Helicobacter pylori is a gram-negative rod associated with gastroduodenal pathologies, such as gastritis, peptic ulcer, and gastric adenocarcinoma. Although most H. pylori infections are clinically silent, the organism is associated with substantial morbidity and mortality. It is unknown why the bacteria are able to produce severe disease in some hosts and be innocuous in others.
Different virulence factors have been described in H. pylori infection, including urease, lipopolysaccharide, adhesins, and vacuolating cytotoxin (2). The formation of vacuoles is associated with a higher degree of virulence, and two genes, cagA and vacA, have been described to be involved in vacuole production. It is assumed that cagA+ strains and those with the s1 allele of vacA are more virulent and produce more severe disease than do strains lacking cagA and vacA s2 strains. Recently, Rudi et al. (5) published a study on cagA and vacA genes and their relationship to the production of several gastroduodenal diseases. They studied 65 strains by PCR (43 from gastritis, 19 from peptic ulcer, and 3 from cancer patients) and found a prevalence of the cagA gene of 84.2% in peptic ulcer and 67.4% in gastritis strains. When the vacA gene was studied, the s1 allele was found to be more prevalent in strains from patients with peptic ulcers (100% showed this allele) and the s2 allele was found only in strains from patients with gastritis. They also found a strong association between the presence of the cagA gene and the s1 allele of vacA.
Recently, we isolated 104 Spanish H. pylori strains, 69 from peptic ulcer patients and 35 from gastritis patients, and aimed to study cagA and vacA status and the relationship with specific pathologies. Antral biopsy specimens were cultured onto blood agar plates (Columbia agar plus 5% horse blood) and incubated for 10 days under microaerobic conditions at 37°C. H. pylori was identified by colony morphology, Gram strain, and positive urease, catalase, and oxidase test results. We performed two PCRs in order to detect the cagA gene and the s1 or s2 allele of the vacA gene. DNA was extracted by the CTAB method (6). cagA was detected by applying a protocol described by Covacci and Rappuoli (3) using primers D008 (5′-ATAATGCTAAATTAGACAACTTGAGCGA-3′) and R008 (5′-TTAGAATAATCAACAAACATCACGCCAT-3′ to detect a 297-bp amplified fragment. For the study of vacA, the protocol of Atherton (1) was performed by using two primers (VA1-F, 5′-ATGGAAATACAACAAACACAC-3′, and VA1-R, 5′-CTGCTTGAATGCGCCAAAC-3′) and two fragments of 259 and 286 bp corresponding to the s1 and s2 alleles were detected. Results were studied by 2% agarose gel electrophoresis and visualized in a UV transilluminator. cagA and vacA detections are shown in Figs. 1 and 2.
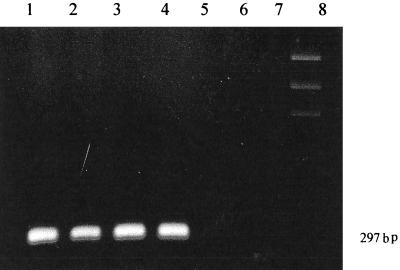
FIG. 1.
cagA detection. Lanes 1, 2, 3, and 4, cagA+; Lanes 5, 6, and 7, cagA absent; Lane 8, marker.
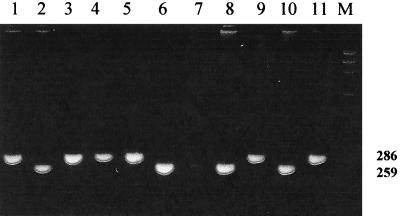
FIG. 2.
Detection of s1 and s2 alleles of the vacA gene. Lanes, 2, 6, 7, 8, and 10, s1 allele; lanes 1, 3, 4, 5, 9, and 11, s2 allele; lane M, DNA marker. Numbers on the right indicate molecular size (in base pairs).
We agree with Rudi et al. about the prevalence of s1 among cagA+ strains; however, we found a smaller percentage (64.1 versus 87.0%) and we detected four strains that lacked cagA and possessed the s1 allele of vacA, a genotype which was not found by those authors. When we studied the relationship between cagA and pathology, we also found no statistically significant differences among strains obtained from patients with ulcers (91% cagA+) and gastritis (83.3% cagA+). Moreover, in our study we found similar percentages of s1 alleles in strains from ulcer patients (62%) and gastritis patients (57%) and we detected the s2 allele in 38% of strains obtained from patients with peptic ulcer disease. These data are not in agreement with those published by Rudi et al., who detected s1 in all ulcer strains and s2 only in gastritis strains. When cagA and vacA genes were combined and were related to the pathologies, cagA+ strains with the vacA s1 allele were found to be more prevalent in ulcer isolates, although differences were not statistically significant.
From our experience, we conclude that, as in other populations (4), cagA and vacA genes cannot be used as predictive markers in H. pylori clinical isolates to identify a particular strain as a gastritis or ulcer producer. Perhaps new virulence factors should be described with more power to discriminate among H. pylori strains.
REFERENCES
- 1.Atherton J C, Cao P, Peek R M, Jr, Tummuru M K R, Blaser M J, Cover T L. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori.. Association of specific vacAtypes with cytotoxin production and peptic ulceration. J Biol Chem. 1995;270:17771–17777. doi: 10.1074/jbc.270.30.17771. [DOI] [PubMed] [Google Scholar]
- 2.Blaser M J. Role of vacA and the cagA locus of Helicobacter pyloriin human disease. Aliment Pharmacol Ther. 1996;10(Suppl. 1):73–77. doi: 10.1046/j.1365-2036.1996.22164008.x. [DOI] [PubMed] [Google Scholar]
- 3.Covacci A, Rappuoli R. PCR amplifications of H. pylori gene sequences. In: Lee A, Megraud F, editors. Helicobacter pylori: techniques for clinical diagnosis. W. B. London, United Kingdom: Saunders Company Ltd.; 1996. pp. 94–111. [Google Scholar]
- 4.Maeda S, Ogura K, Yoshida H, Kanai F, Ikenoue T, Kato N, Shiratori Y, Omata M. Major virulence factors, vacA and cagA, are commonly positive in Helicobacter pyloriisolates in Japan. Gut. 1998;42:338–343. doi: 10.1136/gut.42.3.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rudi R, Kolb C, Maiwald M, Kuck D, Sieg A, Galle P R, Stremmel W. Diversity of Helicobacter pylori vacA and cagAgenes and relationship to VacA and CagA protein expression, cytotoxin production, and associated diseases. J Clin Microbiol. 1998;36:944–948. doi: 10.1128/jcm.36.4.944-948.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilson K. Preparation of genomic DNA from bacteria. In: Asubel R, Brent R, Kingston D D, Moore J G, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Greene Publishing and Wiley Interscience; 1987. pp. 2.4–2.4. [Google Scholar]