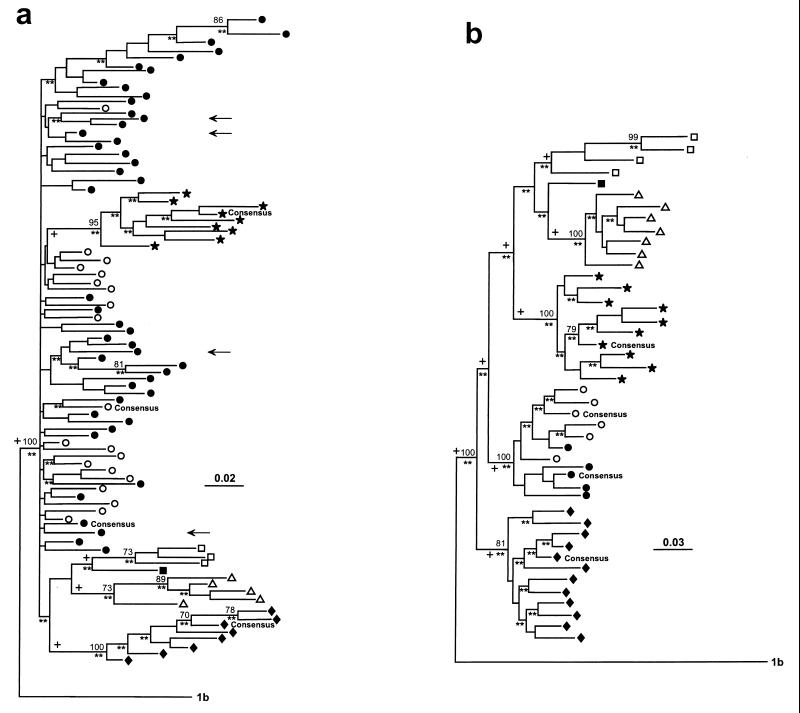
In their recent comparison of two rapid-genotyping methods for hepatitis C virus (HCV) Le Pogam et al. found that type 2 HCV is frequently mistyped by available methods (2). We have reported similar findings (4). By performing a phylogenetic analysis of partial core sequences from 63 type 2 isolates, of which 8 were their own and 55 were found in the literature or in data banks and were representative of subtypes 2a to 2f, Le Pogam et al. also found that subtype 2c, a subtype highly prevalent in Italy (3), is largely paraphyletic. They concluded that any assignment of HCV isolates to subtype 2c is quite weak and that other domains of the viral genome should be investigated for more informative phylogenetic analysis. Following their suggestion, we have reconsidered the phylogenetic position of 45 Italian HCV isolates that we had previously classified as subtype 2c based on comparison with the limited number of sequences then available (3) by performing a new phylogenetic analysis in the core and NS5 regions. In this analysis we also included most of the type 2 sequences presently found in data banks. For the core region analysis a total of 88 sequences (8 of subtype 2a, 7 of subtype 2b, 64 of subtype 2c, 1 of subtype 2d, 5 of subtype 2e, and 3 of subtype 2f) were examined, and for the NS5 region a total of 41 sequences (9 of subtype 2a, 11 of subtype 2b, 9 of subtype 2c, 1 of subtype 2d, 7 of subtype 2e, and 4 of subtype 2f) were examined. The consensus sequences for subtypes 2a, 2b, and 2c, obtained as described previously (1), were also included, while the consensus sequence of subtype 1b served as an outgroup. Phylogenetic analysis was performed by using the Fitch-Margoliash (FM) algorithm with 100 bootstrap replications, the maximum-likelihood (ML) method with a transition/transversion ratio of 2.00, and the DNA parsimony (DNAPARS) method randomizing the data input (Phylogeny Inference Package, version 3.5c).
The phylogenetic reconstructions obtained by the three methods were quite similar although a few sequences assumed slightly different positions within the same clade (not shown). In the core tree, while all the other type 2 subtypes originated from single branches marked by high bootstrap values, subtype 2c isolates did not (Fig. 1a). Thus, in agreement with Le Pogam et al., in the core region subtype 2c proved highly paraphyletic. In the tree shown in Fig. 1a, all the Italian isolates were intermingled with other previously classified subtype 2c isolates. The phylogenetic tree reconstructed by using the NS5 region was, however, quite different. As expected from the genetic distances, in this tree the branches produced were longer, and more importantly, all the 2c isolates, regardless of their position in the core tree, originated from a single branch, thus behaving like the other subtypes (Fig. 1b). We conclude that subtype 2c is not paraphyletic when examined in the NS5 region and that this region should be preferred for the classification of type 2 isolates.
FIG. 1.
Phylogenetic analyses of HCV type 2 isolates based on core sequences (nucleotides 160 to 356) (a) and NS5 sequences (nucleotides 7500 to 7612) (b). The branching pattern of the trees was obtained by the FM algorithm, bootstrap values above 70 out of 100 are shown at the branch points, and double asterisks indicate branches at P < 0.01 as calculated by the ML method. Branches identifying the monophyletic clades also retrieved by the DNAPARS method are indicated by a plus sign. Numbers above the bars indicate the number of substitutions per site calculated by Kimura’s two-parameter model with a transition/transversion ratio of 2.00. (a) Italian isolates examined in both the core and NS5 region are indicated by arrows. In both panels, the subtypes of isolates and consensus sequences are indicated by the following symbols: ★, 2a; ♦, 2b; ○, 2c; ●, 2c (Italian isolates); ■, 2d; ▵, 2e; □, 2f.
Ed. Note: The editor and the authors of the published article did not feel that a response was necessary.
REFERENCES
- 1.Cammarota G, Maggi F, Vatteroni M L, Da Prato L, Barsanti L, Bendinelli M, Pistello M. Partial nucleotide sequencing of six subtype 2c hepatitis C viruses detected in Italy. J Clin Microbiol. 1995;33:2781–2784. doi: 10.1128/jcm.33.10.2781-2784.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Le Pogam S, Dubois F, Christen R, Raby C, Cavicchini A, Goudeau A. Comparison of DNA enzyme immunoassay and line probe assays (Inno-LIPA HCV I and II) for hepatitis C genotyping. J Clin Microbiol. 1998;36:1461–1463. doi: 10.1128/jcm.36.5.1461-1463.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maggi F, Vatteroni M L, Fornai C, Morrica A, Giorgi M, Bendinelli M, Pistello M. Subtype 2c of hepatitis C virus is highly prevalent in Italy and is heterogeneous in the NS5A region. J Clin Microbiol. 1997;35:161–164. doi: 10.1128/jcm.35.1.161-164.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vatteroni M L, Maggi F, Morrica A, Fornai C, Giorgi M, Pistello M, Bendinelli M. Comparative evaluation of five rapid methods for identifying subtype 1b and 2c hepatitis C virus isolates. J Virol Methods. 1997;66:187–194. doi: 10.1016/s0166-0934(97)00054-2. [DOI] [PubMed] [Google Scholar]