Abstract
Papain-like protease (PLpro) is a key enzyme encoded by SARS-CoV-2 that is essential for viral replication and immune evasion. Significant suppression of viral spread and promotion of antiviral immunity can be achieved by inhibition of PLpro, revealing an inspiring strategy for COVID-19 treatment. This study aimed to discover PLpro inhibitors by investigating the national compound library of traditional Chinese medicines (NCLTCMs), a phytochemical library comprising over 9000 TCM-derived compounds. Through virtual screening and enzymatic evaluations, nine natural biflavones were confirmed to be effective PLpro inhibitors with IC50 values ranging from 9.5 to 43.2 μM. Pro-ISG15 cleavage assays further demonstrated that several biflavones exhibited potent inhibitory effects against PLpro-mediated deISGylation, a key process involved in viral immune evasion. Herein, we report the discovery, antiviral evaluation, structure-activity relationship elucidation and molecular docking investigation of biflavones as potent inhibitors of SARS-CoV-2 PLpro.
Keywords: SARS-CoV-2, Papain-like protease, Natural biflavone, deISGylation, Antiviral
Graphical abstract
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of COVID-19, continues to ravage mankind throughout the world. As of July 25, 2021, there have been over 192 million confirmed cases of COVID-19 including 4.1 million deaths reported to the World Health Organization. Nonetheless, there is still a lack of effective treatment for COVID-19, and therapeutic drugs are desperately needed.
PLpro is a key enzyme encoded by SARS-CoV-2 that recognizes and cleaves the LXGG consensus sequence (Leu-X-Gly-Gly, X refers to unspecific amino acids) of both viral and host proteins (Klemm et al., 2020). Cleavage of the viral polyprotein at the LXGG site generates non-structural proteins (nsp1–3) to participate in the assembly of the viral replicase complex, which initiates replication and transcription of the viral genome (Hu et al., 2021a) (Fig. 1 A). Beyond the ability to process viral proteins, ample evidence indicates that PLpro can also manipulate host proteins to evade antiviral responses. For example, recent studies demonstrated that SARS-CoV-2 PLpro could bind to, interact with and finally cleave the interferon stimulated gene product-15 (ISG15) modifier from melanoma differentiation-associated protein 5 (MDA5) to escape immune surveillance. MDA5 is the crucial viral RNA sensor to detect cytosolic SARS-CoV-2 RNA and thereby triggers innate immune response in an ISG15-dependent manner (Liu et al., 2021a; Yin et al., 2021). However, SARS-CoV-2 PLpro is capable of removing the ISG15 modifier from MDA5 by cleaving the LRGG motif at the C-terminus of ISG15 (Fig. 1B), thus terminating MDA5-mediated antiviral signalling. This event is termed deISGylation, a viral evolutionary strategy to evade host innate immunity. Likewise, studies by Shin et al. (2020). Showed that PLpro could facilitate immune evasion by deISGylating interferon regulation factor 3 (IRF3), which in turn disrupts type I interferon-mediated antiviral signalling. In addition to interfering with ISG15 modification, SARS-CoV-2 PLpro could also cleave Lys48 ubiquitin chains from host proteins to promote immune evasion but with much less preference and lower selectivity than ISG15 substrates (Freitas et al., 2020; Klemm et al., 2020).
Fig. 1.
Functions of SARS-CoV-2 PLpro and benefits by targeting PLpro. (A) PLpro proteolyzes viral polyprotein at the LXGG site to generate mature nsp1, nsp2 and nsp3. (B) PLpro removes ISG15 and ubiquitin modifiers from host proteins to evade innate antiviral immunity.
Collectively, PLpro plays a critical role in viral replication and immune evasion of SARS-CoV-2, making it an attractive target for COVID-19 treatment. Encouragingly, recent studies demonstrated that inhibition of PLpro could achieve significant suppression of SARS-CoV-2 viral spread and promotion of antiviral immunity (Shen et al., 2021; Shin et al., 2020). Motivated by this, the present work was devoted to searching for novel PLpro inhibitors, especially those with inhibitory potential against the deISGylation activity of PLpro.
Traditional Chinese medicine (TCM) is an extraordinary reservoir of antiviral agents, from which a wide variety of phytochemicals have been proven to be effective PLpro inhibitors. For instance, tanshinones derived from Salvia miltiorrhiza were found to be specific and selective inhibitors of PLpro (Park et al., 2012b). Furthermore, flavonoids isolated from Angelica keiskei (Park et al., 2016) and Broussonetia papyrifera (Park et al., 2017) displayed significant inhibition of PLpro. In addition, polyphenols from Paulownia tomentosa demonstrated dose-dependent inhibitory effects on both the proteolytic and deubiquitination activities of PLpro (Cho et al., 2013). In addition to the above phytochemicals, previous studies also identified diarylheptanoids from Alnus japonica (Park et al., 2012a) and cinnamic amides from Tribulus terrestris (Song et al., 2014) as potent inhibitors of PLpro.
Enlightened by this, we aimed to discover novel inhibitors of SARS-CoV-2 PLpro by investigating NCLTCMs, a phytochemical library constructed by our group currently possessing over 9000 entities of TCM-derived compounds. For the rapid search and identification of PLpro inhibitors, structure based virtual screening was initially performed to filter potential candidates. The hit compounds were then subjected to enzymatic evaluations to confirm their inhibitory activities. Finally, a panel of natural biflavones (Fig. 2 , 1–9) ranked as the most potent PLpro inhibitors from NCLTCMs, with anti-proteolytic IC50 values ranging from 9.5 to 43.2 μM. Gratifyingly, pro-ISG cleavage assays further demonstrated that several biflavones exhibit significant inhibition of the deISGylation activity of PLpro, offering good prospects in attenuating PLpro-mediated immune evasion. It is also worth mentioning that all biflavones in this study were derived from TCM. Among them, amentoflavone (1), ginkgetin (3), isoginkgetin (4) and sciadopitysin (5) are active ingredients in Ginkgo biloba (Liu et al., 2021b), while podocarpusflavone A (2) is a characteristic chemical constituent of Podocarpus nakaii (Yeh et al., 2012). In addition, morelloflavone (6) exists in Garcinia lateriflora (Ren et al., 2010), while hinokiflavone (7) and cryptomerin B (8) are found in Platycladus orientalis (L.) Franco (Lu et al., 2006). 4′-O-methylochnaflavone (9) is a naturally occurring biflavone from Lonicera japonica Thunb. (Seo et al., 2012; Son et al., 1992). Of note, G. biloba, P. orientalis and L. japonica are frequently used TCMs for antiviral treatment. Remarkably, considerable research has indicated that extracts and formulas of L. japonica are effective for multiple coronaviruses including SARS-CoV-2 (Hu et al., 2021b; Zhang et al., 2020). Our findings suggested that these biflavones might act as antiviral ingredients of the above TCMs by inhibiting PLpro. This article also reports a detailed kinetic investigation, structure-activity relationship elucidation and molecular docking analysis of biflavones.
Fig. 2.
Structures of the biflavones (1–9) from NCLTCMs and the flavone monomers.
2. Results and discussion
2.1. Identification of SARS-CoV-2 PLpro inhibitors from NCLTCMs
Structure based virtual screening is an effective strategy for lead compound discovery (Ghosh et al., 2006). The present study established a molecular docking model based on the existing crystal structure of SARS-CoV-2 PLpro (PDB: 7JRN) and performed virtual screening of all 9032 entries of NCLTCMs. The CDOCKER score (indicated as -CDOCKER interaction energy) was set as the criterion to filter candidates, and molecules that scored over 30 were selected as hit compounds.
The virtual screening process yielded 152 hit compounds, which were subjected to fluorogenic enzymatic assays to corroborate their inhibitory potencies. The assay utilized Z-RLRGG-AMC, a pentapeptide resembling the consensus cleavage sequence of PLpro labelled with 7-amino-4-methylcoumarin (AMC) at the C-terminus. The AMC motif was non-fluorescent when covalently conjugated, but became dramatically fluorescent upon cleavage by PLpro, thus enabling efficient determination of anti-proteolytic activity (Ratia et al., 2008). To eliminate promiscuous inhibitors, the assays were performed in the presence of 10 mM dithiothreitol (DTT) and 100 mM sodium chloride, for which DTT was employed as a reducing agent and electrophile trap, while sodium chloride was added to preclude unspecific electrostatic interactions. Psoralidin, a previously reported natural inhibitor of SARS-CoV PLpro (Kim et al., 2014), was selected as the positive control.
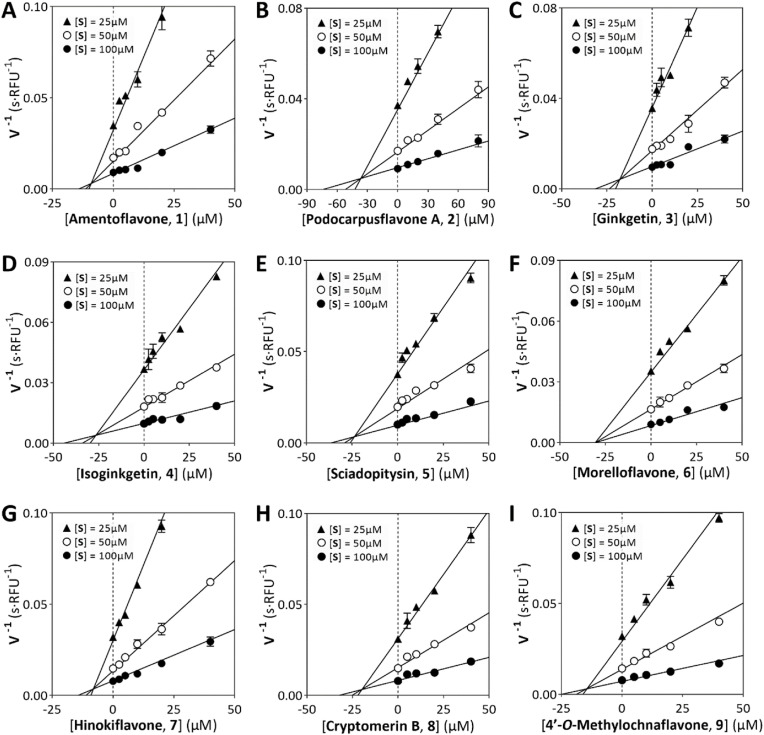
By this method, we determined the anti-proteolytic activities for all hit compounds, 46 of which achieved 50% inhibition at a 50 μM concentration. Among them, a panel of natural biflavones (1–9) were found to be most prominent, with IC50 values ranging from 9.5 to 43.2 μM. Detailed results for virtual screening and experimental evaluation are listed in Table 1 , including CDOCKER scores, binding energies, binding interactions and IC50 values. To gain kinetic insights, inhibition constants (K i) for all biflavones were determined by Dixon plots. As shown in Fig. 3 , reciprocals of initial velocity (V−1) at varying substrate concentrations were plotted as a function of biflavone concentration to produce a family of intersecting lines, and the abscissa of the intersection point was taken as K i. The K i values of the biflavones ranged from 7.8 to 36.5 μM, as listed in Table 1. The intersections also gave clues for inhibition mode, by which upper x-axis intersections indicated competitive or mixed type inhibition, as in the cases of all biflavones except morelloflavone (6). The intersection for 6 fell on the x-axis (Fig. 3F), suggesting that 6 alone manifested non-competitive inhibition. Explanations for this phenomenon will be discussed in depth below.
Table 1.
Virtual screening and anti-proteolytic evaluation results for the biflavones.
| Compound | Docking analysis |
Anti-proteolytic activity |
||||
|---|---|---|---|---|---|---|
| CDOCKER scorea | Binding energyb | Hydrogen bondc | Interacting residues | IC50/μMd | Ki/μMe | |
| Biflavones | ||||||
| 1 | 58.1 | −129.6 | 6 | Lys157, Leu162, Arg166, Glu167, Asn267, Thr301 | 13.0 ± 1.2 | 9.1 ± 0.7 |
| 2 | 50.3 | −66.2 | 4 | Lys157, Leu162, Glu167, Thr301 | 43.2 ± 2.9 | 36.5 ± 4.2 |
| 3 | 55.6 | −117.2 | 5 | Lys157, Leu162, Asp164, Arg166, Gly266 | 29.8 ± 1.5 | 17.7 ± 1.1 |
| 4 | 57.6 | −123.5 | 4 | Lys157, Leu162, Asp164, Arg166 | 31.2 ± 3.4 | 26.5 ± 2.8 |
| 5 | 55.7 | −113.4 | 2 | Lys157, Leu162 | 34.8 ± 3.7 | 23.6 ± 1.3 |
| 6 | 43.1 | −81.6 | 1 | Arg166 | 36.4 ± 4.8 | 30.7 ± 2.9 |
| 7 | 50.9 | −119.1 | 5 | Lys157, Glu161, Asp164, Arg166, Gly266 | 9.5 ± 1.2 | 7.8 ± 0.5 |
| 8 | 47.8 | −87.9 | 3 | Lys157, Glu161, Arg166 | 26.3 ± 3.2 | 14.3 ± 1.0 |
| 9 | 48.1 | −105.2 | 4 | Lys157, Leu162, Arg166, Glu167 | 22.8 ± 0.9 | 19.7 ± 1.6 |
| Flavone monomers | ||||||
| Apigenin | 29.4 | −48.2 | 2 | Ala246, Tyr264 | 75.7 ± 6.4 | n.d.f |
| Acacetin | 28.3 | −38.7 | 1 | Tyr264 | 91.2 ± 11.3 | n.d. |
| Positive control | ||||||
| Psoralidin | 38.7 | −55.8 | 1 | Tyr264 | 27.8 ± 2.2 | n.d. |
Indicated as negative value of CDOCKER interaction energy.
presented in Kcal/mol.
Number of hydrogen bonds formed between inhibitor and PLpro.
Concentrations of 50% inhibition of PLpro anti-proteolytic activity, represented as mean ± SD.
Inhibition constant, determined by Dixon plots.
Not determined.
Fig. 3.
Dixon plots for biflavone 1–9.
In summary, a panel of natural biflavones were identified as the most potent PLpro inhibitors from NCLTCMs via virtual screening and experimental corroboration. Because a variety of flavones were proven to be potent PLpro inhibitors in previous reports (Cho et al., 2013; Park et al., 2016, 2017), the biflavones from NCLTCMs attracted our particular interest and inspired further investigation.
2.2. Structure-activity relationship for biflavones
To correlate the chemical structure of the biflavones with their inhibitory potency against PLpro, we investigated the structure-activity relationship (SAR) for the biflavones. Since biflavones are structurally composed of two flavone monomers connected by C–C bonds (1–6) or C–O–C bonds (7–9), as indicated by the thick red lines in Fig. 2, the influence of connection type was first examined. The C–O–C connections revealed a more pronounced effect on anti-proteolytic activity than the C–C connections. As presented in Tables 1 and 4′-O-6″-connected biflavone 7 displayed the strongest activity with an IC50 value of 9.5 μM. Another biflavone with the same connection type (8) showed a milder yet tolerated IC50 value of 26.3 μM. Meanwhile, 4′-O-3‴-connected biflavone 9 ranked as the third-best inhibitor with an IC50 value of 22.8 μM. In contrast, C–C connections were less tolerated, and only biflavone 1 demonstrated a satisfactory IC50 value of 13.0 μM, whereas the IC50 values for all other C–C-type biflavones (2–6) ranged from 29.8 to 43.2 μM, which were all lower than those of C–O–C-type biflavones. To further explore the influence of the connection bond, the monomeric counterparts of biflavones, apigenin and acacetin, were also assessed for their anti-proteolytic activities. However, dramatic loss of activity was observed. The IC50 values for the monomers decreased to 75.7–91.2 μM, which was 8- to 10-fold weaker than that of dimer 7, suggesting that the dimeric skeleton was indispensable for PLpro inhibition.
We then surveyed the substitution effect of the biflavones, and found that the presence of hydroxy groups was more beneficial than methoxy groups. For instance, of all 8-3‴-connected biflavones (1–5), non-methylated compound 1 conferred the most potent activity with an IC50 value of 13.0 μM, whereas replacement of hydroxy groups with methoxy groups led to a 2.3- to 3.3-fold reduction in anti-proteolytic activity, as evidenced by the IC50 values of 29.8–43.2 μM for 2–5. Similarly, 7 and 8 both characterized a 4′-O-6″-connected skeleton, nonetheless, non-methylated biflavone 7 showed 2.8-fold more potent activity than its di-methylated counterpart (8), with IC50 values of 9.5 μM and 26.3 μM for 7 and 8, respectively.
2.3. Binding analysis of biflavones
We next analysed the binding mode of the biflavones with the aid of molecular docking. The interactions between SARS-CoV-2 PLpro and a representative C–C-type biflavone (1) were first explored. As illustrated in Fig. 4 A, biflavone 1 could dock well into the P3/P4 region of the substrate-binding pocket of PLpro with a “two-winged” pattern, by which the two flavone fragments concurrently occupy the P3 and P4 pockets to block substrate access. Multiple hydrogen bonds dominated the binding interactions. As depicted in Fig. 4D/4G, the 4′-OH and 7″-OH of biflavone 1 served as hydrogen bond donors to interact with the carboxy groups of Asn267 and Glu167, respectively. Meanwhile, 4‴-OH contributed to the hydrogen bond formed with the hydroxy group of Thr301. Additionally, 5″-OH created hydrogen bonds with the backbone of Leu162. In addition to acting as hydrogen bond donors, the 7-O atom and 5″-O atom of 1 functioned as hydrogen bond acceptors toward the guanidine group of Arg166 and amino side chain of Lys157, respectively. The amino acid residues involved in the hydrogen bonds are summarized in Table 1. These intensive polar contacts promised the top docking score of 58.1 as well as the highest binding energy of −129.6 kcal/mol for 1 across all biflavones in this study. Anti-proteolytic evaluation further confirmed the above result. With an IC50 value of 13.0 μM, 1 ranked as the second-best inhibitor among all nine biflavones.
Fig. 4.
Possible interactions of biflavones 1, 7, and 6 with PLpro. (A–C) Binding interactions represented in the Corey-Pauling-Koltun (CPK) model. (D–F) Binding interactions represented in the ball-stick model. The interacting amino acid residues are labelled in black. The hydrogen bonds are indicated by yellow dashed lines with the distances given in Å. The P3 and P4 regions of the substrate binding pocket of PLpro are indicated by corresponding labels. (G–I) 2D diagram of the binding interactions. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Hinokiflavone (7), a representative C–O–C-type biflavone, demonstrated a similar binding pattern to 1 in docking analysis. As shown in Fig. 4E/4H, the 7-OH, 7″-OH and 4‴-OH of 7 engaged in polar contacts with Glu161, Asp164 and Gly266, respectively. The binding of 7 to PLpro was further fortified by hydrogen bonds formed by 7-O atom and 4′-O atom toward Lys157 and Arg166, respectively.
The “two-winged” binding pattern of biflavone revealed a plausible explanation for the dimeric-dependent inhibition, in which the inability to co-occupy the P3 and P4 pockets by flavone monomers accounted for 8- to 10-fold activity loss compared to dimer 7.
In the case of morelloflavone (6), the “two-winged” binding pattern was impaired by a 3–8‴-connection. With the bulky flavone motif substituted at the 3-position, the resulting steric hindrance significantly impeded the binding of 6 with the substrate-binding pocket (Fig. 4C/4 F). As a consequence, 6 only formed one hydrogen bond with PLpro, resulting in a poor docking score of 48.2 and a weak binding energy of −91.2 kcal/mol for 6. In agreement with these findings, 6 exhibited mild anti-proteolytic activity with an IC50 value of 36.4 μM, which was the second least active of all nine biflavones. Moreover, the partial incompetency of 6 for the PLpro substrate-binding pocket was further supported by its non-competitive inhibition profile, as determined by Dixon plot (Fig. 3F).
2.4. Inhibition of PLpro-mediated deISGylation by biflavones
Given the pivotal role of deISGylation in PLpro-mediated immune evasion, we next evaluated whether biflavones could inhibit the deISGylation activity of PLpro by pro-ISG15 cleavage assays. Pro-ISG15 is the precursor of ISG15 harbouring the LRGG↓TEPGGRS sequence at the C-terminus, which could be cleaved by PLpro after the LXGG recognition motif. The cleavage led to a reduction in molecular weight by approximately 0.8 kDa, and the corresponding protein band shift could be efficiently detected and quantified by SDS–PAGE analysis (Klemm et al., 2020; Shin et al., 2020). Accordingly, inhibition of deISGylation could be determined by the proteolysis ratio of pro-ISG15.
Using the above approach, we assessed the inhibitory activities of all biflavones at an initial concentration of 20 μM. In line with recent studies (Klemm et al., 2020; Shin et al., 2020), SARS-CoV-2 PLpro demonstrated robust deISGylation activity, by which the proteolysis ratio of pro-ISG15 exceeded 95% in a 10 min period (Fig. 5 A, second lane). However, the deISGylation process was significantly inhibited in the presence of biflavones, as shown in Fig. 5E. Notably, four biflavones (3, 4, 7 and 9) demonstrated near complete inhibition against PLpro-mediated deISGylation at 20 μM. Another two biflavones (2, 8) displayed milder yet still considerable inhibition rates of 64.9%–74.8% at 20 μM. The inhibition rate of deISGylation was rather weak for 5 and 6 (32.3%–34.1% at 20 μM), but it was indeed consistent with their poor anti-proteolytic activities, with IC50 values ranging from 34.8 to 36.4 μM. Unexpectedly however, despite possessing the second-best anti-proteolytic activity with an IC50 value of 9.5 μM, amentoflavone (1) only demonstrated a mild 48.1% inhibition of deISGylation at 20 μM.
Fig. 5.
pro-ISG15 cleavage assays for the biflavones. (A) Initial screening of all nine biflavones for the inhibitory activities against PLpro-mediated deISGylation at a 20 μM concentration. (B–D) Active biflavones were further evaluated at serially diluted concentrations of 20, 10, 5 and 2.5 μM. (E) Inhibition rates for all 9 biflavones against PLpro-mediated deISGylation at 20 μM. (F) Inhibition rates for C–C-type biflavones (2–4) at concentrations of 20, 10, 5 and 2.5 μM. (G) Inhibition rates for C–O–C-type biflavones (7–9) at concentrations of 20, 10, 5 and 2.5 μM. w/o inb. = without inhibitors.
Biflavones with inhibition rates greater than 50% at 20 μM (2–4 and 7–9) were further investigated for their inhibitory potencies at decreased concentrations of 10 μM, 5 μM and 2.5 μM (Fig. 5B-D). All of the tested compounds displayed dose-dependent inhibition, and the inhibition rate is plotted in Fig. 5F-G. Analogous to anti-proteolytic activity, inhibitory activity against deISGylation also favoured C–O–C connections. As shown in Fig. 5G, at a concentration of 10 μM, C–O–C-type biflavones 7, 8, and 9 achieved 65.6%–98.0% inhibition, whereas the inhibition rate for C–C-type biflavones 2, 3, and 4 was only 53.2%–71.1% (Fig. 5F). When we further reduced the concentration to 5 μM, none of the biflavones except a C–O–C-type biflavone (9) reached 50% inhibition. Surprisingly, 9 even achieved 60.7% inhibition at concentrations down to 2.5 μM, demonstrating promising potential to attenuate PLpro-mediated deISGylation. As mentioned above, 4′-O-methylochnaflavone (9) is a naturally occurring biflavone in Lonicera japonica Thunb. (common name: honeysuckle). L. japonica is not only the most prescribed antiviral TCM, but also a food-medicine herb widely used in daily tea drinks, soft drinks and cosmetics (Shang et al., 2011). Notably, abundant studies indicated that L. japonica is effective for coronaviruses. For instance, cytopathic morphology-based assays by Wu et al. (2004). demonstrated that L. japonica extracts had significant antiviral effects against SARS-CoV. Furthermore, the antiviral efficacy of L. japonica was confirmed by accumulating clinical evidence. During the battle against SARS-CoV-2, greater than 85% of COVID-19 patients in China received TCM treatment and clinical results revealed that TCM could significantly alleviate the symptoms of mild patient (Wang et al., 2021; Yang et al., 2020). Among these TCMs, two L. japonica formulas, namely “Lianhua Qingwen capsule” and “Jinhua Qingan granules”, are listed as recommended medicine for COVID-19 treatment in the “Diagnosis and Treatment Scheme for Novel Coronavirus Pneumonia (Trial fourth edition, Fifth edition, Sixth edition, Seventh edition)”, owing to their obvious curative effect (Hu et al., 2021b; Wei, 2020; Zhang et al., 2020). Remarkably, L. japonica acts as the sovereign drug in both formulas, indicating a principal role in their therapeutic effect for COVID-19 treatment. Nonetheless, the active ingredients and the mechanism underlying the antiviral effect of L. japonica against SARS-CoV-2 remains elusive. In this context, it is of great significance to clarify the active ingredients in L. japonica responsible for SARS-CoV-2 inhibition. This study demonstrated for the first time that 9 exhibited potent inhibition of both proteolytic and deISGylation activities of SARS-CoV-2 PLpro. Our findings suggest that 9 might at least in part contribute to the antiviral activity of L. japonica by inhibiting PLpro, which provide useful information to support the clinical treatment of COVID-19 by L . japonica. In addition, considering the wide applications of L. japonica as a food-medicine herb, further studies regarding the in vivo antiviral activity and pharmacokinetic investigation of 9 will be worthwhile.
3. Conclusion
The involvement of PLpro in SARS-CoV-2 viral replication and immune evasion makes it an important target to combat COVID-19. This study identified natural biflavones as a new class of PLpro inhibitors through investigation of NCLTCMs, a phytochemical library comprising over 9000 TCM-derived compounds. By virtual screening and experimental corroboration, a panel of natural biflavones were confirmed to be potent PLpro inhibitors with IC50 values ranging from 9.5 to 43.2 μM. Moreover, several biflavones demonstrated significant inhibition of PLpro-mediated deISGylation, as indicated by pro-ISG15 assays. Structure-activity relationship analysis revealed that hydroxyl groups contributed to better anti-proteolytic activity, while C–O–C connections were more beneficial. Molecular docking investigation further suggested that the biflavones could occupy the P3/P4 substrate-binding pocket of PLpro to block substrate access. Our findings provide new insight into the active ingredients responsible for the antiviral activity of TCM. In addition, biflavones may represent valuable lead compounds for the development of antiviral agents against SARS-CoV-2 PLpro.
4. Materials and methods
4.1. Materials
All of the tested compounds were retrieved from 4 °C storage of NCLTCMs, which were all pure isolates from natural sources. The NCLTCMs accession numbers for biflavones 1–9 were 3001852, 3000847, 3000889, 3000854, 3000863, 3001926, 3004125, 3004129 and 3003784, respectively. Purities of the biflavones were all greater than 98% as determined by HPLC, and a 1 mg aliquot of each compound was weighed to prepare a 10 mM DMSO stock. Recombinant SARS-CoV-2 papain-like protease (Glu1564-Val1880, Cat. 40593-V07E) and recombinant human pro-ISG15 protein (Met1-Ser165, Cat. 12729-HNAE) were purchased from Sino-Biological Inc (Beijing, China). The fluorogenic substrate Z-RLRGG-AMC was purchased from Bachem (Bubendorf, Switzerland).
4.2. Virtual screening and molecular modelling
The X-ray structure of wild-type PLpro of SARS-CoV-2 complexed with a non-covalent small molecule inhibitor was obtained from the PDB database (PDB ID: 7JRN). The binding site was defined as an 8 Å radius sphere around the centroid of the co-crystallized ligand. The SMILES strings of 9032 NCLTCMs compounds were imported into BIOVIA Discovery Studio 2019 (Dassault Systèmes, San Diego, USA) to build a ligand database, and the ligands were further minimized using the prepare ligands protocol as implemented in Discovery Studio software. The virtual screening process was performed using the CDOCKER algorithm, followed by CHARMM27 force field minimization and binding energy calculations with an explicit solvent model. The hydrogen bond distance was measured using the measure wizard of PyMOL 2.4 (The PyMOL Molecular Graphics System, Schrödinger, LLC). The 3D illustrations of the ligand-protein interactions were rendered in PyMOL.
4.3. Enzymatic assay
A fluorogenic assay was established based on previous protocol with minor modifications. Briefly, 30 μL of serially diluted compounds in 20 mM Tris buffer (pH 8.0) containing 10 mM dithiothreitol (DTT), 50 μM Z-RLRGG-AMC, 150 mM NaCl, and 2% DMSO was transferred to Corning 96-well half area, non-binding black polystyrene plates and incubated at 37 °C for 10 min prior to the proteolysis reaction. The reaction was initiated by the addition of 30 μL of recombinant SARS-CoV-2 PLpro at a final concentration of 50 nM and kept at a constant temperature of 37 °C. The fluorescence intensity at ex/em 365/460 nm was recorded continuously by a BioTek synergy H1 microplate reader. The increase in fluorescence intensity during the initial linear phase of the reaction (first 7 min) was analysed using linear regression to determine the initial velocities. The IC50 values were calculated by the equation vi = v0/(1+[I]/IC50) using the enzyme kinetics module of Sigma plot 14.0, where vi refers to the initial velocities in the presence of inhibitor, v0 refers to the initial velocities in the absence of inhibitor, and [I] is the concentration of inhibitor. The inhibition constant (K i) was determined by Dixon plot analysis, in which reciprocal of initial velocity vs. varying concentrations of biflavones was plotted under three different substrate concentrations (25 μM, 50 μM and 100 μM), and the intersection of the lines was taken as K i.
4.4. Pro-ISG15 cleavage assay
SARS-CoV-2 PLpro at a final concentration of 50 nM was added to recombinant human pro-ISG15 (final concentration 350 nM) solution in the presence or absence of biflavones. The reaction buffer was 20 mM Tris (pH 8.0) containing 10 mM DTT and 150 mM NaCl. After a 10 min incubation at 37 °C, the deISGylation reaction was terminated by the addition of 4X LDS loading buffer, followed by heating at 70 °C for 10 min and finally resolving by SDS-PAGE. The protein bands were stained with eLuminol protein gel stain (Genecopoeia, MD, USA), visualized under a UV transilluminator and quantified by ImageLab 3.0 software (Bio–Rad, CA, USA).
4.5. Statistical analysis
All of the fluorogenic measurements were performed in triplicate, and the blotting experiments were performed in duplicate. The IC50, K i and inhibition rate of pro-ISG15 cleavage are expressed as the mean ± standard error (SD) if not otherwise specified.
Author contributions
Lingyu Li performed all experiments and drafted the manuscript; Liyan Ma performed part of the activity determination work; Yue Hu and Xiaoxue Li participated in the collection and preparation of samples; Meng Yu and Hai Shang participated in the virtual screening process; Zhongmei Zou designed the study and revised the manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors are grateful to the financial support of the National Mega-Project for Innovative Drugs (2019ZX09735002).
References
- Cho J.K., Curtis M.J., Lee K.H., Kim D.W., Ryu H.W., Yuk H.J., Park K.H. Geranylated flavonoids displaying SARS-CoV papain-like protease inhibition from the fruits of Paulownia tomentosa. Bioorg. Med. Chem. 2013;21:3051–3057. doi: 10.1016/j.bmc.2013.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitas B.T., Durie I.A., Murray J., Longo J.E., Miller H.C., Crich D., Hogan R.J., Tripp R.A., Pegan S.D. Characterization and noncovalent inhibition of the deubiquitinase and deISGylase activity of SARS-CoV-2 papain-like protease. ACS Infect. Dis. 2020;6:2099–2109. doi: 10.1021/acsinfecdis.0c00168. [DOI] [PubMed] [Google Scholar]
- Ghosh S., Nie A.H., An J., Huang Z.W. Structure-based virtual screening of chemical libraries for drug discovery. Curr. Opin. Chem. Biol. 2006;10:194–202. doi: 10.1016/j.cbpa.2006.04.002. [DOI] [PubMed] [Google Scholar]
- Hu B., Guo H., Zhou P., Shi Z.L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021;19:141–154. doi: 10.1038/s41579-020-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu K., Guan W.J., Bi Y., Zhang W., Li L.J., Zhang B.L., Liu Q.Q., Song Y.L., Li X.W., Duan Z.P., Zheng Q.S., Yang Z.F., Liang J.Y., Han M.F., Ruan L.G., Wu C.M., Zhang Y.T., Jia Z.H., Zhong N.S. Efficacy and safety of Lianhuaqingwen capsules, a repurposed Chinese herb, in patients with coronavirus disease 2019: a multicenter, prospective, randomized controlled trial. Phytomedicine. 2021;85:153242. doi: 10.1016/j.phymed.2020.153242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.W., Seo K.H., Curtis Long M.J., Oh K.Y., Oh J.W., Cho J.K., Lee K.H., Park K.H. Phenolic phytochemical displaying SARS-CoV papain-like protease inhibition from the seeds of Psoralea corylifolia. J. Enzym. Inhib. Med. Chem. 2014;29:59–63. doi: 10.3109/14756366.2012.753591. [DOI] [PubMed] [Google Scholar]
- Klemm T., Ebert G., Calleja D.J., Allison C.C., Richardson L.W., Bernardini J.P., Lu B.G., Kuchel N.W., Grohmann C., Shibata Y. Mechanism and inhibition of the papain-like protease, PLpro, of SARS-CoV-2. EMBO J. 2020;39 doi: 10.15252/embj.2020106275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G.Q., Lee J.H., Parker Z.M., Acharya D., Chiang J.J., van Gent M., Riedl W., Davis-Gardner M.E., Wies E., Chiang C. ISG15-dependent activation of the sensor MDA5 is antagonized by the SARS-CoV-2 papain-like protease to evade host innate immunity. Nat. Microbiol. 2021;6:467–478. doi: 10.1038/s41564-021-00884-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L.M., Wang Y.T., Zhang J.C., Wang S.F. Advances in the chemical constituents and chemical analysis of Ginkgo biloba leaf, extract, and phytopharmaceuticals. J. Pharmaceut. Biomed. Anal. 2021;193:113704. doi: 10.1016/j.jpba.2020.113704. [DOI] [PubMed] [Google Scholar]
- Lu Y.H., Liu Z.Y., Wang Z.T., Wei D.Z. Quality evaluation of Platycladus orientalis (L.) Franco through simultaneous determination of four bioactive flavonoids by high-performance liquid chromatography. J. Pharmaceut. Biomed. Anal. 2006;41:1186–1190. doi: 10.1016/j.jpba.2006.02.054. [DOI] [PubMed] [Google Scholar]
- Park J.Y., Jeong H.J., Kim J.H., Kim Y.M., Park S.J., Kim D., Park K.H., Lee W.S., Ryu Y.B. Diarylheptanoids from Alnus japonica inhibit papain-Like protease of severe acute respiratory syndrome coronavirus. Biol. Pharm. Bull. 2012;35:2036–2042. doi: 10.1248/bpb.b12-00623. [DOI] [PubMed] [Google Scholar]
- Park J.Y., Kim J.H., Kim Y.M., Jeong H.J., Kim D.W., Park K.H., Kwon H.J., Park S.J., Lee W.S., Ryu Y.B. Tanshinones as selective and slow-binding inhibitors for SARS-CoV cysteine proteases. Bioorg. Med. Chem. 2012;20:5928–5935. doi: 10.1016/j.bmc.2012.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.Y., Ko J.A., Kim D.W., Kim Y.M., Kwon H.J., Jeong H.J., Kim C.Y., Park K.H., Lee W.S., Ryu Y.B. Chalcones isolated from Angelica keiskei inhibit cysteine proteases of SARS-CoV. J. Enzym. Inhib. Med. Chem. 2016;31:23–30. doi: 10.3109/14756366.2014.1003215. [DOI] [PubMed] [Google Scholar]
- Park J.Y., Yuk H.J., Ryu H.W., Lim S.H., Kim K.S., Park K.H., Ryu Y.B., Lee W.S. Evaluation of polyphenols from Broussonetia papyrifera as coronavirus protease inhibitors. J. Enzym. Inhib. Med. Chem. 2017;32:504–512. doi: 10.1080/14756366.2016.1265519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratia K., Pegan S., Takayama J., Sleeman K., Coughlin M., Baliji S., Chaudhuri R., Fu W., Prabhakar B.S., Johnson M.E. A noncovalent class of papain-like protease/deubiquitinase inhibitors blocks SARS virus replication. Proc. Natl. Acad. Sci. U. S. A. 2008;105:16119–16124. doi: 10.1073/pnas.0805240105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y., Lantvit D.D., Carcache de Blanco E.J., Kardono L.B.S., Riswan S., Chai H., Cottrell C.E., Farnsworth N.R., Swanson S.M., Ding Y., Li X., Marais J.P.J., Ferreira D., Kinghorn A.D. Proteasome-inhibitory and cytotoxic constituents of Garcinia lateriflora: absolute configuration of caged xanthones. Tetrahedron. 2010;66:5311–5320. doi: 10.1016/j.tet.2010.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo O.N., Kim G.S., Park S., Lee J.H., Kim Y.H., Lee W.S., Lee S.J., Kim C.Y., Jin J.S., Choi S.K. Determination of polyphenol components of Lonicera japonica Thunb. using liquid chromatography–tandem mass spectrometry: contribution to the overall antioxidant activity. Food Chem. 2012;134:572–577. doi: 10.1016/j.foodchem.2012.02.124. [DOI] [Google Scholar]
- Shang X.F., Pan H., Li M.X., Miao X.L., Ding H. Lonicera japonica Thunb.: ethnopharmacology, phytochemistry and pharmacology of an important traditional Chinese medicine. J. Ethnopharmacol. 2011;138:1–21. doi: 10.1016/j.jep.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z.N., Ratia K., Cooper L., Kong D.Y., Lee H., Kwon Y.J., Li Y.F., Alqarni S., Huang F., Dubrovskyi O. 2021. Potent, Novel SARS-CoV-2 PLpro Inhibitors Block Viral Replication in Monkey and Human Cell Cultures. bioRxiv. [DOI] [Google Scholar]
- Shin D.H., Mukherjee R., Grewe D., Bojkova D., Baek K.W., Bhattacharya A., Schulz L., Widera M., Mehdipour A.R., Tascher G., Geurink P.P., Wilhelm A., van der Heden van Noort G.J., Ovaa H., Müller S., Knobeloch K.P., Rajalingam K., Schulman B.A., Cinatl J., Hummer G., Ciesek S., Dikic I. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature. 2020;587:657–662. doi: 10.1038/s41586-020-2601-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son K.H., Park J.O., Chung K.C., Chang H.W., Kim H.P., Kim J.S., Kang S.S. Flavonoids from the aerial parts of Lonicera japonica. Arch Pharm. Res. (Seoul) 1992;15:365–370. doi: 10.1007/BF02974114. [DOI] [Google Scholar]
- Song Y.H., Kim D.W., Curtis Long M.J., Yuk H.J., Wang Y., Zhuang N., Lee K.H., Jeon K.S., Park K.H. Papain-like protease (PLpro) inhibitory effects of cinnamic amides from Tribulus terrestris fruits. Biol. Pharm. Bull. 2014;37:1021–1028. doi: 10.1248/bpb.b14-00026. [DOI] [PubMed] [Google Scholar]
- Wang H., Xu B., Zhang Y., Duan Y., Gao R., He H., Li X., Li J. Efficacy and safety of traditional Chinese medicine in coronavirus disease 2019 (COVID-19): a systematic review and meta-analysis. Front. Pharmacol. 2021;12 doi: 10.3389/fphar.2021.609213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei P.F. Diagnosis and treatment protocol for novel coronavirus Pneumonia (trial version 7) Chin. Med. J. (Engl.) 2020;133:1087–1095. doi: 10.1097/cm9.0000000000000819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.Y., Jan J.T., Ma S.H., Kuo C.J., Juan H.F., Cheng Y.S.E., Hsu H.H., Huang H.C., Wu D., Brik A., Liang F.S., Liu R.S., Fang J.M., Chen S.T., Liang P.H., Wong C.H. Small molecules targeting severe acute respiratory syndrome human coronavirus. Proc. Natl. Acad. Sci. U.S.A. 2004;101:10012–10017. doi: 10.1073/pnas.0403596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Islam M.S., Wang J., Li Y., Chen X. Traditional Chinese medicine in the treatment of patients infected with 2019-new coronavirus (SARS-CoV-2): a review and perspective. Int. J. Biol. Sci. 2020;16:1708–1717. doi: 10.7150/ijbs.45538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh P., Shieh Y., Hsu L., Kuo L., Lin J., Liaw C., Kuo Y. Naturally occurring cytotoxic [3′→8″]-Biflavonoids from Podocarpus nakaii. J. Tradit. Complement. Med. 2012;2:220–226. doi: 10.1016/S2225-4110(16)30103-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X., Riva L., Pu Y., Martin-Sancho L., Kanamune J., Yamamoto Y., Sakai K., Gotoh S., Miorin L., De Jesus P.D. MDA5 governs the innate immune response to SARS-CoV-2 in lung epithelial cells. Cell Rep. 2021;34:108628. doi: 10.1016/j.celrep.2020.108628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Cao F., Wang Y., Xu X., Sun Y., Li J., Qi X., Sun S., Ji G., Song B. The efficacy and safety of Jinhua Qinggan granule (JHQG) in the treatment of coronavirus disease 2019 (COVID-19): a protocol for systematic review and meta analysis. Medicine (Baltim.) 2020;99 doi: 10.1097/MD.0000000000020531. e20531-e20531. [DOI] [PMC free article] [PubMed] [Google Scholar]