Abstract
For grain crops such as rice (Oryza sativa), grain size substantially affects yield. The histone acetyltransferase GRAIN WEIGHT 6a (GW6a) determines grain size and yield in rice. However, the gene regulatory network underlying GW6a-mediated regulation of grain size has remained elusive. In this study, we show that GW6a interacts with HOMOLOG OF DA1 ON RICE CHROMOSOME 3 (HDR3), a ubiquitin-interacting motif-containing ubiquitin receptor. Transgenic rice plants overexpressing HDR3 produced larger grains, whereas HDR3 knockout lines produce smaller grains compared to the control. Cytological data suggest that HDR3 modulates grain size in a similar manner to GW6a, by altering cell proliferation in spikelet hulls. Mechanistically, HDR3 physically interacts with and stabilizes GW6a in an ubiquitin-dependent manner, delaying protein degradation by the 26S proteasome. The delay in GW6a degradation results in dramatic enhancement of the local acetylation of H3 and H4 histones. Furthermore, RNA sequencing analysis and chromatin immunoprecipitation assays reveal that HDR3 and GW6a bind to the promoters of and modulate a common set of downstream genes. In addition, genetic analysis demonstrates that HDR3 functions in the same genetic pathway as GW6a to regulate the grain size. Therefore, we identified the grain size regulatory module HDR3–GW6a as a potential target for crop yield improvement.
A ubiquitin receptor ubiquitylates and stabilizes a histone acetyltransferase; this module regulates downstream gene expression, altering rice grain size by modulating cell proliferation.
Introduction
In rice (Oryza sativa L.), grain yield is mainly controlled by quantitative trait loci (QTLs) for its component traits, including panicle number, grain number per panicle, and grain size. Notably, a number of QTLs, such as GRAIN LENGTH ON CHROMOSOE 2 (GL2)/GRAIN SIZE ON CHROMOSOME 2 (GS2)/RICE GROWTH-REGULATING FACTOR 4 (OsGRF4), THOUSAND GRAIN WEIGHT ON CHROMOSOME 3 (TGW3)/GRAIN LENGTH 3.3 (GL3.3), THOUSAND-GRAIN WEIGHT 6 (TGW6), GRAIN SIZE ON CHROMOSOME 5 (GS5), GRAIN WEIGHT ON CHROMOSOME 5 (GW5)/GRAIN SIZE ON CHROMOSOME 5 (GSE5), GRAIN LENGTH 3.1 (GL3.1), RICE SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 13 (OsSPL13)/RICE GRAIN LENGTH AND WEIGHT ON CHROMOSOME 7 (GLW7), GRAIN WIDTH ON CHROMOSOME 2 (GW2), GRAIN WEIGHT 6a (GW6a), and GRAIN WIDTH ON CHROMOSOME 8 (GW8)/OsSPL16 for grain size have been identified and characterized as significant players contributing to rice yield (Song et al., 2007, 2015; Li et al., 2011, 2017; Qi et al., 2012; Wang et al., 2012; Ishimaru et al., 2013; Hu et al., 2015, 2018; Che et al., 2016; Si et al., 2016; Duan et al., 2017; Xia et al., 2018; Ying et al., 2018). Furthermore, several signaling pathways including those mediated by plant hormones, G-proteins, the ubiquitin-proteasome, protein kinases, and transcriptional factors have been proposed to play a key role in regulating the grain size (Zuo and Li, 2014; Li et al., 2019).
Protein ubiquitylation has been considered as a central mechanism to modulate various cellular pathways, with profound effects on proteolytic and nonproteolytic functions (e.g. subcellular localization, enzyme activity) (Bonifacino and Traub, 2003; Elsasser and Finley 2005). The process is regulated by the sequential action of an ATP-dependent E1 (ubiquitin-activating)–E2 (ubiquitin-conjugating)–E3 (ubiquitin-ligating) enzyme cascade (Dikic et al., 2009). For example, the causal gene for the above-mentioned QTL GW2 encodes a RING-type E3 ubiquitin ligase that regulates rice grain width and weight, which was proposed to target proteins for degradation by the 26S proteosome (Song et al., 2007). Interestingly, the rice grain size gene WIDE AND THICK GRAIN 1 (WTG1) encodes an otubain-like protease that has deubiquitination activity (Huang et al., 2017). Recently, another positive regulator of grain length and width, rice UBIQUITIN-SPECIFIC PROTEASE 15 (OsUBP15) also possesses deubiquitination activity (Shi et al., 2019). Notably, the grain size QTL GW6a, which encodes a histone acetyltransferase, acts as a positive regulator of grain size and rice yield (Song et al., 2015). However, the downstream or upstream genetic factors of the GW6a-dependent regulation of grain size are currently unknown.
In Arabidopsis thaliana, the ubiquitin receptor DA1 regulates final seed and organ size by limiting the period of cell proliferation (Li et al., 2008). Ubiquitin receptors were reported to interact with ubiquitinated protein substrates of E3 ubiquitin ligases via ubiquitin-interacting motifs (UIMs) and facilitate their degradation by the 26S proteasome (Verma et al., 2004). As expected, DA1 and its close family members DA1-RELATED1 (DAR1) and DAR2 physically interact with and redundantly boost the protein degradation of the plant-specific transcription factors TEOSINTE BRANCHED1/CYCLOIDEA/PCF 14 (TCP14) and TCP15 to regulate cell and organ growth through the alteration of endoreduplication (Peng et al., 2015). Furthermore, DA1 was characterized as a peptidase that is activated by the ubiquitinated modification of the two RING E3 ligases, BIG BROTHER (BB) and DA2 (Arabidopsis homolog of rice GW2), which are subsequently cleaved by the activated DA1 and destabilized (Disch et al., 2006; Dong et al., 2017). However, it remains unknown about whether and how the homologs of DA1 family members control the grain size in rice.
Here, we show that one of the rice homologs of Arabidopsis DA1, HOMOLOG OF DA1 ON RICE CHROMOSOME 3 (HDR3), interacts physically with GW6a. Like GW6a, HDR3 is also a positive regulator of grain size in rice. Unexpectedly, we observed that HDR3 possesses ubiquitin-binding activity and enhanced the ubiquitylation and stability of GW6a in a UIM-dependent manner. The resulting delay of the degradation of GW6a promotes its acetyltransferase activity and increases the expression of its downstream target genes. Thus, our findings reveal a molecular mechanism for grain size control and provide useful information for improvement of grain yield.
Results
Identification of HDR3, a protein homologous to Arabidopsis DA1, that interacts with GW6a
To identify interacting proteins, we fused the GW6a coding sequence to the GAL4 DNA-binding domain (BD-GW6a) and used it as bait for a yeast two-hybrid (Y2H) screening. Ultimately, we paid an especial attention to one identified candidate segment that corresponds to part of the Os03g0267800 gene (we here named HOMOLOG OF DA1 ON RICE CHROMOSOME 3 [HDR3], see below). Consistent with an interaction between HDR3 and GW6a, yeast cells co-transformed with plasmid DNAs containing the GAL4 activation domain (AD)-fused HDR3 (AD-HDR3) and BD-GW6a grew well on the quadruple dropout media, while those transformed with the corresponding control did not (Figure 1A).
Figure 1.
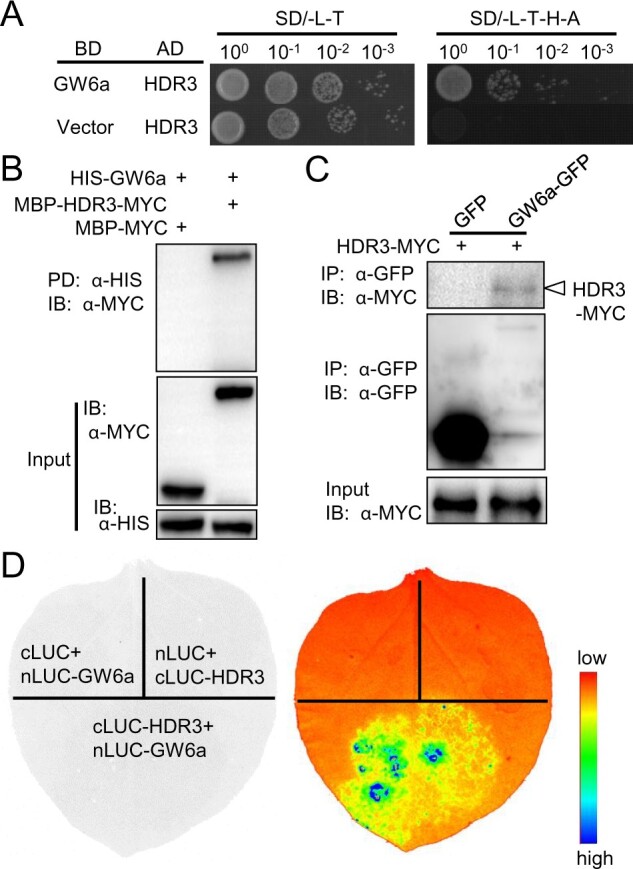
HDR3 associates with GW6a in vitro and in vivo. A, Y2H assays showing that HDR3 interacts with GW6a. SD, yeast drop-out culture medium; L, Leu; T, Trp; H, His; A, Ade. B, HDR3 interacts with GW6a in vitro. MBP-HDR3-MYC and MBP-MYC were pulled down (PD) by HIS-GW6a and analyzed by immunoblotting (IB) using an anti-MYC (α-MYC) antibody. C, HDR3 interacts with GW6a in vivo. Nicotiana benthamiana leaves were transformed by injection of the Agrobacterium EHA105 cells containing 35S::HDR3-MYC and 35S::GW6a-GFP plasmids. Total proteins were immunoprecipitated with GFP-Trap-A, and the immunoblots were probed with the anti-GFP and anti-MYC antibodies, respectively. D, Split firefly luciferase complementation assays showing that HDR3 interacts with GW6a. nLUC-tagged GW6a was co-transformed into tobacco leaves along with cLUC-tagged HDR3.
To verify the HDR3–GW6a interaction by pull-down assays, we produced the fusion proteins HIS-GW6a, MBP-HDR3-MYC, and MBP-MYC in Escherichia coli cells. Following anti-HIS pull-downs, immunoblotting assays using an antibody to MYC detected an evident band indicative of the predicted MBP-HDR3-MYC size, but no band of MBP-MYC in the corresponding control (Figure 1B). Meanwhile, we conducted co-immunoprecipitation (Co-IP) assays to test the interaction in vivo, and inserted HDR3 and GW6a, respectively, into a binary vector to transiently express the fusion proteins HDR3-MYC, GW6a-GFP, and the GFP alone in Nicotiana benthamiana leaves by Agrobacterium-mediated transformation; upon IP using an antibody to GFP, immunoblotblot assays using an anti-MYC antibody detected a band of HDR3-MYC in the GW6a-GFP sample, but no band identified in the GFP sample (Figure 1C). In addition, split firefly luciferase complementation assays were also performed to confirm the interaction, in which activated luciferase reconstitution was indicated by luminescence signals when the fusion constructs cLUC-HDR3 and nLUC-GW6a were present, in contrast to those containing either the fusion construct cLUC-HDR3 and cLUC, or nLUC-GW6a and cLUC (Figure 1D). Thus, we concluded that HDR3 physically interacts with GW6a.
Sequencing analysis suggested that HDR3 is a homolog of DA1 on rice chromosome 3 (hence we named it HDR3) (Li et al., 2008). Database searching identified four homologs of DA1 on the rice genome, and of these homologs, the encoded protein of HDR3 shares the least protein identity (47.5%) with Arabidopsis DA1 (Supplemental Table S1). Nevertheless, we found that only HDR3 could interact with GW6a in yeast cells and the other three homologs (HDR3.1, HDR6, and HDR12) did not (Supplemental Figure S1).
We further mapped the domains of the GW6a–HDR3 interaction using Y2H assays, and observed that the carboxyl terminus of GW6a (GW6a-C) interacted with HDR3 (Supplemental Figure S2, A and B), and the amino terminus of HDR3 (HDR3-N) associated with GW6a (Supplemental Figure S2, C and D). Consistent with these results, amino acid sequence alignment with Arabidopsis DA1, DAR1, DAR2 (Li et al., 2008; Peng et al., 2015), and the four rice HDR homologs revealed a conserved C-terminus containing the LIN-11, ISL-1 AND MEC-3 (LIM) and predicted peptidase domains, and a variable N-terminus (Supplemental Figure S3A). Homology analysis also suggested that HDR3 has a close relationship with Arabidopsis DAR2 (Supplemental Figure S3B). Taking these results together, we identified a member of the DA1 family in rice, HDR3, the distinct N-terminus segment of which interacts with GW6a.
HDR3 functions as a positive regulator of grain size and weight
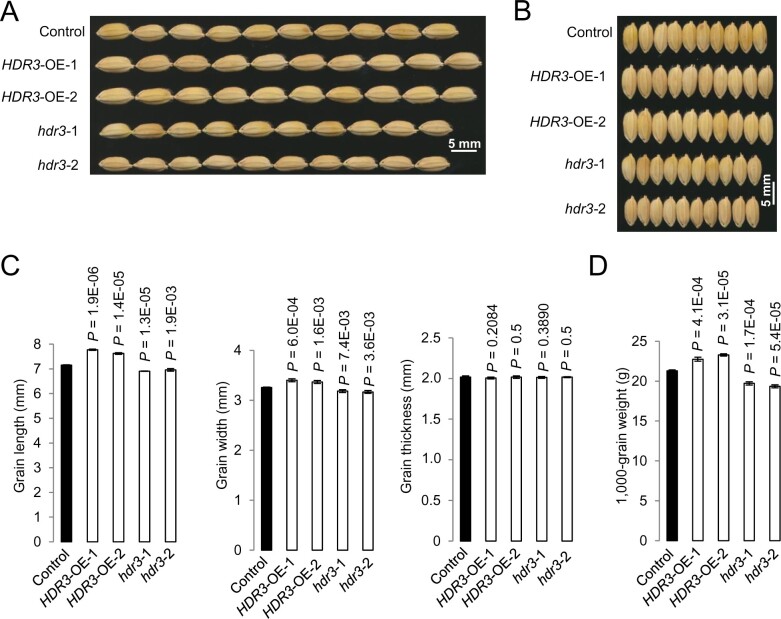
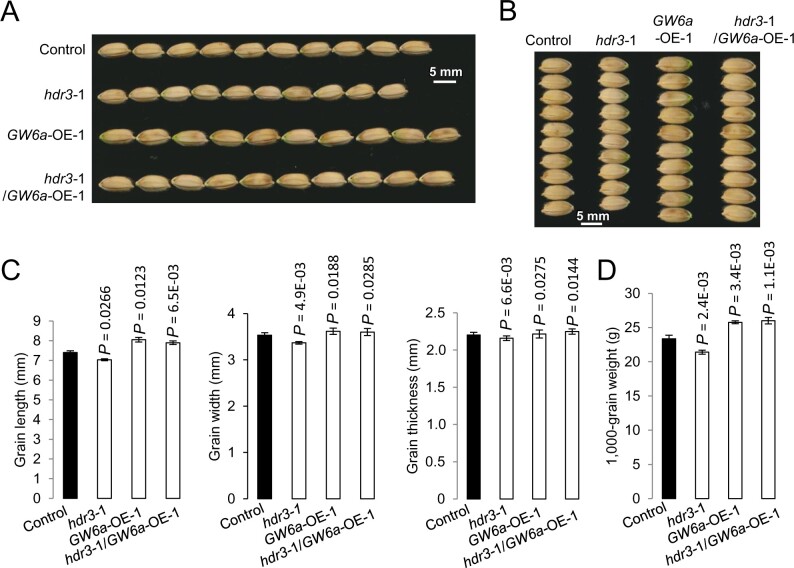
We asked whether HDR3 functions as a regulator of grain size, and produced transgenic rice plants overexpressing the HDR3 gene (HDR3-OE; Supplemental Figure S4A). The HDR3-OE plants set seeds apparently larger than the control did (Figure 2, A and B). Phenotypic measurement and statistical analysis revealed that the HDR3-OE grains were 10% longer and 5% wider, with the grain thickness unchanged, and thus 20% heavier, than the nontransgenic grains (control; Figure 2, C and D).
Figure 2.
HDR3 is a positive modulator of grain length, width, and weight in rice. A, B, Mature grains of the overexpressing rice lines HDR3-OE-1 and -2, and the gene knockout lines hdr3-1 and -2. C, Comparisons of grain length, width, and grain thickness of the transgenic HDR3-OE-1, -2, and hdr3-1, -2, and the corresponding control (Nipponbare). D, Comparisons of 1,000-grain weight of the transgenic HDR3-OE-1, -2, and hdr3-1, -2, and the corresponding control (Nipponbare). Student’s t test was used to generate the P-values in (C) and (D).
In contrast, two independent transgenic plants with gene editing of HDR3 using the CRISPR/Cas9 method (hdr3-1 and hdr3-2) produced seeds obviously smaller than the control (Figure 2, A and B;Supplemental Figure S4, B–D). Accordingly, phenotypic analyses showed that the hdr3 grains were 10% shorter and 5% narrower, with the grain thickness unchanged, and 15% lighter than the control seeds (Figure 2, C and D). Collectively, these results suggested that HDR3 functions as a positive regulator of grain size and weight in rice.
We also examined temporal and spatial expression patterns of HDR3 by reverse transcription quantitative polymerase chain reaction (RT-qPCR). Consistent with the role of HDR3 in the regulation of grain size, we observed that the HDR3 mRNA was preferentially present in panicles and the relative levels rose during panicle development with a peak at the panicle stage of 15 cm in length (Supplemental Figure S5A).
We next investigated the subcellular distribution of HDR3 in rice protoplast cells expressing the HDR3-GFP fusion protein, and immunoblotting using an anti-GFP antibody only detected a conspicuous band indicative of the HDR3-GFP size in the nuclear component (Supplemental Figure S5B); in contrast, the GFP signals were observed using a laser scanning confocal fluorescence microscope suggesting that HDR3-GFP localized in the cytoplasm (Supplemental Figure S5C). We also produced HDR3 and its two derivations fused with eGFP, and these constructs were transiently expressed in N. benthamiana leaf cells; the resultant eGFP signals were observed in the cytoplasm and/or nucleus (Supplemental Figure S5, D and E). These results suggested that HDR3 localizes to the cytoplasm and nucleus.
HDR3 mainly alters the number of cells in spikelet hulls
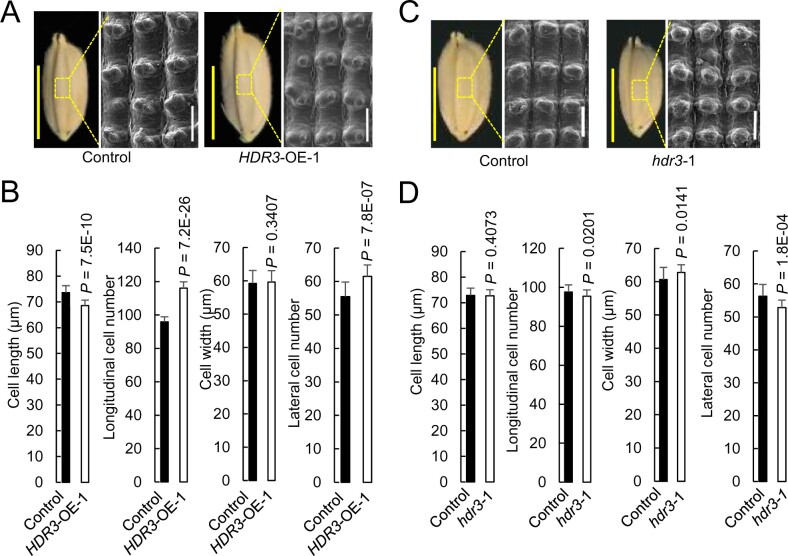
To investigate the cytological basis underlying the HDR3-mediated regulation of grain size, we analyzed cell number and cell size of lemmas in the central parts of mature grains by scanning electron microscopy. We observed that, in the orientation of grain length, the estimated total cell number of spikelet hull of HDR3-OE showed a substantial increase (21% increase) relative to that of the corresponding control, although the cell size showed a slight decrease; similarly, the lateral cell number of spikelet hull of HDR3-OE also increased significantly (11% increase) compared to that of the corresponding control (Figure 3, A and B).
Figure 3.
HDR3 regulates grain size presumably through its positive role in the regulation of cell number in spikelet hulls. A, C, Mature grain and scanning electron microscopic analyses of the outer surfaces of the spikelet hulls of overexpressing rice lines HDR3-OE-1 and the gene knockout lines hdr3-1, and the corresponding control (Nipponbare). White bar = 200 μm; yellow bar = 5 mm. B, D, Comparisons of cell length, and the total of cell number, of the spikelet hulls along the grain length orientation of the transgenic lines HDR3-OE-1 and hdr3-1, respectively, with the corresponding control (Nipponbare). Student’s t test was used to generate the P-values.
We also evaluated cytological features of the hdr3 grains. Consistent with the above results, we observed that, in the orientations of grain length and width, hdr3-1 had a significant decrease of cell number, respectively, of 2.3% and 6.6%, with the cell size unchanged, compared with that of the corresponding control (Figure 3, C and D). These data demonstrated that the regulation of grain size by HDR3 results mainly from the alteration of cell number, but not of cell size, suggesting that HDR3 may function involved in the regulation of cell proliferation.
HDR3 is an active ubiquitin receptor that enhances the ubiquitylation of GW6a
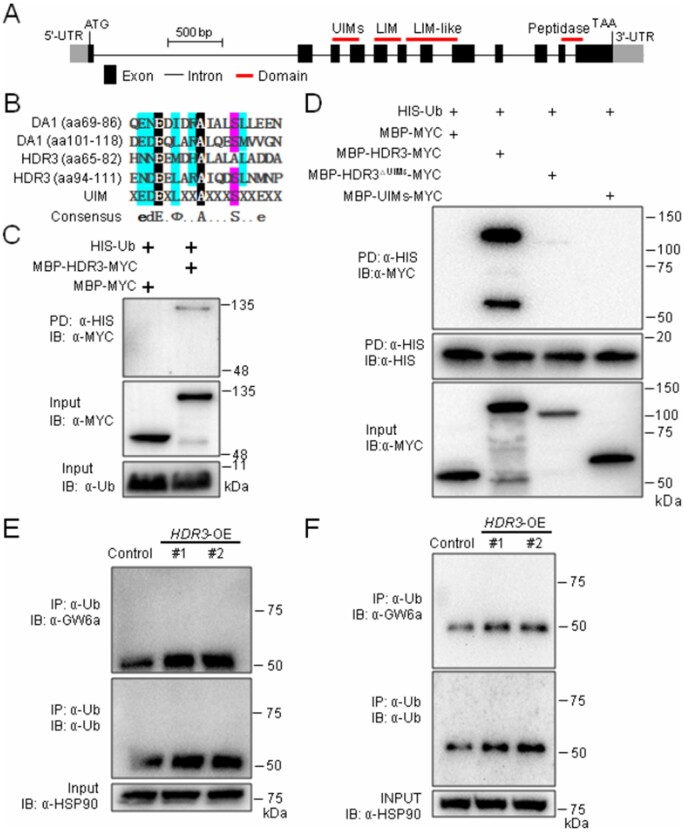
Like DA1, HDR3 presumably possesses four predicted domains: two UIMs (i.e. UIM1 and UIM2; ubiquitin-interacting motifs) in tandem arrangement, one LIM (serves as protein interaction interface), one LIM-like, and one peptidase domain (Dong et al., 2017; Figure 4A). Of these domains, we observed that the amino residues 65–82 and 94–111 of HDR3 make up a relatively conserved core sequence (although the conserved S81 in DA1 was replaced with A77 in HDR3), indicative of functional UIM domains (Figure 4B; Hofmann and Falquet, 2001). To test this hypothesis, we produced the fusion proteins MBP-HDR3-MYC and MBP-MYC, and used them for in vitro ubiquitin-binding assays. These assays revealed that MBP-HDR3-MYC could bind HIS-Ub (pull-downs using an anti-HIS and immunoblotting using an anti-MYC antibody), whereas MBP-MYC could not (Figure 4C). To further test whether UIMs are required for the binding ability of HDR3, we also produced the fusion proteins MBP-HDR3△UIMs-MYC (HDR3△UIMs is the HDR3 allele with a deletion of amino acids 65–111) and MBP-UIMs-MYC for in vitro ubiquitin-binding assays; the results suggested that, in contrast to MBP-HDR3-MYC, MBP-HDR3△UIMs-MYC could not bind HIS-Ub, nor did MBP-UIMs-MYC (Figure 4D). These results suggest that HDR3 is an ubiquitin receptor with ubiquitin-binding activity.
Figure 4.
HDR3 is an UIM-type ubiquitin receptor that promotes the GW6a ubiquitylation. A, The gene structure of HDR3. The translation start codon (ATG) and the stop codon (TAA) are indicated. B, Sequence alignments of the UIM domains of DA1 and HDR3. C, Pull-down assays suggesting that HDR3 has an ubiquitin-binding activity. D, Pull-down assays showing that MBP-HDR3△UIMs-MYC could not bind HIS-Ub, nor did MBP-UIMs-MYC. E, F, Co-IP assays showing that HDR3 acts to enhance the protein ubiquitylation of GW6a. Total proteins were extracted from, respectively, seedlings (E) and young panicles (F), of two independent HDR3-OE plants. Total proteins were immunoprecipitated with Ub-Trap-M, and the immunoblot was probed with α-Ub, α-HSP90 and α-GW6a antibodies, respectively.
In Arabidopsis, DA1 interacts with the E3 ubiquitin ligase DA2 (Xia et al., 2013). We then asked whether HDR3 could associate with GW2 (the rice homolog of DA2; Song et al., 2007; Xia et al., 2013). The following Y2H results showed that the yeast cells co-transformed with plasmid DNAs containing AD-GW2 and BD-HDR3 could grow well on the quadruple dropout media, while those co-transformed with plasmid DNAs containing either AD-GW2 and BD vector, or BD-HDR3 and AD vector, could not (Supplemental Figure S6A). We also verified the HDR3–GW2 interaction using pull-down assays, and observed that after anti-HIS pull-downs, immunoblot experiment using an antibody to MYC could detect the predicted MBP-HDR3-MYC band, but could not detect the corresponding MBP-MYC band (Supplemental Figure S6B). Thus, HDR3 interacts with GW2.
We further investigated whether GW2 could ubiquitinate HDR3. Consistent with our previous observation, in the presence of an ubiquitin-activating enzyme (HIS-E1), an ubiquitin-conjugating enzyme (E2), HIS-ubiquitin (HIS-Ub), and GST-GW2, self-ubiquitylation of GW2 was demonstrated by immunoblot using both anti-GST and anti-Ub antibodies (Supplemental Figure S6C, second lane from the left; Song et al., 2007). Similarly, the ubiquitylation of HDR3 by GW2 was revealed by immunoblotting using an anti-HIS antibody (Supplemental Figure S6C, first lane from the right).
We next tested a direct biochemical relationship between HDR3 and GW6a. Total proteins of HDR3-OE and the control plants were isolated and incubated with Ub-Trap-A rProtein A/G MagPoly beads to immunoprecipitate conjugated ubiquitins. Obviously, higher precipitates were detected in the HDR3-OE extractions than that of the control with anti-Ub and anti-GW6a antibodies (Figure 4, E and F), indicating that the overexpression of HDR3 enhances the ubiquitylation of GW6a.
HDR3 stabilizes GW6a and enhances its histone acetyltransferase activity
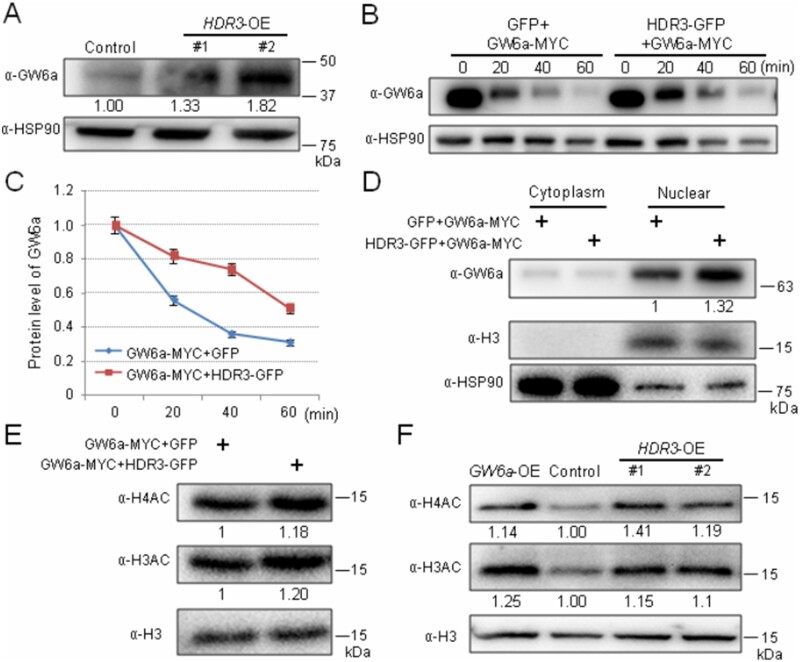
To investigate the biochemical consequences of the HDR3–GW6a interaction, we evaluated the GW6a protein level in transgenic HDR3-OE plants. Surprisingly, immunoblotting using an anti-GW6a antibody showed that the protein amount of GW6a visibly increased compared with the corresponding control (Figure 5A). To confirm the results, we conducted protein degradation experiments in rice protoplasts; we observed that the simultaneous expression of the fusion proteins HDR3-GFP and GW6a-MYC could significantly slow down the GW6a degradation relative to cells expressing just GW6a-MYC (Figure 5, B and C). The addition of the proteasome inhibitor MG132 also clearly inhibited GW6a degradation (Supplemental Figure S7, A and B). In contrast, rice protoplast cells expressing a combination of GW6a-MYC and HDR3△UIMs-GFP exhibited a comparable GW6a level to those expressing GW6a-MYC alone (Supplemental Figure S8, A and B). Furthermore, we observed that, contrary to the effect of HDR3 on the stability of GW6a, the rice protoplasts expressing GW2-GFP dramatically increased the speed of the GW6a protein degradation (Supplemental Figure 8, C and D). In addition, we analyzed the alterations of GW6a amount in cell components, and immunoblotting showed that the protein was predominantly present in the nuclear fraction, where an effect of 30% increase of GW6a level by the HDR3 action was observed (Figure 5D). Thus, we concluded that HDR3 stabilizes GW6a primarily in the nucleus.
Figure 5.
HDR3 stabilizes the protein and enhances the histone acetyltransferase activity of GW6a. A, The GW6a protein levels were elevated in the transgenic HDR3-OE plants compared with the nontransgenic plant (Control). GW6a was detected in protein extract from young panicles by immunoblotting with an anti-GW6a antibody (α-GW6a). B, HDR3 regulates the GW6a stabilization. The GW6a-MYC and HDR3–GFP fusion proteins were produced in rice protoplast cells. Immunoblotting showing the GW6a protein levels detected by an anti-GW6a antibody and anti-HSP90 as a control. C, Quantitative analysis of protein levels showed in (B). D, Immunoblotting analysis of GW6a amount in the cytoplasm and nuclear components extracted from rice protoplasts expressing the indicated fusion proteins. E, HDR3 enhances the histone H4 and H3 acetyltransferase activities by GW6a in rice protoplast cells. F, The overexpression of HDR3 in transgenic rice plants (HDR3-OE) enhances the H4AC and H3AC levels in the young panicles, similar to that of the overexpression of GW6a (GW6a-OE).
GW6a is an active histone H4 acetyltransferase (Song et al., 2015). We assumed that the increase of GW6a protein level by the HDR3 action would elevate histone acetylation. Supporting our idea, we observed that the total proteins extracted from the protoplast cells co-transformed with GW6a-MYC and HDR3-GFP, but not GW6a-MYC and HDR3△UIMs-GFP constructs showed enhanced levels of acetylation at H3 and H4 (H3AC and H4AC; Figure 5E;Supplemental Figure S8E). Furthermore, we found that total proteins extracted from young panicles of the HDR3-OE plants had significantly increased H4AC and H3AC modifications compared with the corresponding control, which were very similar to that of the GW6a-OE plants (Figure 5F).
We next examined the genetic interaction between HDR3 and GW6a. For this purpose, we crossed GW6a-OE-1 (male parent) with hdr3-1 (female parent) to obtain hdr3-1/GW6a-OE-1. As expected, we observed that, the hdr3-1/GW6a-OE-1 plants that are hemizygous for the OE transgene and heterozygous for the hdr3 allele produced grains visually very similar to those of GW6a-OE-1, in a sharp contrast to those of hdr3-1 (Figure 6, A and B). Phenotypic analyses showed that, relative to those of nontransgenic control, the hdr3-1/GW6a-OE-1 grains showed an increase of 6.8% and 10.3%, in grain length and weight, respectively, having a comparable effect to those of GW6a-OE-1 (with 8.8% and 9.9% increase), whereas the hdr3-1 grains exhibited a reduced size and weight (Figure 6, C and D), implying that GW6a-OE could nearly rescue the grain size and weight phenotypes of hdr3. Thus, HDR3 most likely functions in a common genetic pathway as and acts upstream of GW6a to regulate grain size and weight.
Figure 6.
HDR3 functions in a common genetic pathway with and upstream of GW6a to regulate grain size and weight. A, B, Mature grains of hdr3-1, GW6a-OE-1, and hdr3-1/GW6a-OE-1. C, Comparisons of grain length, width, and thickness of hdr3-1, GW6a-OE-1, hdr3-1/GW6a-OE-1 and the corresponding control (nontransgene). D, Comparisons of 1,000-grain weight of hdr3-1, GW6a-OE-1, hdr3-1/GW6a-OE-1, and the corresponding control. Student’s t test was used to generate the P-values in (C) and (D).
The HDR3–GW6a regulatory module alters gene transcription and increases the local acetylation of histone H4 and H3
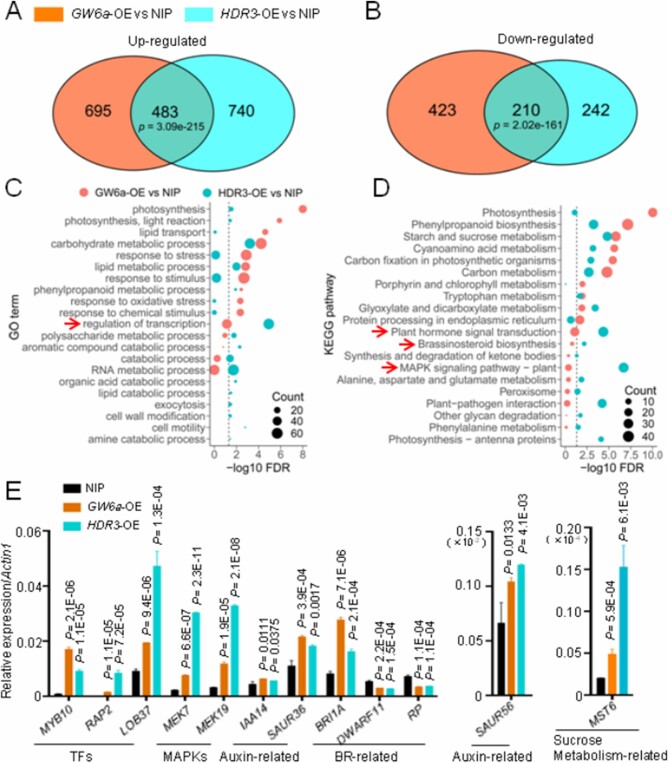
GW6a might function as a transcription regulator (Song et al., 2015). The HDR3–GW6a interaction suggests that this pathway modulates gene transcription. Thus, we compared the transcriptome of the transgenic GW6a-OE and HDR3-OE young panicles to that of the wild-type. The vast majority of genes were upregulated in both GW6a-OE and HDR3-OE panicles (Figure 7, A and B). Notably, 39.5% (483 out of 1,223) of upregulated and 45.5% (210 out of 452) of downregulated genes in HDR3-OE overlapped with those of in GW6a-OE (Figure 7, A and B). Importantly, several gene ontology (GO) terms and kyoto encyclopedia of genes and genomes (KEGG) pathways of the encoded products of the co-regulated genes, such as regulation of transcription, plant hormone signal transduction, brassinosteroid biosynthesis, and MAPK signaling pathway were significantly enriched (Figure 7, C and D). We also confirmed the results by performing RT-qPCR analysis, and found that the selected genes in these pathways have significantly altered transcriptional expression accordingly (Figure 7E). Thus, the HDR3–GW6a pathway regulates the transcription of diverse genes.
Figure 7.
HDR3 and GW6a alter the transcriptional expression of a common set of genes. A, B, Venn diagrams showing overlap of genes up- (A) or downregulated (B) by HDR3 and genes up- (A) or downregulated (B) by GW6a. C, GO enrichment analysis of DE genes for each genetype. GO terms in biological processes significantly enriched (FDR < 0.05) in the DE genes and at the bottom of the GO hierarchical graphs are shown. D, KEGG enrichment analysis of DE genes for each genotype. KEGG pathways significantly enriched (FDR < 0.05) in the DE genes are shown. E, RT-qPCR confirmation of several types of overlapped differentially expressed genes in the young panicles of GW6a-OE and HDR3-OE relative to that of control (NIP). Student’s t test was used to generate the P-values.
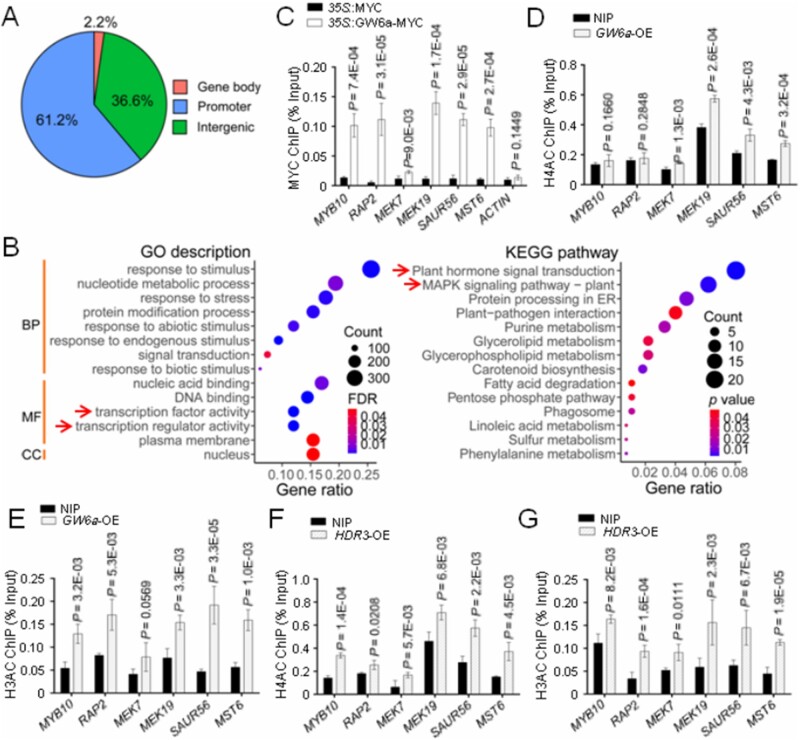
To further investigate how the HDR3–GW6a module alters the expression of the GW6a target genes, we compared chromatin immunoprecipitation sequencing (ChIP-seq) data of rice protoplast cells overexpressing the GW6a-MYC fusion protein with cells overexpressing MYC alone. The ChIP-seq analysis identified 3,336 enriched peaks corresponding to 2,570 genes (a threshold of two-fold enrichment, FDR ˂0.05), the vast majority of which localized at promoter (61.2%) and intergenic (36.6%) regions (Figure 8A). We next defined the genes with MYC enrichment peaks at promoter and/or gene body regions (2,116 peaks, 1,807 genes) as the candidate GW6a target genes. Strikingly, the resulting GO terms and KEGG pathways of the encoded products of these GW6a target genes uncovered several overlapped categories with those of RNA-seq data, such as transcription factor activity, transcription regulator activity, plant hormone signal transduction, and MAPK signaling pathway (Figure 8B). Especially, our chromatin immunoprecipitation quantitative polymerase chain reaction (ChIP-qPCR) results demonstrated that the altered expression of several genes that encode transcription factors (MYB10 and RAP2), MAPK signaling components (MEK7 and MEK19), auxin signaling component (SAUR56), and sucrose metabolism-related regulator (MST6) were most likely due to an elevated binding of GW6a to the corresponding promoter elements (Figures 7, E and 8, C). Consistent with these results, the transgenic young panicles overexpressing GW6a showed an increase of enrichment of H4AC and H3AC at these loci (Figure 8, D and E); similar observations were made in the transgenic young panicles in which HDR3 was significantly overexpressed (Figure 8, F and G).
Figure 8.
HDR3 enhances the local GW6a genomic binding and the corresponding H4AC and H3AC modifications. A, Genomic distribution of GW6a-MYC ChIP-seq peaks. Promoter located at ∼2-kb downstream of the transcriptional starting site (TSS) was analyzed. B, GO and KEGG enrichment analyses for the genes identified to be enriched for GW6a by ChIP-seq. Only the genes that identified to be enriched for GW6a at promoter and gene body regions were used for the analyses. BP, biological process, MF, molecular function, CC, cellular component. C, ChIP-qPCR validation showing fold enrichment of the indicated signal from immuoprecipitation (35S::GW6a-MYC) over the background (35S::MYC) in transformed rice protoplast cells. D, E, ChIP-qPCR analysis showing relative enrichment of the indicated signals in H4AC (D) and H3AC (E) ChIP samples of the GW6a-OE versus the wild-type panicles. F, G, ChIP-qPCR analysis showing relative enrichment of the indicated signals in H4AC (F) and H3AC (G) ChIP samples of the HDR3-OE versus the wild-type panicles. Student’s t test was used to generate the P-values in (B, D, E, F, and G).
Discussion
In this study, we characterized a protein that interacts with GW6a; this protein, HDR3, is homologous to the Arabidopsis DA1. Unexpectedly, we observed that the regulation of grain size by rice HDR3 differs dramatically from that of seed and organ size by Arabidopsis DA1. In Arabidopsis, the R358K mutation in da1-1 increased seed and organ size by elongating the period of cell proliferation in maternal integuments and the mutant protein exerts a negative effect on the function of DA1 and DAR proteins in a dose-dependent manner (Li et al., 2008); at the same time, disruption of the DA1 gene with a T-DNA insertion (loss-of-function da1 mutation) caused increased seed size (Dong et al., 2017), suggesting that DA1 acts as a negative regulator of the seed size trait in Arabidopsis. In contrast, we observed that transgenic rice plants overexpressing HDR3 produced enlarged grains, whereas knocking out the HDR3 gene by the CRISPR/Cas9 method produced significantly smaller grains (Figure 2), supporting the idea that HDR3 positively regulates grain size in rice.
Arabidopsis DA1 controls seed and organ size and has been proposed to be involved in ubiquitin-mediated protein degradation (Li et al., 2008; Xia et al., 2013). Further experimental evidence suggested that DA1 acts as a peptidase whose activity could be activated by its ubiquitylation by DA2 and BB, which are then cleaved by the activated DA1 and decreased stability; in addition to BB and D2, the activated DA1 could cleave and destabilize diverse growth regulatory proteins, including UBP15, the E3 ligase PROTEOLYSIS 1, and the transcription factors TCP15 and TCP22 (Dong et al., 2017). In line with these observations, DA1 could interact with and modulate the stability of UBP15 by the 26S proteasome (Du et al., 2014). Notably, in this study, we found that, among all of the four rice homologs of DA1 (HDRs), GW6a specifically interacts with HDR3 (Figure 1; Supplemental Figures 1 and 2). Surprisingly, our observations suggested that HDR3 functions to stabilize the protein levels of GW6a in an ubiquitin-dependent manner (Figure 5; Supplemental Figures S7 and S8). We therefore uncovered the new HDR3–GW6a regulatory module regulating rice grain size.
The UIM domains have dual roles of binding ubiquitin and promoting ubiquitylation of UIM-containing proteins (Miller et al., 2004). In this study, our in vitro ubiquitin binding assays indicate that the UIM-containing protein HDR3 is indeed a functional ubiquitin receptor (Figure 4, A–D). Furthermore, transient simultaneous expression of GW6a-MYC and HDR3-GFP in rice protoplasts enhanced protein ubiquitylation compared with expressing GW6a-MYC alone (Supplemental Figure S10A). In contrast, the enhancement of ubiquitylation was abolished when expressing GW6a-MYC and HDR3△UIMs-GFP in rice protoplasts (Supplemental Figure S10B), implying that the functional ubiquitin-binding activity of HDR3 is necessary for enhanced protein ubiquitylation. Importantly, the HDR3 upregulation in rice plants results in the enhanced protein ubiquitylation of GW6a (Figure 4, E and F). In contrast to those seen in the case of DA1, the increased GW6a ubiquitylation by HDR3 facilitates the delay of GW6a degradation by the 26S proteasome. Thus, we have revealed a new molecular mechanism where the nonproteolysis function of the UIM-containing protein HDR3 controls seed size.
In the last step of ubiquitylation, the 76-residue protein ubiquitin is catalyzed by an E3 ligase to form an isopeptide bond between its C-terminal glycine residue and a lysine of the target protein (Pickart and Fushman, 2004). Two well-described structurally distinct polyubiquitin chains were generally regarded as functionally different intracellular signals: a single lysine 48 (K48)-linked polyubiquitin chain is sufficient to target a substrate to the 26S proteasome (Chau et al., 1989) and canonical K48-linked chains usually signal proteasome proteolysis; in contrast, K63-linked chains act as nonproteolytic signals in several intracellular pathway, including DNA damage tolerance, the inflammatory response, protein trafficking, and ribosomal protein synthesis (Spence et al., 2000; Ulrich, 2002; Hicke and Dunn, 2003; Sun and Chen, 2004). Notably, our immunoblotting analysis revealed that, relative to those of the nontransgenic control, the levels of K63-linked polyubiquitin chains in the total protein extractions of HDR3-OE young panicles were visibly enhanced, with K48-linked chains unchanged, suggesting that the overexpression of HDR3 has increased its specific binding of K63-linked ubiquitin chains (Supplemental Figure S10C). The zinc finger domain-containing proteins TAB2 and TAB3 bind specific to K63-linked polyubiquitin chains and thereby activate the protein kinase TAK1 and IKK signaling pathway (Kanayama et al., 2004). We assumed that the UIM-type ubiquitin-binding domains may also represent a new class of signaling domain, which deserves future detailed experimental investigations. Based on our experimental data, we summarized a model to explain the potential grain size regulation mechanism by the HDR3–GW6a module in rice (Figure 9).
Figure 9.
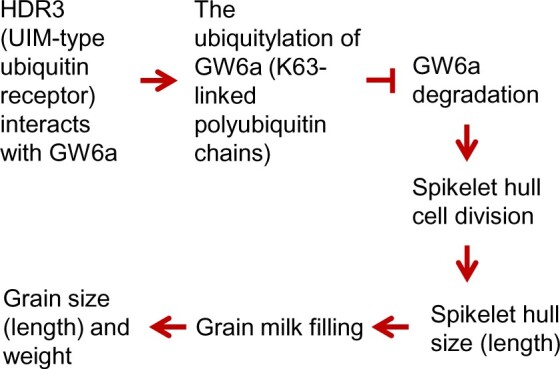
A proposed model for the role of the HDR3–GW6a module’s regulation of grain size (length) and weight in rice. HDR3 acts as an active ubiquitin receptor of UIM type and interacts with the positive grain-size regulator GW6a, leading to the enhanced GW6a ubiquitylation (K63-linked polyubiquitin chains) and delay of its degradation by the 26S proteasome, thereby boosting cell division and spikelet hull size, subsequently increasing milk filling rate and, ultimately, grain size (length) and weight
In addition, it was worth noting that although transiently simultaneous expression in rice protoplasts of a combination of GW6a-MYC, HDR3-GFP, and GW2-GFP showed dramatically stronger protein ubiquitylation than that of GW6a-MYC and GW2-GFP, we did not observe simultaneously a dramatically decreased level of GW6a (Supplemental Figures S8C, S8D, and S10A). This was reasonable given that both GW6a and HDR3 positively regulate grain length (Song et al., 2015), and HDR3 increases GW6a ubiquitylation and protein stability, whereas GW2 is a negative modulator of grain width and length and was thought to function by targeting its substrates to the 26S proteasome for proteolysis (Song et al., 2007). The fact that GW6a suffers the ubiquitin-dependent degradation through the 26S proteasome (Supplemental Figures S7 and S8), hinting that GW2 ubiquitylates and boosts its degradation. In line with this observation, we found that GW2 can interact with GW6a in pull-down assays (Supplemental Figure S11A). We next performed an in vitro E3 ubiquitin ligase activity assay and observed that GW2 can ubiquitylate GW6a revealed by immunoblotting using both anti-GW6a and anti-ubiquitin antibodies (Supplemental Figure S11B). Furthermore, results from protein degradation assays in rice protoplast cells suggested that the degradation of GW6a was promoted by GW2 action; in contrast, the deletion of the RING domain of GW2 (GW2ΔRING) dramatically reduced its promotion effect on GW6a (Supplemental Figure S11, C and D). In this scenario, we assumed that there should be existed another unknown E3 ubiquitin ligase that functions associated with HDR3 to synergistically increase the protein ubiquitylation and entire amount of GW6a in the regulation of grain size.
Materials and methods
Plant materials and growth conditions
The rice (O. sativa) japonica cultivar “Nipponbare” was used for transgenic experiments unless indicated otherwise. The transgenic overexpression lines of GW6a (GW6a-OE, also called OsglHAT1N-OE) were described previously (Song et al., 2015). For each transgenic construct, more than 10 independent positive lines (T0) were produced, and the representative plants of T3 generation showing the typical phenotype were randomly chosen for trait evaluation. The GW6a-OE and HDR3-OE young panicles for transcriptome analyses were harvested from the corresponding plants that were cultivated in greenhouse (12-h light/12-h dark at 30/24�C, light intensity of 30,000 Lux).
Sequence alignment, homology analysis, and conserved domain analysis
The HDR3 homologs were searched against the RAP-DB database using BLAST program (https://rapdb.dna.affrc.go.jp/tools/blast) and the The Arabidopsis Information Resource (TAIR) database (https://www.arabidopsis.org/). Amino acid alignments and the homology analyses of rice and Arabidopsis HDR3 homologs were performed using DNAMAN version of 5.0 (LynnonBiosoft, Canada) based on observed divergency. Protein domains were searched against National Center for Biotechnology Information (NCBI) Conserved Domain Search (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) and SMART programs (http://smart.embl.de/smart/set_mode.cgi?NORMAL=1).
Y2H assays
The Y2H cDNA screening library was constructed using young panicles of Nipponbare (Clontech). The indicated GW6a and HDR3 segments were amplified and cloned into the EcoRI and BamHI restriction sites of the pGBKT7 vector to produce BD-GW6a, BD-GW6a-N, BD-GW6a-C, and BD-HDR3 constructs. Meanwhile, the indicated HDR3 and GW2 fragments were inserted into the EcoRI and BamHI site of pGADT7 to produce the AD-GW2, AD-HDR3, AD-HDR3.1, AD-HDR6, AD-HDR12, AD-HDR3-N, AD-HDR3-C, AD-HDR3-N1, AD-HDR3-N2, AD-HDR3-N3, AD-HDR3-N4, AD-HDR3-N5, and AD-HDR3-N6 constructs. The bait and prey constructs were co-transformed into the yeast cells (strain: Y2HGOLD). Co-transformed yeast clones were serially diluted (1:10) and spotted on the indicated drop-out medium for growth. Primer pairs are listed in Supplemental Table S2.
Co-IP assays
The GW6a and HDR3 coding sequences were cloned into the HindIII and SpeI sites of pSuper1300-GFP and pSuper1300-MYC, respectively, to produce the fusion proteins GW6a-GFP and HDR3-MYC. Each of the fusion constructs was transformed into Agrobacterium EHA105 cells and co-infiltrated into N. benthamiana leaves. After 48–72 h cultivation, these leaves were harvested and ground in liquid nitrogen. Total protein was extracted with extraction buffer (50-mM Tris–MES [pH 8.0], 10-mM EDTA [pH 8.0], 0.5-M Sucrose, 1-mM MgCl2, 1-mM DTT, 1-mM Phenylmethanesulfonyl fluoride (PMSF), 1�Complete protease inhibitor cocktail [Roche]) and incubated with GFP-Trap-A (SA070005, SMART, LIFESCIENCE) for 1–2 h at 4�C. Beads were washed three times with washing buffer (10-mM Tris/HCl [pH 7.5], 0.5-mM EDTA [pH 8.0], 150-mM NaCl, 0.05% SDS, 1% Triton X-100, 1-mM DTT, 1-mM PMSF, 1�Complete protease inhibitor cocktail [Roche]). The immunoprecipitates were separated in 10% sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS–PAGE) and detected by immunoblotting with anti-GFP and anti-MYC antibodies. Primer pairs are listed in Supplemental Table S2.
To verify that HDR3 functions to enhance the GW6a ubiquitylation, 2-week-old seedlings of two independent HDR3-OE plants (-1 and -2) and the wild-type control were harvested and the total proteins of which were extracted. These total proteins were then incubated with anti-Ub-antibody-binding rProtein A/G MagPoly Beads (SM01205, SMART LIFESCIENCES) at 4�C for 2–4 h. The immunoprecipitates were detected by immunoblotting using anti-Ub, anti-GW6a, and anti-HSP90 antibodies.
Split firefly luciferase complementation assays
The coding sequences of GW6a and HDR3 were cloned into the KpnI and SalI sites of pCAMBIA1300-nLUC and pCAMBIA1300-cLUC, respectively, to generate the fusion constructs nLUC-GW6a and cLUC-HDR3. The recombinant constructs were transformed into Agrobacterium EHA105 cells and co-infiltrated into N. benthamiana leaves. The activated luciferase reconstitution signals were imaged using a plant imaging system (Tanon 5200). Primer pairs are listed in Supplemental Table S2.
Pull-down assays
The coding sequences of GW6a and GW2 were cloned into the BamHI and HindIII sites of pET-32a to generate HIS-GW6a and HIS-GW2. The coding sequence of GW6a was also cloned into the BamHI and SalI sites of pGEX4T1 to produce GST-GW6a, and those of HDR3 and HDR3△UIMs were cloned into the SmaI and BamHI sites of pMAL-c2x to generate MBP-HDR3-MYC and MBP-HDR3△UIMs-MYC. The constructs were transformed into Escherichia coli BL21 (DE3), and the recombinant proteins were induced by 1.0-mM IPTG (isopropyl-β-d-thiogalactopyranoside). The indicated fusion proteins were incubated with HIS-Ub at 4�C for 2–3 h. The following eluted proteins were separated by 10% SDS–PAGE and subjected to immunoblotting with anti-HIS, anti-MYC, anti-GST, and anti-Ub antibodies. Primer pairs are listed in Supplemental Table S2.
Transgenic constructs and transformation
We used the vector called AHLG (derived from that of the pBI101-Hm2 vector) for the HDR3-OE constructs, in which a rice ACTIN promoter was used (Ohta et al., 1990; Yamamoto et al., 2007). The rice ACTIN promoter has been described to control rice grain size as shown before (Fang et al., 2016; Xu et al., 2018). The coding sequence of HDR3 was cloned into the ApaI and XbaI sites of AHLG to generate the pActin::HDR3-GFP (HDR3-OE) construct. The HDR3-CR lines were created using the CRISPR/Cas9 system (Ma et al., 2015). Briefly, sgRNA fragment (primers 5′-GCCGGGACGGCGAGGCCAACCGA-3′ and 5′-AAACTCGGTTGGCCTCGCCGTCC-3′ were used) were cloned into the BbsI site of the sgRNA plasmid. The tandem sgRNA cassette was inserted into the pYLCRISPR/Cas9Pubi-H plasmid vector. Transgenic plants were achieved using an Agrobacterium tumefaciens-mediated method. We evaluated the CRISPR/Cas9 mutants using extraction of genomic DNA of the transgenic seedling plants and PCR amplification with a pair of gene-specific primers (5′-GAAGTGTTAGGTGGGATGTTGCTC-3′, and 5′-GTTGCATCAATGGTGCTTAGTG-3′). The amplicons were subjected to DNA sequencing.
Grain-size trait evaluation
Grain size traits described in this study including grain length, width, and thickness were measured using a seed counting and weighing device SC-G (Wanshen, China), and 1,000-grain weight were measured using an electronic analytical balance with three replicates and each replicate contains 10 grains for grain length, width, and thickness measurement, 100 grains for grain weight measurement.
Histological analysis
For cytological observation of cell size of spikelet hulls, the central parts of lemmas of mature seeds were treated and observed using a scanning electron microscope (S-4800; Hitachi). The longitudinal cell length and cell width were analyzed using Image J software. Cell number was determined by grain length and the average cell length.
E3 ubiquitin ligase activity assay
The coding sequences of HDR3 and GW2 were cloned, respectively, into the BamHI and HindIII sites of pET-32a and the BamHI and SalI sites of pGEX4T1 to produce the fusion proteins HIS-HDR3 and GST-GW2. The constructs were transformed into E. coli BL21 (DE3), and the recombinant proteins were induced by 1.0-mM IPTG (isopropyl-β-d-thiogalactopyranoside). Assays for in vitro ubiquitylation were performed as described previously (Song et al., 2007), with some modifications. In brief, 100-ng E1 (BB-E-304-050, Boston Biochem), 200-ng E2 (BB-E2-616-100, Boston Biochem), 5-μg of HIS-ubiquitin (HIS-Ub), and 2-μg purified GST-GW2 and HIS-HDR3, HIS-GW2 and HIS-GW6a fusion proteins were incubated in a 20-μL reaction mix (50-mM Tris–HCl [pH 7.5], 2-mM ATP, 5-mM MgCl2, and 2-mM DTT) for 1 h at 30�C. Polyubiquitinated proteins were detected by immunoblotting with anti-HIS, anti-GST, anti-GW6a, and anti-Ub antibodies. Primer pairs are listed in Supplemental Table S2.
Protoplast isolation and transformation
Rice seeds were surface-sterilized using 70% ethanol and 25% NaClO, and after washing several times were grown in 1/2 Murashige & Skoog (MS) medium containing 0.8% agar at 30�C for 1 week (in the dark). The up-ground tissues of 1-week-old rice seedling were used for protoplast isolation with an enzymatic digestion solution (1.5% cellulose R-10, 0.75% Macerozyme R-10, 0.4-M mannitol, 20-mM MES [pH 5.7], 10-mM CaCl2, 0.1% BSA). The filtrated protoplasts were then washed with W5 solution (154-mM NaCl, 125-mM CaCl2, 5-mM KCl, 2-mM MES [pH 5.7]) and suspended in MMG solution (0.4-M mannitol, 15-mM MgCl2, 4-mM MES [pH 5.7]). Equal volume of 40% PEG solution (40% [w/v] PEG 4000, 0.1-M CaCl2, 0.2-M mannitol) was added to the prepared protoplast cells that contain plasmid DNAs. Finally, the transformed protoplasts were washed with W5 solution and suspended in W1 solution (0.5-M mannitol, 20-mM KCl, 4-mM MES [pH 5.7]) and transferred into a multiwell plate for overnight culture in the dark.
Protein degradation assay
The coding sequences of GW6a, GW2, GW2△RING, HDR3, and HDR3△UIMs were cloned into the HindIII and SpeI sites of pSuper1300-MYC and pSuper1300-GFP to produce the fusion proteins GW6a-MYC, GW2-GFP, GW2△RING-GFP, HDR3-GFP, and HDR3△UIMs-GFP. The corresponding recombinant plasmids were transformed in rice protoplasts and cultured overnight at room temperature, and total proteins were extracted from degradation buffer (25-mM Tris–HCl [pH 7.5], 10-mM NaCl, 10-mM MgCl2, 5-mM DTT, 4-mM PMSF, and 2-mM ATP) with or without MG132 (50 μM). Total proteins were then analyzed by SDS–PAGE and detected by immunoblotting with anti-GW6a and anti-HSP90 antibodies. Image J software was used for the quantitative analysis of relative protein levels. Primer pairs are listed in Supplemental Table S2.
Nuclear/cytoplasmic fractionation
Rice protoplasts transformed with plasmids were cultured overnight and re-suspended in 0.5 mL of Honda buffer (2.5% Ficoll 400, 5% Dextron T40, 0.4-M Sucrose, 25-mM Tris–HCl [pH 7.4], 100-mM MgCl2, 5-mM PMSF, 5-mM DTT, and 1� Complete protease inhibitor cocktail [Roche]). After incubated on ice for 30 min, the re-suspension was centrifuged at 1,500g for 20 min to obtain the crude nuclear and cytoplasmic fractions. The supernatant that contains cytoplasmic fraction was then centrifuged at 16,000g for 15 min to remove the cellular debris. The pellet that contains nuclear fraction was dissolved in the ChIP extraction buffer (0.25-M Sucrose, 10-mM Tris–HCl [pH 8.0], 10-mM MgCl2, 1% Triton X-100, 0.1-mM PMSF, 1-mM DTT, and 1� Complete protease inhibitor cocktail [Roche]) and was centrifuged at 16,000g at 4�C. Finally, the pellet was re-suspended in 200 μL of nuclear lysis buffer (20% glycerol, 200-mM (NH4)2SO4, 25-mM HEPES [pH 8.0], 5-mM MgCl2, 1-mM PMSF, 3-mM DTT, and 1�Complete protease inhibitor cocktail [Roche]). The samples were analyzed by SDS–PAGE and detected by immunoblotting with anti-H3AC, anti-H4AC, anti-GW6a, anti-GFP, anti-H3, and anti-HSP90 antibodies. Image J software was used for quantitative analysis of protein levels.
Subcellular localization
HDR3 and its derivatives were cloned into the BamHI and SalI sites of pCAMBIA2300-eGFP to generate HDR3-FL-eGFP, HDR3-T1-eGFP, and HDR3-T2-eGFP. Each of the constructs was transformed into Agrobacterium EHA105 cells and co-infiltrated into N. benthamiana leaves. After 48–72 h cultivation, the eGFP fluorescence signal was observed using a laser scan confocal microscopy (Leica TCS SP5). Rice protoplasts transformed with plasmid HDR3-GFP (that described earlier) were cultured overnight and were then used to observe the GFP fluorescence accordingly.
RNA extraction and quantitative reverse transcription PCR (RT-qPCR)
Total RNA was extracted using the OminiPlant RNA Kit (CW2598, CWBIO) and 2 μg of which was used to synthesize the first-strand cDNA using the TIANGEN FastQuart RT Kit. The synthesized cDNAs were then diluted five times with RNase-free water and used for RT-qPCR experiments. Rice ACTIN1 and GAPDH were used to normalize the data and the relative expression levels were calculated using 2–ΔΔCt method. Relevant PCR primer pairs are listed in Supplemental Table S3.
RNA-seq analysis
Young panicles (3–5 cm in length) of Nipponbare (NIP), GW6a-OE and HDR3-OE, with three biological replicates were collected and immediately frozen in liquid nitrogen. Total RNA was extracted with TRIzol (ZP401, ZOMANBIO). The cDNA libraries were constructed following Illumina standard protocols and sequenced on the Illumina NovaSeq 6000 platform with 150-bp paired-ends read at Major Bio (Shanghai, China). RNA-seq reads were trimmed using Trim Galore (https://www.bioinformatics. babraham.ac.uk/projects/trim_galore/) to remove library construction adapter and low-quality ends and then aligned to the rice reference genome (IRGSP-1.0) using TopHat (v2.1.1). Then, the Cufflinks was used to assemble the transcripts and gene transcription levels were calculated as FPKM (fragments per kilobase of exon per million mapped reads) for differential expression analysis using Cuffdiff. A criterion of fold-change ≥ 2 and q-value ≤0.05 was used to screen differential expressed genes. GO and KEGG enrichment analysis were performed upon Agrigo webservice (http://bioinfo.cau.edu.cn/agriGO/) and KOBAS 3.0 webservice (http://kobas.cbi.pku.edu.cn/kobas3/?t=1), respectively.
ChIP-seq analysis
For ChIP-seq assays, rice protoplasts were transformed with 35S::MYC and 35S::GW6a-MYC plasmids and cultured overnight at room temperature. The protoplasts were centrifugated and cross-linked with 1% formaldehyde. Chromatin was isolated using sucrose gradient centrifugation and sonicated using a Bioruptor (Bioruptor Plus, Diagenode). The fragmented chromatin was immunoprecipitated by Anti-c-MYC Affinity Beads (SA065001, SMART, LIFESCIENCE). The immunoprecipitated chromatin was then de-cross-linked and DNA was retrieved for library construction following the method in NEXTflex Rapid DNA-Seq Kit (5144-08, BIOO).
The clean reads of ChIP-seq were mapped to the IRGSP-1.0 rice reference genome using the Bowtie 2 program and the unique map reads were used for peak calling with the Model-based analysis of ChIP-Seq (MACS). The peaks were annotated with the ChIPseeker and the GW6a binding sites were defined using the following criteria: (1) the peak was located within −2 kb to 2 kb from transcriptional starting sites of a given gene and (2) fold enrichment was larger than two-fold. Candidate sites were verified by quantitative PCR. Primer pairs are listed in Supplemental Table S3.
Native ChIP-qPCR assay
Two grams of young panicle was ground in liquid nitrogen. Chromatin was extracted and fragmented via Micrococcal Nuclease (M0247S, NEB). Each of the anti-H3AC and anti-H4AC antibodies was incubated with Protein A/G Magnetic Beads (26162, ThermoFisher) for 3–5 h at 4�C. The fragmented chromatin suspension was then incubated with the antibody-binding beads at 4�C overnight. The immunoprecipitated chromatin was retrieved for ChIP-qPCR. The relevant primer pairs are listed in Supplemental Table S3.
Antibodies used in this study
The information of antibodies used in this study is: anti-MYC (HT101-01, TRANS), anti-ubiquitin (sc-8017, SANTA CRUZ), anti-HIS (HT501-01, TRANS), anti-GST (HT601-01, TRANS), anti-H3AC (06-599, Millipore), anti-H4AC (06-06-866, Millipore), anti-H3 (06-755, Millipore), anti-GFP (ab290, Abcam), anti-HSP90 (AbM51099-31-PU, Huada). The anti-GW6a monoclonal antibody was raised against GW6a.
Accession numbers
Data produced in this study are deposited in NCBI sequence Read Archive under accession number PRJNA694519 (https://www.ncbi.nlm.nih.gov/bioproject/694519).
GW6a (Os06g0650300), HDR3 (Os03g0267800), HDR3.1 (Os03g0626600), HDR6 (Os06g0182500), HDR12 (Os12g0596800), GW2 (Os02g0244100), MYB10 (Os09g0401000), RAP2 (Os08g0474000), LOB37 (Os07g0589000), MEK7 (Os01g0699600), MEK19 (Os05g0545400), IAA14 (Os03g0797800), SAUR36 (Os08g0550700), BRI1A (Os06g0274300), DWARF11 (Os04g0469800), RP (Os07g0136800), SAUR56 (Os10g0510500), MST6 (Os07g0559700).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. GW6a interacts with HDR3, but not with its other rice homologs, in Y2H assays (Supports Figure 1).
Supplemental Figure S2. The carboxyl terminus of GW6a interacts with the amino terminus of HDR3 in yeast cells (Supports Figure 1).
Supplemental Figure S3. Sequence alignments and a homology tree of the rice and known Arabidopsis HDR3 homologs (Supports Figure 1).
Supplemental Figure S4. Molecular evidences for the transgenic plants with overexpression and gene edition of HDR3 (Supports Figure 2).
Supplemental Figure S5. The transcriptional expression of HDR3 and the subcellular distribution of its encoded product (Supports Figure 2).
Supplemental Figure S6. GW2 interacts with and ubiquitylates HDR3 in vitro (Supports Figure 4).
Supplemental Figure S7. GW6a suffers ubiquitin-dependent degradation through the 26S proteasome (Supports Figure 5).
Supplemental Figure S8. HDR3, but not GW2, could stabilize GW6a and enhance its activity in a ubiquitin-dependent manner (Supports Figure 5).
Supplemental Figure S9. HDR3 and GW6a alter the transcript abundance of a common set of genes (Supports Figure 7).
Supplemental Figure S10. UIMs of HDR3 is essential for its effects on the boost of protein ubiquitination (Supports Figure 5).
Supplemental Figure S11. GW2 interacts with, ubiquitylates and destabilizes the protein level of GW6a (Supports Figure 5).
Supplemental Table S1. A total of four rice homologs of Arabidopsis DA1 were identified.
Supplemental Table S2. Vectors and primers used for construction.
Supplemental Table S3. Primer pairs used in this study for RT-qPCR.
Supplemental Data Set S1. List of differentially expressed genes identified from comparisons between GW6a-OE/HDR3-OE and the corresponding control (Nipponbare) in young panicles.
Supplemental Data Set S2. GW6a-regulated gene targets enriched in GW6a-MYC ChIP-seq.
Supplemental File S1. Multiple sequence alignment supports Supplemental Figure S3.
Supplementary Material
Acknowledgments
We thank Prof. C.M. Liu in Institute of Botany for sharing the plasmid vector AHLG, and all projects for generating and sharing the data used in this paper.
Funding
This work was supported by grants from the Chinese Academy of Sciences (XDA24010101-2), the National Key Research and Development Program of China (2016YFD0100402), and the National Natural Science Foundation of China (91735302, 91435113, and 31471466).
Conflict of interest statement. The authors declare that they have no competing interests.
X.-J.S. designed the experiments. Q.G. and N.Z. performed most of the experiments; W.-Q.W. performed ChIP-seq and RNA-seq analyses. S.-Y.S. and C.B. provided some technical support for the experiments. X.-J.S. wrote the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) is: Xian-Jun Song (songxj@ibcas.ac.cn).
References
- Bonifacino JS, Traub LM (2003) Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu Rev Biochem 72:395–447 [DOI] [PubMed] [Google Scholar]
- Chau V, Tobias JW, Bachmair A, Marriott D, Ecker DJ, Gonda DK, Varshavsky A (1989) A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science 243:1576–1583 [DOI] [PubMed] [Google Scholar]
- Che RH, Tong HN, Shi BH, Liu YQ, Fang SR, Liu DP, Xiao YH, Hu B, Liu LC, Wang HR, et al. (2016). Control of grain size and rice yield by GL2-mediated brassinosteroid response. Nat Plant 2:15195–15201 [DOI] [PubMed] [Google Scholar]
- Dikic I, Wakatsuki S, Walters KJ (2009) Ubiquitin-binding domains—from structures to functions. Nat Rev Mol Cell Biol 10:659–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disch S, Anastasiou E, Sharma VK, Laux T, Fletcher JC, Lenhard M (2006) The E3 ubiquitin ligase BIG BROTHER controls Arabidopsis organ size in a dosage-dependent manner. Curr Biol 16:272–279 [DOI] [PubMed] [Google Scholar]
- Dong H, Dumenil J, Lu FH, Li N, Vanhaeren H, Naumann C, Klecker M, Prior R, Smith C, McKenzie N, et al. (2017). Ubiquitylation activates a peptidase that promotes cleavage and destabilization of its activating E3 ligases and diverse growth regulatory proteins to limit cell proliferaton in Arabidopsis. Genes Dev 31:197–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L, Li N, Chen LL, Xu YX, Li Y, Zhang YY, Li CY, Li YH (2014) The ubiquitin receptor DA1 regulates seed and organ size by modulating the stability of the ubiquitin-specific protease UBP15/SOD2 in Arabidopsis. Plant Cell 26:665–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan PG, Xu JS, Zeng DL, Zhang BL, Geng MF, Zhang GZ, Huang K, Huang LJ, Xu R, Ge S, et al. (2017) Natural variation in the promoter of GSE5 contributes to grain size diversity in rice. Mol Plant 10:658–694 [DOI] [PubMed] [Google Scholar]
- Fang N, Xu R, Huang LJ, Zhang BL, Duan PG, Li N, Luo YH, Li YH (2016) SMALL GRAIN 11 controls grain size, grain number and grain yield in rice. Rice 9: 64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsasser S, Finley D (2005) Delivery of ubiquitinated substrates to protein-unfolding machines. Nat Cell Biol 7:742–749 [DOI] [PubMed] [Google Scholar]
- Hicke L, Dunn R (2003) Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu Rev Cell Dev Biol 19:141–172 [DOI] [PubMed] [Google Scholar]
- Hofmann K, Falquet L (2001) A ubiquitin-interacting motif conserved in components of the proteasomal and lysosomal protein degradation systems. Trends Biochem Sci 26:347–350 [DOI] [PubMed] [Google Scholar]
- Hu J, Wang YX, Fang YX, Zeng LJ, Xu J, Yu HP, Shi ZY, Pan JJ, Zhang D, Kang SJ, et al. (2015). A rare allele of GS2 enhances grain size and grain yield in rice. Mol Plant 8:1455–1465 [DOI] [PubMed] [Google Scholar]
- Hu ZJ, Lu SJ, Wang MJ, He HH, Sun L, Wang HR, Liu XH, Jiang L, Sun JL, Xin XY, et al. (2018). A novel QTL qTGW3 encodes the GSK3/SHAGGY-like kinase OsGSK5/OsSK41 that interacts with OsARF4 to negatively regulate grain size and weight in rice. Mol Plant 11:736–749 [DOI] [PubMed] [Google Scholar]
- Huang K, Wang DK, Duan PG, Zhang BL, Xu R, Li N, Li YH (2017) WILD AND THICK GRAIN 1, which encodes an otubain-like protease with deubiquitination activity, influence grain size and shape in rice. Plant J 91:849–860 [DOI] [PubMed] [Google Scholar]
- Ishimaru K, Hirotsu N, Madoka Y, Murakami N, Hara N, Onodera H, Kashiwagi T, Ujiie K, Shimizu B, Onishi A, et al. (2013). Loss of function of the IAA-glucose hydrolase gene TGW6 enhances rice grain weight and increases yield. Nat Genet 45:707–711 [DOI] [PubMed] [Google Scholar]
- Kanayama A, Seth RB, Sun LJ, Ea C, Hong M, Shaito A, Chiu Y, Deng L, Chen Z (2004) TAB2 and TAB3 activate the NF-κB pathway through binding to polyubiquitin chains. Mol Cell 15:535–548 [DOI] [PubMed] [Google Scholar]
- Li N, Xu R, Li Y (2019) Molecular networks of seed size control in plants. Annu Rev Plant Biol 70:435–463 [DOI] [PubMed] [Google Scholar]
- Li YH, Zheng LY, Corke F, Smith C, Bevan MW (2008). Control of final seed and organ size by the DA1 gene family in Arabidopsis thaliana. Genes Dev 22:1331–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YB, Fan CC, Xing YZ, Jiang YH, Luo LJ, Sun L, Shao D, Xu CJ, Li XH, Xiao JH, et al. (2011). Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat. Genet. 43:1266–1269 [DOI] [PubMed] [Google Scholar]
- Liu JF, Chen J, Zheng XM, Wu FQ, Lin QB, Heng YQ, Tian P, Cheng ZJ, Yu XW, Zhou KN, et al. (2017). GW5 acts in the brassinosteroid signalling pathway to regulate grain width and weight in rice. Nat Plant 3:17043–17049 [DOI] [PubMed] [Google Scholar]
- Ma XL, Zhang QY, Zhu QL, Liu W, Chen Y, Qiu R, Wang B, Yang ZF, Li HY, Lin YR, et al. (2015). A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant 8:1274–1284 [DOI] [PubMed] [Google Scholar]
- Miller SL, Malotky E, O’Bryan JP (2004) Analysis of the role of ubiquitin-interacting motifs (UIMs) in ubiquitin binding and ubiquitylation. J Biol Chem 279:33528–33537 [DOI] [PubMed] [Google Scholar]
- Ohta N, Masurekar M, Newton A (1990) Cloning and cell cycle-dependent expression of DNA replication gene dnaC from Caulobacter crescentus. J Bacteriol 172: 7027–7034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng YC, Chen LL, Lu YR, Wu YB, Dumenil J, Zhu ZG, Bevan MW, Li YH (2015) The ubiquitin receptors DA1, DAR1, and DAR2 redundantly regulate endoreduplication by modulating the stability of TCP14/15 in Arabidopsis. Plant Cell 27:649–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart CM, Fushman D. (2004). Polyubiquitin chains: polymeric protein signals. Curr Opin Chem Biol 8:610–616 [DOI] [PubMed] [Google Scholar]
- Qi P, Lin YS, Song XJ, Shen JB, Huang W, Shan JX, Zhu MZ, Jiang LW, Gao JP, Lin HX (2012) The novel quantitative trait locus GL3.1 controls rice grain size and yield by regulating cyclin-T1;3. Cell Res 22:1666–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi CL, Ren YL, Liu LL, Wang F, Zhang H, Tian P, Pan T, Wang YF, Jing RN, Liu TZ, et al. (2019). Ubiquitin specific protease 15 has an important role in regulating grain width and size in rice. Plant Physiol 180:381–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si LZ, Chen JY, Huang XH, Gong H, Luo JH, Hou QQ, Zhou TY, Lu TT, Zhu JJ, Shangguan YY, et al. (2016). OsSPL13 controls grain size in cultivated rice. Nat Genet 48:447–456 [DOI] [PubMed] [Google Scholar]
- Song XJ, Huang W, Shi M, Zhu MZ, Lin HX (2007) A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat Genet 39:623–630 [DOI] [PubMed] [Google Scholar]
- Song XJ, Kuroha T, Ayano M, Furuta T, Nagai K, Komeda N, Segami S, Miura K, Ogawa D, Kamura T, et al. (2015). Rare allele of a previously unidentified histone H4 acetyltransferase enhances grain weight, yield, and plant biomass in rice. Proc Natl Acad Sci USA 112:76–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence J, Gali RR, Dittmar G, Sherman F, Karin M, Finley D (2000) Cell cycle-regulated modification of the ribosome by a variant multiubiquitin chain. Cell 102:67–76 [DOI] [PubMed] [Google Scholar]
- Sun L, Chen ZJ (2004) The novel functions of ubiquitination in signaling. Curr Opin Cell Biol 16:119–126 [DOI] [PubMed] [Google Scholar]
- Ulrich HD (2002) Degradation or maintenance: actions of the ubiquitin system on eukaryotic chromatin. Eukaryot Cell 1:1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R, Oania R, Graumann J, Deshaies RJ (2004) Multiubiquitin chain receptors define a layer of substrate selectivity in the ubiquitin-proteasome system. Cell 118:99–110 [DOI] [PubMed] [Google Scholar]
- Wang SK, Wu K, Yuan QB, Liu XY, Liu ZB, Lin XY, Zeng RZ, Zhu HT, Dong GJ, Qian Q, et al. (2012). Control of grain size, shape and quality by OsSPL16 in rice. Nat Genet 44:950–954 [DOI] [PubMed] [Google Scholar]
- Xia D, Zhou H, Liu RJ, Dan WH, Li PB, Wu B, Chen JX, Wang LQ, Gao GJ, Zhang QL, et al. (2018). GL3.3, a novel QTL encoding a GSK3/SHAGGY-like Kinase, epistatically interacts with GS3 to produce extra-long grains. Mol Plant 11:754–756 [DOI] [PubMed] [Google Scholar]
- Xia T, Li N, Dumenil J, Li J, Kamenski A, Benvan MW, Gao F, Li YH (2013) The ubiquitin receptor DA1 interacts with the E3 ubiquitin ligase DA2 to regulate seed and organ size in Arabidopsis. Plant Cell 25:3347–3359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R, Duan PG, Yu HY, Zhou ZK, Zhang BL, Wang RC, Li J, Zhang GZ, Zhuang SS, Lyn J, et al. (2018) Control of grain size and weight by the OsMKKK10-OsMKK4-OsMAPK6 signaling pathway in rice. Mol Plant 11: 860–873 [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Kamiya N, Morinaka Y, Matsuoka M, Sazuka T (2007) Auxin biosynthesis by the YUCCA genes in rice. Plant Physiol 143: 1362–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying JZ, Ma M, Bai C, Huang XH, Liu JL, Song XJ (2018) TGW3, a major QTL that negatively modulates grain length and weight in rice. Mol Plant 11:750–753 [DOI] [PubMed] [Google Scholar]
- Zuo J, Li J (2014) Molecular genetic dissection of quantitative trait loci regulating rice grain size. Annu Rev Genet 48:99–118 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.