Graphical abstract
Keywords: Black soldier fly larvae, Hermetia illucens, Salmonella, Food safety, Insect rearing
Highlights
-
•
BSFL does not show a reducing effect on Salmonella counts on chicken feed.
-
•
The background microbiota is an important factor during inoculation experiments.
-
•
Airborne transmission is possible in laboratory conditions during insect rearing.
-
•
Salmonella free substrate is recommended to avoid the pathogen entry in rearing.
Abstract
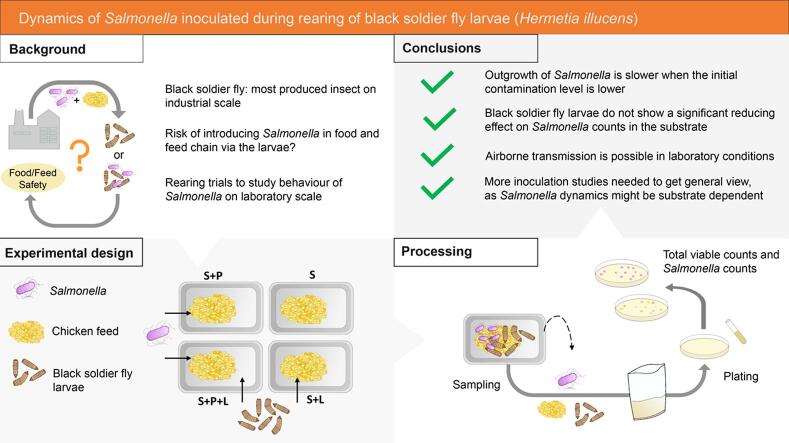
The black soldier fly is currently the most produced edible insect on industrial scale, with its larval stage being processed into animal feed as the main application. As this insect species enters the feed and food chain, good hygiene and monitoring practices are needed to avoid the entrance of foodborne pathogens via the larvae. However, insufficient data on the risk of such introductions via industrial larvae production are available. To address this gap, a range of rearing trials were conducted in which the substrate, chicken feed, was inoculated with different levels of Salmonella and in which total viable counts and Salmonella counts were determined during the following days. The outgrowth of Salmonella was slower in those experiments with a lower initial contamination level than in experiments with a higher level. No significant reducing effect originating from the larvae on the substrate Salmonella counts was observed, in contrast to previous studies using other substrates. Our study also revealed that airborne transmission of Salmonella is possible under rearing conditions corresponding to those applied at industrial production sites. Based on our results, we recommend insect producers to use substrate ingredients free of Salmonella, and not to count on the antimicrobial activities that BSFL may exert in some situations towards food pathogens. More inoculation studies using other Salmonella serotypes, other zoonotic bacteria, other substrates, larvae of other ages and including variations on rearing protocols are needed in order to obtain a general view on the dynamics of food pathogens in this insect species and to support comprehensive risk assessments.
1. Introduction
The mass production of insects is now accepted as an agricultural activity in the Western world. Depending on the insect species, they can be used in human food and animal feed as an alternative source for proteins, but they also deliver other components, such as lipids for biodiesel production and biochemicals for cosmetics. They can also be applied in waste processing to support the circular economy (Sogari et al., 2019). Particularly when insects are processed into food or feed products, safety hazards have to be monitored during rearing and processing to ensure a safe end product (van der Fels-Klerx et al., 2018). The insect species currently produced in the largest volume is the black soldier fly (Hermetia illucens) and the major application of its larval stage is animal feed ingredient (Arru et al., 2019).
In 2015, the European Food Safety Authority (EFSA) published an initial risk profile for the production and consumption of edible insects for food and feed and listed potential safety hazards (EFSA Scientific Committee, 2015). Specific attention was paid to microbiological hazards, including Salmonella. Later, studies focused on the microbial composition of the larvae during rearing and the presence of food pathogens. While most studies do not detect the specific food pathogen Salmonella in the larvae (Osimani et al., 2021), this does not mean this food pathogen cannot be present in the rearing cycle. For example, the presence of Salmonella sp. was observed in the residue of a black soldier fly larvae (BSFL) rearing cycle at an industrial setting, though no Salmonella was found in 25 g samples of the larvae in this study either (Wynants et al., 2019). Hence, even when using only the food- and feed-grade substrates that are currently allowed for insect rearing, good hygiene and monitoring practices are needed to avoid the introduction of this and likely also other foodborne pathogens in the feed and food chain via insects.
In Europe, the use of processed BSFL, or so-called ‘insect meal’, is currently allowed in aquafeed. Authorization in poultry and pig feed is to be expected (Byrne, 2021), and then larvae will enter the feed chain at an even large scale. Hence, monitoring and surveillance programs will have to upscale concomitantly. Information on the killing effect on food pathogens present in the larvae of post-harvest processing is very scarce, even though the aim should be to rear pathogen-free larvae and to avoid any introduction of food pathogens in larvae processing plants. In addition, the legislation in Europe (Regulation (EU) No 2017/893) currently allows the feeding of live insects to poultry, which is shown to benefit poultry welfare (Ipema et al., 2020). Salmonella can asymptomatically colonize the small intestine of poultry, along with the cecum, and therefore broilers and layers belong to the most likely vectors for Salmonella transmission to humans via food consumption (Cosby et al., 2015). Finally, in its brochure called “Three research priorities”, the European insect federation IPIFF (International Platform of Insects for Food and Feed), the first priority mentioned is to explore substrates for insect rearing that are not yet allowed but can further boost the contribution of the sector to a circular economy. Examples of envisaged streams are former foodstuffs containing meat, slaughter waste, etcetera. It goes without saying that in these types of substrates, the surveillance and control of food pathogens such as Salmonella will be of utmost importance. All mentioned facts underpin the high need for more data on the dynamics of food pathogens in BSFL, and in particular in the situation when they enter the rearing cycle via the substrates fed to the insects.
A typical approach to study the behavior of a zoonotic pathogen during rearing of or bioconversion by insects, is to inoculate the substrate with the microorganism, provide it to the insects and follow-up possible colonization of the substrate and insects via classical microbial counts and/or sequencing of the whole microbiota. Some studies were performed in this way for BSFL in combination with a few zoonotic pathogens. The larvae were reported to be able to reduce the load of Escherichia coli, Salmonella spp., and Enterococcus spp. in their substrate by even up to 8 log cycles in some cases (Erickson et al., 2004, Lalander et al., 2013, Lalander et al., 2015, Liu et al., 2008, Lopes et al., 2020). It must be mentioned, though, that the substrate in all aforementioned studies was some type of manure and in one study aquaculture waste, and that the main aim was to find out whether BSFL can be used as bioconversion and sanitizing step in the processing of the waste. These publications, together with an increasing number of reports on the detection and description of a wide range of antimicrobial peptides in BSFL and antimicrobial effects of extracts (Choi et al., 2012, Xu et al., 2020), can lead to the general impression that the presence of food pathogens in whatever substrate of BSFL is not a large risk, since their growth is expected to be suppressed by the larvae. Although BSFL indeed may exert antimicrobial activity against specific micro-organisms and in certain conditions, more data are needed to elucidate the consequences of the presence of food pathogens in the substrate, especially when the larvae are produced as feed ingredient. The production of antimicrobial peptides has proven to be diet-dependent (Vogel et al., 2017)), and so may be the possible pathogen reducing effect. Even for substrates currently allowed and frequently used in industrial BSFL production for animal feed, the interactions between a pathogen such as Salmonella, the larvae and the other micro-organisms present during rearing are not yet uncovered. It is not known for allowed substrates, if and how fast Salmonella can colonize the substrate and/or the larvae, and which factors, such as the contamination level of the pathogen, the type of substrate and other rearing conditions, the background microbiota and the overall hygiene level of the production environment, the age of the larvae, the Salmonella serotype(s) present, influence the interactions.
The aim of this work was to conduct a range of rearing trials with BSFL after inoculating the substrate with Salmonella and to determine total viable counts and Salmonella counts in the days after providing the inoculated substrate. While there are, as mentioned before, many factors that possibly affect the dynamics of Salmonella, we opted to perform all inoculation experiments with the same substrate and in the same rearing conditions. A factor that was varied, however, was the contamination level. The research was started first with trials at a high contamination level, to mimic worst-case scenarios, and then moved on to lower levels, probably implying more realistic scenarios in industry. As substrate chicken feed was chosen, which is frequently used in research as well as in (the first stages of rearing in) industry. The chicken feed was not frozen or autoclaved, so that the endogenous microbiota of the feed was also active during the experiments.
2. Material and methods
2.1. Overview of consecutive series of experiments
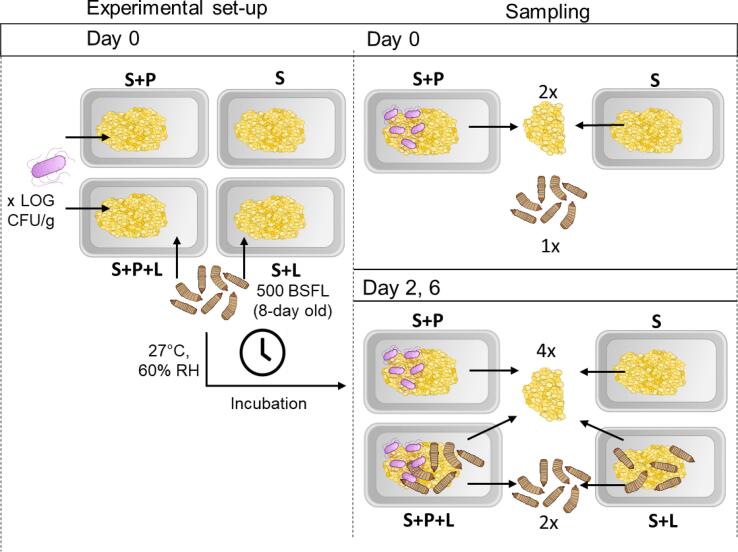
To study the behavior of Salmonella during BSFL rearing, three different experimental set-ups, conducted at laboratory scale, were used. As it is highly likely in insects, that not all strains from a single species are equally able to colonise and survive in insects. Results may well be strain dependent. To account for this strain dependency, it is necessary to work with a mixture of strains. In a first series of experiments, a mixture of three wild-type Salmonella strains was used for inoculation. Since this resulted in the presence of a large quantity of non-specific colonies (background microbiota) on the selective plates (as discussed in detail in the results), two kanamycin resistant mutant strains were used in the second series, and two inoculation levels were included. While the background microbiota issue was resolved in this second series, another issue arose being airborne contamination between the conditions tested (as described further). Therefore, a third series of experiments was conducted in which the conditions were incubated separately to exclude the unwanted impact of this cross-contamination. All experimental set-ups were performed with two or three separate batches of larvae (meaning that the larvae were reared independently during different rearing cycles), and for each repetition, two replicates (i.e. containers with larvae) were included. A general overview of the experimental designs and the varied parameters can be found in Table 1. For each experiment, four different rearing conditions were included, an overview of which is given in Fig. 1: (i) substrate without Salmonella and without larvae (S), (ii) substrate inoculated with pathogen Salmonella but without larvae (S + P), (iii) substrate with larvae but without Salmonella (S + L), and (iv) substrate inoculated with Salmonella and provided with larvae (S + P + L).
Table 1.
Overview of the consecutive series of experiments.
| Series of experiments | Salmonella strains included | Target level for inoculation | Incubation | Number of batches (and of replicates) |
|---|---|---|---|---|
| 1 | S. Typhymurium, S. Infantis, S. Enteritidis | 7 log CFU/g | Inoculated and Uninoculated conditions incubated together | 2 (2) |
| 2 |
S. Typhymurium KANR S. Infantis KANR |
4 log CFU/g 7 log CFU/g |
Inoculated and Uninoculated conditions incubated together | 2 (2) |
| 3 |
S. Typhymurium KANR S. Infantis KANR |
3 log CFU/g | Inoculated and Uninoculated conditions incubated separately | 3 (2) |
Fig. 1.
Schematic overview of experimental set-up and sampling method in a challenge experiment, depicted for one batch of larvae. S = substrate without Salmonella and without larvae, S + P = substrate inoculated with pathogen Salmonella but without larvae, S + L substrate with larvae but without Salmonella, and S + P + L = substrate inoculated with Salmonella and provided with larvae.
2.2. Salmonella cultivation and creation of kanamycin resistant strains
Three different Salmonella strains were used: Salmonella enterica subsp. enterica serovar Enteritidis (LMG 18735), Salmonella enterica subsp. enterica serovar Typhimurium (LMG 18732) and Salmonella enterica subsp. enterica serovar Infantis (LMG 18746), all purchased from the Belgian Coordinated Collection of Microorganisms (BCCM). For experiment series 1, a mixture of all three bacterial strains was used for inoculation. From design 2 onwards, a mixture of kanamycin resistant S. enterica ser. Typhymurium and kanamycin resistant S. enterica ser. Infantis was used.
Antibiotic resistant strains were generated by using a temperature sensitive pHSG415-tnsABCD helper plasmid and a modified mini-Tn7 delivery system as described previously (Shivak et al., 2016). In our study, the delivery plasmid pUC18R6K-mini-Tn7T-pCS26-KmR sig70_c10 LUX was used to incorporate a kanamycin resistance gene into the target bacteria. To achieve chromosomal integration and kanamycin resistance, electrocompetent Salmonella cells were made, as described by Shivak et al. (2016). Next, both the helper plasmid and delivery plasmid were transformed via electroporation. After electroporation, cells were allowed to recover in SOC medium (2% tryptone (Lab M, UK), 0.5% yeast extract (VWR, Belgium), 10 mM NaCl (Acros Organics, Belgium), 2.5 mM KCl (Acros Organics), 10 mM MgCl2 (Acros Organics), 10 mM MgSO4 (Acros Organics), and 20 mM glucose (Acros Organics)) for 2 h at 28 °C. Then, the transformation mix was plated on LB/Kan50 (Luria Bertani, composed of 10.0 g/l peptone (Biokar Diagnostics, France), 5.0 g/l yeast extract, 5.0 g/l NaCl, 15 g/l agar (VWR, Belgium), 50 µg/ml Kanamycin (Thermo Fisher Scientific, Belgium)) and incubated overnight at 37 °C. The correct chromosomal integration of the resistance cassette was checked in the obtained transformants with PCR using two primer pairs (primer pair 1: glmSdetectFor - lux-check and primer pair 2: glmSdetectRev- KmCheck). The conditions for the PCR reaction can be found in Supplementary table 1.
2.3. Inoculation of substrate
The selected Salmonella species were grown overnight at 37 °C in Luria Bertani broth (see LB plates but without agar), and with Kan50 if resistant strains were used (design 2 and 3). Then, the overnight cultures were diluted using LB to a McFarland unit (MFU, DEN-1 McFarland Densitometer, Grant instruments, UK) of 5. Next, the different Salmonella strains were combined in equal volumes to create a suitable Salmonella mixture. For the substrate, chicken starter feed (Startmeel voor Kuikens 259, AVEVE, Belgium) was grinded with a mixer (Espressions EP9800 Powerblender) using two times the “Ice Crush” program. The substrate was then prepared by mixing the grinded chicken starter feed and tap water in a 1:1 ratio (w/w), and 100 g of wetted feed was placed in polypropylene trays (1L). Finally, for all conditions which required a challenge (S + P and S + P + L), an aliquot of the prepared inoculum was added to 100 g of wetted feed to obtain a desired starting concentration of 7 log CFU/g (3.3 ml of a MFU 5 solution) (design 1) or 7 and 4 (1.2 ml of a 1/2000 diluted MFU 2 solution) log CFU/g (design 2) or 3 log CFU/g (0.6 ml of a 1/2000 diluted MFU 2 solution) (design 3). The inoculated feed was homogenized using a sterile spoon. To the uninoculated groups, an volume of sterile LB broth was added equal to their inoculated counterparts.
2.4. Rearing of BSFL and sampling
For this study, BSFL were supplied by and originated from a colony maintained by RADIUS (Thomas More University University of Applied Sciences, Geel, Belgium). They were nursed until day 8 on a mixture of chicken starter feed and tap water in a 1:1 ratio (w/w) in a climate chamber (Pharma 600, Weiss Technik, Belgium) at 27 °C and 60% relative humidity (RH). At that time, the challenge experiment was initiated. Approximately 500 (as determined by the average weight of three times 10 larvae) 8-day-old BSFL were added to the container of each larvae-containing replicate (S + L and S + P + L). The dimensions of the containers used were 10 cm × 15 cm, yielding a density of 3.3 larvae/cm2. The containers were fitted with a lid containing a mesh covered surface (7 cm × 13 cm) to allow air circulation, but prevent larval escape. Next, 100 g of either or not contaminated feed was added to the respective replicates and all containers were placed in a climate chamber (Memmert HPP260, Memmert, Belgium) at 27 °C and 60% relative humidity (RH) until the end of the experiment, which was 6 days after the challenge. On day 2 and day 4, an additional 80 g of uncontaminated feed (chicken starter and tap water) was added to each rearing box. For design 1 and 2, all experimental conditions were incubated together in the same incubator. For design 3, replicates with different conditions were incubated separately to avoid cross-contamination.
Sampling of larvae and substrate took place on day 0, day 2 and day 6 in aseptic conditions (Fig. 1). Larvae were separated from the substrate by sieving. Then they were disinfected prior to sampling in three subsequent washing steps: a first disinfection step with 100 ml of 70% ethanol (1 min at 200 rpm on a laboratory shaking table (Unimax 1010, Germany)) was followed by two rinsing steps with 100 ml of sterile, demineralized water (1 min each at 200 rpm on the shaking table). To monitor the growth of the larvae, the mass of 10 larvae was measured and this was repeated 5 times per replicate.
2.5. Microbiological analysis
For the microbiological analysis, samples of 5 g of disinfected larvae were collected and diluted tenfold in physiological peptone solution (PPS, 0.85% NaCl, 0.01% peptone) before pulverizing the larvae in the solution by using an ethanol sterilized home type mixer (Bosch CNHR 25). Similarly, 5 g of substrate was sampled from each replicate and diluted 1:10 in PPS. Next, both larval and substrate samples were homogenized using a stomacher (BagMixer 400CC, Interscience, France) for 1 min. For each sample, the total aerobic viable count and the specific Salmonella count were determined. All plate counts were performed according to the ISO standards for microbial analyses of food and feed as compiled by Dijk et al. (2015). For the total aerobic viable count, serial dilutions were made in PPS, plated on Plate Count Agar (PCA, Biokar Diagnostics, Beauvais, France), and incubated at 30 °C for 72 h. Salmonella was counted by plating the diluted samples on a chromogenic RAPID′ Salmonella agar (BioRad Laboratories, Belgium) and incubating the plates at 37 °C for 24 h. The cell density of all inocula was also verified by plating a serial dilution on both the RAPID’ Salmonella agar and PCA.
2.6. Statistical analysis
For design 1 to 3, total viable counts as well as Salmonella sp. counts of larvae and substrate samples for each condition were compared between sampling moments using one-way ANOVA, with Tukey HSD as post hoc test in case of equal variances. When the variances were not equal, the Welch’s ANOVA with Steel–Dwass All Pairs post hoc test was used. Furthermore specific pair-wise t-tests were conducted in order to compare total viable counts and/or Salmonella sp. counts between samples at day 6. When counts of a sample were below the detection limit, the detection limit itself was chosen as value to be included for statistical analysis. All these tests were performed using JMP Pro 15.0.0 from SAS. For each test, a significance level of α = 0.05 was considered.
3. Results
3.1. Salmonella dynamics using a high inoculation level
The first goal was to examine a possible worst-case scenario during rearing. This was achieved in experimental design 1, by contaminating the substrate and aiming at an inoculation level of 7 log CFU of Salmonella sp. per g substrate. The actual Salmonella sp. counts reached at day 0 were close to that target level, being 7.4 ± 0.5 log CFU/g (Table 2). No impact on the larval growth was observed from the presence of Salmonella sp. in the substrate (Supplementary Fig. 1), which was to be expected, as no evidence is present in literature that Salmonella sp. would be pathogenic for BSFL (Joosten et al., 2020).
Table 2.
Total aerobic viable counts and Salmonella spp. counts of larvae and substrate samples of experiment series 1, involving a high contamination of wild-type Salmonella strains (7 log CFU/g) and all conditions incubated in the same climate chamber. Values present the mean (±standard deviation) of two batches, each with two replicates per condition (n = 2x2). S = Substrate; S + P = Substrate with pathogen; S + L = Substrate with larvae; and S + P + L = Substrate with pathogen and larvae.
| Experimental condition | Investigated sample | Target Salmonella spp. contamination level in substrate (log CFU/g) |
Total viable count (log CFU/g) |
Salmonella sp. count (log CFU/g) |
||||
|---|---|---|---|---|---|---|---|---|
| Day 0 | Day 2 | Day 6 | Day 0 | Day 2 | Day 6 | |||
| S | substrate |
0* | 6.5 ± 2.3a | 11.1 ± 0.9a | 11.6 ± 0.3a | <2.0 ± 0.0†,a | <2.0 ± 0.1†,a | <2.0 ± 0.0†,a |
| S + P | substrate | 7 | 8.0 ± 0.3a | 11.4 ± 0.5a | 12.1 ± 1.0a | 7.4 ± 0.5a | 8.1 ± 0.4a | 9.5 ± 0.8b |
| S + L | substrate |
0* | 6.5 ± 2.3°,a | >11.5 ± 1.2a | 12.4 ± 1.0a | <2.0 ± 0.0°,†,a | <3.3 ± 2.6ǂ,a | <6.3 ± 5.0ǂ,a |
| S + P + L | substrate | 7 | 8.0 ± 0.3°,a | >11.0 ± 1.7a | 11.9 ± 0.7a | 7.4 ± 0.5°,a | 7.9 ± 1.7a | 8.3 ± 1.7a |
| S + L | larvae |
0* | 9.1 ± 0.5a | 10.3 ± 0.8b | 10.7 ± 0.5b | <2.0 ± 0.0†,a | <3.7 ± 2.0ǂ,a | <5.7 ± 4.2ǂ,a |
| S + P + L | larvae | 7 | 11.0 ± 1.0a | 10.5 ± 0.3a | 6.5 ± 1.2a | 7.3 ± 1.0a | ||
Uninoculated replicates; ° = sample is similar at this timepoint to chicken starter feed without larvae and was not determined a second time;
”<2.0″ indicates that Salmonella sp. was below the detection limit (2 log CFU/g) in every sample;
“<” followed by a value higher than 2.0 log CFU/g indicates that Salmonella sp. was below the detection limit in at least one, but not all samples. abc Average values for total viable counts and Salmonella sp. counts within each row that share a letter in superscript did not significantly (p ≥ 0.05) increase or decrease between sampling days.
A clear increase was observed in the total aerobic viable count from day 0 till day 6 for all substrate samples, irrespective of the presence of larvae, with counts increasing from 6.5 to 8.0 log CFU/g to 11.6–12.4 log CFU/g (Table 2). The difference between the inoculated substrate (S + P and S + P + L; 8 log CFU/g) and the uninoculated samples (S and S + L; 6.5 log CFU/g) at day 0 can be attributed to the supplemented Salmonella in the inoculated samples. Indeed Salmonella is capable of growing on the Plate Count Agar and the addition of 7 log CFU/g will thus impact the initial TVC. For the substrate without larvae (S and S + P), this is in contrast to observations made in earlier challenge experiments reported for Tenebrio molitor, where the total viable counts did not significantly change over a same period of time for that type of sample (Wynants et al., 2019, Wynants et al., 2019). A plausible explanation is that the higher moisture content of the substrate for BSFL (approximately 50%) is more suited for microbial growth compared to that of the substrate for Tenebrio molitor, that consisted of dry wheat bran with carrot pieces as moisture source. Another observation is that the presence of larvae does not seem to influence the microbial load (at least, in quantitative terms) of the substrate in any way (Table 2).
At day 0, the total aerobic viable count of the larvae (in S + L as well as in S + P + L) was higher than that of the substrate, and with 9.1 ± 0.5 log CFU/g, this number is in the range of earlier reports on the total viable count present in larvae (Wynants et al., 2018). The increase over time in their interior microbial load was also much less pronounced than the increase in their substrate. Final numbers, ranging from 10.5 to 10.7 log CFU/g, remained approximately one log CFU lower than the microbial load in the substrate. This likely can be explained by the larval interior being completely occupied as ecological niche, in terms of nutrient availability and/or it can point towards the presence of control mechanisms restricting the microbial load inside the larvae.
From the Salmonella sp. counts, it is clear that the pathogen can thrive well in the substrate as their number increased significantly in the inoculated, larvae-free condition (S + P) from 7.4 ± 0.5 log CFU/g at day 0 to 9.5 ± 0.8 log CFU/g at day 6 (Table 2). At the same time, Salmonella was not detected (detection limit is 2 log CFU/g) in the uninoculated substrate without larvae (S) at day 0, 2 and 6. Interestingly, this was different in the uninoculated substrate samples with larvae (S + L). Here, Salmonella was detected from day 2 in two out of four replicates and its counts increased further in these replicates to reach counts close to those of the substrates of the inoculated experiments (S + P and S + P + L, Table 2). Two explanations are possible here. First, all larval samples showed Salmonella levels below the detection limit at day 0, but it cannot be excluded that it was present below the detection limit and started growing to detectable levels during the test. Secondly, and according to us more likely, cross-contamination occurred in the climate chamber due to airborne transmission of Salmonella, as will be further addressed in the discussion. In contrast to the studies mentioned in the Introduction Section, no significant decreases in Salmonella sp. counts were observed over time in the presence of larvae, neither in the substrate samples nor in the larval samples (Table 2). Overall, the Salmonella counts in the larvae were approximately between 1 and 2 log CFU/g lower than in the corresponding substrate sample.
The aforementioned observations were hindered by a background growth of micro-organisms on the selective medium for Salmonella count (Supplementary Figure 2). Though distinction with Salmonella and proper counting of Salmonella was still possible due to the colony morphology, and in particular the color, the large abundances of the background microbiota were unwanted. Indeed, the presence of the background microbiota could hinder the next step in our study to explore the impact of lower, more realistic, inoculation levels on the microbiological safety of BSFL. To circumvent this problem, all three Salmonella sp. used were genetically engineered to express a kanamycin resistance gene. The procedure was successful for S. Typhimurium and S. Infantis, so these two strains were mixed and used as inoculum in experimental design 2 and 3 (Table 1). The use of kanamycin, at a concentration of 50 µg/ml, indeed had a significant impact on the background growth (Supplementary Figure 2).
3.2. Salmonella dynamics using a high and low inoculation level and kanamycin-resistant strains
Using a mixture of the two resistant Salmonella strains, a second set of challenge experiments was executed (design 2). This design included both a low (4 log CFU/g), as well as the previous high (7 log CFU/g) inoculation level. No impact of the inoculations was observed on larval growth (data not shown), as in design 1. The microbiological results are shown in Table 3. The total viable counts were slightly lower over the whole design than the counts in design 1. However, the trends were comparable, showing a 4 to 5 log CFU/g increase in the substrate samples over the 6-day time frame and a higher initial count in the substrate inoculated with 7 log CFU/g. The initial total viable count in the uninoculated larvae (S + L) at day 0 was 8.2 ± 0.4 log CFU/g. This count only slightly increased to, on average, 9.2 ± 0.5 log CFU/g at day 6 and a similar observation was made for the inoculated larvae (S + P + L). As in design 1, the total viable count in the larvae was between 1 and 2 log CFU/g lower than the count reported in their corresponding substrates, which adds weight to the hypothesis that the ecological niche is fully occupied and/or that larvae control to some extent the total microbial load in their interior.
Table 3.
Total aerobic viable counts and Salmonella sp. counts of larvae and substrate samples of experiment series 2, involving both a low (4 log CFU/g) and high (7 log CFU/g) contamination of resistant Salmonella strains and all conditions incubated in the same climate chamber. Values represent the mean (±standard deviation) of two batches, each with two replicates per condition (n = 2x2). S = Substrate; S + P = Substrate with pathogen; S + L = Substrate with larvae; and S + P + L = Substrate with pathogen and larvae.
| Experimental condition | Investigated sample | Target Salmonella sp. contamination level in substrate (log CFU/g) |
Total viable count (log CFU/g) |
Salmonella sp. count (log CFU/g) |
|||||
|---|---|---|---|---|---|---|---|---|---|
| Day 0 | Day 2 | Day 6 | Day 0 | Day 2 | Day 6 | ||||
| S | Substrate | 0* | 5.2 ± 0.2a | 9.6 ± 0.3b | 10.0 ± 0.2b | <2.0 ± 0.0†a | <2.0 ± 0.0†,a | 4.0 ± 1.3a | |
| S + P | Substrate | 4 | 5.0 ± 0.1a | 9.5 ± 0.3b | 10.0 ± 0.1b | 3.6 ± 0.4a | <2.6 ± 0.7ǂ,a | 5.9 ± 0.8a | |
| S + P | Substrate | 7 | 7.8 ± 0.2a | 9.5 ± 0.5b | 10.4 ± 0.2c | 7.6 ± 0.1a | 7.5 ± 1.1a | 6.8 ± 1.3a | |
| S + L | Substrate | 0* | 5.2 ± 0.2°,a | 10.2 ± 0.0b | 10.9 ± 0.4c | <2.0 ± 0.0°,†,a | <4.5 ± 2.5ǂ,a | 6.2 ± 0.5a | |
| S + P + L | Substrate | 4 | 5.0 ± 0.1°,a | 9.9 ± 0.2b | 10.7 ± 0.3c | 3.6 ± 0.4°,a | <4.7 ± 2.3ǂ,a | 6.9 ± 0.7a | |
| S + P + L | Substrate | 7 | 7.8 ± 0.2°,a | 10.4 ± 0.1a | 11.7 ± 0.8a | 7.6 ± 0.1°,a | 7.8 ± 0.3a | 7.5 ± 0.3a | |
| S + L | Larvae | 0* | 8.2 ± 0.4a | 8.6 ± 0.5a,b | 9.2 ± 0.5b | <2.0 ± 0.0†,a | <4.2 ± 2.5ǂ,a | 5.1 ± 1.6a | |
| S + P + L | Larvae | 4 | 8.2 ± 0.4a | 8.8 ± 0.5a,b | 9.1 ± 0.1b | <2.0 ± 0.0†,a | <4.3 ± 2.2ǂ,a | 5.9 ± 0.4a | |
| S + P + L | Larvae | 7 | 9.0 ± 0.3a | 9.1 ± 0.3a | 9.6 ± 0.3a | <2.0 ± 0.0†,a | 6.5 ± 0.3a | 5.7 ± 0.1a | |
Uninoculated replicates; ° = sample is similar at this timepoint to chicken starter feed without larvae and was not determined a second time;
“<2.0” indicates that Salmonella sp. count was below the detection limit (2 log CFU/g) in every sample;
“<” followed by a value higher than 2.0 log CFU/g indicates that Salmonella sp. count was below the detection limit in at least one, but not all samples; abc Average values for total viable counts and Salmonella sp. counts within each row that share a letter in superscript did not significantly (p ≥ 0.05) increase or decrease between sampling days.
For Salmonella (Table 3), the counts in the substrate (S + P) at day 0 were respectively 3.6 ± 0.4 log CFU/g and 7.6 ± 0.1 for the log 4 and log 7 challenge. At the high inoculation level, Salmonella counts remained fairly constant over time in the substrate, both in the absence (S + P) and presence (S + P + L) of larvae. In contrast, at the low inoculation level, an increase was observed over the 6-day period to a count of 5.9 ± 0.8 log CFU/g and 6.9 ± 0.7 log CFU/g in the absence (S + P) or presence (S + P + L) of larvae respectively. At day 2, considerable variation between replicates was observed in the substrate for both conditions (S + P and S + P + L), with Salmonella counts even below the detection limit in respectively two and one out of the four replicates. This indicates that at lower inoculation levels, the initial colonization speed of the substrate by Salmonella sp. is more variable and can even lead to an apparent removal of this pathogen. Nevertheless, the pathogen managed to successfully colonize the substrate over time. This observation is not impacted by the presence of larvae, as a similar trend is observed in both conditions and S + P and S + P + L at the 4 log CFU/g inoculation level.
Focusing on the larvae, for the 7 log CFU/g challenge test we observed a colonization evolution to 5.7 ± 0.1 CFU/g at day 6, which was a lower end value than in series 1 where a Salmonella count of 7.3 ± 1.0 was reached. Yet a similarity with series 1 was that the counts in the larvae both at day 2 and day 6 were about 1.5 log CFU/g and significantly (p < 0.001) lower than in the corresponding substrates. It should not be interpreted as a specific reduction of Salmonella numbers by the larvae during the rearing process, because this difference between larvae and their substrate is also observed for the total viable count. Moreover, after a challenge with 4 log CFU/g, at day 6 Salmonella reached even the same count (5.9 ± 0.4 CFU/g) as after the 7 log CFU/g challenge.
Extreme care that was taken in all series of experiments to avoid cross-contamination during manipulations of containers and samples. Therefore, the most intriguing observation from experiment series 2 was the presence of the Salmonella in the various uninoculated samples (S and S + L). Since kanamycin was used in the medium, the colonies found on the plates originated from resistant strains. All four replicates of the uninoculated samples (S + L), both in the substrate and larvae, were contaminated at day 6. With respectively 6.2 ± 0.5 and 5.1 ± 1.6 log CFU/g, these counts are comparable to the counts observed in the inoculated samples. All four replicates of the substrate (S) were also contaminated at day 6, while these replicates had a count below the detection limit at day 2. A possible hypothesis for these contaminations is that a small amount of naturally resistant Salmonella sp. were present below the detection limit and reached sufficient levels to be detected over time. Another possible explanation is the occurrence of airborne transmission of Salmonella in the climate chamber, which will be explored in more detail in the discussion. Though additional research is needed to confirm the following finding, the larvae seem to aggravate this transmission, as contaminations manifested themselves earlier if larvae were present (three out of four replicates of substrate contaminated at day 2 for S + L) compared to substrate without larvae, and the contaminations also reached a higher level in presence of larvae. Their movement through the substrate via their feeding behavior might contribute to airborne distribution via aerosols and/or dust particles.
3.3. Salmonella dynamics using a low inoculation level, kanamycin-resistant strains and prevention of airborne transmission
A third series of experiments with the same resistant strains was performed, and conditions were incubated separately from each other (i.e. consecutively), so that airborne transmission between the different conditions was completely excluded. An even lower contamination level than in the previous series of tests was applied, i.e. a target level of 3 log CFU/g Salmonella, to cover more potential situations in the insect industry related to Salmonella-infected substrates. The microbiological counts can be found in Table 4. As for the previous two series of tests, no impact on larval growth was observed (data not shown). The data for the total viable counts were similar to those found in the previous series, with the total viable counts increasing in all substrates, regardless of larval presence, to levels over 10 log CFU/g. At the same time, the total viable counts showed a smaller increase in the larvae than in the substrate and reached a level that was about 1 log CFU/g lower than that of the substrate.
Table 4.
Total aerobic viable counts and Salmonella spp. counts from larvae and substrate samples of experiment series 3 involving a low contamination of resistant Salmonella strains (3 log CFU/g) and all conditions incubated separately in climate chamber. Values represent the mean (±standard deviation) of three batches, each with two replicates per condition (n = 3x2). S = Substrate; S + P = Substrate with pathogen; S + L = Substrate with larvae; and S + P + L = Substrate with pathogen and larvae.
| Experimental condition | Investigated sample | Target Salmonella spp. contamination level in substrate (log CFU/g) |
Total viable count (log CFU/g) |
Salmonella spp. count (log CFU/g) |
|||||
|---|---|---|---|---|---|---|---|---|---|
| Day 0 | Day 2 | Day 6 | Day 0 | Day 2 | Day 6 | ||||
| S | Substrate | 0* | 4.7 ± 0.6a | 9.1 ± 0.2b | 10.1 ± 0.2c | <2.0 ± 0.0†,a | <2.0 ± 0.0†,a | <2.0 ± 0.0†,a | |
| S + P | Substrate | 3 | 4.4 ± 0.1a | 9.6 ± 0.1b | 10.4 ± 0.2c | <3.3 ± 0.3a | <2.3 ± 0.2b | <4.4 ± 1.9a,b | |
| S + L | Substrate | 0* | 4.7a | 10.3 ± 0.1b | 10.7 ± 0.2c | <2.0†,a | <2.0 ± 0.0†,a | <2.0 ± 0.0†,a | |
| S + P + L | Substrate | 3 | 5.9a | 10 ± 0.2b | 10.8 ± 0.2c | 3.5a | <2.0 ± 0.1ǂ,a | <3.4 ± 1.5ǂ,a | |
| S + L | Larvae | 0* | 8.2a | 9.3 ± 0.2a | 9.6 ± 0.9a | <2.0†,a | <2.1 ± 0.1,a,† | <2.0 ± 0.0†,a | |
| S + P + L | Larvae | 3 | 6.9a,b | 8.6 ± 0.1a | 9.3 ± 0.4b | <2.0†,a | <2.0 ± 0.0ǂ,a | 5.2 ± 0.6b | |
Uninoculated replicates;
“<2.0 or < 2.1″ indicates that Salmonella spp. was below the detection limit (2 to 2.1 log CFU/g) in every sample;
“<” followed by a value higher than 2.1 log CFU/g indicates that Salmonella spp. was below the detection limit in at least one, but not all samples; abc Average values for total viable counts and Salmonella sp. counts within each row that share a letter in superscript did not significantly (p ≥ 0.05) increase or decrease between sampling days.
No Salmonella was detected over the course of the challenge test in any of the uninoculated samples (S and S + L). In this third series of experiments, it can be excluded that Salmonella was present in the larvae under the detection limit at the start of the trial. In the same way as the 4 log CFU/g inoculation in series 2, for the inoculated substrate (S + P) a reduction occurred at day 2 to a level of < 2.3 ± 0.2 log CFU/g (with two out of six replicates below the detection limit). At day 6, the Salmonella counts were higher again, as they reached a value of < 4.4 ± 1.9 log CFU/g. Interestingly, in this design the two replicates that were below the detection limit at day 2 remained below the limit at day 6. In addition, at this low inoculation level, the larvae seemed to hinder colonization, since all six replicates of the substrate with pathogen and larvae (S + P + L) were below the detection limit at day 2. At day 6, four of the six replicates had a Salmonella count below detection, leading to an average count of < 3.4 ± 1.5 log CFU/g. This means that no significant increase in counts occurred in the substrate. This was not the case in the larvae, however. While no Salmonella was detected in the larvae on day 2, all replicates had detectable counts on day 6 with an average of 5.2 ± 0.6 log CFU/g. This number does not differ much from the final counts observed in experiment series 2 (Table 3) for the larvae at either of the two inoculation levels, indicating that the intestines of the larvae offer a habitable niche for Salmonella to colonize the larvae to a certain extent. That niche might even be more suited for Salmonella growth than the surrounding substrate, when only a low amount of Salmonella cells is present. The latter phenomenon might be substrate dependent, however, and needs further investigation.
4. Discussion
In the industrial practice of BSFL rearing for feed purposes, Salmonella can be present in ingredients used to prepare the substrate mixture. Typical (and currently allowed) ingredients include cereals and cereal-based materials such as distillers dried grains with solubles, fruit and vegetables and derived products, former foodstuffs (purely vegetal or containing eggs or milk) and compound feed. Evidence is present in literature for each of these ingredient categories that they can contain Salmonella (Berghofer et al., 2003, Centers for Disease Control and Prevention, 1998, Gosling et al., 2021, Jongen and (Ed.), 2005, Lee et al., 2016, Eglezos, 2010), which is why it was selected in this study. It is worth noting that Salmonella is only one of a number of foodborne pathogens that have been found in insects, including S. aureus, C. perfringens and the B. cereus group (Vandeweyer, De Smet, Van Looveren, & Van Campenhout, 2021). Future research will be needed to explore how their presence in the substrate affects the microbiological quality of BSFL.
The contamination level of Salmonella in a substrate mixture ready to provide to BSFL can vary, because (1) the load in individual contaminated ingredients can vary, and (2) the ingredients and/or the finished feed may be stored before feeding to the insects, causing Salmonella counts to increase or reduce during storage. Since the Salmonella load of the substrate can vary and since it is not known whether or this load determines the fate of the pathogen in contact with larvae in their substrate, several inoculation levels were included in this research. While a lower inoculation level indeed pointed at a slower colonization by Salmonella, or a longer suppression of Salmonella by the larvae, in none of the cases investigated, the larvae were able to eradicate the pathogen completely, or even to reduce its counts over time. In trials with inoculated substrate and larvae (S + P + L), both substrate and larvae were still contaminated after 6 days and Salmonella counts were as high as or sometimes several log cycles higher than values at day 0. These results are in strong contrast with the reports mentioned earlier (Erickson et al., 2004, Lalander et al., 2013, Lalander et al., 2015, Liu et al., 2008, Lopes et al., 2020), that generally describe a pronounced reduction of Salmonella (and some other zoonotic bacteria) by BSFL treatment of the waste, investigated as a possible hygienization step. Several possible explanations can be put forward for this difference. The manure types and aquaculture waste can be expected to substantially differ from the substrate used in our study, both in terms of chemical and microbiological composition. Salmonella cells inoculated in manure or aquaculture waste may be faced with another and potentially less favorable nutrient profile and a larger background microbiota than the chicken feed in our study. Therefore, in this potentially tougher environment, the cells may have less competitive advantage and/or may be less fit, and in this way they may be more vulnerable for the antimicrobial mechanisms exposed by BSFL, such as antimicrobial peptides, lysozymes, other antimicrobial components and the low pH encountered during intestinal passage (Bonelli et al., 2019). The fact that the substrate is different, can also have an impact on the larvae, which may be more triggered by the chemical and microbiological composition of manure or aquaculture waste than that of chicken feed to activate their immune system and exert their antimicrobial activities (Vogel et al., 2017).
A difficulty encountered in our inoculation experiments was the fact that, when trying to specifically count the Salmonella inoculated in the experiments, colonies from the background microbiota were also present on the selective plates. This indicates that micro-organisms with properties very close to the pathogen are present in the background microbiota. In our study, we eliminated these organisms by using antimicrobial resistance as an additional selection mechanism in the plates. In our previous work on inoculation experiments with Salmonella during yellow mealworm rearing (Wynants et al., 2019a), no substantial background microbiota hindered the Salmonella counts, and an additional selection mechanism was therefore not necessary. The other studies on BSFL already cited (with the exception of Erickson et al., 2004) did not use additional selection mechanisms in their media, and from their results, it is not clear whether background organisms are included in the counts or not. It can be advised for future inoculation experiments with BSFL, with authorized or not (yet?) authorized substrates, to anticipate on the abundance of organisms closely related to the wildtype target organism and include an extra selective or elective aid.
In our experimental set-up, we incorporated two types of uninoculated conditions, being the substrate alone and the substrate containing larvae. In the first two experiment series, we discovered that the uninoculated samples did not remain free of Salmonella. This was true for the two series and for both substrate and larvae. This finding, and taking into account our precautions to avoid cross-contamination during manipulations, urged us to conclude that the infection must have happened via transmission in the air (with 60% RH) in the ventilated climate chamber. This was confirmed by the fact that cross contamination did not occur when incubation of uninoculated and inoculated containers were incubated separately. While airborne transmission is generally not associated with Salmonella as a route for spreading in the food industry, multiple reports document on airborne transmission of Salmonella between animals in poultry (Adell et al., 2014, Gast et al., 1998, Holt et al., 1998, Kallapura et al., 2014, Lever and Williams, 1996, Richardson et al., 2003) and pig (Ikeguchi et al., 2005, Oliveira et al., 2006) houses. In a previous study by our research group on the dynamics of Salmonella sp. in mealworm rearing (Wynants et al., 2019, Wynants et al., 2019), uninoculated and inoculates replicates were incubated together (in containers that were not covered and that were placed at the same distance from each other as in this study and using in the same climate chamber (at 28 °C and 65% RH) as in this study), and there was no cross-contamination. The mealworms were reared in wheat bran (without water addition), however, which is expected to have a much lower moisture content than the moistened chicken feed in the current study. It is assumed that the moistened substrate in this study facilitates airborne transmission, viability during transmission and growth upon arrival on a new location (e.g. a neighboring container) of Salmonella cells. It is reasonable to extrapolate these findings and possible explanations to large scale BSFL rearing in stacked, open crates in a production facility with moistened and circulated air. If one crate contains substrate and crawling larvae that are highly contaminated, a rapid spread to other containers via the air throughout the rearing facility may take place. Even when harvested larvae are further processed with treatments that may reduce or eradicate Salmonella, a massive outbreak in the rearing facility should be avoided.
5. Conclusions
Our study revealed that, when reared on chicken feed, BSFL does not show a reducing effect on Salmonella counts in the substrate. It can be concluded though, that outgrowth of Salmonella is slower when the initial contamination level is lower. This contrasts a number of studies stating a reduction of Salmonella counts in BSFL, when reared on manure, which indicates a substrate-dependency in the fate of food pathogens in BSFL. In addition, our study demonstrates that airborne transmission is possible in laboratory conditions and we expect that it also may occur in industrial production facilities. Altogether, these observations lead to the general recommendation for insect producers to use substrate ingredients free of Salmonella, to avoid the entrance of the pathogen in the rearing and post-harvest processing line by any other route, and not to count on the antimicrobial activities that BSFL exert in some situations to eradicate the food safety risk. Future inoculation experiments are needed, using other Salmonella serotypes and other zoonotic bacteria, other substrates, larvae of other ages and variations on the rearing protocols to further elaborate on the dynamics of this pathogen and to support risk assessments. From our study, we can advise on the use of antibiotic resistant organisms to allow a proper monitoring of the inoculated strain. PCR technology can also assist in pathogen monitoring, provided proper primers for the target organism are available and background interference of the matrix can be excluded.
CRediT authorship contribution statement
J. De Smet: Conceptualization, Methodology, Investigation, Writing – original draft. D. Vandeweyer: Methodology, Data curation. L. Van Moll: Investigation, Writing – original draft, Visualization. D. Lachi: Investigation. L. Van Campenhout: Conceptualization, Writing – review & editing, Supervision, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
DV is financed by the Research Foundation - Flanders (FWO) via the SBO project ENTOBIOTA (S008519N) as well as by the European Union's Horizon 2020 Research and Innovation program via the H2020 project SUSINCHAIN (grant agreement number 861976). LVM and DL are funded by the former and latter project, respectively. JDS holds a postdoctoral fellowship grant (grant number 12V5219N) of the FWO. We would like to thank RADIUS for providing the 8-day old BSF larvae to start the rearing experiments and Dr. Aaron White for sharing his plasmids to generate Kanamycine resistant Salmonella strains.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.foodres.2021.110692.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Adell E., Moset V., Zhao Y., Jiménez-Belenguer A., Cerisuelo A., Cambra-López M. Comparative performance of three sampling techniques to detect airborne Salmonella species in poultry farms. Annals of Agricultural and Environmental Medicine. 2014;21(1):15–24. [PubMed] [Google Scholar]
- Arru B., Furesi R., Gasco L., Madau F.A., Pulina P. The introduction of insect meal into fish diet: The first economic analysis on European sea bass farming. Sustainability. 2019;11(6):1697. doi: 10.3390/su11061697. [DOI] [Google Scholar]
- Berghofer L.K., Hocking A.D., Miskelly D., Jansson E. Microbiology of wheat and flour milling in Australia. International Journal of Food Microbiology. 2003;85(1):137–149. doi: 10.1016/S0168-1605(02)00507-X. [DOI] [PubMed] [Google Scholar]
- Bonelli M., Bruno D., Caccia S., Sgambetterra G., Cappellozza S., Jucker C.…Casartelli M. Structural and functional characterization of Hermetia illucens larval midgut. Frontiers in Physiology. 2019;10 doi: 10.3389/fphys.2019.0020410.3389/fphys.2019.00204.s00110.3389/fphys.2019.00204.s002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne, J. (2021). Insects as feed: EU legislative developments ahead. https://www.feednavigator.com/Article/2020/11/27/Insects-as-feed-EU-legislative-developments-ahead.
- Centers for Disease Control and Prevention Multistate outbreak of Salmonella serotype Agona infections linked to toasted oats cereal. Morbidity and Mortality Weekly Report. 1998;47(22):462–464. [PubMed] [Google Scholar]
- Choi W.-H., Yun J.-H., Chu J.-P., Chu K.-B. Antibacterial effect of extracts of Hermetia illucens (Diptera: Stratiomyidae) larvae against Gram-negative bacteria. Entomological Research. 2012;42(5):219–226. doi: 10.1111/j.1748-5967.2012.00465.x. [DOI] [Google Scholar]
- Cosby D.E., Cox N.A., Harrison M.A., Wilson J.L., Jeff Buhr R., Fedorka-Cray P.J. Salmonella and antimicrobial resistance in broilers: A review. Journal of Applied Poultry Research. 2015;24(3):408–426. doi: 10.3382/japr/pfv038. [DOI] [Google Scholar]
- Dijk, R., van den Berg, D., Beumer, R. R., De Boer, E., Dijkstra, A. F., Mout, L., Stegeman, H., Uyttendaele, M., & Veld, S. In ’t. (2015). Microbiologie van Voedingsmiddelen: Methoden, principes en criteria. MYbusinessmedia.
- EFSA Scientific Committee Risk profile related to production and consumption of insects as food and feed. EFSA Journal. 2015;13(10):4257. doi: 10.2903/j.efsa.2015.4257. [DOI] [Google Scholar]
- Eglezos S. Microbiological quality of wheat grain and flour from two mills in Queensland Australia. Journal of Food Protection. 2010;73(8):1533–1536. doi: 10.4315/0362-028X-73.8.1533. [DOI] [PubMed] [Google Scholar]
- Erickson M.C., Islam M., Sheppard C., Liao J., Doyle M.P. Reduction of Escherichia coli O157:H7 and Salmonella enterica serovar Enteritidis in chicken manure by larvae of the black soldier fly. Journal of Food Protection. 2004;67(4):685–690. doi: 10.4315/0362-028x-67.4.685. [DOI] [PubMed] [Google Scholar]
- Gast, R. K., Mitchell, B. W., & Holt, P. S. (1998). Airborne transmission of Salmonella Enteritidis infection between groups of chicks in controlled-environment isolation cabinets. Avian Diseases, 42(2), 315–320. https://doi.org/https://doi.org/10.2307/1592482. [PubMed]
- Gosling R.J., Mawhinney I., Richardson K., Wales A., Davies R. Control of Salmonella and pathogenic E. coli contamination of animal feed using alternatives to formaldehyde-based treatments. Microorganisms. 2021;9:263. doi: 10.3390/microorganisms9020263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt, P. S., Mitchell, B. W., & Gast, R. K. (1998). Airborne horizontal transmission of Salmonella Enteritidis in molted laying chickens. Avian Diseases, 42(1), 45–52. https://doi.org/https://doi.org/10.2307/1592575. [PubMed]
- Ikeguchi, A., Okushima, L., Zhang, G., & Strom, J. S. (2005). Contaminant air propagation between naturally ventilated scale model pig buildings under steady-state conditions. Biosystems Engineering, 90(2), 217–226. https://doi.org/10.1016/j.biosystemseng.2004.10.011.
- Ipema Allyson F., Gerrits Walter J.J., Bokkers Eddie A.M., Kemp Bas, Bolhuis J. Elizabeth. Provisioning of live black soldier fly larvae (Hermetia illucens) benefits broiler activity and leg health in a frequency- and dose-dependent manner. Applied Animal Behaviour Science. 2020;230:105082. doi: 10.1016/j.applanim.2020.105082. [DOI] [Google Scholar]
- Jongen, W. (Ed.). (2005). Improving the safety of fresh fruit and Vegetables (1st ed.). Woodhead Publishing.
- Joosten Lotte, Lecocq Antoine, Jensen Annette Bruun, Haenen Olga, Schmitt Eric, Eilenberg Jørgen. Review of insect pathogen risks for the black soldier fly (Hermetia illucens) and guidelines for reliable production. Entomologia Experimentalis et Applicata. 2020;168(6-7):432–447. doi: 10.1111/eea.v168.6-710.1111/eea.12916. [DOI] [Google Scholar]
- Kallapura G., Morgan M.J., Pumford N.R., Bielke L.R., Wolfenden A.D., Faulkner O.B.…Tellez G. Evaluation of the respiratory route as a viable portal of entry for Salmonella in poultry via intratracheal challenge of Salmonella Enteritidis and Salmonella Typhimurium1. Poultry Science. 2014;93(2):340–346. doi: 10.3382/ps.2013-03602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalander C., Diener S., Magri M.E., Zurbrügg C., Lindström A., Vinnerås B. Faecal sludge management with the larvae of the black soldier fly (Hermetia illucens) — From a hygiene aspect. Science of The Total Environment. 2013;458–460:312–318. doi: 10.1016/j.scitotenv.2013.04.033. [DOI] [PubMed] [Google Scholar]
- Lalander C.H., Fidjeland J., Diener S., Eriksson S., Vinnerås B. High waste-to-biomass conversion and efficient Salmonella spp. reduction using black soldier fly for waste recycling. Agronomy for Sustainable Development. 2015;35(1):261–271. doi: 10.1007/s13593-014-0235-4. [DOI] [Google Scholar]
- Lee K.-M., Herrman T.J., Post L. Evaluation of selected nutrients and contaminants in distillers grains from ethanol production in Texas. Journal of Food Protection. 2016;79(9):1562–1571. doi: 10.4315/0362-028X.JFP-16-072. [DOI] [PubMed] [Google Scholar]
- Lever M.S., Williams A. Cross-infection of chicks by airborne transmission of Salmonella Enteritidis PT4. Letters in Applied Microbiology. 1996;23(5):347–349. doi: 10.1111/j.1472-765x.1996.tb00205.x. [DOI] [PubMed] [Google Scholar]
- Liu Q., Tomberlin J.K., Brady J.A., Sanford M.R., Yu Z. Black soldier fly (Diptera: Stratiomyidae) larvae reduce Escherichia coli in dairy manure. Environmental Entomology. 2008;37(6):1525–1530. doi: 10.1603/0046-225X-37.6.1525. [DOI] [PubMed] [Google Scholar]
- Lopes I.G., Lalander C., Vidotti R.M., Vinnerås B. Reduction of bacteria in relation to feeding regimes when treating aquaculture waste in fly larvae composting. Frontiers in Microbiology. 2020;11:1616. doi: 10.3389/fmicb.2020.01616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira C.J.B., Carvalho L.F.O.S., Garcia T.B. Experimental airborne transmission of Salmonella Agona and Salmonella Typhimurium in weaned pigs. Epidemiology and Infection. 2006;134(1):199–209. doi: 10.1017/S0950268805004668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osimani A., Ferrocino I., Corvaglia M.R., Roncolini A., Milanović V., Garofalo C.…Clementi F. Microbial dynamics in rearing trials of Hermetia illucens larvae fed coffee silverskin and microalgae. Food Research International. 2021;140 doi: 10.1016/j.foodres.2020.110028. [DOI] [PubMed] [Google Scholar]
- Richardson L.J., Hofacre C.L., Mitchell B.W., Wilson J.L. Effect of electrostatic space charge on reduction of airborne transmission of Salmonella and other bacteria in broiler breeders in production and their progeny. Avian Diseases. 2003;47(4):1352–1361. doi: 10.1637/7013. [DOI] [PubMed] [Google Scholar]
- Shivak Dylan J., MacKenzie Keith D., Watson Nikole L., Pasternak J. Alex, Jones Brian D., Wang Yejun.…Kivisaar M. A modular, Tn7-based system for making bioluminescent or fluorescent Salmonella and Escherichia coli strains. Applied and Environmental Microbiology. 2016;82(16):4931–4943. doi: 10.1128/AEM.01346-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogari G., Amato M., Biasato I., Chiesa S., Gasco L. The potential role of insects as feed: A multi-perspective review. Animals. 2019;9(4):119. doi: 10.3390/ani9040119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Fels-Klerx, H. J., Camenzuli, L., Belluco, S., Meijer, N., & Ricci, A. (2018). Food safety issues related to uses of insects for feeds and foods. Comprehensive Reviews in Food Science and Food Safety, 17(5), 1172–1183. https://doi.org/10.1111/1541-4337.12385. [DOI] [PubMed]
- Vandeweyer D., De Smet J., Van Looveren N., Van Campenhout L. Biological contaminants in insects as food and feed. Journal of Insects as Food and Feed. 2021:1–16. [Google Scholar]
- Vogel H., Müller A., Heckel D., Gutzeit H., Vilcinskas A. Nutritional immunology: Diversification and diet-dependent expression of antimicrobial peptides in the black soldier fly Hermetia illucens. Developmental & Comparative Immunology. 2017;78:141–148. doi: 10.1016/j.dci.2017.09.008. [DOI] [PubMed] [Google Scholar]
- Wynants E., Frooninckx L., Crauwels S., Verreth C., De Smet J., Sandrock C.…Van Campenhout L. Assessing the microbiota of Black Soldier Fly larvae (Hermetia illucens) reared on organic waste streams on four different locations at laboratory and large Scale. Microbial Ecology. 2019;77(4):913–930. doi: 10.1007/s00248-018-1286-x. [DOI] [PubMed] [Google Scholar]
- Wynants E., Frooninckx L., Van Miert S., Geeraerd A., Claes J., Van Campenhout L. Risks related to the presence of Salmonella sp. during rearing of mealworms (Tenebrio molitor) for food or feed: Survival in the substrate and transmission to the larvae. Food Control. 2019;100:227–234. doi: 10.1016/j.foodcont.2019.01.026. [DOI] [Google Scholar]
- Xu, J., Luo, X., Fang, G., Zhan, S., Wu, J., Wang, D., & Huang, Y. (2020). Transgenic expression of antimicrobial peptides from black soldier fly enhance resistance against entomopathogenic bacteria in the silkworm, Bombyx mori. Insect Biochemistry and Molecular Biology, 127, 103487. https://doi.org/https://doi.org/10.1016/j.ibmb.2020.103487. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.