Abstract
Background
Osteosarcoma (OS) is a rare cancer with a bimodal age distribution that peaks in children and young adults. It has been shown that the expression of programmed cell death protein 1 (PD-1) and programmed death ligand-1 (PD-L1) on tumor-infiltrating immune cells negatively correlates with prognosis of OS patients. However, a comprehensive assessment of the tumor-infiltrating immune cells in OS and their function has not been performed.
Methods
CD8+ T cells were isolated from biopsy tissue samples collected from OS patients and control subjects. Mass cytometry, Treg suppression assay, mixed lymphocyte reaction assay, and effector T cell functional assay were performed to analyze the function of tumor-infiltrating T cells. A xenograft metastasis model was established in BALB/c nude mice.
Results
Macrophages and CD3+ T cells comprised most of the tumor-infiltrating immune cells in OS, with a disproportionately higher number of helper CD4+ T cells than effector CD8+ T cells. Whereas the tumor-infiltrating regulatory T cells were functionally intact, the CD8+ T cells showed increased expression of the immune checkpoint receptor (ICR) PD-1 and T cell immunoglobulin and mucin-domain containing 3 (TIM3) and were functionally inactive. TIM3 blockade using a monoclonal antibody restored the T cell alloreactive function of the CD8+ T cells ex vivo. TIM3 blockade in a xenograft model of OS impaired tumor growth in vivo. TIM3 blockade decreased the number of tumor-infiltrating CD4+ T cells while increasing the numbers and functional activation of tumor-infiltrating CD8+ T cells in vivo.
Conclusions
These results highlight that TIM3 blockade might be a viable therapeutic option and should be tested in additional preclinical models.
Keywords: Osteosarcoma, tumor microenvironment (TME), T cell immunoglobulin and mucin-domain containing 3 (TIM3), exhausted T cells
Introduction
Osteosarcoma (OS) is a rare cancer but the most common bone neoplasm (1). It originates as a localized aggressive tumor with a high incidence of metastasis to distant organs (1,2). Preoperative neoadjuvant therapy, followed by surgical excision, and postoperative chemotherapy with high-dose methotrexate, ifosfamide, and etoposide is a typical regimen for patients with OS (3-6). However, such treatments are not efficacious for all patients (1,7-9), requiring the identification of alternative therapeutic targets.
The bone microenvironment in which OS grows is a dynamic and specialized compartment composed of the malignant bone cells (osteocytes, osteoclasts, and osteoblasts), vascular cells, stromal cells (mesenchymal stem cells and fibroblasts), mineralized extracellular matrix, and immune cells (10-12). Recent studies have highlighted the importance of the tumor microenvironment (TME) in OS pathogenesis and progression (13-15), especially the role of tumor-infiltrating macrophages, which express an M2-like phenotype.
OS cells can control the balance between M1 and M2 macrophages, in turn regulating the T cell response within the TME via programmed cell death protein 1 (PD-1) and programmed death ligand-1 (PD-L1) (15). Indeed, different studies using mouse models of OS or clinical trials in OS patients have shown objective responses on survival and metastatic progression using immune checkpoint receptors (ICRs) (16-22). The expression of both PD-L1 and PD-1 negatively correlates with prognosis in OS patients (22). However, a comprehensive analysis of the TME in OS is yet to be done.
In the present study, we investigated the TME in OS patients and the function of tumor-infiltrating T cells. The novel idea of our work was to explore the effect of inhibiting T cell immunoglobulin and mucin-domain containing 3 (TIM3) on the function of tumor-infiltrating cytotoxic T cells and tumor progression in a mouse model of OS.
We present the following article in accordance with the ARRIVE reporting checklist (available at https://dx.doi.org/10.21037/atm-21-3928).
Methods
Patients and sample processing
Biopsy tissue samples were collected from OS patients. Control samples were derived from to-be-discarded bone fragments obtained from patients undergoing debridement surgery for traumatic injury to the long-bones. The study protocol was approved by the Institutional Review Board of the Fourth Affiliated Hospital of China Medical University. All patients involved in the study provided signed informed consent. The handling of clinical samples was in accordance with the Declaration of Helsinki (as revised in 2013). For mass cytometry, solid tissue cells from the patient biopsies (n=15; 7 males and 8 females, mean age 17.3 years) or healthy subjects (n=5) were used. Tissue samples were processed to single cells over a 70-µm mesh (Greiner) and then stored viably in a Recovery-Cell culture medium (Thermo Fisher Scientific).
Cell culture and transduction
The 143B OS cell line was purchased from American Type Culture Collection (ATCC, Manassas, USA), and grown in Dulbecco’s Modified Eagle Medium (Invitrogen, Carlsbad, CA, USA). The medium was supplemented with 10% fetal bovine serum (Invitrogen) and 1% penicillin-streptomycin solution. Cells were incubated in a 5% CO2 environment at 37 °C. Exponentially growing cells were transduced with firefly luciferase lentivirus (BPS Bioscience, San Diego, CA, USA) using polybrene (10 µg/mL; Sigma-Aldrich, St. Louis, MO, USA). Transduced cells were selected using puromycin (1 µg/mL; Sigma-Aldrich), and successful expression of firefly luciferase was verified using the Luciferase Assay Kit (Promega).
Cell isolation, culture, and treatment
CD8+ T cells were isolated using positive selection from OS biopsy samples, control biopsy samples, or peripheral blood mononuclear cells (PBMCs) from healthy subjects using magnetic-activated cell sorting CD3 and CD8 microbeads (Miltenyi Biotec). The cells were cultured in 6-well plates coated with OKT3 (Ortho Biotech; 1 µg/mL) and anti-CD28 (clone CD28.2, Becton Dickinson; 1 µg/mL) antibodies.
Spontaneous xenograft metastasis model
All animal procedures were performed under the approval of the Laboratory Animal Welfare and Ethics Committee of the Fourth Affiliated Hospital of China Medical University and in accordance with the NIH Guide for the Care and Use of Laboratory Animals. All mice were housed in a controlled environment. A protocol was prepared before the study without registration. 143B cells (0.5×106) stably expressing firefly luciferase were trypsinized, washed with PBS, and resuspended in a 1:1 solution of PBS and Matrigel (phenol red-free and reduced growth factors; BD Biosciences) and injected into the lateral tail vein of 6-week-old female BALB/c nude mice (n=20). The mice were randomly divided into two experimental groups (n=10/group); the control group received intraperitoneal (i.p.) injection of isotype control, whereas the treatment group received i.p. injection of 100 µg of anti-mouse TIM-3 monoclonal antibody (clone B8.2C12; Bio X Cell, Lebanon, USA) on day 0 and 2 after tumor cell inoculation. Bodyweight and tumor size was measured every 7 days. Mice (n=5/group) were humanely killed at 4-week post-inoculation. The other 5 mice/group were kept until all mice in one group died, and the data were used for survival rate analysis. The tumors were surgically removed and processed similarly to patients’ samples, and stored at −80 °C.
Mass cytometry by time-of-flight (CyTOF) staining
Staining with cisplatin (DVS Sciences) was done to differentiate between live and dead cells. Cells were then incubated with 19 antibodies, all metal-tagged, followed by an intercalator (DVS Sciences). Staining followed the guidelines of the Maxpar cell staining protocol (Fluidigm Corporation) (23). A CyTOF-II mass cytometer (Fluidigm) was used for sample runs. Post-run, the EQ Four Element Calibration Beads (EQ Beads, 201078; Fluidigm) were used to normalize the signal. Events were initially gated for viability using Ir191 versus Ir193, and double-positive cells were used in downstream analyses to identify different subpopulations post-dynamic downsampling.
Treg suppression assay
The in vitro suppressive capacity of isolated PBMCs of healthy subjects (nTregs), patients with chronic tonsillitis undergoing tonsillectomy (tonsil-Tregs), OS Tregs (OS-Treg), and CD25– was assessed using a carboxyfluorescein succinimidyl ester (CFSE)-based inhibition assay. The CFSE (ThermoFisher Scientific, Carlsbad, CA, USA)-labeled PBMCs were stimulated with OKT3 (0.5 µg/mL) and irradiated (40 Gy) allogeneic PBMC (at a 4:1 effector: feeder ratio). CFSE dilution was assessed after 7 days, and suppression was defined as the Treg-mediated percent inhibition of T cells’ proliferation.
Mixed lymphocyte reaction (MLR) assay
Responders (CD3+CD8+ T cells) were purified from tumor-infiltrating immune cells and stained with CFSE as described above. CFSE-labeled responder CD3+CD8+ T cells were seeded in 96-well plate format in RPMI 10% AB serum either alone or with irradiated (40 Gy) allogeneic PBMCs (stimulators) at 1:1, 2:1, and 5:1 ratio. CD14+ antigen was present in all wells. T cell proliferation was calculated by analyzing the CFSE dilution of the responders after 72 h. Where indicated, CD3+CD8+ T cells were treated with anti-human TIM3 monoclonal antibody (TSR-022; Creative Biolabs, Shirley, USA).
Effector T cell functional assay
Sorted tumor-infiltrating CD3+CD8+ cells were cultured in medium ± phorbol 12-myristate 13-acetate (PMA, 20 ng/mL; Sigma-Aldrich) and Ionomycin (1 µg/mL; Sigma-Aldrich). Luminex (Millipore) was used to quantify the levels of indicated cytokines in the supernatants collected before or after 3 days of PMA/ionomycin stimulation.
Statistical analysis
Quantitative data are expressed as the mean ± standard deviation (SD). Student’s t-test was used for two-group comparison, and the differences between more than two groups were analyzed using ANOVA. Kaplan-Meier curves were computed to analyze survival rates and the log-rank (Mantel-Cox) test used to evaluate statistical significance. P<0.05 was considered statistically significant.
Results
Phenotypic analysis of TME in OS patients
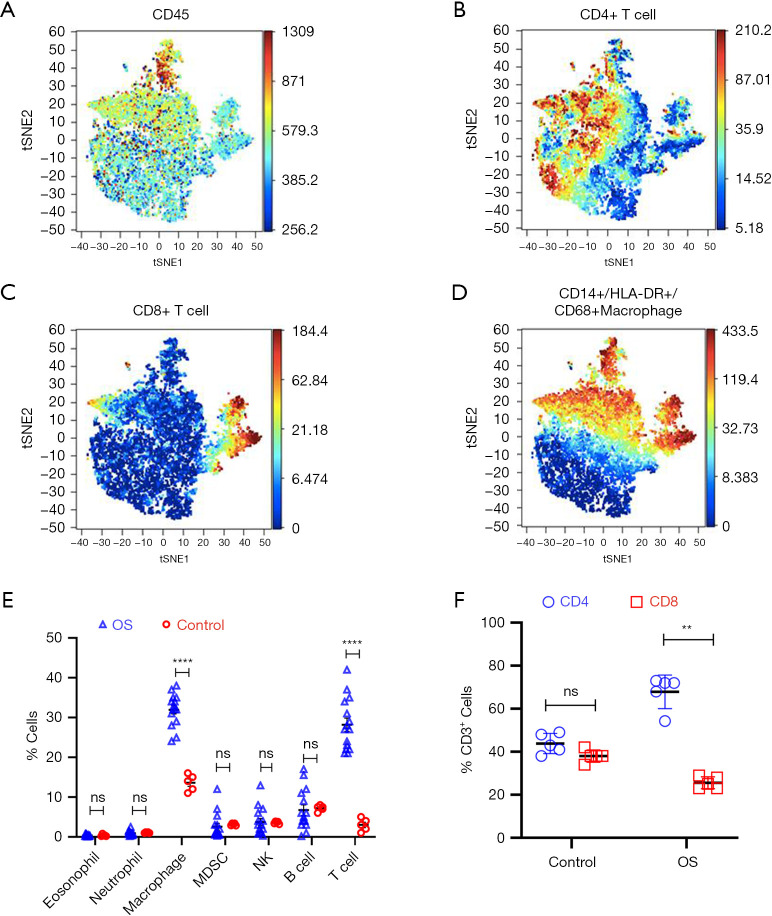
We first analyzed the immune cell composition using CyTOF of patient biopsies (n=15 and controls (n=5) (Figure 1A-1E). The immune infiltrate in OS varied between 33% and 49%, with macrophages (CD45+CD14+HLA-DR+CD68+) and CD45+CD3+ T cells as the most abundant populations (Figure 1E).
Figure 1.
Phenotypic analysis of tumor-infiltrating immune cells in OS patients. (A-D) CyTOF (viSNE plot) analysis of live CD45+ cells from representative tumors. (E) Quantification of tumor-infiltrating cells shown in (A-D) (OS, n=15; Control, n=5). (F) Relative percent of CD4+ and CD8+ T cells in tumor-infiltrating CD3+ T cells in OS and controls (OS, n=15; Control, n=5). For (E and F), data are mean ± SD; **, P<0.01; ****, P<0.0001, ns, not significant (P>0.05)—one-way ANOVA with Tukey’s post hoc test. CyTOF, mass cytometry by time-of-flight; OS, osteosarcoma.
The tumor-infiltrating CD3+ T cells were further analyzed using automatic gating via FlowSOM (24). This algorithm does multivariate clustering of cells, intuitively using the self-organized map (SOM) algorithm, and categorizes cells into relevant meta-clusters based on their surface markers. Within OS, CD3+CD4+ helper T cells were significantly higher in number than effector CD3+CD8+ T cells compared with control (Figure 1F).
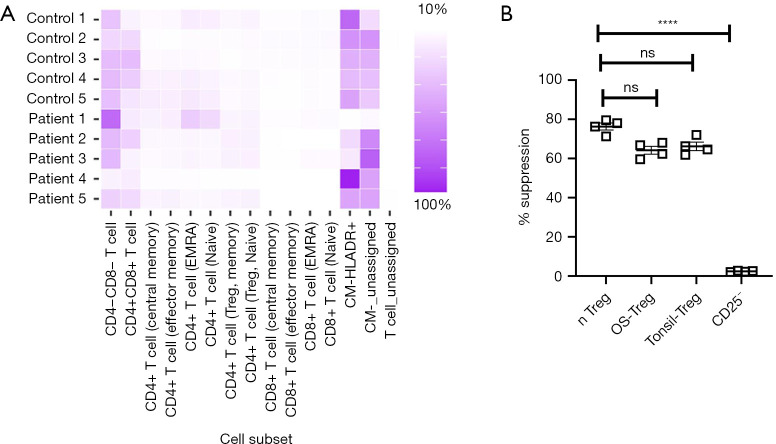
We did not observe significant enrichment of T cell subtypes (Figure 2A). Tregs isolated from OS demonstrated intact suppressive activity, similar to that observed for nTregs isolated from the PBMCs of healthy controls or from tonsils (Figure 2B). Taken together, the results indicated that the tumor-infiltrating immune cells in OS are enriched with macrophages and T cells, with disproportionately higher numbers of CD4+ T cells compared with CD8+ T cells.
Figure 2.
Intact suppression activity of tumor-infiltrating Tregs in OS. (A) Relative indicated T cell subtypes among tumor-infiltrating T cells was done using a self-organizing map (FlowSOM) and visualized as pie charts (MST plots). No enrichment of specific T cell subtypes was observed. (B) Relative suppressor capacity of Tregs isolated from OS patients, freshly isolated ‘natural’ Tregs [nTregs (CD3+CD4+CD25Bright)], tonsil, or CD25– cells (n=4 each). Data are mean ± SD. ****, P<0.0001, ns, not significant (P>0.05)—one-way ANOVA with Turkey’s post hoc test. Tregs, regulator T cells; OS, osteosarcoma; MST, minimal spanning tree.
Characteristics of infiltrating T cells in OS
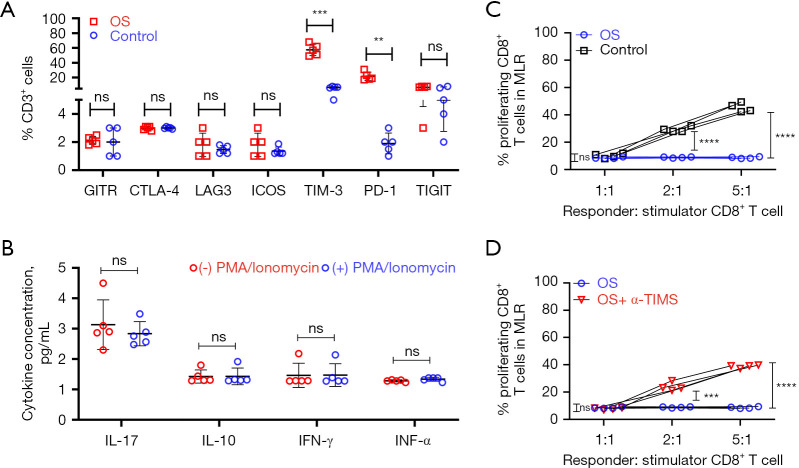
Given that the enhanced expression of ICRs can induce T cell exhaustion (25) and prior literature reports high expression of ICRs in OS (16,18,19,21,22), we determined the expression of inhibitory receptors on tumor-infiltrating CD8+ T cells in OS. CD8+ T cells infiltrating the TME of OS patients showed higher expression of the inhibitory receptors TIM3 and PD-1, but not glucocorticoid-induced TNFR-related protein (GITR), cytotoxic T lymphocyte antigen-4 (CTLA-4), lymphocyte activation gene 3 (LAG-3), inducible T cell costimulator (ICOS), or T cell immunoreceptor with Ig and ITIM domain (TIGIT) compared with controls (Figure 3A).
Figure 3.
Infiltrating T lymphocytes in OS patients are functionally and phenotypically exhausted, rescued by TIM3 blockade ex vivo. (A) CyTOF analysis of immune checkpoint receptors’ expression in OS tumor tissue and controls were performed. Quantification of data shown is shown. Data are represented as mean ± standard deviation (n=5 each group). (B) Expression of cytokines in CD8+ T cells isolated from OS patients pre-and post-activation with PMA and ionomycin. Data are mean ± SD, n=5. (C) A MLR assay showed that CD8+ T cells isolated from OS patients were dysfunctional compared to controls (n=4 each). (D) Same as C, but CD8+ T cells from OS patients were treated for 24 hours with 20 µg/mL anti-TIM3 monoclonal antibody (TSR-022) or isotype control before the MLR assay (n=4 each). For (A-D), **, P<0.01; ***, P<0.001; ****, P<0.0001, ns, not significant (P>0.05)—one-way ANOVA with Tukey’s post hoc test. CyTOF, mass cytometry by time-of-flight; OS, osteosarcoma; MLR, mixed lymphocyte reaction; PMA, phorbol 12-myristate 13-acetate.
Because the density of tumor-infiltrating CD8+ T cells can be associated with disease prognosis, as is observed in other cancers (26), we next assessed the functional capacity of CD8+ T cells within the TME of OS patients. CD8+ T cells isolated from OS patients demonstrated attenuated expression of Th1/Th2 cytokines, interleukin (IL)-17, IL-10 interferon-gamma (IFN-γ), and tumor necrosis factor-alpha (TNFα), both in the native state and after polyclonal stimulation (Figure 3B). CD8+ T cells within the TME of OS had impaired alloreactivity compared with control (Figure 3C), consistent with altered effector function. Because previous work by others has shown that inhibition of PD-1 might inhibit OS pathogenesis in mice models (16,18,19,21,22), we decided to focus on TIM3. Thus, we next evaluated if TIM3 blockade would restore T cell function in OS in an ex vivo setting and found that treatment of patient-derived OS CD8+ T cells with anti-human TIM3 antibody (TSR-022) for 24 h significantly rescued their effector functions (Figure 3D). Taken together, these results suggested that TIM3 blockade could influence the immune dysfunction observed in tumor-infiltrating CD8+ T cells of OS patients.
Effect of TIM3 blockade in an in vivo model of OS
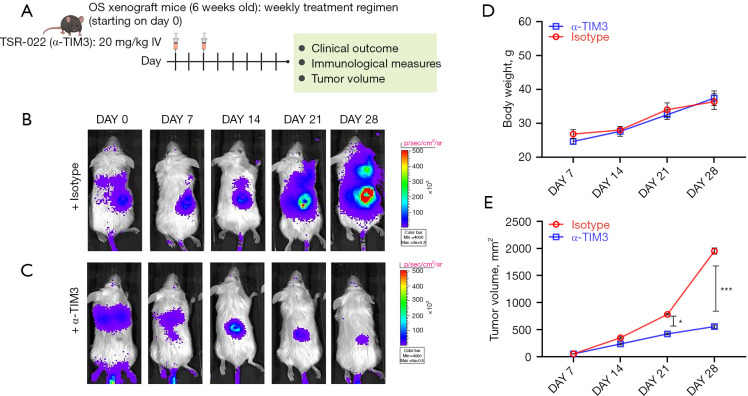
A spontaneous metastasis model was established by injecting 143B OS cells stably expressing firefly luciferase in the lateral tail vein of BALB/c nude mice, which were then randomly divided into two groups (n=10/group: anti-mouse TIM3 antibody and isotype control) and injected on days 0 and 2 after tumor cell inoculation (Figure 4A). Bodyweight and tumor growth were monitored up to 4 weeks after the initial injection, and survival was monitored until all mice in one group had died. Tissues were harvested when either mice were dead or when tumor volume reached 3,000 mm3.
Figure 4.
TIM3 blockade slows OS tumor progression in vivo. (A) Schematic representation of OS xenograft experiment. BALB/c nude mice were injected with 143B cells stably expressing firefly luciferase into the lateral tail vein. Mice either received i.p. injection of isotype control or anti-mouse TIM-3 antibody (clone B8.2C12) after tumor cell inoculation. Tumors were imaged live in vivo (B,C). Shown are images at the end of 4 weeks (n=10/group). (D) Change in bodyweight of mice in the different experimental groups over 4 weeks (n=5/group). (E) Tumor growth rate in the two experimental groups (n=5/group). TIM3, T cell immunoglobulin and mucin-domain containing 3; OS, osteosarcoma. *, P<0.05; ***, P<0.001.
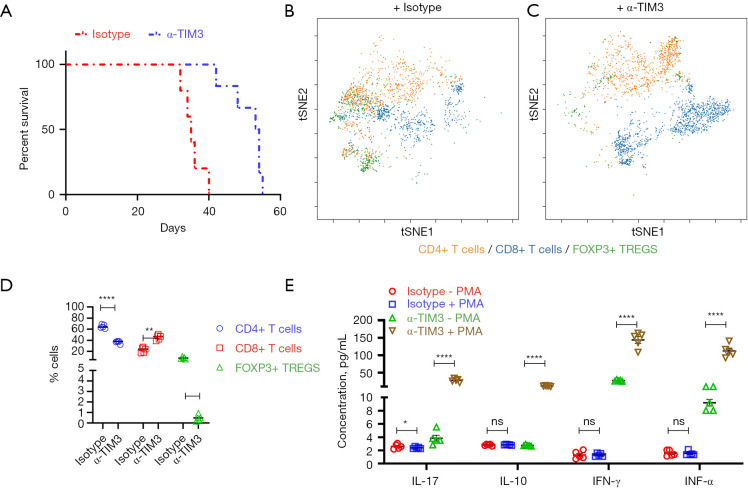
Compared with the isotype control group (Figure 4B), treatment with anti-mouse TIM-3 antibody prevented tumor growth (Figure 4C). No significant difference in bodyweight was observed in the animals from the different experimental groups (Figure 4D). The tumor growth rate was significantly higher in the isotype control group than the anti-mouse TIM-3 antibody group (Figure 4E). The anti-TIM-3 antibody therapy significantly improved survival compared with the isotype control group (Figure 5A). Taken together, these results showed potent anti-tumorigenic activity of the anti-mouse TIM-3 antibody.
Figure 5.
TIM3 blockade restores the function of tumor-infiltrating CD8+ T cells in vivo. (A) Kaplan-Meier survival curve showing significantly improved survival in mice treated with anti-mouse TIM-3 antibody compared with isotype control (P=0.0016). Of note, all mice in the vehicle group died by week 6. (B,C) CyTOF (viSNE plot) analysis of live CD45+ cells from representative tumors from mice treated with isotype control (B) or anti-mouse TIM-3 antibody (C). (D) Quantification of tumor-infiltrating cells shown in (B,C) (n=5 each). (E) Expression of cytokines in CD8+ T cells isolated from representative tumors from mice treated with isotype control or anti-mouse TIM-3 antibody pre- and post-activation with PMA and ionomycin. Data are mean ± SD, n=5. For (D,E), *, P<0.05; **, P<0.01; ****, P<0.0001, ns, not significant (P>0.05)—one-way ANOVA with Tukey’s post hoc test. TIM3, T cell immunoglobulin and mucin-domain containing 3; CyTOF, mass cytometry by time-of-flight; OS, osteosarcoma; PMA, phorbol 12-myristate 13-acetate.
Effect of TIM3 blockade on function of tumor-infiltrating CD8+ T cells
We next evaluated the tumor-infiltrating CD4+ T cells, CD8+ T cells, and FOXP3+ Tregs in mice treated with isotype control or anti-mouse TIM-3 antibody. Compared with the isotype control, TIM3 blockade significantly decreased the number of tumor-infiltrating CD4+ T cells and FOXP3+ Tregs while increasing the number of tumor-infiltrating CD8+ T cells (Figure 5B-5D). This was in striking contrast to the disproportionately higher tumor-infiltrating CD4+/CD8+ T cell ratio observed in OS patients (Figure 1F) or mice treated with isotype control (Figure 5B,5D).
Given that overexpression of TIM3 is associated with tumor exhaustion and functional impairment, we hypothesized that TIM3 blockade would restore the effector function of the observed high number of tumor-infiltrating CD8+ T cells. Hence, the functional capacity of tumor-infiltrating CD8+ T cells in mice treated with isotype control or anti-mouse TIM-3 antibody was assessed. CD8+ T cells isolated from mice treated with isotype control demonstrated attenuated expression of Th1/Th2 cytokines, IL-17, IL-10 IFN-γ, and TNFα, both in the native state and after polyclonal stimulation with PMA and ionomycin (Figure 5E), as was observed in OS patients (Figure 3B). In comparison, expression of all four cytokines was higher in tumor-infiltrating T cells isolated from mice treated with anti-mouse TIM-3 antibody in the native state, and significantly induced after polyclonal stimulation (Figure 5E). Taken together, these results showed that TIM3 blockade can inhibit tumor progression in OS by increasing functionally active CD8+ T cells in the TME.
Discussion
TIM3 is a negative regulator of lymphocyte function and survival that, like PD-1, is a marker of CD4 and CD8 T cell exhaustion (27). PD-1+TIM3+ CD8 T cells have been identified in different cancers and represent a more severely impaired cytotoxic lymphocyte population (compared with PD-1 or TIM3 only) as measured by inflammatory cytokine production and proliferation capacity (28,29). Here, we did not find PD-1+TIM-3+ CD8 T cells in either the OS patients or in the mouse model. Therefore, it remains to be determined if TIM3 blockade might increase expression of PD-1 in these cells, because adaptive resistance to one ICR blockade is often associated with redundant overexpression of other ICRs (30-32). In such a scenario, dual blockade of both PD-1 and TIM3 will have to be tested. This is especially important given the encouraging results obtained for other ICR inhibitors. Although combining multiple coinhibitory blocking antibodies in the clinical setting should be approached with great caution, previous studies of different cancer types have suggested that blocking a single inhibitory receptor only leads to activation of the unblocked pathway (33), necessitating multiple blockades to circumvent the adaptive resistance.
We observed that CD4+ helper T cells were present in a higher proportion than effector CD8+ T cells within the OS TME. TIM3 blockade decreased the percentage of tumor-infiltrating CD4+ T cells and increased that of the CD8+ T cells. It remains to be determined whether the observed change in CD4+/CD8+ T cell ratio is due to apoptosis of CD4+ T cells and functional restoration of CD8+ T cells or directed by increased infiltration of functionally effective CD8+ T cells from the peripheral blood to the tumor milieu.
There are several limitations in this study that should be considered when interpreting the results. Firstly, due to the small sample size, additional verification in a larger number of OS patients is necessary. Secondly, due to the high heterogeneity of OS, attenuation of tumor progression observed in mice might not be replicated in humans. Hence, additional experiments with TIM3 blockade are necessary using patient-derived xenograft models that are much closer to replicating OS in humans than the xenograft with 143B cells we used in the current study. Finally, due to rarity of OS and the limited availability of OS samples, data integration from multiple datasets, be it gene expression or protein arrays, is being used as an essential source of data to study and characterize OS. However, such a strategy has an inherent bias towards batch effects due to the lack of rigorous statistical controls.
In summary, the TME in OS is enriched for macrophages and T cells with comparatively higher numbers of helper T cells than effector T cells. The tumor-infiltrating effector T cells exhibit increased expression of the ICRs, TIM3, and PD-1, and are functionally exhausted. Blockade of TIM3 can restore the alloreactivity of these effector T cells ex vivo, whereas in vivo it inhibits tumor progression and increases infiltration of functionally active effector T cells within the TME.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: This study was supported by the Science and Technology Research Project of Liaoning Provincial Department of Education (No. LK201628) and the Scientific Research Funding Project of the Education Department of Liaoning Province (No. ZF2019025) and the Project of School of Nursing, China Medical University (No. 2017HL-26).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study protocol was approved by the Institutional Review Board of the Fourth Affiliated Hospital of China Medical University. All patients involved in the study provided signed informed consent. The handling of clinical samples was in accordance with the Declaration of Helsinki (as revised in 2013). All animal procedures were performed under the approval of the Laboratory Animal Welfare and Ethics Committee of the Fourth Affiliated Hospital of China Medical University and in accordance with the NIH Guide for the Care and Use of Laboratory Animals.
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://dx.doi.org/10.21037/atm-21-3928
Data Sharing Statement: Available at https://dx.doi.org/10.21037/atm-21-3928
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/atm-21-3928). Dr. WL reports that this study was supported by the Science and Technology Research Project of Liaoning Provincial Department of Education (No. LK201628) and the Scientific Research Funding Project of the Education Department of Liaoning Province (No. ZF2019025). The other authors have no conflicts of interest to declare.
(English Language Editor: K. Brown)
References
- 1.Arndt CA, Rose PS, Folpe AL, et al. Common musculoskeletal tumors of childhood and adolescence. Mayo Clin Proc 2012;87:475-87. 10.1016/j.mayocp.2012.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kumar R, Kumar M, Malhotra K, et al. Primary Osteosarcoma in the Elderly Revisited: Current Concepts in Diagnosis and Treatment. Curr Oncol Rep 2018;20:13. 10.1007/s11912-018-0658-1 [DOI] [PubMed] [Google Scholar]
- 3.Harrison DJ, Geller DS, Gill JD, et al. Current and future therapeutic approaches for osteosarcoma. Expert Rev Anticancer Ther 2018;18:39-50. 10.1080/14737140.2018.1413939 [DOI] [PubMed] [Google Scholar]
- 4.Kager L, Tamamyan G, Bielack S. Novel insights and therapeutic interventions for pediatric osteosarcoma. Future Oncol 2017;13:357-68. 10.2217/fon-2016-0261 [DOI] [PubMed] [Google Scholar]
- 5.O'Reilly R, Cheung NK, Bowman L, et al. NCCN pediatric neuroblastoma practice guidelines. The National Comprehensive Cancer Network. Oncology (Williston Park) 1996;10:1813-22. [PubMed] [Google Scholar]
- 6.Ward WG, Mikaelian K, Dorey F, et al. Pulmonary metastases of stage IIB extremity osteosarcoma and subsequent pulmonary metastases. J Clin Oncol 1994;12:1849-58. 10.1200/JCO.1994.12.9.1849 [DOI] [PubMed] [Google Scholar]
- 7.Bacci G, Ferrari S, Longhi A, et al. Pattern of relapse in patients with osteosarcoma of the extremities treated with neoadjuvant chemotherapy. Eur J Cancer 2001;37:32-8. 10.1016/S0959-8049(00)00361-0 [DOI] [PubMed] [Google Scholar]
- 8.Gorlick R, Anderson P, Andrulis I, et al. Biology of childhood osteogenic sarcoma and potential targets for therapeutic development: meeting summary. Clin Cancer Res 2003;9:5442-53. [PubMed] [Google Scholar]
- 9.Kempf-Bielack B, Bielack SS, Jürgens H, et al. Osteosarcoma relapse after combined modality therapy: an analysis of unselected patients in the Cooperative Osteosarcoma Study Group (COSS). J Clin Oncol 2005;23:559-68. 10.1200/JCO.2005.04.063 [DOI] [PubMed] [Google Scholar]
- 10.Alfranca A, Martinez-Cruzado L, Tornin J, et al. Bone microenvironment signals in osteosarcoma development. Cell Mol Life Sci 2015;72:3097-113. 10.1007/s00018-015-1918-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mathieu M, Martin-Jaular L, Lavieu G, et al. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat Cell Biol 2019;21:9-17. 10.1038/s41556-018-0250-9 [DOI] [PubMed] [Google Scholar]
- 12.Paget S. The distribution of secondary growths in cancer of the breast. 1889. Cancer Metastasis Rev 1989;8:98-101. [PubMed] [Google Scholar]
- 13.Bolli E, Movahedi K, Laoui D, et al. Novel insights in the regulation and function of macrophages in the tumor microenvironment. Curr Opin Oncol 2017;29:55-61. 10.1097/CCO.0000000000000344 [DOI] [PubMed] [Google Scholar]
- 14.Heymann MF, Lézot F, Heymann D. The contribution of immune infiltrates and the local microenvironment in the pathogenesis of osteosarcoma. Cell Immunol 2019;343:103711. 10.1016/j.cellimm.2017.10.011 [DOI] [PubMed] [Google Scholar]
- 15.Miwa S, Shirai T, Yamamoto N, et al. Current and Emerging Targets in Immunotherapy for Osteosarcoma. J Oncol 2019;2019:7035045. 10.1155/2019/7035045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hingorani P, Maas ML, Gustafson MP, et al. Increased CTLA-4(+) T cells and an increased ratio of monocytes with loss of class II (CD14(+) HLA-DR(lo/neg)) found in aggressive pediatric sarcoma patients. J Immunother Cancer 2015;3:35. 10.1186/s40425-015-0082-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liao Y, Chen L, Feng Y, et al. Targeting programmed cell death ligand 1 by CRISPR/Cas9 in osteosarcoma cells. Oncotarget 2017;8:30276-87. 10.18632/oncotarget.16326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lussier DM, Johnson JL, Hingorani P, et al. Combination immunotherapy with α-CTLA-4 and α-PD-L1 antibody blockade prevents immune escape and leads to complete control of metastatic osteosarcoma. J Immunother Cancer 2015;3:21. 10.1186/s40425-015-0067-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen JK, Cote GM, Choy E, et al. Programmed cell death ligand 1 expression in osteosarcoma. Cancer Immunol Res 2014;2:690-8. 10.1158/2326-6066.CIR-13-0224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tawbi HA, Burgess M, Bolejack V, et al. Pembrolizumab in advanced soft-tissue sarcoma and bone sarcoma (SARC028): a multicentre, two-cohort, single-arm, open-label, phase 2 trial. Lancet Oncol 2017;18:1493-501. 10.1016/S1470-2045(17)30624-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng B, Ren T, Huang Y, et al. PD-1 axis expression in musculoskeletal tumors and antitumor effect of nivolumab in osteosarcoma model of humanized mouse. J Hematol Oncol 2018;11:16. 10.1186/s13045-018-0560-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng W, Xiao H, Liu H, et al. Expression of programmed death 1 is correlated with progression of osteosarcoma. APMIS 2015;123:102-7. 10.1111/apm.12311 [DOI] [PubMed] [Google Scholar]
- 23.Finck R, Simonds EF, Jager A, et al. Normalization of mass cytometry data with bead standards. Cytometry A 2013;83:483-94. 10.1002/cyto.a.22271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van Gassen S, Callebaut B, Van Helden MJ, et al. FlowSOM: Using self-organizing maps for visualization and interpretation of cytometry data. Cytometry A 2015;87:636-45. 10.1002/cyto.a.22625 [DOI] [PubMed] [Google Scholar]
- 25.Wherry EJ, Kurachi M. Molecular and cellular insights into T cell exhaustion. Nat Rev Immunol 2015;15:486-99. 10.1038/nri3862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reiser J, Banerjee A. Effector, Memory, and Dysfunctional CD8(+) T Cell Fates in the Antitumor Immune Response. J Immunol Res 2016;2016:8941260. 10.1155/2016/8941260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu C, Anderson AC, Kuchroo VK. TIM-3 and its regulatory role in immune responses. Curr Top Microbiol Immunol 2011;350:1-15. [DOI] [PubMed] [Google Scholar]
- 28.Fourcade J, Sun Z, Benallaoua M, et al. Upregulation of Tim-3 and PD-1 expression is associated with tumor antigen-specific CD8+ T cell dysfunction in melanoma patients. J Exp Med 2010;207:2175-86. 10.1084/jem.20100637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sakuishi K, Apetoh L, Sullivan JM, et al. Targeting Tim-3 and PD-1 pathways to reverse T cell exhaustion and restore anti-tumor immunity. J Exp Med 2010;207:2187-94. 10.1084/jem.20100643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koyama S, Akbay EA, Li YY, et al. Adaptive resistance to therapeutic PD-1 blockade is associated with upregulation of alternative immune checkpoints. Nat Commun 2016;7:10501. 10.1038/ncomms10501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Romero D. Immunotherapy: PD-1 says goodbye, TIM-3 says hello. Nat Rev Clin Oncol 2016;13:202-3. 10.1038/nrclinonc.2016.40 [DOI] [PubMed] [Google Scholar]
- 32.Shayan G, Srivastava R, Li J, et al. Adaptive resistance to anti-PD1 therapy by Tim-3 upregulation is mediated by the PI3K-Akt pathway in head and neck cancer. Oncoimmunology 2017;6:e1261779. 10.1080/2162402X.2016.1261779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Curran MA, Montalvo W, Yagita H, et al. PD-1 and CTLA-4 combination blockade expands infiltrating T cells and reduces regulatory T and myeloid cells within B16 melanoma tumors. Proc Natl Acad Sci U S A 2010;107:4275-80. 10.1073/pnas.0915174107 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as