Abstract
The cotranscriptional placement of the 7-methylguanosine cap on pre-mRNA is mediated by recruitment of capping enzyme to the phosphorylated carboxy-terminal domain (CTD) of RNA polymerase II. Immunoblotting suggests that the capping enzyme guanylyltransferase (Ceg1) is stabilized in vivo by its interaction with the CTD and that serine 5, the major site of phosphorylation within the CTD heptamer consensus YSPTSPS, is particularly important. We sought to identify the CTD kinase responsible for capping enzyme targeting. The candidate kinases Kin28-Ccl1, CTDK1, and Srb10-Srb11 can each phosphorylate a glutathione S-transferase–CTD fusion protein such that capping enzyme can bind in vitro. However, kin28 mutant alleles cause reduced Ceg1 levels in vivo and exhibit genetic interactions with a mutant ceg1 allele, while srb10 or ctk1 deletions do not. Therefore, only the TFIIH-associated CTD kinase Kin28 appears necessary for proper capping enzyme targeting in vivo. Interestingly, levels of the polyadenylation factor Pta1 are also reduced in kin28 mutants, while several other polyadenylation factors remain stable. Pta1 in yeast extracts binds specifically to the phosphorylated CTD, suggesting that this interaction may mediate coupling of polyadenylation and transcription.
Eukaryotic pre-mRNAs are transcribed by RNA polymerase II (Pol II) and undergo several processing events before maturing into mRNA. Soon after initiation of transcription, pre-mRNA is capped at its 5′ terminus (27, 48). Transcripts are further processed by the splicing and polyadenylation machineries before translocation to the cytoplasm for translation. Cotranscriptional mRNA processing is facilitated by the recruitment of mRNA processing factors to the carboxy-terminal domain (CTD) of the Pol II large subunit (8, 22, 23, 26, 36, 37, 56).
The CTD is composed of a tandemly repeated heptad with the consensus sequence YSPTSPS (1, 10). Mammalian Pol II CTD has 52 repeats, whereas the yeast Saccharomyces cerevisiae CTD has only 26 (12). Deletion of the mouse (4), Drosophila (58), or yeast (2, 43) CTD is lethal, and partial deletions result in conditional phenotypes, reducing transcription and response to activators (5, 19, 38, 49). The CTD is phosphorylated in vivo, primarily at serine 2 and serine 5 of the heptapeptide consensus repeat (12). Hyperphosphorylation of the CTD appears to be coordinated with transcription initiation and elongation in vivo (45, 54). Phosphorylation is mediated by one or more CTD kinase activities, but the timing and role of specific kinases are not clearly defined.
Several putative CTD kinases are members of the cyclin-dependent kinase (CDK) family. These kinases typically consist of a catalytic subunit bound to a regulatory cyclin subunit. The Kin28-Ccl1 (Cdk7-cyclin H) kinase complex associated with the general transcription factor TFIIH can phosphorylate the CTD after preinitiation complex (PIC) formation, thereby positively regulating transcription (16, 21). The Srb10-Srb11 (Cdk8-cyclin C) kinase complex is associated with the RNA Pol II holoenzyme and may negatively regulate initiation of transcription by phosphorylating the CTD before PIC formation (21, 35) or by phosphorylating upstream activator complexes (24). CTD kinase 1 (CTDK1) is necessary for proper CTD phosphorylation in vivo (33) and may also be involved in transcriptional repression (32). The Ctk1 subunit is most similar to the Cdk9 subunit of mammalian CTD kinase and elongation factor pTEFb, suggesting a possible role for Ctk1 in elongation (62). Of these three CTD kinases, only Kin28-Ccl1 is essential for viability, and the functions of Srb10 and Ctk1 are not redundant. Phosphorylation of different sites within the consensus CTD repeat and temporal and spatial regulation of the kinases are likely to play crucial roles in the interplay between the CTD and the many factors that bind to it.
Placement of a cap structure on the 5′ end of a nascent pre-mRNA is the first detectable mRNA processing event. The reaction occurs in three steps: removal of the gamma phosphate from the pre-mRNA by RNA triphosphatase, transfer of GMP by guanylyltransferase, and methylation of the N7 position of the new guanosine cap (for review, see references 41 and 51). Capping is restricted to Pol II transcripts by capping enzyme recruitment to a phosphorylated CTD. This interaction is mediated by a direct association of the capping enzyme guanylyltransferase Ceg1 with the phosphorylated CTD (8, 36, 56). Interestingly, Ceg1 guanylyltransferase activity on the CTD is allosterically regulated by its association with the mRNA triphosphatase subunit Cet1 (7).
The CTD is also required for efficient splicing and polyadenylation in mammalian cells (37). Certain splicing factors can be coimmunoprecipitated with hyperphosphorylated Pol II (30, 40, 57). Polyadenylation factors can bind to a CTD affinity column, yet demonstrate no apparent preference for the phosphorylation state of the CTD (37). In addition, the CTD has been shown to be an essential cofactor in mRNA polyadenylation (22). While either unphosphorylated or hyperphosphorylated CTD stimulates the 3′ cleavage reaction, the ability of creatine phosphate or phosphoserine to also stimulate cleavage suggests that a phosphorylated CTD may be the relevant in vivo cofactor.
We sought to further characterize the CTD phosphorylation event responsible for capping enzyme recruitment. Genetic experiments with S. cerevisiae suggest that the CTD kinase Kin28, but neither Srb10 nor CTDK1, is necessary for capping enzyme targeting. While any of these kinases can phosphorylate a glutathione S-transferase (GST)-CTD fusion protein to allow capping enzyme binding in vitro, only kin28 mutant alleles exhibit genetic interactions with ceg1-250 in vivo. Ceg1 levels are reduced in cells carrying Kin28 mutants or a partial CTD truncation. Furthermore, conditional mutants in the serine 5, but not serine 2, position of the CTD consensus heptapeptide repeat YSPTSPS are lethal in combination with ceg1-250. These data support the model that Kin28 phosphorylates the CTD at the serine 5 position to mediate cotranscriptional recruiting of the capping enzyme. It was also observed that levels of the 3′ RNA processing factor Pta1 are decreased in kin28 mutants and that Pta1 could bind specifically to a phosphorylated CTD. Therefore, CTD phosphorylation by Kin28 may also mediate coupling of transcription and polyadenylation.
MATERIALS AND METHODS
Plasmid construction.
The plasmids used in this study are summarized in Table 1. To generate pRS426-KIN28, the 1.3-kb HindIII-BamHI fragment from YCplac22-KIN28 was ligated into the HindIII and BamHI sites of pRS426. pRS314-hakin28(T17D), pRS314-hakin28(K36A), and pRS314-hakin28(T162A) will be described by Keogh et al. (unpublished data). The remaining plasmids were constructed as previously described (7, 9, 17, 33, 35, 44, 50, 52, 55). DNA manipulations and transformation into bacteria were performed by standard techniques (3).
TABLE 1.
Characteristics of plasmids used in this study
| Plasmid | Relevant features | Source or reference |
|---|---|---|
| pRS315-CEG1 | CEN/ARS LEU2 CEG1 | 17 |
| pRS316-CEG1 | CEN/ARS URA3 CEG1 | 17 |
| pRS316-CET1 | CEN/ARS URA3 CET1 | 7 |
| YCplac22-KIN28 | CEN/ARS TRP1 KIN28 | 9 |
| YCplac22-kin28-16 | CEN/ARS TRP1 kin28-16(N123D, P206L, V232A, L293S) | 9 |
| pRS426-KIN28 | 2 μm URA3 KIN28 | This study |
| pRS314-hakin28(T17D) | CEN/ARS TRP1 kin28(T17D), C-terminal HAa tag | This study |
| pRS314-hakin28(K36A) | CEN/ARS TRP1 kin28(K36A), C-terminal HA tag | This study |
| pRS314-hakin28(T162A) | CEN/ARS TRP1 kin28(T162A), C-terminal HA tag | This study |
| pDJ29 | srb10Δ::HIS3 | 35 |
| RY2973 | CEN/ARS URA3 SRB10 | 35 |
| pSZH | ctk1Δ::HIS3 | 52 |
| pJYC1513 | CEN/ARS URA3 CTK1 | 33 |
| pRP1-101 | CEN/ARS LEU2 rpb1Δ101 (11 wild-type heptapeptide repeats) | 44 |
| pRP112 | CEN/ARS URA3 RPB1 | 44 |
| pRP114 | CEN/ARS LEU2 RPB1 | 44 |
| pY1WT(10) | CEN/ARS LEU2 rpb1 (10 wild-type repeats) C-terminal HA tag | 55 |
| pY1A2(8)WT(7) | CEN/ARS LEU2 rpb1 (8 S2A, 7 wild-type repeats) C-terminal HA tag | 55 |
| pY1A5(5)WT(7) | CEN/ARS LEU2 rpb1 (5 S5A, 7 wild-type repeats) C-terminal HA tag | 55 |
| rpb1-15 | CEN/ARS LEU2 rpb1-15(T4292A) | 49, 50 |
| rpb1-18 | CEN/ARS LEU2 rpb1-18(G808A) | 49, 50 |
| rpb1-19 | CEN/ARS LEU2 rpb1-19(G4031A) | 49, 50 |
HA, hemagglutinin.
Yeast strains.
The yeast strains used in this study are summarized in Table 2. YSB625 was generated by mating YSB491 with FY834. Ade+ Lys+ diploids were selected, sporulated, and dissected. YSB625 was identified as an Ade+ Lys− Ts− spore, whose Ts− phenotype could be complemented by pRS315-CEG1, but not by pRS315. YSB626 and YSB627 were generated by mating 24-1.1A with YSB517. Leu+ Trp+ diploids were selected, sporulated, and dissected. YSB626 was identified as a Leu+ Trp+ Ts+ spore. YSB627 was identified as a Leu+ Trp+ Ts− spore; the Ts− phenotype was complemented by pRS316-CEG1 but not by pRS316. YSB626 and YSB627 were transformed with pRS426-KIN28, and the Trp+ YCplac22-KIN28 was shuffled out, resulting in Leu+ Ura+ Trp− strains. To generate an srb10Δ strain, pRS316-CEG1 was transformed into YSB625. The resulting strain was transformed with SalI-linearized pDJ29, and His+ Ura+ transformants were selected to generate YSB652. The cold sensitivity phenotype associated with srb10Δ was observed in YSB652 and could be complemented by RY2973. To generate a ctk1Δ strain, pRS316-CEG1 was transformed into YSB625. The resulting strain was transformed with the 2.9-kb SnaBI-VspI fragment of pSZH/ctk1Δ::HIS3, and His+ Ura+ transformants were selected, to generate YSB653. The cold and caffeine sensitivity phenotypes associated with ctk1Δ were observed in YSB653 and could be complemented by pRS316-CTK1.
TABLE 2.
S. cerevisiae strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| PY469 | MATa ura3 leu2 trp1 his3 ade2 ade3 can1 | D. Pellman |
| YSB491 | MATa ura3 leu2 trp1 his3 ade2 ade3 can1 ceg1-250 | 8 |
| YSB625 | MATα ura3 leu2 trp1 his3 lys2 can1 ceg1-250 | This study |
| FY834 | MATα ura3 leu2 trp1 his3 lys2 | F. Winston |
| N398 | MATa ura3 leu2 TRP1 his3 rpb1Δ187::HIS3 (pRP1-101) | R. Young |
| N418 | MATa ura3 leu2 TRP1 his3 rpb1Δ187::HIS3 (pRP112) | R. Young |
| YSB516 | MATα ura3 leu2 TRP1 his3 ceg1-250 rpb1Δ187::HIS3 (pRP112) | 8 |
| YSB626 | MATa ura3 leu2 trp1 his3 ade2 ade3 can1 kin28Δ::LEU2 (pRS426-KIN28) | This study |
| YSB627 | MATα ura3 leu2 trp1 his3 ade2 ade3 can1 ceg1-250 kin28Δ::LEU2 (pRS426-KIN28) | This study |
| 24-1.1A | MATa ura3 leu2 trp1 his3 ade2 ade3 can1 kin28Δ::LEU2 (YCplac22-KIN28) | 9 |
| YSB652 | MATα ura3 leu2 trp1 his3 lys2 srb10Δ::HIS3 ceg1-250 (pRS316-CEG1) | This study |
| YSB653 | MATα ura3 leu2 trp1 his3 lys2 ctk1Δ::HIS3 ceg1-250 (pRS316-CEG1) | This study |
In order to compare growth of yeast strains, the strains were grown overnight at 30°C in synthetic complete minimal medium. Cultures were normalized to an optical density at 600 nm (OD600) of 0.2, and three serial dilutions of 1:8 were prepared. Aliquots of the four dilutions were then spotted on minimal medium plates and incubated for 3 days at 30°C. Medium preparation, yeast transformations, and other yeast manipulations were performed by standard methods as described previously (20).
CTD kinase and CEG1 genetic analyses.
kin28 mutants were analyzed by plasmid shuffling of YCplac22-KIN28, pRS314-hakin28(T17D), pRS314-hakin28(K36A), YCplac22-kin28-16, or pRS314-hakin28(T162A) into YSB626 (CEG1+) or YSB627 (ceg1-250) and growth on −Leu −Trp +fluoroorotic acid (FOA) synthetic complete medium plates for 3 days at 30°C. To generate a pta1 kin28 strain, FY1283 was mated with YSB626, and a spore was identified which was Ura+, Leu+, FOAS, and Ts−. This pta1 kin28 strain, YSB688, and YSB626 and FY1283 were analyzed by plasmid shuffling of YCplac22-KIN28, pRS314-hakin28(T17D), pRS314-hakin28(K36A), YCplac22-kin28-16, or pRS314-hakin28(T162A) and growth on −Trp +FOA synthetic complete medium plates for 3 days at 30°C. The wild type (YSB625) and srb10Δ (YSB652) and ctk1Δ (YSB653) mutants were analyzed in combination with CEG1 and ceg1-250 by comparing the growth levels of strains transformed with pRS316-CEG1 on −His −Ura plates and −His +FOA plates for 3 days at 30°C.
Yeast extract preparation and immunoblotting analysis.
Yeast whole-cell extracts were prepared as described previously (14). Lysis buffer contained 20 mM HEPES (pH 7.6), 10% glycerol, 200 mM KoAc, 1 mM EDTA, 1 mM phenylmethyl sulfonyl fluoride and the phosphatase inhibitors NaF (10 mM) and Na3VO4 (0.1 mM). Protein levels were detected by standard Western blotting procedures (3). Antibodies against Ceg1 (18) and polyadenylation factors (28, 29, 53) (antibody 1664 [53]) have been described previously. Monoclonal antibody B3, which recognizes the phosphorylated CTD (42, 46), was generously provided by B. Blencowe, and the monoclonal antibody against Pta1 was a gift of P. O'Connor. Anti-Cet1 antibody was prepared by T. Takagi and will be described elsewhere.
In vitro CTD interaction experiments.
GST-CTD interaction experiments were performed as described previously (8) with some modifications. GST-CTD was bound to glutathione agarose (2 mg of protein/ml of beads). GST-CTD-agarose (∼200 ng of protein per reaction) was phosphorylated for 1 hour with the following different kinases: recombinant Kin28-Ccl1 (0.9 μg; 20 mM HEPES-KOH [pH 7.3], 15 mM magnesium acetate, 100 mM potassium acetate, 1 mM dithiothreitol, 2.5 mM EGTA, 10% glycerol [generously provided by S. Koh, C. Hengartner, and R. Young]), recombinant Srb10-Srb11 (0.8 μg; same buffer as Kin28-Ccl1 [also provided by S. Koh, C. Hengartner, and R. Young]), CTDK1 (75 ng; 25 mM Tris-Cl [pH 7.9] [purified as in reference 33], 10 mM MgCl2), and casein kinase I (500 U; manufacturer's buffer; New England Biolabs). Each buffer contained 200 μM ATP and 3 μCi of [γ-32P]ATP (3,000 Ci/mmol). At the end of the reaction, 20 μl of glutathione agarose was added to each tube as a carrier, and the beads were washed.
While the phosphorylation reaction was carried out, recombinant Ceg1 and Cet1 (50 and 100 ng per reaction, respectively) were incubated with GST-agarose in buffer A containing 150 μg of bovine serum albumin per ml, 1× phosphatase inhibitors (1 mM NaN3, 1 mM NaF, 0.4 mM Na3VO4), 0.01% NP-40, and 0.05% Triton X-100 for 1 h at room temperature. The GST-agarose was then removed by centrifugation. The precleared Ceg1-Cet1 mixture was then added to nonphosphorylated and phosphorylated GST-CTD and incubated for 1 h. Beads were precipitated, washed extensively, and used for an enzyme-GMP formation assay and immunoblotting as described previously (8). Phosphorylated GST-CTD was detected by immunoblotting with H14 monoclonal antibody (BAbCO, Richmond, Calif.). GST-CTD was detected by immunoblotting with anti-GST monoclonal antibody (Santa Cruz Biotechnology, Inc., Santa Cruz, Calif.).
For interaction studies with polyadenylation factors, the GST fusion proteins were incubated with the first column fractions from the factor purification (29). Binding and analysis were carried out as described above.
RESULTS
In vitro CTD phosphorylation by various kinases is sufficient to recruit capping enzyme.
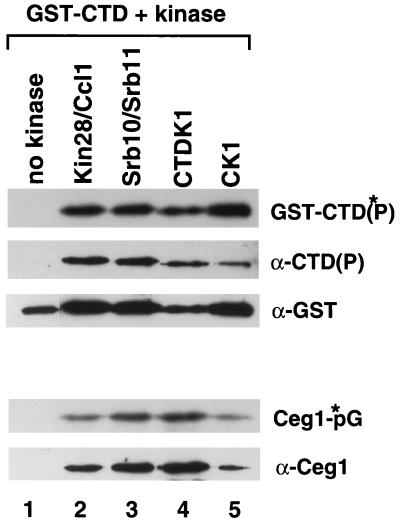
Many different kinases have been shown to phosphorylate the CTD in vitro. The particular kinases necessary for capping enzyme to bind Pol II have not been identified. Therefore, we decided to test the ability of individual CTD kinases to phosphorylate a GST-CTD fusion protein and thereby recruit capping enzyme. We picked the three kinases with clear in vivo connections to Pol II: the TFIIH-associated Kin28-Ccl1 complex, the Pol II holoenzyme-associated Srb10-Srb11 complex, and CTDK1, which is necessary for normal levels of CTD phosphorylation in vivo. A GST-CTD fusion protein was incubated with no kinase (GST-CTD), Kin28-Ccl1, Srb10-Srb11, CTDK1, or the control protein casein kinase 1 [GST-CTD(P)]. Both Ceg1 and Cet1 capping enzyme subunits were then mixed with the GST-CTD beads, and the complexes were pelleted. No capping enzyme was detected in the unphosphorylated GST-CTD pellet (Fig. 1, lane 1). However, each of the four kinases tested was able to phosphorylate the GST-CTD sufficiently to recruit Ceg1 (lanes 2 to 5, α-Ceg1). Because we have previously found that the guanylyltransferase is allosterically regulated by the CTD and Cet1 (7), we tested the ability of the bound Ceg1 to form a covalent complex with GMP. No obvious differences were observed between CTD phosphorylated with different kinases (Ceg1-*pG). Therefore, there are no apparent differences between kinases for in vitro CTD phosphorylation and capping enzyme recruitment.
FIG. 1.
In vitro CTD phosphorylation by various kinases allows binding of Ceg1. Glutathione-agarose carrying GST-CTD was phosphorylated with [γ-32P]ATP by various kinases. Lanes: 1, no kinase; 2, Kin28-Ccl1; 3, Srb10-Srb11; 4, CTDK1; 5, casein kinase 1. Phosphorylated GST-CTD [GST-CTD(P)] and nonphosphorylated GST-CTD glutathione-agarose beads were incubated with Ceg1 and Cet1. The beads were pelleted and washed extensively. Phosphorylation of GST-CTD was detected by autoradiogram [GST-CTD(*P)] and immunoblotting [α-CTD(P)] with the H14 monoclonal antibody. GST-CTD and GST-CTD(P) were detected by immunoblotting with anti-GST antibody. Capping enzyme in the pellet was detected by both on autoradiogram of enzyme-GMP formation (Ceg1-*pG) and immunoblotting (α-Ceg1) with anti-Ceg1 antibody.
Kin28 is the CTD kinase necessary for capping enzyme recruitment in vivo.
Whereas various kinases can phosphorylate the CTD in a manner sufficient to recruit capping enzyme in vitro, these kinases are likely to function at different times or locations in vivo. For example, Srb10 is able to phosphorylate the CTD before PIC formation, whereas Kin28 phosphorylates the CTD after PIC formation (21). To test the in vivo role of specific kinases in CTD phosphorylation and capping enzyme recruitment, a genetic approach was taken. Previously, we found that a truncated CTD mutant (rpb1Δ101, 11 repeats) and the ceg1-250 capping enzyme mutant, both of which are viable at 30°C, are lethal in combination (8). Here, we analyzed the combination of ceg1-250 with different CTD kinase mutants.
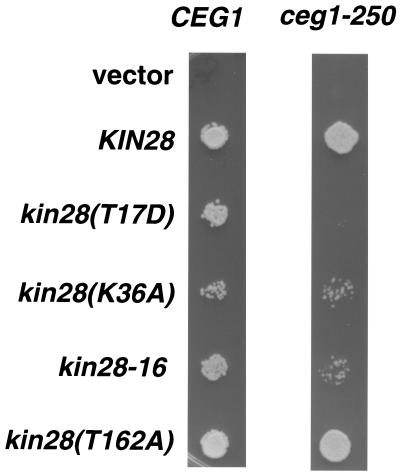
A summary of genetic interactions between ceg1-250 and several CTD kinase mutants is shown in Table 3. The srb10Δ ceg1-250 double mutant does not display any combined growth phenotypes different from that of either single mutant alone. Similarly, a ctk1Δ ceg1-250 double mutant grew no worse than either single mutant. However, the combination of certain kin28 mutants with ceg1-250 (Table 3 and Fig. 2) resulted in either synthetic lethality (T17D) or slower growth (kin28-16). The kin28(K36A) mutant, which has a mild effect on Kin28 activity (data not shown), showed only a modest reduction in growth rate when combined with ceg1-250. In contrast, another kin28 mutant (T162A) that does not reduce CTD phosphorylation in vivo (data not shown and see below) displayed no growth defect in combination with ceg1-250 (Fig. 2). In conclusion, in the three likely CTD kinases, only kin28 exhibits genetic interactions with ceg1.
TABLE 3.
Summary of genetic analyses between CTD kinase mutants and ceg1-250
| CTD kinase genotypea | Result with growth at 30°C for 3 daysb
|
|
|---|---|---|
| CEG1 | ceg1-250 | |
| KIN28 | ++++ | ++++ |
| kin28(T17D) | ++ | − |
| kin28(K36A) | ++ | ++/+ |
| kin28-16 | ++ | + |
| SRB10 | ++++ | ++++ |
| srb10Δ | ++ | ++ |
| CTK1 | ++++ | ++++ |
| ctk1Δ | ++ | ++ |
Construction and analyses of double mutant combinations are described in Materials and Methods.
++++, wild-type growth after 3 days; ++, 50% reduced colony size; +, 75% reduced colony size; −, no apparent colonies.
FIG. 2.
kin28 mutants display synthetic mutant phenotypes in combination with ceg1-250. kin28 mutants were analyzed in combination with CEG1 and ceg1-250 upon shuffling of pRS314, YCplac22-KIN28, pRS314-hakin28(T17D), pRS314-hakin28(K36A), YCplac22-kin28-16, and pRS314-hakin28(T162A) into YSB626 and YSB627, respectively. Growth of double mutants was compared by spotting a 1:8 dilution of an OD600 = 0.2 culture onto −Leu −Trp +FOA synthetic complete medium plates for 2 days at 30°C.
Genetic interactions with the ceg1-250 mutant suggest that kin28(T17D), and -16 mutants are defective for the CTD kinase activity necessary for capping enzyme recruitment. This is in contrast to the kin28(T162A) mutant, which includes a mutated threonine thought to be phosphorylated by Cak1, the cyclin-dependent kinase-activating kinase (15). Our laboratory and others have recently demonstrated that the kin28(T162A) mutant, while still viable in S. cerevisiae, is not phosphorylated at this site by Cak1 and is reduced in its kinase activity (31; Keogh et al., unpublished data). Our genetic results here suggest that this T162A mutation has no effect on the Kin28 activity necessary for the CTD phosphorylation event that recruits capping enzyme (Table 3 and Fig. 2). This was investigated further by genetic analyses with CAK1 conditional mutants and CDC28 mutants that allowed for the deletion of CAK1 (gifts of F. Cross and D. O. Morgan [11, 15]). Mutants were viable and displayed no additional phenotypes in the case of cak1-22 ceg1-250, as well as cdc28-169-43244 cak1Δ ceg1-250 (data not shown). Thus, even if Kin28 fails to receive an activating phosphorylation at T162, it retains sufficient CTD kinase activity to recruit capping enzyme, despite its overall reduced kinase activity.
We previously reported a synthetic lethal combination of a partially truncated CTD and ceg1-250 (8) and now find that the double mutant kin28(T17D) ceg1-250 is also a lethal combination. Tetrad analysis of a cross between rpb1Δ101 and kin28(T17D) reveals that the double mutant is also inviable (data not shown). This contrasts with loss-of-function alleles in srb10, which improve growth of CTD truncation mutants (21). Our data indicate that the Pol II CTD and the TFIIH-associated CTD kinase Kin28 interact genetically with each other and with the capping enzyme.
CTD phosphorylation and Ceg1 protein levels are reduced in CTD truncation and Kin28 mutants.
Disruption of either Kin28 or Ctk1 activity results in a decrease in CTD phosphorylation (12). To examine whether such a decrease affects levels of capping enzyme components, immunoblotting was performed with a variety of CTD kinase mutants (Fig. 3). By using the B3 monoclonal antibody that recognizes phosphoepitopes on the CTD (45), a decrease in CTD phosphorylation [CTD(P)] was seen in ctk1Δ and CTD truncation strains, but not in an srb10Δ strain. CTD phosphorylation is also decreased in the kin28 mutant kin28(T17D), kin28(K36A), and kin28-16 strains, but not in the kin28(T162A) strain. The decrease in CTD phosphorylation in specific kin28 mutants parallels those in strains that exhibit synthetic phenotypes in combination with ceg1-250 (Fig. 2).
FIG. 3.
CTD phosphorylation and Ceg1 are affected in CTD truncation and CTD kinase mutants. Whole-cell extracts were prepared from strains grown for 6 h at 30°C. Eighty micrograms of extract from each strain was assayed by immunoblotting with B3 [α-CTD(P)], anti-Ceg1, and anti-Cet1 antibodies. Lanes: 1, wild type, PY469; 2, ceg1-250, YSB491; 3, ctk1Δ, YSB653; 4, srb10Δ, YSB652; 5, rpb1Δ101 (CTD truncation, 11 wild-type heptapeptide repeats), N398; 6 to 9, FOAR strains yielded from shuffling of kin28 mutants (T17D, K36A, -16, and T162A, respectively) into YSB626, as described in the legend to Fig. 2.
Wild-type Ceg1 levels were reduced in the CTD truncation strain (Fig. 3, lane 5). Ceg1 was also reduced in the kin28(T17D) and kin28-16 mutants, but was relatively unaffected in kin28(K36A) and kin28(T162A) mutants (lanes 6 to 9). The reduction in Ceg1 levels correlates well with the genetic interactions with ceg1-250. The most severe reductions are caused by CTD truncation and kin28(T17D); when combined with the further reduction in guanylyltransferase levels caused by the ceg-250 mutation (lane 2), they are synthetically lethal. kin28-16 is more affected by combination with ceg1-250 than kin28(K36A) and has correspondingly reduced levels of Ceg1. kin28(T162A) does not affect phosphorylated CTD or Ceg1 levels and shows no genetic interactions with ceg1-250.
Interestingly, Ceg1 was unaffected in the ctk1Δ strain, despite the decrease in CTD phosphorylation (lane 3). Therefore, an overall decrease in CTD phosphorylation alone is not sufficient to reduce Ceg1 levels. It is likely that the reduction in CTD phosphorylation caused by ctk1Δ reflects a defect different from that caused by the kin28 mutations. In an srb10Δ strain, Ceg1 levels were actually slightly increased (lane 4). The increase is not due to an increase in Ceg1 mRNA levels (data not shown). Although the mechanism is not understood, it may be a reflection of the competition between Kin28 and Srb10 for CTD phosphorylation as proposed by Hengartner et al. (21). Surprisingly, levels of the triphosphatase subunit Cet1 remained largely unaffected in all CTD kinase mutants (Fig. 3, α-Cet1 panel), even when Ceg1 was reduced. Therefore, Cet1 is likely to be stable when present in excess over Ceg1.
Since Ceg1 protein levels are decreased in a CTD truncation mutant, we tested whether they could be rescued by a wild-type polymerase. The rpb1Δ101 mutant strain was transformed with plasmids containing RPB1, CEG1, or CET1, and whole-cell extracts were prepared. Immunoblotting (Fig. 4) shows that addition of an RPB1 gene with full-length CTD restores levels of Ceg1 protein (lane 4). An additional copy of CEG1 also increases overall levels of Ceg1 protein (lane 5). We previously observed that an additional copy of CET1 raises Ceg1 protein levels of a ceg1-250 mutant (7) in the context of a wild-type Pol II CTD, but additional copies of CET1 fail to rescue Ceg1 levels caused by the CTD truncation (lane 6). The change in levels of Ceg1 is mediated at the protein level (probably stability), since RNA analysis showed that Ceg1 mRNA levels were unaffected in the CTD truncation mutant (data not shown).
FIG. 4.
Ceg1 protein is restored with wild-type Rpb1. Whole-cell extracts were prepared from strains grown for 6 h at 30°C. Eighty micrograms of extract from each strain was assayed by immunoblotting with B3 [α-CTD(P)], anti-Ceg1, and anti-Cet1 antibodies. Lanes: 1, wild type, PY469; 2, ceg1-250, YSB491; 3 to 6, rpb1Δ101 (CTD truncation, 11 wild-type heptapeptide repeats), N398 transformed with vector alone (pRS316), RPB1 (pRP112), pRS316-CEG1, and pRS316-CET1, respectively.
The effects of CTD and Kin28 mutations on Ceg1 levels provide further in vivo evidence for their functional interactions. We previously showed that capping enzyme is recruited to the hyperphosphorylated CTD in vitro (7, 8). The immunoblotting results suggest that capping enzyme guanylyltransferase levels are posttranslationally regulated. Ceg1 bound to the phosphorylated CTD levels may be stabilized relative to unbound Ceg1. This could provide a mechanism for keeping capping enzyme levels correlated with the amount of actively transcribing RNA Pol II.
Serine 5 of the heptapeptide repeat is critical for capping enzyme recruitment.
The primary phosphorylation sites of the CTD repeat YSPTSPS are serine 2 and serine 5 (59). During active growth, the yeast CTD is predominantly phosphorylated on serine 5, while serine 2 phosphorylation increases upon heat shock or diauxic shift (46). Mutant CTDs in which every serine 2 or every serine 5 is replaced by alanine do not support viability (55). However, conditional mutants have been generated in which the amino- or carboxy-terminal half of the CTD is wild type and the other half changes all serine 2 positions [rpb1(S2A)] or serine 5 positions [rpb1(S5A)] to alanine (55). To examine the effect of such a mutated CTD on capping enzyme levels, whole-cell extracts were prepared and subjected to immunoblot analysis (Fig. 5A). Whereas Cet1 remained unaffected in rpb1(S2A) and rpb1(S5A) extracts, Ceg1 levels were reduced in both mutants, although not to the extent seen with the CTD truncation mutant.
FIG. 5.
Serine 5 is critical for capping enzyme recruitment. rpb1 mutants were analyzed in combination with CEG1 and ceg1-250 upon shuffling of Leu2-marked vector alone (pRS315), RPB1 (wild-type CTD, 26 repeats; pRP114), rpb1(CTDΔ) [a CTD truncation mutant, 10 repeats; pY1WT(10)], rpb1(S2A) [pY1A2(8)WT(7)], rpb1(S5A) [pY1A5(5)WT(7)], rpb-15, rpb1-18, and rpb1-19 into N418 and YSB516, respectively. rpb1 mutants were isolated by growth on −Leu +FOA media. (A) Whole-cell extracts were prepared from strains grown for 6 h at 30°C. Eighty micrograms of extract from each strain was assayed by immunoblotting with B3 [α-CTD(P)], anti-Ceg1, and anti-Cet1 antibodies. Lanes: 1, wild type, PY469; 2, ceg1-250, YSB491; 3 to 5, FOAR CEG1 rpb1 shuffled mutants rpb1(CTDΔ) [10 repeats, pY1WT(10)], rpb1(S2A) [pY1A2(8)WT(7)], and rpb1(S5A) [pY1A5(5)WT(7)], respectively. (B) Growth of rpb1 mutants was compared by spotting them onto −Leu +FOA synthetic complete medium plates for 2 days at 30°C.
We also analyzed the growth effects of RPB1 mutants in the presence of ceg1-250. Both CEG1 and ceg1-250 strains were generated which allowed plasmid shuffling of RPB1, and various conditional mutants were tested for the ability to support viability at the normally permissive temperature of 30°C (Fig. 5B). Mutations in regions of RPB1 outside of the CTD had no deleterious effects in combination with ceg1-250 (rpb1-15, -18, and -19). As observed previously, a partially truncated CTD (10 wild-type consensus repeats) is synthetically lethal in combination with ceg1-250. The rpb1(S5A) ceg1-250 double mutant is inviable. This contrasts with the serine 2 mutant, which displays no significant growth reduction in combination with ceg1-250.
The rpb1(S2A) and rpb1(S5A) mutants tested as shown in Fig. 5B were mutated in the amino-terminal half of the CTD. S2A and S5A mutants in the carboxy-terminal half of the CTD (55) were also tested to see whether capping enzyme was more dependent on one particular half of the CTD. We observed lethality for both S5A mutants in combination with ceg1-250 (data not shown). Similarly, both S2A mutants were viable but slower growing in combination with ceg1-250 (data not shown). These data suggest that both halves of the CTD contribute to recruitment of capping enzyme. Furthermore, phosphorylation of serine 5, the site modified by the Kin28 kinase, appears to play a particularly critical role in capping enzyme recruitment to Pol II.
The 3′ processing factor Pta1 is affected by kin28 mutants.
Mammalian mRNA 3′ processing factors, such as the cleavage and polyadenylation specificity factor (CPSF) and cleavage stimulatory factor (CstF), also are linked to Pol II transcription via the CTD, although it remains unclear whether CTD phosphorylation is required for the interaction. Previous experiments have not shown a clear preference by CPSF and the CstF complex for phosphorylated CTD by using a CTD affinity column (37). It has also been suggested that CPSF may be initially recruited to promoters by TFIID and then transferred to the CTD at the start of transcription (13). By immunoblotting, we examined whether the levels of the 3′ processing machinery in yeast are affected by CTD kinase mutants.
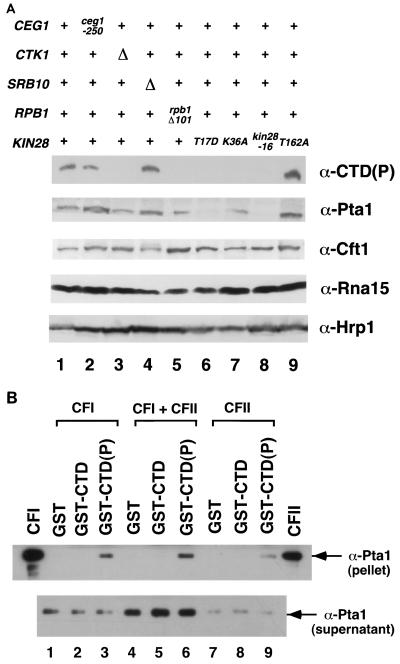
Four separable factors are required for 3′ end formation in yeast; cleavage-polyadenylation factor IA (CFIA), CFIB, and CFII are required for 3′ cleavage, while CFIA, CFIB, polyadenylation factor I (PFI), and poly(A) polymerase (PAP) perform the poly(A) addition (6, 29). Analysis of several members of the 3′ polyadenylation machinery is shown in Fig. 6A. Pta1 and Cft1 are components of both PFI and CFII (47, 53, 60, 61), Rna15 is a member of CFIA (29, 39), and Hrp1 constitutes CFIB (28). Cft1, Rna15, and Hrp1 levels were not changed even when there was a decrease in CTD phosphorylation. In contrast, Pta1 levels were significantly reduced in the two kin28 mutants most defective for CTD phosphorylation (T17D and kin28-16), the same mutants that had the strongest effect on Ceg1 levels. The interaction between polyadenylation factors and Kin28 was further supported by genetic analysis. The conditional pta1-2 allele displays synthetic lethality in combination with kin28(T17D), kin28(K36A), and kin28-16, but no reduced growth phenotype in combination with kin28(T162A) (data not shown). Northern blotting showed that levels of PTA1 mRNA were not reduced in the kin28 mutants, consistent with a defective interaction at the protein level (data not shown). Surprisingly, the partial CTD truncation (rpb1Δ101) did not appear to have a strong effect on wild-type Pta1 protein levels, although synthetic lethal interactions were observed between pta1-2 and several rpb1 mutants (data not shown). These data suggest that CTD phosphorylation by Kin28 plays a role in recruitment of 3′ processing machinery, possibly through an interaction with PFI and/or CFII.
FIG. 6.
Interactions between the 3′ processing factor Pta1 and the phosphorylated CTD. (A) Pta1 protein levels are reduced in kin28 mutant strains. Whole-cell extracts were prepared from strains grown for 6 h at 30°C. Eighty micrograms of extract from each strain was assayed by immunoblotting with B3 [α-CTD(P)], anti-Cft1, anti-Pta1, anti-Rna15, and anti-Hrp1 antibodies. Lanes: 1, wild type, PY469; 2, ceg1-250, YSB491; 3, ctk1Δ, YSB653; 4, srb10Δ, YSB 652; 5, rpb1Δ101 (CTD truncation, 11 wild-type heptapeptide repeats), N398; 6 to 9, FOAR strains yielded from shuffling kin28 mutants (T17D, K36A, -16, and T162A, respectively) into YSB626, as described in the legend to Fig. 2. (B) Pta1 is specifically retained on the phosphorylated CTD. Partially purified polyadenylation factors were incubated with GST (lanes 1, 4, and 7), unphosphorylated GST-CTD fusion protein (lanes 2, 5, and 8), or phosphorylated GST-CTD(P) (lanes 3, 6, and 9). Beads were pelleted and washed, and bound proteins were assayed by immunoblotting with anti-Pta1 antibodies. The CFI and CFII lanes are positive controls.
To test for the interactions in vitro, a GST-CTD fusion protein was phosphorylated and incubated with partially purified CFI and CFII (these are early fractions that both contain Pta1). No association of Pta1 with either GST or unphosphorylated GST-CTD was observed (Fig. 6B, lanes 1, 2, 4, 5, 7, and 8). In contrast, Pta1 was effectively retained on phosphorylated GST-CTD(P) beads (lanes 3, 6, and 9). We also observed preferential retention of Cft1 on the phosphorylated GST-CTD column (data not shown). At this point it is not clear if Pta1 and Cft1 directly contact the phosphorylated CTD or are recruited indirectly as part of a larger complex.
DISCUSSION
Capping enzyme does not use specific RNA sequences to recognize mRNAs transcribed by Pol II. Rather, it is targeted to the Pol II initiation complex and caps mRNAs cotranscriptionally. Capping enzyme is only one of a myriad of factors that bind to the CTD at various stages of the transcription cycle, but has been the only factor to show clear specificity for the phosphorylated form of polymerase. Several CTD kinases are candidates for mediating CTD phosphorylation and capping enzyme recruitment. Here, we present in vivo evidence that it is the TFIIH-associated CTD kinase Kin28 and its CTD phosphorylation site serine 5 that are necessary for capping enzyme recruitment. Furthermore, we find that the yeast 3′-processing factor Pta1 also binds specifically to the phosphorylated CTD in vitro and is stabilized by Kin28 activity in vivo.
Several lines of evidence indicate that Kin28 is the kinase responsible for capping enzyme recruitment. First, combination of a capping enzyme guanylyltransferase mutation with mutant kin28 alleles results in exacerbated phenotypes, and the severity of the synthetic phenotype (from lethality to slower growth to no effect) correlates with severity of the kin28 allele (Table 3 and Fig. 2). This finding parallels the synthetic lethality observed between the capping enzyme mutant and a partial CTD truncation (7). We also observed synthetic lethality between the kin28(T17D) allele and the CTD truncation (data not shown), completing the triangle of interactions between these three components of the capping enzyme recruitment mechanism. These synthetic lethal interactions suggest that KIN28, the Pol II CTD, and CEG1 all interact within the same pathway, because compromising any two members of the pathway reduces cell viability. Based on the genetic analysis alone, it is impossible to determine whether these interactions are direct or indirect. We cannot rule out the possibility that the kin28 mutant alleles affect expression of some other factor required for Ceg1 and Pta1 stability or function. However, in combination with the published biochemical experiments (7, 8, 21, 26, 36, 37), it seems most likely that Kin28 phosphorylation of the CTD mediates recruitment of the capping and polyadenylation factors.
A second indication that Kin28 is the relevant kinase comes from the observation that the CTD truncation and kin28 mutants exhibit reduced levels of Ceg1 protein (Fig. 3 and 4), but not mRNA (data not shown). This suggests that Ceg1 bound to the phosphorylated CTD is stabilized relative to unbound Ceg1. This differential stability, in combination with the allosteric interactions observed between capping enzyme subunits and the CTD (7, 25), may be important for preventing untargeted capping enzyme activity.
Kin28 phosphorylates serine 5 of the CTD consensus repeat. We find that rpb1 alleles carrying mutations of serine 5 to alanine in either the first or second half of the CTD are synthetically lethal in combination with a ceg1 mutant. Similar rpb1 alleles that change serine 2 to alanine are viable in the presence of ceg1-250, but the double mutant grows more slowly than either mutant alone. Therefore, both serines 2 and 5 may contribute to binding of capping enzyme to the CTD, but the contribution of serine 5 is likely to be more important. This is in good agreement with studies of the mammalian capping enzyme reporting that capping enzyme could bind to a CTD peptide phosphorylated at either serine 2 or 5, but that only the serine 5 phosphorylation provided the allosteric activation of guanylyltransferase activity (25).
The in vivo specificity of capping enzyme interaction with the Kin28 CTD kinase contrasts markedly with the absence of interactions seen with the srb10 and ctk1 deletions. All three kinases can phosphorylate the CTD in vitro to allow binding of capping enzyme. Like the kin28 mutants, a ctk1Δ mutant is decreased in bulk CTD phosphorylation, yet no genetic or biochemical perturbation of capping enzyme is seen. Therefore, although both Kin28 and Ctk1 are necessary for CTD phosphorylation in vivo, it is likely that the CTDK1 phosphorylation occurs at a location or time that is not relevant to capping enzyme recruitment. Whereas Kin28 is present as a component of TFIIH in the promoter-bound transcription complex, Ctk1 is not (E. J. Cho, unpublished results). It appears that the Kin28 kinase only functions in the context of the promoter (21). Since capping enzyme can be recruited directly to the initiation complex (8) and capping occurs after only 20 to 30 nucleotides have been transcribed (27, 48), it makes good sense that Kin28 should be the kinase responsible for capping enzyme recruitment. CTDK1 may phosphorylate the CTD at a later phase in the transcription cycle, perhaps as a modulator of transcriptional elongation efficiency by RNA Pol II (34).
The CTD also appears to mediate coupling of transcription to mRNA splicing and 3′ processing (37). In some cases, the phosphorylated CTD appears to preferentially interact with these RNA processing machineries, while other experiments show less of a difference between the CTD species (see references 22, 23, and 37 and references therein). We find that in kin28 mutants defective for CTD phosphorylation, the polyadenylation factor Pta1 is notably less abundant and that Pta1 in crude fractions can bind specifically to the phosphorylated CTD. Pta1 is proposed to be a component of both PFI and CFII in yeast, and both complexes contain homologues of several subunits of the mammalian CPSF (47, 60, 61). Further in vivo studies with Pta1, as well as other yeast polyadenylation and splicing components, may reveal whether the function of these factors requires or is simply enhanced by a particular CTD phosphorylation state.
Our data support and extend the emerging model for the cotranscriptional processing of RNA Pol II transcripts. Hyperphosphorylation of the CTD by Kin28, a component of the general transcription factor TFIIH, is coordinated with the transition from transcription initiation to elongation. Specifically, recruitment of mRNA processing machinery to the CTD structure unique to RNA Pol II provides an elegant means of targeting cap placement, splicing, and cleavage or polyadenylation to the proper RNA substrate. Several studies have suggested coordinated activities between the different mRNA processing machineries, and their close proximity on the CTD provides a means for these interactions. Future studies of the cross-talk between processing machineries and the transcription complex should prove insightful when considering the association and subsequent dissociation of factors depending upon the CTD phosphorylation state.
ACKNOWLEDGMENTS
We thank Jeff Corden, Rick Young, Mark Solomon, Fred Winston, David Pellman, David Morgan, and Fred Cross for the gifts of plasmids and yeast strains. We are particularly grateful to Rick Young, Christoph Hengartner, and Sang Seok Koh for Srb10-Srb11 and Kin28-Ccl1 protein preparations. We also thank Toshimitsu Takagi and Yasutaka Takase for the generation of Cet1 antibody, Ben Blancowe for the B3 antibody, and Patrick O'Connor for Pta1 antibody.
This work was supported by grants from NIH to S.B., C.L.M., and A.G. S.B. gratefully acknowledges support from the Pew Scholars Program and an American Cancer Society Junior Faculty Research Award.
REFERENCES
- 1.Allison L A, Moyle M, Shales M, Ingles C J. Extensive homology among the largest subunits of eukaryotic and prokaryotic RNA polymerases. Cell. 1985;42:599–610. doi: 10.1016/0092-8674(85)90117-5. [DOI] [PubMed] [Google Scholar]
- 2.Allison L A, Wong J K-C, Fitzpatrick V D, Moyle M, Ingles C J. The C-terminal domain of the largest subunit of RNA polymerase II of Saccharomyces cerevisiae, Drosophila melanogaster, and mammals: a conserved structure with an essential function. Mol Cell Biol. 1988;8:321–329. doi: 10.1128/mcb.8.1.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. 1 and 2. New York, N.Y: Greene Publishing Associates and Wiley-Interscience; 1991. [Google Scholar]
- 4.Bartolomei M S, Halden N F, Cullen C R, Corden J L. Genetic analysis of the repetitive carboxyl-terminal domain of the largest subunit of RNA polymerase II. Mol Cell Biol. 1988;8:330–339. doi: 10.1128/mcb.8.1.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carlson M. Genetics of transcriptional regulation in yeast: connections to the RNA polymerase II CTD. Annu Rev Cell Dev Biol. 1997;13:1–23. doi: 10.1146/annurev.cellbio.13.1.1. [DOI] [PubMed] [Google Scholar]
- 6.Chen J, Moore C. Separation of factors required for cleavage and polyadenylation of yeast pre-mRNA. Mol Cell Biol. 1992;12:3470–3481. doi: 10.1128/mcb.12.8.3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho E J, Rodriguez C R, Takagi T, Buratowski S. Allosteric interactions between capping enzyme subunits and the RNA polymerase II carboxy-terminal domain. Genes Dev. 1998;12:3482–3487. doi: 10.1101/gad.12.22.3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cho E J, Takagi T, Moore C R, Buratowski S. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 1997;11:3319–3326. doi: 10.1101/gad.11.24.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cismowski M J, Laff G M, Solomon M J, Reed S I. KIN28 encodes a C-terminal domain kinase that controls mRNA transcription in Saccharomyces cerevisiae but lacks cyclin-dependent kinase-activating kinase (CAK) activity. Mol Cell Biol. 1995;15:2983–2992. doi: 10.1128/mcb.15.6.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corden J L, Cadena D L, Ahearn J J, Dahmus M E. A unique structure at the carboxyl terminus of the largest subunit of eukaryotic RNA polymerase II. Proc Natl Acad Sci USA. 1985;82:7934–7938. doi: 10.1073/pnas.82.23.7934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cross F R, Levine K. Molecular evolution allows bypass of the requirement for activation loop phosphorylation of the Cdc28 cyclin-dependent kinase. Mol Cell Biol. 1998;18:2923–2931. doi: 10.1128/mcb.18.5.2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dahmus M E. Reversible phosphorylation of the C-terminal domain of RNA polymerase II. J Biol Chem. 1996;271:19009–19012. doi: 10.1074/jbc.271.32.19009. [DOI] [PubMed] [Google Scholar]
- 13.Dantonel J C, Murthy K G, Manley J L, Tora L. Transcription factor TFIID recruits factor CPSF for formation of 3′ end of mRNA. Nature. 1997;389:399–402. doi: 10.1038/38763. [DOI] [PubMed] [Google Scholar]
- 14.Eisenmann D M, Arndt K M, Ricupero S L, Rooney J W, Winston F. SPT3 interacts with TFIID to allow normal transcription in Saccharomyces cerevisiae. Genes Dev. 1992;6:1319–1331. doi: 10.1101/gad.6.7.1319. [DOI] [PubMed] [Google Scholar]
- 15.Espinoza F H, Farrell A, Nourse J L, Chamberlin H M, Gileadi O, Morgan D O. Cak1 is required for Kin28 phosphorylation and activation in vivo. Mol Cell Biol. 1998;18:6365–6373. doi: 10.1128/mcb.18.11.6365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feaver W J, Svejstrup J Q, Henry N L, Kornberg R D. Relationship of CDK-activating kinase and RNA polymerase II CTD kinase TFIIH/TFIIK. Cell. 1994;79:1103–1109. doi: 10.1016/0092-8674(94)90040-x. [DOI] [PubMed] [Google Scholar]
- 17.Fresco L D, Buratowski S. Active site of the mRNA capping enzyme guanylyltransferase from Saccharomyces cerevisiae: similarity to the nucleotidyl attachment motif of DNA and RNA ligases. Proc Natl Acad Sci USA. 1994;91:6624–6628. doi: 10.1073/pnas.91.14.6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fresco L D, Buratowski S. Conditional mutants in the yeast mRNA capping enzyme show that the cap enhances, but is not required for, mRNA splicing. RNA. 1996;2:584–596. [PMC free article] [PubMed] [Google Scholar]
- 19.Gerber H P, Hagmann M, Seipel K, Georgiev O, West M A, Litingtung Y, Schaffner W, Corden J L. RNA polymerase II C-terminal domain required for enhancer-driven transcription. Nature. 1995;374:660–662. doi: 10.1038/374660a0. [DOI] [PubMed] [Google Scholar]
- 20.Guthrie C, Fink G R, editors. Guide to yeast genetics and molecular biology. Vol. 194. Boston, Mass: Academic Press, Inc.; 1991. [Google Scholar]
- 21.Hengartner C J, Myer V E, Liao S M, Wilson C J, Koh S S, Young R A. Temporal regulation of RNA polymerase II by Srb10 and Kin28 cyclin-dependent kinases. Mol Cell. 1998;2:43–53. doi: 10.1016/s1097-2765(00)80112-4. [DOI] [PubMed] [Google Scholar]
- 22.Hirose Y, Manley J L. RNA polymerase II is an essential mRNA polyadenylation factor. Nature. 1998;395:93–96. doi: 10.1038/25786. [DOI] [PubMed] [Google Scholar]
- 23.Hirose Y, Tacke R, Manley J L. Phosphorylated RNA polymerase II stimulates pre-mRNA splicing. Genes Dev. 1999;13:1234–1239. doi: 10.1101/gad.13.10.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hirst M, Kobor M S, Kuriakose N, Greenblatt J, Sadowski I. GAL4 is regulated by the RNA polymerase II holoenzyme-associated cyclin-dependent protein kinase SRB10/CDK8. Mol Cell. 1999;3:673–678. doi: 10.1016/s1097-2765(00)80360-3. [DOI] [PubMed] [Google Scholar]
- 25.Ho C K, Shuman S. Distinct roles for CTD Ser-2 and Ser-5 phosphorylation in the recruitment and allosteric activation of mammalian mRNA capping enzyme. Mol Cell. 1999;3:405–411. doi: 10.1016/s1097-2765(00)80468-2. [DOI] [PubMed] [Google Scholar]
- 26.Ho C K, Sriskanda V, McCracken S, Bentley D, Schwer B, Shuman S. The guanylyltransferase domain of mammalian mRNA capping enzyme binds to the phosphorylated carboxyl-terminal domain of RNA polymerase II. J Biol Chem. 1998;273:9577–9585. doi: 10.1074/jbc.273.16.9577. [DOI] [PubMed] [Google Scholar]
- 27.Jove R, Manley J L. In vitro transcription from the adenovirus 2 Major Late Promoter utilizing templates truncated at promoter-proximal sites. J Biol Chem. 1984;259:8513–8521. [PubMed] [Google Scholar]
- 28.Kessler M M, Henry M F, Shen E, Zhao J, Gross S, Silver P A, Moore C L. Hrp1, a sequence-specific RNA-binding protein that shuttles between the nucleus and the cytoplasm, is required for mRNA 3′-end formation in yeast. Genes Dev. 1997;11:2545–2556. doi: 10.1101/gad.11.19.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kessler M M, Zhao J, Moore C L. Purification of the Saccharomyces cerevisiae cleavage/polyadenylation factor I. Separation into two components that are required for both cleavage and polyadenylation of mRNA 3′ ends. J Biol Chem. 1996;271:27167–27175. doi: 10.1074/jbc.271.43.27167. [DOI] [PubMed] [Google Scholar]
- 30.Kim E, Du L, Bregman D B, Warren S L. Splicing factors associate with hyperphosphorylated RNA polymerase II in the absence of pre-mRNA. J Cell Biol. 1997;136:19–28. doi: 10.1083/jcb.136.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kimmelman J, Kaldis P, Hengartner C J, Laff G M, Koh S S, Young R A, Solomon M J. Activating phosphorylation of the Kin28p subunit of yeast TFIIH by Cak1p. Mol Cell Biol. 1999;19:4774–4787. doi: 10.1128/mcb.19.7.4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuchin S, Carlson M. Functional relationships of Srb10-Srb11 kinase, carboxy-terminal domain kinase CTDK-I, and transcriptional corepressor Ssn6-Tup1. Mol Cell Biol. 1998;18:1163–1171. doi: 10.1128/mcb.18.3.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee J M, Greenleaf A L. CTD kinase large subunit is encoded by CTK1, a gene required for normal growth of Saccharomyces cerevisiae. Gene Expr. 1991;1:149–167. [PMC free article] [PubMed] [Google Scholar]
- 34.Lee J M, Greenleaf A L. Modulation of RNA polymerase II elongation efficiency by C-terminal heptapeptide repeat domain kinase I. J Biol Chem. 1997;272:10990–10993. doi: 10.1074/jbc.272.17.10990. [DOI] [PubMed] [Google Scholar]
- 35.Liao S M, Zhang J, Jeffery D A, Koleske A J, Thompson C M, Chao D M, Viljoen M, van Vuuren H J J, Young R A. A kinase-cyclin pair in the RNA polymerase II holoenzyme. Nature. 1995;374:193–196. doi: 10.1038/374193a0. [DOI] [PubMed] [Google Scholar]
- 36.McCracken S, Fong N, Rosonina E, Yankulov K, Brothers G, Siderovski D, Hessel A, Foster S, Shuman S, Bentley D L. 5′-Capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes Dev. 1997;11:3306–3318. doi: 10.1101/gad.11.24.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson S D, Wickens M, Bentley D L. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature. 1997;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- 38.Meisels E, Gileadi O, Corden J L. Partial truncation of the yeast RNA polymerase II carboxyl-terminal domain preferentially reduces expression of glycolytic genes. J Biol Chem. 1995;270:31255–31261. doi: 10.1074/jbc.270.52.31255. [DOI] [PubMed] [Google Scholar]
- 39.Minvielle S L, Preker P J, Keller W. RNA14 and RNA15 proteins as components of a yeast pre-mRNA 3′-end processing factor. Science. 1994;266:1702–1705. doi: 10.1126/science.7992054. [DOI] [PubMed] [Google Scholar]
- 40.Misteli T, Spector D L. RNA polymerase II targets pre-mRNA splicing factors to transcription sites in vivo. Mol Cell. 1999;3:697–705. doi: 10.1016/s1097-2765(01)80002-2. [DOI] [PubMed] [Google Scholar]
- 41.Mizumoto K, Kaziro Y. Messenger RNA capping enzymes from eukaryotic cells. Prog Nucleic Acid Res. 1987;34:1–28. doi: 10.1016/s0079-6603(08)60491-2. [DOI] [PubMed] [Google Scholar]
- 42.Mortillaro M J, Blencowe B J, Wei X, Nakayasu H, Du L, Warren S L, Sharp P A, Berezney R. A hyperphosphorylated form of the large subunit of RNA polymerase II is associated with splicing complexes and the nuclear matrix. Proc Natl Acad Sci USA. 1996;93:8253–8257. doi: 10.1073/pnas.93.16.8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nonet M, Scafe C, Sexton J, Young R. Eukaryotic RNA polymerase conditional mutant that rapidly ceases mRNA synthesis. Mol Cell Biol. 1987;7:1602–1611. doi: 10.1128/mcb.7.5.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nonet M L, Young R A. Intragenic and extragenic suppressors of mutations in the heptapeptide repeat domain of Saccharomyces cerevisiae RNA polymerase II. Genetics. 1989;123:715–724. doi: 10.1093/genetics/123.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O'Brien T, Hardin S, Greenleaf A, Lis J T. Phosphorylation of RNA polymerase II C-terminal domain and transcriptional elongation. Nature. 1994;370:75–77. doi: 10.1038/370075a0. [DOI] [PubMed] [Google Scholar]
- 46.Patturajan M, Schulte R J, Sefton B M, Berezney R, Vincent M, Bensaude O, Warren S L, Corden J L. Growth-related changes in phosphorylation of yeast RNA polymerase II. J Biol Chem. 1998;273:4689–4694. doi: 10.1074/jbc.273.8.4689. [DOI] [PubMed] [Google Scholar]
- 47.Preker P J, Ohnacker M, Minvielle S L, Keller W. A multisubunit 3′ end processing factor from yeast containing poly(A) polymerase and homologues of the subunits of mammalian cleavage and polyadenylation specificity factor. EMBO J. 1997;16:4727–4737. doi: 10.1093/emboj/16.15.4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rasmussen E B, Lis J T. In vivo transcriptional pausing and cap formation on three Drosophila heat shock genes. Proc Natl Acad Sci USA. 1993;90:7923–7927. doi: 10.1073/pnas.90.17.7923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scafe C, Chao D, Lopes J, Hirsch J P, Henry S, Young R A. RNA polymerase II C-terminal repeat influences response to transcriptional enhancer signals. Nature. 1990;347:491–494. doi: 10.1038/347491a0. [DOI] [PubMed] [Google Scholar]
- 50.Scafe C, Martin C, Nonet M, Podos S, Okamura S, Young R A. Conditional mutations occur predominantly in highly conserved residues of RNA polymerase II subunits. Mol Cell Biol. 1990;10:1270–1275. doi: 10.1128/mcb.10.3.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shuman S. Capping enzyme in eukaryotic mRNA synthesis. Prog Nucleic Acid Res. 1995;50:101–129. doi: 10.1016/s0079-6603(08)60812-0. [DOI] [PubMed] [Google Scholar]
- 52.Sterner D E, Lee J M, Hardin S E, Greenleaf A L. The yeast carboxyl-terminal repeat domain kinase CTDK-I is a divergent cyclin–cyclin-dependent kinase complex. Mol Cell Biol. 1995;15:5716–5724. doi: 10.1128/mcb.15.10.5716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stumpf G, Domdey H. Dependence of yeast pre-mRNA 3′-end processing on CFT1: a sequence homolog of the mammalian AAUAAA binding factor. Science. 1996;274:1517–1520. doi: 10.1126/science.274.5292.1517. [DOI] [PubMed] [Google Scholar]
- 54.Weeks J R, Hardin S E, Shen J, Lee J M, Greenleaf A L. Locus-specific variation in phosphorylation state of RNA polymerase II in vivo: correlations with gene activity and transcript processing. Genes Dev. 1993;7:2329–2344. doi: 10.1101/gad.7.12a.2329. [DOI] [PubMed] [Google Scholar]
- 55.West M L, Corden J L. Construction and analysis of yeast RNA polymerase II CTD deletion and substitution mutations. Genetics. 1995;140:1223–1233. doi: 10.1093/genetics/140.4.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yue Z, Maldonado E, Pillutla R, Cho H, Reinberg D, Shatkin A J. Mammalian capping enzyme complements mutant Saccharomyces cerevisiae lacking mRNA guanylyltransferase and selectively binds the elongating form of RNA polymerase II. Proc Natl Acad Sci USA. 1997;94:12898–12903. doi: 10.1073/pnas.94.24.12898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yuryev A, Patturajan M, Litingtung Y, Joshi R V, Gentile C, Gebara M, Corden J L. The C-terminal domain of the largest subunit of RNA polymerase II interacts with a novel set of serine/arginine-rich proteins. Proc Natl Acad Sci USA. 1996;93:6975–6980. doi: 10.1073/pnas.93.14.6975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zehring W A, Lee J M, Weeks J R, Jokerst R S, Greenleaf A L. The C-terminal repeat domain of RNA polymerase II largest subunit is essential in vivo but is not required for accurate transcription initiation in vitro. Proc Natl Acad Sci USA. 1988;85:3698–3702. doi: 10.1073/pnas.85.11.3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang J, Corden J L. Identification of phosphorylation sites in the repetitive carboxyl-terminal domain of the mouse RNA polymerase II largest subunit. J Biol Chem. 1991;266:2290–2296. [PubMed] [Google Scholar]
- 60.Zhao J, Kessler M, Moore C. Cleavage factor II of Saccharomyces cerevisiae contains homologues to subunits of the mammalian cleavage/polyadenylation specificity factor and exhibits sequence-specific, ATP-dependent interaction with precursor RNA. J Biol Chem. 1997;272:10831–10838. doi: 10.1074/jbc.272.16.10831. [DOI] [PubMed] [Google Scholar]
- 61.Zhao J, Kessler M, Helmling S, O'Connor J P, Moore C L. Pta1, a component of yeast CFII, is required for both cleavage and poly(A) addition of mRNA precursor. Mol Cell Biol. 1999;19:7733–7740. doi: 10.1128/mcb.19.11.7733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhu Y, Pe'ery T, Peng J, Ramanathan Y, Marshall N, Marshall T, Amendt B, Mathews M B, Price D H. Transcription elongation factor P-TEFb is required for HIV-1 tat transactivation in vitro. Genes Dev. 1997;11:2622–2632. doi: 10.1101/gad.11.20.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]