for the development of natural killer cells in mice. Immunity. 1998;9:555–563. doi: 10.1016/s1074-7613(00)80638-x. [DOI] [PubMed] [Google Scholar]
- 5.Batchelor A, Piper D, Charles de la Brousse F, McKnight S, Wolberger C. The structure of GABPα/β: an ETS domain-ankryin repeat heterodimer bound to DNA. Science. 1998;279:1037–1041. doi: 10.1126/science.279.5353.1037. [DOI] [PubMed] [Google Scholar]
- 6.Berardi M, Sun C, Zehr M, Abildgard F, Peng J, Speck N, Bushweller J. The core binding factor Runt domain is a structural and functional homologue of the Ig domains of the Rel homology region of NF-κB. Structure. 1999;7:1247–1256. doi: 10.1016/s0969-2126(00)80058-1. [DOI] [PubMed] [Google Scholar]
- 7.Bhat N K, Fisher R J, Fujiwara S, Ascione R, Papas T S. Temporal and tissue-specific expression of mouse ets genes. Proc Natl Acad Sci USA. 1987;84:3161–3165. doi: 10.1073/pnas.84.10.3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bories J-C, Willerford D M, Grevin D, Davidson L, Camus A, Martin P, Stehelin D, Alt F W. Increased T-cell apoptosis and terminal B-cell differentiation induced by inactivation of the Ets-1 proto-oncogene. Nature. 1995;377:635–638. doi: 10.1038/377635a0. [DOI] [PubMed] [Google Scholar]
- 9.Brass A L, Kehrli E, Eisenbeis C F, Storb U, Singh H. Pip, a lymphoid-restricted IRF, contains a regulatory domain that is important for autoinhibition and ternary complex formation with the ets factor PU.1. Genes Dev. 1996;10:2335–2347. doi: 10.1101/gad.10.18.2335. [DOI] [PubMed] [Google Scholar]
- 10.Chan S K, Mann R S. A structural model for a homeotic protein-extradenticle-DNA complex accounts for the choice of HOX protein in the heterodimer. Proc Natl Acad Sci USA. 1996;93:5223–5228. doi: 10.1073/pnas.93.11.5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan S K, Popperl H, Krumlauf R, Mann R S. An extradenticle-induced conformational change in a HOX protein overcomes an inhibitory function of the conserved hexapeptide motif. EMBO J. 1996;15:2476–2487. [PMC free article] [PubMed] [Google Scholar]
- 12.Dickinson L A, Trauger J W, Baird E E, Dervan P B, Graves B J, Gottesfeld J M. Inhibition of Ets-1 DNA binding and ternary complex formation between Ets-1, NF-κB and DNA by a designed DNA-binding ligand. J Biol Chem. 1999;274:12765–12773. doi: 10.1074/jbc.274.18.12765. [DOI] [PubMed] [Google Scholar]
- 13.Dombroski A J, Walter W A, Gross C A. Amino-terminal amino acids modulate sigma-factor DNA-binding activity. Genes Dev. 1993;7:2446–2455. doi: 10.1101/gad.7.12a.2446. [DOI] [PubMed] [Google Scholar]
- 14.Donaldson L W, Petersen J M, Graves B J, McIntosh L P. Secondary structure of the ETS domain places murine Ets-1 in the superfamily of winged helix-turn-helix DNA-binding proteins. Biochemistry. 1994;33:13509–13516. doi: 10.1021/bi00250a001. [DOI] [PubMed] [Google Scholar]
- 15.Donaldson L W, Petersen J M, Graves B J, McIntosh L P. Solution structure of the ETS domain from murine Ets-1: a winged helix-turn-helix DNA binding motif. EMBO J. 1996;15:125–134. [PMC free article] [PubMed] [Google Scholar]
- 16.Erman B, Cortes M, Nikolajczyk B S, Speck N A, Sen R. ETS-core binding factor: a common composite motif in antigen receptor gene enhancers. Mol Cell Biol. 1998;18:1322–1330. doi: 10.1128/mcb.18.3.1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fleischman L F, Holtzclaw L, Russell J T, Mavrothalassitis G, Fisher R J. ets-1 in astrocytes: expression and transmitter-evoked phosphorylation. Mol Cell Biol. 1995;15:925–931. doi: 10.1128/mcb.15.2.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fleischman L F, Pilaro A M, Murakami K, Kondoh A, Fisher R J, Papas T S. c-Ets-1 protein is hyperphosphorylated during mitosis. Oncogene. 1993;8:771–780. [PubMed] [Google Scholar]
- 19.Giese K, Kingsley C, Kirshner J R, Grosschedl R. Assembly and function of a TCRα enhancer complex is dependent on LEF-1 induced DNA bending and multiple protein-protein interactions. Genes Dev. 1995;9:995–1008. doi: 10.1101/gad.9.8.995. [DOI] [PubMed] [Google Scholar]
- 20.Granger S W, Fan H. In vivo footprinting of the enhancer sequences in the upstream long terminal repeat of Moloney murine leukemia virus: differential binding of nuclear factors in different cell types. J Virol. 1998;72:8961–8970. doi: 10.1128/jvi.72.11.8961-8970.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Graves B J, Cowley D O, Goetz T L, Petersen J M, Jonsen M D, Gillespie M E. Autoinhibition as a transcriptional regulatory mechanism. Cold Spring Harbor Symp Quant Biol. 1998;63:621–629. doi: 10.1101/sqb.1998.63.621. [DOI] [PubMed] [Google Scholar]
- 22.Graves B J, Petersen J M. Specificity within the ets family of transcription factors. Adv Cancer Res. 1998;75:1–55. doi: 10.1016/s0065-230x(08)60738-1. [DOI] [PubMed] [Google Scholar]
- 23.Gu T-L, Goetz T L, Graves B J, Speck N A. Auto-inhibition and partner proteins core-binding factor β (CBFβ) and Ets-1, modulate DNA binding by CBFα2 (AML1) Mol Cell Biol. 2000;20:91–103. doi: 10.1128/mcb.20.1.91-103.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gunther C V, Graves B J. Identification of ETS domain proteins in murine T lymphocytes that interact with the Moloney murine leukemia virus enhancer. Mol Cell Biol. 1994;14:7569–7580. doi: 10.1128/mcb.14.11.7569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hagman J, Grosschedl R. An inhibitory carboxyl terminal domain in Ets-1 and Ets-2 mediates differential binding of ETS family factors to promoter sequences of the mb-1 gene. Proc Natl Acad Sci USA. 1992;89:8889–8893. doi: 10.1073/pnas.89.19.8889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hernandez-Munain C, Roberts J L, Krangel M S. Cooperation among multiple transcription factors is required for access to minimal T-cell receptor alpha-enhancer chromatin in vivo. Mol Cell Biol. 1998;18:3223–3233. doi: 10.1128/mcb.18.6.3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jonsen M D, Petersen J M, Xu Q, Graves B J. Characterization of the cooperative function of inhibitory sequences of Ets-1. Mol Cell Biol. 1996;16:2065–2073. doi: 10.1128/mcb.16.5.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kagoshima H, Shigesada K, Satake M, Ito Y, Miyoshi H, Ohki M, Pepling M, Gergen P. The Runt domain identifies a new family of heteromeric transcriptional regulators. Trends Genet. 1993;9:338–341. doi: 10.1016/0168-9525(93)90026-e. [DOI] [PubMed] [Google Scholar]
- 29.Kamachi Y, Ogawa E, Asano M, Ishida S, Murakami Y, Satake M, Ito Y. Purification of a mouse nuclear factor that binds to both the A and B cores of the polyomavirus enhancer. J Virol. 1990;64:4808–4819. doi: 10.1128/jvi.64.10.4808-4819.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanno T, Kanno Y, Chen L F, Ogawa E, Kim W Y, Ito Y. Intrinsic transcriptional activation-inhibition domains of the polyomavirus enhancer binding protein 2/core binding factor α subunit revealed in the presence of the β subunit. Mol Cell Biol. 1998;18:2444–2454. doi: 10.1128/mcb.18.5.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim W-Y, Sieweke M, Ogawa D, Wee H-J, Englemeier U, Graf T, Ito Y. Mutual activation of Ets-1 and AML1 DNA binding by direct interaction of their autoinhibitory domains. EMBO J. 1999;18:1609–1620. doi: 10.1093/emboj/18.6.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kodandapani R, Pio F, Ni C-Z, Piccialli G, Klemsz M, McKercher S, Maki R A, Ely K R. A new pattern for helix-turn-helix recognition revealed by the PU.1 ETS-domain-DNA complex. Nature. 1996;380:456–460. doi: 10.1038/380456a0. [DOI] [PubMed] [Google Scholar]
- 33.Kola I, Brookes S, Green A R, Garber R, Tymms M, Papas T S, Seth A. The Ets-1 transcription factor is widely expressed during murine embryo development and is associated with mesodermal cells involved in morphogenetic processes such an organ formation. Proc Natl Acad Sci USA. 1993;90:7588–7592. doi: 10.1073/pnas.90.16.7588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Levanon D, Negreanu V, Bernstein Y, Bar-Am I, Avivi L, Groner Y. AML1, AML2, AML3, the human members of the runt domain gene-family: cDNA structure, expression, and chromosomal localization. Genomics. 1994;23:425–432. doi: 10.1006/geno.1994.1519. [DOI] [PubMed] [Google Scholar]
- 35.Lim F, Kraut N, Frampton J, Graf T. DNA binding by c-Ets-1, but not v-Ets, is repressed by an intramolecular mechanism. EMBO J. 1992;11:643–652. doi: 10.1002/j.1460-2075.1992.tb05096.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ling Y, Lakey J H, Roberts C E, Sharrocks A D. Molecular characterization of the B-box protein-protein interaction motif of the ETS-domain transcription factor Elk-1. EMBO J. 1997;16:2431–2440. doi: 10.1093/emboj/16.9.2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Logan S K, Garabedian M J, Campbell C E, Werb Z. Synergistic transcriptional activation of the tissue inhibitor of metalloproteinases-1 promoter via functional interaction of AP-1 and Ets-1 transcription factors. J Biol Chem. 1996;271:774–782. doi: 10.1074/jbc.271.2.774. [DOI] [PubMed] [Google Scholar]
- 38.Mao S, Frank R C, Zhang J, Miyazaki Y, Nimer S D. Functional and physical interactions between AML1 proteins and an ETS protein, MEF: implications for the pathogenesis of t(8;21)-positive leukemias. Mol Cell Biol. 1999;19:3635–3644. doi: 10.1128/mcb.19.5.3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mayall T P, Sheridan P L, Montminy M R, Jones K A. Distinct roles for P-CREB and LEF-1 in TCRα enhancer assembly and activation on chromatin templates in vitro. Genes Dev. 1997;11:887–899. doi: 10.1101/gad.11.7.887. [DOI] [PubMed] [Google Scholar]
- 40.Mittal V, Hernandez N. Role for the amino-terminal region of human TBP in U6 snRNA transcription. Science. 1997;275:1136–1140. doi: 10.1126/science.275.5303.1136. [DOI] [PubMed] [Google Scholar]
- 41.Mo Y, Vaessen B, Johnston K, Marmorstein R. Structures of SAP-1 bound to DNA targets from the E74 and c-fos promoters: insights into DNA sequence discrimination by Ets proteins. Mol Cell. 1998;2:201–212. doi: 10.1016/s1097-2765(00)80130-6. [DOI] [PubMed] [Google Scholar]
- 42.Muthusamy N, Barton K, Leiden J M. Defective activation and survival of T-cells lacking the Ets-1 transcription factor. Nature. 1995;377:639–642. doi: 10.1038/377639a0. [DOI] [PubMed] [Google Scholar]
- 43.Myszka D G, Jonsen M D, Graves B J. Equilibrium analysis of high affinity interactions using BIACORE. Anal Biochem. 1998;265:326–330. doi: 10.1006/abio.1998.2937. [DOI] [PubMed] [Google Scholar]
- 44.Nagata T, Gupta V, Sorce D, Kim W Y, Sali A, Chait B T, Shigesada K, Ito Y, Werner M H. Immunoglobulin motif DNA recognition and heterodimerization of the PEBP2/CBF Runt domain. Nat Struct Biol. 1999;6:615–619. doi: 10.1038/10658. [DOI] [PubMed] [Google Scholar]
- 45.Niki M, Okada H, Takano H, Kuno J, Tani K, Hibino H, Asano S, Ito Y, Satake M, Noda T. Hematopoiesis in the fetal liver is impaired by targeted mutagenesis of a gene encoding a non-DNA binding subunit of the transcription factor, polyomavirus enhancer binding protein 2/core binding factor. Proc Natl Acad Sci USA. 1997;94:5697–5702. doi: 10.1073/pnas.94.11.5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.North T E, Gu T-L, Stacy T, Wang Q, Howard L, Binder M, Marin-Padilla M, Speck N A. Cbfa2 is required for the formation of intra-aortic hematopoietic clusters. Development. 1999;126:2563–2575. doi: 10.1242/dev.126.11.2563. [DOI] [PubMed] [Google Scholar]
- 47.Nye J A, Petersen J M, Gunther C V, Jonsen M D, Graves B J. Interaction of murine Ets-1 with GGA-binding sites establishes the ETS domain as a new DNA-binding motif. Genes Dev. 1992;6:975–990. doi: 10.1101/gad.6.6.975. [DOI] [PubMed] [Google Scholar]
- 48.Ogawa E, Inuzuka M, Maruyama M, Satake M, Naito-Fujimoto M, Ito Y, Shigesada K. Molecular cloning and characterization of PEBP2β, the heterodimeric partner of a novel drosophila runt-related DNA binding protein PEBP2α. Virology. 1993;194:314–331. doi: 10.1006/viro.1993.1262. [DOI] [PubMed] [Google Scholar]
- 49.Ogawa E, Maruyama M, Kagoshima H, Inuzuka M, Lu J, Satake M, Shigesada K, Ito Y. PEBP2/PEA2 represents a new family of transcription factor homologous to the products of the Drosophila runt and the human AML1 gene. Proc Natl Acad Sci USA. 1993;90:6859–6863. doi: 10.1073/pnas.90.14.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Okuda T, van Deursen J, Hiebert S W, Grosveld G, Downing J R. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 1996;84:321–330. doi: 10.1016/s0092-8674(00)80986-1. [DOI] [PubMed] [Google Scholar]
- 51.Petersen J M, Skalicky J J, Donaldson L W, McIntosh L P, Alber T, Graves B J. Modulation of transcription factor Ets-1 DNA binding: DNA-induced unfolding of an alpha helix. Science. 1995;269:1866–1869. doi: 10.1126/science.7569926. [DOI] [PubMed] [Google Scholar]
- 52.Petrovick M S, Hiebert S W, Friedman A D, Hetherington C J, Tenen D G, Zhang D E. Multiple functional domains of AML1: PU.1 and C/EBPα synergize with different regions of AML1. Mol Cell Biol. 1998;18:3915–3925. doi: 10.1128/mcb.18.7.3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sasaki K, Yahi H, Bronson R T, Tominaga K, Matsunashi T, Deguchi K, Tani Y, Kishimoto T, Komori T. Absence of fetal liver hematopoiesis in transcriptional co-activator, core binding factor β (Cbfβ) deficient mice. Proc Natl Acad Sci USA. 1996;93:12359–12363. doi: 10.1073/pnas.93.22.12359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Satake M, Nomura S, Yamaguchi-Iwai Y, Takahama Y, Hashimoto Y, Niki M, Kitamura Y, Ito Y. Expression of the Runt domain-encoding PEBP2α genes in T cells during thymic development. Mol Cell Biol. 1995;15:1662–1670. doi: 10.1128/mcb.15.3.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sato M, Morii E, Komori T, Kawahata H, Sugimoto M, Terai K, Shimizu H, Yasui T, Ogihara H, Yasui N, Ochi T, Kitamura Y, Ito Y, Nomura S. Transcriptional regulation of osteopontin gene in vivo by PEBP2αA/CBFA1 and ETS1 in the skeletal tissues. Oncogene. 1998;17:1517–1525. doi: 10.1038/sj.onc.1202064. [DOI] [PubMed] [Google Scholar]
- 56.Sharrocks A, Brown A, Ling Y, Yates P. The ETS domain transcription factor family. Int J Biochem Cell Biol. 1997;29:1371–1387. doi: 10.1016/s1357-2725(97)00086-1. [DOI] [PubMed] [Google Scholar]
- 57.Sheridan P L, Sheline C T, Cannon K, Voz M L, Pazin M J, Kadonaga J T, Jones K A. Activation of the HIV-1 enhancer by the LEF-1 HMG protein on nucleosome assembled DNA in vitro. Genes Dev. 1995;9:2090–2104. doi: 10.1101/gad.9.17.2090. [DOI] [PubMed] [Google Scholar]
- 58.Sieweke M H, Tekotte H, Jarosch U, Graf T. Cooperative interaction of Ets-1 with USF-1 required for HIV-1 enhancer activity in T cells. EMBO J. 1998;17:1728–1739. doi: 10.1093/emboj/17.6.1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Skalicky J J, Donaldson L W, Petersen J M, Graves B J, McIntosh L P. Structural coupling of the inhibitory regions flanking the ETS domain of murine Ets-1. Protein Sci. 1996;5:296–309. doi: 10.1002/pro.5560050214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Speck N A, Renjifo B, Golemis E, Fredrickson T N, Hartley J W, Hopkins N. Mutation of the core or adjacent LVb elements of the Moloney murine leukemia virus enhancer alters disease specificity. Genes Dev. 1990;4:233–242. doi: 10.1101/gad.4.2.233. [DOI] [PubMed] [Google Scholar]
- 61.Speck N A, Renjifo B, Hopkins N. Point mutations in the Moloney murine leukemia virus enhancer identify a lymphoid-specific viral core motif and 1,3-phorbol myristate acetate-inducible element. J Virol. 1990;64:543–550. doi: 10.1128/jvi.64.2.543-550.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Speck N A, Stacy T. A new transcription factor family associated with human leukemias. Crit Rev Eukaryotic Gene Expr. 1995;5:337–364. doi: 10.1615/critreveukargeneexpr.v5.i3-4.60. [DOI] [PubMed] [Google Scholar]
- 63.Stark M R, Escher D, Johnson A D. A trans-acting peptide activates the yeast a1 repressor by raising its DNA-binding affinity. EMBO J. 1999;18:1621–1629. doi: 10.1093/emboj/18.6.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Steger D J, Workman J L. Stable co-occupancy of transcription factors and histones at the HIV-1 enhancer. EMBO J. 1997;16:2463–2472. doi: 10.1093/emboj/16.9.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sun W, Graves B J, Speck N A. Transactivation of the Moloney murine leukemia virus and T-cell receptor β-chain enhancers by cbf and ets requires intact binding sites for both proteins. J Virol. 1995;69:4941–4949. doi: 10.1128/jvi.69.8.4941-4949.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tian G, Erman B, Ishii H, Gangopadhyay S S, Sen R. Transcriptional activation by ETS and leucine zipper-containing basic helix-loop-helix proteins. Mol Cell Biol. 1999;4:2946–2957. doi: 10.1128/mcb.19.4.2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Treisman R, Marais R, Wynne J. Spatial flexibility in ternary complexes between SRF and its accessory proteins. EMBO J. 1992;11:4631–4640. doi: 10.1002/j.1460-2075.1992.tb05565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang L C, Kuo F, Fujiwara Y, Gilliland D G, Golub T R, Orkin S H. Yolk sac angiogenic defect and intra-embryonic apoptosis in mice lacking the Ets-related factor TEL. EMBO J. 1997;16:4374–4383. doi: 10.1093/emboj/16.14.4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang Q, Stacy T, Miller J D, Lewis A F, Gu T L, Huang X, Bushweller J H, Bories J C, Alt F W, Ryan G, Liu P P, Wynshaw-Boris A, Binder M, Marin-Padilla M, Sharpe A H, Speck N A. The CBFβ subunit is essential for CBFα2 (AML1) function in vivo. Cell. 1996;87:697–708. doi: 10.1016/s0092-8674(00)81389-6. [DOI] [PubMed] [Google Scholar]
- 70.Wang S, Wang Q, Crute B E, Melnikova I N, Keller S R, Speck N A. Cloning and characterization of subunits of the T-cell receptor and murine leukemia virus enhancer core-binding factor. Mol Cell Biol. 1993;13:3324–3339. doi: 10.1128/mcb.13.6.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wasylyk C, Kerckaert J-P, Wasylyk B. A novel modulator domain of Ets transcription factors. Genes Dev. 1992;6:965–974. doi: 10.1101/gad.6.6.965. [DOI] [PubMed] [Google Scholar]
- 72.Werner M H, Clore G M, Fisher C L, Fisher R J, Trinh L, Shiloach J, Gronenborn A M. Correction of the NMR structure of the ETS1/DNA complex. J Biomol NMR. 1997;10:317–328. doi: 10.1023/a:1018399711996. [DOI] [PubMed] [Google Scholar]
- 73.Wernert N, Raes M B, Lassalle P, Dehouck M P, Gosselin B, Vandenbunder B, Stehelin D. c-ets1 proto-oncogene is a transcription factor expressed in endothelial cells during tumor vascularization and other forms of angiogenesis in humans. Am J Pathol. 1992;140:119–127. [PMC free article] [PubMed] [Google Scholar]
- 74.Wotton D, Ghysdael J, Wang S, Speck N A, Owen M J. Cooperative binding of Ets-1 and core binding factor to DNA. Mol Cell Biol. 1994;14:840–850. doi: 10.1128/mcb.14.1.840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yanagi K, Prive G G, Dickerson R E. Analysis of local helix geometry in three B-DNA decamers and eight dodecamers. J Mol Biol. 1991;217:201–214. doi: 10.1016/0022-2836(91)90620-l. [DOI] [PubMed] [Google Scholar]