Abstract
Background
Gallbladder carcinoma (GBC) remains a highly lethal disease worldwide. MiR-552 family members promote the malignant progression of a variety of digestive system tumors, but the role of miR-552-3p in GBC has not been elucidated. miR-552-3p was predicted to target the 3'-untranslated region (3'UTR) of the mRNA for the tumor suppressor gene “repulsive guidance molecule BMP co-receptor a” (RGMA). The aim of the present study was to clarify the roles and mechanisms of miR-552-3p targeting RGMA in the malignant progression of GBC.
Methods
In vitro: expression of miR-552-3p was detected by real-time quantitative PCR (qRT-PCR) in tumor and non-tumor adjacent tissues (NATs). Lentivirus-miR-552-3p was employed to knockdown this miRNA in GBC cell lines. Stem cell-related transcription factors and markers were assessed by qRT-PCR. Cell Counting Kit-8 (CCK-8), sphere formation and transwell assays were used to determine the malignant phenotypes of GBC cells. Targeting the 3'UTR of RGMA by miR-552-3p was verified by integrated analysis including bioinformatics prediction, luciferase assays, measures of changes of gene expression and rescue experiments. In vivo: mouse models of subcutaneous tumors and lung metastases were established to observe the effect of miR-552-3p on tumorigenesis and organ metastasis, respectively.
Results
MiR-552-3p was abnormally highly expressed in GBC tissues and cancer stem cells. Interference with miR-552-3p in SGC-996 and GBC-SD cells significantly inhibited GBC stem cell expansion. Reciprocally, miR-552-3p promoted GBC cell proliferation, migration and invasion both in vitro and in vivo; hence, interference with this miRNA impeded the malignant progression of GBC. Furthermore, the important tumor suppressor gene RGMA was identified as a target of miR-552-3p. The effects of miR-552-3p on cell proliferation and metastasis were abrogated or enhanced by gain or loss of RGMA function, respectively. Mechanistically, miR-552-3p promoted GBC progression by reactivating the Akt/β-catenin pathway and epithelial-mesenchymal transformation (EMT). Clinically, miR-552-3p correlated with multi-malignant characteristics of GBC and acted as a prognostic marker for GBC outcome.
Conclusions
MiR-552-3p promotes the malignant progression of GBC by inhibiting the mRNA of the tumor suppressor gene RGMA, resulting in reactivation of the Akt/β-catenin signaling pathway.
Keywords: Gallbladder carcinoma (GBC), stemness, miR-552-3p, repulsive guidance molecule BMP co-receptor a (RGMA), epithelial-mesenchymal transformation (EMT)
Introduction
GBC is the most common cancer of the gallbladder, the incidence of which has been rising rapidly recently. It has become a major malignant disease affecting people’s health (1). Unfortunately, most patients with GBC are at advanced stage at the time of diagnosis (2). The most effective treatment is surgical removal of the tumor, but unfortunately, 40–75% of those patients have metastases at diagnosis, with only 10% of patients have resectable tumors (1,3). The early clinical features of gallbladder carcinoma are similar to cholecystitis and the low incidence rate of this tumor has resulted in insufficient attention being paid to this cancer type. In addition, a lack of gallbladder serosa adjacent to the liver facilitates hepatic invasion and metastatic progression, a leading cause of its dismal outcome (4). Because research into this disease has long been limited, studying its biological mechanisms is essential for improving means of prevention, early detection, and treatment of this fatal disease.
Cancer stem cells (CSCs) are cells with self-renewing capacity and multilineage differentiation potential, which are closely related to malignant biological behavior such as invasion, metastasis, and chemoresistance (5,6). Some studies have shown that GBC CSCs can be recognized on the basis of their expression of several defined surface markers such as CD44 and CD133 (7,8). Silencing CD44 in vitro has been reported to inhibit GBC proliferation, migration and invasion, and attenuate CSC function (9). CD133-positive GBC cells represent a subpopulation of tumor cells that initiate and sustain tumor development, having higher self-renewing capacity leading to higher tumorigenicity and chemoresistance (10). CD44+ and CD133+ populations exhibit CSC-like characteristics in human GBC (11). Accumulating evidence indicates that recurrence, metastasis, tumor grade, and chemoresistance of GBC patients are all closely associated with the expression of CSC markers such as CD44 and CD133 on GBC cells (12,13).
MicroRNAs (miRNAs) are small noncoding RNAs 19–25 nt in length which negatively regulate the expression of target genes by specifically binding to their mRNA 3'-untranslated region (UTR). It has been known for many years that miRNAs are critical regulators of tumor biology, which makes them attractive tools and targets for cancer treatment. They have been the subject of intensive research for the past 20 years (14). Several studies have reported important roles for miRNAs in CSC regulation. CD133 is involved in signaling pathways and miRNA regulation in CSCs (15). MiR-106b was reported to modulate CSC characteristics via TGF-β/Smad signaling in CD44-positive gastric cancer cells (16). MiR-136 enhances the antitumor effect of paclitaxel in chemoresistant ovarian cancer cells by inhibiting tumor stem cell activity via targeting Notch3 (17). MiR-135a was found to inhibit CSC-driven medulloblastoma development by directly repressing Arhgef6 expression (18).
Previously, miR-552 was reported to act as an oncogene in various different tumors, including hepatocellular carcinoma, gastric cancer, colon cancer and laryngeal cancer by promoting cell cycle progression, proliferation, invasion, and migration (19-22). MiR-552-3p is a mature form of miR-552, highly expressed in gastric adenocarcinoma, colon adenocarcinoma, and esophageal cancer according to the results of a pan-cancer screening study using the Illumina hi-seq system (23). Wei et al. analyzed miRNA-seq data and found that miR-552-3p expression was up-regulated nearly 3.6-fold in gastric cancer tissue compared with normal tissues (24). However, whether miR-552-3p is involved in the development of GBC remains unknown.
In the present study, we found that miR-552-3p is highly expressed in gallbladder CSCs and GBC tumor tissues. Using gain-and loss-of function analysis in GBC cell line, we demonstrated that miR-552-3p promotes stemness, tumorigenicity, malignant proliferation and metastasis of GBC cells. Intriguingly, gene enrichment pathway analysis of miR-552-3p targets showed that the Wnt and cadherin signaling pathways were most affected. Combined with bioinformatics analysis, expression detection, and target identification, we confirmed that the important tumor suppressor gene repulsive guidance molecule BMP co-receptor a (RGMA) is a target gene of miR-552-3p in GBC. Further mechanistic studies revealed that by inhibiting the expression of RGMA, miR-552-3p reactivates Akt/β-catenin and epithelial-mesenchymal transformation (EMT) signaling pathways, and finally promotes the occurrence and development of GBC.
We present the following article in accordance with the ARRIVE reporting checklist (available at https://dx.doi.org/10.21037/atm-21-2013).
Methods
Cell lines and cell culture
The human GBC cell lines GBC-SD (RRID:CVCL_6903) and SGC-996 (RRID:CVCL_M737) were purchased from Chinese Academy of Sciences (Shanghai, China). The GBC-SD and SGC-996 cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) (Gibco, NY, USA) and 2 mM L-glutamine, and 25 µg/mL of gentamicin and maintained at 37 °C in a humidified atmosphere with 5% CO2. According to the cell line verification test recommendations, cell lines were validated by microscopy, growth curve analysis and mycoplasma detection before the experiments.
Patients and tissue specimens
We collected a total of 41 GBC patients’ tissue samples versus non-tumor adjacent tissues (NATs) samples from the Eastern Hepatobiliary Surgery Hospital (Shanghai, China). Freshly resected tissues were frozen at −80 °C for subsequent protein extraction and RNA extraction. Formalin-immersed tissues were stored at 4 °C for Immunohistochemistry production. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethic Committee of Eastern Hepatobiliary Surgery Hospital (No. EHBHKY2020-K-016) and individual consent for this retrospective analysis was waived.
Cell transfection
Lentivirus-miR-552-3p mimic and lentivirus-miR-552-3p sponge and lentivirus miR-GFP (negative control) were purchased from Genepharma (Shanghai, China). Has-miR-552-3p mimics and negative control RNA were purchased from Riobio (Guangzhou, China). Additionally, RGMA overexpression plasmid, RGMA negative control plasmid, RGMA small interfering RNA and negative control small interfering RNA were purchased from Generalbiol (Chuzhou, China) for the rescue experiments. The miRNA mimics, miRNA plasmids and inhibitor were transfected into SGC-996 and GBC-SD cells using transfection reagent (Polyplus Transfection, USA). The sequences of miR-552-3p mimic and RGMA siRNA are showed in Table S1.
Quantitative real-time PCR
Total RNA was extracted from the above tissue samples or cultured cells with TRIzol reagent (Invitrogen, Carlsbad, CA, USA), and the complementary DNA template was prepared with oligo (dT) random primers or miRNA RT primers and M-MLV (Moloney murine leukemia virus) reverse transcriptase (Promega) according to the manufacturer’s protocol. RNA expression was measured by qRT-PCR with SYBR® Green (Takara, Dalian, China). GAPDH and U6 served as internal controls for the mRNA levels and miRNA levels respectively. Relative RNA expression levels were quantified with the 2−ΔΔCt method. The sequences of the primers used here are listed in Table S2.
Flow cytometric analysis
The GBC cells were incubated with the primary anti-CD44 (Proteintech Cat#15675-1-AP, RRID:AB_2076198) or anti-CD133 (Proteintech Cat# 18470-1-AP, RRID:AB_2172859) for 30 min at room temperature, then washed 3 times with PBS. Diluted secondary antibody (BioLegend, Cat#406414, RRID:AB_2563202) was added to the samples, and then incubated for 45 min at room temperature. One milliliter washing buffer was added and centrifuged at 1,000 rpm/min for 5 min, and repeat the washing once. The cells were resuspended with 300 µL 1× PBS buffer, and the results were analyzed by a MoFlo XDP cell sorter (Beckman Coulter, Indianapolis, IN, USA) according to the manufacturer’s instructions.
Spheroid formation assay
GBC-SD and SGC-996 cells were cultured in a 6-well ultra-low attachment culture plate (Invitrogen, Carlsbad, CA, USA) for 7 days, and the total numbers of spheres were counted under the microscope.
In vitro limiting dilution assay
Different numbers of GBC-SD or SGC-996 miR-552-3p sponge and their control cells were seeded into 96 well ultra-low adhesion plates and cultured in a 5% CO2 incubator for 7 days. CSC proportions were analyzed using Poisson distribution statistics and the L-Calc version 1.1 software program (Stem Cell Technologies, Inc., Vancouver, Canada) as described (25).
Cell proliferation assays
For cell proliferation analysis, GBC cells were seeded in 96-well plates (3×103 cells per well). ATP activity was measured by a Cell Counting Kit-8 (CCK-8) at indicated time points. For cell EdU immunofluorescence staining, GBC cells were seeded into 96-well plates (3×103 cells) and performed using the EdU Kit (RiboBio). The results were quantified with a Zeiss axiophot photomicroscope (Carl Zeiss) and Image-Pro plus 6.0 software. For colony formation assay, the GBC cells were seeded in 12-well plates (0.8×103 cells/well), and then incubated at 37 °C for 10 days. After that the cells were fixed with 10% neutral formalin for 15 min and stained with crystal violet (Beyotime, Haimen, China) for 15 min. Finally, the results were photographed under a digital camera.
Wound-healing assay
For the wound-healing assay, GBC-SD and SGC-996 cells were seeded in 6-well plates. When the cells had reached 90% confluence, a 20-µL sterile micropipette tip was used to form a 2-mm wide strip across the well. The cells migrating into the wounded areas were visualized and photographed at a time during 0 to 72 h after washing the well with PBS.
Cell migration assay
For cell migration experiments, GBC cells (GBC-SD and SGC-996 are 1×105 and 2×105 cells /mL, 200 µL per well, respectively) were seeded into the upper chamber of a polycarbonate transwell in serum-free DMEM. The lower chamber was added with DMEM containing 10% FBS as chemoattractant. The cells were incubated for 24 h and the chamber was fixed. Cell count is expressed as the average number of the cells in each field.
Cell invasion assay
For cell invasion experiments, GBC cells (GBC-SD and SGC-996 are 1×105 and 2×105 cells/mL, 200 µL per well, respectively) were seeded into the upper Matrigel. The concentration of working solution is 200 µg/mL (Coring® Matrigel Matrix, #354234). A total of 100 µL per well of the diluted Matrigel matrix was carefully added to the center of each upper chamber. Next, coated the chamber of polycarbonate transwell in serum-free DMEM. The lower chamber was added with DMEM containing 10% FBS as chemoattractant. The cells were incubated for 24 h and the chamber was fixed. Cell count is expressed as the average number of the cells in each field.
Western blotting assay
The GBC cells were collected with cell lysis buffer, then disposed like we described before (26). Thirty micrograms protein of cell extracts were subjected to Western blot with the indicated antibodies. The protein band, specifically bound to the primary antibody, was detected using an IRDye 800CW-conjugated secondary antibody and LI-COR imaging system (LI-COR Biosciences, Lincoln, NE, USA). The primary antibodies were RGMA (Abcam, Cat# ab169761, RRID:AB_2885097), p-Akt (Cell Signaling Technology, Cat# 4060, RRID:AB_2315049), Akt (Cell Signaling Technology, Cat# 4685, RRID:AB_2225340), E-cadherin (Proteintech, Cat# 60335-1-Ig, RRID:AB_2881444), vimentin (Proteintech, Cat# 10366-1-AP, RRID: AB_2273020), β-actin (Cell Signaling Technology, Cat# 3700, RRID:AB_2242334) and GAPDH (Cell Signaling Technology, Cat# 5174, RRID:AB_10622025).
Luciferase reporter assay
A 1,000-bp fragment of the RGMA 3'-UTR containing miR-552-3p-binding sites was inserted into a luciferase reporter plasmid (OBIO technology, Shanghai, China), and a synthetic RGMA 3'-UTR mutant fragment was inserted into an equivalent reporter plasmid. For the luciferase assays, HEK-293T (ATCC, Cat# CRL-3216, RRID:CVCL_0063) cells were seeded in 6-well plates in triplicate and allowed to settle for 24 h. The cells were then co-transfected with 10 ng of firefly luciferase reporter plasmid and an equal amount (50 pmol) of miR-552-3p mimics or scrambled negative control RNA using transfection reagent (Polyplus Transfection, USA). Forty-eight hours after transfection, the cells were assayed using a luciferase assay kit (Promega, Madison, WI, USA).
Immunohistochemistry
Immunohistochemistry was performed using the anti-RGMA (1:200; R&D Systems, Cat# AF2459, RRID:AB_355273). The sections were then rinsed three times in PBS before blocking with 10% normal goat serum (15 min at room temperature). The sections were rinsed in PBS and incubated with a primary antibody overnight at 4 °C. Afterwards, the tissue sections were rinsed three times in PBS and incubated with the secondary antibody for 60 min at room temperature. The sections were then immersed in DAB for 5–10 min and counterstained with 10% Mayer’s hematoxylin.
Immunofluorescence
After transfections with the mimics or inhibitors, GBC cells were seeded into 24-well plates. Two days later, the cells were washed with PBS and fixed in 4% paraformaldehyde for 15 min at room temperature. Afterwards, the cells were incubated with blocking buffer (5% BSA, 0.1% Triton X-100) alone for 1 h and then incubated with the primary antibodies overnight at 4 °C. The primary antibody was removed, and cells were incubated with the secondary fluorescent antibody for 1 h at room temperature. After 3 times of PBS washing, the cells were stained with DAPI and photographed under a fluorescence microscope (Olympus, Japan).
Nude mouse subcutaneous and lung metastasis tumor models
The subcutaneous xenograft model was established using 4 to 6 weeks male BALB/C NU/NU mice. The mice were purchased from the experimental animal center of Naval Medical University (Shanghai, China). The mice were housed under specific pathogen-free conditions and fed a regular autoclaved chow diet with water ad libitum. A total of 2×106 GBC-SD cells stably expression miR-552-3p sponge and negative control (Lv-miR-552-3p sponge and Lv-miR-NC) were inoculated subcutaneously into the ventral areas of the mice (n=6 per group). The sizes of the tumors were measured every 4 days after inoculation. The sizes were evaluated using the following formula:
Tumor volume = [tumor length × (tumor width)2] ×0.5. To establish the lung metastasis model, we injected 2×106 GBC-SD miR-552-3p sponge and negative control cells via the tail vein. The mice were killed 3 months after injection, at which time the lungs were harvested, and the number of metastatic nodules in each lung was counted. Experiments were performed under a project license (NO.: EHBHKY2020-K-016) granted by the Ethic Committee of Eastern Hepatobiliary Surgery Hospital, in compliance with The Second Military Medical University guidelines for the care and use of animals.
Statistical analysis
GraphPad Prism (GraphPad Software, Inc., La Jolla, USA) was used for all statistical analyses. Data were shown as mean ± standard deviation (SD). For continuous variables, if which obey the normal distribution, Student’s t-test was used to evaluate differences between two groups of data. Otherwise, variables were compared using nonparametric test for which with an abnormal distribution. Differences between multi-groups were compared using analysis of variance (ANOVA) when applicable or a nonparametric test. The Kaplan-Meier method and log-rank test was used to evaluate the correlation between miRNA expression and patient survival. Differences in proportions were compared with the Chi-square test. For all cytological experiments, there were three replicates in each group. There were 6 replicates and 5 replicates per group in nude mouse subcutaneous and lung metastasis tumor models, respectively (*, P<0.05; **, P<0.01; ***, P<0.001; ****, P<0.0001). P values <0.05 were considered statistically significant.
Results
MiR-552-3p is highly expressed in gallbladder carcinoma tissues and CSCs
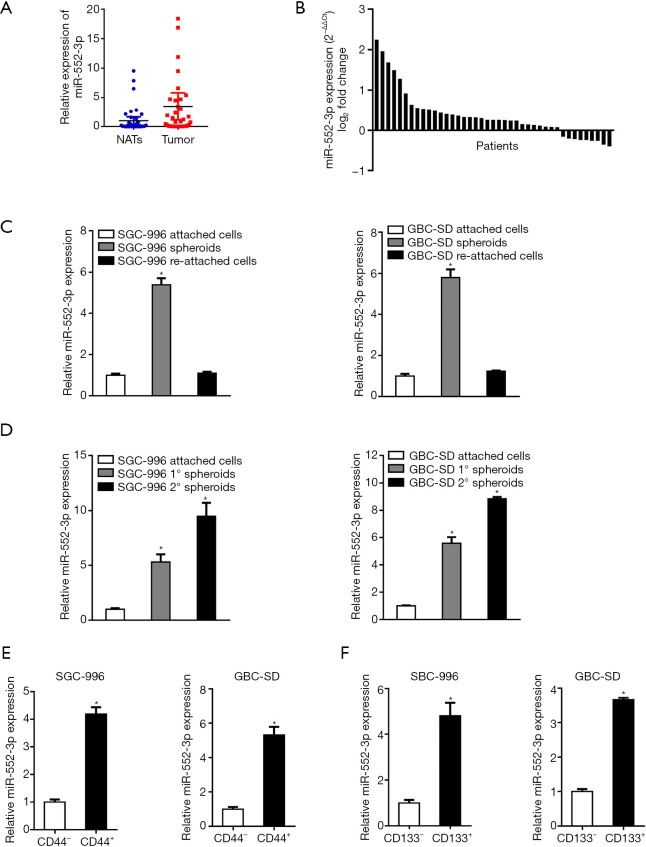
According to qRT-PCR results, miR-552-3p is markedly upregulated in GBC tissues compared with NATs (Figure 1A) with relatively high expression in 32 out of a total of 41 cases (P<0.05; Figure 1B). Furthermore, levels of miR-552-3p expression were elevated in spheroids formed by SGC-996 and GBC-SD cells relative to 2D-cultured cells (Figure S1A), indicating that miR-552-3p might correlate with the stemness of GBC cells. Next, we enriched CSCs by sphere formation assays. As shown in Figure1C, the expression of miR-552-3p was upregulated in the self-renewing spheroids, but then down-regulated again when GBC cells re-attached to a flat surface. Moreover, upon continued serial passaging of GBC spheroids, the expression of miR-552-3p gradually increased with the number of passages (Figure 1D).
Figure 1.
MiR-552-3p is highly expressed in GBC tissues and CSCs (n=41). (A) The relative miR-552-3p expression levels in GBC tissues and non-tumor adjacent tissues (NATs); (B) the relative expression level of miR-552-3p was detected in 41 patients; (C) the expression of miR-552-3p in spheroids and reattached cells was compared by q-PCR; (D) real-time PCR was used to detect the expression of miR-552-3p in passaged GBC spheroids; (E) real-time PCR was used to analyze the expression of miR-552-3p in CD44+ GBC cells sorted by flow cytometry relative to CD44− GBC cells; (F) real-time PCR was used to analyze the expression of miR-552-3p in CD133+ GBC cells sorted by flow cytometry relative to CD133− GBC cells. *, P<0.05. GBC, gallbladder cancer; CSCs, cancer stem cells; NATs, non-tumor adjacent tissues.
To further explore the expression of miR-552-3p in gallbladder CSCs, CD44+/CD133+ GBC cells were sorted by flow cytometry. As shown in Figure 1E,1F, the expression of miR-552-3p in the CD44+/CD133+ GBC CSC population was higher than that in the corresponding CD44−/CD133− cells.
In summary, the above results indicate that miR-552-3p is an abnormally highly-expressed miRNA in GBC tissues and CSCs.
MiR-552-3p promotes GBC CSC expansion
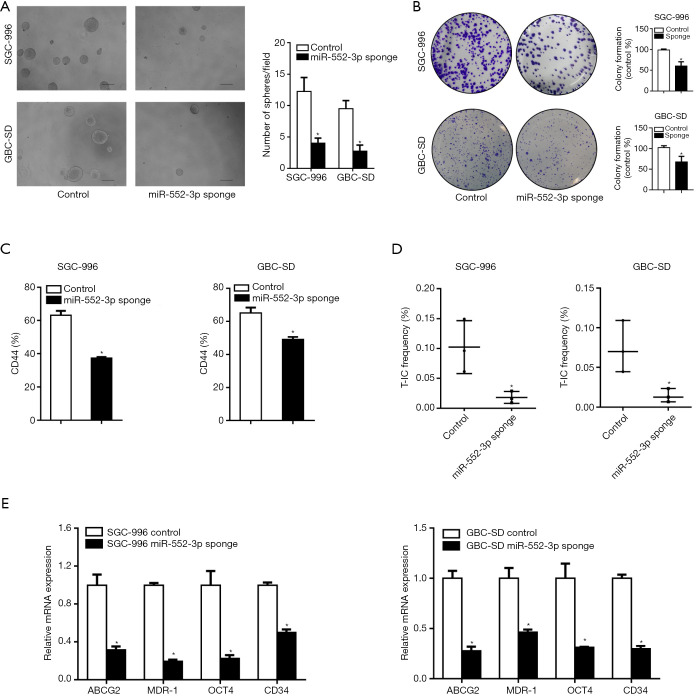
To determine the function of miR-552-3p in GBC CSCs, we constructed GBC cell lines in which miR-552-3p was stably affected, using a lentiviral system (Figure S1B). Fewer spheroids were formed by GBC cells in which miR-552-3p expression was interfered, relative to controls (Figure 2A). Colony formation assays also showed that interference with miR-552-3p expression significantly reduced colony formation by GBC cells (Figure 2B). Moreover, interference with miR-552-3p expression decreased the proportion of CD44+ cells detected by flow cytometry (Figure 2C). Additionally, in vitro limiting dilution assays showed that suppression of miR-552-3p significantly reduced CSC frequency (Figure 2D). Consistently, the expression of GBC stemness-associated transcription factors and CSC markers was also suppressed in cells in which miR-552-3p had been interfered (Figure 2E).
Figure 2.
MiR-552-3p promotes GBC stem cells expansion. (A) The representative images of spheres formed by miR-552-3p knockdown and miR-552-3p control GBC cells (scale bar =100 μm); (B) cloning ability of miR-552-3p knockdown and control GBC cells (dyeing with crystal violet); (C) the percentage of CD44+ in miR-552-3p knockdown and control GBC cells was analyzed by flow cytometry; (D) the proportion of CSC was determined by in vitro limited dilution assay; (E) the expression of stem cell related transcription factors and CSCs markers in miR-552-3p knockdown and control GBC cells were detected by real-time PCR. *, P<0.05. GBC, gallbladder cancer; CSCs, cancer stem cells; T-IC, tumor-initiating cell.
Thus, the above results indicate that miR-552-3p promotes GBC CSC expansion.
MiR-552-3p acts as a key regulator of GBC malignant phenotypes in vitro and in vivo
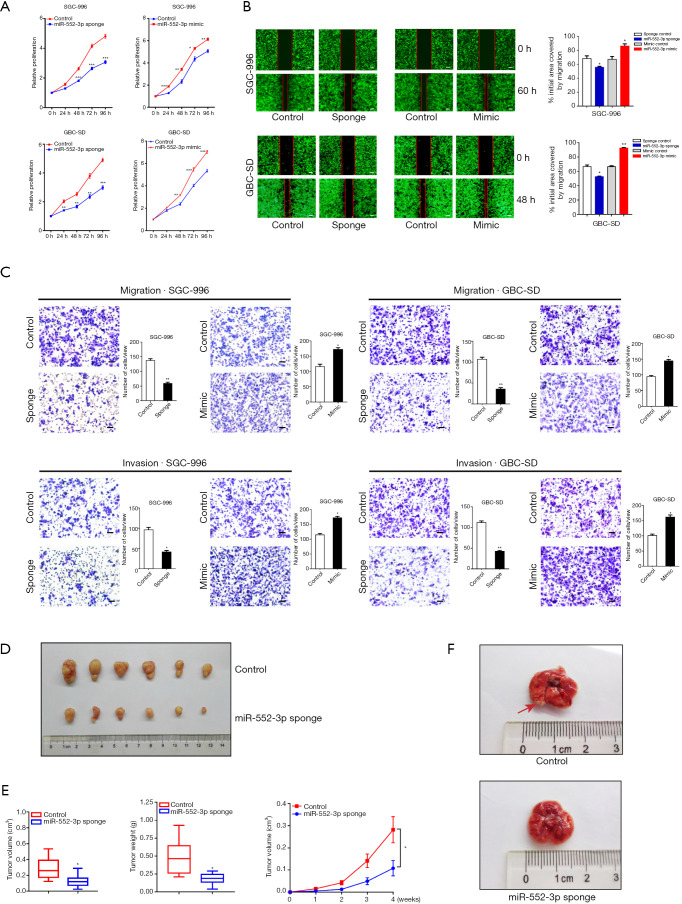
To elucidate the effects of miR-552-3p on GBC cell function in more detail, a series of experiments were performed to determine its effects on malignant phenotypes. Proliferation assays using the CCK-8 showed that interference with miR-552-3p expression reduced GBC cell proliferation. Conversely, its overexpression increased the number of viable cells, and increased proliferation (Figure 3A). Consistent with this, 5-ethynyl-2'-deoxyuridine (EdU) staining confirmed that miR-552-3p interference inhibited GBC cell proliferation (Figure S1C).
Figure 3.
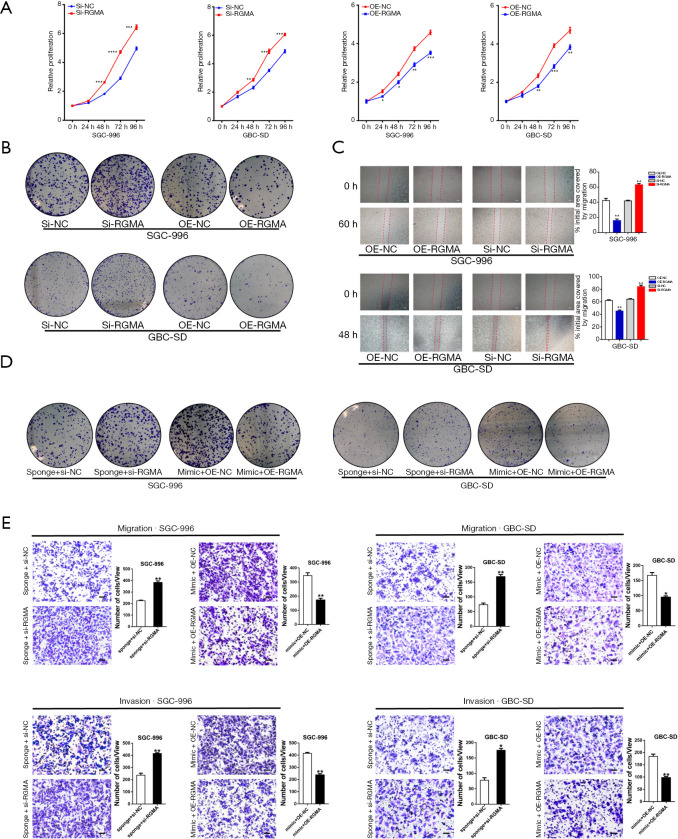
MiR-552-3p acts as a key regulator for GBC malignant phenotypes in vitro and in vivo. (A) The proliferation of miR-552-3p knockdown or overexpression in GBC cells was measured by CCK-8 assay; (B) the migratory ability of miR-552-3p knockdown or overexpression in GBC cells was measured by wound-healing assay (scale bar =500 μm); (C) cell migration and invasion were evaluated in GBC cells with miR-552-3p knockdown or overexpression (dyeing with crystal violet; scale bar =100 μm); (D) the GBC-SD cells stably expressing miR-552-3p sponge or sponge NC (Lv-miR-NC) were subcutaneously injected into nude mice (n=6). Xenografted tumor weight was measured 4 weeks later; (E) the size and weight and production situation of subcutaneous tumor were measured; (F) the representative image of tumor metastasis images of nude mice was injected with miR-552-3p sponge (Lv-miR-552-3p sponge) or sponge NC (Lv-miR-NC) cells. The red arrow indicates a metastatic gallbladder carcinoma in the lung. Compared with the control group, *, P<0.05; **, P<0.01; ***, P<0.001. GBC, gallbladder cancer; CCK-8, Cell Counting Kit-8.
To explore the effects of miR-552-3p on GBC cell migration and metastatic capability, the migratory potential of cells in which miR-552-3p was either silenced or overexpressed was analyzed in the wound healing assay. The degree of wound closure in this assay with cells overexpressing miR-552-3p was significantly greater than in controls at 48 and 60 h, whereas inhibition of miR-552-3p expression using a miRNA sponge had the reverse effect (Figure 3B). To further clarify the effect of proliferation on migration and metastatic capability, we complemented the wound-healing assay using mitomycin C to inhibit GBC cell proliferation. We found that under the background of inhibiting cell growth, miR-552-3p can effectively promote the migration ability of GBC cells (Figure S1D). Additionally, transwell migration and invasion assays showed that downregulation of miR-552-3p significantly decreased the migration and invasion capabilities of GBC cells. Conversely, overexpression of miR-552-3p significantly promoted GBC cell migration and invasion (Figure 3C).
To further determine the effects of miR-552-3p on GBC cell proliferation, migration and invasion in vivo, we used GBC-SD cells with stably decreased expression of miR-552-3p in a subcutaneous tumor-bearing mouse model and in a lung metastasis model following caudal vein tumor cell injection in nude mice. In the first model, the size of subcutaneous tumors was measured and recorded every week. After 4 weeks, the mice were sacrificed and the volumes and weights of xenografted tumors assessed. This revealed that tumors were smaller and of lower weight in animals with tumors lacking miR-552-3p (Figure 3D,3E). In the second model, more lung metastases appeared in the miR-552-3p controls compared with the miR-552-3p sponge-treated mice (Figure 3F).
Collectively, the above results indicate that miR-552-3p has a positive regulatory effect on GBC cell migration and invasion in vitro and in vivo.
RGMA is direct target of miR-552-3p in GBC
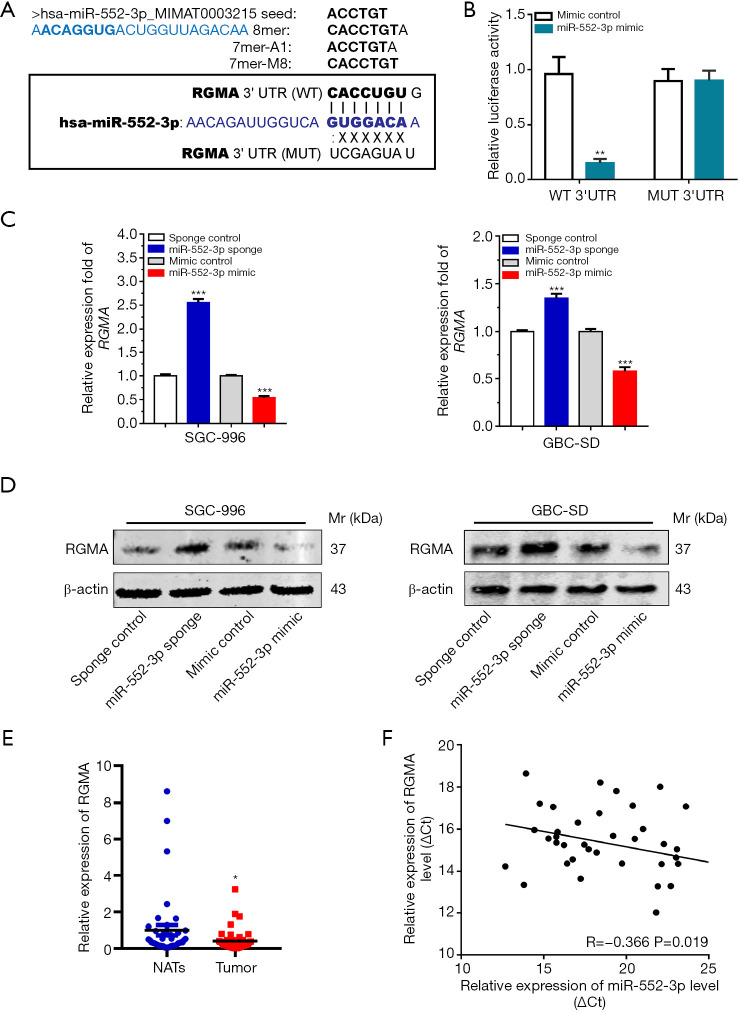
MiRNAs generally function by binding to the 3'UTR of their target gene products, leading to their degradation or otherwise inhibiting translation (27). Therefore, we analyzed miR-552-3p target genes and verified their functions. First, we identified potential target genes of miR-552-3p by searching the TargetScan database (http://www.targetscan.org) and the RNA22 database (https://cm.jefferson.edu/rna22/Interactive/) (28). We then analyzed the function of the possible target genes using the GeneCard database (www.genecards.org). We found that, among the top ten possible target genes scored, there was an important tumor suppressor gene called RGMA (Figure 4A).
Figure 4.
RGMA is direct target of miR-552-3p in GBC. (A) Prediction of miRNA target genes based on TargetScan database and RNA22 tool; (B) luciferase reporter assay was performed in 293T cells co-transfected with miR-552-3p mimics and pMIR-REPORT luciferase-RGMA 3'UTR (WT) or pMIR-REPORT luciferase-RGMA 3'UTR (MUT); (C) qRT-PCR analyses of RGMA expression levels in SGC-996 and GBC-SD cells transfected with miR-sponge-NC and miR-552-3p sponge or miR-mimic-NC and miR-552-3p mimic; (D) Western blotting analyses of RGMA expression levels in SGC-996 and GBC-SD cells transfected with miR-sponge-NC and miR-552-3p sponge or miR-mimic-NC and miR-552-3p mimc; (E) real-time PCR analysis of the relative expression of RGMA between nontumor and tumor group; (F) the correlation between miR-552-3p expression levels and RGMA expression levels was determined by linear regression analysis (P=0.019, R=−0.366, Pearson’s correlation coefficient). Compared with the control or NATs group, *, P<0.05; **, P<0.01; ***, P<0.001. GBC, gallbladder cancer; RGMA, repulsive guidance molecule BMP co-receptor a; UTR, untranslated region; Mr, molecular mass.
We then cloned RGMA 3'-UTRs containing wild-type or mutant miR-552-3p-binding sites (Figure 4A) into luciferase reporter plasmids. Dual luciferase reporter assays showed that cells transfected with miR-552-3p mimics specifically inhibited RGMA-3'-UTR-WT luciferase reporter activity but not RGMA-3'-UTR-MUT reporter activity (Figure 4B). Moreover, qRT-PCR and Western blotting results also showed that miR-552-3p suppressed RGMA mRNA and protein expression in GBC cells (Figure 4C,4D).
Subsequently, RGMA was found to be more weakly expressed in cancer tissues than in NATs (Figure 4E). Moreover, miR-552-3p expression and RGMA expression were significantly negatively correlated with one another in GBC tissues (Figure 4F). In addition, immunohistochemistry of GBC tissues and NATs also confirmed that RGMA was under-expressed in GBC tissues (Figure S2A). As further evidence for the relationship between miR-552-3p and RGMA, significant negative correlations between the two were also found in esophageal carcinoma, colon adenocarcinoma, stomach adenocarcinoma and lung adenocarcinoma tumor samples (Figure S2B).
Taken together, these results indicate that miR-552-3p negatively regulates RGMA expression in GBC by directly targeting its 3'-UTR region.
Gain or loss of RGMA function abrogates or enhances the impact of miR-552-3p in GBC cells, respectively
To determine whether RGMA plays a crucial role in miR-552-3p-induced alterations in GBC cell proliferation and metastasis, we introduced the RGMA gene or siRNA into SGC-996 and GBC-SD cells, and then evaluated functional alterations in these cells.
The CCK-8 assay showed that RGMA overexpression attenuated GBC cell proliferation relative to the control group. Conversely, inhibiting RGMA expression with siRNA increased the number of viable cells and increased their proliferative capacity (Figure 5A). RGMA overexpression prevented tumor cell colony formation, whereas RGMA interference induced it (Figure 5B). The effect of silencing or overexpressing RGMA on the migratory potential of these GBC cells was also analyzed by the wound healing assay with similar results (Figure 5C).
Figure 5.
Gain and loss of RGMA function abrogates and enhance the impact of miR-552-3p on GBC cell, respectively. (A) The proliferation of RGMA knockdown and overexpression GBC cells was measured by CCK-8 assay; (B) the proliferation of RGMA knockdown and overexpression GBC cells was performed by colony formation assay (dyeing with crystal violet); (C) the migratory ability of RGMA knockdown and overexpression GBC cells was measured by wound-healing assay (scale bar =500 μm); (D) cell proliferation ability was assayed in GBC cells transfected with miR-552-3p sponge and RGMA siRNA or miR-552-3p mimic and RGMA-overexpression vectors by colony formation assay (dyeing with crystal violet); (E) cell migration and invasion were evaluated in GBC cells transfected with miR-552-3p sponge and RGMA siRNA or miR-552-3p mimic and RGMA overexpression vectors (dyeing with crystal violet; scale bar =100 μm). Compared with the NC group, *, P<0.05; **, P<0.01; ***, P<0.001; ****, P<0.0001. GBC, gallbladder cancer; RGMA, repulsive guidance molecule BMP co-receptor a; CCK-8, Cell Counting Kit-8.
Subsequently, we co-transfected GBC cells with either miR-552-3p sponge together with si-RGMA, or an miR-552-3p mimic together with pcDNA3.1(+)-RGMA. This functional rescue experiment showed that restoration of RGMA expression significantly reversed the promoting effects of the miR-552-3p mimic on colony formation, whereas inhibition of RGMA expression significantly restored the inhibitory effect of miR-552-3p (Figure 5D). In addition, the recovery of RGMA expression significantly reversed the promoting effects of miR-552-3p on cell migration and invasion, and inhibition of its expression significantly restored the inhibitory effect of miR-552-3p interference (Figure 5E).
The above results indicate that RGMA plays an important role in the mechanisms underlying the tumor-promoting functions of miR-552-3p in GBC.
MiR-552-3p promotes GBC malignant progression by activating the Akt/β-catenin and EMT pathways
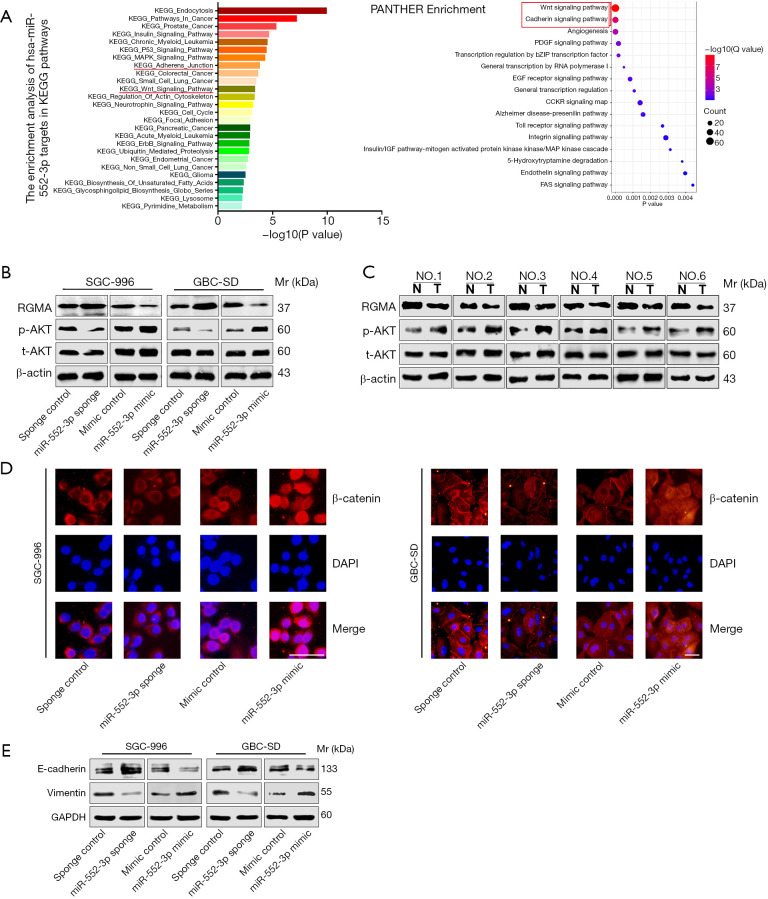
In order to further explore the downstream signal transduction pathways of miR-552-3p for promoting GBC progression, we used bioinformatics methods (29,30) to investigate signal pathways involved by target gene enrichment analysis. Interestingly, tumor-related Wnt and cadherin signaling pathways were enriched, underlying miR-552-3p function (Figure 6A). Based on this, we then further analyzed whether miR-552-3p promotes GBC cell malignant progression via the Wnt and cadherin pathways.
Figure 6.
MiR-552-3p promotes GBC malignant progression by reactivating Akt/β-catenin pathway and EMT. (A) KEGG and PANTHER enrichment pathway analysis for miR-552-3p’s target gene; (B) protein expression levels of RGMA and the activation vs. inhibition situations of Akt after miR-552-3p knockdown or overexpression in GBC cells; (C) protein expression levels of RGMA and the activation vs. inhibition situations of Akt after miR-552-3p knockdown or overexpression in GBC tissues vs. nontumor tissues; (D) the nuclear translocation of β-catenin was visualized by immunofluorescence assay in GBC cells (scale bar =50 μm); (E) E-cadherin and vimentin protein expression levels in the indicated cells were examined by Western blotting. GBC, gallbladder cancer; RGMA, repulsive guidance molecule BMP co-receptor a; EMT, epithelial-mesenchymal transformation; Mr, molecular mass.
It has been reported that RGMA plays a critical role in tumor cell migration and adhesion (31). RGMA combined with its receptor neogenin-1 (NEO1) can inactivate the downstream protein tyrosine kinase 2 (PTK2)-Akt signal transduction pathway (32). Activated Akt in turn activates β-catenin and prevents its degradation. This initiates nuclear translocation, which then activates transcription of a cascade of downstream proliferation and invasion-related genes (33,34). According to these previous studies, we hypothesized that miR-552-3p would affect the Wnt and cadherin pathways through inhibiting RGMA. We found that miR-552-3p sponge increased the expression of RGMA and inhibited the phosphorylation of Akt in two GBC cell lines. Conversely, cells treated with miR-552-3p mimic reduced expression of RGMA and activated Akt (Figure 6B). Consistent with this, we found that RGMA expression was decreased and Akt phosphorylation increased in GBC tumor tissues compared to normal tissues (Figure 6C). Next, cellular immunofluorescence showed that inhibiting miR-552-3p reduced the translocation of β-catenin in GBC cells, while its overexpression promoted the entry of β-catenin into the nucleus (Figure 6D). In terms of cell adhesion, miR-552-3p sponge increased E-cadherin protein while it decreased the expression of vimentin protein. In contrast, overexpression of miR-552-3p induced the opposite effects (Figure 6E).
Thus, we found that miR-552-3p promotes GBC cell malignant progression via inhibition of RGMA expression and reactivation of the Akt/β-catenin pathway and the EMT pathway.
MiR-552-3p is a significant predictor of GBC prognosis
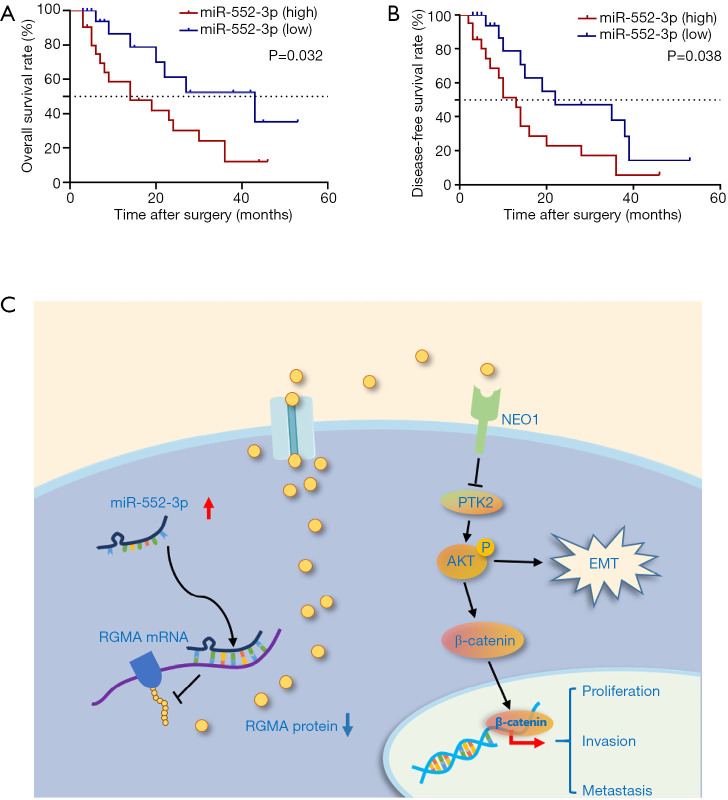
To explore the clinical significance of miR-552-3p, we stratified 41 GBC patients into two groups according to their tumor’s high or low miR-552-3p expression using the median value as the cut-off. We found that high miR-552-3p expression correlated with larger tumors and liver metastasis (Table S3). Furthermore, Kaplan-Meier analysis showed shorter disease-free survival (DFS) and overall survival (OS) of patients with high miR-552-3p expression, relative to patients with low miR-552-3p expression (Figure 7A,7B).
Figure 7.
MiR-552-3p acts as a significant prognosis predictor of GBC. (A) Kaplan-Meier overall survival curve of GBC patients based on miR-552-3p expression; (B) Kaplan-Meier disease-free survival curve of GBC patients based on miR-552-3p expression; (C) miR-552-3p promotes malignant progression of gallbladder carcinoma by reactivating the Akt/β-catenin signaling pathway due to inhibition of the tumor suppressor gene RGMA. GBC, gallbladder cancer; RGMA, repulsive guidance molecule BMP co-receptor a.
Discussion
GBC is a great challenge for health care because of its high metastatic potential and great heterogeneity. The OS of GBC patients is worse than for other anatomic biliary cancer primary sites. A study reported that the overall median survival time was about 8.5 months in 385 GBC patients (35). Moreover, GBC is one of the few cancers with a higher incidence in women than in men (36). Tumorigenesis is the genetic result of the imbalance between the regulation of pro-oncogenes and tumor suppressor genes, accompanied by uncontrolled cell-proliferate and invasion, resulting in the disruption of normal tissue structure, homeostatic imbalance of the organic environment, eventually exceeding the body’s compensatory ability.
Accumulated studies have shown that miRNAs play roles similar to oncogenes and tumor suppressor genes in tumors (16,17,37). MiRNAs constitute a highly conserved class of small non-protein-coding RNAs which maintain cell homeostasis by negative gene regulation. Modulation of the function of CSCs, as a class of tumor-initiating cells, will directly affect cancer cell proliferation and invasive potential. However, few miRNAs involved in CSC regulation have thus far been reported. On account of the fact that miR-552 was highly expressed in some tumors of the digestive system relative to normal tissue or adjacent non-tumor tissues (19,20), we analyzed levels of expression of the miR-552-3p form in GBC tissues and CSCs by qRT-PCR. We found that miR-552-3p is up-regulated in GBC tissues when compared with non-tumor tissues. More importantly, miR-552-3p was mostly expressed in CD44+ and CD133+ gallbladder carcinoma stem cells, as well as in stem cell-enriched spheroids.
To date, the biological functions of the miR-552 family have been mainly reported in the field of proliferation and metastasis in some tumors, while the effects of miR-552-3p on CSCs are still unclear, especially in gallbladder carcinoma. In this study, we focused on GBC CSCs and performed functional analyses. As expected, interference with miR-552-3p suppressed gallbladder CSC expansion, self-renewal and stemness-related gene expression. In addition, effects on the cancer cell phenotype after miR-552-3p reduction or overexpression were explored. miR-552-3p interference inhibited GBC cell proliferation, migration and invasion in vitro. In vivo experiments also confirmed that interference with miR-552-3p expression inhibited the growth of subcutaneous tumors and lung metastases of gallbladder carcinoma. In contrast, overexpression of miR-552-3p had the opposite effect.
The precise molecular mechanism by which miR-552-3p affects various pathological processes involved in GBC remains unclear. Bioinformatic approaches concluded that RGMA may be the target gene of miR-552-3p. RGMA encodes a member of the repulsive guidance molecule family and acts as a critical tumor suppressor gene (38-40). To confirm this pathway, we first ascertained that RGMA is a target of miR-552-3p using luciferase reporter assays. Second, we determined that miR-552-3p could regulate the expression of RGMA at the mRNA and protein levels in GBC cells and that knockdown of RGMA could promote the GBC cell malignant phenotype. Third, we documented that expression of miR-552-3p and RGMA was negatively correlated in GBC tissues. In addition, considering that GBCs are mainly adenocarcinomas, the correlation of miR-552-3p with RGMA expression in several other digestive system adenocarcinomas and even lung adenocarcinoma, is consistent with the conclusion that miR-552-3p negatively regulates RGMA. The most important finding here is that gain and loss of RGMA function abrogate and enhance the impact of miR-552-3p on cell proliferation and metastasis, respectively. All these results confirm that miR-552-3p influences the malignant phenotype of GBC by regulating the target gene RGMA.
To further clarify the downstream signaling pathway of miR-552-3p in regulating GBC CSCs and the malignant phenotype through RGMA, we performed enrichment pathway analysis of miR-552-3p target genes. We found that Wnt signaling and cadherin signaling pathways were enriched. Interestingly, a previous study has demonstrated that binding of RGMA to its receptor NEO1 can inactivate the downstream PTK2-Akt signal transduction pathway (32). Considering the effect of Akt activation on the Wnt and cadherin signaling pathways, we further explored whether miR-552-3p could affect the activity of these two pathways by inhibiting RGMA. As expected, activation of Akt and nuclear localization of β-catenin were induced by miR-552-3p overexpression and RGMA inhibition. At the same time, we found that miR-552-3p could also regulate EMT progression.
In the clinical context, compared with GBC patients whose tumors expressed low levels of miR-552-3p, those with high-expression had larger tumors and more liver metastasis. Moreover, GBC patients with high expression of miR-552-3p had worse OS and DFS than those with low expression of miR-552-3p. Thus, miR-552-3p expression levels can be used as a new potential prognostic marker for gallbladder carcinoma.
In summary, our present study documented tumorigenic effects of miR-552-3p in GBC. We first reported that miR-552-3p was upregulated in GBC CSCs and tissues, and promoted self-renewal, malignant proliferation, tumorigenicity and metastasis of GBC cells. Mechanistically, we found that miR-552-3p promotes the malignant progression of GBC by inhibiting the tumor suppressor gene RGMA, leading to reactivation of the Akt/β-catenin and EMT signaling pathways (Figures 6B,6E,7C). Moreover, miR-552-3p may serve as a predictive marker for prognosis, as well as a therapeutic target for GBC patients.
Supplementary
The article’s supplementary files as
Acknowledgments
We would like to thank Yanting Yu, Shanhua Tang, Linna Guo, Dan Cao, Shanna Huang, Shennian Ge, Qi Yang and Liang Tang for technical assistance.
Funding: We gratefully acknowledge the support from the State Key Project on Infectious Diseases of China (grant No.2018ZX10723204-002-002), National Natural Science Foundation of China (grant No. 91859205, 81988101, 81830054, 81672777, 81902940, 81902942), Natural Science Foundation of Shanghai (grant No. 19ZR1400300), Shanghai Rising-Star Program (grant No. 17QA1405700), Shanghai Top Young Talents Program, Foundation of Shanghai Shenkang Hospital Development Center (grant No. SHDC2020CR2011A, SHDC12016127).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethic Committee of Eastern Hepatobiliary Surgery Hospital (No.: EHBHKY2020-K-016) and individual consent for this retrospective analysis was waived. Animal experiments were performed under a project license (No.: EHBHKY2020-K-016) granted by the Ethic Committee of Eastern Hepatobiliary Surgery Hospital, in compliance with Second Military Medical University guidelines for the care and use of animals.
Footnotes
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://dx.doi.org/10.21037/atm-21-2013
Data Sharing Statement: Available at https://dx.doi.org/10.21037/atm-21-2013
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/atm-21-2013). The authors have no conflicts of interest to declare.
References
- 1.Kanthan R, Senger JL, Ahmed S, et al. Gallbladder Cancer in the 21st Century. J Oncol 2015;2015:967472. 10.1155/2015/967472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ramachandran A, Srivastava DN, Madhusudhan KS. Gallbladder cancer revisited: the evolving role of a radiologist. Br J Radiol 2021;94:20200726. 10.1259/bjr.20200726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zaidi MY, Maithel SK. Updates on Gallbladder Cancer Management. Curr Oncol Rep 2018;20:21. 10.1007/s11912-018-0664-3 [DOI] [PubMed] [Google Scholar]
- 4.Hundal R, Shaffer EA. Gallbladder cancer: epidemiology and outcome. Clin Epidemiol 2014;6:99-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ruiu R, Tarone L, Rolih V, et al. Cancer stem cell immunology and immunotherapy: Harnessing the immune system against cancer's source. Prog Mol Biol Transl Sci 2019;164:119-88. 10.1016/bs.pmbts.2019.03.008 [DOI] [PubMed] [Google Scholar]
- 6.Garcia-Mayea Y, Mir C, Masson F, et al. Insights into new mechanisms and models of cancer stem cell multidrug resistance. Semin Cancer Biol 2020;60:166-80. 10.1016/j.semcancer.2019.07.022 [DOI] [PubMed] [Google Scholar]
- 7.Sharma KL, Yadav A, Gupta A, et al. Association of genetic variants of cancer stem cell gene CD44 haplotypes with gallbladder cancer susceptibility in North Indian population. Tumour Biol 2014;35:2583-9. 10.1007/s13277-013-1340-8 [DOI] [PubMed] [Google Scholar]
- 8.Mizukami T, Kamachi H, Mitsuhashi T, et al. Cytoplasmic CD133 expression correlates with histologic differentiation and is a significant prognostic factor in extrahepatic bile duct cancer and gallbladder cancer. Oncol Lett 2018;16:6423-30. 10.3892/ol.2018.9499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He Y, Xue C, Yu Y, et al. CD44 is overexpressed and correlated with tumor progression in gallbladder cancer. Cancer Manag Res 2018;10:3857-65. 10.2147/CMAR.S175681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi CJ, Gao J, Wang M, et al. CD133(+) gallbladder carcinoma cells exhibit self-renewal ability and tumorigenicity. World J Gastroenterol 2011;17:2965-71. 10.3748/wjg.v17.i24.2965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi C, Tian R, Wang M, et al. CD44+ CD133+ population exhibits cancer stem cell-like characteristics in human gallbladder carcinoma. Cancer Biol Ther 2010;10:1182-90. 10.4161/cbt.10.11.13664 [DOI] [PubMed] [Google Scholar]
- 12.Yadav A, Gupta A, Rastogi N, et al. Association of cancer stem cell markers genetic variants with gallbladder cancer susceptibility, prognosis, and survival. Tumour Biol 2016;37:1835-44. 10.1007/s13277-015-3929-6 [DOI] [PubMed] [Google Scholar]
- 13.Fatima N, Srivastava AN, Nigam J, et al. Clinicopathological correlation of cancer stem cell markers Oct-4 and CD133 expression as prognostic factor in malignant lesions of gallbladder: An immunohistochemical study. Indian J Pathol Microbiol 2019;62:384-90. 10.4103/IJPM.IJPM_134_19 [DOI] [PubMed] [Google Scholar]
- 14.Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov 2017;16:203-22. 10.1038/nrd.2016.246 [DOI] [PubMed] [Google Scholar]
- 15.Aghajani M, Mansoori B, Mohammadi A, et al. New emerging roles of CD133 in cancer stem cell: Signaling pathway and miRNA regulation. J Cell Physiol 2019;234:21642-61. 10.1002/jcp.28824 [DOI] [PubMed] [Google Scholar]
- 16.Yu D, Shin HS, Lee YS, et al. miR-106b modulates cancer stem cell characteristics through TGF-β/Smad signaling in CD44-positive gastric cancer cells. Lab Invest 2014;94:1370-81. 10.1038/labinvest.2014.125 [DOI] [PubMed] [Google Scholar]
- 17.Jeong JY, Kang H, Kim TH, et al. MicroRNA-136 inhibits cancer stem cell activity and enhances the anti-tumor effect of paclitaxel against chemoresistant ovarian cancer cells by targeting Notch3. Cancer Lett 2017;386:168-78. 10.1016/j.canlet.2016.11.017 [DOI] [PubMed] [Google Scholar]
- 18.Hemmesi K, Squadrito ML, Mestdagh P, et al. miR-135a Inhibits Cancer Stem Cell-Driven Medulloblastoma Development by Directly Repressing Arhgef6 Expression. Stem Cells 2015;33:1377-89. 10.1002/stem.1958 [DOI] [PubMed] [Google Scholar]
- 19.Qu W, Wen X, Su K, et al. MiR-552 promotes the proliferation, migration and EMT of hepatocellular carcinoma cells by inhibiting AJAP1 expression. J Cell Mol Med 2019;23:1541-52. 10.1111/jcmm.14062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feng X, Zhu M, Liao B, et al. Upregulation of miR-552 Predicts Unfavorable Prognosis of Gastric Cancer and Promotes the Proliferation, Migration, and Invasion of Gastric Cancer Cells. Oncol Res Treat 2020;43:103-11. 10.1159/000505377 [DOI] [PubMed] [Google Scholar]
- 21.Cao J, Yan XR, Liu T, et al. MicroRNA-552 promotes tumor cell proliferation and migration by directly targeting DACH1 via the Wnt/β-catenin signaling pathway in colorectal cancer. Oncol Lett 2017;14:3795-802. 10.3892/ol.2017.6600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gu J, Han T, Sun L, et al. miR-552 promotes laryngocarcinoma cells proliferation and metastasis by targeting p53 pathway. Cell Cycle 2020;19:1012-21. 10.1080/15384101.2020.1743910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lai CH, Liang XZ, Liang XY, et al. Study on miRNAs in Pan-Cancer of the Digestive Tract Based on the Illumina HiSeq System Data Sequencing. Biomed Res Int 2019;2019:8016120. 10.1155/2019/8016120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wei C, Gao JJ. Downregulated miR-383-5p contributes to the proliferation and migration of gastric cancer cells and is associated with poor prognosis. PeerJ 2019;7:e7882. 10.7717/peerj.7882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li XF, Chen C, Xiang DM, et al. Chronic inflammation-elicited liver progenitor cell conversion to liver cancer stem cell with clinical significance. Hepatology 2017;66:1934-51. 10.1002/hep.29372 [DOI] [PubMed] [Google Scholar]
- 26.Xiang DM, Sun W, Ning BF, et al. The HLF/IL-6/STAT3 feedforward circuit drives hepatic stellate cell activation to promote liver fibrosis. Gut 2018;67:1704-15. 10.1136/gutjnl-2016-313392 [DOI] [PubMed] [Google Scholar]
- 27.McGeary SE, Lin KS, Shi CY, et al. The biochemical basis of microRNA targeting efficacy. Science 2019;366:eaav1741. [DOI] [PMC free article] [PubMed]
- 28.Miranda KC, Huynh T, Tay Y, et al. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell 2006;126:1203-17. 10.1016/j.cell.2006.07.031 [DOI] [PubMed] [Google Scholar]
- 29.Li JH, Liu S, Zhou H, et al. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res 2014;42:D92-7. 10.1093/nar/gkt1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xie C, Mao X, Huang J, et al. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res 2011;39:W316-22. 10.1093/nar/gkr483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lah GJ, Key B. Novel roles of the chemorepellent axon guidance molecule RGMa in cell migration and adhesion. Mol Cell Biol 2012;32:968-80. 10.1128/MCB.06128-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Endo M, Yamashita T. Inactivation of Ras by p120GAP via focal adhesion kinase dephosphorylation mediates RGMa-induced growth cone collapse. J Neurosci 2009;29:6649-62. 10.1523/JNEUROSCI.0927-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Borhani S, Corciulo C, Larranaga-Vera A, et al. Adenosine A2A receptor (A2AR) activation triggers Akt signaling and enhances nuclear localization of β-catenin in osteoblasts. FASEB J 2019;33:7555-62. 10.1096/fj.201900014R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fang D, Hawke D, Zheng Y, et al. Phosphorylation of beta-catenin by AKT promotes beta-catenin transcriptional activity. J Biol Chem 2007;282:11221-9. 10.1074/jbc.M611871200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McNamara MG, Lopes A, Wasan H, et al. Landmark survival analysis and impact of anatomic site of origin in prospective clinical trials of biliary tract cancer. J Hepatol 2020;73:1109-17. 10.1016/j.jhep.2020.05.014 [DOI] [PubMed] [Google Scholar]
- 36.Siegel RL, Miller KD, Fuchs HE, et al. Cancer Statistics, 2021. CA Cancer J Clin 2021;71:7-33. 10.3322/caac.21654 [DOI] [PubMed] [Google Scholar]
- 37.Mishra S, Yadav T, Rani V. Exploring miRNA based approaches in cancer diagnostics and therapeutics. Crit Rev Oncol Hematol 2016;98:12-23. 10.1016/j.critrevonc.2015.10.003 [DOI] [PubMed] [Google Scholar]
- 38.Li VS, Yuen ST, Chan TL, et al. Frequent inactivation of axon guidance molecule RGMA in human colon cancer through genetic and epigenetic mechanisms. Gastroenterology 2009;137:176-87. 10.1053/j.gastro.2009.03.005 [DOI] [PubMed] [Google Scholar]
- 39.Li J, Ye L, Kynaston HG, et al. Repulsive guidance molecules, novel bone morphogenetic protein co-receptors, are key regulators of the growth and aggressiveness of prostate cancer cells. Int J Oncol 2012;40:544-50. [DOI] [PubMed] [Google Scholar]
- 40.Harada K, Fujita Y, Yamashita T. Repulsive guidance molecule A suppresses angiogenesis. Biochem Biophys Res Commun 2016;469:993-9. 10.1016/j.bbrc.2015.12.073 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as