Abstract
To examine the neuroprotective roles of lncRNA-MIAT in Parkinson’s disease (PD). RNA sequencing expression profiles were utilized to screen the dysregulated lncRNAs in patients with PD and to explore the underlying molecular mechanisms by which the lncRNAs regulate the pathogenesis of PD. 6-hydroxydopamine-induced SH-SY5Y cell lines and a PD mouse model were used to prove how the overexpressing or knocking-down of MIAT produce a marked effect in both in vitro and in vivo experiments. Subsequently, the subcellular localization of MIAT was detected via RNA fluorescence in situ hybridization (FISH) assays. Quantitative PCR, as well as western blotting, were used to determine the expression levels of the associated genes and proteins. We utilized Cell Counting Kit-8 (CCK8) assays to measure the viability of the cells, and the apoptotic rate was determined using Annexin V-FITC/PI double staining. The expressions of tyrosine hydroxylase (TH) and Parkin were quantified in the substantia nigra using immunohistochemical staining. Also, TUNEL staining was performed to visualize the apoptotic cells in the substantia nigra. Compared with the normal rats, the downregulation of MIAT was observed in the cortex, hippocampus, substantia nigra, and striatum of the PD rats. Overexpression of MIAT exhibited a neuroprotective effect on the SH-SY5Y cells. Through RNA-sequencing of the PD mice treated with an overexpression of MIAT and through a differentially expressed genes analysis, it was hypothesized that MIAT could upregulate the expression of synaptotagmin-1 (SYT1) through sponging of miR-34-5p. Interactions between MIAT, miR-34-5p, and SYT1 were confirmed using RIP and dual-luciferase reporter assays. At the same time, the MIAT overexpression group exhibited elevated Parkin and TH protein levels, increased cell viability but a decreased apoptosis rate of the SH-SY5Y cells in contrast with the negative control (NC) group. In vivo, compared with the NC group, the overexpression of MIAT resulted in an increase in the positive rates of Parkin and TH, and the apoptosis was decreased in the PD mice. The behavioral test results showed that the motor coordination and autonomous activity of the mice were enhanced in the MIAT overexpression group compared with the NC group. LncRNA-MIAT regulates the growth of SHSY5Y cells by sponging miR-34-5p which targets SYT1 and exerts a neuroprotective effect in a mouse model of PD.
Keywords: Long non-coding RNA-myocardial infarction-associated transcript, synaptotagmin-1, microRNA-34-5p, SHSY5Y cells, Parkinson’s disease
Introduction
Parkinson’s disease (PD), with the most noteworthy feature of progressive loss of neurons in the substantia nigra pars compacta, has been confirmed as the second most common neurodegenerative disorder. The pathogenesis of PD is closely associated with oxidative stress, dysfunctional mitochondria, the accumulation of α-synuclein aggregates, and the dysregulation of anti-oxidant mechanisms [1,2]. Resting tremor, bradykinesia, high myodynamia, and postural instability are all principal manifestations of PD [3]. Although substantial improvement has been made in understanding the pathophysiology of PD, there are still no effective measures for preventing the progression of the disease. Furthermore, the cellular and molecular mechanisms with regard to the development of PD remain unclear.
In the current study, a microarray was carried out to screen for differentially expressed genes in patients with PD. Based on the results, synaptotagmin-1 (SYT1) was further assessed. SYT1, a member of the synaptotagmin family of proteins, can function as a calcium ion sensor located in synaptic vesicles and regulate neuron exocytosis [4]. It has been demonstrated that the loss of regulation of the vesicle cycle can serve an essential role in the degeneration of dopaminergic neurons and that SYT1 participates in the pathogenesis of PD [5,6].
MicroRNAs (miRNAs) are non-coding RNAs with endogenous competitive features, typically 21-23 nucleotides in length [7]. A growing body of research has shown that miRNAs can exert essential roles in gene expressions at various stages and in genome rearrangement [8,9]. Dysregulated miRNA expression has been confirmed to underlie several types of nervous system diseases, showing their essential roles in the treatment of disorders of the nervous system [10,11]. Past research has confirmed that miR-34 expression is dysregulated in a variety of neurodegenerative diseases [12]. Furthermore, miR-34a may offset the protective function of schisandrin in animal models of PD by regulating the Nrf2 signaling pathway [13].
Long non-coding RNAs (lncRNAs) are a group of RNAs with more than 200 nucleotides in length. LncRNAs do not participate in protein coding [14]. Many studies have revealed that lncRNA scan exert a great impact on diverse biological processes, such as nucleus-cytoplasm transport, genome imprinting, the regulation of gene expression, as well as RNA splicing and translational regulation [15]. Additionally, lncRNAs can affect the expression of RNA targets by competitively binding to miRNAs through miRNA response elements according to the competitive endogenous RNA (ceRNA) hypothesis [16,17]. LncRNA myocardial infarction-associated transcript (MIAT), also known as Gomafu, exhibits dysregulated expression in a variety of cardiovascular diseases [18-20]. Recently, it has been suggested that the cardiovascular system shares several similar mechanisms with the nervous system in terms of function, so lncRNA-MIAT may increase the risk of diseases associated with the nervous system, such as schizophrenia and Alzheimer’s disease [21,22]. Nevertheless, the role of lncRNA-MIAT in PD has not been studied before. Therefore, we aimed to determine the specific mechanism of lncRNA-MIAT in PD and the particular interactions of lncRNA-MIAT with miR-34-5p and SYT1. The results demonstrated that lncRNA MIAT acts as a ceRNA of miR-34-5p, resulting in the upregulation of SYT1, which reveals a possible mechanism underlying the effects of lncRNA-MIAT in the pathogenesis of PD.
Materials and methods
Ethics statements
The present research was examined and verified by the Animal Care and Use Committees of the First Affiliated Hospital, Harbin Medical University. All experimental operations were carried out in accordance with the International Convention on Laboratory Animal Ethics and the Guide for the Care and Use of Laboratory Animals.
Animal surgery
All animal experiments were conducted in accordance with the Regulations for the Administration of Affairs Concerning Experimental Animals. A total of 8 PD and 9 wild-type (WT) BALB/c mice (50 days old, half of them male and half of them female, weighing 22-30 g, and purchased from the Southern Model Biological Research Center) were housed at 18-22°C, a relative humidity of 50-60%, and a noise level below 60 dB. Before the surgery, the mice had ad libitum access to drinking and eating. The MIAT transcript sequence and the MIAT shRNA sequence were inserted using an adeno-associated virus (AAV; Invitrogen; Thermo Fisher Scientific, Inc.). The mice were given 2% pentobarbital sodium (16 mg/kg) for anesthesia. The solution containing the recombinant virus (10 μL, 1 × 1012/mL) was precisely injected into the rostral pole of the right substantia nigra pars compacta at three different locations three times [13]. The injection was administrated once a day and three times in succession. After carrying out the ethological experiments, all the mice were euthanized with highly concentrated pentobarbital sodium and their entire brains were collected.
Total RNA extraction, sequencing, and differentially expressed gene analysis
Total RNA from the mice’s whole brains (3 PD mice with MIAT-overexpression vs. 3 untreated PD mice) was extracted according to the TRIzol® Reagent protocol (Thermo Fisher) and quantified using the NanoDrop2000 spectrophotometer (Thermo Fisher Scientific). The total RNA integrity was assessed using RNA 6000 Nano Kits and Agilent Bioanalyzer 2100 (Agilent Biotechnologies, Santa Clara, CA, USA). Sequencing libraries were generated using a Colibri Stranded RNA Library Prep Kits for Illumina (NEB, MA, USA). Subsequently, the mRNAs purified from 2 µg of RNA per brain by oligo (dT) magnetic beads were randomly fragmented into pieces of approximately 200 base pairs in length using the fragmentation buffer. The fragmented mRNAs were then used for the first-strand cDNA synthesis using reverse transcriptase and random hexamer primers. The second-strand cDNA was synthesized using DNA polymerase I and RNase H. The DNA fragments were ligated to adaptors and barcodes for the polymerase chain reaction (PCR) templates. The libraries were sequenced using an IlluminaHiSeq 2500 platform (Illumina, CA, USA). FastQC was used to assess the sequencing quality. The sequences of pair-end Fastq files were aligned with the Musmusculus reference genome (mm10) using Bowtie (1.0.1). The BAM files were sorted, indexed, and qualified using SAMtools and HT-SEQ. The R/Bioconductor package edgeR was used to obtain and annotate differentially expressed genes in MIAT-overexpressed PD mice based on the threshold value (|log2FC|>2, FDR<0.05). The multiple testing corrections were performed using the Benjamini-Hochberg procedure to reduce the false discovery rate. The volcano map of differential gene expression was plotted using ggplot2. The DAVID tool was used to carry out the GO enrichment analysis. The MicroRNA.org database (microrna.org/microrna/home.do), miRDB (mirdb.org/), and TargetScan Human version 7.2 (targetscan.org/vert_72/) were used to predict the genes targeted by the miRNAs. Venn diagrams were plotted using the online website Venny (bioinformatics.psb.ugent.be/webtools/Venn/) to compare and analyze the data of different sets.
Cell culture and treatment
Neuroblastoma SH-SY5Y cells offered by Hunan Key Laboratory of Medical Genetics of Central South University (Changsha, Hunan, China) and 293T cells (CL-0005, Procell, Co., Ltd.) were cultivated in Minimum Essential Medium (MEM) supplemented with 10% Fetal bovine serum (FBS) and 1% penicillin and streptomycin (100 U/ml and 100 μg/ml respectively) at 37°C with 5% CO2. The cell lines were identified by 21-short tandem repeat loci authentication performed by Procell, Co., Ltd. The cell passaging was performed with a cell number to the medium volume ratio of 1:4. Before the transfection, SH-SY5Y cells were serum freed and co-incubated for 24 h with 40 μM of 6-hydroxydopamine (6-OHDA) dissolved in normal saline. The MIAT overexpression plasmid, the MIAT short hairpin (sh) RNA plasmid, the SYT1 overexpression plasmid, and the SYT1 shRNA plasmids were all generated using Biovector. The miR-34-5p mimic and miR-34-5p inhibitor were purchased from Thermo Fisher Scientific, Inc. SH-SY5Y cells were seeded in 96 well plates and grown to 70-90% confluence. Subsequently, 5 μL of Opti-MEM was mixed with 0.15 μL of Lipofectamine® 3000 (Cat. No. L3000015; Thermo Fisher Scientific, Inc.). Next, 10 μL of Opti-MEM, 0.4 μL of P3000™ Reagent, and 0.15 μg of DNA were gently mixed. A total of 5 µl from both solutions was mixed and incubated for 15 min. The aforementioned procedures were performed at room temperature. Ultimately, the DNA-liposome complex was added to the cells, and the cells were routinely cultured. The cells were transfected for 48 h.
RNA-fluorescence in situ hybridization (FISH)
A Ribo™ fluorescence in situ hybridization kit (Cat. No. C10910; Guangzhou RiboBio Co., Ltd.) was utilized for the subcellular localization test. The cell slides were prepared and subsequently fixed with 4% paraformaldehyde. The hybrid buffer was incubated with the fluorescently labeled MIAT probes (Sigma-Aldrich; Merck KGaA). The scrambled probe was set as NC. The immobilized cells were incubated with a hybrid buffer/probe mixture and then counterstained with DAPI. Anti-fluorescence quenching was added, and nail polish was used to seal the cells. The cells were then observed under a laser confocal microscope (ImageXpress® Nano; Molecular Devices, Inc.).
Dual-luciferase reporter assay
The predicted binding sites of miR-34-5p on MIAT and SYT1 were further confirmed using dual-luciferase reporter assays. The DNA fragments containing WT or mutant (MUT) binding sites with 50 bp flanking sequences were synthesized and cloned into the pMIR-GLO luciferase vector (Cat. No. E1330; Promega Corporation). The recombinant plasmids were isolated and sequenced. The MiR-34-5p mimic and the mimic NC sequences were synthesized by Beijing Genomics institution (BGI) and co-transfected with WT/MUT target gene fragments into 293T cells using Lipofectamine® 3000 according to the manufacturer’s instructions. After 4 h of transfection, the medium was changed, and the cells were cultured in an incubator at 37°C with 5% CO2 for 48 h. Subsequently, the medium was discarded, and PBS was used to wash the cells three times. The dual-luciferase kit (Cat. No. E2920; Promega Corporation) and an automatic chemiluminescence analyzer (DXI800, Beckman Kurt) were used to detect the fluorescence. The relative value of firefly luciferase activity value/renilla luciferase activity value was considered as luciferase activity.
RNA binding protein immunoprecipitation (RIP) assay
An RIP kit (Cat. No. 17-702; EMD Millipore) was used to detect the binding of MIAT and SYT1 to the Ago2 protein. After washing the 293T cells with pre-cooled PBS, the supernatant was discarded. Afterward, the pellet was placed in an ice bath for 5 min with a proper RIPA lysis buffer (Cat. No. P0013B; Biyuntian). The solution was then centrifuged at 4°C, and the content of the cell extract was co-precipitated with the antibody as a part of the input. Each co-precipitation reaction system consisted of 50 μL of magnetic beads and was re-suspended with 100 μL of RIP washing buffer. A total of 5 μL of antibody was added to the treatment groups. After washing, the magnetic bead antibody complex was re-suspended in 900 μL of RIP washing buffer and incubated overnight in 100 μL of cell extract at 4°C. The sample was placed in a magnetic pedestal to separate the magnetic bead-protein complex. Then protease K was used to digest the sample and input. RNA was then extracted and the amount of binding RNA was measured using quantitative (q) PCR. The antibodies used for the RIP assay were anti-Ago2 (Cat. No. ab186733; Abcam) and anti-immunoglobulin G (IgG; Cat. No. ab109489; Abcam).
Reverse transcription-qPCR
TRIzol® reagent (Cat. No. 15596; Thermo Fisher Scientific, Inc.) was used to extract the total RNA from the substantia nigra. NanoDrop 2000 (Thermo Fisher Scientific, Inc.) was used to determine the concentration and purity of the RNA. Reverse transcription was performed using an iScriptcDNA Synthesis kit (Cat. No. RR047A; Takara Bio, Inc.) according to the manufacturer’s protocol. SYBR Green Master mix (Cat. No. HRR081B; Takara Bio, Inc.) was used for the qPCR. Each reaction consisted of 25 μL of SYBR® Premix ExTaqTMII (2 ×), 2 μL of PCR forward and reverse primers, 1 μL of ROX Reference Dye (50 ×), 4 μL of DNA template, and 16 μL of ddH2O. The ABI PRISM® 7300 system (Applied Biosystems; Thermo Fisher Scientific, Inc.) was used to accomplish the PCR reaction. The thermocycling parameters were: pre-denaturation at 95°C for 4 min; followed by 40 cycles of 94°C for 30 sec, annealing at 58°C, and extension at 72°C for 1 min. GAPDH was selected as the reference gene. All the primers were purchased from BGI, and the 2-ΔΔCq method was used for the calculation of the genes’ relative expression levels [23]. The sequences are presented in Table 1.
Table 1.
PCR primers
| Gene | Forward primer (5’-3’) | Reverse primer (5’-3’) |
|---|---|---|
| MIAT | GCTCACACCTCCTATTCCT | CTTCACCAACTCTCCCACT |
| SYT1 | GAACTGCATAAAATCCCATTGCC | CAGCAGGTCACGACTAGAAGG |
| GAPDH | GATGCTGGTGCTGAGTATGCG | GTGGTGCAGGATGCATTGCTCTGA |
Western blotting
After washing with PBS twice, the SH-SY5Y cells (4*107) were pre-treated with 1 mL of cell lysis buffer (0.1% sodium dodecyl sulfate, 1% Triton X 100, 2 mM EDTA, 50 mM Tris, and 150 mM NaCl) on ice. Five minutes later, at 4°C, the supernatant of the cell lysates was extracted by centrifuging at 12,000 g for 5 min. A bicinchoninic acid protein assay kit (Cat. No. 23225; Thermo Fisher Scientific, Inc.) was utilized to measure the concentration of protein. SDS-PAGE was performed, and the resolved proteins were transferred to PVDF membranes. PBS containing 0.2% Tween-20 and 5% non-fat milk (PBS-T) was used to block the non-specific binding at room temperature on a shaker for 1 h. After the blocking solution was removed, the membranes were incubated with anti-Parkin antibodies (Cat. No. ab77924; Abcam, diluted to 5 μg/mL) or anti-tyrosine hydroxylase (TH) antibodies (Cat. No. ab112; Abcam; 1:200) at 4°C overnight. GAPDH was used as the loading control. Subsequently, the PVDF membrane was washed with PBS-T and incubated with diluted goat anti-mouse IgG (Cat. No. ab97040; Abcam, 1:5,000) secondary antibodies at 4°C for 4-6 h. The signals were visualized using enhanced chemiluminescence for 1 min at room temperature. The ratio of the gray value of the protein band to the gray value of the internal reference band was considered the relative protein level.
Cell Counting Kit-8 (CCK-8) assays
CCK-8 (Cat. No. C0038; Beyotime Institute of Biotechnology) assays were used to measure the cell viability of the SH-SY5Y cells. Initially, the cells were seeded into 96-well plates at 2*103 cells/well and cultured for 0, 24, 48, and 72 h, respectively. Afterward, 10 μL of CCK-8 solution was added into each well at 37°C for 2 h incubation. A microplate reader (Varioskan LUX; Thermo Fisher Scientific, Inc.) was used to measure the optical density (OD) at 450 nm.
Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) dual label staining
An annexin-V-FITC apoptosis detection kit (Cat. No. C1062M, Beyotime Institute of Biotechnology) was employed to determine the cells’ apoptosis rates. Briefly, 200 μL of Annexin V-FITC binding solution was added to the cells and the cells were incubated for 15 min. Subsequently, the cells were centrifuged at 1,000 r/min for 5 min. Afterward, the precipitate was collected and resuspended in 180 μL of Annexin V-FITC binding solution and 20 μL of iodine propidium (PI) staining solution (Cat. No. P8080-10 mg; Solvay), and then incubated on ice for 5 min in the dark. The apoptosis was measured in the treated cells within 30 min using a FACSCalibur flow cytometer (BD Biosciences).
Immunohistochemical (IHC) staining
The substantia nigra was embedded in paraffin and sectioned. The sections were placed in a 60°C for 1 h, de-waxed with xylene I and II for 10 min, and dehydrated using a gradient of alcohol solutions (100%, 90%, 80%, 70%, 60%, and 50%). The slices were washed with H2O2 and ddH2O for 1 min, respectively. Next, the antigen retrieval was conducted via immersing sections in 0.01 M citrate buffer and microwaving. The slices were then treated with 5% BSA for incubation at room temperature to block the non-specific binding. Subsequently, 20 minutes later, the slices were incubated with anti-Parkin antibodies (Cat. No. ab77924, Abcam; 1:1,000) or anti-TH antibodies (Cat. No. ab112; Abcam; 1:750) at 37°C for 1 h followed PBS rinsing. An anti-IgG antibody (Cat. No. ab97040; Abcam; 1:1,000) was used to enhance the immunoreactive signal. The slices were incubated with an appropriate quantity of SABC at 37°C for 45 min and rinsed with PBS. Diaminobenzidine and hematoxylin were used to develop the antibody staining and stain the nuclei. After rehydration with a gradient of ethanol solutions, neutral gum was used to seal the tissues. ImageJ version 2.0 (National Institutes of Health) was used for determining the positive cell rate.
TUNEL assay
The substantia nigra was fixed with 10% formaldehyde for 24 h and the tissues were embedded in paraffin, sliced, baked, dewaxed, and dehydrated as described above. At room temperature, the slices were treated with 10 mmol/L Tris-HCl containing 1 μg/mL proteinase K for degradation. Subsequently, the slices were washed with PBS to block the endogenous peroxidase activity. A TUNEL kit (Cat. No. 11684817910, Roche Diagnostics) was used to measure the apoptosis in the tissues. The tissues were incubated with 50 μL of TUNEL solution and a Converter-POD for 60 min in the dark. Subsequently, the nuclei were stained using hematoxylin and the slices were sealed using neutral resin. The slices were visualized under an optical microscope (CKX53, Olympus Corporation). The apoptotic rate (%) in each visual field was calculated as the number of positive apoptotic cells/total number of cells) *100%.
Behavioral assays
The rotating rod system (BW-MED-RRS, Shanghai Ruanlong, China) was used for the rotating rod test. A rotating rod was rotated at 30 rpm. The times when the mice fell off the rod were recorded in each group on days 7, 14, 21, and 28 after the injection.
Open field test: Referring to the method of Kawai et al. [24], a wooden open field chamber (100 cm L × 100 cm W × 40 cm H) was equipped with a bottom divided into 25 squares (20 cm × 20 cm). The inner part was painted black, the grid on the sidewall was defined as the peripheral area, and the rest was defined as the central area. We placed the mice in the central field for ambulation for 5 min and recorded the number of times that the mice crossed the grid (horizontal exercise ability) and rearing (vertical exercise ability).
The pole test: First, the mice were positioned upward on the rough-surfaced ligneous pole (70 cm in length and 1 cm diameter) with a 2-5 cm diameter foam shell stuck to the top and two layers of the plaster wrapped around. The mice were placed on the top of the ball and we recorded the time it took the mice to climb the top half of the pole, the time required for the mice to climb the downward half of the pole, and the full length of the pole. Points were awarded according to the following criteria: 3 points for completing one of the above actions within 3 seconds; 2 points for completing it within 6 seconds; 1 point for exceeding 6 seconds.
Statistical analysis
All the data analyses were carried out using SPSS version 21.0 (IBM, Corp.). The data were expressed as the mean ± standard deviation of three independent repeats. The comparisons between the two groups were performed using Student’s t-tests. The comparisons among multiple groups were analyzed using ANOVA followed by a Least-Significant Difference post-hoc test. P<0.05 was considered to indicate a significant difference.
Results
The MIAT expressions were downregulated in various regions of the brain
qPCR quantified the expression levels of MIAT in a variety of the brain regions of the PD mice and the 6-OHDA induced SH-SY5Y cells, and the results showed varying degrees of MIAT downregulation in the hippocampus, substantia nigra, striatum, and the whole brain (all P<0.05), but there was not a statistically significant inexpression of the cortex (P>0.05). Among the regions assessed, the decreases in the expression of MIAT were most prominent in the substantia nigra (Figure 1A). Additionally, the in vitro expression of MIAT 48 h after induction was significantly decreased compared with the normal saline group (Figure 1B; P<0.05). Thus, it was hypothesized that the MIAT expression was associated with the pathogenesis of PD.
Figure 1.
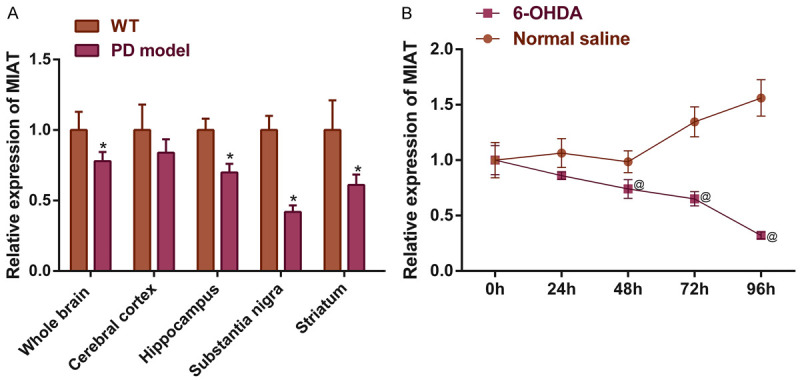
Aberrant MIAT expressions in different brain regions in a PD mouse model and in SH-SY5Y cells. MIAT expressions in (A) different brain regions of a PD mouse model, and (B) in 6-OHDA induced SH-SY5Y cells. *P<0.05 vs. WT; @ P<0.05 vs. normal saline. MIAT, myocardial infarction-associated transcript; PD, Parkinson’s disease, OHDA, hydroxydopamine.
MIAT protected the neuronal function of the SH-SY5Y cells
To investigate the effect of MIAT on the SH-SY5Y cell growth and function by overexpressing or knocking down the expression of MIAT, MIAT shRNA and exon recombinant plasmids were successfully constructed (all P<0.05; Figure 2A). Subsequently, the Parkin and TH protein levels were quantified by performing western blotting. TH is a neuron-specific biomarker and Parkin is a protein associated with the pathogenesis of PD that plays a protective role in dopaminergic neurons. The cell viability was measured using CCK-8 assays, and the results indicated that MIAT overexpression significantly increased cell growth 48 h after induction and enhanced the protein expression levels of Parkin and TH in comparison with the negative control group. The knockdown of MIAT achieved the opposite effects (all P<0.05; Figure 2B-D). Thus, MIAT improves the growth and function of SH-SY5Y cells.
Figure 2.
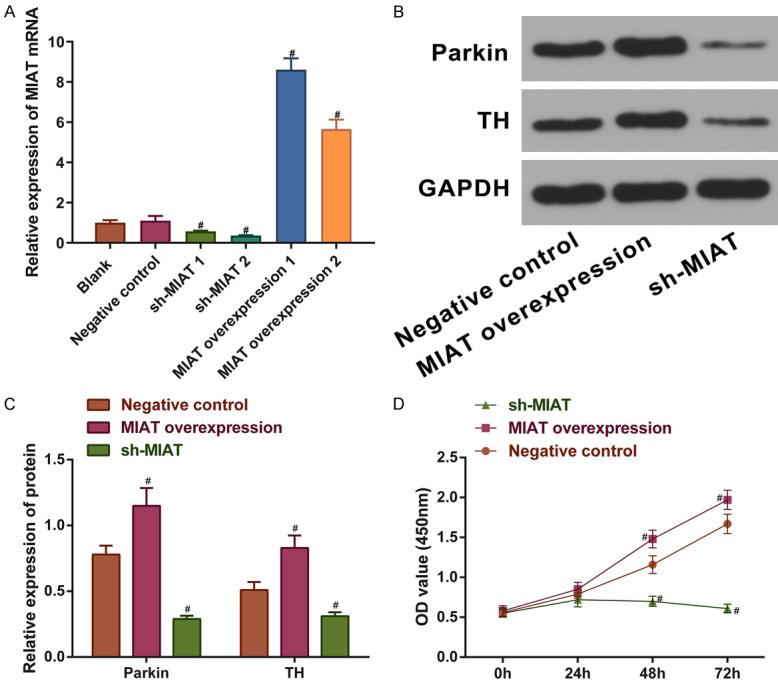
Neuroprotective effects of MIAT on SH-SY5Y cells. A. MIAT mRNA levels in SH-SY5Y cells with MIAT knocked down or overexpressed. B and C. Protein expression levels of Parkin and TH in SH-SY5Y cells with MIAT knocked down or overexpressed. D. Cell viability of treated SH-SY5Y cells. # P<0.05 vs. negative control. MIAT, myocardial infarction-associated transcript; TH, tyrosine hydroxylase.
Determining the molecular mechanism underlying the MIAT activity in PD
To determine the detailed mechanism of MIAT in PD, we up-regulated the expression of MIAT in the PD mice. The whole brains from 3 MIAT-overexpression PD mice and 3 untreated PD mice were used for the RNA-sequencing to find a MIAT target gene. Compared with the PD mice, the MIAT expression showed a marked increase, and 10 and 24 genes were found to be significantly up-regulated and down-regulated, respectively (Figure 3A and Table S1). The results from the GO enrichment analysis of the down-regulated genes suggested MIAT plays a potential regulatory role in dopaminergic neuron differentiation (GO: 1904339, fold enrichment = 508.84, FDR = 0.006, Table 2). But we failed to find a significant enrichment in any GO term for the up-regulated genes. For the fold change of genes in our analysis, even if it was not statistically significant, it might still have biological significance. Therefore, we focused on SYT1 (log2FC = 2.19, FDR = 0.052), a member of the synaptotagmin family, which serves as the calcium ion sensor located in the synaptic vesicles and that regulates neuronal exocytosis [25,26]. SYT1 serves an important regulatory role in dopamine release, and the dysregulation of SYT1 activity may underlie PD [6]. The miRNAs that bind to SYT1 were predicted. The common miRNAs in various prediction tools were included, and hsa-miR-429, hsa-miR-449a, hsa-miR-34-5p, hsa-miR-133b, and hsa-miR-146b-5p were used as consensus miRNAs (Figure 3B). Early research had shown that miR-34 expression is dysregulated in multiple neurodegenerative diseases [12]. In addition, miR-34a may reverse the neuroprotective effects of schisandrin in the PD model by negatively regulating the Nrf2 signaling pathway [13]. Thus, miR-34-5p may be a potential target in PD molecular therapy. LncRNAs may serve as ceRNA of coding genes to bind to miRNAs, resulting in an increased expression of the protein coded by the target gene. LncRNA-MIAT expression is dysregulated in various diseases, such as schizophrenia and myocardial infarction [21,27-29]. However, the mechanism of lncRNA-MIAT in PD is not well understood. Combined with the study results of MIAT function in the SH-SY5Y cells (Figures 1 and 2), it was hypothesized that MIAT could upregulate SYT1 expression by adsorbing miR-34-5p, thus exerting neuroprotective effects in PD.
Figure 3.
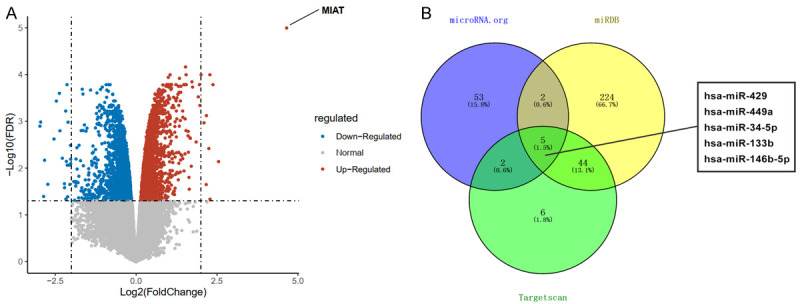
Differentially expressed genes in PD mice treated with MIAT over-expression. A. A volcano map of the differentially expressed genes. The blue dots indicate down-regulated genes, and the red dots indicate up-regulated genes. The dashed line parallel to the Y-axis is log2FC = |2|, and the dashed line parallel to the X-axis is FDR = 0.05 (MIAT over-expression PD mice vs PD mice). B. The common miRNAs targeting SYT1 were determined using a bioinformatics analysis. PD, Parkinson’s disease; SYT1, synaptotagmin-1; miRNA, microRNA.
Table 2.
A GO enrichment analysis of the differentially expressed genes
| Category | Term | Count | % | P | Genes | List Total | Pop Hits | Pop Total | Fold Enrichment | Bonferroni | Benjamini | FDR |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GOTERM_BP_DIRECT | GO: 1904339~negative regulation of dopaminergic neuron differentiation | 3 | 8.571429 | 1.05E-05 | GSK3B, SHH, WNT3A | 33 | 3 | 16792 | 508.8485 | 0.006693 | 0.006705 | 0.006621 |
| GOTERM_BP_DIRECT | GO: 0001938~positive regulation of endothelial cell proliferation | 4 | 11.42857 | 3.02E-04 | NRP1, NF1, THBS4, VEGFA | 33 | 69 | 16792 | 29.49846 | 0.175234 | 0.096162 | 0.094952 |
| GOTERM_BP_DIRECT | GO: 0048844~artery morphogenesis | 3 | 8.571429 | 7.22E-04 | NRP1, NF1, VEGFA | 33 | 21 | 16792 | 72.69264 | 0.368893 | 0.1436 | 0.141794 |
| GOTERM_BP_DIRECT | GO: 0001569~patterning of blood vessels | 3 | 8.571429 | 0.001289 | NRP1, SHH, VEGFA | 33 | 28 | 16792 | 54.51948 | 0.560391 | 0.1436 | 0.141794 |
| GOTERM_BP_DIRECT | GO: 0010628~positive regulation of gene expression | 5 | 14.28571 | 0.001477 | WNT3A, ERBB2, ALOX12, ACTG2, VEGFA | 33 | 262 | 16792 | 9.710849 | 0.609936 | 0.1436 | 0.141794 |
| GOTERM_BP_DIRECT | GO: 0043066~negative regulation of apoptotic process | 6 | 17.14286 | 0.001572 | GSK3B, SHH, PROK2, ALOX12, LIMS2, VEGFA | 33 | 455 | 16792 | 6.71009 | 0.632961 | 0.1436 | 0.141794 |
| GOTERM_BP_DIRECT | GO: 0045765~regulation of angiogenesis | 3 | 8.571429 | 0.001581 | ERBB2, NF1, PROK2 | 33 | 31 | 16792 | 49.2434 | 0.634898 | 0.1436 | 0.141794 |
| GOTERM_BP_DIRECT | GO: 0045785~positive regulation of cell adhesion | 3 | 8.571429 | 0.003026 | ERBB2, ALOX12, VEGFA | 33 | 43 | 16792 | 35.50106 | 0.854905 | 0.18629 | 0.183947 |
| GOTERM_BP_DIRECT | GO: 0010595~positive regulation of endothelial cell migration | 3 | 8.571429 | 0.003456 | NRP1, ALOX12, VEGFA | 33 | 46 | 16792 | 33.18577 | 0.889767 | 0.18629 | 0.183947 |
| GOTERM_BP_DIRECT | GO: 0051781~positive regulation of cell division | 3 | 8.571429 | 0.003605 | SHH, THBS4, VEGFA | 33 | 47 | 16792 | 32.47969 | 0.899802 | 0.18629 | 0.183947 |
| GOTERM_BP_DIRECT | GO: 0038190~VEGF-activated neuropilin signaling pathway | 2 | 5.714286 | 0.003808 | NRP1, VEGFA | 33 | 2 | 16792 | 508.8485 | 0.911982 | 0.18629 | 0.183947 |
| GOTERM_BP_DIRECT | GO: 1902336~positive regulation of retinal ganglion cell axon guidance | 2 | 5.714286 | 0.003808 | NRP1, VEGFA | 33 | 2 | 16792 | 508.8485 | 0.911982 | 0.18629 | 0.183947 |
| GOTERM_BP_DIRECT | GO: 0090259~regulation of retinal ganglion cell axon guidance | 2 | 5.714286 | 0.003808 | NRP1, VEGFA | 33 | 2 | 16792 | 508.8485 | 0.911982 | 0.18629 | 0.183947 |
| GOTERM_BP_DIRECT | GO: 0045964~positive regulation of dopamine metabolic process | 2 | 5.714286 | 0.007602 | GPR37, HPRT1 | 33 | 4 | 16792 | 254.4242 | 0.992255 | 0.322865 | 0.318804 |
| GOTERM_BP_DIRECT | GO: 0033138~positive regulation of peptidyl-serine phosphorylation | 3 | 8.571429 | 0.007837 | GSK3B, WNT3A, VEGFA | 33 | 70 | 16792 | 21.80779 | 0.993343 | 0.322865 | 0.318804 |
| GOTERM_BP_DIRECT | GO: 0048010~vascular endothelial growth factor receptor signaling pathway | 3 | 8.571429 | 0.008275 | NCKAP1, NRP1, VEGFA | 33 | 72 | 16792 | 21.20202 | 0.994975 | 0.322865 | 0.318804 |
| GOTERM_BP_DIRECT | GO: 0001525~angiogenesis | 4 | 11.42857 | 0.00863 | NRP1, PROK2, NRCAM, VEGFA | 33 | 223 | 16792 | 9.127327 | 0.995999 | 0.322865 | 0.318804 |
| GOTERM_BP_DIRECT | GO: 0045944~positive regulation of transcription from RNA polymerase II promoter | 7 | 20 | 0.009595 | NPAS4, GSK3B, SHH, DMRT1, WNT3A, NEUROG1, VEGFA | 33 | 981 | 16792 | 3.630927 | 0.997849 | 0.339039 | 0.334774 |
| GOTERM_BP_DIRECT | GO: 0050731~positive regulation of peptidyl-tyrosine phosphorylation | 3 | 8.571429 | 0.010626 | NRP1, THBS4, VEGFA | 33 | 82 | 16792 | 18.61641 | 0.998892 | 0.344723 | 0.340387 |
| GOTERM_BP_DIRECT | GO: 0060070~canonical Wnt signaling pathway | 3 | 8.571429 | 0.010875 | GSK3B, SHH, WNT3A | 33 | 83 | 16792 | 18.39211 | 0.999056 | 0.344723 | 0.340387 |
| GOTERM_BP_DIRECT | GO: 0008284~positive regulation of cell proliferation | 5 | 14.28571 | 0.011382 | SHH, WNT3A, PROK2, ALOX12, VEGFA | 33 | 466 | 16792 | 5.459748 | 0.999319 | 0.344723 | 0.340387 |
| GOTERM_BP_DIRECT | GO: 0071679~commissural neuron axon guidance | 2 | 5.714286 | 0.013266 | NRP1, VEGFA | 33 | 7 | 16792 | 145.3853 | 0.999798 | 0.35155 | 0.347128 |
| GOTERM_BP_DIRECT | GO: 0048842~positive regulation of axon extension involved in axon guidance | 2 | 5.714286 | 0.013266 | NRP1, VEGFA | 33 | 7 | 16792 | 145.3853 | 0.999798 | 0.35155 | 0.347128 |
| GOTERM_BP_DIRECT | GO: 0060982~coronary artery morphogenesis | 2 | 5.714286 | 0.013266 | NRP1, VEGFA | 33 | 7 | 16792 | 145.3853 | 0.999798 | 0.35155 | 0.347128 |
| GOTERM_BP_DIRECT | GO: 0035767~endothelial cell chemotaxis | 2 | 5.714286 | 0.017025 | NRP1, VEGFA | 33 | 9 | 16792 | 113.0774 | 0.999982 | 0.416455 | 0.411217 |
| GOTERM_BP_DIRECT | GO: 0032886~regulation of microtubule-based process | 2 | 5.714286 | 0.017025 | GSK3B, ERBB2 | 33 | 9 | 16792 | 113.0774 | 0.999982 | 0.416455 | 0.411217 |
| GOTERM_BP_DIRECT | GO: 0048643~positive regulation of skeletal muscle tissue development | 2 | 5.714286 | 0.018899 | SHH, WNT3A | 33 | 10 | 16792 | 101.7697 | 0.999995 | 0.42928 | 0.42388 |
| GOTERM_BP_DIRECT | GO: 2000727~positive regulation of cardiac muscle cell differentiation | 2 | 5.714286 | 0.018899 | GSK3B, WNT3A | 33 | 10 | 16792 | 101.7697 | 0.999995 | 0.42928 | 0.42388 |
| GOTERM_BP_DIRECT | GO: 0021904~dorsal/ventral neural tube patterning | 2 | 5.714286 | 0.02077 | SHH, WNT3A | 33 | 11 | 16792 | 92.51791 | 0.999998 | 0.455505 | 0.449776 |
| GOTERM_BP_DIRECT | GO: 0007417~central nervous system development | 3 | 8.571429 | 0.021844 | NCKAP1, SHH, NRCAM | 33 | 120 | 16792 | 12.72121 | 0.999999 | 0.463093 | 0.457268 |
| GOTERM_CC_DIRECT | GO: 0043235~receptor complex | 3 | 8.571429 | 0.022097 | NRP1, GPR37, ERBB2 | 34 | 127 | 18224 | 12.66142 | 0.872 | 0.89956 | 0.89956 |
| GOTERM_BP_DIRECT | GO: 0032228~regulation of synaptic transmission, GABAergic | 2 | 5.714286 | 0.022637 | NPAS4, NF1 | 33 | 12 | 16792 | 84.80808 | 1 | 0.464428 | 0.458586 |
| GOTERM_CC_DIRECT | GO: 0005576~extracellular region | 8 | 22.85714 | 0.022961 | SHH, WNT3A, APOF, PROK2, NRCAM, TRH, THBS4, VEGFA | 34 | 1610 | 18224 | 2.663354 | 0.881996 | 0.89956 | 0.89956 |
| GOTERM_BP_DIRECT | GO: 0001934~positive regulation of protein phosphorylation | 3 | 8.571429 | 0.024277 | WNT3A, ERBB2, VEGFA | 33 | 127 | 16792 | 12.02004 | 1 | 0.482514 | 0.476444 |
| GOTERM_BP_DIRECT | GO: 0035729~cellular response to hepatocyte growth factor stimulus | 2 | 5.714286 | 0.026361 | GSK3B, NRP1 | 33 | 14 | 16792 | 72.69264 | 1 | 0.508058 | 0.501667 |
| GOTERM_BP_DIRECT | GO: 0002042~cell migration involved in sprouting angiogenesis | 2 | 5.714286 | 0.028218 | NRP1, VEGFA | 33 | 15 | 16792 | 67.84646 | 1 | 0.527851 | 0.521212 |
| GOTERM_MF_DIRECT | GO: 0048018~receptor agonist activity | 2 | 5.714286 | 0.029916 | WNT3A, VEGFA | 33 | 16 | 16881 | 63.94318 | 0.980714 | 1 | 1 |
| GOTERM_BP_DIRECT | GO: 0048593~camera-type eye morphogenesis | 2 | 5.714286 | 0.030072 | NF1, VEGFA | 33 | 16 | 16792 | 63.60606 | 1 | 0.546451 | 0.539578 |
| GOTERM_CC_DIRECT | GO: 0005925~focal adhesion | 4 | 11.42857 | 0.033247 | CNN1, NCKAP1, NRP1, LIMS2 | 34 | 391 | 18224 | 5.483376 | 0.95543 | 0.89956 | 0.89956 |
| GOTERM_BP_DIRECT | GO: 0007413~axonal fasciculation | 2 | 5.714286 | 0.033769 | NRP1, NRCAM | 33 | 18 | 16792 | 56.53872 | 1 | 0.596583 | 0.589079 |
| GOTERM_BP_DIRECT | GO: 0031290~retinal ganglion cell axon guidance | 2 | 5.714286 | 0.035612 | NRP1, NRCAM | 33 | 19 | 16792 | 53.563 | 1 | 0.605927 | 0.598305 |
| GOTERM_BP_DIRECT | GO: 0007411~axon guidance | 3 | 8.571429 | 0.036709 | NRP1, SHH, WNT3A | 33 | 159 | 16792 | 9.600915 | 1 | 0.605927 | 0.598305 |
| GOTERM_MF_DIRECT | GO: 0008201~heparin binding | 3 | 8.571429 | 0.036775 | NRP1, THBS4, VEGFA | 33 | 160 | 16881 | 9.591477 | 0.992334 | 1 | 1 |
| GOTERM_BP_DIRECT | GO: 0002052~positive regulation of neuroblast proliferation | 2 | 5.714286 | 0.037452 | SHH, VEGFA | 33 | 20 | 16792 | 50.88485 | 1 | 0.605927 | 0.598305 |
| GOTERM_CC_DIRECT | GO: 0098794~postsynapse | 2 | 5.714286 | 0.039111 | NPAS4, GSK3B | 34 | 22 | 18224 | 48.72727 | 0.974536 | 0.89956 | 0.89956 |
| GOTERM_BP_DIRECT | GO: 1904886~beta-catenin destruction complex disassembly | 2 | 5.714286 | 0.041122 | GSK3B, WNT3A | 33 | 22 | 16792 | 46.25895 | 1 | 0.605927 | 0.598305 |
| GOTERM_BP_DIRECT | GO: 0071542~dopaminergic neuron differentiation | 2 | 5.714286 | 0.041122 | SHH, VEGFA | 33 | 22 | 16792 | 46.25895 | 1 | 0.605927 | 0.598305 |
| GOTERM_BP_DIRECT | GO: 0031334~positive regulation of protein complex assembly | 2 | 5.714286 | 0.041122 | GSK3B, VEGFA | 33 | 22 | 16792 | 46.25895 | 1 | 0.605927 | 0.598305 |
| GOTERM_BP_DIRECT | GO: 0001666~response to hypoxia | 3 | 8.571429 | 0.042328 | NF1, TRH, VEGFA | 33 | 172 | 16792 | 8.875264 | 1 | 0.605927 | 0.598305 |
| GOTERM_BP_DIRECT | GO: 0048754~branching morphogenesis of an epithelial tube | 2 | 5.714286 | 0.042951 | SHH, VEGFA | 33 | 23 | 16792 | 44.24769 | 1 | 0.605927 | 0.598305 |
| GOTERM_BP_DIRECT | GO: 0035924~cellular response to vascular endothelial growth factor stimulus | 2 | 5.714286 | 0.042951 | NRP1, VEGFA | 33 | 23 | 16792 | 44.24769 | 1 | 0.605927 | 0.598305 |
| GOTERM_BP_DIRECT | GO: 0002092~positive regulation of receptor internalization | 2 | 5.714286 | 0.044778 | WNT3A, VEGFA | 33 | 24 | 16792 | 42.40404 | 1 | 0.605927 | 0.598305 |
| GOTERM_BP_DIRECT | GO: 0007422~peripheral nervous system development | 2 | 5.714286 | 0.044778 | ERBB2, NF1 | 33 | 24 | 16792 | 42.40404 | 1 | 0.605927 | 0.598305 |
| GOTERM_BP_DIRECT | GO: 0007507~heart development | 3 | 8.571429 | 0.04732 | SHH, ERBB2, NF1 | 33 | 183 | 16792 | 8.341778 | 1 | 0.607254 | 0.599616 |
| GOTERM_BP_DIRECT | GO: 0048843~negative regulation of axon extension involved in axon guidance | 2 | 5.714286 | 0.04842 | NRP1, WNT3A | 33 | 26 | 16792 | 39.14219 | 1 | 0.607254 | 0.599616 |
| GOTERM_MF_DIRECT | GO: 0019838~growth factor binding | 2 | 5.714286 | 0.049978 | NRP1, ERBB2 | 33 | 27 | 16881 | 37.89226 | 0.998725 | 1 | 1 |
| GOTERM_BP_DIRECT | GO: 0033077~T cell differentiation in thymus | 2 | 5.714286 | 0.050236 | SHH, RAG2 | 33 | 27 | 16792 | 37.69248 | 1 | 0.607254 | 0.599616 |
| GOTERM_BP_DIRECT | GO: 0014065~phosphatidylinositol 3-kinase signaling | 2 | 5.714286 | 0.050236 | ERBB2, NF1 | 33 | 27 | 16792 | 37.69248 | 1 | 0.607254 | 0.599616 |
| GOTERM_BP_DIRECT | GO: 0001656~metanephros development | 2 | 5.714286 | 0.050236 | SHH, NF1 | 33 | 27 | 16792 | 37.69248 | 1 | 0.607254 | 0.599616 |
| GOTERM_BP_DIRECT | GO: 0007420~brain development | 3 | 8.571429 | 0.050605 | SYT1, FOXG1, NF1 | 33 | 190 | 16792 | 8.03445 | 1 | 0.607254 | 0.599616 |
| GOTERM_CC_DIRECT | GO: 0005886~plasma membrane | 13 | 37.14286 | 0.051555 | GSK3B, NRP1, GPR37, SYT1, KCNE3, WNT3A, TRH, NMBR, SHH, OR10W1, ERBB2, NRCAM, LIMS2 | 34 | 4121 | 18224 | 1.690852 | 0.992324 | 0.917167 | 0.917167 |
| GOTERM_MF_DIRECT | GO: 0042802~identical protein binding | 5 | 14.28571 | 0.051659 | SYT1, ERBB2, HPRT1, ETNPPL, VEGFA | 33 | 749 | 16881 | 3.414856 | 0.998988 | 1 | 1 |
| GOTERM_MF_DIRECT | GO: 0046982~protein heterodimerization activity | 4 | 11.42857 | 0.057132 | NPAS4, DMRT1, ERBB2, VEGFA | 33 | 465 | 16881 | 4.400391 | 0.999523 | 1 | 1 |
| GOTERM_CC_DIRECT | GO: 0016529~sarcoplasmic reticulum | 2 | 5.714286 | 0.059815 | MRVI1, THBS4 | 34 | 34 | 18224 | 31.52941 | 0.996567 | 0.917167 | 0.917167 |
| GOTERM_BP_DIRECT | GO: 0050918~positive chemotaxis | 2 | 5.714286 | 0.064646 | NRP1, VEGFA | 33 | 35 | 16792 | 29.07706 | 1 | 0.761382 | 0.751805 |
| GOTERM_BP_DIRECT | GO: 0043280~positive regulation of cysteine-type endopeptidase activity involved in apoptotic process | 2 | 5.714286 | 0.073544 | WNT3A, ALOX12 | 33 | 40 | 16792 | 25.44242 | 1 | 0.850432 | 0.839735 |
| GOTERM_CC_DIRECT | GO: 0009986~cell surface | 4 | 11.42857 | 0.073944 | NRP1, SHH, WNT3A, VEGFA | 34 | 542 | 18224 | 3.95572 | 0.999148 | 0.948781 | 0.948781 |
| GOTERM_BP_DIRECT | GO: 0010977~negative regulation of neuron projection development | 2 | 5.714286 | 0.080603 | GSK3B, WNT3A | 33 | 44 | 16792 | 23.12948 | 1 | 0.915419 | 0.903904 |
| GOTERM_BP_DIRECT | GO: 0007267~cell-cell signaling | 3 | 8.571429 | 0.084064 | NRP1, SHH, TRH | 33 | 254 | 16792 | 6.010021 | 1 | 0.937977 | 0.926179 |
| GOTERM_CC_DIRECT | GO: 0005578~proteinaceous extracellular matrix | 3 | 8.571429 | 0.084481 | SHH, WNT3A, VEGFA | 34 | 268 | 18224 | 6 | 0.999703 | 0.948781 | 0.948781 |
| GOTERM_CC_DIRECT | GO: 0005615~extracellular space | 6 | 17.14286 | 0.092816 | NRP1, SHH, WNT3A, THBS4, ACTG2, VEGFA | 34 | 1347 | 18224 | 2.387528 | 0.999872 | 0.948781 | 0.948781 |
| GOTERM_BP_DIRECT | GO: 0042472~inner ear morphogenesis | 2 | 5.714286 | 0.094566 | WNT3A, NEUROG1 | 33 | 52 | 16792 | 19.5711 | 1 | 1 | 0.988976 |
MIAT upregulated SYT1 expression by adsorbing miR-34-5p expression
To explore the regulatory mechanism of MIAT in SH-SY5Y cells, the MIAT subcellular localization was determined using a FISH assay. The results obtained from the FISH assay demonstrated that MIAT was distributed in the nuclei and cytoplasms, suggesting that MIAT may regulate gene expression by serving as a molecular sponge (Figure 4A). Subsequently, a dual-luciferase reporter assay was carried out to confirm the associations between miR-34-5p, MIAT, and SYT1 predicted by MicroRNA.org, miRDB, and TargetScanHuman 7.2 (Figure 4B and 4C). In comparison with the negative control group, the miR-34-5p mimic bound to the MIAT wild-type fragment and the SYT1 wild-type fragment, led to significant reductions in the luciferase activity (all P<0.05; Figure 4D).
Figure 4.
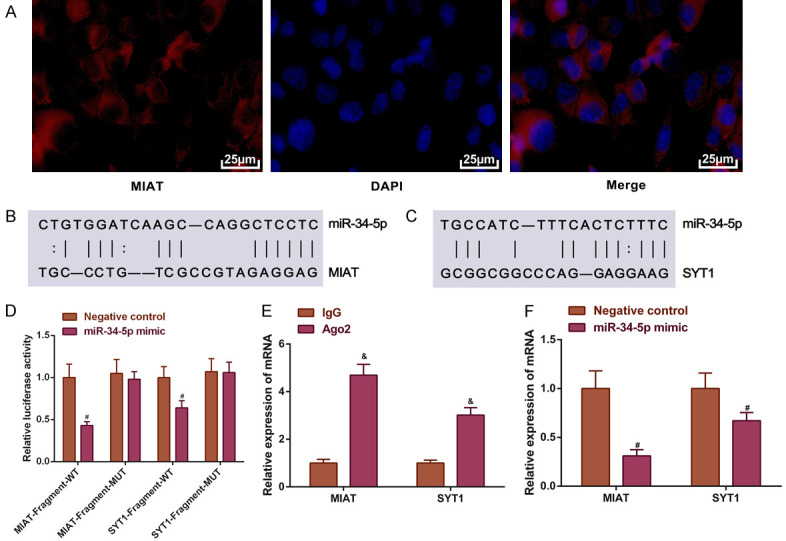
MIAT upregulates the expression of SYT1 by sponging miR-34-5p. A. The subcellular localization of MIAT. Magnification, × 200; B and C. The predicted binding sites of miR-34-5p on MIAT and SYT1; D. The relative luciferase activity; E. The relative mRNA expression levels of MIAT and SYT1 enriched by Ago2; F. The relative mRNA expression levels of MIAT and SYT1 in SH-SY5Y cells transfected with miR-34-5p mimic. # P<0.05 vs. control; & P<0.05 vs. IgG. MIAT, myocardial infarction-associated transcript; SYT1, synaptotagmin-1; miR, microRNA; IgG, immunoglobulin G.
The Ago2 protein and the miRNAs participate in the formation of an RNA-induced silencing complex, thereby binding to the target RNAs and inducing the degradation of target RNAs. RIP was used to determine whether Ago2 recruited MIAT or SYT1. The results showed that Ago2 antibodies pulled down the MIAT or the SYT1 mRNA (P<0.05; Figure 4E).
The effects of miR-34-5p on MIAT and SYT1 mRNA expressions were determined by overexpression and knockdown of miR-34-5p. Compared to the negative control, the decreased levels of MIAT and SYT1 mRNA expressions were observed in the miR-34-5p mimic group (all P<0.05; Figure 4F). These results demonstrated that MIAT can upregulate SYT1 expression by adsorbing miR-34-5p.
MIAT adsorbed miR-34-5p to promote Parkin and TH protein expressions in the SH-SY5Y cells
Consistent with the results in Figure 4, we found that the SYT1 protein level was up-regulated by MIAT but inhibited by miR-34-5p (all P<0.05; Figure 5). To ascertain whether MIAT regulated the levels of Parkin and TH proteins by adsorbing miR-34-5p in vitro, MIAT was overexpressed in the SH-SY5Y cells, and the miR-34-5p expression was decreased. Western blotting showed that higher levels of the Parkin and TH proteins were observed in the MIAT overexpression group and the miR-34-5p inhibitor group in comparison with the negative control group (all P<0.05; Figure 5A and 5B). In the miR-34-5p mimic group, the protein expression levels of Parkin and TH were significantly decreased (all P<0.05; Figure 5A and 5B). MIAT partially reversed the effects of miR-34-5p on the protein levels of Parkin and TH (P<0.05; Figure 5A and 5B).
Figure 5.
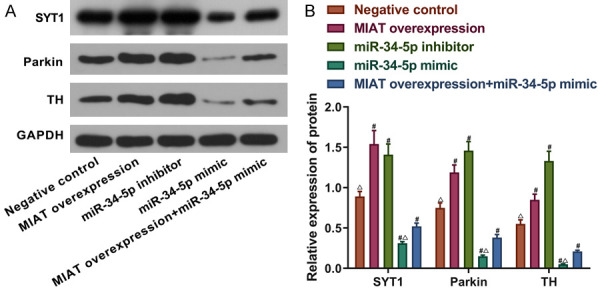
MIAT adsorbs miR-34-5p and increases the Parkin and TH protein expression levels in SH-SY5Y cells. A and B. The relative protein expression levels of SYT1, Parkin and TH in the SH-SY5Y cells with miR-34-5p or MIAT knocked down or overexpressed. # P<0.05 vs. the negative control; Δ P<0.05 vs. MIAT overexpression + miR-34-5p simulation. MIAT, myocardial infarction-associated transcript; miR, microRNA; TH, tyrosine hydroxylase; SYT1, synaptotagmin-1.
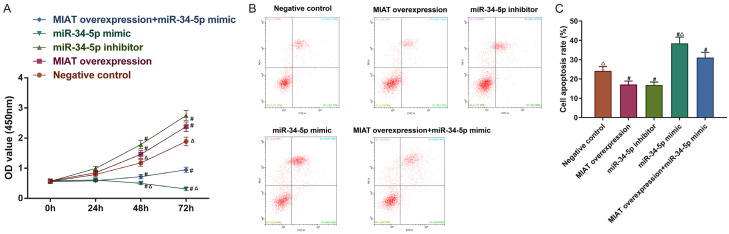
MIAT adsorbed miR-34-5p to increase cell viability and decrease the apoptosis of the SH-SY5Y cells
To determine whether MIAT regulated cell viability by targeting miR-34-5p, the MIAT overexpression vector, the miR-34-5p inhibitors, and the miR-34-5p mimic were transfected into the SH-SY5Y cells. CCK-8 assays and annexin V-FITC/PI dual-label staining were performed, and the results demonstrated that compared with the negative control group, the cell viability was increased but the apoptosis was decreased in the MIAT overexpression group and the miR-34-5p inhibitor group. Conversely, the opposite results were discovered in the miR-34-5p mimic group (all P<0.05; Figure 6A-C). MIAT overexpression partially reversed the effects of the miR-34-5p mimics on cell viability as well as the apoptosis (P<0.05; Figure 6A-C).
Figure 6.
MIAT adsorbs miR-34-5p, increases the SH-SY5Y cell viability, and reduces apoptosis. A. The SH-SY5Y cell viability. B. The early and late phase apoptosis. C. Quantification of the apoptosis. # P<0.05 vs. negative control; Δ P<0.05 vs. MIAT overexpression + miR-34-5p simulation. MIAT, myocardial infarction-associated transcript; miR, microRNA.
MIAT regulated the SYT1 expression to promote the expressions of Parkin and TH in the SH-SY5Y cells by absorbing miR-34-5p
We then explored the effects of the MIAT/SYT1/miR-34-5p regulatory network on the protein expressions of Parkin and TH. MIAT overexpression, SYT1 overexpression, and a miR-34-5p mimic transfection were performed in the SH-SY5Y cells. The Western blot results demonstrated that in comparison with the negative control group, the Parkin and TH’s protein expressions were increased by the overexpression of MIAT and SYT1, but the miR-34-5p mimic resulted in a decrease in their expressions (all P<0.05; Figure 7A and 7B). Notably, overexpressing SYT1 partially reversed the effects of miR-34-5p overexpression on the expressions of Parkin and TH (P<0.05; Figure 7A and 7B).
Figure 7.
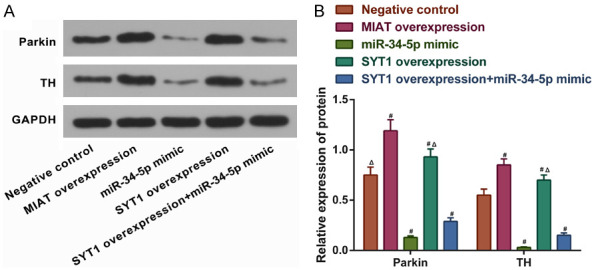
MIAT regulates SYT1 to increase the protein expression levels of Parkin and TH in SH-SY5Y cells by adsorbing miR-34-5p. A and B. The relative protein expression levels of Parkin and TH in SH-SY5Y cells with miR-34-5p or MIAT knocked down or overexpressed. # P<0.05 vs. negative control; ▲ P<0.05 vs. SYT1 overexpression + miR-34-5p mimic. MIAT, myocardial infarction-associated transcript; SYT1, synaptotagmin-1; miR, microRNA; TH, tyrosine hydroxylase.
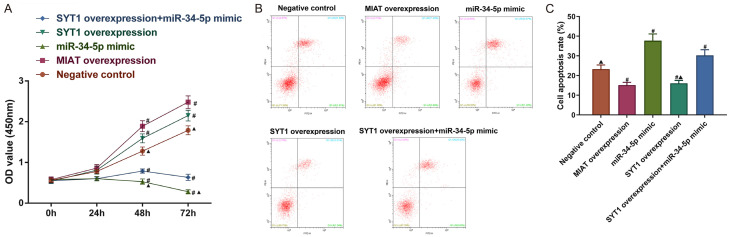
MIAT mediated SYT1 to promote the viability and inhibit the apoptosis of the SH-SY5Y cells by adsorbing miR-34-5p
To further investigate the regulatory mechanisms of the MIAT/SYT1/miR-34-5p network on viability and apoptosis, the SH-SY5Y cells were transfected with a MIAT overexpression vector, a SYT1 overexpression vector, or a miR-34-5p mimic. from the CCK-8 assay and Annexin V-FITC/PI dual-label staining results illustrated that in comparison with the negative control group, the cell viability was increased and the apoptosis was decreased in the cells overexpressing MIAT or SYT1 (all P<0.05; Figure 8A-C). Conversely, the transfection of the miR-34-5p mimic resulted in a decreased cell viability and increased apoptosis (all P<0.05; Figure 8A-C). The SYT1 overexpression partly reversed the effects of the miR-34-5p mimic on the cell viability and apoptosis (P<0.05; Figure 8A-C).
Figure 8.
MIAT regulates SYT1 to increase the viability and reduce the apoptosis of SH-SY5Y cells by adsorbing miR-34-5p. A. SH-SY5Y cell viability. B. Early and late phase apoptosis. C. Quantification of the apoptosis. # P<0.05 vs. negative control. ▲ P<0.05 vs. SYT1 overexpression + miR-34-5p mimic. MIAT, myocardial infarction-associated transcript; SYT1, synaptotagmin-1; miR, microRNA.
MIAT increased the Parkin and TH expressions in the substantia nigra of the PD mice
Whether MIAT increased the Parkin and TH expressions in vivo as well as in vitro was determined. An AAV containing MIAT transcripts or MIAT shRNA was used to infect the substantia nigra of the PD mice. Afterward, immunohistochemistry was performed to quantify the proportions of the TH and Parkin-positive cells. In the normal substantia nigra, TH and Parkin were highly expressed, and the neuronal cells were morphologically normal and structurally intact in the dense zone, with a large number of regularly arranged cells (Figure 9A). Following the MIAT overexpression, the number of Parkin or TH positive neurons in the substantia nigra was upregulated, and the cell body outlines and synapses were visible. Cell body shrinkage was observed in the sh-MIAT group. The TH and Parkin proteins were co-localized both in the cytoplasms and the nuclei (Figure 9A and 9B). At the same time, the number of TH and Parkin-positive neurons observed in the MIAT overexpression group was increased in comparison with the negative control group (all P<0.05; Figure 9C). Conversely, transfection with sh-MIAT reduced the number of TH and Parkin-positive neurons in the PD mice (all P<0.05; Figure 9C).
Figure 9.
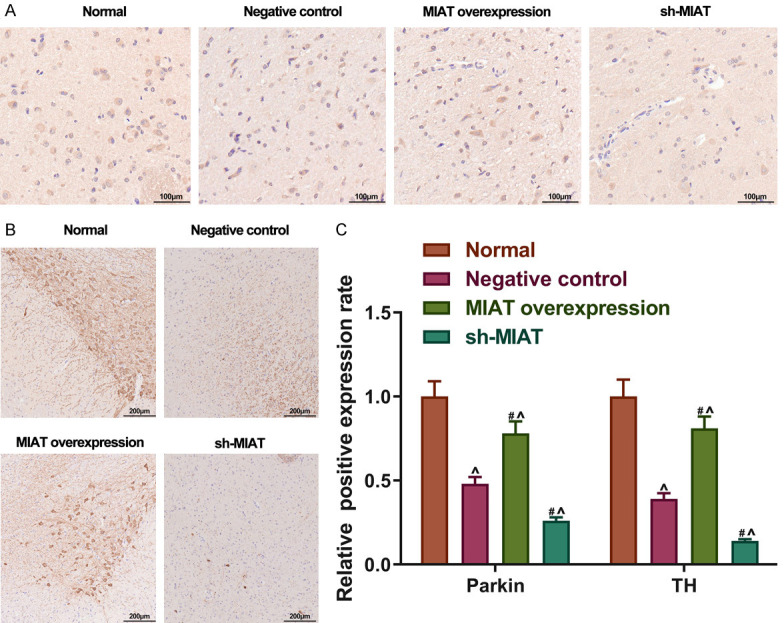
MIAT increases Parkin and TH expression in the substantia nigra of PD mice. A and B. The Parkin and TH-positive cells measured using immunohistochemistry staining in vivo. Magnification, × 200. C. Relative positive expression rate of Parkin and TH in each group. # P<0.05 vs. the negative control. ^P<0.05 vs. normal. MIAT, myocardial infarction-associated transcript; PD, Parkinson’s disease; TH, tyrosine hydroxylase.
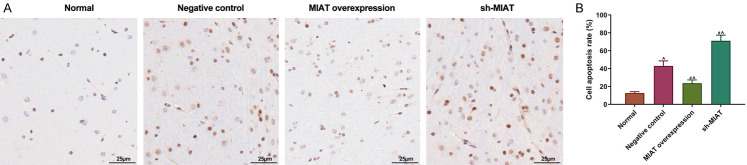
MIAT exerted a neuroprotective effect on the substantia nigra of PD mice
We further investigated the neuroprotective effects of MIAT. TUNEL staining was used to observe the neuron damage in the substantia nigra of the PD mice. In comparison with normal substantia nigra, the apoptotic neurons in the substantia nigra increased significantly and were damaged more severely in the NC group. In addition, the apoptosis of the neurons was strikingly reduced in the MIAT overexpression group, whereas MIAT shRNA significantly increased the apoptotic neurons compared with the NC group (all P<0.05; Figure 10A and 10B).
Figure 10.
MIAT has a neuroprotective effect on the substantia nigra of PD mice. A and B. Apoptosis was detected using TUNEL staining in vivo. Magnification, × 400. # P<0.05 vs. negative control. ^P<0.05 vs. normal. MIAT, myocardial infarction-associated transcript; PD, Parkinson’s disease.
Combined with the results of the in vitro experiments, the data suggest that MIAT has a neuroprotective effect in PD.
MIAT partially rescued behavioral phenotypes of PD mice
To investigate the effects of MIAT on the locomotor activity and the motor coordination ability in the PD group, rotation tests, open field tests, and the pole climbing tests were performed. Compared to the normal group, the remaining groups of mice showed significantly increased dyskinesia. In addition, compared to the negative control, PD mice treated with MIAT overexpression showed that the time it took for the mice to fall was prolonged on the 7th, 14th, and 21st days. Also, the ambulation and rearing scores were significantly increased on the 21st and 28th days, respectively. But after being transfected with MIAT shRNA, it took a much shorter time for the PD mice to fall off the rod on the 21st and 28th days. The rearing and ambulation scores (7 d, 14 d, and 21 d) decreased on the same conditions, compared to the negative controls (all P<0.05; Figure 11). Thus, the overexpression of MIAT can partially improve the motor performance of PD mice.
Figure 11.
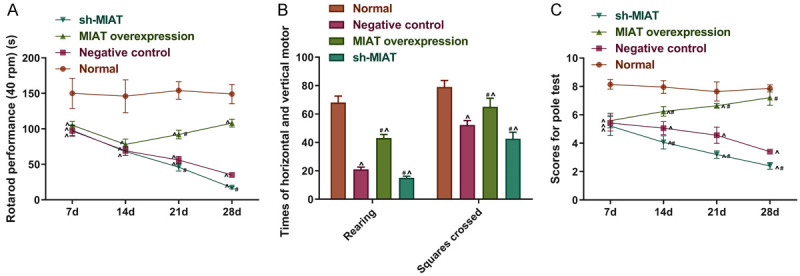
MIAT partly rescued the motor disability of PD mice. A-C. Autonomous activities and physical coordination were measured using rotating tests, open field tests and pole tests. # P<0.05 vs. the negative control. ^P<0.05 vs. normal. MIAT, myocardial infarction-associated transcript; PD, Parkinson’s disease.
Discussion
With several biological mechanisms underlying the diseases of the nervous system being revealed in recent years, some specific genes have been shown to exert significant effects on the occurrence and development of PD. In the present study, the cellular model and the animal model were used to explore the pathological functions of lncRNA-MIAT throughout PD’s progression. The results suggested that MIAT expression is significantly downregulated in the regions of the substantia nigra and corpus striatum in animal model. Additionally, it was shown that lncRNA MIAT can function as a ceRNA to regulate SYT1 by sponging miR-34-5p, which consequently participates in the progression of PD and exerts a neuroprotective effect.
A genome-wide association study indicated that lncRNA-MIAT is associated with the occurrence of myocardial infarction [27]. Accumulating evidence shows that lncRNA-MIAT potentially participated in multiple cellular processes and disease progressions, such as myocardial infarction, hypoxic-ischemic injury, age-related cataracts, and microvascular dysfunction [18,30,31]. In recent years, the effects of lncRNA-MIAT on several diseases associated with the nervous system have gained increasing attention. A previous study revealed that lncRNA-MIAT is a high-risk gene for paranoid schizophrenia and participates in the pathophysiology of schizophrenia [29]. Barry et al. determined that lncRNA-MIAT expression is remarkably decreased in the cortex and grey matter in patients with schizophrenia after death, implying that the dysregulation of MIAT contributes to neurological disorders [21]. In the current study, MIAT was found to be upregulated in the PD mice’s substantia nigra and corpus striatum. Overexpressing MIAT promoted the expression levels of Parkin and TH in SH-SY5Y cells. Furthermore, the cell viability of SH-SY5Y cells was enhanced. All the results demonstrated the neuroprotective role of MIAT in PD.
From the perspective of the ceRNA hypothesis, the present study found that lncRNA-MIAT could act as a ceRNA in regulating the SYT1 expression by absorbing miR-34-5p. Preceding research has shown that lncRNA-MIAT can affect lung cancer progression by epigenetically regulating miR-34a [32]. Another study reported that the expression level of miR-34 is elevated in patients with Alzheimer’s disease, and that it may serve as a novel target to treat certain cognitive disorders [33]. Hu et al. confirmed that miR-34c downregulation can ameliorate neurodegeneration and memory impairments [34]. Furthermore, the dysregulation of miR-34 has been demonstrated in PD, Alzheimer’s disease, and Huntington’s disease, suggesting its significant role in several neurodegenerative disorders [12]. The present study is the first study to investigate the regulatory relationship between lncRNA-MIAT and miR-34-5p in PD. Our results state clearly that MIAT can increase the concentration of Parkin and TH in SH-SY5Y cells by sponging miR-34-5p to promote the cell viability of SH-SY5Y cells significantly.
SYT1 modulates the release of the calcium-dependent neurotransmitter as a pivotal intercessor and is tightly associated with physiological cognitive development [35]. The local accumulation of amyloid β peptide is hypothesized to underlie neuronal degeneration, memory deficits, synapse loss, and dysfunction. SYT1 regulates synaptic amyloid β, so it may be a potential target in treating nervous system disorders [36]. In our study, MIAT and SYT1 overexpressions boosted the Parkin and TH levels and particularly affected the cell viability and apoptotic rate. The downregulation of Parkin and TH caused by increasing the miR-34-5p expression was partially reversed by SYT1, suggesting that MIAT may regulate SYT1 by absorbing miR-34-5p, which subsequently affects the pathogenic process of PD.
In summary, the ceRNA regulatory network, lncRNA-MIAT/miR-34-5p/SYT1, may play an important part in the pathogenesis of PD. MIAT may pose as a ceRNA in modulating the SYT1 expression by sponging miR-34-5p, which consequently exerts neuroprotective effects in PD. The molecular regulation mechanism of the present research is shown in Figure 12.
Figure 12.
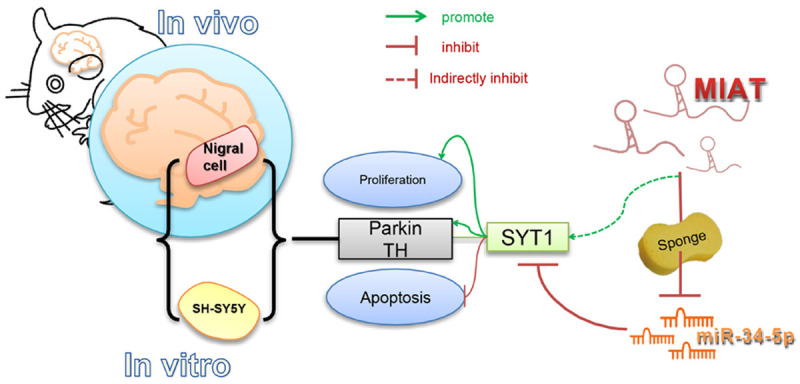
The molecular regulation mechanism by which LncRNA-MIAT regulates the growth of SHSY5Y cells via the miR-34-5p-SYT1 axis, showing neuroprotective effects in a mouse model of Parkinson’s disease.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Kalia LV, Lang AE. Parkinson’s disease. Lancet. 2015;386:896–912. doi: 10.1016/S0140-6736(14)61393-3. [DOI] [PubMed] [Google Scholar]
- 2.Falkenburger BH, Saridaki T, Dinter E. Cellular models for Parkinson’s disease. J Neurochem. 2016;139(Suppl 1):121–130. doi: 10.1111/jnc.13618. [DOI] [PubMed] [Google Scholar]
- 3.Jankovic J. Parkinson’s disease: clinical features and diagnosis. J Neurol Neurosurg Psychiatry. 2008;79:368–376. doi: 10.1136/jnnp.2007.131045. [DOI] [PubMed] [Google Scholar]
- 4.Xu J, Mashimo T, Sudhof TC. Synaptotagmin-1, -2, and -9: Ca(2+) sensors for fast release that specify distinct presynaptic properties in subsets of neurons. Neuron. 2007;54:567–581. doi: 10.1016/j.neuron.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 5.Mingazov ER, Ugrumov MV. Gene expression of proteins of the vesicle cycle in dopaminergic neurons in modeling of Parkinson’s disease. Dokl Biochem Biophys. 2016;468:206–208. doi: 10.1134/S1607672916030133. [DOI] [PubMed] [Google Scholar]
- 6.Liu C, Kaeser PS. Mechanisms and regulation of dopamine release. Curr Opin Neurobiol. 2019;57:46–53. doi: 10.1016/j.conb.2019.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fan Y, Siklenka K, Arora SK, Ribeiro P, Kimmins S, Xia J. miRNet-dissecting miRNA-target interactions and functional associations through network-based visual analysis. Nucleic Acids Res. 2016;44:W135–141. doi: 10.1093/nar/gkw288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 9.Ameres SL, Zamore PD. Diversifying microRNA sequence and function. Nat Rev Mol Cell Biol. 2013;14:475–488. doi: 10.1038/nrm3611. [DOI] [PubMed] [Google Scholar]
- 10.Codocedo JF, Inestrosa NC. Environmental control of microRNAs in the nervous system: implications in plasticity and behavior. Neurosci Biobehav Rev. 2016;60:121–138. doi: 10.1016/j.neubiorev.2015.10.010. [DOI] [PubMed] [Google Scholar]
- 11.Shafi G, Aliya N, Munshi A. MicroRNA signatures in neurological disorders. Can J Neurol Sci. 2010;37:177–185. doi: 10.1017/s0317167100009902. [DOI] [PubMed] [Google Scholar]
- 12.Hernandez-Rapp J, Rainone S, Hebert SS. MicroRNAs underlying memory deficits in neurodegenerative disorders. Prog Neuropsychopharmacol Biol Psychiatry. 2017;73:79–86. doi: 10.1016/j.pnpbp.2016.04.011. [DOI] [PubMed] [Google Scholar]
- 13.Ba Q, Cui C, Wen L, Feng S, Zhou J, Yang K. Schisandrin B shows neuroprotective effect in 6-OHDA-induced Parkinson’s disease via inhibiting the negative modulation of miR-34a on Nrf2 pathway. Biomed Pharmacother. 2015;75:165–172. doi: 10.1016/j.biopha.2015.07.034. [DOI] [PubMed] [Google Scholar]
- 14.Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172:393–407. doi: 10.1016/j.cell.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boon RA, Jae N, Holdt L, Dimmeler S. Long noncoding RNAs: from clinical genetics to therapeutic targets? J Am Coll Cardiol. 2016;67:1214–1226. doi: 10.1016/j.jacc.2015.12.051. [DOI] [PubMed] [Google Scholar]
- 16.Sui J, Li YH, Zhang YQ, Li CY, Shen X, Yao WZ, Peng H, Hong WW, Yin LH, Pu YP, Liang GY. Integrated analysis of long non-coding RNAassociated ceRNA network reveals potential lncRNA biomarkers in human lung adenocarcinoma. Int J Oncol. 2016;49:2023–2036. doi: 10.3892/ijo.2016.3716. [DOI] [PubMed] [Google Scholar]
- 17.Song YX, Sun JX, Zhao JH, Yang YC, Shi JX, Wu ZH, Chen XW, Gao P, Miao ZF, Wang ZN. Non-coding RNAs participate in the regulatory network of CLDN4 via ceRNA mediated miRNA evasion. Nat Commun. 2017;8:289. doi: 10.1038/s41467-017-00304-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan B, Yao J, Liu JY, Li XM, Wang XQ, Li YJ, Tao ZF, Song YC, Chen Q, Jiang Q. lncRNA-MIAT regulates microvascular dysfunction by functioning as a competing endogenous RNA. Circ Res. 2015;116:1143–1156. doi: 10.1161/CIRCRESAHA.116.305510. [DOI] [PubMed] [Google Scholar]
- 19.Li Y, Wang J, Sun L, Zhu S. LncRNA myocardial infarction-associated transcript (MIAT) contributed to cardiac hypertrophy by regulating TLR4 via miR-93. Eur J Pharmacol. 2018;818:508–517. doi: 10.1016/j.ejphar.2017.11.031. [DOI] [PubMed] [Google Scholar]
- 20.Zhou X, Zhang W, Jin M, Chen J, Xu W, Kong X. lncRNA MIAT functions as a competing endogenous RNA to upregulate DAPK2 by sponging miR-22-3p in diabetic cardiomyopathy. Cell Death Dis. 2017;8:e2929. doi: 10.1038/cddis.2017.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barry G, Briggs JA, Vanichkina DP, Poth EM, Beveridge NJ, Ratnu VS, Nayler SP, Nones K, Hu J, Bredy TW, Nakagawa S, Rigo F, Taft RJ, Cairns MJ, Blackshaw S, Wolvetang EJ, Mattick JS. The long non-coding RNA Gomafu is acutely regulated in response to neuronal activation and involved in schizophrenia-associated alternative splicing. Mol Psychiatry. 2014;19:486–494. doi: 10.1038/mp.2013.45. [DOI] [PubMed] [Google Scholar]
- 22.Jiang Q, Shan K, Qun-Wang X, Zhou RM, Yang H, Liu C, Li YJ, Yao J, Li XM, Shen Y, Cheng H, Yuan J, Zhang YY, Yan B. Long non-coding RNA-MIAT promotes neurovascular remodeling in the eye and brain. Oncotarget. 2016;7:49688–49698. doi: 10.18632/oncotarget.10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pagani IS, Spinelli O, Mattarucchi E, Pirrone C, Pigni D, Amelotti E, Lilliu S, Boroni C, Intermesoli T, Giussani U, Caimi L, Bolda F, Baffelli R, Candi E, Pasquali F, Lo Curto F, Lanfranchi A, Porta F, Rambaldi A, Porta G. Genomic quantitative real-time PCR proves residual disease positivity in more than 30% samples with negative mRNA-based qRT-PCR in chronic myeloid leukemia. Oncoscience. 2014;1:510–521. doi: 10.18632/oncoscience.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kawai H, Makino Y, Hirobe M. Novel endogenous 1,2,3,4-tetrahydroisoquinoline derivatives: uptake by dopamine transporter and activity to induce parkinsonism. J Neurochem. 1998;70:745–751. doi: 10.1046/j.1471-4159.1998.70020745.x. [DOI] [PubMed] [Google Scholar]
- 25.Fernandez-Chacon R, Konigstorfer A, Gerber SH, Garcia J, Matos MF, Stevens CF, Brose N, Rizo J, Rosenmund C, Sudhof TC. Synaptotagmin I functions as a calcium regulator of release probability. Nature. 2001;410:41–49. doi: 10.1038/35065004. [DOI] [PubMed] [Google Scholar]
- 26.Fukuda M, Aruga J, Niinobe M, Aimoto S, Mikoshiba K. Inositol-1,3,4,5-tetrakisphosphate binding to C2B domain of IP4BP/synaptotagmin II. J Biol Chem. 1994;269:29206–29211. [PubMed] [Google Scholar]
- 27.Ishii N, Ozaki K, Sato H, Mizuno H, Susumu S, Takahashi A, Miyamoto Y, Ikegawa S, Kamatani N, Hori M, Satoshi S, Nakamura Y, Tanaka T. Identification of a novel non-coding RNA, MIAT, that confers risk of myocardial infarction. J Hum Genet. 2006;51:1087–1099. doi: 10.1007/s10038-006-0070-9. [DOI] [PubMed] [Google Scholar]
- 28.Iaconetti C, Gareri C, Polimeni A, Indolfi C. Non-coding RNAs: the “dark matter” of cardiovascular pathophysiology. Int J Mol Sci. 2013;14:19987–20018. doi: 10.3390/ijms141019987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rao SQ, Hu HL, Ye N, Shen Y, Xu Q. Genetic variants in long non-coding RNA MIAT contribute to risk of paranoid schizophrenia in a Chinese Han population. Schizophr Res. 2015;166:125–130. doi: 10.1016/j.schres.2015.04.032. [DOI] [PubMed] [Google Scholar]
- 30.Li EY, Zhao PJ, Jian J, Yin BQ, Sun ZY, Xu CX, Tang YC, Wu H. LncRNA MIAT overexpression reduced neuron apoptosis in a neonatal rat model of hypoxic-ischemic injury through miR-211/GDNF. Cell Cycle. 2019;18:156–166. doi: 10.1080/15384101.2018.1560202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen Y, Dong LF, Zhou RM, Yao J, Song YC, Yang H, Jiang Q, Yan B. Role of long non-coding RNA MIAT in proliferation, apoptosis and migration of lens epithelial cells: a clinical and in vitro study. J Cell Mol Med. 2016;20:537–548. doi: 10.1111/jcmm.12755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fu Y, Li C, Luo Y, Li L, Liu J, Gui R. Silencing of long non-coding RNA MIAT sensitizes lung cancer cells to gefitinib by epigenetically regulating miR-34a. Front Pharmacol. 2018;9:82. doi: 10.3389/fphar.2018.00082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zovoilis A, Agbemenyah HY, Agis-Balboa RC, Stilling RM, Edbauer D, Rao P, Farinelli L, Delalle I, Schmitt A, Falkai P, Bahari-Javan S, Burkhardt S, Sananbenesi F, Fischer A. microRNA-34c is a novel target to treat dementias. EMBO J. 2011;30:4299–4308. doi: 10.1038/emboj.2011.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu S, Wang H, Chen K, Cheng P, Gao S, Liu J, Li X, Sun X. MicroRNA-34c downregulation ameliorates amyloid-beta-induced synaptic failure and memory deficits by targeting VAMP2. J Alzheimers Dis. 2015;48:673–686. doi: 10.3233/JAD-150432. [DOI] [PubMed] [Google Scholar]
- 35.Baker K, Gordon SL, Melland H, Bumbak F, Scott DJ, Jiang TJ, Owen D, Turner BJ, Boyd SG, Rossi M, Al-Raqad M, Elpeleg O, Peck D, Mancini GMS, Wilke M, Zollino M, Marangi G, Weigand H, Borggraefe I, Haack T, Stark Z, Sadedin S Broad Center for Mendelian Genomics. Tan TY, Jiang Y, Gibbs RA, Ellingwood S, Amaral M, Kelley W, Kurian MA, Cousin MA, Raymond FL. SYT1-associated neurodevelopmental disorder: a case series. Brain. 2018;141:2576–2591. doi: 10.1093/brain/awy209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuzuya A, Zoltowska KM, Post KL, Arimon M, Li X, Svirsky S, Maesako M, Muzikansky A, Gautam V, Kovacs D, Hyman BT, Berezovska O. Identification of the novel activity-driven interaction between synaptotagmin 1 and presenilin 1 links calcium, synapse, and amyloid beta. BMC Biol. 2016;14:25. doi: 10.1186/s12915-016-0248-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.