Abstract
Protease-digested lactoferrin fragments often exhibit improved therapeutic properties. However, there are limited studies investigating the anticancer properties of these fragments. The fragment with improved anticancer activities is an attractive alternative to chemotherapeutic drugs—presenting severe side effects. Herein, we report the isolation and characterization of recombinant engineered-lactoferrin (rtHLF4), exhibiting up to 100-fold improved anticancer activity compared to the full-length lactoferrin (flHLF). Further, rtHLF4 exerts its anticancer effect in a shorter duration. Through transcriptomic analysis of various cancer biomarkers, rtHLF4 was found to upregulate various pro-apoptotic markers and downregulate signaling proteins involved in angiogenesis and metastasis. We further determined that rtHLF4 showed no hemolytic activity at high concentrations. We believe that this anticancer protein can be further developed as a cancer treatment.
Keywords: Lactoferrin, Anticancer, Tryptic-digestion
Cancer kills approximately 10 million patients each year, accounting for over 15% of deaths worldwide.1 Thus, there is an urgency to search for new anticancer treatments. Current anticancer treatments often have undesirable side effects, whereas natural anticancer remedies have shown to be less efficacious, requiring a high local concentration to elicit the anticancer properties. The lactoferrin protein exerts multiple therapeutic properties, including anticancer, antimicrobial, antiparasitic, antiviral, and antifungal activity.2 It was recently found that lactoferrin can hasten the recovery of patients infected with COVID-19.3,4 This transferrin-class protein exhibits these therapeutic activities by recruiting immune-related cytokines and depleting nutrients in the diseased area through metal ion sequestration.5
Protease-digested lactoferrin often results in fragments that exhibit improved therapeutic activities. Lactoferricin (peptic-digested lactoferrin) has improved antimicrobial and antifungal properties compared to the full-length lactoferrin (flHLF).6 These therapeutic activities are mainly attributed to the lactoferricin’s amphipathic and cationic properties that are similar to bacteriocins and pyocins.6 Other products such as lactoferrampin showed increased killing efficacy against certain fungal strains.7 However, there are limited studies that suggest that protease-digested lactoferrin fragments show improved anticancer activities.
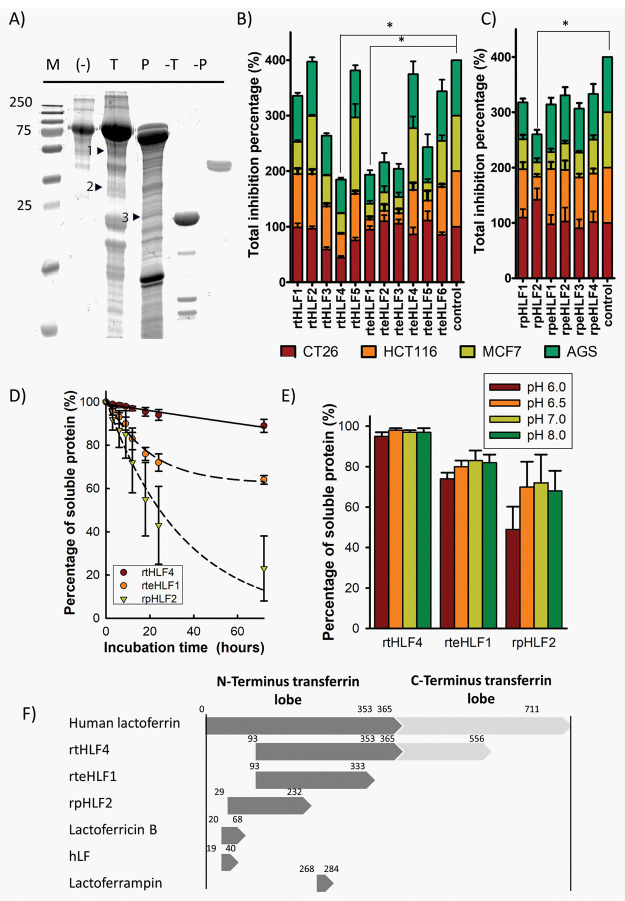
This study adopts a rational approach to investigate protease-digested lactoferrin fragments with improved anticancer properties. Previous studies found that lactoferrin is highly resistant to proteolytic degradation, resulting in incomplete digested fragments that exhibit different bioactivities.8 Protease-digested recombinantly expressed human lactoferrin, sorted by fragment size (Figure 1A), were screened for anticancer activity against gastric (AGS), breast (MCF7), and colorectal cancer (HCT116 and CT26) cell lines (Figure 1B,C). Three promising candidates (rtHLF4, rteHLF1, and rpHLF1) were found to have improved inhibition against the four cancer cell lines, inhibiting over 50% of the cultured cancer cells. Out of the three fragments, only fragment rtHLF4 showed improved stability under human physiological temperature (Figure 1D) and pH (Figure 1E). Protein identification using MALDI-TOF of these fragments revealed that all three peptides are truncated at the N- and C-terminus (Figure 1F).
Figure 1.
Isolation of anticancer lactoferrin fragments. (A) SDS-PAGE of trypsin (T) and pepsin (P) digested lactoferrin. Fragments with positive anticancer activity are indicated with arrows (M, marker; T, Trypsin; P, Pepsin; 1, rtHLF4; 2, rteHLF1; 3, rpHLF2). (B) Anticancer activity of trypsin-digested lactoferrin fragments. (C) Anticancer activity of pepsin-digested lactoferrin fragments. (D) Isolated lactoferrin fragment stability at 37 °C. (E) Purified lactoferrin fragment stability under different pH after 12 h. (F) The sequence length of isolated fragments compared to lactoferrin and other lactoferrin-derived fragments (*P ≤ 0.0083 after Bonferroni correction; each group performed in triplicate).
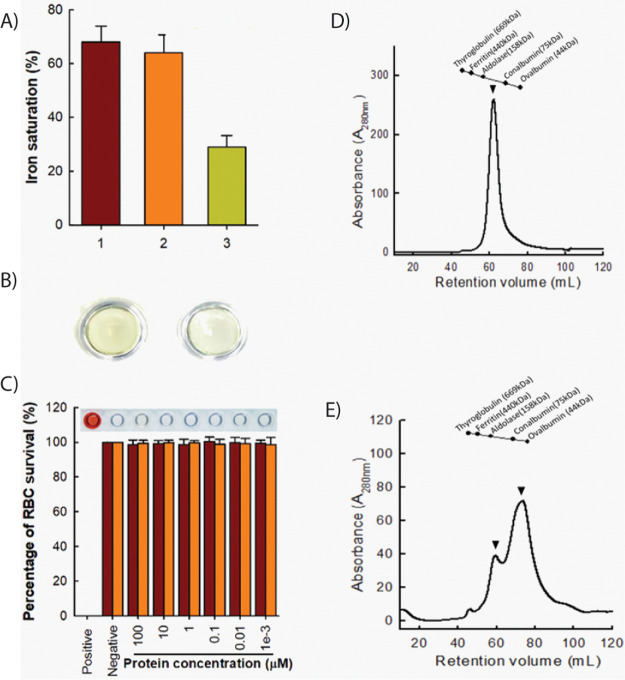
We focused our studies on rtHLF4 due to its stability exhibited under human-physiological conditions and its comparable anticancer properties to rteHLF1 and rpHLF2 (Figure 1B,C). We hypothesize that the rtHLF4 truncated C-terminus lobe functions to stabilize the lactoferrin protein, where rteHLF1 and rpHLF2 lacking the C-terminus domain are less stable under different pH and temperature changes. Recombinant expression of rtHLF4 showed that the level of ferric occupancy in the binding pockets is 3-fold lower than that of flHLF (Figure 2A), leading to the color of rtHLF4 being much lighter than that of flHLF under the same concentration (Figure 2B). The flHLF comprises two globular lobes, each with one iron-binding site.9 The 3-fold lower ferric ion occupancy in rtHLF4 suggests that the truncated N-terminus and disrupted C-terminus lobe cannot retain the ferric-binding activity due to the loss of its conserved ferric-binding residues. The modeled rtHLF4 structure showed perturbation of its C-terminus lobe affecting the ferric binding pocket (Figure S1b)10 as compared to the flHLF structure (Figure S1c). Additionally, rtHLF4 can exist in both the monomeric and dimeric conformation (Figure 2E), whereas flHLF has the preference of being in the dimeric state (Figure 2D). These findings indicate that the truncated C-terminus lobe helps in protein stability, but incurs the loss of ferric-binding and interferes with the dimerization interface (Figure S1a).
Figure 2.
Structural and biosafety analysis of rtHLF4. (A) Percentage of ferric ion saturation of lactoferrin and rtHLF4 (1, commercial lactoferrin; 2, flHLF; 3, rtHLF4; each group performed in triplicate). (B) Color of recombinant lactoferrin (left) and rtHLF4 (right; samples prepared at 0.1 mM). (C) Hemolytic assay of purified lactoferrin (maroon) and rtHLF4 (orange) on human red blood cells (inset: hemolytic results using the corresponding rtHLF4 concentrations; n = 10, positive control, 0.1 mg/mL indomethacin). (D) Size exclusion chromatographic separation of flHLF (protein weight, 80 kDa) (arrows indicate dimeric conformation of the isolated protein). (E) Size exclusion separation of rtHLF4 (protein weight, 51 kDa; arrows indicate dimeric and monomeric conformation of the isolated protein).
We conducted a hemolytic toxicity assay to evaluate the safety of using recombinant rtHLF4 on healthy cells. The hemolytic toxicity assay using blood acquired from 10 donors (aged between 25–40 years old) found no hemolytic activities in the erythrocytes treated with 100 μM of recombinantly expressed flHLF or rtHLF4 (Figure 2C). These results suggest that the observed anticancer activities result from direct rtHLF4 interactions with the cancer cells and are not the result of cell toxicity.
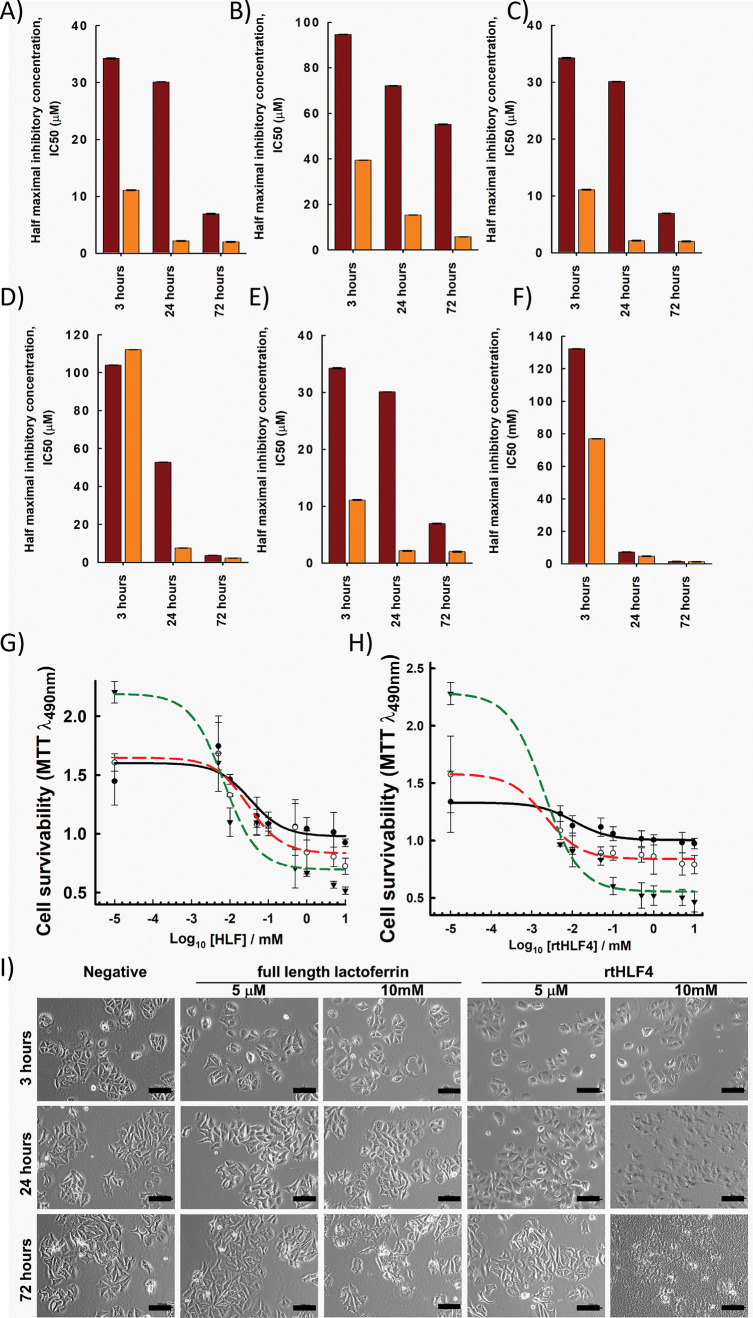
We compared the anticancer activities of both rtHFL4 and flHLF against human cancer cell lines including breast (MCF7), stomach (HGC27 and AGS), and colorectal (Caco2, LoVo, and HCT116) cancer cell lines. We evaluated the anticancer activity by observing the treatment time (3, 24, and 72 h) needed to elicit complete inhibition of the cancer cells. rtHLF4 is more potent against cancer cells by 1 to 2 orders of magnitude when compared to flHLF. Additionally, rtHLF4 elicits complete cancer cell inhibition within 24 h of treatment (Figure 3A–F) compared to the flHLF that needs a longer incubation time. The dose-dependent-cell viability assay of rtHLF4 against MCF-7 (Figure3G,H) and other cancer cells (Figures S2–S6) revealed improved efficacy and sensitivity against these cancer cell lines. Similarly, the cell morphology of MCF7 (Figure 3I) and other cancer cells (Figures S2c–S6c) showed an indication of observable cell death annotated by detachment and cellular apoptosis within 24 h of treatment with rtHLF4, while flHLF requires a higher concentration and longer incubation time to elicit a similar level of cellular death. Studies have shown that flHLF’s anticancer activity results from the ferric ion sequestration, causing gradual killing activity arising from nutrient starvation.8 As rtHLF4 has lesser ferric ion binding ability, we hypothesize that the mechanism of action attributing to rtHLF4’s improved anticancer properties differs from that of flHLF.
Figure 3.
Anticancer activity of lactoferrin and rtHLF4. (A) The half maximal inhibitory concentration, IC50, of flHLF (maroon) and rtHLF4 (orange) tested against breast cancer (A) MCF7; gastric cancer (B) HGC27 and (C) AGS; and colorectal cancer (D) Caco2, (E) LoVo, and (F) HCT116. Dose–response of MCF7 against varying concentrations of (G) flHLF and (H) rtHLF4. (I) Cell morphology of MCF7 cell lines treated with varying concentrations of flHLF and rtHLF4 (bar = 100 μm).
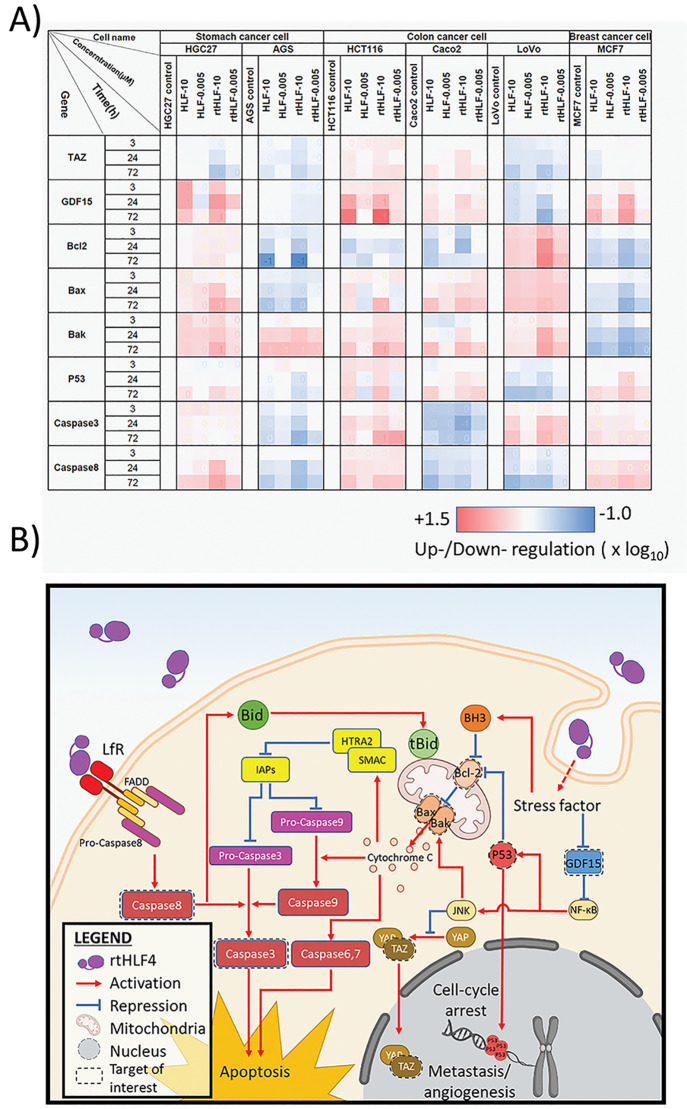
In order to understand the mechanism of action, we investigated the gene expression of anticancer-associated biomarkers in the cancer cell cultures treated with rtHLF4 and flHLF. The gene expression of pro-apoptotic, angiogenesis, and metastatic proteins was monitored using quantitative PCR and Western blot (Figure 4A). In comparison to flHLF, millimolar and micromolar rtHLF4 concentration administered to the cancer cells revealed changes in protein expression level involved in the death receptor pathway (caspase3 and caspase8), mitochondrial outer membrane permeabilization (MOMP; Bak/Bax, and Bcl2), and P53-related pathway (P53, GDF15, and TAZ).
Figure 4.
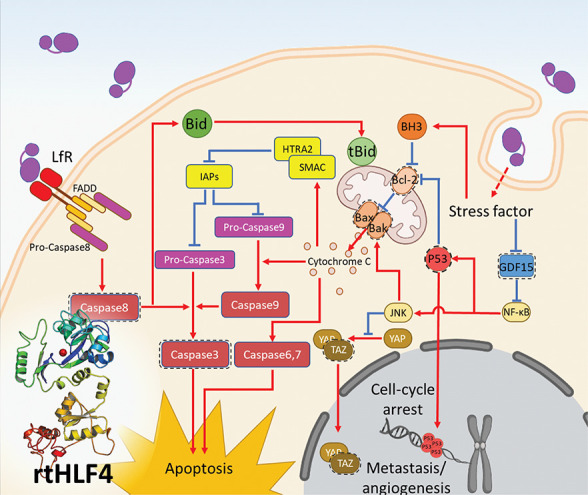
Anticancer mechanisms in gastric, colon, and breast cancer cells following treatment with flHLF and rtHLF4. (A) quantitative PCR results of different genes in gastric, colon, and breast cancer cells. Expression levels were compared against housekeeping gene GADPH (flHLF/rtHLF4–10:10 μM of flHLF/rtHLF4; flHLF/rtHLF4–0.005:0.005 μM of flHLF/rtHLF4). (B) The different anticancer pathway in cancer cells triggered by rtHLF4 treatment.
rtHLF-treated HGC27, HCT116, and MCF7 showed elevated caspase3 and caspase8 expression, suggesting that rtHLF4 triggers activation of caspase8, leading to the proteolytic cleavage of caspase3 (Figures 4a, S7, S8, S12). Studies have shown that lactoferrin-binding receptors with Fas-associated death domain (FADD) trigger the activation of caspase8, triggering the death receptor pathway (Figure 4b).11,12 Caspase8 then activates caspase3 that subsequently cleaves critical cellular protein, leading to cancer cell apoptosis. These results are consistent with previous studies that indicated that the presence of FADD-associated lactoferrin-binding receptors such as CD91, TLR2, TLR4, and intelectin-1 activates the death receptor pathway upon lactoferrin binding.13 It was found that these receptors are present in various cancer cell lines including colorectal epithelial cancer, breast cancer, and gastric cancer.14 We hypothesize that the improved upregulation of the caspase8 can be attributed to the predominant monomeric state of rtHLF, allowing better receptor–ligand interactions.
rtHLF4 also triggers cellular apoptosis via the upregulation of the mitochondrial outer membrane permeabilization (MOMP) proteins, Bax/Bak in HGC27, AGS, Caco2, LoVo, and HCT116 cells (Figures 4a, S8–S12). Upregulated MOMP releases cytochrome C, triggering the caspase6, -7, and -9 activation.15 Caspase9 activates caspase3, leading to cellular apoptosis. The Bax/Bak activation is triggered through various factors including Bcl-2 suppression, P53 activation, and JNK pathway activation (Figure 4b). We observed improved Bcl-2 suppression and upregulated Bak/Bax in AGS, Caco2, and HCT116 cancer cells treated with rtHLF4. Upregulated P53 was found in HGC27, Caco2, and HCT116 treated with rtHLF4 that similarly observed elevated levels of Bak/Bax. These findings suggest that Bax/Bak activation results from the internalization of rtHLF4, leading to increased stress factors within the cell. This activates the Bcl-2 homology 3 (BH3) that represses Bcl-2, thus alleviating the repression of the Bax/Bak proteins.15 Additionally, these stress factors also contribute to the upregulation of P53 and JNK proteins that repress Bcl-2 and upregulate Bak/Bax activities, respectively.16,17 Evidence shows that the cytochrome C released from the mitochondria following MOMP triggers the upregulation of HTRA2/SMAC activation that represses Survivin, a major inhibitor of apoptosis protein (IAP).18 IAP blocks the effect of caspases 3, 7, and 9, preventing cellular apoptosis. Previous studies revealed that bovine lactoferrin significantly decreases IAP expression in colorectal cancer cell lines while observing the upregulation of IAP inhibitors SMAC and HTRA2.18 Our results reveal similar traits of caspase3 activation in both flHLF and rtHLF4, where the latter showed an approximately 2-fold increase of caspase3 in four cancer cell lines (MCF7, HGC27, AGS, and LoVo). The rtHLF4-treated LoVo cells showed upregulated caspase3 and Bax/Bak proteins, while showing downregulation of GDF15, suggesting that the repression of GDF15 relieves the NF-κB activating JNK, leading to Bax/Bak activation. Through the Bak/Bax activation, the caspase3 proteolytic cleavage is triggered by cytochrome C–caspase9 activation.19
In addition, in the experiment of rtHLF4’s anticancer activities, we found that rtHLF4 slows cancer cell growth better than flHLF (Figure 3A–F) by 1 to 2 orders of magnitude. We hypothesize that rtHLF4 additionally inhibits the PDZ-binding motif (TAZ) and growth differentiation factor 15 (GDF15). rtHLF4 showed better inhibition of TAZ in HGC27, AGS, and LoVo cells and improved inhibition of GDF15 in AGS and LoVo (Figures 4a, S8, S9, S11). TAZ conventionally regulates mesenchymal stem cell differentiation, but it is also a known coactivator yes-associated protein (YAP) of the Hippo-pathway genes that upregulate multiple tumorigenic genes including cell migration, invasion, and anchorage-independent growth.20,21 The TAZ/YAP activation plays an essential role in cancer initiation and solid tumor formation through the activation of Survivin expression (Figure 4b).22 Thus, rtHLF4’s suppression of TAZ prevents both the cancer metastatic potential and angiogenesis. GDF15, a protein from the transforming growth factor beta (TGF-β) superfamily, is upregulated and secreted by various metastatic cancers including glioblastoma, ovarian, prostate, breast, and colorectal cancers. While the corresponding role in cancer metastasis is not fully understood, upregulated GDF15 is associated with poor patient survival after cancer remission.23 The ability of rtHLF4 to inhibit GDF15 indicates the potential use of the lactoferrin fragment for the prevention of cancer metastasis and upregulation of P53 mediated cell apoptosis (Figure 4b).
In summary, we identified three protease-digested human lactoferrin fragments that present improved anticancer properties as compared to flHLF. Among the three, rtHLF4 showed the best stability and tolerance to human physiological conditions. Upon closer inspection on rtHLF4’s mode of mechanism, we found that rtHLF4 elicits a wide range of anticancer effects on various cancer cell types via lactoferrin receptor interactions, internalization of rtHLF4, and inhibition of metastatic genes, thus indicating the potential use of this protein in future cancer therapy. Further investigation would be needed to better understand the anticancer mechanism prior to development of rtHLF4 as a viable treatment. These include a closer investigation of receptor proteins and binding proteins involved in the internalization of rtHLF4 to the cytoplasm. We intend to further investigate such interactions using thermal proximity coaggregation to profile the protein complex dynamics within these cancer cells.24 The rtHLF4 potentially can also function as an output mechanism for synthetic biology-driven cellular engineering for anticancer therapy.25,26
Acknowledgments
The authors wish to thank Xinyi Chen and Jun Chen for their comments on the manuscript. The authors also wish to acknowledge the advice provided by Professor Dai Lei and Assistant Professor Tan Yang from the Shenzhen Institute of Advanced Technology (SIAT). The authors recognize Zeyang Qu, Jingyu Xiao, and Juliana Rizal for their help in the experimental designs.
Glossary
Abbreviations
- rtHLF4
recombinant engineered-lactoferrin
- flHLF
full-length lactoferrin
- MALDI-TOF
matrix assisted laser desorption ionization-time-of-flight
- FADD
Fas-associated death domain
- TLR
toll-like receptor
- BH3
Bcl-2 homology 3
- MOMP
mitochondrial outer membrane permeabilization
- TAZ
PDZ-binding motif
- GDF15
growth differentiation factor 15
- YAP
yes-associated protein
- TGF-β
transforming growth beta
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsptsci.1c00134.
Experimental details and supporting figures and tables; details of the materials and methods involved in the experiments, including the list of RT-PCR primer sequences; the sequence and structure of flHLF and rtHLF4, including all anticancer activities of the protein against the different cancer cells (PDF)
Author Contributions
§ These authors contributed equally. C.L.H. designed the study. Y.P., N.C. and K.L. performed the experiments. Y.P., N.C., and C.L.H. analyzed the data. C.L.H. wrote the manuscript. C.L.H. supervised the project. All authors discussed the results and commented on the manuscript.
This work was supported by the Shenzhen Institutes of Advanced Technology External Funds (DWKF20190001), National Natural Science Foundation of China’s Research Fund for International Young Scientists (22050410270), Guangdong Innovative Projects for the Characteristics of General Colleges and Universities (2018KTSCX200), and Guangdong Innovative and Entrepreneurial Research Team Program (2019ZT08Y191)
The authors declare no competing financial interest.
Supplementary Material
References
- Ritchie H., and Roser M.. Causes of Death. https://ourworldindata.org/causes-of-death#citation. [Google Scholar]
- González-Chávez S. A.; Arévalo-Gallegos S.; Rascón-Cruz Q. (2009) Lactoferrin: structure, function and applications. Int. J. Antimicrob. Agents 33 (4), 301.e1. 10.1016/j.ijantimicag.2008.07.020. [DOI] [PubMed] [Google Scholar]
- Wang Y.; Wang P.; Wang H.; Luo Y.; Wan L.; Jiang M.; Chu Y. (2020) Lactoferrin for the treatment of COVID-19 (Review). Exp. Ther. Med. 20 (6), 272. 10.3892/etm.2020.9402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salaris C.; Scarpa M.; Elli M.; Bertolini A.; Guglielmetti S.; Pregliasco F.; Blandizzi C.; Brun P.; Castagliuolo I. (2021) Protective Effects of Lactoferrin against SARS-CoV-2 Infection In Vitro. Nutrients 13 (2), 328. 10.3390/nu13020328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel H. J. (2012) Lactoferrin, a bird’s eye view. Biochem. Cell Biol. 90 (3), 233–44. 10.1139/o2012-016. [DOI] [PubMed] [Google Scholar]
- Gifford J. L.; Hunter H. N.; Vogel H. J. (2005) Lactoferricin: a lactoferrin-derived peptide with antimicrobial, antiviral, antitumor and immunological properties. Cell. Mol. Life Sci. 62 (22), 2588–98. 10.1007/s00018-005-5373-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Kraan M. I. A.; Groenink J.; Nazmi K.; Veerman E. C. I.; Bolscher J. G. M.; Nieuw Amerongen A. V. (2004) Lactoferrampin: a novel antimicrobial peptide in the N1-domain of bovine lactoferrin. Peptides 25 (2), 177–183. 10.1016/j.peptides.2003.12.006. [DOI] [PubMed] [Google Scholar]
- Levay P. F.; Viljoen M. (1995) Lactoferrin: a general review. Haematologica 80 (3), 252–267. [PubMed] [Google Scholar]
- Lönnerdal B.; Iyer S. (1995) Lactoferrin: Molecular Structure and Biological Function. Annu. Rev. Nutr. 15 (1), 93–110. 10.1146/annurev.nu.15.070195.000521. [DOI] [PubMed] [Google Scholar]
- Yang J.; Zhang Y. (2015) I-TASSER server: new development for protein structure and function predictions. Nucleic Acids Res. 43 (W1), W174–W181. 10.1093/nar/gkv342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita K.-i.; Matsuda E.; Sekine K.; Iigo M.; Tsuda H. (2004) Lactoferrin enhances Fas expression and apoptosis in the colon mucosa of azoxymethane-treated rats. Carcinogenesis 25 (10), 1961–1966. 10.1093/carcin/bgh205. [DOI] [PubMed] [Google Scholar]
- Ho C. L.; Hwang I. Y.; Loh K.; Chang M. W. (2015) Matrix-immobilized yeast for large-scale production of recombinant human lactoferrin. MedChemComm 6 (3), 486–491. 10.1039/C4MD00537F. [DOI] [Google Scholar]
- Kell D. B.; Heyden E. L.; Pretorius E. (2020) The Biology of Lactoferrin, an Iron-Binding Protein That Can Help Defend Against Viruses and Bacteria. Front. Immunol. 2020 (1221), 11. 10.3389/fimmu.2020.01221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amarante-Mendes G. P.; Adjemian S.; Branco L. M.; Zanetti L. C.; Weinlich R.; Bortoluci K. R. (2018) Pattern Recognition Receptors and the Host Cell Death Molecular Machinery. Front. Immunol. 9, 2379. 10.3389/fimmu.2018.02379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill K. L.; Huang K.; Zhang J.; Chen Y.; Luo X. (2016) Inactivation of prosurvival Bcl-2 proteins activates Bax/Bak through the outer mitochondrial membrane. Genes Dev. 30 (8), 973–88. 10.1101/gad.276725.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemann M. T.; Lowe S. W. (2006) The p53–Bcl-2 connection. Cell Death Differ. 13 (8), 1256–1259. 10.1038/sj.cdd.4401962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blatt N. B.; Boitano A. E.; Lyssiotis C. A.; Opipari A. W. Jr.; Glick G. D. (2008) Bz-423 superoxide signals apoptosis via selective activation of JNK, Bak, and Bax. Free Radical Biol. Med. 45 (9), 1232–42. 10.1016/j.freeradbiomed.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons J. A.; Kanwar J. R.; Kanwar R. K. (2015) Iron-free and iron-saturated bovine lactoferrin inhibit survivin expression and differentially modulate apoptosis in breast cancer. BMC Cancer 15 (1), 425. 10.1186/s12885-015-1441-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L.; Xu G.; Li J.; Liu W.; Jia W.; Ma J.; Wei D. (2017) Bovine lactoferricin P13 triggers ROS-mediated caspase-dependent apoptosis in SMMC7721 cells. Oncol. Lett. 13 (1), 511–517. 10.3892/ol.2016.5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S. W.; Lim C. J.; Guo K.; Ng C. P.; Lee I.; Hunziker W.; Zeng Q.; Hong W. (2008) A Role for TAZ. in Migration, Invasion, and Tumorigenesis of Breast Cancer Cells. Cancer Res. 68 (8), 2592. 10.1158/0008-5472.CAN-07-2696. [DOI] [PubMed] [Google Scholar]
- Dey A.; Varelas X.; Guan K.-L. (2020) Targeting the Hippo pathway in cancer, fibrosis, wound healing and regenerative medicine. Nat. Rev. Drug Discovery 19 (7), 480–494. 10.1038/s41573-020-0070-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatani K.; Maehama T.; Nishio M.; Goto H.; Kato W.; Omori H.; Miyachi Y.; Togashi H.; Shimono Y.; Suzuki A. (2016) Targeting the Hippo signalling pathway for cancer treatment. J. Biochem. 161 (3), 237–244. 10.1093/jb/mvw074. [DOI] [PubMed] [Google Scholar]
- Spanopoulou A.; Gkretsi V. (2020) Growth differentiation factor 15 (GDF15) in cancer cell metastasis: from the cells to the patients. Clin. Exp. Metastasis 37 (4), 451–464. 10.1007/s10585-020-10041-3. [DOI] [PubMed] [Google Scholar]
- Tan C. S. H.; Go K. D.; Bisteau X.; Dai L.; Yong C. H.; Prabhu N.; Ozturk M. B.; Lim Y. T.; Sreekumar L.; Lengqvist J.; Tergaonkar V.; Kaldis P.; Sobota R. M.; Nordlund P. (2018) Thermeal proximigy coaggregation for system-wide profiling of protein complex dynamics in cells. Science 359 (6380), 1170–1177. 10.1126/science.aan0346. [DOI] [PubMed] [Google Scholar]
- Ho C. L.; Tan H. Q.; Chua K. J.; Kang A.; Lim K. H.; Ling K. L.; Yew W. S.; Lee Y. S.; Thiery J. P.; Chang M. W. (2018) Engineered commensal microbes for diet-mediated colorectal-cancer chemoprevention. Nature Biomedical Engineering 2 (1), 27–37. 10.1038/s41551-017-0181-y. [DOI] [PubMed] [Google Scholar]
- Lubkowicz D.; Ho C. L.; Hwang I. Y.; Yew W. S.; Lee Y. S.; Chang M. W. (2018) Reprogramming Probiotic Lactobacillus reuteri as a Biosensor for Staphylococcus aureus Derived AIP-I Detection. ACS Synth. Biol. 7 (5), 1229–1237. 10.1021/acssynbio.8b00063. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.