Abstract
Transcription in human papillomaviruses (HPVs) is mainly regulated by cellular transcription factors and virus-encoded E2 proteins that act as sequence-specific DNA-binding proteins. Although the functions of E2 as a transcriptional activator and a repressor have been well documented, the role of cellular factors involved in E2-mediated regulation of the HPV promoters and the mechanism by which E2 modulates viral gene expression remain unclear. Using reconstituted cell-free transcription systems, we found that cellular enhancer-binding factors and general cofactors, such as TAFIIs, TFIIA, Mediator, and PC4, are not required for E2-mediated repression. Unlike other transcriptional repressors that function through recruitment of histone deacetylase or corepressor complexes, HPV E2 is able to directly target components of the general transcription machinery to exert its repressor activity on the natural HPV E6 promoter. Interestingly, preincubation of TATA binding protein (TBP) or TFIID with HPV template is not sufficient to overcome E2-mediated repression, which can be alleviated only via formation of a minimal TBP (or TFIID)-TFIIB-RNA polymerase II-TFIIF preinitiation complex. Our data therefore indicate that E2 does not simply work by displacing TBP or TFIID from binding to the adjacent TATA box. Instead, E2 appears to function as an active repressor that directly inhibits HPV transcription at steps after TATA recognition by TBP or TFIID.
Transcription in eukaryotes is often regulated by extracellular molecules that act through distinct signal transduction pathways to modulate specific gene expression via controlling the activity of gene-specific transcription factors. These gene-specific transcription factors then work in conjunction with general transcription factors (GTFs) and cofactors to enhance or inhibit the level of transcription. Although many studies have been conducted to elucidate the mechanisms of transcriptional activation in eukaryotes, relatively little is known about the mechanisms of repression. In general, transcriptional repressors can work either passively to antagonize the activator function or actively to inhibit the activity of the general transcription machinery (30). Counteraction of the activator function by passive repressors can be achieved by direct competition of the same DNA-binding sites (36, 37, 41, 54, 55), interference of overlapping or neighboring activator-binding sites (21, 24, 38, 58), modification of the DNA-binding property of the activators (60), titrating away limiting protein factors required for activator function (15, 31), or masking and/or altering the function of the activation domain or blocking the DNA-binding activity of the activators through protein-protein interactions (3, 24, 46, 61). In contrast, active repressors are able to directly inhibit the activity or the assembly of the general transcription machinery, with or without the help of corepressors (2, 23, 27, 29, 40, 43, 45, 51). The recruitment of histone deacetylase complexes by some repressors or corepressors represents another level of gene regulation via alteration of chromatin structure (48).
To decipher the mechanisms of transcriptional repression, we use human papillomaviruses (HPVs) as a model system, since repression of HPV transcription plays a critical role in regulating the expression of viral oncoproteins that accounts for the pathogenic nature of many HPV-induced human diseases including benign genital warts and cervical cancer. E2, encoded by HPVs, is a sequence-specific transcription factor that recognizes a consensus sequence, ACCN6GGT, found in the upstream regulatory region (URR or long control region) of all papillomaviruses so far identified (41, 55). Depending on the sequence context of the E2-binding sites, E2 can function as a transcriptional activator or as a repressor. Four consensus E2-binding sites, whose positions are highly conserved among HPVs, are located in the URR (44). Binding of E2 to the promoter-distal sites enhances, whereas E2 binding to the promoter-proximal elements inhibits, E6 promoter activity. Although activation of the E6 promoter, which controls the expression of many viral gene products involved in HPV DNA replication, transcription, and transformation, requires E2 working in conjunction with URR enhancer-binding factors (14, 16, 20, 21, 32, 59, 63, 72), E2-mediated repression of the E6 promoter is believed to occur in the promoter-proximal region by preventing the binding of adjacent cellular factors such as Sp1 and TATA binding protein (TBP) that are critical for preinitiation complex (PIC) assembly (17, 21, 22, 59). Indeed, binding of Sp1 to its cognate sequence is displaced by increasing amounts of E2 as demonstrated by electrophoretic mobility shift assays (17, 21, 59), correlating with the observation that mutations on the Sp1-binding site partially relieve E2-mediated repression while mutations on the promoter-proximal E2-binding sites enhance E6 promoter activity during transient transfection assays (17, 21, 50, 59). In addition to passive repression, E2 has been suggested to function as an active repressor by preventing the assembly of a functional PIC based on in vitro transcription studies performed with HeLa nuclear extracts (22). Although preincubation of HPV DNA templates with HeLa nuclear extracts apparently alleviates E2-mediated repression, it is not clear whether the inhibitory activity of E2 is overcome solely by components of the GTFs or by other cellular proteins, such as Sp1, present in HeLa nuclear extracts. Therefore, the role of individual cellular factors, including both enhancer-binding proteins and GTFs, in HPV transcription remains to be investigated.
To elucidate the mechanism of E2-mediated regulation of the E6 promoter and to define the role of cellular transcription factors in HPV transcription, we have established E2-dependent cell-free transcription systems reconstituted either with individually purified GTFs and cofactors (68) or with a TATA-binding factor (TBP or TFIID) and preassembled TFIID-deficient RNA polymerase II (pol II) holoenzyme that contains pol II, a subset of GTFs (TFIIB, TFIIE, TFIIF, and TFIIH), SRBs (suppressors of RNA polymerase B mutations), and chromatin remodeling factors (67, 69). Using a combination of these in vitro transcription systems, we demonstrate that E2 protein, derived from HPV type 11 (HPV-11), which induces benign genital warts and laryngeal papillomatosis in infected patients, can suppress the homologous E6 promoter independently of cellular enhancer-binding factors, general cofactors such as TAFIIs in TFIID, TFIIA, Mediator, and USA-derived components, and protein phosphorylation on E2. The N-terminal domain of E2, which is important for transactivation, is not required for E2-mediated repression. Only a subset of GTFs, including TBP, TFIIB, pol II, and TFIIF, are needed and sufficient for E2-mediated repression of the HPV-11 E6 promoter. Surprisingly, using order-of-addition and Sarkosyl to manipulate the sequence of factor assembly in the transcription assays, we find that preincubation of TBP (or TFIID, which is a multiprotein complex comprising TBP and approximately a dozen TAFIIs) with DNA templates cannot overcome E2-mediated repression, indicating that steric hindrance of TBP (or TFIID) binding to the TATA box may not be the sole mechanism by which E2 inhibits E6 promoter activity. Interestingly, alleviation of E2-mediated repression can be achieved via formation of a minimal TBP (or TFIID)-TFIIB-pol II-TFIIF PIC. These findings suggest that, unlike other transcriptional repressors that function through recruitment of histone deacetylase or corepressor complexes, HPV E2 is able to directly target components of the general transcription machinery to exert its repressor activity on the natural HPV E6 promoter.
MATERIALS AND METHODS
Plasmid constructions.
Bacterial expression plasmids pF:E1-11d, pF:E2-11d, pF:E4-11d, pF:CM-11d, pF:CM2-11d, and pF:CM4-11d, used for the expression of FLAG-tagged HPV-11 E1, E2, E4, E1M ^E2C, E1Ma ^E4, and double-spliced E2C (ds-E2C) (5, 7), were constructed from p6HisF:E1-11d, p6HisF:E2-11d (11), p6HisF:E4-11d, p6HisF:CM-11d, p6HisF:CM2-11d, and p6HisF:CM4-11d, respectively, by PCR amplification with an NcoI site-containing sense primer (5′ CAAGGGAATTCGCCATGGACTACAAA 3′) that anneals to the FLAG sequence and an antisense primer (5′ TGCTAGTTATTGCTCAGCGG 3′) located downstream of the BamHI site in p6HisF-11d (11), after cloning individual PCR products into the NcoI and BamHI sites of pET-11d (Novagen). The plasmid p6HisF:E1-11d was generated by cloning the NdeI (created at the initiation codon by PCR)-BglII fragment of the HPV-11 E1 cDNA from pVL-E1 (6) into the NdeI and BamHI sites of p6HisF-11d. Plasmids p6HisF:E4-11d, p6HisF:CM-11d, p6HisF:CM2-11d, and p6HisF:CM4-11d were made by cloning the N-terminal fragment of each cDNA from pRSE4 (12), pMT2-CM-M849 (7), pMT2-CM2-M849 (same as pMT2-CM-M849 except containing the E1Ma ^E4 splice sites [5]), and pMT2-CM4 (7), respectively, between NdeI (created at the initiation codon) and NarI sites together with a common C-terminal fragment between NarI and XhoI sites of pCMV-E2 (8) into the NdeI and XhoI sites of p6HisF-11d.
The transfer plasmids pVL-F:CM, pVL-F:CM2, pVL-F:CM4, and pVL-F:E4, used for baculoviral expression of FLAG-tagged HPV-11 E1M ^E2C, E1Ma ^E4, ds-E2C, and E4, were constructed by substituting the NcoI-BamHI fragments of HPV inserts isolated from pF:CM-11d, pF:CM2-11d, pF:CM4-11d, and pF:E4-11d, respectively, for the HPV-11 E2 insert in pVL-F°:E2 (69) between the NcoI and BamHI sites. The transfer plasmid pVL-F°:E1, used for baculoviral expression of FLAG-tagged HPV-11 E1 without a heart muscle kinase phosphorylation site linked to the FLAG epitope, was made by first cloning the NdeI-EcoRI fragment of the full-length E1 cDNA from p6HisF:E1-11d, following partial NdeI and EcoRI digestion, into the NdeI-EcoRI-linearized pFLAG°(AS)-7 vector (66) to create pF°:E1-7. The FLAG-tagged HPV-11 E1-coding region was then isolated from pF°:E1-7 and cloned into pVL1392 between the BglII and EcoRI sites to generate pVL-F°:E1.
The transcriptional template pGL7072-161, containing the HPV-11 URR spanning nucleotides 7072 to 7933/1 to 161 linked to a luciferase reporter gene, was constructed by cloning the HPV-11 insert, amplified from pSVO10/HPV-11 (5) with a KpnI site-containing sense primer (5′ TTCCCGGGTACCGGATCCCTATAAGGATATG 3′) and a BglII site-containing antisense primer (5′ CCAAGCTTAGATCTAAACGTCTTGCACAAC 3′) by PCR, into pGL2-Basic (Promega) between the KpnI and BglII sites. The G-less cassette templates p7072-70GLess/I+, p7072-70GLess/I−, p7862-70GLess/I+, and p7862-70GLess/I− (with numbers corresponding to the first and last nucleotides of the HPV-11 inserts) used for in vitro transcription were constructed by cloning the PCR products, generated from HPV-11 URR-containing plasmid pUR23-3 (13) with an EcoRI site-containing sense primer (5′ TTGGTACCGAATTCGGATCCCTATAAGGATATG 3′ for the cloning of p7072-70GLess/I+ and p7072-70GLess/I− and 5′ GGAGAGGGTACCGAATTCATGAGTAACCTAAGGTCA 3′ for the cloning of p7862-70GLess/I+ and p7862-70GLess/I−) and a SacI site-containing antisense primer (5′ GAGAGGAGATCTGAGCTCTTATATATAACCGTTTTC 3′), into either pIGL (65), which contains the adenovirus major late promoter (MLP) initiator linked to a G-less cassette of 388 nucleotides, or pGL (65), which contains a G-less cassette of 377 nucleotides without the MLP initiator, between EcoRI and SacI sites. The transcription templates, p7862-70(2M)GLess/I−, p7862-70(3M)GLess/I−, p7862-70(4M)GLess/I−, p7862-70(23M)GLess/I−, p7862-70(24M)GLess/I−, p7862-70(34M)GLess/I−, and p7862-70(234M)GLess/I−, which contain mutations on E2-binding sites 2, 3, 4, 2 and 3, 2 and 4, 3 and 4, and 2 and 3 and 4, respectively, were constructed similarly to p7862-70GLess/I−, except that DNA templates containing various E2-binding site mutations in the backbone of 24-N (21) were used for PCR amplification.
Protein purification.
Bacterially expressed FLAG-tagged HPV-11 proteins E1, E2, E4, E1M ^E2C, E1Ma ^E4, and ds-E2C were purified from BL21(DE3)pLysS strains harboring pF:E1-11d, pF:E2-11d, pF:E4-11d, pF:CM-11d, pF:CM2-11d, and pF:CM4-11d, respectively, according to the published protocol (10). Purification of FLAG-tagged HPV-11 E1, E2, E4, E1M ^E2C, and ds-E2C from insect Sf9 cells was performed by using pVL-F°:E1, pVL-F°:E2, pVL-F:E4, pVL-F:CM, and pVL-F:CM4, individually, as described elsewhere (69). Isolation of recombinant FLAG-tagged human GTFs (TFIIB, TBP, TFIIEα, and TFIIEβ), FLAG-tagged multiprotein complexes (TFIID, TFIIH, and pol II), six-histidine-tagged TFIIF subunits (RAP30 and RAP74), recombinant PC4, and TFIID-deficient pol II holoenzyme was conducted as described previously (9, 10, 34, 67, 68).
In vitro transcription with HeLa nuclear extracts.
In vitro transcription performed with HeLa nuclear extracts and analyzed by primer extension was carried out in a 25-μl reaction mixture containing 250 ng of pGL7072-161, 5 ng of pHIV+58 (33), 40 mM HEPES-Na (pH 8.4), 0.2 mg of bovine serum albumin per ml, 60 mM KCl, 3 mM MgCl2, 4 mM dithiothreitol (DTT), 0.6 mM nucleoside triphosphates (NTPs), 4 U of RNasin (Promega), and 5 μl of HeLa nuclear extracts, prepared as previously described (18), in the absence or presence of increasing amounts of baculovirus-expressed HPV-11 E2 protein. After incubation of the reaction mixture at 30°C for 60 min, the RNA synthesized was extracted by acid phenol and precipitated with ethanol. Purified RNA was incubated with 0.06 pmol of each 32P-labeled primer specific for the luciferase [Luc(AS)-5 primer, 5′ CTCTTCATAGCCTTATGCAG 3′] and the chloramphenicol acetyltransferase (CAT) gene (5′ CAACGGTGGTATATCCAGTG 3′) in 10 μl of buffer containing 8.8 mM Tris-HCl (pH 7.5), 0.88 mM EDTA, and 0.25 M KCl, at 85°C for 2 min, 65°C for 1 h, and finally at room temperature for 10 min. Primer extension was then performed by adding 25 μl of reverse transcriptase cocktail containing 5 U of avian myeloblastosis virus reverse transcriptase (Promega), 10 mM DTT, 10 mM MgCl2, 20 mM Tris-HCl (pH 8.7), 1 mM dNTPs, 1.5 μg of actinomycin D, and 20 U of RNasin at 42°C for 45 min. The reverse transcriptase product was then ethanol precipitated, resolved by 8% polyacrylamide gels containing 50% urea, and detected by autoradiography. Unless otherwise specified, relative intensity in each set of reactions is defined as the signal intensity quantitated by PhosphorImager (Molecular Dynamics) from the HPV templates relative to that performed in the absence of E2.
In vitro transcription performed with G-less cassette templates in HeLa nuclear extracts was similarly conducted in a 25-μl reaction mixture containing 50 ng of HPV-11 G-less cassette template, 50 ng of pMLΔ53 (42), 25 mM HEPES-Na (pH 8.4), 0.5 mg of bovine serum albumin per ml, 72 mM KCl, 4 mM MgCl2, 5 mM DTT, 0.5 mM ATP, 0.5 mM UTP, 25 μM CTP, 1 μl of [α-32P]CTP (3,000 Ci/mmol; Amersham Pharmacia Biotech), 0.1 mM 3′-O-methyl-GTP, 20 U of RNasin, 10 U of RNase T1 (Amersham Pharmacia Biotech), and 1 μl of HeLa nuclear extracts, in the absence or presence of increasing amounts of baculovirus-expressed HPV-11 E2 protein. Reactions were then performed and analyzed as described elsewhere (67).
Reconstituted in vitro transcription assays.
In vitro transcription performed with a TATA-binding factor and TFIID-deficient pol II holoenzyme (f:pol II [67]) was conducted with 50 ng of HPV-11 G-less cassette template, 50 ng of pMLΔ53, 1 ng of TBP or an equivalent amount of FLAG-tagged TFIID (normalized by Western blotting with anti-TBP antibodies), and 3 μl (∼90 ng) of f:pol II, in the absence or presence of HPV-11 proteins as specified in the figures. Reactions were then conducted and analyzed as described elsewhere (67). The order-of-addition experiments outlined in Fig. 6A were similarly conducted as described elsewhere (67) by preincubating 50 ng of p7072-70GLess/I− and 50 ng of pMLΔ53 with TBP and f:pol II, individually or together, at 30°C for 30 min, in the absence or presence of baculovirus-expressed HPV-11 E2. The remaining transcription components and ribonucleoside triphosphates were then added, with or without 0.015% Sarkosyl, to initiate transcription. E2, if included (50 ng), was also added at different time points as indicated.
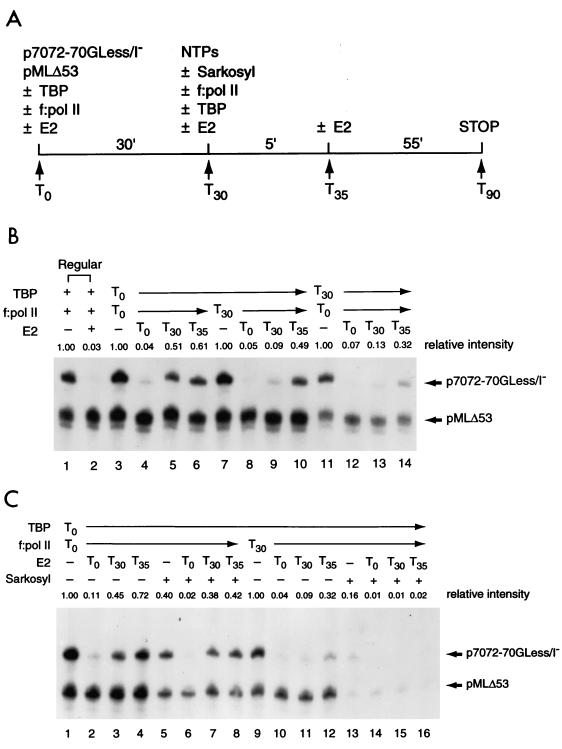
FIG. 6.
Preincubation of TBP with transcriptional templates cannot overcome E2-mediated repression. (A) Outline of the order-of-addition experiment. Transcriptional templates (p7072-70GLess/I− and pMLΔ53) were incubated with TBP and TFIID-deficient pol II holoenzyme (f:pol II), individually or together, at 30°C for 30 min. The remaining transcription components and ribonucleoside triphosphates (NTPs) were then added, in the absence (−) or presence (+) of 0.015% Sarkosyl, to initiate transcription. E2, if included (50 ng), was added at different time points (T0, T30, or T35) during the incubation. Reactions were then processed as described in Materials and Methods. (B) Preincubation of TBP alone with the transcriptional templates is unable to overcome E2-mediated repression. The order-of-addition experiment was conducted as described for panel A. Two regular reactions performed in the absence (−) or presence (+) of E2 were also conducted (lanes 1 and 2) for comparison. (C) E2 inhibits multiple rounds of transcription. The order-of-addition experiment was carried out as described for panel B, with (+) or without (−) 0.015% Sarkosyl added at T30.
Unless otherwise specified, in vitro transcription reconstituted with recombinant GTFs (TFIIB, TBP, TFIIE, and TFIIF) and highly purified epitope-tagged multiprotein complexes (TFIIH and pol II) was performed with 50 ng of HPV template, 50 ng of pMLΔ53, 1 ng of TBP, 10 ng of TFIIB, 5 ng of TFIIEα, 5 ng of TFIIEβ, 28 ng of reconstituted TFIIF (20 ng of RAP74 and 8 ng of RAP30), 15 ng of TFIIH, and 30 ng of pol II, in the absence or presence of 50 ng of E2. The reactions were then processed as described previously (68). The order-of-addition experiment performed with a minimal transcription system containing TBP, TFIIB, pol II, and TFIIF is outlined at the bottom of Fig. 8A. Briefly, transcriptional templates (p7072-70GLess/I+ or p7072-70GLess/I− and pMLΔ53) were incubated first with TBP, TBP-TFIIB, TBP-TFIIB-pol II, or TBP-TFIIB-pol II-TFIIF individually in different tubes at 30°C for 30 min. The remaining transcriptional components and NTPs were then added, in the absence or presence of E2, to initiate transcription and processed as described above.
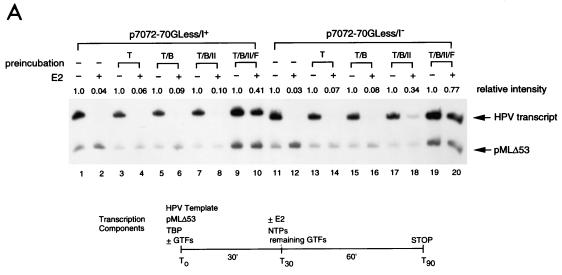
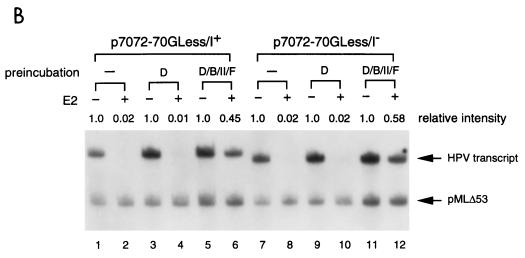
FIG. 8.
E2-mediated repression of the HPV-11 E6 promoter can be alleviated by forming a promoter-bound TBP (or TFIID)-TFIIB-pol II-TFIIF complex. (A) TBP as the TATA-binding factor for PIC assembly. In vitro transcription was performed as outlined at the bottom by preincubating transcriptional templates with TBP (T), TBP-TFIIB (T/B), TBP-TFIIB-pol II (T/B/II), or TBP-TFIIB-pol II-TFIIF (T/B/II/F), in different reaction tubes at 30°C for 30 min. The remaining components and NTPs were then added, in the absence (−) or presence (+) of HPV-11 E2, to initiate transcription. The reactions were processed as described in Materials and Methods. Regular E2-mediated reactions with all four protein components but without a two-step incubation were also conducted for comparison (lanes 1 and 2 and lanes 11 and 12). (B) TFIID as the TATA-binding factor for PIC assembly. In vitro transcription was performed as outlined in panel A, except that TFIID (D) was used as the TATA-binding factor.
Mapping of transcription start sites.
The transcription start site from pGL7072-161 was mapped by first performing an in vitro transcription reaction with 50 ng of pGL7072-161 and 5 μl of HeLa nuclear extracts, in the absence or presence of different amounts of α-amanitin, as described above except that primer extension was carried out with a different luciferase primer [Luc(AS)-4, 5′ ACCAACAGTACCGGAATGCCA 3′]. The same primer was also used to generate DNA sequencing markers for mapping of the transcription start site, which was visualized by autoradiography after separation on a DNA sequencing gel.
For mapping of the transcription start sites from HPV-11 G-less cassette templates (p7072-70GLess/I+, p7862-70GLess/I+, p7072-70GLess/I−, and p7862-70GLess/I−) and pMLΔ53, 250 ng of each DNA template was incubated with 5 μl of HeLa nuclear extracts. In vitro transcription and primer extension were similarly performed as described above except for the use of a primer (5′ TGAGAGTGAATGATGATAGATTTGGG 3′) that anneals to the G-less cassettes. This primer was also used to create DNA sequencing markers for mapping of the transcription start sites.
DNase I footprinting.
The DNA fragment, spanning nucleotides 7902 to 7933/1 to 161 of HPV-11, used for DNase I footprinting was generated by PCR amplification of pGL7072-161 with a KpnI-containing sense primer (5′ TTCCCGGGTACCGGTACATATTGCCCTG 3′) and a BglII-containing antisense primer (5′ CCAAGCTTAGATCTAAACGTCTTGCACAAC 3′). The PCR product was end labeled with 32P by T4 polynucleotide kinase. The top-strand specifically end-labeled template was generated by BglII digestion and further purified by passage through a MicroSpin G-25 column (Amersham Pharmacia Biotech). Approximately 3 fmol of labeled DNA fragment (∼50,000 cpm) was used for DNase I footprinting analysis according to the same protocol as described previously (9).
RESULTS
Establishment of cell-free transcription systems regulated by HPV-11 E2.
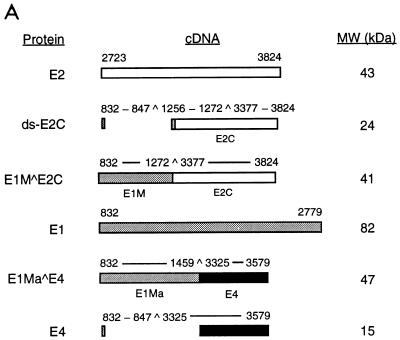
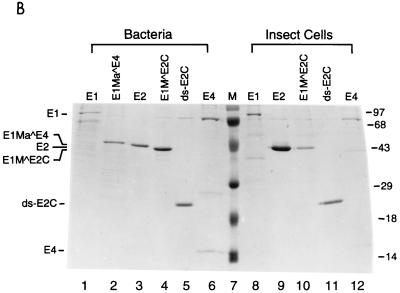
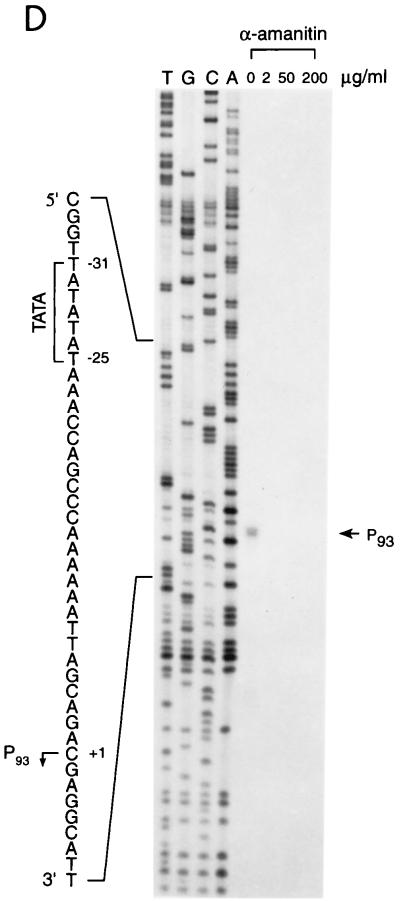
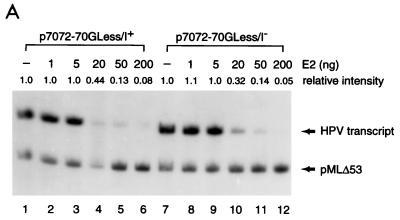
To dissect the mechanisms of E2-mediated regulation of the natural HPV E6 promoter, we decided to use HPV-11 as a model system, since multiple forms of HPV-11 E2 proteins that have the same DNA-binding and dimerization domain but differ in their N-terminal amino acid residues have been identified and shown to play distinct roles in viral DNA replication and transcription (5, 7). Three of the HPV-11 E2 family proteins (E2, ds-E2C, and E1M ^E2C) as well as the E1 family proteins (E1, E1M ^E2C, and E1Ma ^E4) and E4 (Fig. 1A) to be used as controls for in vitro transcription assays were purified from both bacteria and insect cells via FLAG-epitope tagging and peptide elution methods (Fig. 1B). To test whether our purified FLAG-tagged full-length E2 protein (simply referred to as E2 throughout the text) was transcriptionally active and was capable of recapitulating E2-mediated regulation of HPV transcription in cell-free systems, we first performed a transcription assay with HeLa nuclear extracts, which provide all the cellular transcription factors and cofactors necessary for E2-mediated transcription in vitro (22, 56, 63). A DNA template, pGL7072-161, containing the HPV-11 URR spanning nucleotides 7072 to 7933/1 to 161 linked to a luciferase reporter gene was used for initial characterization. A human immunodeficiency virus type 1 (HIV-1) promoter-driven CAT reporter plasmid, pHIV+58, with the HIV-1 upstream regulatory sequence up to −167 (33) was also included in the transcription reaction as a negative control for E2-mediated response. The amounts of transcripts formed in the nuclear extract from both DNA templates were measured by primer extension with 32P-labeled luciferase and CAT primers and analyzed on an 8% polyacrylamide–urea gel. The expected transcript size is approximately 200 nucleotides for pGL7072-161 and 129 nucleotides for pHIV+58. As shown in Fig. 1C, transcription from the HPV-11 template is enhanced up to 2-fold at low concentrations of E2 but inhibited up to 13-fold at high amounts of E2, as observed previously for transfected cells (21). Two minor transcripts also responsive to E2 are caused by strong pausing sites found in the HPV-11 transcript during reverse transcriptase reactions (data not shown). Transcription from the internal HIV control template remains constant throughout the experiment (Fig. 1C, lanes 1 to 6). This result not only confirms a previous hypothesis for E2 regulation of the HPV-11 E6 promoter (13) and is consistent with a result similarly obtained for HeLa nuclear extracts with HPV-18 E2 protein (56) but also demonstrates that our FLAG-tagged E2 protein, which shows sequence-specific binding activity as assayed by DNase I footprinting (see below), indeed has transcriptional activity as expected from a natural untagged E2 protein. Since the transcription start site of the HPV-11 E6 promoter has been estimated thus far by RNase protection assays only to nucleotides between 90 and 99 (13, 19, 53), we have now precisely mapped the E6 start site to nucleotide 93 (P93) by primer extension and defined the location of the TATA box (TATATAT, −31 to −25, corresponding to HPV-11 nucleotides 62 to 68) used for specifying the transcription start site of the E6 promoter (Fig. 1D). Initiation at P93 represents an authentic pol II-specific transcript, as transcription from P93 is completely abolished at low concentrations of α-amanitin (Fig. 1D).
FIG. 1.
Recapitulation of E2-mediated transcriptional regulation of the homologous E6 promoter in vitro. (A) HPV-11 E2, E1, and E4 family proteins. Numbers next to carets indicate the nucleotide positions of exon boundaries adjacent to splice donors and acceptors, whereas numbers at the beginning and end of each box are the first and last nucleotides of individual open reading frames. The E2, E1, and E4 open reading frames are shown as white, gray, and black boxes, respectively. The apparent molecular mass (MW) of each protein, estimated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, is also listed on the right. (B) Purified recombinant HPV-11 proteins. Coomassie blue staining of FLAG-tagged HPV-11 proteins purified from either bacteria (lanes 1 to 6) or Sf9 insect cells (lanes 8 to 12) by immunoaffinity purification and peptide elution methods (see Materials and Methods) is shown. The positions of HPV-11 proteins separated by sodium dodecyl sulfate–15% polyacrylamide gel electrophoresis are indicated on the left. Prestained protein size markers (in kilodaltons; from GIBCO-BRL) are depicted on the right. (C) Transcriptional regulation of the HPV-11 E6 promoter by homologous E2 protein. In vitro transcription was performed in HeLa nuclear extracts (N.E.) with pGL7072-161, which contains the HPV-11 URR spanning nucleotides 7072 to 7933/1 to 161, and an HIV-1 internal control template, pHIV+58, in the absence (−) or presence of increasing amounts of baculovirus-expressed HPV-11 E2 protein. Transcripts derived from the HPV-11 and HIV-1 templates are indicated, respectively, by arrows. Relative intensity is defined as the signal intensity quantitated by PhosphorImager (Molecular Dynamics) from the HPV template relative to that performed in the absence of E2. (D) Mapping of the HPV-11 E6 promoter start site. In vitro transcription was performed with HeLa nuclear extracts with pGL7072-161, and the RNA synthesized was subjected to primer extension analysis as described in Materials and Methods. Different amounts of α-amanitin (in micrograms per milliliter) were also included in the reactions to score for pol II-specific transcripts. The DNA sequencing marker, prepared from the same primer and DNA template as employed for in vitro transcription, was used for the assignment of the transcription initiation site. The DNA sequence surrounding the transcription start site (+1) at P93, indicated by the arrow, and the TATA box of the E6 promoter are shown on the left.
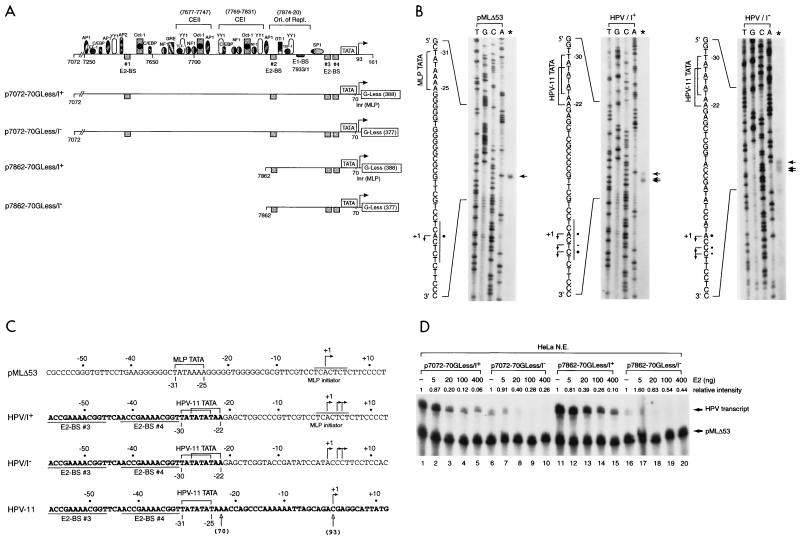
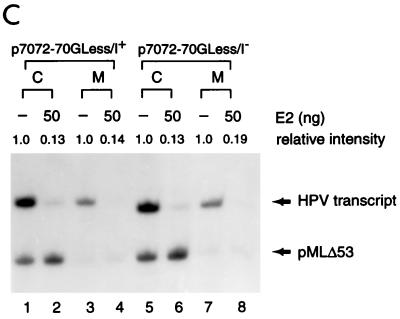
To facilitate the in vitro analysis of HPV transcription, we also created four DNA templates (Fig. 2A) containing HPV-11 URR and the E6 TATA box linked to G-less cassettes with or without an initiator element (Inr) derived from the adenovirus MLP. The inclusion of the MLP Inr in some of the G-less cassette templates (p7072-70GLess/I+ and p7862-70GLess/I+) may strengthen E6 core promoter activity and further enhance the effect of E2-mediated regulation of the E6 promoter, which does not have an apparent consensus initiator sequence. To ensure that transcription from these HPV-11 G-less templates is indeed driven by the homologous E6 promoter, we first mapped the transcription start sites from each of these DNA templates by in vitro transcription-primer extension performed with HeLa nuclear extracts. As shown in Fig. 2B, a cluster of three start sites was found in the HPV-11 templates, irrespective of the absence or presence of the MLP Inr sequence (middle and right panels). Although the relative intensities of these start sites vary slightly among these HPV templates, the correct spacing of the start sites from the E6 TATA box (Fig. 2C) indicates that transcription from these G-less cassettes is indeed driven by the E6 promoter. Interestingly, the entire AT-rich sequence of the E6 promoter, which spans 10 nucleotides, is apparently used as TATA boxes. In contrast, only one start site is detected from the adenovirus MLP in pMLΔ53 (42), which contains only a single TATA box (TATAAAA) surrounded by GC-rich sequences (Fig. 2B, left panel). This analysis demonstrates that the HPV-11 E6 promoter is fully active and correctly specifies the transcription start sites in the G-less cassette templates. These four HPV-11 G-less cassette templates and pMLΔ53, used as an internal control, were then tested for their response to HPV-11 E2 protein by in vitro transcription performed with HeLa nuclear extracts. Under a condition with limiting amounts of cellular factors provided by HeLa nuclear extracts, we clearly detected E2-mediated repression from all four HPV templates, although inhibition of the E6 promoter seems to be stronger in the presence of the MLP Inr sequence (Fig. 2D, compare lanes 1 to 5 with lanes 6 to 10 and lanes 11 to 15 with lanes 16 to 20), whose presence indeed strengthens the effect of E2-mediated repression presumably due to the enhancement of basal transcription from the E6 promoter by some Inr-binding proteins present in HeLa nuclear extracts (Fig. 2D, compare lanes 1 and 6 and lanes 11 and 16). Moreover, since the inclusion of constitutive enhancer elements I and II (CEI and CEII [Fig. 2A]), which are functional in vivo (13, 28), does not enhance E6 promoter activity (Fig. 2D, compare lanes 1 and 11 and lanes 6 and 16), this result also suggests that the HPV-11 enhancer elements are not active in vitro when transcriptional assays are performed on naked DNA templates with HeLa nuclear extracts. Nevertheless, the experiment shows that E2-mediated repression of the HPV E6 promoter can indeed be observed with G-less cassette templates, which can be used for mechanistic studies of HPV transcription (see below).
FIG. 2.
Use of G-less cassette templates for transcriptional analysis of HPV-11 E6 promoter activity. (A) HPV-11 G-less cassettes. Plasmids p7072-70GLess/I+ and p7072-70GLess/I− contain the HPV-11 URR spanning nucleotides 7072 to 7933/1 to 70 linked to either a G-less cassette of 388 nucleotides preceded by the adenovirus MLP Inr element (I+) or another G-less cassette of 377 nucleotides without the MLP Inr (I−). Plasmids p7862-70GLess/I+ and p7862-70GLess/I− containing a truncated HPV-11 URR from nucleotide 7862 to nucleotide 70 without CEI and CEII were similarly linked to G-less cassettes with or without the MLP Inr. The compilation of cis-acting elements and trans-acting factors in the HPV-11 URR is mainly based on published information (28, 44, 72). The boundaries of CEI (28) and CEII (13) and the origin of replication (7) are indicated by brackets. (B) Start site mapping of HPV-11 and MLP G-less cassette templates. In vitro transcription reactions were performed with HeLa nuclear extracts with either an adenovirus MLP-driven G-less cassette template (pMLΔ53 [42]), HPV-11 p7072-70GLess/I+ and p7862-70GLess/I+ templates (both give the same start sites and are thus indicated as HPV/I+), or p7072-70GLess/I− and p7862-70GLess/I− templates (both give the same start sites and are thus indicated as HPV/I−). The transcription start sites (arrows), MLP Inr (underlined), and TATA boxes (brackets) are shown on the left of each panel with intensities of relative start sites denoted by dots. (C) Comparison of nucleotide sequences surrounding the core promoter elements of the MLP and HPV templates. HPV-11 nucleotides are in boldface while vector or MLP sequences are in regular type. Brackets indicate the boundaries of the TATA boxes with lines above the sequence demarking the MLP initiator element. The promoter-proximal E2-binding sites (E2-BS 3 and E2-BS 4) are underlined, and the nucleotides (open arrows) corresponding to HPV-11 number designations are indicated in parentheses. (D) E2-mediated repression of the homologous E6 promoter. In vitro transcription was performed with HeLa nuclear extracts with 50 ng of each HPV and pMLΔ53 templates, in the absence (−) or presence of increasing amounts of baculovirus-expressed E2 protein.
General cofactors, such as TAFIIs and PC4, and phosphorylation on E2 are not required for E2-mediated repression of the E6 promoter.
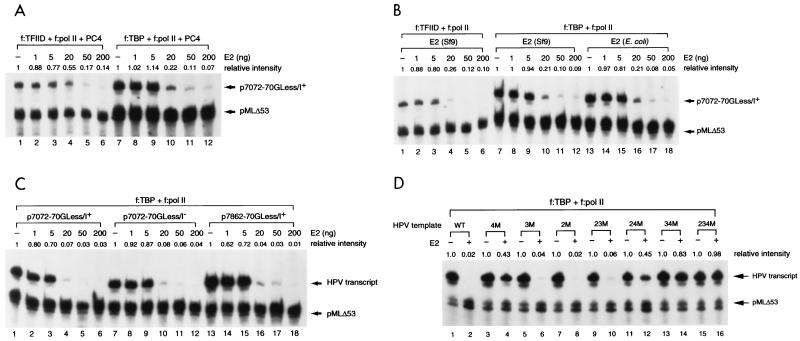
Although in vitro transcription performed with nuclear extracts provides a valuable analysis to determine the domains and concentrations of E2 involved in papillomaviral transcription (22, 56, 57, 63), the molecular mechanisms by which E2 works in conjunction with cellular proteins and the role of cellular factors involved in E2-mediated repression of the E6 promoter cannot be easily addressed in transcription assays with crude nuclear extracts. To overcome this problem, we resorted to the use of a cell-free transcription system reconstituted with a TATA-binding factor (TBP or TFIID) and a preassembled TFIID-deficient pol II holoenzyme complex (67, 69). This system allows us to address the requirement of general transcription cofactors, such as the TAFII components of TFIID and USA-derived cofactor PC4, that are known to possess both stimulatory and inhibitory activities for E2-mediated repression. As shown in Fig. 3A, repression of the E6 promoter could indeed be observed in this two-component transcription system in a dose-dependent and E2-binding site-specific manner (Fig. 3A, lanes 1 to 6). Repression of the E6 promoter persisted when TBP was used in place of TFIID, indicating that TAFIIs are not required for E2-mediated repression (Fig. 3A, lanes 7 to 12). In agreement with our previous report (67), the level of basal transcription conducted with TBP is, in general, higher than that of transcription performed with TFIID (Fig. 3A, compare lanes 1 and 7), implicating a suppressing role of TAFIIs in basal transcription (4). To examine whether PC4 is also dispensable for E2-mediated repression and to investigate whether phosphorylation on E2 is critical for E2 function, we conducted a similar experiment by leaving PC4 out of the reaction and using E2 proteins purified from either bacteria (Escherichia coli) or insect cells (Sf9) for comparison. As shown in Fig. 3B, PC4 and phosphorylation on E2 are not needed for E2-mediated repression (compare Fig. 3A and B, lanes 1 to 12, and Fig. 3B, lanes 7 to 12 with lanes 13 to 18), as bacterially expressed E2 protein is able to inhibit E6 promoter activity in the absence of PC4. This result is consistent with a previous report indicating that mutations on the phosphorylation sites of bovine papillomavirus type 1 (BPV-1) E2 do not affect the transcriptional activity of E2 (39). However, it is important to note that the ability of bacterially expressed E2 protein to inhibit E6 promoter activity does not rule out the possibility that phosphorylation may further contribute to the regulatory property of E2. This issue remains to be further investigated.
FIG. 3.
E2-mediated repression of the HPV-11 E6 promoter in a reconstituted two-component transcription system. (A) TAFIIs are not required for E2-mediated repression. In vitro transcription was performed with TFIID-deficient pol II holoenzyme (f:pol II [67]) and either FLAG-tagged TFIID (f:TFIID [9]) or FLAG-tagged TBP (f:TBP), in conjunction with general cofactor PC4 (25, 35), in the absence (−) or presence of different amounts of E2. Unless otherwise specified, baculovirus-expressed FLAG-tagged HPV-11 E2 was used in the assay. (B) PC4 and phosphorylation on E2 are not required for E2-mediated repression. In vitro transcription was performed as described for panel A, except that E2 purified from either insect cells (Sf9) or bacteria (E. coli) was used and no PC4 was included in the experiment. (C) The initiator element and CEI and CEII are not required for E2-mediated repression. In vitro transcription was performed as described for panel A, except that different HPV-11 DNA templates were used. (D) E2 mainly acts through the no. 4 E2-binding site to inhibit E6 promoter activity. DNA templates containing either wild-type (WT) or mutated E2-binding sites were constructed in the backbone of p7862-70GLess/I− as described in Materials and Methods. 4M, 3M, 2M, 23M, 24M, 34M, and 234M are DNA templates containing mutations on the no. 4, no. 3, no. 2, no. 2 and 3, no. 2 and 4, no. 3 and 4, and no. 2 and 3 and 4 E2-binding sites, respectively. In vitro transcription was performed as described for panel A, except that different HPV-11 DNA templates were used.
Since E2 appears to repress E6 promoter activity by excluding TBP or TFIID from binding to the TATA box (17, 21, 22, 59), we speculated that the initiator and enhancer elements might not be involved in E2-mediated repression. Indeed, suppression of the E6 promoter could be observed on all HPV-11 URR-containing templates with or without the MLP initiator element (Fig. 3C, compare lanes 1 to 6 and lanes 7 to 12) and in the presence or absence of CEI and CEII (Fig. 3C, compare lanes 1 to 6 and lanes 13 to 18). The observation that the levels of basal transcription are the same between those two DNA templates with or without the initiator element (Fig. 3C, compare lanes 1 and 7) suggests that initiator-binding proteins which are present in nuclear extracts accounting for the enhanced basal transcription from the Inr-containing templates (Fig. 2D) are absent in our reconstituted two-component transcription system. This analysis indicates that cellular proteins that bind to the initiator element and CEI and CEII are not absolutely required for E2-mediated repression.
E2-mediated repression of the E6 promoter mainly functions through the promoter-proximal no. 4 E2-binding site.
To directly demonstrate that E2-mediated repression indeed acts through its cognate binding sites and to further evaluate the relative contributions of individual E2-binding sites in E2-mediated repression, we performed an in vitro transcription assay with G-less cassette templates containing individually mutated E2-binding sites introduced in the backbone of p7862-70GLess/I−. The same set of mutations linked to a CAT reporter gene was used previously for the studies of individual E2-binding sites in E2-mediated repression in transfected cells (21). As shown in Fig. 3D, mutations at all three E2-binding sites (2, 3, and 4) completely relieve E2-mediated repression, suggesting that E2 indeed acts through its cognate binding sites in a sequence-specific manner (compare lanes 1 and 2 and lanes 15 and 16). Interestingly, the no. 4 E2-binding site plays a predominant role in E2-mediated repression, as single-site mutations in no. 4, but not no. 2 and 3 sites, alleviate most of the repressing activity (Fig. 3D, lanes 1 to 8). The transcriptional result with pairwise mutations further suggests that the no. 2 E2-binding site has little or no effect on E2-mediated repression, whereas the no. 3 E2-binding site contributes to E2-mediated repression in our two-component transcription system (Fig. 3D, lanes 9 to 14). The difference in the contributions of no. 2 and 3 E2-binding sites between our in vitro assay and previous in vivo transfection studies (21) is likely due to the absence of cellular proteins binding to the sequence motifs adjacent to no. 2 and 3 E2-binding sites in our two-component transcription system (67). A hierarchy of E2-binding sites in contributing to E2-mediated repression apparently occurs in the order of no. 4 > no. 3 > no. 2, which likely reflects their relative distance from the TATA box, as indicated by the facts that 3 and 4 have an identical consensus sequence and affinity for E2 recognition (1) and that the DNase I footprint exhibited by TBP clearly extends to the no. 4 but not the no. 3 E2-binding site (see below). It is also likely that binding of an E2 dimer to the no. 3 E2-binding site enhances the stability of the E2 dimer binding to the no. 4 E2-binding site. This may explain a contributory role of the no. 3 E2-binding site in E2-mediated repression. The same results obtained here with TBP were also obtained with TFIID as the TATA-binding factor and with the same set of E2-binding site mutations introduced in the initiator-containing p7862-70GLess/I+ template (data not shown).
The N-terminal domain of E2 is not directly involved in E2-mediated repression.
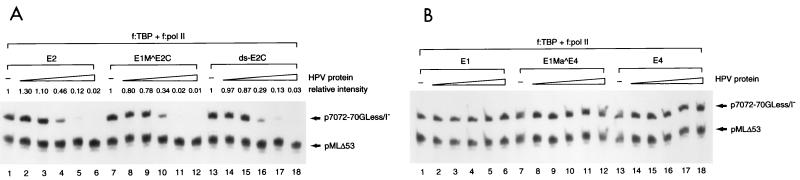
To define the protein domain responsible for E2-mediated repression, we tested the efficiency of transcriptional repression by three forms of HPV-11 E2 proteins that have the same DNA-binding and dimerization domain but differ in their N-terminal amino acid residues (Fig. 1A). Clearly, all three forms of E2 proteins, normalized by Western blotting with anti-FLAG antibodies (data not shown), have comparable activity in suppressing the E6 promoter (Fig. 4A). In contrast, equivalent amounts of other HPV-11 proteins such as E1, E1Ma ^E4, and E4 proteins, also normalized by Western blotting (data not shown), do not show any effect on E6 promoter activity (Fig. 4B). This result indicates that the N-terminal domain of E2 is not required for E2-mediated repression and that the C-terminal domain of E2 is sufficient for E2-mediated repression.
FIG. 4.
Effect of HPV-11 proteins in regulating the homologous E6 promoter. (A) The N-terminal domain of E2 is not required for E2-mediated repression. In vitro transcription was performed with p7072-70GLess/I− template, as described in the legend to Fig. 3A, in the absence (−) or presence of increasing amounts (0.02, 0.10, 0.45, 1.0, and 4.5 pmol) of E2, E1M ^E2C, or ds-E2C. (B) E1 family and E4 proteins individually have no effect on HPV-11 E6 promoter activity. In vitro transcription was performed as described for panel A, in the absence (−) or presence of increasing amounts (0.02, 0.10, 0.45, 1.0, and 4.5 pmol) of E1, E1Ma ^E4, or E4 proteins.
Preincubation of HPV templates with TBP or TFIID cannot alleviate E2-mediated repression of the E6 promoter.
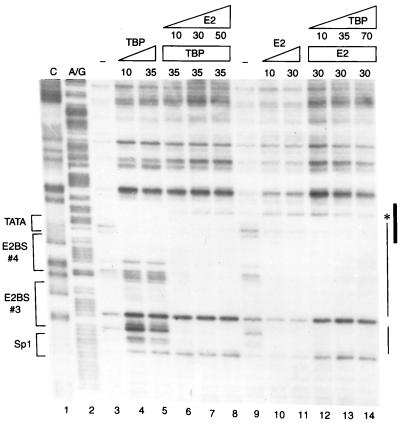
To define the mechanism of E2-mediated repression, we would like to first confirm by DNase I footprinting whether binding of HPV-11 E2 and human TBP to the homologous E6 promoter is indeed mutually exclusive, similar to a previous result showing that binding of TBP could be displaced by an increasing amount of BPV-1 E2 in an electrophoretic mobility shift assay with an oligonucleotide containing the TATA box and an E2-binding site derived from the HPV-18 E6 promoter (22). As shown in Fig. 5, the DNase I footprints generated by E2 and TBP, individually, indeed overlap (compare lanes 4 and 5 with lanes 10 and 11). But interestingly, E2 apparently has higher affinity toward its two binding sites than that of TBP for the TATA box in the E6 promoter, as demonstrated by an efficient displacement of TBP by increasing amounts of E2 (Fig. 5, lanes 4 to 8) yet an inefficient exclusion of E2 by molar excess of TBP (Fig. 5, lanes 10 to 14). Given the observation that an efficient repression of E6 promoter activity occurs only at high concentrations (at least 20 to 50 ng) of E2, relative to 1 ng of TBP used in the transcription assays (Fig. 3), the footprinting data also indicate that steric hindrance of TBP binding by E2 is unlikely to be the sole mechanism for E2-mediated repression. This viewpoint has indeed been further supported by order-of-addition experiments (see below).
FIG. 5.
Binding of TBP and HPV-11 E2 to the HPV-11 E6 promoter is mutually exclusive. DNase I footprinting of the HPV-11 E6 promoter was performed with different amounts (in nanograms) of baculovirus-expressed FLAG-tagged E2 and bacterially expressed FLAG-tagged TBP as described in Materials and Methods. The Maxam-Gilbert sequencing method (52) was used to prepare the C and A/G footprinting markers (lanes 1 and 2). No protein (lanes 3 and 9) or increasing amounts of TBP or E2 or of a combination of these two proteins were included in the footprinting reactions. Sequence motifs in the E6 promoter-proximal region recognized by TBP (TATA), E2 (E2BS 3 and E2BS 4), and Sp1 (Sp1) are marked on the left, whereas the actual DNase I footprints observed by TBP (thick line) and E2 (thin line) are indicated on the right. The asterisk denotes a hypersensitive site induced by E2 binding.
If exclusion of TBP or TFIID binding to the adjacent TATA box by E2 is indeed the mechanism of E2-mediated repression, we speculated that preincubation of DNA template with TBP or TFIID would relieve E2-mediated repression. To address this important issue, we performed an order-of-addition experiment as outlined in Fig. 6A. In this experiment, transcriptional templates, p7072-70GLess/I− and pMLΔ53, were preincubated with TBP and TFIID-deficient pol II-holoenzyme (f:pol II), individually or together, at 30°C for 30 min. The remaining transcription components and NTPs were then added to initiate transcription. E2 was included at different steps of the reaction to evaluate its repressive activity prior to or after PIC assembly. As expected, PIC formation prior to the addition of E2 almost completely overcomes E2-mediated repression (Fig. 6B, lanes 3 to 6). A residual twofold repression observed by E2 added after PIC formation (Fig. 6B, compare lane 3 with lanes 5 and 6) was likely due to inhibition of reinitiation of transcription, since approximately two rounds of transcription were typically observed in this two-component transcription system (67) (see also below). Surprisingly, preincubation of TBP alone with transcriptional templates was not sufficient to alleviate E2-mediated repression (Fig. 6B, compare lane 7 with lanes 8 and 9), indicating that TBP binding to the TATA box could not prevent E2 from binding to its cognate sequence, consistent with the result of DNase I footprinting (Fig. 5). Preincubation of f:pol II with DNA templates could not relieve E2-mediated repression (Fig. 6B, lanes 11 to 14), consistent with our previous finding that pol II holoenzyme alone could not stably bind to the DNA template in the absence of a TATA-binding factor (67, 69). The decreased levels of overall basal transcription when pol II holoenzyme was preincubated with DNA templates (Fig. 6B, compare lanes 3 to 6 with lanes 11 to 14) are likely due to its nonspecific DNA-binding activity that appears to titrate out pol II holoenzyme available for promoter-specific transcription. Interestingly, when E2 was added 5 min after including the remaining transcription component and NTPs, most of the E2-mediated repression was alleviated (Fig. 6B, compare lanes 9 and 10 and lanes 13 and 14). This observation suggests that PIC formation can readily occur within 5 min. The same result was also obtained when TFIID was used in place of TBP as the TATA-binding factor (data not shown).
If E2 indeed inhibits the assembly of PIC, it should be capable of suppressing reinitiation of transcription, which requires the reassembly of transcriptional components. As predicted, when 0.015% Sarkosyl was added 30 min after PIC assembly to prevent reinitiation of transcription (67), E2 no longer inhibited the E6 promoter after PIC formation (Fig. 6C, lanes 5 to 8). Again, preincubation of TBP alone could not overcome E2-mediated repression (Fig. 6C, lanes 9 to 16). In this experiment, approximately 2.5 rounds of transcription were observed (Fig. 6C, compare lanes 1 and 5).
E2-mediated repression requires only components of the general transcription machinery.
The findings that corepressors are often implicated in repressor function and that pol II holoenzyme apparently contains many undefined components that may be functionally involved in E2-mediated repression prompted us to use a more refined in vitro transcription system to further define the role of cellular factors in E2-mediated repression. To this end, we performed an in vitro transcription assay with only recombinant GTFs (TBP, TFIIB, TFIIE, and TFIIF) and highly purified epitope-tagged multiprotein TFIIH and pol II complexes (34, 68), considering the fact that enhancer-binding factors and general cofactors such as TAFIIs and PC4 are not required for E2-mediated repression in our two-component transcription system (Fig. 3). Remarkably, we are able to detect for the first time that E2-mediated repression on the natural HPV-11 promoter can be observed in a completely defined cell-free transcription system containing only TBP, TFIIB, TFIIE, TFIIF, TFIIH, and pol II in a dose-dependent manner, irrespective of the presence or absence of an initiator element (Fig. 7A) and similar to the levels of repression detected in the two-component transcription system (Fig. 3) and in HeLa nuclear extracts (Fig. 2D). This finding suggests that E2-mediated repression does not require corepressors, nor general cofactors TFIIA and Mediator, which are not present in our highly purified in vitro transcription system (68), and that E2 can indeed act as an active repressor to directly inhibit the activity or the assembly of the general transcription machinery.
FIG. 7.
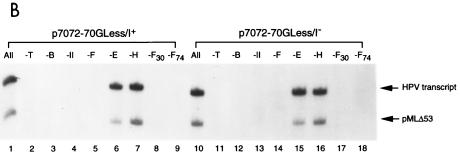
Requirement of GTFs for HPV-11 E2-mediated repression of the homologous E6 promoter. (A) E2-mediated repression in a highly purified in vitro transcription system. In vitro transcription was conducted with recombinant TFIIB, TBP, TFIIE, TFIIF, and FLAG-tagged TFIIH and FLAG-tagged pol II as described in Materials and Methods, in the absence (−) or presence of increasing amounts of E2. Relative intensity in each set of reactions is defined as the ratio of the HPV signal, which is quantitated first by PhosphorImager and then normalized with the internal control, obtained in each reaction to that performed in the absence of E2 (i.e., the first lane of each reaction set). (B) TFIIE and TFIIH are not required for transcription from supercoiled HPV DNA templates. Transcription reactions were performed as described for panel A. The transcription components indicated above the lanes were then left out from the complete reaction (All). The subunits of TFIIF, RAP30 (F30) and RAP74 (F74), were also selectively left out from the complete reaction. (C) E2-mediated repression of the E6 promoter can be observed in a minimal transcription system containing only TBP, TFIIB, TFIIF, and pol II. In vitro transcription reactions were performed with TBP, TFIIB, TFIIF, and pol II, with or without (−) 50 ng of E2 and in the absence (M) or presence (C) of both TFIIE and TFIIH.
To define which components of the general transcription machinery are essential for transcription from the HPV-11 E6 promoter, we conducted a leave-out experiment by omitting individual transcription components from a complete reaction that contained TBP, TFIIB, TFIIE, TFIIF, TFIIH, and pol II. As shown in Fig. 7B, TFIIE and TFIIH are not absolutely required for HPV transcription (compare lane 1 with lanes 6 and 7 and lane 10 with lanes 15 and 16), consistent with the observation that TFIIE and TFIIH are dispensable for transcription from supercoiled DNA templates in our highly purified in vitro transcription system (34, 68). In contrast, TBP, TFIIB, pol II, and both subunits of TFIIF (RAP30 and RAP74) are essential for HPV transcription (Fig. 7B). To see whether a minimal transcription system containing only TBP, TFIIB, pol II, and TFIIF is indeed sufficient to support HPV transcription and whether E2-mediated repression of the E6 promoter can still occur with only four components of the general transcription machinery, we directly compared the transcription activity of the HPV template in a minimal system with that in a complete system that additionally contained TFIIE and TFIIH. As shown in Fig. 7C, although the overall level of basal transcription was lower in the minimal system than in the complete system (compare lanes 1 and 3 and lanes 5 and 7), E2-mediated repression in the minimal system still persisted to an extent similar to that observed in the complete system (compare lanes 2 and 4 and lanes 6 and 8). This result suggests that E2-mediated repression requires only four components of the cellular proteins, TBP, TFIIB, pol II, and TFIIF.
Formation of a minimal PIC composed of TBP (or TFIID), TFIIB, pol II, and TFIIF is necessary to overcome E2-mediated repression.
To define the steps of PIC assembly targeted by E2, we conducted a two-step order-of-addition experiment as outlined at the bottom of Fig. 8A. In this experiment, DNA templates were preincubated respectively with TBP, TBP plus TFIIB, TBP plus TFIIB and pol II, and TBP plus TFIIB/pol II and TFIIF, following the order of PIC assembly, in separate tubes. After a 30-min incubation, the remaining GTFs and NTPs were added, with or without E2, to initiate transcription. Reactions were then continued for an hour before they were analyzed for RNA synthesis. As shown in Fig. 8A, preincubation of TBP with TFIIB or TBP with TFIIB and pol II, which further stabilized TBP binding to the TATA box, was not sufficient to overcome E2-mediated repression (lanes 3 to 8 and 13 to 18). Only when a minimal transcription complex was formed on the E6 promoter was E2-mediated repression alleviated (Fig. 8A, compare lanes 9 and 10 and lanes 19 and 20). The same result was also obtained with TFIID as the TATA-binding factor (Fig. 8B). This study further demonstrates that E2-mediated repression does not simply work by displacing TBP (or TFIID) binding to the TATA box. Other mechanisms affecting the stability of the assembled PIC are also involved in E2-mediated repression of the E6 promoter.
DISCUSSION
Using reconstituted cell-free transcription systems, we are able to identify cellular proteins directly involved in E2-mediated repression of the HPV E6 promoter that regulates the expression of many viral gene products involved in DNA replication, transcription, and transformation. Surprisingly, E2-mediated repression does not require either gene-specific or general transcription cofactors, such as TAFIIs, TFIIA, Mediator, and USA-derived cofactors, normally implicated in activator-dependent transcription. This inhibitory activity of E2 is also independent of transcriptional corepressors, sometimes required for repressor function, and does not simply work by displacing adjacent Sp1 and TBP from binding to their cognate sequences. Instead, E2 appears to inhibit multiple steps of PIC assembly until the formation of a minimal TBP (or TFIID)-TFIIB-pol II-TFIIF complex.
Multiple tiers of transcriptional regulation.
Our finding that a common C-terminal domain present in E2, E1M ^E2C, and ds-E2C is sufficient for E2-mediated repression suggests that the N-terminal domain of E2, which is important for transactivation, is not essential for E2-mediated repression. This conclusion is distinct from a previous result indicating that the N-terminal domain of BPV-1 E2 also plays a role in E2-mediated repression of the HPV-18 E6 promoter experimentally conducted with virus-infected or transiently transfected HeLa cells (26). It is likely that the endogenous HPV-18 E6 promoter in HeLa cells and the introduced HPV-18 promoter during transient transfection assays are packaged into chromatin. Therefore, E2 may need to work in conjunction with chromatin remodeling factors that either help mobilize the promoter-proximal nucleosomes, thereby allowing E2 to bind to its cognate binding sites, or in turn recruit histone deacetylase complexes to further enhance nucleosome-mediated repression of the HPV promoter. This possibility is supported by the recent findings that only the N-terminal, not the C-terminal, domain of HPV-11 E2 can efficiently interact with SWI/SNF chromatin remodeling factor (S.-Y. Wu, S. Y. Hou, and C.-M. Chiang, unpublished data) and that some chromatin remodeling factors can associate with histone deacetylase complexes (62, 64, 70, 71). These observations may explain why mutations at the N-terminal domain of BPV-1 E2 affect E2-mediated repression of the HPV-18 promoter in HeLa cells. It thus seems to be a common theme for both transcriptional activators and repressors to work in conjunction with chromatin remodeling factors to regulate promoter activity in a chromatin context. Once the promoter region is exposed, many cellular proteins, including both positive and negative regulatory factors and cofactors, then constitute another level of gene regulation. Frequently, gene activity reflects a counterbalance between positive and negative regulatory factors in a complicated cellular environment or in a crude cell extract that has multiple protein activities. Therefore, it is of vital importance to define the role of individual transcription factors at each tier of gene regulation. The development of highly purified and well-defined in vitro transcription systems allows us to pinpoint the role of transcriptional regulators directly involved in the transcriptional process. We found that only four components of the general transcription machinery, TBP, TFIIB, pol II, and TFIIF, are absolutely required for E2-mediated repression on the homologous E6 promoter.
Active repression mediated by E2.
An active role of E2-mediated repression does not work simply by displacing TBP and Sp1 from binding to their cognate sequences adjacent to the promoter-proximal E2-binding sites. Despite the fact that an equivalent amount of HPV-11 E2 efficiently prevents TBP from binding to the TATA box (Fig. 5), inhibition of the homologous E6 promoter can be achieved only at high concentrations (20 to 50 ng) of E2, relative to 1 ng of TBP used in the transcription assays (Fig. 3), indicating that steric exclusion of TBP binding by E2 is not the sole mechanism for E2-mediated repression. This argument is further supported by the finding that preincubation of TBP (or TFIID) alone with transcriptional templates is not sufficient to overcome E2-mediated repression (Fig. 6B). The inability of TBP-bound templates to relieve E2-mediated repression is not due to the instability of TBP binding to the transcriptional templates, since inclusion of TFIIB, which has been shown to stabilize TBP binding to the TATA box (47, 49), during the preincubation period is still unable to alleviate E2-mediated repression (Fig. 8A). It is likely that E2 may target a protein surface that is masked only when a minimal TBP-TFIIB-pol II-TFIIF complex has been formed. Alternatively, E2 binding may cause conformational change on the DNA template, which can be reversed only by forming a TBP-TFIIB-pol II-TFIIF complex, not by intermediate PICs. These interesting possibilities are currently under investigation.
Role of cellular proteins in E2-mediated repression.
In transcriptional assays performed with HeLa nuclear extracts, efficient repression of the HPV E6 promoter could be observed only when limiting amounts of cellular factors, provided by nuclear extracts, were used in the assay (Fig. 2D). Inhibition of the E6 promoter by E2 was not detected when an excess amount of nuclear extracts was used (data not shown), suggesting that some cellular proteins present in HeLa nuclear extracts may functionally antagonize E2-mediated repression. This notion is further supported by the observation that only E2-mediated repression, not transactivation, was observed in our reconstituted cell-free transcription systems with TBP as the TATA-binding factor, presumably due to the lack of cellular cofactors critical for E2-mediated activation or required to counteract E2-mediated repression. Although cellular enhancer-binding factors and initiator-binding proteins are not directly involved in E2-mediated repression, they may potentiate PIC assembly, when reaching a critical concentration in vitro or in vivo, to counteract E2-mediated repression. The observations that deletion of the HPV-11 CEII allows homologous E2 to function as a repressor in cervical carcinoma cell line C-33A (13) and that overexpression of HPV-11 E2 can antagonize the nullifying effect of CEII on E2-mediated repression in transient transfection assays (21) further indicate a complicated regulatory circuit between viral E2 and cellular enhancer-binding proteins. The current challenge is to recapitulate the enhancer effect in our highly purified in vitro transcription system by introducing either nucleosome-assembled HPV chromatin templates or enhanceosome-like transcription complexes to dissect the regulatory circuits exerted by various viral and cellular proteins.
ACKNOWLEDGMENTS
We are grateful to G. Dong, T. R. Broker, and L. T. Chow for DNA templates containing various E2-binding site mutations; M. W. Van Dyke for pIGL and pGL plasmids; E. Kershnar for FLAG-tagged TFIIH; and J. Kim for advice on DNase I footprinting.
C.-M.C. is a Pew Scholar in the Biomedical Sciences. This research is supported in part by the American Cancer Society Research Project Grant RPG-97-135-01-MBC and in part by National Institutes of Health grant CA81017.
REFERENCES
- 1.Alexander K A, Phelps W C. A fluorescence anisotropy study of DNA binding by HPV-11 E2C protein: a hierarchy of E2-binding sites. Biochemistry. 1996;35:9864–9872. doi: 10.1021/bi960447d. [DOI] [PubMed] [Google Scholar]
- 2.Auble D T, Hansen K E, Mueller C G F, Lane W S, Thorner J, Hahn S. Mot1, a global repressor of RNA polymerase II transcription, inhibits TBP binding to DNA by an ATP-dependent mechanism. Genes Dev. 1994;8:1920–1934. doi: 10.1101/gad.8.16.1920. [DOI] [PubMed] [Google Scholar]
- 3.Barsoum J, Prakash S S, Han P, Androphy E J. Mechanism of action of the papillomavirus E2 repressors: repression in the absence of DNA binding. J Virol. 1992;66:3941–3945. doi: 10.1128/jvi.66.6.3941-3945.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burley S K, Roeder R G. TATA box mimicry by TFIID: autoinhibition of pol II transcription. Cell. 1998;94:551–553. doi: 10.1016/s0092-8674(00)81596-2. [DOI] [PubMed] [Google Scholar]
- 5.Chiang C-M, Broker T R, Chow L T. An E1M̂E2C fusion protein encoded by human papillomavirus type 11 is a sequence-specific transcription repressor. J Virol. 1991;65:3317–3329. doi: 10.1128/jvi.65.6.3317-3329.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiang C-M, Broker T R, Chow L T. Properties of bovine papillomavirus E1 mutants. Virology. 1992;191:964–967. doi: 10.1016/0042-6822(92)90273-r. [DOI] [PubMed] [Google Scholar]
- 7.Chiang C-M, Dong G, Broker T R, Chow L T. Control of human papillomavirus type 11 origin of replication by the E2 family of transcription regulatory proteins. J Virol. 1992;66:5224–5231. doi: 10.1128/jvi.66.9.5224-5231.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiang C-M, Ustav M, Stenlund A, Ho T F, Broker T R, Chow L T. Viral E1 and E2 proteins support replication of homologous and heterologous papillomaviral origins. Proc Natl Acad Sci USA. 1992;89:5799–5803. doi: 10.1073/pnas.89.13.5799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiang C-M, Ge H, Wang Z, Hoffmann A, Roeder R G. Unique TATA-binding protein-containing complexes and cofactors involved in transcription by RNA polymerases II and III. EMBO J. 1993;12:2749–2762. doi: 10.1002/j.1460-2075.1993.tb05936.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chiang C-M, Roeder R G. Expression and purification of general transcription factors by FLAG epitope-tagging and peptide elution. Peptide Res. 1993;6:62–64. [PubMed] [Google Scholar]
- 11.Chiang C-M, Roeder R G. Cloning of an intrinsic human TFIID subunit that interacts with multiple transcriptional activators. Science. 1995;267:531–536. doi: 10.1126/science.7824954. [DOI] [PubMed] [Google Scholar]
- 12.Chin M T, Hirochika R, Hirochika H, Broker T R, Chow L T. Regulation of human papillomavirus type 11 enhancer and E6 promoter by activating and repressing proteins from the E2 open reading frame: functional and biochemical studies. J Virol. 1988;62:2994–3002. doi: 10.1128/jvi.62.8.2994-3002.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chin M T, Broker T R, Chow L T. Identification of a novel constitutive enhancer element and an associated binding protein: implications for human papillomavirus type 11 enhancer regulation. J Virol. 1989;63:2967–2976. doi: 10.1128/jvi.63.7.2967-2976.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chong T, Apt D, Gloss B, Isa M, Bernard H-U. The enhancer of human papillomavirus type 16: binding sites for the ubiquitous transcription factors oct-1, NFA, TEF-2, NF1, and AP-1 participate in epithelial cell-specific transcription. J Virol. 1991;65:5933–5943. doi: 10.1128/jvi.65.11.5933-5943.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cooney A J, Leng X, Tsai S Y, O'Malley B W, Tsai M-J. Multiple mechanisms of chicken ovalbumin upstream promoter transcription factor-dependent repression of transactivation by the vitamin D, thyroid hormone, and retinoic acid receptors. J Biol Chem. 1993;268:4152–4160. [PubMed] [Google Scholar]
- 16.Cripe T P, Alderborn A, Anderson R D, Parkkinen S, Bergman P, Haugen T H, Pettersson U, Turek L P. Transcriptional activation of the human papillomavirus 16 P97 promoter by an 88-nucleotide enhancer containing distinct cell-dependent and AP-1 responsive modules. New Biol. 1990;2:450–463. [PubMed] [Google Scholar]
- 17.Demeret C, Yaniv M, Thierry F. The E2 transcriptional repressor can compensate for Sp1 activation of the human papillomavirus type 18 early promoter. J Virol. 1994;68:7075–7082. doi: 10.1128/jvi.68.11.7075-7082.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DiLorenzo T P, Steinberg B M. Differential regulation of human papillomavirus type 6 and 11 early promoters in cultured cells derived from laryngeal papillomas. J Virol. 1995;69:6865–6872. doi: 10.1128/jvi.69.11.6865-6872.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dollard S C, Broker T R, Chow L T. Regulation of the human papillomavirus type 11 E6 promoter by viral and host transcription factors in primary human keratinocytes. J Virol. 1993;67:1721–1726. doi: 10.1128/jvi.67.3.1721-1726.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dong G, Broker T R, Chow L T. Human papillomavirus type 11 E2 proteins repress the homologous E6 promoter by interfering with the binding of host transcription factors to adjacent elements. J Virol. 1994;68:1115–1127. doi: 10.1128/jvi.68.2.1115-1127.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dostatni N, Lambert P F, Sousa R, Ham J, Howley P M, Yaniv M. The functional BPV-1 E2 trans-activating protein can act as a repressor by preventing formation of the initiation complex. Genes Dev. 1991;5:1657–1671. doi: 10.1101/gad.5.9.1657. [DOI] [PubMed] [Google Scholar]
- 23.Fondell J D, Brunel F, Hisatake K, Roeder R G. Unliganded thyroid hormone receptor α can target TATA-binding protein for transcriptional repression. Mol Cell Biol. 1996;16:281–287. doi: 10.1128/mcb.16.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gaubatz S, Imhof A, Dosch R, Werner O, Mitchell P, Buettner R, Eilers M. Transcriptional activation by Myc is under negative control by the transcription factor AP-2. EMBO J. 1995;14:1508–1519. doi: 10.1002/j.1460-2075.1995.tb07137.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ge H, Roeder R G. Purification, cloning, and characterization of a human coactivator, PC4, that mediates transcriptional activation of class II genes. Cell. 1994;78:513–523. doi: 10.1016/0092-8674(94)90428-6. [DOI] [PubMed] [Google Scholar]
- 26.Goodwin E C, Naeger L K, Breiding D E, Androphy E J, DiMaio D. Transactivation-competent bovine papillomavirus E2 protein is specifically required for efficient repression of human papillomavirus oncogene expression and for acute growth inhibition of cervical carcinoma cell lines. J Virol. 1998;72:3925–3934. doi: 10.1128/jvi.72.5.3925-3934.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goppelt A, Stelzer G, Lottspeich F, Meisterernst M. A mechanism for repression of class II gene transcription through specific binding of NC2 to TBP-promoter complexes via heterodimeric histone fold domains. EMBO J. 1996;15:3105–3116. [PMC free article] [PubMed] [Google Scholar]
- 28.Hirochika H, Hirochika R, Broker T R, Chow L T. Functional mapping of the human papillomavirus type 11 transcriptional enhancer and its interaction with the trans-acting E2 proteins. Genes Dev. 1988;2:54–67. doi: 10.1101/gad.2.1.54. [DOI] [PubMed] [Google Scholar]
- 29.Ikeda K, Halle J-P, Stelzer G, Meisterernst M, Kawakami K. Involvement of negative cofactor NC2 in active repression by zinc finger-homeodomain transcription factor AREB6. Mol Cell Biol. 1998;18:10–18. doi: 10.1128/mcb.18.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson A D. The price of repression. Cell. 1995;81:655–658. doi: 10.1016/0092-8674(95)90524-3. [DOI] [PubMed] [Google Scholar]
- 31.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin S-C, Heyman R A, Rose D W, Glass C K, Rosenfeld M G. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 32.Kanaya T, Kyo S, Laimins L A. The 5′ region of the human papillomavirus type 31 upstream regulatory region acts as an enhancer which augments viral early expression through the action of YY1. Virology. 1997;237:159–169. doi: 10.1006/viro.1997.8771. [DOI] [PubMed] [Google Scholar]
- 33.Kato H, Horikoshi M, Roeder R G. Repression of HIV-1 transcription by a cellular protein. Science. 1991;251:1476–1479. doi: 10.1126/science.2006421. [DOI] [PubMed] [Google Scholar]
- 34.Kershnar E, Wu S-Y, Chiang C-M. Immunoaffinity purification and functional characterization of human transcription factor IIH and RNA polymerase II from clonal cell lines that conditionally express epitope-tagged subunits of the multiprotein complexes. J Biol Chem. 1998;273:34444–34453. doi: 10.1074/jbc.273.51.34444. [DOI] [PubMed] [Google Scholar]
- 35.Kretzschmar M, Kaiser K, Lottspeich F, Meisterernst M. A novel mediator of class II gene transcription with homology to viral immediate-early transcriptional regulators. Cell. 1994;78:525–534. doi: 10.1016/0092-8674(94)90429-4. [DOI] [PubMed] [Google Scholar]
- 36.Labbé E, Silvestri C, Hoodless P A, Wrana J L, Attisano L. Smad2 and Smad3 positively and negatively regulate TGFβ-dependent transcription through the forkhead DNA-binding protein FAST2. Mol Cell. 1998;2:109–120. doi: 10.1016/s1097-2765(00)80119-7. [DOI] [PubMed] [Google Scholar]
- 37.Lambert P F, Spalholz B A, Howley P M. A transcriptional repressor encoded by BPV-1 shares a common carboxy-terminal domain with the E2 transactivator. Cell. 1987;50:69–78. doi: 10.1016/0092-8674(87)90663-5. [DOI] [PubMed] [Google Scholar]
- 38.Lee K C, Crowe A J, Barton M C. p53-mediated repression of alpha-fetoprotein gene expression by specific DNA binding. Mol Cell Biol. 1999;19:1279–1288. doi: 10.1128/mcb.19.2.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lehman C W, King D S, Botchan M R. A papillomavirus E2 phosphorylation mutant exhibits normal transient replication and transcription but is defective in transformation and plasmid retention. J Virol. 1997;71:3652–3665. doi: 10.1128/jvi.71.5.3652-3665.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li C, Manley J L. Even-skipped represses transcription by binding TATA binding protein and blocking the TFIID-TATA box interaction. Mol Cell Biol. 1998;18:3771–3781. doi: 10.1128/mcb.18.7.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McBride A A, Romanczuk H, Howley P M. The papillomavirus E2 regulatory proteins. J Biol Chem. 1991;266:18411–18414. [PubMed] [Google Scholar]
- 42.Meisterernst M, Roy A L, Lieu H M, Roeder R G. Activation of class II gene transcription by regulatory factors is potentiated by a novel activity. Cell. 1991;66:981–993. doi: 10.1016/0092-8674(91)90443-3. [DOI] [PubMed] [Google Scholar]
- 43.Mermelstein F, Yeung K, Cao J, Inostroza J A, Erdjument-Bromage H, Eagelson K, Landsman D, Levitt P, Tempst P, Reinberg D. Requirement of a corepressor for Dr1-mediated repression of transcription. Genes Dev. 1996;10:1033–1048. doi: 10.1101/gad.10.8.1033. [DOI] [PubMed] [Google Scholar]
- 44.O'Connor M, Chan S Y, Bernard H-U. Transcription factor binding sites in the long control regions of genital HPVs. In: Myers G, Bernard H-U, Delius H, Baker C, Icenogle J, Halpern A, Wheeler C, editors. Human papillomaviruses 1995 compendium, part III-A. Los Alamos, N.Mex: Los Alamos National Laboratory; 1995. pp. 21–40. [Google Scholar]
- 45.Olave I, Reinberg D, Vales L D. The mammalian transcriptional repressor RBP (CBF1) targets TFIID and TFIIA to prevent activated transcription. Genes Dev. 1998;12:1621–1637. doi: 10.1101/gad.12.11.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oliner J D, Pietenpol J A, Thiagalingam S, Gyuris J, Kinzler K W, Vogelstein B. Oncoprotein MDM2 conceals the activation domain of tumour suppressor p53. Nature. 1993;362:857–860. doi: 10.1038/362857a0. [DOI] [PubMed] [Google Scholar]
- 47.Orphanides G, Lagrange T, Reinberg D. The general transcription factors of RNA polymerase II. Genes Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- 48.Pazin M J, Kadonaga J T. What's up and down with histone deacetylation and transcription? Cell. 1997;89:325–328. doi: 10.1016/s0092-8674(00)80211-1. [DOI] [PubMed] [Google Scholar]
- 49.Roeder R G. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem Sci. 1996;21:327–335. [PubMed] [Google Scholar]
- 50.Romanczuk H, Thierry F, Howley P M. Mutational analysis of cis elements involved in E2 modulation of human papillomavirus type 16 P97 and type 18 P105 promoters. J Virol. 1990;64:2849–2859. doi: 10.1128/jvi.64.6.2849-2859.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ross J F, Liu X, Dynlacht B D. Mechanism of transcriptional repression of E2F by the retinoblastoma tumor suppressor protein. Mol Cell. 1999;3:195–205. doi: 10.1016/s1097-2765(00)80310-x. [DOI] [PubMed] [Google Scholar]
- 52.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 53.Smotkin D, Prokoph H, Wettstein F O. Oncogenic and nononcogenic human genital papillomaviruses generate the E7 mRNA by different mechanisms. J Virol. 1989;63:1441–1447. doi: 10.1128/jvi.63.3.1441-1447.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sommer A, Bousset K, Kremmer E, Austen M, Lüscher B. Identification and characterization of specific DNA-binding complexes containing members of the Myc/Max/Mad network of transcriptional regulators. J Biol Chem. 1998;273:6632–6642. doi: 10.1074/jbc.273.12.6632. [DOI] [PubMed] [Google Scholar]
- 55.Sousa R, Dostatni N, Yaniv M. Control of papillomavirus gene expression. Biochim Biophys Acta. 1990;1032:19–37. doi: 10.1016/0304-419x(90)90010-x. [DOI] [PubMed] [Google Scholar]
- 56.Steger G, Corbach S. Dose-dependent regulation of the early promoter of human papillomavirus type 18 by the viral E2 protein. J Virol. 1997;71:50–58. doi: 10.1128/jvi.71.1.50-58.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Steger G, Ham J, Lefebvre O, Yaniv M. The bovine papillomavirus 1 E2 protein contains two activation domains: one that interacts with TBP and another that functions after TBP binding. EMBO J. 1995;14:329–340. doi: 10.1002/j.1460-2075.1995.tb07007.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stenlund A, Botchan M R. The E2 trans-activator can act as a repressor by interfering with a cellular transcription factor. Genes Dev. 1990;4:123–136. doi: 10.1101/gad.4.1.123. [DOI] [PubMed] [Google Scholar]
- 59.Tan S-H, Leong L E-C, Walker P A, Bernard H-U. The human papillomavirus type 16 E2 transcription factor binds with low cooperativity to two flanking sites and represses the E6 promoter through displacement of Sp1 and TFIID. J Virol. 1994;68:6411–6420. doi: 10.1128/jvi.68.10.6411-6420.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tao Y, Kassatly R F, Cress W D, Horowitz J M. Subunit composition determines E2F DNA-binding site specificity. Mol Cell Biol. 1997;17:6994–7007. doi: 10.1128/mcb.17.12.6994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thut C J, Goodrich J A, Tjian R. Repression of p53-mediated transcription by MDM2: a dual mechanism. Genes Dev. 1997;11:1974–1986. doi: 10.1101/gad.11.15.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tong J K, Hassig C A, Schnitzler G R, Kingston R E, Schreiber S L. Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature. 1998;395:917–921. doi: 10.1038/27699. [DOI] [PubMed] [Google Scholar]
- 63.Ushikai M, Lace M J, Yamakawa Y, Kono M, Anson J, Ishiji T, Parkkinen S, Wicker N, Valentine M-E, Davidson I, Turek L P, Haugen T H. trans activation by the full-length E2 proteins of human papillomavirus type 16 and bovine papillomavirus type 1 in vitro and in vivo: cooperation with activation domains of cellular transcription factors. J Virol. 1994;68:6655–6666. doi: 10.1128/jvi.68.10.6655-6666.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wade P A, Jones P L, Vermaak D, Wolffe A P. A multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated Snf2 superfamily ATPase. Curr Biol. 1998;8:843–846. doi: 10.1016/s0960-9822(98)70328-8. [DOI] [PubMed] [Google Scholar]
- 65.Wang J C, Sawadogo M, Van Dyke M W. Plasmids for the in vitro analysis of RNA polymerase II-dependent transcription based on a G-free template. Biochim Biophys Acta. 1998;1397:141–145. doi: 10.1016/s0167-4781(98)00012-8. [DOI] [PubMed] [Google Scholar]
- 66.Wu S-Y, Chiang C-M. Establishment of stable cell lines expressing potentially toxic proteins by tetracycline-regulated and epitope-tagging methods. BioTechniques. 1996;21:718–725. doi: 10.2144/96214rr05. [DOI] [PubMed] [Google Scholar]
- 67.Wu S-Y, Chiang C-M. Properties of PC4 and an RNA polymerase II complex in directing activated and basal transcription in vitro. J Biol Chem. 1998;273:12492–12498. doi: 10.1074/jbc.273.20.12492. [DOI] [PubMed] [Google Scholar]
- 68.Wu S-Y, Kershnar E, Chiang C-M. TAFII-independent activation mediated by human TBP in the presence of the positive cofactor PC4. EMBO J. 1998;17:4478–4490. doi: 10.1093/emboj/17.15.4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu S-Y, Thomas M C, Hou S Y, Likhite V, Chiang C-M. Isolation of mouse TFIID and functional characterization of TBP and TFIID in mediating estrogen receptor and chromatin transcription. J Biol Chem. 1999;274:23480–23490. doi: 10.1074/jbc.274.33.23480. [DOI] [PubMed] [Google Scholar]
- 70.Xue Y, Wong J, Moreno G T, Young M K, Côté J, Wang W. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell. 1998;2:851–861. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- 71.Zhang Y, LeRoy G, Seelig H-P, Lane W S, Reinberg D. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell. 1998;95:279–289. doi: 10.1016/s0092-8674(00)81758-4. [DOI] [PubMed] [Google Scholar]
- 72.Zhao W, Chow L T, Broker T R. A distal element in the HPV-11 upstream regulatory region contributes to promoter repression in basal keratinocytes in squamous epithelium. Virology. 1999;253:219–229. doi: 10.1006/viro.1998.9478. [DOI] [PubMed] [Google Scholar]