Abstract
Folates are indispensable for plant development, but their molecular mode of action remains elusive. We synthesized a probe, “5-F-THF-Dayne,” comprising 5-formyl-tetrahydrofolate (THF) coupled to a photoaffinity tag. Exploiting this probe in an affinity proteomics study in Arabidopsis thaliana, we retrieved 51 hits. Thirty interactions were independently validated with in vitro expressed proteins to bind 5-F-THF with high or low affinity. Interestingly, the interactors reveal associations beyond one-carbon metabolism, covering also connections to nitrogen (N) metabolism, carbohydrate metabolism/photosynthesis, and proteostasis. Two of the interactions, one with the folate biosynthetic enzyme DIHYDROFOLATE REDUCTASE-THYMIDYLATE SYNTHASE 1 (AtDHFR-TS1) and another with N metabolism-associated glutamine synthetase 1;4 (AtGLN1;4), were further characterized. In silico and experimental analyses revealed G35/K36 and E330 as key residues for the binding of 5-F-THF in AtDHFR-TS1 and AtGLN1;4, respectively. Site-directed mutagenesis of AtGLN1;4 E330, which co-localizes with the ATP-binding pocket, abolished 5-F-THF binding as well as AtGLN1;4 activity. Furthermore, 5-F-THF was noted to competitively inhibit the activities of AtDHFR-TS1 and AtGLN1;4. In summary, we demonstrated a regulatory role for 5-F-THF in N metabolism, revealed 5-F-THF-mediated feedback regulation of folate biosynthesis, and identified a total of 14 previously unknown high-affinity binding cellular targets of 5-F-THF. Together, this sets a landmark toward understanding the role of folates in plant development.
Exploration of proteins that interact or bind with folates reveals folate-modulated growth and development in Arabidopsis.
Introduction
Plants produce a variety of endogenous small molecules to modulate their growth and development in concert with internal signals and/or external cues. Among those, vitamins form a diverse group of organic small molecules which, despite being effective at low concentrations, are vital for development. Folates, a collective term for tetrahydrofolate (THF) and its derivatives, also known as vitamin B9, are essential for central metabolism. Structurally, folates are tripartite molecules comprising pteridine, p-aminobenzoate (PABA), and glutamate moieties (Figure 1). THF derivatives contain one-carbon (C1) units at various oxidation states attached to their N5 and/or N10 positions (Blancquaert et al., 2014; Strobbe and Van Der Straeten, 2017). By virtue of this, folates function as donors or acceptors of C1 units in C1 transfer reactions, serving as essential cofactors for enzymes catalyzing synthesis of many vital biomolecules, such as nucleotides (purines and thymidylate), amino acids (methionine, glycine, and serine), and vitamin B5 (pantothenate; Cossins, 2000). Folates are also implicated in synthesis and/or modification of chlorophyll, lignin, lipids, and proteins (Van Wilder et al., 2009; Henkel et al., 2012; Tang et al., 2014; Gorelova et al., 2017a).
Figure 1.
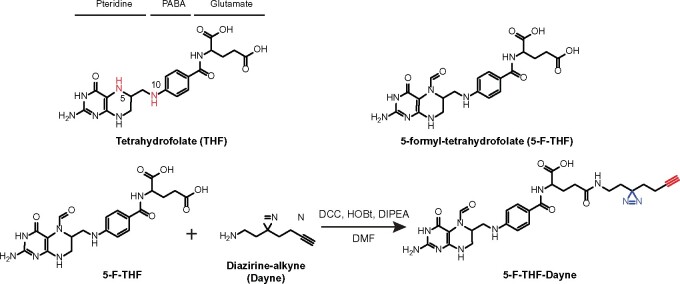
Schematic structure of THF, 5-F-THF, and 5-F-THF-Dayne probe. THF consists of three moieties: pteridine, PABA, and glutamate. The N5 and N10 positions in THF are marked in red. In the 5-F-THF-Dayne probe, a “diazirine-alkyne” (“Dayne”) photoaffinity tag was coupled to the γ-carboxyl group. The diazo part “N=N” is indicated in blue and the alkyne moiety in red. For detailed synthesis procedure see “Methods” section. HOBt, hydroxyl-benzotriazole; DIPEA, N,N-diisopropyl-ethylamine.
In plants, folates have been closely linked with a myriad of physiological processes including photorespiration (Rebeille et al., 1994; Li et al., 2003), plastid biogenesis (Bouvier et al., 2006; Van Wilder et al., 2009), epigenetic regulation of gene expression, and immunity (Zhang et al., 2012; Zhou et al., 2013; Gonzalez and Vera, 2019). Defects in folate biosynthesis and metabolism have been shown to cause multiple aberrations in plant growth and development. For instance, mutation in 5-F-THF cycloligase (5FCL) results in reduced growth and delayed flowering in Arabidopsis thaliana (Goyer et al., 2005). In the gla1 mutant, a knockout for the folate biosynthetic gene DIHYDROFOLATE SYNTHETASE, embryo development arrests at the transition from globular to heart stage (Ishikawa et al., 2003). Further, the double homozygous mutation dhfr-ts1 � dhfr-ts2 (encoding two isoforms of dihydrofolate reductase-thymidylate synthase [TS]) is embryo-lethal (Gorelova et al., 2017b). Mutation in the gene for plastidial FOLYLPOLYGLUTAMATE SYNTHETASE 1 (FPGS1, dfb mutant) leads to altered root development, seedling establishment in darkness, and seed reserve accumulation, including higher nitrogen (N) in seeds (Meng et al., 2014). In addition, dfb displays a shorter hypocotyl under low N supply (Meng et al., 2014). Similarly, fpgs2 exhibits reduced primary root growth and lateral root development under low-N supply, as well as several metabolic changes typical of plant responses to low N stress (Jiang et al., 2013). Mutation of both the FPGSs in the fpgs1�fpgs2 double mutant causes multiple defects, including reduced fertility and embryo or seedling death (Mehrshahi et al., 2010). In maize, the study of bm2 and bm4 demonstrated the roles of two folate biosynthesis enzymes methylenetetrahydrofolate reductase and FPGS in lignin biosynthesis (Tang et al., 2014; Li et al., 2015). Despite the above advances, the molecular mechanism of action of folates remains ill-defined. Furthermore, knowledge regarding the targets of folates, in terms of enzymes and/or other regulatory proteins, remains limited.
Previous studies in mammals suggest that cellular folates do not exist in isolation. Hence, all folate forms are assumed to be bound to folate-binding proteins (FBPs; Luka, 2008). In mammals, FBPs are well investigated and comprised folate receptors, carriers, and transporters, besides the major folate-dependent C1 metabolism enzymes (Luka, 2008; Luka et al., 2011). In contrast, in plants, outside of a few folate-dependent C1 metabolism enzymes (Li et al., 2003), three folate transporters (Hanson and Gregory, 2011), and one recently reported Arabidopsis FBP (Puthusseri et al., 2018a, 2018b), most plant FBPs are yet to be identified. Considering the fundamental roles of folates in living organisms, the number, identity, and functions of FBPs are likely comparable between plants and mammals. Indeed, FBP of bovine origin was successfully expressed to stabilize folates in folate-biofortified rice (Blancquaert et al., 2015).
We reasoned that determining the folate-interacting proteome could serve as the first milestone toward the broader goal of understanding the role of folates in plant growth and development. Chemical strategies have been well established to enrich subsets of the functional proteome and have been empowered with mass spectrometry-based methods to delve deeply and precisely into the biochemical state of organisms and their perturbations by small molecules (Parker and Pratt, 2020; Louis et al., 2021). Here, we utilized a small molecular probe-based affinity proteomics approach to scout the FBPs in A. thaliana. We first coupled 5-formyl-THF (5-F-THF) with diazirine-alkyne (Dayne) to synthesize the novel “5-F-THF-Dayne” probe (Figure 1). Then, after confirming the bioactivity of the probe, it was used as a bait to fish for 5-F-THF-binding proteins (FFBPs) in Arabidopsis. Fifty-one proteins covering candidates with diverse biological functions, including C/N metabolism, photosynthesis, and proteostasis, were identified. Binding of 5-F-THF with two targeted FFBPs, DHFR-TS1, and GLUTAMINE SYNTHETASE 1 (GS1);4 (AtGLN1;4) was studied in detail. The binding sites for 5-F-THF in AtDHFR-TS1 and AtGLN1;4 were determined and the effect of 5-F-THF binding on AtDHFR-TS1 and AtGLN1;4 enzyme activities were revealed. This study significantly expands our knowledge on fundamental folate biology and holds promise to facilitate further research on the action mechanism of folates in plants.
Results
Synthesis of a 5-F-THF-derived photo-affinity probe, 5-F-THF-Dayne
Folic acid and 5-F-THF are the two most stable folate forms (Gorelova et al., 2017a). 5-F-THF is the natural folate derivative in plants with least defined function. In this study, monoglutamated 5-F-THF was coupled to a Dayne photo-affinity tag to generate the 5-F-THF-Dayne probe (Figure 1; for synthesis procedure see “Materials and Methods”). The probe contains three essential structural components: (1) the core scaffold of 5-F-THF as “spearhead”, which determines the binding specificity and directs the probe to the target proteins; (2) a photo-reactive diazarine moiety, which forms a covalent bond with the bound protein under ultraviolet (UV) irradiation; and (3) a small terminal alkyne handle, which can be conjugated to suitable reporters (tetramethylrhodamine [TAMRA]-N3 or biotin-N3) via copper-catalyzed alkyne-azide cycloaddition reaction, allowing subsequent cell- and in gel-based target detection as well as mass spectrometry-based target identification.
In vivo and in vitro validation of the bioactivity of 5-F-THF-Dayne
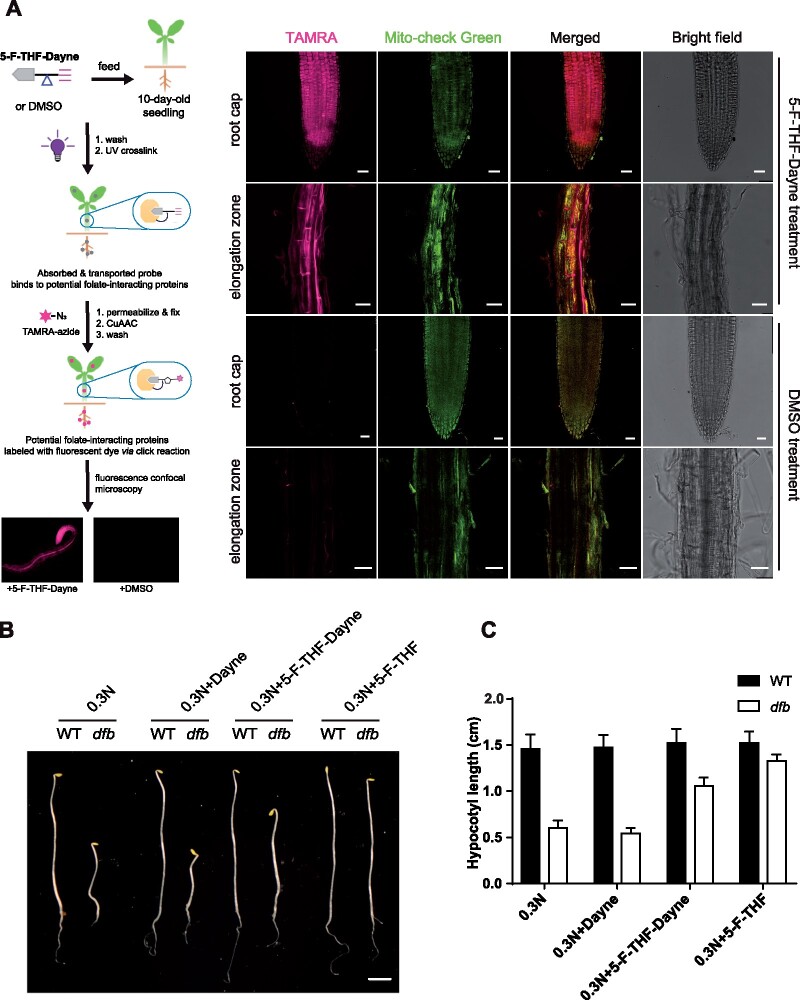
Prior to using the 5-F-THF-Dayne probe in affinity proteomics, we tested whether it retained the biological activity of the 5-F-THF molecule. For this, probe-fed Arabidopsis seedlings were analyzed by in situ fluorescence microscopy (see Supplemental Methods for more details). After 3-h feeding, significant red fluorescence of TAMRA was observed in the optical section of the roots, both in the root cap and elongation zone (Figure 2A). A proportion of these signals also overlapped with the green signals of the mitochondrial marker (Figure 2A). This suggested that Arabidopsis roots can easily uptake 5-F-THF-Dayne and that the probe is distributed across the cell. Signals were also visible in stem tissues (Supplemental Figure S1A), where they were more concentrated in the vasculature (Supplemental Figure S1B). After a feeding time of 6 h, red fluorescence was also observed in leaf scans (Supplemental Figure S1D), indicating active translocation of the probe across the plant body. Next, we checked if 5-F-THF-Dayne is interconverted/metabolized to other folate derivatives. Liquid chromatography- mass spectrometry (LC–MS) analysis with probe-fed seedlings showed peaks representing, besides 5-F-THF-Dayne, 5,10-methenyltetrahydrofolate (5,10-CH=THF)-Dayne and 5-methyl(M)-THF-Dayne (Supplemental Figure S2). This suggested that 5-F-THF-Dayne can be metabolized by plants to other folate derivatives with intact Dayne-tag. Additionally, the mere fact of the conversion of probe (carrying monoglutamated 5-F-THF) to other forms in vivo demonstrates that the metabolic enzymes in the folate biosynthesis pathway do interact with monoglutamated folates and metabolize them. Finally, we performed a pharmacological assay using the Arabidopsis dfb mutant to test whether plants can functionally use 5-F-THF-Dayne. In a previous study, 5-F-THF was shown to rescue the short hypocotyl phenotype of the dfb mutant under low N availability (Meng et al., 2014). In this study, 5-F-THF-Dayne was noted to restore the hypocotyl growth of dfb (Figure 2, B and C). Collectively, these results established that 5-F-THF-Dayne was absorbed, metabolized, and used by Arabidopsis seedlings.
Figure 2.
Confirmation of the in vivo biological activity of the 5-F-THF-Dayne probe. A, In vivo fluorescence imaging of 5-F-THF-Dayne in root tips of 10-day-old Arabidopsis WT seedlings. The stepwise workflow is shown at the left. Briefly, root tips were fed with 5-F-THF-Dayne probe for 3 h, followed by a click reaction with the TAMRA azide fluorophore. The tissue was then analyzed under a confocal microscope. Confocal images at the right show active uptake of the probe by Arabidopsis roots. The red signal of TAMRA (shown in magenta) shows the distribution of the probe in the root cap and elongation zone. Root tips incubated with DMSO as a control showed no fluorescence postclick reaction with TAMRA. Mito-check green, mitochondrion marker. Bar = 25 μm. B and C, Complementation of the dfb mutant showing active utilization of 5-F-THF-Dayne by the mutant seedlings. B, Hypocotyl growth (bar = 0.2 cm) and (C) length of 6-day-old Arabidopsis WT and dfb mutant grown in darkness on low N (0.3 N) medium, supplemented with Dayne, 5-F-THF (50 μM) and 5-F-THF-Dayne (50 μM), respectively.
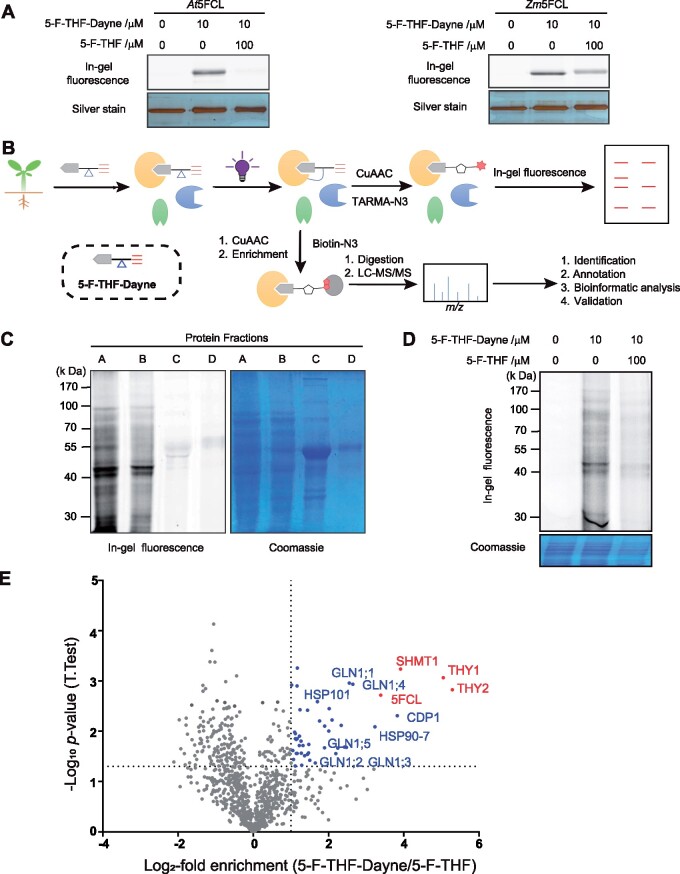
Next, the binding competence of 5-F-THF-Dayne was further verified by assessing its binding with the known 5-F-THF metabolism enzyme 5FCL, using a previously described in-gel fluorescence assay (Li et al., 2018). Upon 5-F-THF-Dayne treatment, a clear fluorescent band was observed, suggesting an effective labeling of the 5FCL protein by 5-F-THF-Dayne (Figure 3A). Alongside, it demonstrated the feasibility of protein labeling in vitro. Incubation with excess 5-F-THF together with the addition of probe (5-F-THF-Dayne) weakened the fluorescence/labeling, especially in the At5FCL sample, confirming the specificity of interaction (Figure 3A).
Figure 3.
Confirmation of in vitro activity of the 5-F-THF-Dayne and chemoproteomics profiling of 5-F-THF-binding proteins in Arabidopsis. A, In vitro interaction of 5-F-THF-Dayne with 5FCL from Arabidopsis (At5FCL) and maize (Zm5FCL) as indicated by in-gel fluorescence imaging. 5-F-THF-Dayne, 5-F-THF probe. B, Workflow for profiling of 5-F-THF-Dyane interacting proteins. The irregular grapes in yellow, blue, and green colors represent extracted proteins. In brief, 5-F-THF-Dayne probe was incubated with the Arabidopsis protein extract; subsequently, the photoreactive diazirine moiety was covalently linked with the interacting proteins after irradiation by UV light. Eventually, the bound proteins were visualized by in-gel fluorescence detection or enriched for LC–MS/MS identification through a copper-catalyzed azide–alkyne cycloaddition reaction with TAMRA-N3 or biotin-N3, respectively. c, In-gel fluorescence labeling of four protein fractions (A, B, C, and D) of the Arabidopsis proteome by 5-F-THF-Dayne (left) and Coomassie staining of the gel (right). D, Competitive labeling of the Arabidopsis proteome by 5-F-THF-Dayne. E, Quantitative mass spectrometry-based profiling of the 5-F-THF-Dayne binding proteins. Blue and red dots depict selected targets that were outcompeted by 5-F-THF (criteria: t-test difference log2-fold enrichment ≥1 and − log10 (P-value) ≥1.33). Red dots represent the identified enzymes involved in C1 metabolism.
These results confirmed the potential of this probe, thus we used it for affinity proteomics, applying the workflow shown in Figure 3B.
Mass spectrometry-based profiling of the folate proteome of Arabidopsis leaves
The four protein fractions (A, B, C, and D; see “Materials and Methods”) obtained from Arabidopsis leaves were incubated with 5-F-THF-Dayne. Fractions A and B showed a strong fluorescent band upon in-gel fluorescence scanning, unlike fractions C and D, which contained the majority of Ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCo; Figure 3C). Hence, fractions A and B were combined and used for the subsequent pull-down steps. To ensure the specificity of probe interaction with cellular proteins, three reactions were set up in parallel (details in “Materials and Methods”): (1) the blank control, protein samples incubated with dimethyl sulfoxide (DMSO); (2) the experimental run, where protein samples were incubated with 5-F-THF-Dayne (10 μM); and (3) competition binding, where protein samples were incubated with both 5-F-THF-Dayne (10 μM) and monoglutamated 5-F-THF (100 μM, 10 folds higher than the probe). The reaction complex was analyzed by in-gel fluorescence assay. Obvious protein signals were observed in the experimental lane with the incubation of 5-F-THF-Dayne, while being greatly decreased in the competition lane by the treatment with excess 5-F-THF (Figure 3D). These results suggested that protein labeling by the 5-F-THF-Dayne probe was highly specific. The bound proteins were identified using mass spectrometry (Supplemental Data Set S1).
To reduce false positives, identified proteins were ranked in the corresponding volcano plots as a log2 of competition ratio (5-F-THF-Dayne versus 5-F-THF) against statistical significance (−log10P-value). Fifty-one proteins competed by a factor >1 with P <0.05 were considered significant probe hits (Figure 3E;Supplemental Data Set S2).
Validation of affinity proteomics data
Among the shortlisted 51 proteins, two members were known 5-F-THF metabolic enzymes, that is, serine hydroxymethyltransferase 1 (SHMT1) and 5FCL. As described above, interaction of 5-F-THF with 5FCL was confirmed through an in-gel labeling assay (Figure 3A). The in vivo interaction of 5-F-THF with SHMT1 was reported previously (Roje et al., 2002). In this study, their binding was verified in vitro through MicroScale Thermophoresis (MST; Supplemental Data Set S2). Besides these two known FFBPs, the binding of an additional 28 putative recombinant FFBPs with 5-F-THF was quantified by MST analysis (Supplemental Data Set S2). Based on the dissociation constant (Kd) values, these proteins were categorized into two classes: (1) High-affinity FFBPs—comprising 14 proteins with Kd < 100 �M (Supplemental Data Set S2)—and (2) low-affinity FFBPs—the remaining 14 proteins with Kd > 100 �M. Furthermore, 10 of the high-affinity bindings were also verified by the Thermo-Shift Assay (Supplemental Figure S3). Notably, the Kds of the low-affinity FFBPs are likely to be above the level of 5-F-THF in vivo and their physiological relevance is more speculative than those with lower, more physiologically relevant Kds. Nevertheless, future detailed characterization of these low-affinity interactions may shed light on their physiological relevance. The remaining 21 candidates from the shortlisted 51 proteins have not yet been tested and independently verified for 5-F-THF binding and are henceforth referred to as putative FFBPs. While some of these putative candidates may prove to be bona fide FBBPs in future, they were not further considered in this study. For the ease of information, however, their identities and functions are listed in Supplemental Data Set S2. We focused only on the 30 independently verified high- and low-affinity targets for further analysis. In addition, the binding of two high-affinity FFBPs, AtDHFR-TS1 and AtGLN1;4, was scrutinized in depth.
Bioinformatics analysis of the 30 verified proteins and their predicted functions
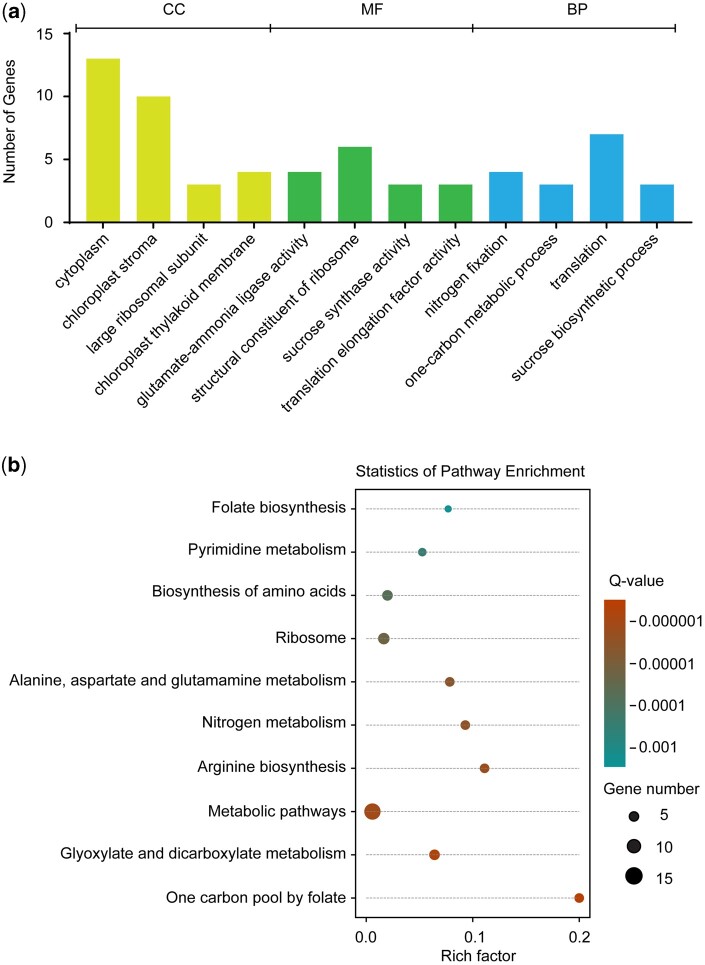
The online Gene Ontology (GO) platform (The Gene Ontology Consortium, 2019) was used for the bioinformatics analysis of the 30 verified proteins. GO enrichment analysis revealed their localization, mainly linked to cytoplasm and plastids/chloroplasts, and associated with the large ribosomal subunits (Figure 4A). Furthermore, the proteins were noted to constitute four groups based on their molecular function (MF). Consistent with the prevailing evidence that 5-F-THF functions as an enzyme regulator (Roje et al., 2002), three groups contained catalytic proteins/enzymes. The fourth group was represented by structural constituents of ribosomes (Figure 4A). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis revealed broad enrichment of the identified proteins mainly in metabolic pathways, C1 metabolism, N fixation, sucrose biosynthesis, and translation (Figure 4B). This indicated a possible connection of folates with other cellular processes besides catalysis of C1-related conversions. Finally, the MapMan annotation of the plant proteome database (Sun et al., 2009) was used and the proteins were categorized into the following broad biological processes (BPs).
Figure 4.
Bioinformatics analysis of the identified 5-F-THF-binding proteins utilizing GO platform. A, Gene function enrichment of 5-F-THF-binding proteins. CC, cellular component; B, KEGG pathway analysis of the identified 5-F-THF-binding proteins.
C1 metabolism
Noticeably, 4 of the 30 proteins had direct links with C1 metabolism (Supplemental Data Set S2), including the two known FFBPs SHMT1 and 5FCL, and the two previously unknown high-affinity FFBPs DHFR-TS1 and DHFR-TS2. SHMT1, a mitochondrial enzyme, catalyzes the reversible conversion of THF and serine to 5,10-CH2-THF and glycine, and provides a major bulk of C1 groups for the synthesis of thymidylate, methionine, and purines (Mouillon et al., 1999). In a previous report, 5-F-THF was revealed to be a potential inhibitor of the SHMT1 activity (Roje et al., 2002). 5FCL was reported to catalyze the conversion of 5-F-THF to 5,10-CH = THF during the folate-mediated C1 metabolism process (Goyer et al., 2005). The binding of 5-F-THF with DHFR-TS1 and DHFR-TS2 has not previously been reported. DHFR-TSs catalyze the conversion of dihydrofolate to THF, in the penultimate step of folate biosynthesis (Gorelova et al., 2017b). Noticeably, out of the three DHFR-TS proteins (AtDHFR-TS1–3) encoded by the Arabidopsis genome, only AtDHFR-TS1 and AtDHFR-TS2 show catalytic activity (Gorelova et al., 2017b). In the light of this, their identity as high-affinity FFBPs hints toward a regulatory role of 5-F-THF on these two folate biosynthetic isoforms. Being bifunctional enzymes, their TS unit catalyzes the conversion of deoxyuridine monophosphate to deoxythymidine monophosphate using 5,10-CH2-THF as a C1 donor and connects C1-metabolism with nucleotide metabolism. Another low-affinity FFBP, uracil phosphoribosyltransferase, is not a part of C1-metabolism sensu stricto, but is involved in nucleotide metabolism (Mainguet et al., 2009). Folates are closely connected to nucleotide synthesis through C1 metabolism (Gorelova et al., 2017a). The above interactions thus indicated that besides assisting as co-factors in nucleotide synthesis, folates may also regulate the kinetics of these reactions through 5-F-THF-mediated regulation.
Translational control and proteostasis
Proteostasis is of utmost importance to maintain a functional cellular state, which ultimately contributes to development. Our 30 verified probe hits included 12 proteins, 6 high-affinity, and 6 low-affinity, connected to proteostasis (Supplemental Data Set S2). The six high-affinity FFBPs included two ribosomal proteins ribosomal protein S18 (RPS18) and L13 family protein (RPL13AD; representing the 30S and 60S subunits, respectively), two translation elongation factor EF1B gamma chains—EF1Bγ1 and 2, a chloroplastic ribosome recycling factor, and a chaperonin containing TCP1 subunit 4 (CCT4). Furthermore, the six FFBPs belonging to the low-affinity group were the four ribosomal proteins—RPS14C, S7e family protein (RPS7A), RPL12-C, and RPL12-A (representing the 40S and 50S subunits), a translation elongation factor EMB2726, and a chaperone protein HSP101. Overall, these interactions indicated that folates may modulate not only the synthesis, but also the folding of a broad range of cellular proteins into their functional states.
N metabolism
Lately, a link between folate metabolism and N assimilation was proposed (Jiang et al., 2013; Meng et al., 2014). However, the molecular connection between the two processes is yet unknown. Here, four cytosolic GS1 isoforms, GLN1;1, GLN1;2, GLN1;4, and GLN1;5, were identified as interactors of 5-F-THF. GLN1;1 and GLN1;4 were confirmed to be high-affinity FFBPs, while GLN1;2 and GLN1;5 demonstrated low-affinity binding (Supplemental Data Set S2). GS1 catalyzes the most important step in N assimilation, that is, incorporation of into amino acids. The interaction of 5-F-THF with GLN1s will help to explore the molecular connection between folates and N metabolism.
Carbohydrate metabolism
This group comprises four proteins (Supplemental Data Set S2), including one high-affinity FFBP, the Photosystem II Subunit Q-1 (PSBQ1), and the three low-affinity FFBPs—sucrose synthases 1, 2, and 3 (SUS1, SUS2, and SUS3). PSBQ1 is required for the assembly and stability of photosystem II and is thus directly associated with photosynthesis (Zhang et al., 2016). SUS1, 2, and 3 catalyze cleavage of sucrose intoUridine diphosphate (UDP)—glucose, and fructose, which serve as energy sources, structural components, and signaling molecules (Stein and Granot, 2019). Interestingly, an interaction between sucrose signaling and folate biosynthesis has already been reported (Stokes et al., 2013). Further, SUS is also implicated in the biosynthesis of sugar-derived structural polymers, like cellulose, necessary for cell wall development, hence cell division (Stein and Granot, 2019). All-inclusive, these interactions provide clues to investigate the relationship of folates with C metabolism in the future.
Miscellaneous
Several verified proteins function in varying yet important BPs, like (1) stress response: the low-affinity FFBP HSP101. HSP101 is one of the most important chaperones involved in acquisition of thermotolerance in plants (Mishra and Grover, 2016); (2) cell division, cytoskeleton, and cell wall organization: one high-affinity FFBP—CHLOROPLASTIC DIVISION PROTEIN 1 (CDP1) and two low-affinity FFBPs—PECTIN ACETYLESTERASE 11 (PAE11) and PROFILIN 1 (PRF1). CDP1 is an essential component of the chloroplast division machinery (Zhang et al., 2009), and PRF1 affects the structure of the cytoskeleton by binding with actin (Cao et al., 2016). PAE11 regulates the physical properties of the cell wall by hydrolyzing acetyl esters in homogalacturonan regions of pectin (Gou et al., 2012); (3) lipid metabolism: the high-affinity FFBP, SUPPRESSOR OF SA INSENSITIVE 2 (SSI2). SSI2, a stearoyl-ACP desaturase, is involved in fatty acid desaturation and has been implicated in salicylic acid-mediated systemic-acquired resistance (SAR) signaling (Kachroo et al., 2005). The interaction of 5-F-THF with SSI2 warrants dedicated future research, as it may answer how folic acid induces SAR in Arabidopsis (Wittek et al., 2015).
Computational modeling and molecular docking analysis of 5-F-THF against the 16 selected FFBPs
Chemically, folates contain a hydrophobic head, comprising pterin and PABA moieties, and a hydrophilic glutamate tail, a negatively charged and rotatable group. Further, at the structural level, 5-F-THF has a high level of flexibility, with up to 11 rotatable bonds (Petrova et al., 2019; Figure 5A), and both the conjunction of pterin and PABA in the head as well as the glutamate tail possess great flexibility of torsion. This implies that the 5-F-THF molecule may easily acquire different conformations adapting to different sizes or shapes of binding pockets, hinting toward a diversity in the identity of the interactors.
Figure 5.
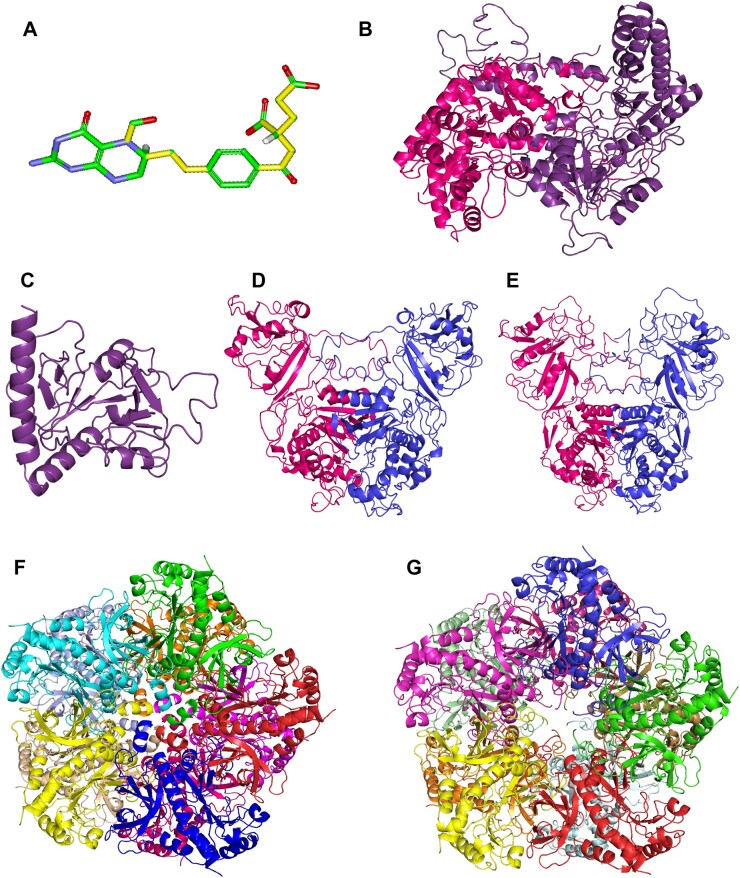
Structure of the ligand 5-F-THF and homology models of the selected FFBPs. 3D structure of 5-F-THF (A) and homology models of the two known FFBPs—SHMT1 (B) and 5FCL (C), and four high-affinity FFBPs –AtDHFR-TS1 (D), AtDHFR-TS2 (E), AtGLN1;1 (F) and AtGLN1;4 (G). The 5-F-THF molecule shown in stick model and its 11 rotatable bonds are presented in yellow (color of atoms: green for C, red for oxygen, blue for N, and white for hydrogen).
To further characterize the binding attributes of the 16 selected proteins (14 high-affinity + 2 known FFBPs) with the ligand 5-F-THF, we performed a docking analysis and studied the interactions. First, the homology models for all the 16 proteins were constructed by a combination of multiple template modeling and ab initio folding simulation (independent of the availability of the crystal structures, in order to reduce artifacts in the process). After optimization and construction of the multimeric structures (see “Materials and Methods” for detailed steps), the quality of the models was examined by checking the confidence level provided by the homology modeling software Phyre2, the Root-mean square deviation (RMSD) between the generated model and the main template, and the obtained Z-score (Supplemental Table S1; Supplemental Figure S4; see “Materials and Methods” for more details). Thirteen of the 16 models were of sufficient quality—(1) confidence level >60%, (2) RMSD < 4.5 �, and (3) Z-scores situated within the cloud of Z-scores of all the experimentally evaluated proteins available in the Protein Data Bank (PDB)—to perform docking analysis (Supplemental Table S1; Supplemental Figure S4). The three remaining models were of low quality. In two cases, this was due to the lack of appropriate templates for their construction (CDP1 and ENODL2, with model confidence levels <40%); and for the third (CCT4), the reason was the high structural differences between the model and crystal structure.
With the set of 13 models that passed our quality controls, a docking analysis was performed. Taking into account the inherent difficulties associated with the docking of highly flexible molecules, like folates (Petrova et al., 2019), a Lamarckian genetic algorithm (LGA)—a method suitable for highly flexible ligands (Morris et al., 2009)—was used for the docking analysis. Further, two positive and two negative controls (see “Materials and Methods”) were added to assess the quality of the modeling procedures used in this study. The complexes between the 13 homology models and the ligand 5-F-THF showed binding energies lower than the negative controls and in a range equivalent to the positive controls (Supplemental Table S2). This finding supports interaction of our protein models with the 5-F-THF ligand. Moreover, the positive interactions of 5-F-THF with the known FFBPs 5FCL and SHMT1 (interaction energies of −10.9 � 0.071 kcal and −9 � 0.127 kcal, respectively) noted in this study, further support the robustness of our modeling and docking methodology. Notably, the results of the docking analysis largely corroborated the ones obtained with the MST assay. The three best binding energies, −11.1 � 0.262 kcal, −9.9 � 0.962 kcal, and −9.44 � 0.283 kcal, obtained in the docking analysis for EF1Bγ1, DHFR-TS1, and RPL13AD, respectively (Supplemental Table S2), were well in accordance with the lowest Kd values in the high-affinity FFBPs group (0.34, 0.90, and 0.17 �M, respectively; Supplemental Data Set S2). Furthermore, the slightly higher interaction energies for GLN1;1 and GLN1;4 (−7.71 � 0.028 kcal and −7.75 � 0.417 kcal, respectively) corresponded with their highest Kd values in the high-affinity FFBPs group (23.20 and 55 �M, respectively; Supplemental Data Set S2). Altogether, these results are supportive of the structural interaction of 5-F-THF with high-affinity FFBPs.
Confirmation of the binding of 5-F-THF with recombinant AtDHFR-TS1 and AtGLN1;4 in vitro
First, binding of 5-F-THF-Dayne with recombinant AtDHFR-TS1 and AtGLN1;4 was confirmed by a gel-based labeling experiment. Clear fluorescent bands were observed for the 5-F-THF-Dayne-labeled protein (Figures 6, A and 7, A, Lanes 1 and 2). Addition of excess 5-F-THF (100 μM, 10 folds) in the protein sample together with the probe (5-F-THF-Dayne, 10 μM) weakened the fluorescence/labeling, highlighting the specificity of the interaction. A competitive binding assay using other folate analogs indicated that dihydrofolic acid (DHF) and 5-formyl-tetrahydropteroyltriglutamate had more or less the same competition effect as 5-F-THF (Supplemental Figure S5).
Figure 6.
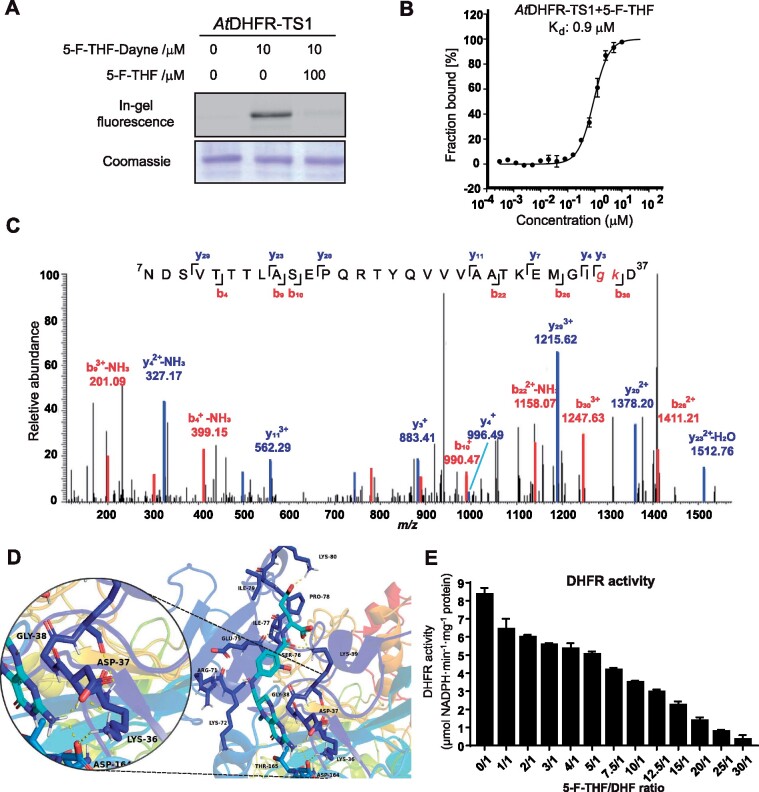
In vitro characterization of the binding of 5-F-THF with the recombinant Arabidopsis AtDHFR-TS1. A, Gel-based labeling results showing the binding of 5-F-THF-Dayne (10 μM) with AtDHFR-TS1. B, Dose-response curve reflecting binding of the recombinant AtDHFR-TS1 with 5-F-THF as assessed by MST analysis. The curve was generated by NanoTemper Analysis version 1.2.231. Normalized fluorescence (hot fluorescence/initial fluorescence) was plotted as a function of 5-F-THF concentration. Three independent thermophoresis measurements were performed; results are shown as mean � SE of the three repetitions. C and D, Identification of the binding site of 5-F-THF in AtDHFR-TS1. C, LC–MS/MS revealed that G35 or K36 is the potential binding position of 5-F-THF-Dayne in AtDHFR-TS1. G, glycine; K, lysine. D, Molecular docking prediction of the binding of 5-F-THF with AtDHFR-TS1. 5-F-THF is represented in light blue and hydrogen bonds are represented with dotted yellow lines. E, Enzymatic activity of the native AtDHFR-TS1 under conditions of varying 5-F-THF/ DHF ratio (50 �M DHF was used). DHFR activity values are means � se of three independent reactions. For the corresponding enzyme kinetics curve showing NADPH absorbance at 340 nm, see Supplemental Figure S8.
Figure 7.
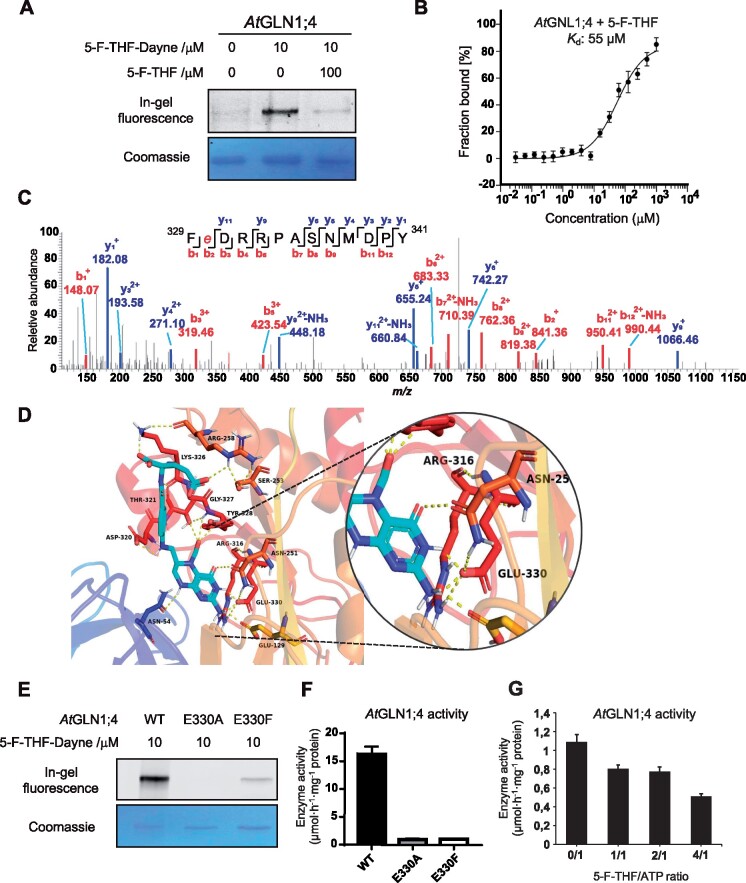
In vitro characterization of the binding of 5-F-THF with the recombinant Arabidopsis AtGLN1;4. A, Gel-based labeling results showing the binding of 5-F-THF-Dayne (10 μM) with AtGLN1;4. B, Dose-response curve of the recombinant AtGLN1;4 with 5-F-THF obtained by MST analysis. The binding curve was generated by NanoTemper Analysis version 1.2.231. Normalized fluorescence (hot fluorescence/initial fluorescence) was plotted as a function of 5-F-THF concentration. Three independent thermophoresis measurements were performed; results are shown as mean � SE of the three repetitions. C and D, Identification of the binding site of 5-F-THF in AtGLN1;4. C, LC–MS/MS identification of E330 as the potential binding position of 5-F-THF-Dayne in AtGLN1;4. E, glutamate. D, Molecular docking prediction of the binding of 5-F-THF with AtGLN1;4. 5-F-THF is represented in light blue and hydrogen bonds are represented with dotted yellow lines. E, The gel-based labeling of native and mutant AtGLN1;4s (E330A/F) by 5-F-THF-Dayne. F, Enzymatic activities of native and mutated (E330A/F) AtGLN1;4s. G, Enzymatic activity of native AtGLN1;4 under conditions of varying 5-F-THF/ATP ratio (400 �M ATP was used, Km value as indicated by Ishiyama et al., 2004).
Further, as discussed above, binding of 5-F-THF with AtDHFR-TS1 and AtGLN1;4 was quantified through MST and supported by docking analysis (Figures 6, B and 7, B; Supplemental Table S2). Interaction of AtDHFR-TS1 with DHF and AtGLN1;4 with ATP served as positive controls (Supplemental Figure S6). The Kd values of AtDHFR-TS1 for 5-F-THF and DHF were noted to be 0.90 and 1.12 μM, respectively, which were in the same order of magnitude (Figure 6B;Supplemental Figure S6A). This suggested that AtDHFR-TS1 had roughly similar affinities for 5-F-THF and DHF (AtDHFR-TS1’s substrate). Likewise, the Kd of AtGLN1;4 for 5-F-THF and ATP was similar, that is, 55 and 50 μM, respectively (Figure 7B;Supplemental Figure S6B), which pointed toward similar affinities. Biochemically, 5-F-THF contains a glutamic acid tail and given that glutamate acts as a substrate of GLN1, we hypothesized that 5-F-THF could also serve as an alternative substrate to AtGLN1;4, explaining their interaction. However, AtGLN1;4 enzyme assays, using glutamate and 5-F-THF as substrates, rejected this hypothesis, as a sharp peak depicting glutamine production was noted for glutamate, but not for 5-F-THF (Supplemental Figure S7).
Characterization of 5-F-THF binding sites in recombinant AtDHFR-TS1 and AtGLN1;4 and the effect of 5-F-THF binding on their activities
To unearth the binding attributes of 5-F-THF with AtDHFR-TS1 and AtGLN1;4, we investigated the 5-F-THF-binding pockets in the two proteins through tandem mass spectrometry analysis. To refine and detect the residues involved in the ligand-receptor complex, a second docking analysis limiting the gridbox to the previously defined binding pocket was performed. Recombinant AtDHFR-TS1 was photo-labeled by 5-F-THF-Dayne, digested, and subjected to LC–MS/MS analysis. The peptide 7NDSVTTTLASEPQRTYQVVVAATKEMGIGKD35 was found to exhibit a mass increase of 564.25 Da, corresponding to 5-F-THF-Dayne with loss of N2, while the MS2 spectrum of the peptide localized the labeling to G35 or K36 (Figure 6C). The second docking analysis showed a location of the ligand 5-F-THF near Gly38 (Figure 6D), similar to the co-crystal result of Toxoplasma gondii TS-DHFR and folate (Sharma et al., 2013). Furthermore, it interacted with residues D39, K36, and D164 forming a hydrogen bond network, matching with the results obtained by LC–MS/MS analysis (Figure 6d). Taken together, these data demonstrated that AtDHFR-TS1 could interact with 5-F-THF. This prompted us to study the effect of 5-F-THF binding on the DHFR activity of the AtDHFR-TS1 enzyme for a possible regulatory role. Addition of 5-F-THF reduced the DHFR activity in a competitive manner (Figure 6E;Supplemental Figure S8). When the 5-F-THF concentration was kept the same as the substrate (DHF) in the assay (50 �M), the DHFR activity was reduced by ∼25%. Further increase in 5-F-THF concentration led to more pronounced reduction of the activity (Figure 6E).
Next, we determined the binding site of 5-F-THF in AtGLN1;4 using the same strategy as above. In this case, the peptide 329FEDRRPASNMDPY341 was found to exhibit a mass increase of 564.25 Da and the labeling localized to E330 (Figure 7C). From the refined molecular docking analysis, the ligand 5-F-THF was found to interact with E330, the same as that revealed by LC–MS/MS analysis (Figure 7D); this residue corresponds to the binding pocket of adenosine (ATP/ADP) (Unno et al., 2006). This finding prompted us to further characterize this interaction, to unveil a possible regulatory role of 5-F-THF in AtGLN1;4 enzyme action and decipher the connection between C1 and N metabolisms. To further test the involvement of E330 in the binding, the effect of the E330F/A mutation on the interaction of 5-F-THF-Dayne with AtGLN1;4 was investigated. Mutating E330 greatly prevented the interaction of the probe with AtGLN1;4 (Figure 7E). In addition, it completely abolished AtGLN1;4 enzyme activity (Figure 7F). These observations hinted toward a possible 5-F-THF-mediated regulation of AtGLN1;4 activity. To test this, the effect of 5-F-THF on AtGLN1;4 enzyme activity was determined in vitro. As enzymatic activity was noted also for the AtGLN1;1 isoform (Supplemental Figure S9), we included both AtGLN1;4 and AtGLN1;1 in this assay. As was the case for AtDHFR-TS1, addition of 5-F-THF was noted to decrease the enzymatic activity of both AtGLN1;4 and AtGLN1;1 in a competitive manner: the higher the 5-F-THF/ATP ratio, the greater the inhibition (Figure 7G;Supplemental Figure S9).
Discussion
Despite the paramount importance of folates in plant growth and development, the understanding of their action mechanism remains poor. We thus aimed at identifying folate-binding/interacting proteins in Arabidopsis to (1) pinpoint the metabolic steps that folates may regulate through these interactions and (2) generate a broader picture of folate-mediated metabolic pathways in plants.
Folates are small molecules and therefore tracing their interacting proteins is challenging. Nevertheless, motivated by the identification of artemisinin-binding proteins using artemisinin-derived small molecular probe in HeLa cells (Zhou et al., 2016), we attempted to synthesize and test a folate-derived probe as the first step in our study. To design the probe, 5-F-THF was chosen as a bait for the following merits and reasons. First and foremost, 5-F-THF, being the most stable natural folate, promised the probe stability and thus efficacy of the strategy. Second, despite constituting majority of the mitochondrial folate pool (Orsomando et al., 2005), little is known regarding the role of 5-F-THF in plants. It is the only derivative that does not function as a C1 donor. Exploring the 5-F-THF-interacting proteome would shed light on its function. Third, the role of 5-F-THF in modulation of enzyme activities was revealed (Roje et al., 2002; Goyer et al., 2005), which hinted toward its regulatory function and thus made it a promising candidate. For the foregoing reasons, a novel 5-F-THF-Dayne probe was customized (Figure 1). Encouragingly, as desired, tagging had little effect on the biological activity of 5-F-THF (Figure 2). The active uptake of the 5-F-THF-Dayne, its effective translocation across the seedling and most importantly, the conversion of 5-F-THF-Dayne to other folate-Dayne derivates by plants, is promising (Figure 2; Supplemental Figures S1 and S2). The in vitro binding activity of 5-F-THF-Dayne with the known folate-associated enzyme 5FCL was also demonstrated (Figure 3A). Encouraged by this, the probe was exploited in an affinity proteomics workflow (Figure 3, B–D) and 51 putative FFBPs in Arabidopsis were revealed through mass-spectrometric profiling. This work establishes that a folate-derived probe can be used to determine the interacting proteome in plants. Moreover, this study opens a door for use of the tagged folates for in vivo experiments in plants as well as other organisms. Folates are prone to oxidation and thus highly unstable, which makes them difficult to be monitored spatially/temporarily. Tagged folates can conveniently be traced in vivo, and thus can contribute tremendously to folate biology in plants.
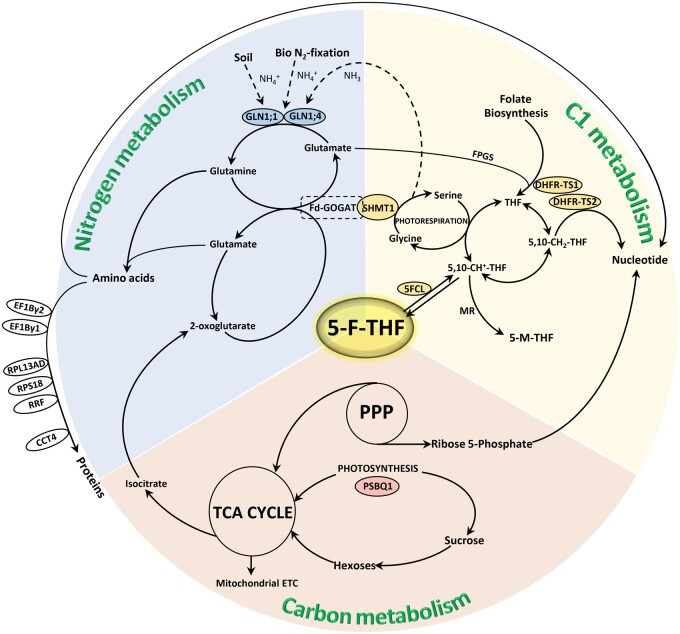
The credibility of the use of 5-F-THF-Dayne in affinity proteomics was guaranteed by confirming the binding of several of the putative FFBPs with 5-F-THF (Supplemental Data Set S2; Supplemental Figure S3). Fourteen high-affinity FFBPs were identified. In silico docking analysis was supportive of the structural interaction of 5-F-THF with the high-affinity FFBPs (Supplemental Table S2). Surveying the connection of these high-affinity FFBPs with plant metabolism, a broad schematic plan depicting C1, N, and C metabolisms with the metabolic steps possibly regulated by 5-F-THF is suggested (Figure 8). The proposed plan would serve as a prototype for future characterization of these bindings as well as functional relevance of folates therein.
Figure 8.
Broad schematic representation of the C1, N, and C metabolism, reflecting 5-F-THF as a nexus between the three metabolisms. Known- and high-affinity FFBPs/enzymes are depicted. The two N metabolism-related high-affinity FFBPs, the GSs—GLN1;1 and GLN1;4 are encircled with blue fill, the C metabolism-related high-affinity FFBP, PSBQ1, with pink fill, and the four C1 metabolism-related FFBPs, DHFR-TS1 and 2( high-affinity), SHMT1 (known) and 5FCL (known), with yellow fill. The encircled high-affinity FFBPs indicated on the left hand of the scheme have broad association with N metabolism, more specifically in amino acid and protein metabolism—RPS18, RPL12-A, RPL12-C, S11 family protein (RPS14C), RPS7A, RPL13AD, translation elongation factor gamma chain (EF1Bγ1 and EF1Bγ2), RRF, and CCT4. The scheme is proposed as a reference road map for future studies involving characterization of these bindings as well as functional relevance in folate-mediated plant development.
In this study, the detailed characterization of 5-F-THF binding with AtDHFR-TS1 (C1 metabolism/folate biosynthesis) and AtGLN1;4 (N metabolism) was undertaken. In view of the modulation of folate biosynthesis and the worldwide focus toward folate biofortification, 5-F-THF binding with the folate biosynthetic enzyme AtDHFR-TS1 was of much interest. An inhibitory effect of 5-F-THF on few folate-dependent mammalian enzymes was reported in 1990s (Stover and Schirch, 1993). Likewise in plants, 5-F-THF was noted to inhibit SHMT1 activity (Goyer et al., 2005). Here, we explored the functional relevance of the interaction of 5-F-THF with AtDHFR-TS1 by examining the effect of 5-F-THF on AtDHFR-TS1 enzyme activity. 5-F-THF was revealed to slow down AtDHFR-TS1 activity in a competitive manner: the higher the concentration of 5-F-THF over the substrate’s (DHF’s), the lower the DHFR activity (Figure 6E;Supplemental Figure S8). It represents a possible feedback regulatory module, thus far unexplored for this step in the folate biosynthetic pathway. It is much conceivable that plants employ 5-F-THF as an additional layer of regulation over folate biosynthesis to maintain the physiological level within the cell. This discovery not only enlightens us further on folate homeostasis and metabolism but also warrants consideration in future folate biofortification attempts, or in multi-biofortification approaches (Van Der Straeten et al., 2020). We propose that boosting folate biosynthesis could be combined with sequestration of 5-F-THF away from the target tissue to avoid feedback inhibition.
Interaction of 5-F-THF with AtGLN1;4 uncovered the molecular link between folates and N metabolism. As previously reported, metabolic changes typical of low-N stress, including significant reduction in glutamine levels, were observed in the fpgs2 mutant, and the GLN1 enzyme activity was noted to be significantly altered (Jiang et al., 2013). These observations suggested that the regulatory role of folates on N metabolism might impact the GLN1 function. In this study, the 5-F-THF—GLN1;4 interaction was supported by multiple lines of evidence. We found that 5-F-THF shares the binding site of ATP (E330) in GLN1;4, which is decisive for the activity of GLN1;4 enzyme. Mutation of the E330 residue not only prevented 5-F-THF binding with GLN1;4 but also abolished its enzymatic activity (Figure 7, E and F). Furthermore, we revealed that 5-F-THF competitively inhibits the activities of the GLN1;4 as well as GLN1;1, the two cytosolic GS1s reported to have high affinity to ammonium and glutamate (Ishiyama et al., 2004; Figure 7g;Supplemental Figure S9). Based on these results, we propose that the 5-F-THF–AtGLN1s interaction may present an important nexus between folate and N metabolism, and call for future genetic studies to substantiate this link. Moreover, GLN1 is the entry point into N metabolism, and the flux then goes to ferredoxin-dependent glutamine oxoglutarate aminotransferase (Fd-GOGAT), which catalyzes the reductive conversion of 2-oxoglutarate plus glutamine yielding two molecules of glutamate (Figure 8). Strikingly, Fd-GOGAT has itself been reported to physically interact with the C1-metabolism-related protein/enzyme SHMT (Figure 8; Jamai et al., 2009). In view of this, the 5-F-THF–GLN1 module is even more appealing, as it closes the circle of C1–N metabolism.
Thus far, 5-F-THF remained the most enigmatic folate derivative; except for its inhibitory action over SHMT1, there was no other information regarding 5-F-THF function in plants. Owing to this knowledge gap and the fact that 5-F-THF can catalytically be converted to other folate derivatives and enter C1 metabolism (Gorelova et al., 2017a, 2017b), it was largely conceived as a storage form. By revealing the 5-F-THF interacting proteome, we bridge this knowledge gap and bring forth several interesting facets of 5-F-THF function in plants. Our results reveal 5-F-THF-interacting proteins involved in C and N metabolism, reiterating the function of folates beyond being just a C1 donor. This was further substantiated by 5-F-THF-mediated regulation of AtDHFR-TS1 and AtGLN1;4 activity. Based on these arguments and exhaustive engagement of other folate species in housekeeping C1 metabolism, we propose that 5-F-THF, being the most stable form, acts as a regulator of diverse metabolic pathways. Future comparison of our current data with those corresponding to other folate derivatives will yield more detailed insight into the control of cellular function by individual folate forms.
In conclusion, this pioneering work (1) adds an important tool (a Dayne-tagged folate probe) in the existing tool box for plant biology, (2) reveals one of the regulatory modules that connect folates with N metabolism, (3) demonstrates 5-F-THF-mediated feedback regulation on folate biosynthesis, and (4) proposes additional cellular targets of 5-F-THF. Several of these interactions may prove to be the missing molecular links substantiating previous reports connecting folates with the plethora of their developmental roles in plants. Hence, this study opens an avenue toward approaching the question of how folates modulate plant growth and development.
Materials and methods
Synthesis of photoaffinity probe 5-F-THF-Dayne
5-F-THF (30 mg) was dissolved in anhydrous N,N-dimethylformamide (DMF; 3.0 mL), and then dicyclohexylcarbodiimide (DCC; 20 mg), hydroxyl-benzotriazole(15 mg), and N,N-diisopropyl-ethylamine (40 �L) were added into the solution. After 1-h stirring, 2-(2-azidoethyl)-2-(but-3-ynyl)-1,3-dioxolane (4 mg) was added to the reaction mixture followed by stirring at room temperature for another 8 h in the dark. The mixture was washed with saturated Na2CO3, 1 mmol L−1 HCl, and brine (a saturated solution of sodium chloride in distilled water). The organic layer was dried by anhydrous Na2SO4, filtered, and concentrated. The crude product was purified by high-performance liquid chromatography (HPLC) to obtain the desired product—5-F-THF-Dayne (see Supplemental methods).
Chromatographic separation was carried out at 30�C on a Waters Acquity ultra performance liquid chromatography (UPLC) BEH C18 column (1.7 μm, 50 mm � 2.1 mm, 130 �; Waters, Milford, MA, USA). Mobile Phase A (0.1% formic acid in H2O) and mobile Phase B (0.1% formic acid in acetonitrile) were used to establish a 21-min gradient elution, 16 min of 20−95% B, and 3 min of 95% B, 2 min of 95−20% B followed by re-equilibrating at 20% B for 2.5 min. The flow rate was 300 �L min−1. Compound 2-(2-azidoethyl)-2-(but-3-ynyl)-1,3-dioxolane was synthesized as previously described (Li et al., 2013).
Chemicals and equipment
For information on different chemicals and equipment used in this study, see Supplemental methods.
Plants and culture conditions
The A. thaliana ecotype Columbia-0 was used in this study, and plants were grown at 22�C with light intensity maintained at white light 300 μmol m−2 s−1 under a 16-h light/8-h dark cycle. Arabidopsis T-DNA insertion mutant dfb (SALK_015472) was ordered from the ABRC seed stock center. For hypocotyl growth, the wild-type (WT) and dfb mutant seeds were sterilized, cultured, and assessed as previously described (Meng et al., 2014).
Arabidopsis proteome extraction and fraction isolation
Fresh Arabidopsis leaves from 4-week-old seedlings were harvested and protein samples were prepared as previously described (Li et al., 2018). About 200 g of fresh tissue was homogenized and extracted in potassium phosphate (KPi) buffer (50 mM KPi pH 8.0, 5.0 mM MgCl2, 1.0 mM EDTA, 10 mM DTT, 10 g PVPP, 10 g PVP, and protease inhibitor cocktail [Roche, Basel, Switzerland]). Three biological repeats with independent pools of leaves were used. Proteins from the clear extracts were precipitated using ammonium persulfate (30%–75% saturation) and re-dissolved in HEPES buffer (50 mM HEPES, 5 mM MgCl2, 10 mM EDTA, 2 mM DTT, pH 8.0). Dissolved proteins were then separated via a DEAE column (2 � 15 cm; Aogma Biosciences, Burlington, MA, USA) and eluted by sodium chloride solution at different concentration range (50–400 mM). Four fractions (A, B, C, and D) were collected and subsequently concentrated with a 10-kDa molecular weight cut-off column (Millipore, Burlington, MA, USA) for further analysis. This enabled separation of the highly abundant proteins, particularly RuBisCo, which avoided interference in probe labeling.
In-gel and in vivo fluorescence labeling
For in-gel fluorescence labeling, Arabidopsis proteome fractions A, B, C, and D (50 μL) were transferred to a 96-well plate and incubated with 5-F-THF-Dayne (10 μM) on ice for 30 min. After irradiation with 365 nm UV light (8 W) for 20 min, 20 μL protein-probe complexes were transferred to 0.6-mL tubes and 1% SDS was added, followed by addition of freshly prepared click cocktail [containing TAMRA-N3 (0.1 mM), CuSO4 (1 mM), tris 3-hydroxypropyl-triazolylmethyl amine (THPTA; 0.1 mM) and ascorbic sodium (1 mM)] at room temperature for 1 h. Finally, SDS loading buffer was added and the samples were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE) (12% gel). Gels were imaged by FUJIFILM FLA 9000 plus DAGE fluorescence scanner and stained by Coomassie blue. For the in vivo fluorescence labeling protocol, see Supplementary materials and methods.
Photoaffinity pull down of 5-F-THF-Dayne binding proteins
About 0.3 mL of concentrated Arabidopsis protein extracts (3.0 mg mL−1) were incubated with 100 μM of 5-F-THF or DMSO together with 5-F-THF-Dayne (10 μM) for 1 h. All samples were photo-irradiated in a 6-well plate on ice, using three 365 nm UV lamps (8 W) for 20 min. The resulting samples were collected in separate tubes and precipitated with CH3OH/CHCl3 as described previously (Li et al., 2018). The precipitates were re-suspended in 220 μL of click buffer (50 mM HEPES, pH8.0, 1% SDS) and subsequently 200 μL of the resulting suspensions were subjected to click reaction with Biotin-N3 (500 μM; Biomatrick Inc., San Diego, CA, USA), CuSO4 (1 mM; Sigma, St. Louis, MO, USA), THPTA, 100 μM; Sigma), and ascorbic sodium (1 mM; Sigma). The remaining 20 μL samples were subjected to click reaction with TAMRA-N3. Biotin-clicked samples were precipitated with CH3OH/CHCl3, washed with CH3OH, and re-dissolved in 200 μL of click buffer by sonication.
On-beads digestion and protein identification
An aliquot of 50 �L of streptavidin-sepharose (GE Healthcare, Chicago, IL, USA) beads were added to each sample and incubated at room temperature with continuous mixing for 1 h. The beads were washed sequentially 3 times each with 1� PBS buffer containing 1% SDS (w/v) and 1� PBS buffer containing 0.5 M NaCl, twice with 4 M Urea in 50 mM triethylammonium bicarbonate (TEAB), and 5 times with 50 mM TEAB. Each wash was performed on a rotator for 15 min. The bound proteins were subjected to on-beads reductive alkylation with 200 �L of 10 mM dithiothreitol at 56�C for 30 min and 200 �L of 55 mM iodoacetamide at 37�C in dark for another 30 min, followed by washing with 50 mM TEAB for 3 times. Bound proteins then underwent on-beads digestion with 0.25 �g of sequencing grade modified trypsin (Promega, Madison, WI, USA) reconstituted in 50 �L of 50 mM TEAB overnight at 37�C. The digests were labeled with respective TMT-2plex™ Isobaric Label Reagents (Thermo Scientific, Waltham, MA, USA), according to the manufacturer’s procedures. The labeled peptides were desalted by Pierce C18 spin columns and evaporated to dryness on a SpeedVac. The dried peptides were resuspended in 20 �L of formic acid/acetonitrile/H2O (v:v:v = 0.5%/2%/97.5%) with sonication. The proteomics experiment was carried out in biological triplicates. For the identification of the proteins, Orbitrap Fusion Lumos Mass Spectrometry was exploited. For technicalities involved, see Supplementary Methods.
Protein purification and competitive photolabeling of recombinant proteins
The AtGLN1;4, At5FCL, and Zm5FCL cDNAs were cloned into a pET28a expression vector, while the cDNAs of other proteins (including AtGLN1;1), except AtDHFR-TS1, were cloned in pLM303 expression vector. Constructs were transformed into BL21 (DE3) Escherichia coli to obtain His-tag and His-MBP-tag fusions for pET28a and pLM303 constructs, respectively (for primer details, see Supplemental Table S3). For production of recombinant AtDHFR-TS1 protein and to assure its biological activity, the protocol described previously (Gorelova et al., 2017b) was used with minor modification (see Supplementary methods). A detailed description of the steps involved in the purification of the recombinant proteins is provided in Supplemental Methods. Recombinant AtDHFR-TS1 or AtGLN1;4 was diluted to a final concentration of 0.1 mg mL−1 in PBS. AtDHFR-TS1 was incubated with 5-F-THF-Dayne (10 μM) and NAPDH (2 μM) in the presence or absence of seven substrates on ice. AtGLN1;4 was incubated with 5-F-THF-Dayne (10 μM) in the presence or absence of 5-F-THF (100 μM) on ice. The following steps were the same as in the in-gel fluorescence labeling.
MST analysis
For validation of the affinity proteomics data, the binding affinities of 29 probe hits/proteins (showing expression in the soluble fraction) were quantified by MST. For AtGLN1;4 and AtDHFR-TS1, purified recombinant proteins (10 �M) were labeled with a RED fluorescent dye NT-647-NHS according to the user manual of the Monolith NT™ Protein Labeling Kit RED-NHS (NanoTemper, M�nchen, Germany; MO-L001). For the rest 27, the soluble recombinant proteins in the supernatant were analyzed by SDS–PAGE, quantified by Bradford assay, and labeled at their His-Tag with RED fluorescence as specified by the kit MO-L018 (Nano temper). The 5-F-THF ligand in a range of concentration (0.03 �M to 5 mM) was incubated with the labeled proteins (0.4 �M) at room temperature for 5 min, and then the samples were loaded into the NanoTemper glass capillaries (NanoTemper Technologies) and microthermophoresis was performed using 40% LED power on the MST instrument (Monolith NT.115). The binding curve was generated by the NanoTemper Analysis version 1.2.231 software. Normalized fluorescence (hot fluorescence/initial fluorescence) was plotted as a function of each ligand concentration.
GO enrichment and KEGG enrichment analysis
GO enrichment analysis of the identified proteins was subsequently performed using R-based hyper geometric test. KEGG enrichment analysis was carried out using KEGG Orthology and KOBAS version 3.0 (Available online: http://kobas.cbi.pku.edu.cn) based on the hyper geometric test.
Homology modeling and binding analysis of 5-F-THF with high-affinity FFBPs
For homology modeling, the primary structure of the 16 high-affinity FFBPs (including 2 known FFBPs, 5FCL, and SHMT1) were downloaded from the SwissProt Protein Database (See UniProt accession numbers in Supplemental Data Set S2). The homology modeling was carried out under the intensive mode of Phyre2 (http://www.sbg.bio.ic.ac.uk/~phyre2), applying a combination of multiple template modeling and ab initio folding simulation (Kelley et al., 2015). The complete list of templates used for each model is detailed in Supplemental Table S1. To assess the quality of the models, both the confidence of the model provided by the Phyre2 software and the Z-score on proSAweb (https://prosa.services.came.sbg.ac.at/prosa.php) were considered (Wiederstein and Sippl, 2007). Furthermore, in order to confirm the robustness of the obtained models, the root-mean-square deviation (RMSD)of the tridimensional models and the best template were determined by superimposition of both structures using PyMOL software version 2.3.0 (http://www.pymol.org/pymol). Finally, the models were geometrically optimized by a two-step energy minimization protocol using NAMD software (Phillips et al., 2020) to accommodate, first, the lateral chains and, subsequently, the backbone of the proteins, using a dielectric constant of 80 and 2,000 steps of 1fs per timestep. For multimeric proteins, crystal structures showing their quaternary structure (i.e. the biological assembly) were used as templates and energy minimization of the complex was performed with the aforementioned parameters (see templates in Supplemental Table S1).
The 2D structure of the ligand 5-F-THF was geometrically optimized to a 3D model using ACD/ChemSketch software version 2020.1.1 (Advanced Chemistry Development, Inc., Toronto, ON, Canada; Figure 1). The charges of both the models and the ligand were adjusted by the PDB2PQR online server (https://server.poissonboltzmann.org/pdb2pqr; Dolinsky et al., 2004) using the CHARMM force field (Vanommeslaeghe et al., 2010) depending on the annotation of the different cellular environments of the proteins. For the detection/characterization of the 5-F-THF binding pockets and determining the interaction energies associated with 5-F-THF binding to the 14 high-affinity and the two known FFBPs (5FCL and SHMT1), a molecular docking analysis was performed by AutoDock version 4 (AutoDock suite; Morris et al., 2009). In order to remove the bias, the whole protein was covered by a grid (126 � � 126 � � 126 � for x, y, and z) with a space of 0.5 � between grid points. All the receptors were considered rigid bodies and the 11 rotatable bonds of the ligand were considered flexible (representing the high flexibility of the 5-F-THF molecule; Petrova et al., 2019). The LGA was used as the docking search method for its suitability with highly flexible ligands, with the generation of 100 conformations, an initial population size of 300, a maximum number of evaluations of 25,000,000, and a maximum number of generations of 27,000. In order to maximize accuracy of binding predictions, two positive, as well as two negative controls, were analyzed using the same docking approach. As positive controls, the known folate-binding enzymes 5FCL (origin—Bacillus anthracis; PDB ID: 2JCB) and SHMT (Geobacillus stearothermophilus; PDB ID: 1KL2) were selected, whereas the nonfolate binding enzyme JAR1 and the structural model of the actin gene (A. thaliana; PDB ID: 4EPL and UniProt accession number: P0CJ46, respectively) were chosen as negative controls. To refine the docking analysis of the structural models of DHFR-TS1 and GLN1;4, a second docking analysis was performed with a gridbox centered on the previously defined binding pocket and with a size 80 � � 80 � � 80 � for x, y, and z and the space between grid points reduced to 0.375 �. The search parameters were as follows: 20 conformations generated, population size of 300, a maximum number of evaluations of 25,000,000, and a maximum number of generations of 27,000. In order to reflect the stability of the binding affinity energies, all the results are expressed as the average of the first and second-best scores of the docking analysis.
Binding site identification
Identification of the 5-F-THF-binding sites in AtDHFR-TS1 or AtGLN1;4 involved as many steps as incubation of the recombinant proteins with 5-F-THF-Dayne, UV irradiation for covalent linking, SDS–PAGE, and in-gel chemical modification. Thereafter, protein in the gel was subjected to trypsin/chymotrypsin digestion, extraction, followed by analysis on a Thermo Orbitrap Fusion Lumos proteomic mass spectrometer. For detailed protocol, see Supplementary methods.
In vitro enzymatic assays of AtDHFR-TS1, AtGLN1;1, and AtGLN1;4 to study the effect of 5-F-THF
To determine the effect of 5-F-THF on AtDHFR-TS1, the DHFR enzyme activity assay was performed as described previously (Gorelova et al., 2017a, 2017b) with minor modifications. Briefly, the reaction mixture of a final volume of 0.5 mL contained 0.2 mM NADPH, 50 μM 7,8-DHF (Schircks laboratories, Z�rich, Switzerland) and 0.5 pmol purified AtDHFR-TS1 in 50 mM phosphate buffer (pH 7.4; 5 mM β-mercaptoethanol and 1 mM DTT). The concentration of 5-F-THF in the reaction was in the range of 0–1,500 �M. The reaction was run at room temperature. The protocol estimates exhaustion/oxidation of NADPH during the AtDHFR-TS1-catalyzed DHF-to-THF conversion by detecting A340 spectrophotometrically using IMPLEN NanoPhotometer NP80 Mobile.
To determine the effect of 5-F-THF on the enzymatic activity of AtGLN1;1 and AtGLN1;4, 5-F-THF was added with a concentration varying from 0 to 1.6 mM to a 600-�L enzymatic reaction mixture (37.5 mM imidazole buffer, pH 7.0; 10 mM L-Glutamate sodium; 20 mM MgSO4; 0.4 mM ATP; 10 mM NH2OH, 0.2 mg AtGLN1;4 protein). After incubation at 30�C for 30 min, 200 �L termination buffer (88 mM FeCl3, 670 mM HCl, and 200 mM trichloroacetic acid) was added and the reaction mixture was allowed to rest at room temperature for 10 min. After centrifugation at 4,000 g at room temperature for 10 min, the supernatant was used for detection of A540 in a spectrophotometer. GS enzyme activity was expressed as micromol of Glu-γ-monohydroxamate formed per hour per milligram of protein.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. In vivo fluorescence imaging of 5-F-THF-Dayne in shoots and leaves of 10-day-old Arabidopsis WT seedlings.
Supplemental Figure S2. Representative LC–MS chromatogram of 5-F-THF-Dayne-fed Arabidopsis seedlings extracts showing peaks for 5-F-THF-Dayne, 5-10-CH=THF-Dayne and 5-M-THF-Dayne.
Supplemental Figure S3. Thermal Shift Assay confirming the binding of 5-F-THF with 10 high-affinity FFBPs.
Supplemental Figure S4. Z-scores for the homology models of two known and 14 high-affinity FFBPs.
Supplemental Figure S5. Competitive labeling of AtDHFR-TS1 with 5-F-THF-Dayne in the presence of different folate derivatives and analogs.
Supplemental Figure S6. Quantification of binding affinities of AtDHFR-TS1 and AtGLN1;4 for DHF and ATP, respectively, through in vitro MST
Supplemental Figure S7. Characterization of the transamination activity of AtGLN1;4 by HR-ESI-MS analysis in the presence of glutamate and 5-F-THF, respectively, as substrates.
Supplemental Figure S8. DHFR enzyme kinetics curve showing the effect of different concentrations of 5-F-THF on the reaction.
Supplementary Figure S9. Enzymatic activity of AtGLN1;1 under conditions of varying 5-F-THF/ATP ratio (concentration).
Supplemental Data Set S1 (Excel sheet) . Quantitative proteomic profiling of 5-F-THF-Dayne against 5-F-THF.
Supplemental Data Set S2 (Excel sheet). Significantly enriched 5-F-THF-Dayne hits (51 proteins) from the Arabidopsis proteome.
Supplemental Table S1 List of templates used for each FFBP homology model, confidence level of the model, and RMSD between the model and the main template.
Supplemental Table S2. Binding energies and ligand efficiencies obtained in the docking analysis between the constructed homology models and the ligand 5-F-THF.
Supplemental Table S3. Primers used in this study.
Supplementary Material
Acknowledgments
We thank Dr W. Yuan and Mr S. Bu in the Core Facility Center of the Institute of Plant Physiology and Ecology for mass spectrometry and NMR assistance.
Funding
This work was supported by the National Natural Science Foundation of China (NSFC) (31970326, 21807102, and 91856112), Chinese Academy of Sciences (Grant Nos. XDB27020203, 153D31KYSB20170121, and 153D31KYSB20160074), and the Agricultural Science and Technology Innovation Program (ASTIP, CAAS-ZDRW202004), and by grants from Ghent University (BOF-GOA2018) and the Research Foundation Flanders (FWO; 3G012609 and 35963) to DVDS. RCM is indebted to FWO (grant number 42316) and the Ghent University Special Research Fund (BOF—grant number BOF20/PDO/040) for a postdoctoral fellowship.
Conflict of interest statement. The authors declare no conflict of interest.
Y.X., D.V.D.S., and C.Z. designed and supervised the research. W.L. performed probe design, probe biological assessment in vitro, in vivo absorption, and affinity proteomics. Q.L. performed biological assessment of the probe in vivo, high-throughput assessment of 30 interactions through MST experiments, GLN1 activity tests, and 5-F-THF inhibition assays. R.C.M., together with Q.L., performed assessment of the high-affinity bindings through Thermo-Shift Assay experiments. R.C.M. performed DHFR-TS1 purification, enzyme activity tests, and 5-F-THF inhibition assays. R.S.M. performed the computational modeling and docking analysis. Q.L., R.C.M., W.L., R.S.M., H.W., and X.C. conducted the data analysis. R.C.M., Q.L., W.L., R.S.M., D.V.D.S., C.Z., and Y.X. prepared the manuscript. R.C.M. drafted the entire discussion. All authors have read and approved the final manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) are: Chunyi Zhang (zhangchunyi@caas.cn) and Dominique Van Der Straeten (Dominique.VanDerStraeten@UGent.be).
References
- Blancquaert D, De Steur H, Gellynck X, Van Der Straeten D (2014) Present and future of folate biofortification of crop plants. J Exp Bot 65: 895–906 [DOI] [PubMed] [Google Scholar]
- Blancquaert D, Van Daele J, Strobbe S, Kiekens F, Storozhenko S, De Steur H, Gellynck X, Lambert W, Stove C, Van Der Straeten D (2015) Improving folate (vitamin B-9) stability in biofortified rice through metabolic engineering. Nat Biotechnol 33: 1076. [DOI] [PubMed] [Google Scholar]
- Bouvier F, Linka N, Isner JC, Mutterer J, Weber APM, Camara B (2006) Arabidopsis SAMT1 defines a plastid transporter regulating plastid biogenesis and plant development. Plant Cell 18: 3088–3105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L, Henty-Ridilla JL, Blanchoin L, Staiger CJ (2016) Profilin-dependent nucleation and assembly of actin filaments controls cell elongation in Arabidopsis. Plant Physiol 170: 220–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway LP, Li WC, Parker CG, , (2021) Chemoproteomic-enabled phenotypic screening. Cell Chem Biol 28: 371–393 [DOI] [PubMed] [Google Scholar]
- Cossins E (2000) The fascinating world of folate and one-carbon metabolism. Can J Bot 78: 691–708 [Google Scholar]
- Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA (2004) PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res 32: W665– W66–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez B, Vera P (2019) Folate metabolism interferes with plant immunity through 1C methionine synthase-directed genome-wide DNA methylation enhancement. Mol Plant 12: 1227–1242 [DOI] [PubMed] [Google Scholar]
- Gorelova V, Ambach L, Rebeille F, Stove C, Van Der Straeten D (2017a) Folates in plants: research advances and progress in crop biofortification. Front Chem 5: 21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorelova V, De Lepeleire J, Van Daele J, Pluim D, Mei C, Cuypers A, Leroux O, Rebeille F, Schellens JHM, Blancquaert D, et al. (2017b) Dihydrofolate reductase/thymidylate synthase fine-tunes the folate status and controls redox homeostasis in plants. Plant Cell 29: 2831–2853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou JY, Miller LM, Hou G, Yu XH, Chen XY, Liu CJ (2012) Acetylesterase-mediated deacetylation of pectin impairs cell elongation, pollen germination, and plant reproduction. Plant cell 24: 50–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyer A, Collakova E, de la Garza RD, Quinlivan EP, Williamson J, Gregory JF, Shachar-Hill Y, Hanson AD (2005) 5-Formyltetrahydrofolate is an inhibitory but well tolerated metabolite in Arabidopsis leaves. J Biol Chem 280: 26137–26142 [DOI] [PubMed] [Google Scholar]
- Hanson AD, Gregory JF (2011) Folate biosynthesis, turnover, and transport in plants. Annu Rev Plant Biol 62: 105–125 [DOI] [PubMed] [Google Scholar]
- Henkel AS, Dewey AM, Anderson KA, Olivares S, Green RM (2012) Reducing endoplasmic reticulum stress does not improve steatohepatitis in mice fed a methionine- and choline-deficient diet. Am J Physiol-Gastr L 303: G54–G59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa T, Machida C, Yoshioka Y, Kitano H, Machida Y (2003) The GLOBULAR ARREST1 gene, which is involved in the biosynthesis of folates, is essential for embryogenesis in Arabidopsis thaliana. Plant J 33: 235–244 [DOI] [PubMed] [Google Scholar]
- Ishiyama K, Inoue E, Watanabe-Takahashi A, Obara M, Yamaya T, Takahashi H (2004) Kinetic properties and ammonium-dependent regulation of cytosolic isoenzymes of glutamine synthetase in Arabidopsis. J Biol Chem 279: 16598–16605 [DOI] [PubMed] [Google Scholar]
- Jamai A, Salome PA, Schilling SH, Weber APM, McClung CR (2009) Arabidopsis photorespiratory serine hydroxymethyltransferase activity requires the mitochondrial accumulation of ferredoxin-dependent glutamate synthase. Plant Cell 21: 595–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Liu YY, Sun H, Han YT, Li JL, Li CK, Guo WZ, Meng HY, Li S, Fan YL, et al. (2013) The mitochondrial folylpolyglutamate synthetase gene is required for nitrogen utilization during early seedling development in Arabidopsis. Plant Physiol 161: 971–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachroo P, Venugopal SC, Navarre DA, Lapchyk L, Kachroo A (2005) Role of salicylic acid and fatty acid desaturation pathways in ssi2-mediated signaling. Plant Physiol 139: 1717–1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protocol 10: 845–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Hill-Skinner S, Liu SZ, Beuchle D, Tang HM, Yeh CT, Nettleton D, Schnable PS (2015) The maize brown midrib4 (bm4) gene encodes a functional folylpolyglutamate synthase. Plant J 81: 493–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Moore M, King J (2003) Investigating the regulation of one-carbon metabolism in Arabidopsis thaliana. Plant Cell Physiol 44: 233–241 [DOI] [PubMed] [Google Scholar]
- Li Z, Hao P, Li L, Tan CY, Cheng X, Chen GY, Sze SK, Shen HM, Yao SQ (2013) Design and synthesis of minimalist terminal alkyne-containing diazirine photo-crosslinkers and their incorporation into kinase inhibitors for cell- and tissue-based proteome profiling. Angewandte Chem 52: 8551–8556 [DOI] [PubMed] [Google Scholar]
- Li WC, Zhou YQ, You WJ, Yang MQ, Ma YR, Wang ML, Wang Y, Yuan SG, Xiao YL (2018) Development of photoaffinity probe for the discovery of steviol glycosides biosynthesis pathway in Stevia rebuadiana and rapid substrate screening. Acs Chem Biol 13: 1944–1949 [DOI] [PubMed] [Google Scholar]
- Luka Z (2008) Methyltetrahydrofolate in folate-binding protein glycine N-methyltransferase. Vitam Horm 79: 325–345 [DOI] [PubMed] [Google Scholar]
- Luka Z, Moss F, Loukachevitch LV, Bornhop DJ, Wagner C (2011) Histone demethylase LSD1 is a folate-binding protein. Biochemistry 50: 4750–4756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mainguet SE, Gakiere B, Majira A, Pelletier S, Bringel F, Guerard F, Caboche M, Berthome R, Renou JP (2009) Uracil salvage is necessary for early Arabidopsis development. Plant J 60: 280–291 [DOI] [PubMed] [Google Scholar]
- Mehrshahi P, Gonzalez-Jorge S, Akhtar TA, Ward JL, Santoyo-Castelazo A, Marcus SE, Lara-Nunez A, Ravanel S, Hawkins ND, Beale MH, et al. (2010) Functional analysis of folate polyglutamylation and its essential role in plant metabolism and development. Plant J 64: 267–279 [DOI] [PubMed] [Google Scholar]
- Meng HY, Jiang L, Xu BS, Guo WZ, Li JL, Zhu XQ, Qi XQ, Duan LX, Meng XB, Fan YL, et al. (2014) Arabidopsis plastidial folylpolyglutamate synthetase is required for seed reserve accumulation and seedling establishment in darkness. PLoS One 9: e101905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra RC, Grover A (2016) ClpB/Hsp100 proteins and heat stress tolerance in plants. Crit Rev Biotechnol 36: 862–874 [DOI] [PubMed] [Google Scholar]
- Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ (2009) AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 30: 2785–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouillon JM, Aubert S, Bourguignon J, Gout E, Douce R, Rebeille F (1999) Glycine and serine catabolism in non-photosynthetic higher plant cells: their role in C1 metabolism. Plant J 20: 197–205 [DOI] [PubMed] [Google Scholar]
- Orsomando G, de la Garza RD, Green BJ, Peng M, Rea PA, Ryan TJ, Gregory JF, Hanson AD (2005) Plant g-Glutamyl hydrolases and folate Polyglutamates characterization, compartmentation, and co-occurrence in vacuoles. J Biol Chem 280: 28877–28884 [DOI] [PubMed] [Google Scholar]
- Parker CG, Pratt MR (2020) Click chemistry in proteomic investigations. Cell 180: 605–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova J, Gocheva G, Ivanova N, Iliev S, Atanasova B, Madjarova G, Ivanova A (2019) Molecular simulation of the structure of folate and antifolates at physiological conditions. J Mol Graph Modell 87: 172–184 [DOI] [PubMed] [Google Scholar]
- Phillips JC, Hardy DJ, Maia JDC, Stone JE, Ribeiro JV, Bernardi RC, Buch R, Fiorin G, Henin J, Jiang W, et al. (2020) Scalable molecular dynamics on CPU and GPU architectures with NAMD. J Chem Phys 153: 044130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthusseri B, Divya P, Veeresh L, Kumar G, Neelwarne B (2018a) Evaluation of folate-binding proteins and stability of folates in plant foliages. Food Chem 242: 555–559 [DOI] [PubMed] [Google Scholar]
- Puthusseri B, Divya P, Lokesh V, Kumar G, Savanur MA, Neelwarne B (2018b) Novel folate binding protein in Arabidopsis expressed during salicylic acid-induced folate accumulation. J Agr Food Chem 66: 505–511 [DOI] [PubMed] [Google Scholar]
- Rebeille F, Neuburger M, Douce R (1994) Interaction between glycine decarboxylase, serine hydroxymethyltransferase and tetrahydrofolate polyglutamates in pea leaf mitochondria. Biochem J 302: 223–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roje S, Janave MT, Ziemak MJ, Hanson AD (2002) Cloning and characterization of mitochondrial 5-formyltetrahydrofolate cycloligase from higher plants. J Biol Chem 277: 42748–42754 [DOI] [PubMed] [Google Scholar]
- Sharma H, Landau MJ, Vargo MA, Spasov KA, Anderson KS (2013) First three-dimensional structure of toxoplasma gondii thymidylate synthase-dihydrofolate reductase: insights for catalysis, interdomain interactions, and substrate channeling. Biochemistry 52: 7305–7317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein O, Granot D (2019) An overview of sucrose synthases in plants. Front Plant Sci 10: 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes ME, Chattopadhyay A, Wilkins O, Nambara E, Campbell MM (2013) Interplay between sucrose and folate modulates auxin signaling in Arabidopsis. Plant Physiol 162: 1552–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stover P, Schirch V (1993) The metabolic role of leucovorin. Trends Biochem Sci 18: 102–106 [DOI] [PubMed] [Google Scholar]
- Strobbe S, Van Der Straeten D (2017) Folate biofortification in food crops. Curr Opin Biotechnol 44: 202–211 [DOI] [PubMed] [Google Scholar]
- Sun Q, Zybailov B, Majeran W, Friso G, Olinares PDB, van Wijk KJ (2009) PPDB, the plant proteomics database at Cornell. Nucleic Acids Res 37: D969–D974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang HM, Liu SZ, Hill-Skinner S, Wu W, Reed D, Yeh CT, Nettleton D, Schnable PS (2014) The maize brown midrib2 (bm2) gene encodes a methylenetetrahydrofolate reductase that contributes to lignin accumulation. Plant J 77: 380–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Ontology Consortium (2019) The gene ontology resource: 20 years and still going strong. Nucleic Acids Res 47: D330–D338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unno H, Uchida T, Sugawara H, Kurisu G, Sugiyama T, Yamaya T, Sakakibara H, Hase T, Kusunoki M (2006) Atomic structure of plant glutamine synthetase: a key enzyme for plant productivity. J Biol Chem 281: 29287– 3367–3385. [DOI] [PubMed] [Google Scholar]
- Van Der Straeten D, Bhullar N, De Steur H, Gruissem W, Mackenzie D, Pfeiffer W, Qaim M, Slamet-Loedin I, Strobbe S, Tohme J, et al. (2020) Multiplying the efficiency and impact of biofortification through metabolic engineering. Nat Commun 11: 5203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Wilder V, De Brouwer V, Loizeau K, Gambonnet B, Albrieux C, Van Der Straeten D, Lambert WE, Douce R, Block MA, Rebeille F, et al. (2009) C1 metabolism and chlorophyll synthesis: the Mg-protoporphyrin IX methyltransferase activity is dependent on the folate status. New Phytol 182: 137–145 [DOI] [PubMed] [Google Scholar]
- Vanommeslaeghe K, Hatcher E, Acharya C, Kundu S, Zhong S, Shim J, Darian E, Guvench O, Lopes P, Vorobyov I, et al. (2010) CHARMM general force field: a force field for drug‐like molecules compatible with the CHARMM all‐atom additive biological force fields. J Comput Chem 31: 671–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederstein M, Sippl MJ (2007) ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res 35: W407–W410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittek F, Kanawati B, Wenig M, Hoffmann T, Franz-Oberdorf K, Schwab W, Schmitt-Kopplin P, Vlot AC (2015) Folic acid induces salicylic acid-dependent immunity in Arabidopsis and enhances susceptibility to Alternaria brassicicola. Mol Plant Pathol 16: 616–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Hu Y, Jia J, Li D, Zhang R, Gao H, He Y (2009) CDP1, a novel component of chloroplast division site positioning system in Arabidopsis. Cell Res 19: 877–886. [DOI] [PubMed] [Google Scholar]
- Zhang HM, Deng XY, Miki D, Cutler S, La HG, Hou YJ, Oh J, Zhu JK. (2012) Sulfamethazine suppresses epigenetic silencing in Arabidopsis by impairing folate synthesis. Plant Cell 24: 1230–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Duan Z, Zhang J, Peng L (2016) BIOGENESIS FACTOR REQUIRED FOR ATP SYNTHASE 3 facilitates assembly of the chloroplast ATP synthase complex. Plant Physiol 171: 1291–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Li W, Xiao Y (2016) Profiling of multiple targets of artemisinin activated by hemin in cancer cell proteome. ACS Chem Biol 11: 882–888 [DOI] [PubMed] [Google Scholar]
- Zhou HR, Zhang FF, Ma ZY, Huang HW, Jiang L, Cai T, Zhu JK, Zhang CY, He XJ (2013) Folate polyglutamylation is involved in chromatin silencing by maintaining global DNA methylation and histone H3K9 dimethylation in Arabidopsis. Plant Cell 25: 2545–2559 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.