Abstract
Objective: To detect the expression pattern of TRIM11 in CC and to investigate the effect of TRIM11 knockdown on CC progression. Methods: First, the expression pattern and function of TRIM11 in CC were inferred by GEPIA database. The expression of TRIM11 in CC tissues and cells was verified by RT-qPCR and Western blot assays. TRIM11 shRNA was transfected into CC cells. The effect of TRIM11 knockdown on CC cell proliferation and cell apoptosis was assessed by CCK-8 assay, clone formation test and flow cytometry (FCM). Furthermore, the effect of TRIM11 knockdown on cell migration and invasion was measured by Transwell assay. The apoptosis-related proteins (Bax, Bcl-2 and Cleaved caspase 3) and PI3K/AKT pathway-related proteins (PI3K, p-PI3K, AKT and p-Akt) were examined by Western blot assay. Results: TRIM11 was found to be dramatically upregulated in CC tissues and cells. Besides this, TRIM11 was closely related to FIGO stage, infiltration depth and metastasis. Moreover, high expression of TRIM11 was found to be associated with poor prognosis of CC patients. Functional experiments showed that knockdown of TRIM11 obviously suppressed CC cell proliferation, migration and invasion, while accelerated cell apoptosis. In addition, the PI3K inhibitor (LY294002) was used to assess the connection between PI3K/AKT pathway and TRIM11 in CC cells. We found that TRIM11 overexpression suppressed the phosphorylation of PI3K and AKT in CC cells. Conclusion: Hence, TRIM11 regulates CC cell progression by modulating PI3K/AKT pathway. The results suggested that TRIM11 might be a possible target for the diagnosis and prognosis of CC patients.
Keywords: TRIM11, cervical cancer, PI3K/AKT pathway
Introduction
Cervical cancer (CC), the primary malignant tumor of the cervix, is one of the most common malignant tumors in Gynecology. CC ranks second only to breast cancer in morbidity and mortality [1]. The incidence of CC is relatively high in China, and the age of onset has become younger in recent years [2]. At present, scholars generally believe that CC is related to HPV, especially to HR-HPV persistent infection [3,4]. However, in recent years, studies have found that some CC patients have negative HPV screening results [5]. The results indicate that CC lesions are not only caused by virus infection, but the result of multiple factors. It is generally believed in modern oncology that the occurrence and development of tumors, including CC, are closely related to environmental factors and living habits. At present, the apparent causes of CC include history of fertility, smoking, unclean sexual intercourse, premature sexual intercourse, and the history of multiple artificial terminations of pregnancy [6,7]. Notably, searching effective molecular markers for clinical diagnosis and prognosis prediction of CC has become a hot spot for medical researchers.
Tripartite-motif protein (TRIM) family exists in almost all multicellular animals, and plays a major role in the innate immune response as immunoregulatory proteins and E3 ubiquitin ligases [8]. Recent studies have found that TRIM family can participate in various biological processes [9]. TRIM proteins can also participate in the post-transcriptional modification of corresponding proteins of proto-oncogenes and tumor suppressor genes, cell proliferation and apoptosis, and can regulate nuclear receptors in a displacement way [10-12]. TRIM11 is a member of the TRIM protein family, and its expression level varies in different tissues and types of cells in the human body. Recent studies have revealed that TRIM11 is a novel HIV-1 capsid binding protein, and TRIM11 overexpression facilitated HIV-1 uncoating [13]. Moreover, TRIM11 was found to facilitate cell proliferation and cell invasion in hepatocellular carcinoma, and is closely associated with poor prognosis of patients with hepatocellular carcinoma [14,15]. Besides, TRIM11 facilitates lung cancer cell multiplication and metastasis, suggesting that TRIM11 plays an oncogenic role in lung cancer [16]. Strikingly, TRIM11 was found to interfere with different stages of MLV or HIV-1 replication. However, the role of TRIM11 in CC is not clear.
In this study, we detected the expression pattern of TRIM11 in CC. The effect of TRIM11 knockdown in CC progression and its diagnostic and prognostic value on CC was also investigated.
Materials and methods
CC patients and clinical tissues samples
The specimens were collected from 52 cervical cancer patients who were surgically treated at the Jinan City People’s Hospital. Fifty-two cases of normal cervical tissues were obtained at the same period of total hysterectomy for uterine fibroids. The age of patients was ranged from 26 to 76 years old, with a mean age of 46 years. All CC patients were home visits and had not received chemotherapy, radiotherapy or other treatment before surgery. All patients were independently diagnosed by at least two pathologists. Our experiments were approved by the ethics committee of the Jinan City People’s Hospital (NO: 2017-Gy0031). And all patients provided the written informed consent.
Cell culture
CC cell lines Hela, Siha, C4-1, C-33A, and human cervical endometrial epithelial cells End1/E6E7 were obtained from Shanghai JingKang Bioengineering CO., LTD., (Shanghai, China). All cells were cultured with DMEM medium (Hyclone, USA) with 10% FBS (Gibco, USA), 100 U/ml penicillin and 100 μg/ml streptomycin (Beyotime Institute of Biotechnology, China), and conventionally cultured in a 5% CO2 incubator at 37°C. The cells were digested with trypsin after reaching the logarithmic growth phase.
Cell transfection
After being cultured with 5% CO2 at 37°C for 24 h, TRIM11 shRNA was transfected into Siha and Hela cells according to the instructions of Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). After transfection for 48 h, the transfection efficiency was detected by RT-qPCR assay. TRIM11 shRNA (5’-GGAGAAGTCACTGGAGCAT-3’) was obtained from Shanghai Jima Pharmaceutical Technology Co., Ltd., (Shanghai, China).
RT-qPCR assay
Total RAN was extracted from CC tissues and cells by the TRIzol (Invitrogen, USA) method. Subsequently, the concentration and purity of RNA were explored by NanoDrop. Next, cDNA was prepared by reverse transcription. 2 μL reverse transcriptions were taken for PCR detection. 20 μL PCR reaction system was established according to the instructions (2 μL reverse transcriptions, 10 μL SYBR Green Mix (Thermo Fisher, USA), 0.5 μL upstream primer, and 0.5 μL downstream primer). RT-PCR was carried out by ABI 7300 Real-Time PCR system. The reaction program was 95°C for 10 min followed by 40 cycles of 95°C for 15 s, 60°C for 45 s. GAPDH was used as internal reference of TRIM1. 2-ΔΔCq method was used to analyze the mRNA expression of TRIM11. The primers were as followed: TRIM11, forward: 5’- 5’-CGAAGACCTCAAGGCGAAGC-3’, reverse: 5’-CAGCAAACGGCGAAGACG-3’; GAPDH, forward: 5’-AATCCCATCACCATCTTC-3’, reverse: 5’-AGGCTGTTGTCATACTTC-3’.
Western blot assay
Western blot analysis was performed to detect the protein expressions of TRIM11, cell apoptosis-related proteins (Bax, Bcl-2 and caspase 3) and PI3K/AKT pathway-related proteins (PI3K, p-PI3K, AKT and p-AKT). The electrophoresis voltage was controlled in the range of 80~120 V and lasted for 45~70 min. After membrane transferring, the proteins were sealed in 5% bovine serum protein for 1 h. Then added with the primary antibodies (TRIM11, 1:1500, cwbiotech, China; Bax, 1:1500, Beyotime Institute of Biotechnology, China; Bcl-2, 1:1500, Beyotime Institute of Biotechnology, China; Cleaved caspase 3, 1:1500, Beyotime Institute of Biotechnology, China; β-actin, 1:1500, SantaCruz, USA; GAPDH, 1:3000, SantaCruz, USA), and cultured at 4°C overnight. The proteins were rinsed with TBST buffer, then added with the goat anti-rabbit secondary antibody (1:500, cwbiotech, China) for 1 h. Then chemiluminescence reagent was added to develop the proteins. Bio-rad Gel DolEZ imaging device was used to image the proteins, and Image J software was used to analyze the gray level of the target bands.
Immunohistochemical (IHC) staining
Paraffin sections were baked at 60°C for 2 h, and then recovered in sodium citrate buffer. Then, 3% H2O2 was added to quench the activity of endogenous peroxidase. After sealed with PBS solution with 10% BSA for 60 min, the sections were incubated with primary antibody at 4°C overnight. After washing three times with PBS, the sections were incubated with secondary antibody. The staining was performed using DA-Chromogen (Dako, Glostrup, Denmark) complexes according to the manufacturer’s instructions. The nucleus was then counterstained with hematoxylin. Finally, the images were analyzed by the Image J software.
CCK-8 assay
Siha and Hela cells in the logarithmic growth phase were cultured in 96-well plates (1×104 cells/well). After incubation for 24, 48, 72 and 96 h, 100 μL serum-free medium and 10 μL CCK-8 solution (Dojindo, Japan) were added. After being incubated for 1-4 h, the absorbance value was measured at 450 nm by the enzyme plate analyzer.
Clone formation test
The transfected cells were digested and prepared for cell suspension. Cell suspension was injected into a 6-well plate (200 cells/well). The inoculated cells were put into the incubator for further culture, and the medium was changed every three days. The formation of clone cells was stopped when the number of most single clone cells was greater than 50. 1 mL paraformaldehyde (ZhongShan GoldBridge Biotechnology Co., Ltd., China) was added to each well for 30 min to fix the cells. Then, 500 μL Gimassa solutions were added to each well for 20 min. After carefully absorbing the Gimassa dye, the fixed clones were washed and soaked repeatedly until the background was clean. The 6-well plate was dried and photographed under a microscope.
Transwell assay
Transwell assay was used to measure CC cell migration and invasion. The cells transfected with TRIM1 shRNA were assigned as the experimental group, and untransfected cells as the control cells. After digested with trypsin, cells were put in a 24-well plate with Transwell chamber. 100 μl cell suspension (2×105 cells/ml) was added to the upper chamber, 250 μL DMEM medium with 10% FBS was added to the lower chamber. In contrast to the migration experiment, the upper Transwell chamber was coated with Matrigel (Corning, USA). After incubated at 37°C and 5% CO2 for 48 h, the cells in the lower chamber were fixed with 4% paraformaldehyde (ZhongShan GoldBridge Biotechnology Co., Ltd., China) for 15 min, and stained with 1% crystal violet (Beijing Solarbio Science & Technology Co., Ltd., China) for 15 min. The chamber was rinsed with PBS (Beijing Solarbio Science & Technology Co., Ltd., China), and observed under a microscope.
Flow cytometery (FCM) analysis
Cell apoptosis of Hela and Siha cells was assessed by FCM. The cells (1×106 cells/ml) were suspended by 0.3 ml PBS containing 10% FBS. Then, the cells were added with 0.7 mL anhydrous ethanol to fix at -20°C for more than 24 h. Next, the cells were centrifuged at 3000 r/min for 30 s, discard the supernatant, resuspended the cells with 1 mL PBS. After discarding the supernatant, the precipitated cells were suspended with 100 μl RNase A (1 mg/mL) at 37°C for 30 min. After adding Annexin V-FITC and 400 μL (50 μg/mL) PI (Thermo Fisher Scientific, USA), the cells were cultured in the dark for 10 min. Finally, Flow cytometer (BD, USA) was used to detect the cell apoptosis.
Statistical analysis
Data were analyzed by SPSS19.0 software. The values were presented as mean ± SEM. The differences between the two groups were compared by Student’s t test. Tukey’s post hoc test was used for comparisons between multiple groups. Kaplan Meier survival analysis was used to analyze the relationship between TRIM11 levels and clinicopathological features. Cox proportional risk models were used for univariate and multivariate analyses. P<0.05 was considered as statistically significant. All the experiments were repeated three times.
Results
TRIM11 was obviously upregulated in CC samples
GEPIA database revealed that TRIM11 was dramatically upregulated in CC tissues (n = 306) compared to the non-tumor tissues (Figure 1A). In our study, TRIM11 showed the same upregulation trend in 52 CC patients (Figure 1B). Next, IHC straining was utilized to confirm the expression level of TRIM11 in CC tissues. We noticed that the intensity of TRIM11 staining in CC tissues was obviously increased compared to that in non-tumor tissues (Figure 1C). Strikingly, GEPIA database displayed that CC patients with high TRIM11 expression had worse overall survival than the patients with low TRIM11 expression (Figure 1D). Similarly to previous data, we found that CC patient with high TRIM11 expression had shorter overall survival time (Figure 1E). Furthermore, Kaplan Meier survival analysis was used to detect the relationship between TRIM11 levels and clinicopathological features of 52 CC patients. The results revealed that TRIM11 expression was closely correlated with FIGO stage, infiltration depth and metastasis (Table 1). Next, Cox proportional risk results verified that TRIM11 expression was independent factor with poor OS in patients with CC (Table 2). All data demonstrated that TRIM11 is involved in the progression of CC, and closely associated with poor prognosis of CC patients.
Figure 1.
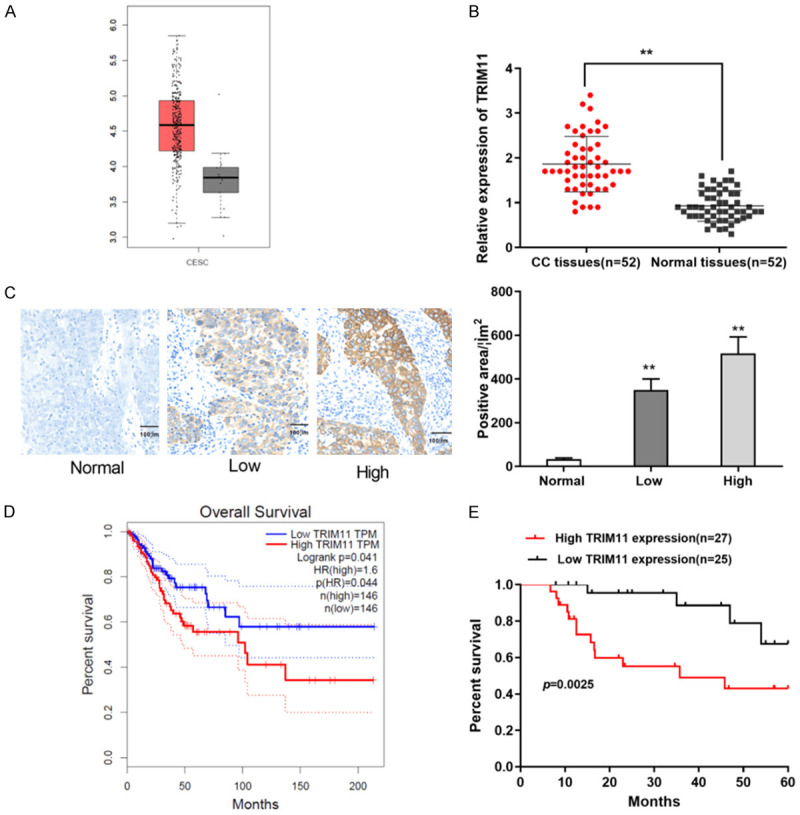
TRIM11 was obviously upregulated in CC samples. A: GEPIA database showed that TRIM11 was significantly upregulated in CC tissues (n = 306) compared with non-tumor tissues (n = 13); B: TRIM11 was obviously upregulated in CC tissues (n = 52) compared with non-tumor tissues (n = 52); C: IHC straining showed that the intensity of TRIM11 staining was significantly increased in CC tissues (n = 52) (scale bar = 100 μm, 100×); D: GEPIA database displayed that patients with high TRIM11 expression (n = 146) had worse overall survival than the patients with low TRIM11 expression (n = 146); E: Patients with high TRIM11 expression (n = 27) had worse overall survival than the patients with low TRIM11 expression (n = 25). **P<0.01. CC: cervical cancer.
Table 1.
Association between TRIM11 expression and clinicopathological characteristics of patients with CC
| n = 52 | High expression (27) | Low expression (25) | х2 value | P value | |
|---|---|---|---|---|---|
| Age (n) | 0.624 | 0.430 | |||
| <45 years | 20 | 9 | 11 | ||
| ≥ea years | 32 | 18 | 14 | ||
| Tumor size (n) | 1.997 | 0.158 | |||
| <4 cm | 24 | 15 | 9 | ||
| ≥5 cm | 28 | 12 | 16 | ||
| FIGO stage (n) | 6.429 | 0.011 | |||
| Ib1-Ib2 | 34 | 22 | 12 | ||
| IIa1-IIb | 18 | 5 | 13 | ||
| Infiltration depth (n) | 9.282 | 0.002 | |||
| <1/2 | 22 | 6 | 16 | ||
| ≥6/2 | 30 | 21 | 9 | ||
| Metastasis (n) | 4.854 | 0.028 | |||
| Present | 29 | 19 | 10 | ||
| Absent | 23 | 8 | 15 |
Note: CC: cervical cancer.
Table 2.
Univariate and multivariate Cox regression models for estimating the overall survival
| Variable | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|
|
|
|
|||
| HR (95% CI) | P-value | HR (95% CI) | P-value | |
| Age (n) | 0.628 | 0.284 | ||
| <45 years | 1.225 (0.539-2.782) | reference | ||
| ≥ef years | 1.630 (0.666-3.986) | |||
| Tumor size (n) | 0.254 | 0.088 | ||
| <4 cm | 1.345 (0.426-3.242) | reference | ||
| ≥e cm | 2.321 (0.882-6.107) | |||
| FIGO stage (n) | 0.374 | 0.055 | ||
| Ib1-Ib2 | 0.659 (0.263-1.654) | reference | ||
| IIa1-IIb | 0.355 (0.123-1.021) | |||
| Infiltration depth (n) | 0.128 | 0.043 | ||
| <1/2 | 1.847 (0.838-4.070) | reference | ||
| ≥efe | 2.388 (1.028-5.543) | |||
| Metastasis(n) | 0.034 | 0.028 | ||
| Present | 2.362 (1.067-5.230) | reference | ||
| Absent | 2.852 (1.122-7.246) | |||
| TRIM11 (n) | 0.011 | 0.004 | ||
| High | 3.032 (1.290-7.122) | reference | ||
| Low | 3.103 (1.551-7.853) | |||
Note: HR: hazard ratio; CI: confidence interval.
Knockdown of TRIM11 blocked cell proliferation of CC cells
Western blot assay was utilized to measure the protein expression of TRIM11 in CC cells. As shown in Figure 2A, TRIM11 was remarkably upregulated in Hela, Siha, C4-1 and C-33A cells compared with End1/E6E7 cells. Notably, the expressions of TRIM11 in Hela and Siha cells was higher than that in other cell lines, so we selected Siha and Hela cells for the subsequent experiments. To explore the effect of TRIM11 on the progression of CC, TRIM11 sh-RNA was transfected into Siha and Hela cells. We found that the expression of TRIM11 was obviously reduced by TRIM11 sh-RNA transfection (Figure 2B). Cell proliferative potential was detected by CCK-8 assay and Clone formation test. Distinctly, the cell proliferation of Siha and Hela cells was dramatically inhibited by TRIM11 knockdown (Figure 2C, 2D). As expected, knockdown of TRIM11 lessened the number of colonies of Siha and Hela cells (Figure 2E). Our results indicated that knockdown of TRIM11 blocked cell proliferation in CC cells.
Figure 2.
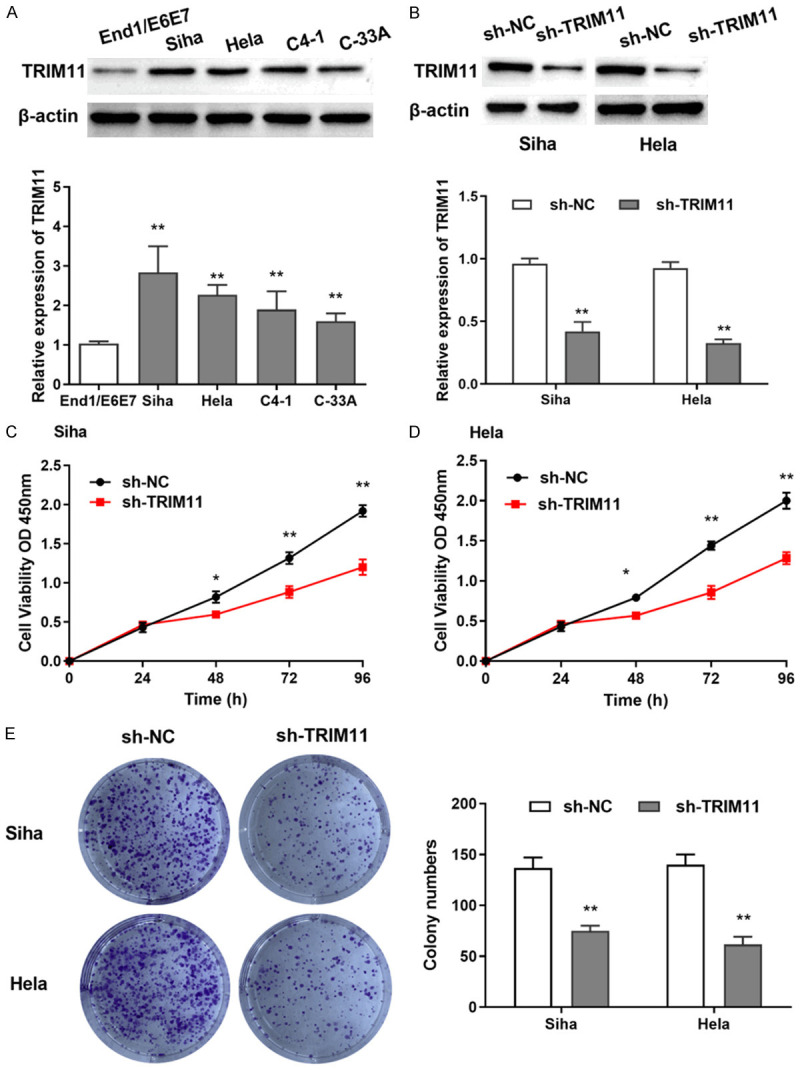
Knockdown of TRIM11 blocked cell proliferation of CC cells. A: The expression of TRIM11 was significantly higher in Siha, Hela, C4-1 and C-33A cells than in End1/E6E7 cells; B: The expression of TRIM11 was significantly reduced by TRIM11 shRNA; C: TRIM11 knockdown inhibited cell proliferation of Siha cells; D: Cell proliferation of Hela cells was inhibited by TRIM11 knockdown; E: TRIM11 knockdown inhibited the colony formation in Siha and Hela cells. *P<0.05, **P<0.01.
Knockdown of TRIM11 suppressed cell migration and invasion of CC cells
Transwell assay was used to detect cell migration and invasion in Siha and Hela cells. Our results displayed that knockdown of TRIM11 reduced the number of migrated cells of Siha and Hela cells (Figure 3A). In the same way, the number of invaded cells in Siha and Hela cells was dramatically reduced by TRIM11 knockdown (Figure 3B). All data displayed that TRIM11 knockdown suppressed cell migration and invasion in CC cells.
Figure 3.
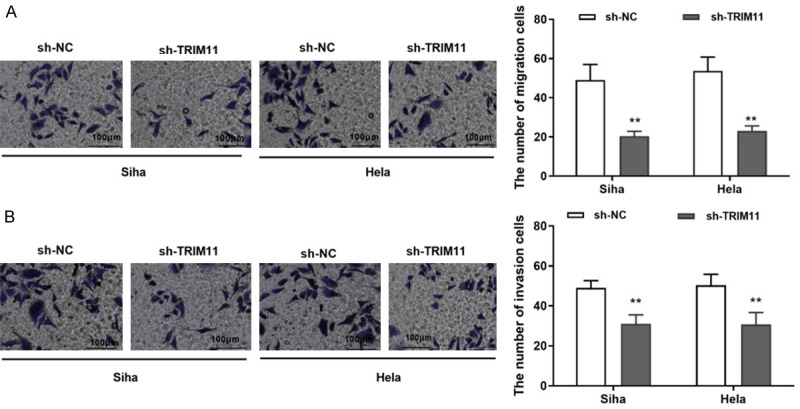
Knockdown of TRIM11 suppressed cell migration and invasion of CC cells. A: TRIM11 knockdown suppressed cell migration in Siha and Hela cells (scale bar = 100 μm, 100×); B: TRIM11 knockdown suppressed cell invasion in Siha and Hela cells (scale bar = 100 μm, 100×). **P<0.01. CC: cervical cancer.
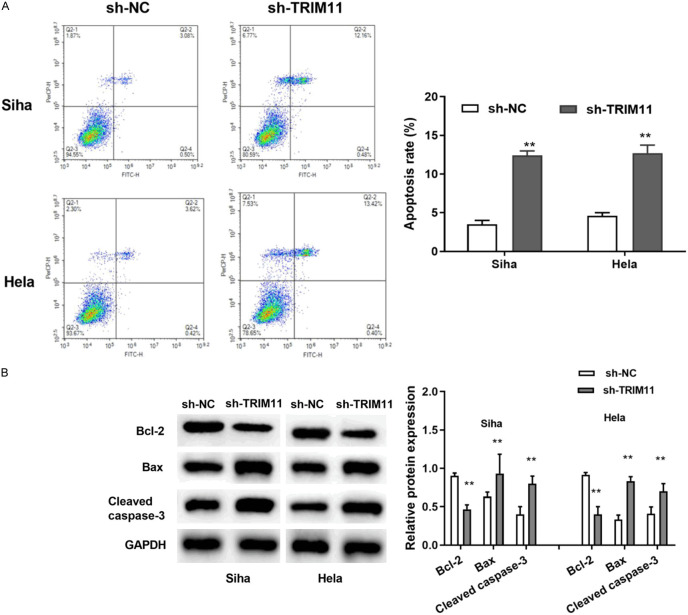
Knockdown of TRIM11 accelerated cell apoptosis of CC cells
As shown in Figure 4A, cell apoptosis of Siha cells was obviously increased by TRIM11 knockdown. Similarly, TRIM11 knockdown induced cell apoptosis in Hela cells (Figure 4A). Next, the cell apoptosis-related proteins (Bax, Bcl-2 and Cleaved caspase 3) were detected by Western blot assay. The results indicated that TRIM11 knockdown obviously ascended the expression of Bax and Cleaved caspase 3, while decreased the expression of Bcl-2 (Figure 4B). These results indicated that TRIM11 knockdown could promote cell apoptosis of CC cells.
Figure 4.
Knockdown of TRIM11 induced cell apoptosis of CC cells. A: TRIM11 knockdown promoted cell apoptosis in Siha and Hela cells; B: TRIM11 silencing increased the expression of Bax and Cleaved caspase 3, while reduced the expression of Bcl-2. **P<0.01. CC: cervical cancer.
TRIM11 regulated CC cell progression by modulating PI3K/AKT pathway
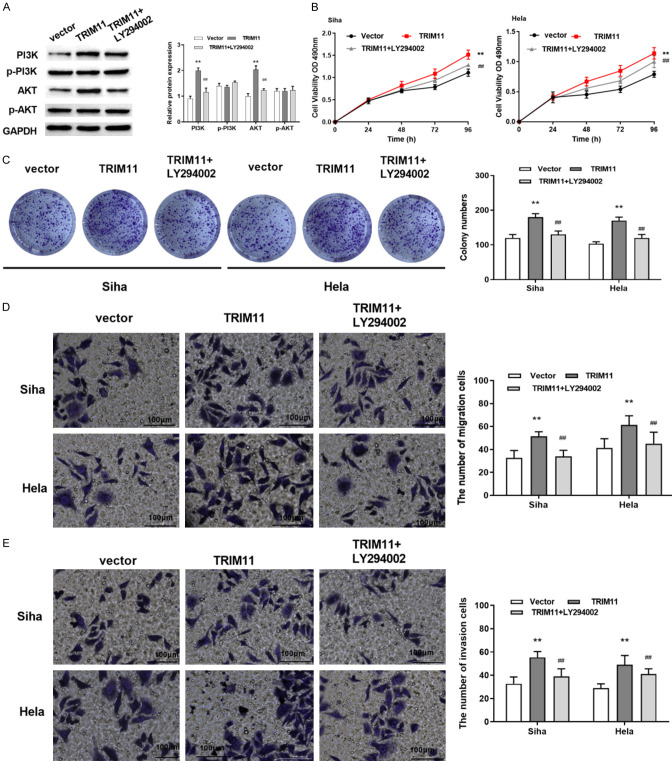
To explore the effect of TRIM11 on CC progression, TRIM11 overexpression was transfected into Hela cells. Western blot assay was performed to assess the expressions of PI3K/AKT pathway related proteins. TRIM11 overexpression was found to suppress the phosphorylation of PI3K and AKT in CC cells (Figure 5A). Then, PI3K pathway inhibitor (LY294002) was added to Hela and Siha cells with TRIM11 overexpression. We found that TRIM11 promoted cell proliferation, while LY294002 reversed the effect of TRIM11 (Figure 5B, 5C). Similarly, LY294002 suppressed the facilitation of TRIM11 overexpression on cell migration and invasion (Figure 5D, 5E). Our findings verified that TRIM11 regulated CC cell progression by modulating PI3K/AKT pathway.
Figure 5.
TRIM11 regulated CC cell progression by modulating PI3K/AKT pathway. A: TRIM11 overexpression suppressed the phosphorylation of PI3K and AKT in CC cells; B, C: TRIM11 overexpression promoted cell proliferation, while LY294002 reversed the effect of TRIM11 in Siha and Hela cells; D, E: TRIM11 overexpression promoted cell migration and invasion, while LY294002 suppressed the facilitation of TRIM11 in Siha and Hela cells (scale bar = 100 μm, 100×). Compared with the vector group, **P<0.01; compared with the TRIM11 group, ##P<0.01. CC: cervical cancer.
Discussion
It has been reported that the TRIM protein family can participate in many important biological processes, such as cell differentiation, cell cycle, cell apoptosis, and influence virus infection [17]. TRIM59 was found to act as a carcinogen in bladder cancer cells by regulating TGF-β/Smad2/3 signaling pathway [18]. Aierken et al. reported that TRIM59 was overexpressed in CC tissues, and knockdown of TRIM59 blocked cell growth, invasion and migration of CC cells [19]. Moreover, TRIM14 was reported to be overexpressed in CC tissues and cells, and TRIM14 overexpression facilitated cell growth and suppressed cell apoptosis by activating the AKT pathway [20]. Nevertheless, TRIM3 was found to inhibit cell proliferation by inactivating the p38 pathway in CC, suggesting that TRIM3 acted as a tumor suppressor in CC [21]. In recent years, the role of TRIM11 in the progression of several tumors has been increasingly recognized, but its molecular mechanism in CC remains unknown. In this study, we highlighted the clinical significance and role of TRIM11 in the evolution of CC carcinogenesis.
In the current study, we showed that TRIM11 was obviously elevated in CC tissues and cells compared with the non-tumor group. Moreover, the patients with high TRIM11 expression had a shorter survival time than patients with low TRIM11 expression. In parallel with our results, TRIM11 was revealed to be overexpressed in breast cancer, and was associated with the poor prognosis of patients with breast cancer [22,23]. Furthermore, TRIM11 was found to be upregulated in lung adenocarcinoma, chordoma and gastric cancer [24-26]. Besides that, TRIM11 was confirmed to be closely correlated with FIGO stage, infiltration depth and metastasis. Hence, our results suggested that TRIM11 might be involved in CC progression.
Functionally, TRIM11 knockdown was found to block cell proliferation, migration and invasion, while promoted cell apoptosis in CC cells. Our findings indicated that TRIM11 might be a carcinogen in the progression of CC. Consistent with our results, TRIM11 knockdown was reported to restrain cell growth and EMT progression in hepatocellular carcinoma cells [15]. Moreover, Chen et al. found that TRIM11 knockdown restrained cell proliferation and cell invasion, but facilitated cell apoptosis in ovarian cancer cells [27].
Studies have shown that TRIM11 may participate in multiple signaling pathways in cancer progression. For example, TRIM11 facilitates cell progression in lymphomas by inducing the β-catenin signaling [28]. Moreover, Huang et al. reported that TRIM11 accelerated tumor angiogenesis via promoting the STAT3/VEGFA pathway in lung adenocarcinoma [24]. PI3K/AKT pathway is reported to be closely related to the occurrence and development of human tumors. Abnormal activity of PI3K/AKT pathway can not only regulate cell proliferation and survival of tumor cells, but also participate in cell migration, invasion, adhesion and angiogenesis of tumor cells. In our study, we explored the effect of PI3K inhibitor (LY294002) on TRIM11 in CC progression. Our results showed that TRIM11 knockdown restrained CC cell progression by inactivating PI3K/AKT pathway. In line with our findings, knockdown of TRIM11 was reported to block cell progression by inhibiting PI3K/AKT pathway in lung cancer [16] and hepatocellular carcinoma [15].
In summary, our results demonstrated that knockdown of TRIM11 suppressed CC cell multiplication, migration and invasion through the PI3K/AKT pathway. Our finding suggested that TRIM11 may be a possible target marker and therapeutic target for the diagnosis and prognosis of patients with CC. However, the specific mechanism of TRIM11 and its effect on tumor formation in vitro still need to be further studied.
Disclosure of conflict of interest
None.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Wei M, Zhou W, Bi Y, Wang H, Liu Y, Zhang ZJ. Rising mortality rate of cervical cancer in younger women in urban China. J Gen Intern Med. 2019;34:281–284. doi: 10.1007/s11606-018-4732-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen X, Wei S, Ma H, Jin G, Hu Z, Suping H, Li D, Hang D, Wu X, Li N. Telomere length in cervical exfoliated cells, interaction with HPV genotype, and cervical cancer occurrence among high-risk HPV-positive women. Cancer Med. 2019;8:4845–4851. doi: 10.1002/cam4.2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Torres-Poveda K, Ruiz-Fraga I, Madrid-Marina V, Chavez M, Richardson V. High risk HPV infection prevalence and associated cofactors: a population-based study in female ISSSTE beneficiaries attending the HPV screening and early detection of cervical cancer program. BMC Cancer. 2019;19:1205. doi: 10.1186/s12885-019-6388-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yin XH, Wang ZQ, Yang SZ, Jia HY, Shi M. Clinical observation of laparoscopic radical hysterectomy for cervical cancer. Int J Clin Exp Med. 2014;7:1373–1377. [PMC free article] [PubMed] [Google Scholar]
- 6.Clarke MA, Fetterman B, Cheung LC, Wentzensen N, Gage JC, Katki HA, Befano B, Demarco M, Schussler J, Kinney WK, Raine-Bennett TR, Lorey TS, Poitras NE, Castle PE, Schiffman M. Epidemiologic evidence that excess body weight increases risk of cervical cancer by decreased detection of precancer. J. Clin. Oncol. 2018;36:1184–1191. doi: 10.1200/JCO.2017.75.3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hindin P, Btoush R, Carmody DP. History of childhood abuse and risk for cervical cancer among women in low-income areas. J Womens Health (Larchmt) 2019;28:23–29. doi: 10.1089/jwh.2018.6926. [DOI] [PubMed] [Google Scholar]
- 8.Ikeda K, Inoue S. TRIM proteins as RING finger E3 ubiquitin ligases. Adv Exp Med Biol. 2012;770:27–37. doi: 10.1007/978-1-4614-5398-7_3. [DOI] [PubMed] [Google Scholar]
- 9.Kato K, Ahmad S, Zhu Z, Young JM, Mu X, Park S, Malik HS, Hur S. Structural analysis of RIG-I-like receptors reveals ancient rules of engagement between diverse RNA helicases and TRIM ubiquitin ligases. Mol Cell. 2021;81:599–613. e8. doi: 10.1016/j.molcel.2020.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jaworska AM, Wlodarczyk NA, Mackiewicz A, Czerwinska P. The role of TRIM family proteins in the regulation of cancer stem cell self-renewal. Stem Cells. 2020;38:165–173. doi: 10.1002/stem.3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eberhardt W, Haeussler K, Nasrullah U, Pfeilschifter J. Multifaceted roles of TRIM proteins in colorectal carcinoma. Int J Mol Sci. 2020;21:7532. doi: 10.3390/ijms21207532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li YJ, Zhang GP, Zhao F, Li RQ, Liu SJ, Zhao ZR, Wang X. Target therapy of TRIM-14 inhibits osteosarcoma aggressiveness through the nuclear factor-κB signaling pathway. Exp Ther Med. 2018;15:2365–2373. doi: 10.3892/etm.2017.5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yuan T, Yao W, Tokunaga K, Yang R, Sun B. An HIV-1 capsid binding protein TRIM11 accelerates viral uncoating. Retrovirology. 2016;13:72. doi: 10.1186/s12977-016-0306-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Y, Li L, Qian X, Ge Y, Xu G. High expression of TRIM11 correlates with poor prognosis in patients with hepatocellular carcinoma. Clin Res Hepatol Gastroenterol. 2017;41:190–196. doi: 10.1016/j.clinre.2016.09.010. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Z, Xu C, Zhang X, Huang L, Zheng C, Chen H, Wang Y, Ju H, Yao Q. TRIM11 upregulation contributes to proliferation, invasion, and EMT of hepatocellular carcinoma cells. Oncol Res. 2017;25:691–699. doi: 10.3727/096504016X14774897404770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang X, Shi W, Shi H, Lu S, Wang K, Sun C, He J, Jin W, Lv X, Zou H, Shu Y. TRIM11 overexpression promotes proliferation, migration and invasion of lung cancer cells. J Exp Clin Cancer Res. 2016;35:100. doi: 10.1186/s13046-016-0379-y. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Watanabe M, Hatakeyama S. TRIM proteins and diseases. J Biochem. 2017;161:135–144. doi: 10.1093/jb/mvw087. [DOI] [PubMed] [Google Scholar]
- 18.Chen W, Zhao K, Miao C, Xu A, Zhang J, Zhu J, Su S, Wang Z. Silencing Trim59 inhibits invasion/migration and epithelial-to-mesenchymal transition via TGF-β/Smad2/3 signaling pathway in bladder cancer cells. Onco Targets Ther. 2017;10:1503–1512. doi: 10.2147/OTT.S130139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aierken G, Seyiti A, Alifu M, Kuerban G. Knockdown of tripartite-59 (TRIM59) inhibits cellular proliferation and migration in human cervical cancer cells. Oncol Res. 2017;25:381–388. doi: 10.3727/096504016X14741511303522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Diao W, Zhu C, Guo Q, Cao Y, Song Y, Feng H, Li J, Xue X, Lu P. Tripartite motif-containing 14 regulates cell proliferation and apoptosis in cervical cancer via the Akt signaling pathway. Mol Med Rep. 2020;22:5145–5154. doi: 10.3892/mmr.2020.11634. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Song Y, Guo Q, Gao S, Hua K. Tripartite motif-containing protein 3 plays a role of tumor inhibitor in cervical cancer. Biochem Biophys Res Commun. 2018;498:686–692. doi: 10.1016/j.bbrc.2018.03.046. [DOI] [PubMed] [Google Scholar]
- 22.Tang J, Luo Y, Tian Z, Liao X, Cui Q, Yang Q, Wu G. TRIM11 promotes breast cancer cell proliferation by stabilizing estrogen receptor α. Neoplasia. 2020;22:343–351. doi: 10.1016/j.neo.2020.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dai X, Geng F, Li M, Liu M. Tripartite motif-containing 11 regulates the proliferation and apoptosis of breast cancer cells. Oncol Rep. 2019;41:2567–2574. doi: 10.3892/or.2019.7015. [DOI] [PubMed] [Google Scholar]
- 24.Huang J, Tang L, Zhao Y, Ding W. TRIM11 promotes tumor angiogenesis via activation of STAT3/VEGFA signaling in lung adenocarcinoma. Am J Cancer Res. 2019;9:2019–2027. [PMC free article] [PubMed] [Google Scholar]
- 25.Wang B, Wang G, Wang Q, Zhu Z, Wang Y, Chen K, Yang H. Silencing of TRIM11 suppresses the tumorigenicity of chordoma cells through improving the activity of PHLPP1/AKT. Cancer Cell Int. 2019;19:284. doi: 10.1186/s12935-019-1007-7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Luo N, Wang Z. TRIM11 stimulates the proliferation of gastric cancer through targeting CPEB3/EGFR axis. J BUON. 2020;25:2097–2104. [PubMed] [Google Scholar]
- 27.Chen Y, Sun J, Ma J. Proliferation and invasion of ovarian cancer cells are suppressed by knockdown of TRIM11. Oncol Lett. 2017;14:2125–2130. doi: 10.3892/ol.2017.6432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hou Y, Ding M, Wang C, Yang X, Ye T, Yu H. TRIM11 promotes lymphomas by activating the β-catenin signaling and Axin1 ubiquitination degradation. Exp Cell Res. 2020;387:111750. doi: 10.1016/j.yexcr.2019.111750. [DOI] [PubMed] [Google Scholar]