Abstract
Cancer-associated fibroblasts (CAFs) serve as a predominant regulator in the tumor microenvironment. However, the crosstalk between CAFs and OS cells remains mostly unclear. Recent studies explored that long non-coding RNA (LncRNAs) involved in regulating osteosarcoma (OS) formation and development, but their functions in CAFs are unknown. Here, we first investigated the SNHG17 was upregulated in OS tissues and correlated with the poor prognosis through the integrating clinical data. We then evaluated the function of SNHG17 in vitro using the stable SNHG17-depleted OS cells. HOS cells with SNHG17 knocked down were performed to generate the OS xenograft model. Through immunohistochemistry assay and TUNEL apoptosis assay, the role of SNHG17 on OS development was assessed in vivo. We then examined the SNHG17 expression in exosomes derived from CAFs, normal fibroblasts (NFs), and tumor tissues from the OS clinical samples. The interaction among SNHG17, miR-2861, and MMP2 was predicted by bioinformatics analysis and identified by RIP and luciferase assays. The cell proliferation, migration, and apoptosis of SJSA-1 and HOS cells co-cultured with CAFs-derived exosomes were assessed by CCK-8 and colony formation assays. We found that SNHG17 was upregulated in the tumor tissues and presented a pro-tumorigenic effect on OS both in vitro and in vivo. It also was an essential exosomal cargo of CAFs and could affect OS cell proliferation and migration in vitro. CAFs-released exosomal SNHG17 acted as an essential molecular sponge for miR-2861 in OS cells. Moreover, MMP2 was a direct target of miR-2861 and was regulated by SNHG17. Overall, our findings identified that SNHG17 was an essential exosomal cargo of OS-related CAFs that contributes to proliferation and metastasis of OS, supporting the therapeutic potency of targeting the crosstalk between cancer cells and CAFs.
Keywords: Cancer-associated fibroblasts, osteosarcoma, LncRNA SNHG17, exosomes
Introduction
Osteosarcoma (OS) is a kind of malignant bone tumor that usually occurs in children and young people [1,2]. It presented severe local pain and swelling, and exhibits a predilection to aggressive and metastasize to pulmonary at early stage [3]. The introduction of several therapeutic strategies, including chemotherapy, surgery, and radiotherapy, achieving a great improvement for patients who develop localized OS [4]. However, the prognosis is still dismal for patients with metastasis and relapse disease [5]. Besides, the acquisition of resistance to such therapeutic options remains the main clinical problem for OS, which might cause treatment failure, recurrence, and even death [6,7]. Therefore, it is of considerable significance to further understand the underlying mechanism of OS progression and to identify a novel approach for OS diagnosis and prognosis.
Emerging evidence reported that cancer-associated fibroblasts (CAFs) serve as a predominant regulator for OS formation and drug response [8-10]. The communication between CAFs and tumor cells has been revealed to rely on the release of some mediators, including exosomes. Currently, interest in the molecular crosstalk between cancer cells and CAFs has been a renaissance. Exosomes are a type of cystic vehicle with a 40-150 nm diameter, which emerged as a key mediator for multiple diseases [11,12]. By transferring molecules including microRNAs (miRNAs), long non-coding RNAs (LncRNAs) and proteins, exosomes involve in the cell-cell communication among different cells [13]. The link between CAFs-derived exosomes and OS was supported by Jiang et al., they showed that CAFs could transfer exosomes into OS cells to promote cell proliferation and metastasis in vitro. Based on the data of sequence, the enriched miR-1228 in exosomes were required for achieving these functions [8]. Unfortunately, the role of exosomal LncRNAs in OS proliferation and metastasis remains unknown.
LncRNAs are a class of non-coding RNAs with more than 200 nucleotides. Recent studies have pointed out deregulation of LncRNAs plays an essential role in the progression of several cancer types. For example, Li et al. revealed that CAFs-derived exosomal LncRNA SNHG3 controlled the metabolic process in breast cancer cells and exerted an oncogenic function [14]. Besides, increased LncRNA H19 in CAFs-derived exosomes had also been reported to regulate the stemness of cancer stem cells and to enhance the chemoresistance of colon cancer cells [15]. Of interest, LncRNA small nucleolar RNA Host Gene 17(SNHG17), a critical lncRNA located on 20q11.23 with 1186 bp length, is markedly correlated with the proliferation, invasion, and metastasis of several cancer types, such as non-small cell lung cancer, colorectal cancer, prostate cancer and prostate cancer [16-19]. Nevertheless, the biological effect of SNHG17 in the progression of OS remains elusive.
In this study, we aimed to determine the expression pattern of SNHG17 in osteosarcoma CAFs-derived exosomes and investigate the underlying mechanism of its function. We found that SNHG17 was enriched in the CAFs-derived exosomes and promoted OS proliferation and metastasis. Moreover, we discovered that SNHG17 functions as a competing endogenous RNA (ceRNA) to control the expression of MMP2 through binding with miR-2861. Our finding confirmed that SNHG17 served as an oncogenic regulator for OS development and is a possible biomarker for OS diagnosis and prognosis.
Methods
Clinical samples and fibroblasts isolation
Five pairs of primary OS adjacent tumor-free tissues and tumor tissues were collected from the Second Affiliated Hospital of Inner Mongolia Medical University. After washed by PBS supplemented with 20% antibiotics (Thermo, USA), the normal fibroblasts (NFs) and cancer related fibroblasts (CAFs) were isolated according to the previous study [20]. In brief, the tissues were digested by collagenase type II (1 mg/mL, Thermo, USA) for 2 h at 37°C. The cells were then plated on the culture flask for 30 min to remove the non-adherent cells according to the different adherent ability. The non-adherent cells, which were considered as the fibroblasts, were collected and maintained with Dulbecco’s modified Eagle’s medium/F12 (DMEM/F12, Gibco, USA) supplemented with 10% fetal bovine serum (FBS, Gibco, USA). The CAFs were isolated from the tumor tissue. The human study was approved by the ethical committee of the Second Affiliated Hospital of Inner Mongolia Medical University (No. Y K D2017142).
Conditioned medium collection
CAFs and NFs were cultured in the 10 cm dish for 48 h until 80% confluence. The culture medium was changed by DMEM/F12 with 10% exosome-depleted FBS (Thermo, USA). After 48 h, the medium was collected, followed by centrifugation for 30 min at 300×g at 4°C. The supernatant was collected and be used to treat the OS cells for 4 days. For one group, in order to inhibit exosome generation, CAFs (106 cells per well in the 6-well plate) were treated by 15 µM GW4869 (Sigma, USA) for 48 h.
Cell culture and transfection
The HBO, HOS, SJSA-1, MG63, and HEK293T cell lines were purchased from the American Type Culture Collection (USA) and maintained in Dulbecco’s modified Eagle’s medium (Gibco, USA) supplemented with 10% fetal bovine serum (Gibco, USA).
For cell transfections, the HOS and MG63 cells were plated in the 6-well plate at the density of 106 cells per well. The recombinant lentiviruses expressing the sh-SNHG17#1, sh-SNHG17#2, and sh-NC were obtained from Sangon Biotech Company (Shanghai, China) and were transfected into cells using polybrene (Genechem, MOI = 30) for 48 h. The miR-2861 mimic, inhibitor, and related control were designed by GenePharma (Shanghai, China). Cells were transfected with these molecules for 24 h (5 nM for all) using the Lipofectamine 2000 reagent (Invitrogen, USA) following the manufacturer’s instructions.
RT-PCR
Total RNA was extracted from the cells or tissues using TRIzol reagent (Thermo, USA) and revere-transcribed into cDNA using PrimeScript RT Master Mix Kit or Prime-Script miRNA cDNA Synthesis Kit (TaKaRa, Japan). Next, gene expression was performed on the qRT-PCR system (Bio-Rad, USA) using SYBR Green reagent (Bio-Rad, USA). U6 and GAPDH were used as the controls. The data were calculated by the 2-ΔΔCT method. The sequence of primers was showed as follows: miR-2861 (human): forward 5’-GCGGGGGCCTGGCGGT-3’, reverse 5’-AGTGCAGGGTCCGAGGTATT-3’; SNHG17 forward: 5’-AGAGAATGGAGAGTGAGGCTACC-3’, reverse: 5’-CCAGGCATGGACAGAGGGAT-3’; miR-4700-3p (human): forward 5’-GCACAGGACTGACTCCTCACC-3’, reverse 5’-AGTGCAGGGTCCGAGGTATT-3’; VEGF (human): forward 5’-TGCGGATCAAACCTCACCAA-3’, reverse 5’-GCTTAACCCTGGCACAGATCA-3’; BCL2L (human): forward 5’-ACTCTTCCGGGATGGGGTAA-3’, reverse 5’-AAGGGCTGTTGGGGATCTCT-3’; HIF1A (human): forward 5’-AGAGGTTGAGGGACGGAGAT-3’, reverse 5’-CAACATGAAATGTCCTGCGT-3’; BIRC5 (human): forward 5’-GGACTGCCGCTTTAATCCCT-3’, reverse 5’-TGAACAGGGTTTGAGCAGTTC-3’; SOX4 (human): forward 5’-CAGCAAACCAACAATGCCGA-3’, reverse 5’-GATCTGCGACCACACCATGA-3’; TWIST1 (human): forward 5’-GGAGGGAGGGGGCACTAATA-3’, reverse 5’-ACATGCTTGTGCCTGTCAGT-3’; MYC (human): forward 5’-ACTCTGGTAAGCGAAGCCC-3’, reverse 5’-ACATGGGCAGTCTAAGGGGA-3’; TIAM (human): forward 5’-TGCCTTCGCTGGATGGAAAT-3’, reverse 5’-GGGACCGAGACGCAAACATA-3’; KIAA (human): forward 5’-TCAACAGAGCTATGGTGGGAC-3’, reverse 5’-CATCGCCCCTTCACATCCAA-3’; CCND1 (human): forward 5’-ACACCTAGTGCCACGGAAAT-3’, reverse 5’-CACCATCACCACACAGACCA-3’; CCND2 (human): forward 5’-CTGTCTCTGATCCGCAAGCA-3’, reverse 5’-GTTCCTACGGCCTTTTGCCT-3’; MMP2 (human): forward 5’-ACGCTAAGACCCAGTGTGTG-3’, reverse 5’-TTGGGGTGGAAAGTCTTGGG-3’; FN14 (human): forward 5’-CTGACGACCCCACCTCTTATC-3’, reverse 5’-ACCTTGGAAGGTTCCCCTGA-3’; TCF1 (human): forward 5’-AAGACTTCACGCCACCCATC-3’, reverse 5’-GTCCCCCTTCCCAGCAACTA-3’; HES6 (human): forward 5’-TAACCCCTGCCAGACGGAG-3’, reverse 5’-GCAATTTGGGCTGTGGTCAG-3’; HEY1 (human): forward 5’-AGAGAACGGTGTGTGGTGTG-3’, reverse 5’-ATGCACTAGCTCAATCCGCA-3’; HES1 (human): forward 5’-GATAATGCTTGCGCTCCGTG-3’, reverse 5’-TCTGGAAGAAATCACCGCGA-3’; NRARP (human): forward 5’-TACCTTCCCGCCAACTACCT-3’, reverse 5’-TGCTGGTTGCGAGACTCAAA-3’; GLI1 (human): forward 5’-AGCCCCTACCCAAGTCCATT-3’, reverse 5’-TAGTGGTTGAGGCAGTCCCA-3’; PATCHED (human): forward 5’-AAAGCAGCAGACAAATGGGGA-3’, reverse 5’-GCCGTACAGGAGTTTAGGCT-3’; GLI3 (human): forward 5’-TGTCAAAAGGCGCCCAGATT-3’, reverse 5’-TCGCGGAGTTCTTCTGAACC-3’; GAPDH (human): forward 5’-GCACCGTCAAGGCTGAGAAC-3’, reverse 5’-TGGTGAAGACGCCAGTGGA-3’; U6 (human): forward 5’-GAAGCGCGGCCACGAG-3’, reverse 5’-AGTGCAGGGTCCGAGGTATT-3’.
Cell proliferation
According to the manufacturer’s instructions, cell viability was evaluated using the cell counting kit-8 (CCK-8) (Sigma, USA). Briefly, after different treatments, the cells were plated in the 96-well plates at a density of 5000 cells per well. After 24 h, the CCK-8 solution was added and incubated for another 4 h. Finally, each well’s absorbance was determined using a microplate reader (Bio-Tek, USA) at 450 nm.
Colony assay
104 cells were seeded on the 3-cm dish and co-cultured with the exosomes or condition medium for 48 h. Then the medium was changed and washed by PBS. In one group, cells were plated after different transfections. The cells were maintained in the fresh medium for 2 weeks. Finally, the colonies were fixed and dyed with crystal violet (Beyotime, China). The images were observed under a light microscope (Olympus, Japan), and the visible colonies were recorded.
Migration assay
Transwell assay was performed to evaluate cell migration. In brief, cells were plated to upper transwell chambers (BD Biosciences, USA). Cells were co-cultured with the exosomes or condition medium for 48 h. After washed by PBS, migration cells were fixed with methanol and incubated with 0.1% crystal violet for 10 min. The visible cells were counted at five randomly selected fields in each well under a light microscope (Olympus, Japan).
Flow cytometry analysis
The apoptosis rates were assessed by flow cytometry via an Annexin V/PI kit (BD Biosciences, USA). Briefly, stably transfected cells were seeded in 6-well plates (105 cells per well). After washed by PBS several times, cells were resuspended in binding buffer, incubated with 5 μL FITC-Annexin V and 5 μL PI for 15 min in the dark at room temperature. Cells were then analyzed by the FACScan flow cytometry system (Becton Dickinson, San Diego, CA, USA).
Western blot
Cells were collected and homogenized to extracted total proteins using radioimmunoprecipitation assay buffer (Beyotime, China). Next, equal protein (20 µg/lane) was separated and transferred onto the polyvinylidene fluoride membrane (Bio-Rad, USA). Subsequently, the membranes were blocked by 5% BSA solution for 2 h at room temperature, followed by incubated with primary antibodies against cleaved caspase-3 (1:1000, Abcam, USA), cleaved caspase-9 (1:1000, Abcam, USA), caspase-3 (1:1000, Abcam, USA), caspase-9 (1:1000, Abcam, USA), MMP2 (1:2000, Abcam, USA), Bax (1:1000, Abcam, USA), GAPDH (1:5000, Abcam, USA), Fibronectin (1:2000, Abcam, USA), a-SMA (1:1000, Abcam, USA) and Tublin (1:5000, Abcam, USA) at 4°C overnight. After hybridized with related secondary antibodies (1:5000, Abcam, USA) for 2 h at room temperature, the bands were developed by the ECL kit (Thermo, USA) and analyzed by Image J software. GAPDH and Tublin were used as the internal control.
Animals and transfection
The BALB/c nude mice (4-6 weeks) were obtained from the Animal Centre of Nanjing University. After different transfection, 106 HOS cells that were stably expressing sh-NC or sh-SNHG17#1 were injected subcutaneously into the left axilla of mice. The tumor volume was recorded every week and measured according to the formula as follows: tumor size (mm3) = (length × width2)/2. At the end of this experiment, the mice were sacrificed, and the tumor tissues were removed, weighed, and fixed by 10% formalin solution. All animal experiments were approved by the Ethics Committee of The Second Affiliated Hospital of Inner Mongolia Medical University (No. YKD2017142).
H&E and IHC
The fixed tumor tissues of mice were embedded in paraffin. Five-micrometer-thickness sections were cut using a microtome (Leica Biosystems, Germany) and stained with hematoxylin/eosin solution (Beyotime, China) following the manufacturer’s protocol. The images were observed under a light microscope.
The sections were then incubated by the primary antibody against Ki-67 (1:500, Abcam, USA) overnight at 4°C. After stained using a secondary antibody (30-40 μL, Abcam) at room temperature for 30 min, the positive signals of Ki-67 were developed using a commercial kit (Thermo, USA) according to the manufacturer’s instructions. The nuclei were stained by hematoxylin solution. The images were determined under light microscopy (Olympus, Japan).
TUNEL assay
TUNEL staining was performed to evaluate the apoptosis of tumor tissues using the In-Situ Cell Death Detection Kit (Roche, Germany) following the manufacturer’s protocol. In brief, the sections were deparaffinized, rehydrated, and blocked by proteinase K (20 mg/mL). Next, the sections were incubated with 3% hydrogen peroxide in methanol for 10 min, followed by treatment with a TUNEL solution for 60 min at 37°C. The cells were then incubated with fluorescein-dUTP, and the nucleus was stained by DAPI. Staining cells were observed under the fluorescence microscope.
Exosome isolation and identification
The cell supernatants were collected as described above, and centrifuged at 300×g for 15 min, 2000×g for 30 min and 10000×g for 30 min at 4°C. The supernatants were then filtered with a 0.22-µm filter (Millipore, USA), followed by centrifuging at 100000×g for 4 h. After washed by PBS, the exosomes were resuspended in 200 µL PBS for further study.
Nanoparticle tracking analysis (NTA) was used to determine the concentration of CAFs-exosomes and NFs-exosomes using the Nanosight (Merkel Technologies Ltd., Israel). The particle size distribution plot relayed on the diameter (x-axis), and intensity (y-axis) was generated. The biomarkers of exosomes, including CD9 (Abcam, 1:1000) and CD63 (Abcam, 1:2000), were determined using western blot as described above.
Exosome treatment
Cells were plated in a 6-cm dish and incubated with 30 ng/mL exosomes (in 50 µL PBS) derived from normal fibroblasts or cancer-associated fibroblasts (with or without GW4869 treatment). Cells were collected for further study after 48 h.
Bioinformatics analysis
The target miRNAs of LncRNA SNHG17 was predicted by two online databases: LncBase (http://carolina.imis.athena-innovation.gr/) and RegRNA (http://regrna2.mbc.nctu.edu.tw/). The potential miRNAs targeting MMP2 was assessed by miRDB (http://www.mirdb.org/), and the miRWalk database (http://www.mirwalk.umm.uni-heidelberg.de/).
RNA binding protein immunoprecipitation (RIP)
The RIP experiment was performed as the previous study. The HOS cells were collected and lysed using the RIP lysis buffer supplemented with protease and RNase inhibitor. The lysates were then incubated with the magnetic beads containing the Ago2 antibody (Abcam, USA) or IgG for 4 h at 4°C. The beads were then washed by PBS for several times, and the immunoprecipitated RNA was extracted by TRIzol reagent and subjected to RT-qPCR. The Ago2 was assessed by western blot.
Luciferase assay
The wildtype or mutant 3’-UTR of MMP2 or LncSNHG17 have generated the pmirGLO vector (Promega, USA) and obtained from GenePharma (Shanghai, China). Mutations were established by a QuikChange Site-Directed Mutagenesis Kit (Stratagene, USA). HEK293T cells were plated on the 6-well plate at a density of 105 cells per well. The vector was co-transfected into cells with miR-2861 mimic, inhibitor, or related controls. After 48 h, the cells were collected to detect the luciferase activity by the dual-luciferase reporter assay system (Promega). The ratio of firefly and renilla luciferase activity was analyzed.
Statistical analysis
All data were analyzed by Prism version 8.0 (La Jolla, USA) and represented as mean ± SD from three independent experiments. Differences between two groups were analyzed by two-tailed Student’s t-test. Kaplan-Meier method was used to evaluate the overall survival rate. The correlation between SNHG17 levels and MMP2 expression was measured by spearman’s correlation coefficient. The P<0.05 was considered statistically significant.
Results
LncRNA SNHG17 was upregulated in the OS tissues and corresponding with the poor prognosis of OS
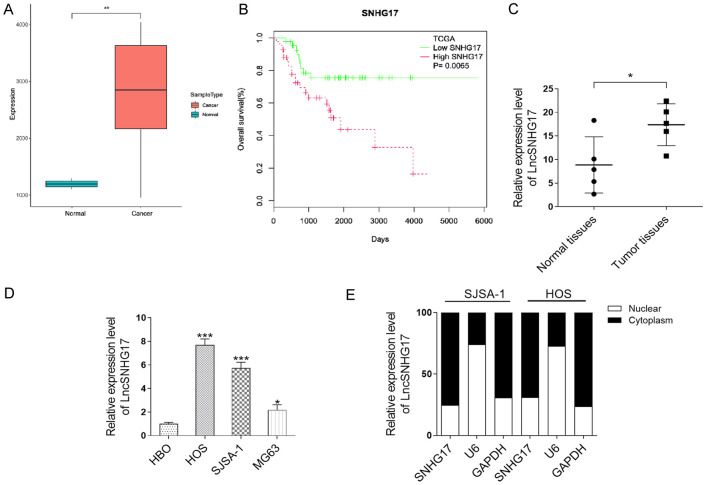
The expression level of SNHG17 in the OS tissues and related normal tissues was determined using the TCGA database. As shown in Figure 1A, the transcript level of SNHG17 was about 3 times higher in the tumor tissues than that in the normal tissues. The relationship between overall survival of OS and SNHG17 expression was also determined through Kaplan-Meier analysis. The results showed that OS patients with high-expression SNHG17 were associated with poor prognosis (P = 0.0065) (Figure 1B). Further, through qRT-PCR analysis, we found that the level of SNHG17 was higher expressed in our OS samples (n = 5) than the normal tissues (n = 5) (Figure 1C). To evaluate the role of this lncRNA, we checked the expression of SNHG17 in a panel of OS cells. As shown in Figure 1D, compared with the normal cell (HBO), SNHG17 expression was almost 8, 6, and 3 folds higher in HOS, SJSA-1, and MG63 cells, respectively. Besides, we further examined the subcellular localization of SNHG17 in HOS and SJSA-1 cells. The qRT-PCR analysis showed that SNHG17 was mainly enriched in the cytoplasm of both cells rather than the nuclear (Figure 1E).
Figure 1.
LncRNA SNHG17 is upregulated in the OS tissues and corresponding with the poor prognosis of OS. A. Relative expression of SNHG17 in human OS tissues compared with adjacent normal tissue via TCGA data analysis. B. Kaplan-Meier overall survival curves were performed based on the LncRNA SNHG17 expression level. C. The expression level of LncRNA SNHG17 in OS clinical samples (n = 5) were measured by qRT-PCR. *P<0.05 vs. the normal tissues. D. LncRNA SNHG17 expression was determined by qRT-PCR analysis in a normal cell line (HBO) and OS cell lines (MG63, HOS, and SJSA-1). *P<0.05, **P<0.01, ***P<0.001 vs. the HBO group. E. Localization of LncRNA SNHG17 in SJSA-1 and HOS cells.
LncRNA SNHG17 promotes the proliferation and migration in vitro
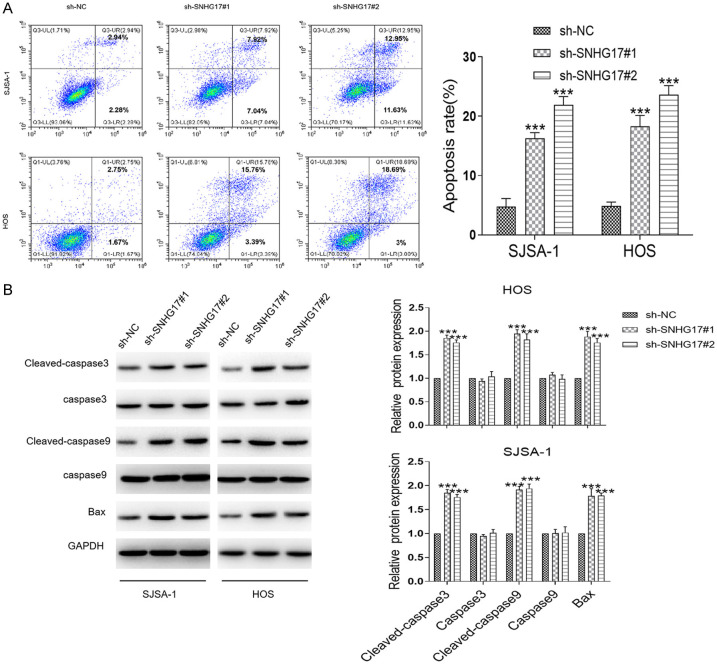
Changes in SNHG17 expression in OS cells (HOS and SJSA-1) transfected with two shRNA against SNHG17 (sh-SNHG17#1 and sh-SNHG17#2), or related control (sh-NC) were assessed by qRT-PCR. As shown in Figure 2A, transfection with the sh-SNHG17#1 or sh-SNHG17#2 presented about 70% and 50% reduction of the SNHG17 level in SJSA-1 cells compared with the control group, respectively. Besides, we investigated similar results in HOS cells, where these shRNAs transfection induced about 75% and 60% decrease in the SNHG17 levels than the si-NC group. Consistently, the CCK-8 assay showed that the silence of SNHG17 led to a marked inhibition of cell viability in these cells (Figure 2B). Consistently, inhibition of SNHG17 suppressed the colony formation of HOS and SJSA-1 cells (Figure 2C). Besides, we found down-regulation of SNHG17 remarkably decreased the migration of OS cells and increased the apoptotic index in tumor cells (Figures 2D and 3A). Besides, western blot analysis demonstrated that compared with the sh-NC group, SJSA-1 and HOS cells with sh-SNHG17 transfection had higher expression levels of the apoptotic genes cleaved-caspase 3, cleaved caspase 9, and Bax (Figure 3B). Together, SNHG17 promoted OS cell proliferation and migration in vitro.
Figure 2.
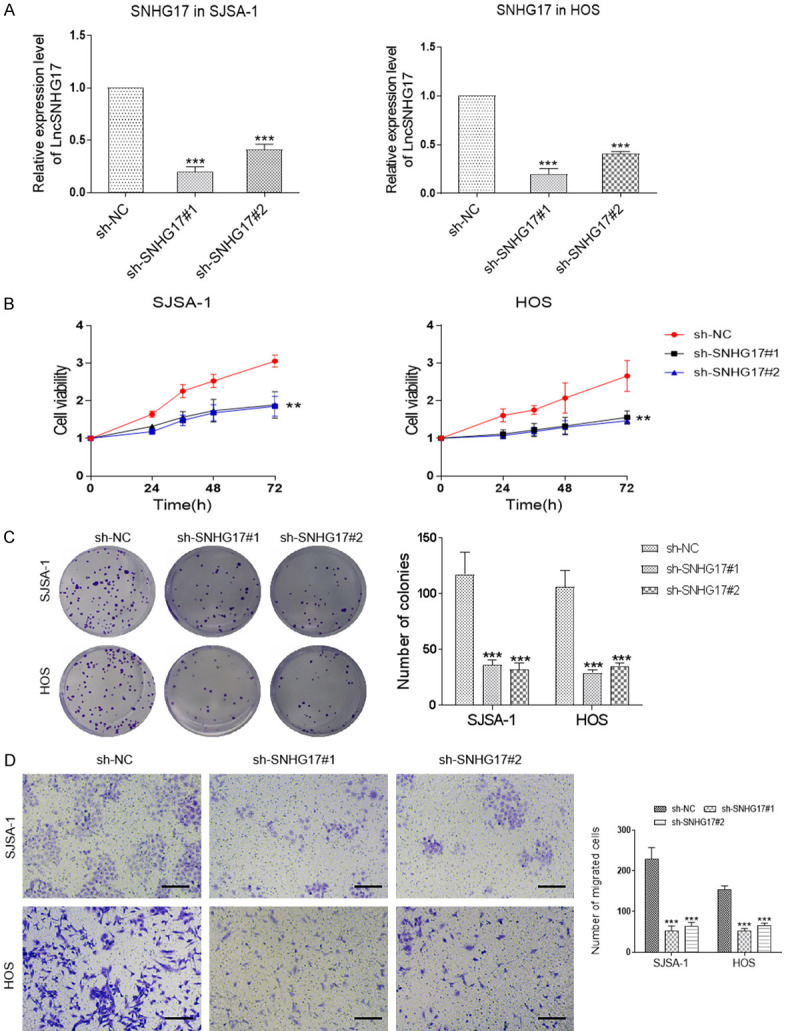
LncRNA SNHG17 promotes the proliferation and migration of osteosarcoma in vitro. A. LncRNA SNHG17 expression levels were assessed in the SJSA-1 and HOS cells with shRNAs treatment. ***P<0.001 vs. the sh-NC group. B. The cell viability was performed in SJSA-1 and HOS cells following transfection with sh-NC or sh-SNHG17. C. Representative images of colony-formation assays for sh-SNHG17-transfected SJSA-1 and HOS cells. D. Transwell assays were applied to measure the sh-SNHG17-transfected SJSA-1 and HOS cells migration capacity. Bar = 20 μm. *p<0.05, **p<0.01, ***p<0.001 vs. the sh-NC group.
Figure 3.
LncRNA SNHG17 promotes cell apoptosis in vitro. A. The rate of cell apoptosis was performed in SJSA-1 and HOS cells following transfection with sh-NC or sh-SNHG17. B. Representative blots and statistical analysis for cleaved caspase-3, caspase-3, cleaved caspase-9, caspase-9, and Bax in sh-SNHG17-transfected SJSA-1 and HOS cells were performed. *P<0.05, **P<0.01, ***P<0.001 vs. the sh-NC group.
LncRNA SNHG17 promotes the proliferation and inhibits apoptosis in vivo
To further investigate the role of SNHG17 in vivo, the SNHG17-depleted HOS cells were subcutaneously injected to establish the xenograft model. As shown in Figure 4A, the expression of SNHG17 was dropped by almost 50% by sh-SNHG17#1 transfection compared with the control one. Accordingly, the volume of tumor in the sh-SNHG17 group was smaller than the sh-NC transfected mice (Figure 4B). Besides, as shown in Figure 4C, tumor growth of mice was delayed by SNHG17 inhibition. Four weeks after inoculation, the tumor tissues were isolated. A significant decrease also presented in the tumor weight, with 2.94 ± 0.08 g in the sh-NC group and 0.44 ± 0.07 g in the sh-SNHG17 group, respectively (Figure 4D). Besides, IHC staining demonstrated mice with sh-SNHG17#1 presented more positive Ki-67 signals, a marker of cell proliferation. Conversely, the TUNEL assay resulted in more apoptotic cells in the sh-NC transfected mice than in the sh-SNHG17 group (Figure 4E). Accordingly, we considered that SNHG17 enhanced tumorigenicity of OS in vivo.
Figure 4.
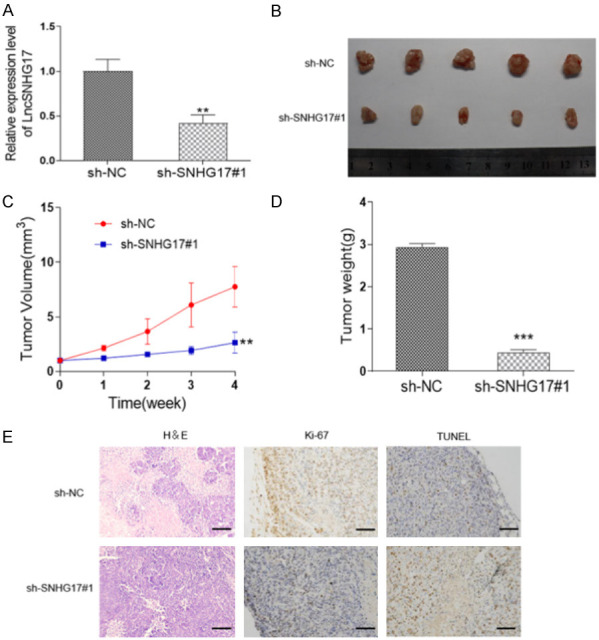
LncRNA SNHG17 promotes the proliferation and inhibits apoptosis in vivo. HOS cells were transfected with sh-NC or sh-SNHG17#1 and then injected into nude mice. A. Transfection efficiency in HOS cells was determined using qRT-PCR. B. The picture of tumors in each group. C. Tumor volume was recorded every week after injection. D. The tumor weight of each group was detected. E. Representative images of H&E staining, TUNEL assay, and IHC staining of Ki-67 were performed. Bar = 50 μm. *p<0.05, **p<0.01, ***p<0.001 vs. the sh-NC group.
CAF-derived exosomes are involved in the enhanced expression of LncRNA SNHG17 in osteosarcoma
Next, as shown in Figure 5A, the result of western blot demonstrated that both NFs and CAFs exhibited fibronectin and a-SMA expression, suggesting that we had successfully isolated the fibroblasts form clinical tissues. Besides, compared with the normal fibroblasts, we found about 30 folds higher level of SNHG17 in CAFs derived from our samples (Figure 5B). Hence, we hypothesized whether the CAFs derived SNHG17 could regulate tumor cell proliferation. To achieve this purpose, we treated the tumor cells with condition medium of NFs and CAFs. The CCK-8 analysis showed that the cell viability of SJSA-1 and HOS cells were both notably increased after CAFs-medium treatment compared with the NFs-medium administration (Figure 5C). Similarly, the same treatment also promoted the colony formation and cell migration of HOS and SJSA-1 cells (Figure 5D and 5E). Interestingly, when we treated the tumor cells with a medium derived from the GW4869-treated CAFs, lower cell viability, and less cell migration were displayed in both cells than the control group (Figure 5). As an inhibitor of exosome secretion, we then hypothesized CAFs might affect tumor proliferation through transferring exosomes.
Figure 5.
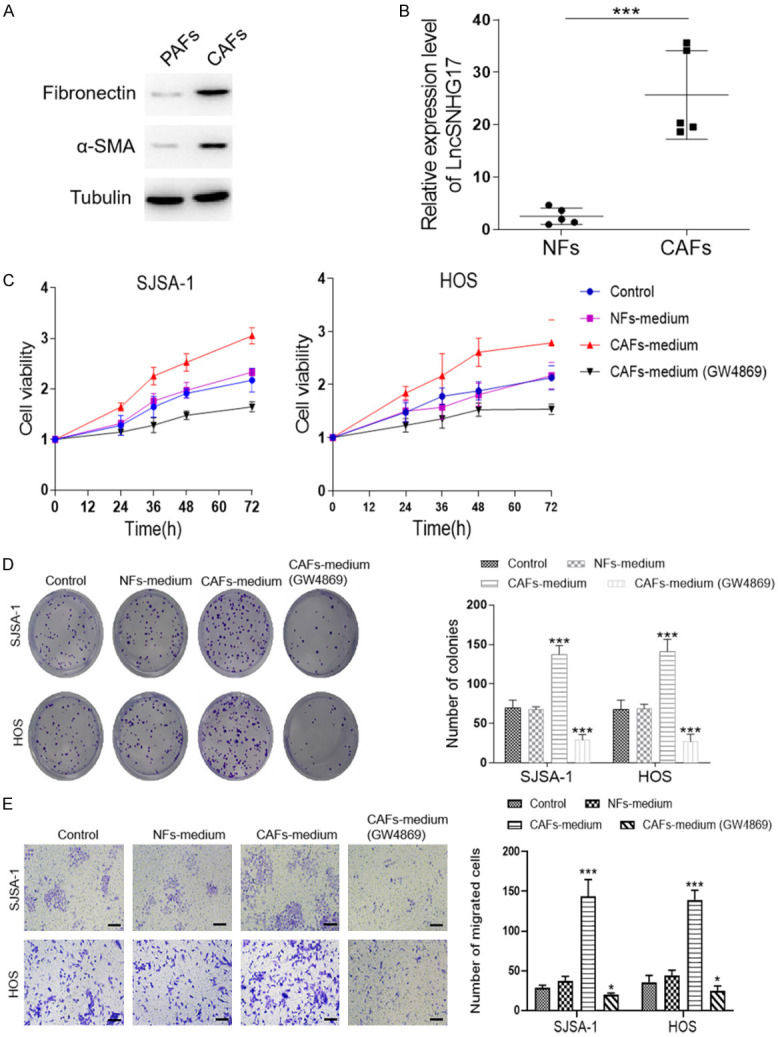
CAFs-derived exosomes are involved in the enhanced expression of LncRNA SNHG17 in osteosarcoma. (A) Western blot analysis of fibroblast-related markers (fibronectin, a-SMA) in the NFs and CAFs. (B) The expression levels of LncRNA SNHG17 in normal fibroblasts and cancer-associated fibroblasts isolated from OS patients were detected by qRT-PCR, n = 5. (C) The cell viability of SJSA-1 and HOS cells were determined after different condition medium treatments. (D) The colony assay and (E) migration assay of SJSA-1 and HOS cells after different treatments were performed. Bar = 20 μm. *P<0.05, **P<0.01, ***P<0.001 vs. the sh-NC group. #P<0.05, ##P<0.01, ###P<0.001 vs. the CAF-medium group.
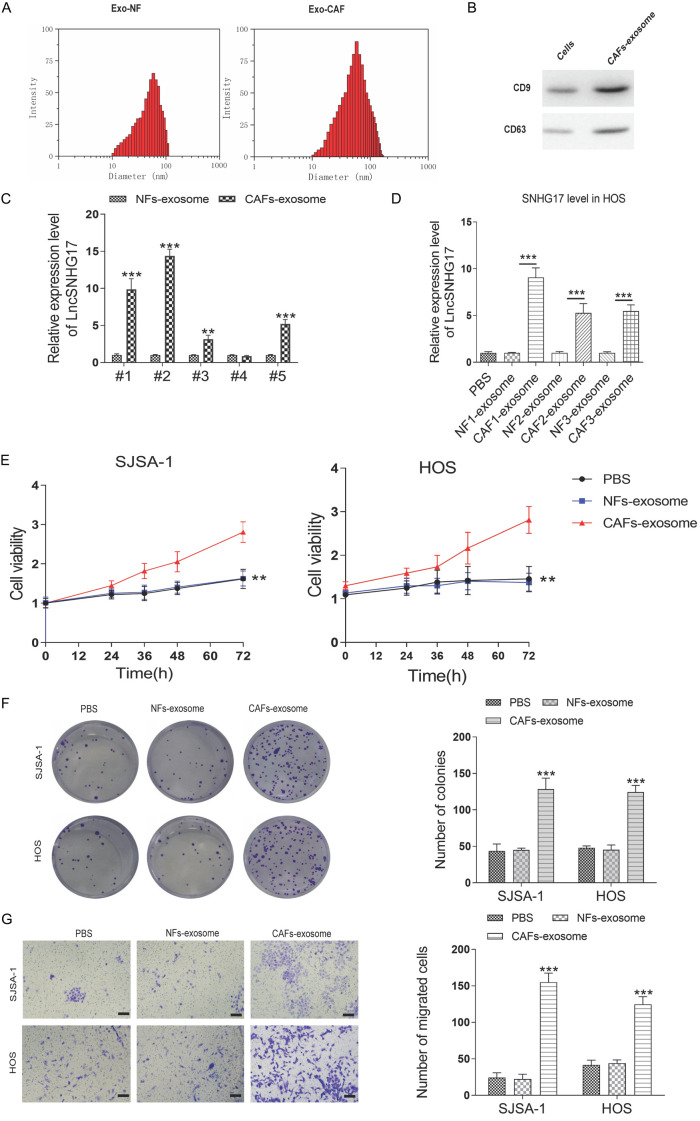
We then isolated the exosomes from the conditioned medium of CAFs and NFs using series ultracentrifugation. The diameter of such exosomes was observed using NTA. These exosomes presented a typical cystic structure with 100 nm diameter, which was consistent with previous studies [12,13] (Figure 6A). Two positive markers of exosomes, including CD9 and CD63, were both obviously enriched in CAFs-derived exosomes (Figure 6B). Since there were no standard internal controls for exosome proteins, we did not detect the expression of β-actin or GAPDH in this study. Few previous studies determined that the expression of β-actin in the exosomes, but in some cases, its abundance was low [21-23]. We further checked the SNHG17 expression level in the CAFs of our OS samples. As shown in Figure 6C, SNHG17 was highly expressed in the CAFs-exosomes derived from the patients than that in the NFs-exosomes.
Figure 6.
CAFs-derived exosomes transfer SNHG17 to osteosarcoma cells. (A) The size distribution of the isolated exosomes was detected by NTA. (B) Western blotting analysis for exosomal markers CD63 and CD9 of tumor cells and exosomes derived from CAFs. (C) The expression level of LncRNA SNHG17 in the exosomes derived from normal fibroblast and CAF of five pairs of clinical samples. **P<0.01, ***P<0.001 vs. the NFs-exosomes group. (D) The expression level of LncRNA SNHG17 in the HOS cells after treatment with exosomes derived from NFs or CAFs of three pairs of clinical samples. ***P<0.001. (E) Cells were treated by PBS, NFs-exosomes, or CAFs-exosome, and the cell viability was determined by CCK-8 assay. (F) The colony assay and (G) migration assay of SJSA-1 and HOS cells after different treatments were performed. Bar = 20 μm. *P<0.05, **P<0.01, ***P<0.001 vs. the sh-NC group. #P<0.05, ##P<0.01, ###P<0.001 vs. the CAFs-exosome group.
To investigate the function of CAFs-released exosomes on tumor cell proliferation, the exosomes were used to treat HOS and SJSA-1 cells. As expected, incubation with exosomes released by CAFs markedly increased the SNHG17 expression in HOS cells. However, exosomes derived from the corresponding NFs displayed no effect on the SNHG17 level (Figure 6D). Accordingly, CAFs-derived exosomes significantly enhanced the proliferation and migration of SJSA-1 and HOS cells (Figure 6E-G). Together, these data revealed that CAFs-derived exosomes were involved in the enhanced expression of SNHG17, proliferation, and migration in OS cells.
CAFs-derived exosomes transfer SNHG17 promotes the progression of osteosarcoma via inhibiting MMP2
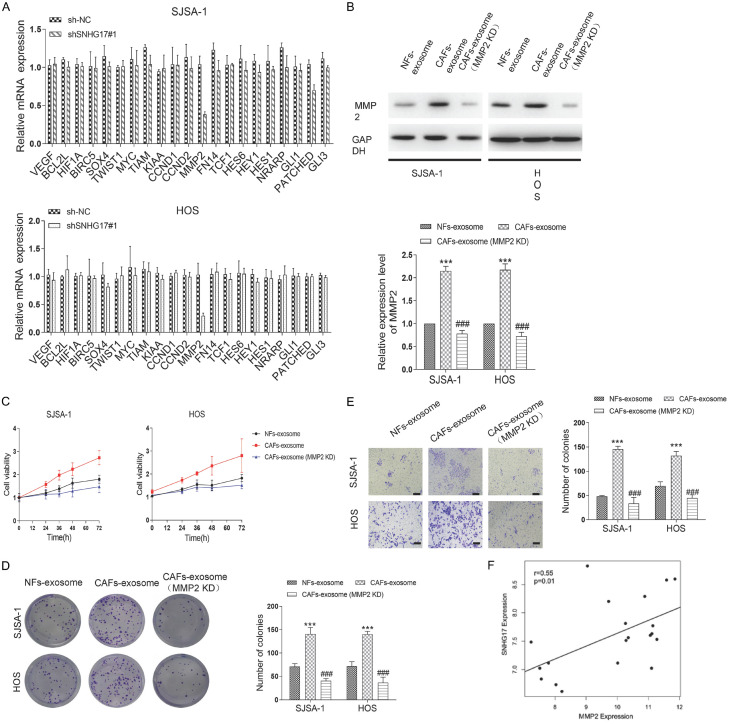
Given that several pathways, including NOTCH signaling, Wnt pathway, NF-κB signaling, and Hedgehog signaling, are critical for tumorigenesis, we sought whether exosomal SNHG17 could regulate these pathways. The results showed that silence of SNHG17 with sh-SNHG17#1 in SJSA-1 and HOS cells only significantly resulted in decreased expression of MMP2 (Figure 7A). Using western blot analysis, we found a 2.2-folds increase, and a 2.3-folds upregulation of MMP2 expression in CAFs-exosomes treated SJSA-1 and HOS cells, respectively. However, the reverse experiment where MMP2 was inhibited before exosome isolation re-downregulated the protein level of the MMP2 (Figure 7B). Accordingly, through CCK-8, colony formation assay, and migration assay, we found downregulation of MMP2 reversed the effect of CAFs-exosomes in both cells (Figure 7C-E). Next, we analyzed the SNHG17 and MMP2 in OS tissues using the TCGA database and found that the level of SNHG17 positively correlated with the level of MMP2 (r = 0.55, P = 0.01) (Figure 7F).
Figure 7.
CAFs-derived exosomes transfer SNHG17 promotes the progression of osteosarcoma via inhibiting MMP2. (A) The expression of NOTCH signaling, Wnt pathway, NF-κB signaling, and Hedgehog signaling related genes in SJSA-1 and HOS cells after SNHG17 depletion were performed by qRT-PCR analysis. (B) Representative blots and statistical analysis for MMP2 in SJSA-1 and HOS cells after different treatments were performed. (C) Cells were treated by NFs-exosomes, CAFs-exosome, or CAFs-exosome (MMP2 KD), and the cell viability was determined by CCK-8 assay. (D) The colony assay and (E) migration assay of SJSA-1 and HOS cells after different treatments were performed. Bar = 20 μm. (F) Spearman correlation analysis determined the relationship between the levels of SNHG17 and MMP2 in OS cancer tissues from TCGA database. *P<0.05, **P<0.01, ***P<0.001 vs. the sh-NC group. #p<0.05, ##P<0.01, ###P<0.001 vs. the CAFs-exosome group.
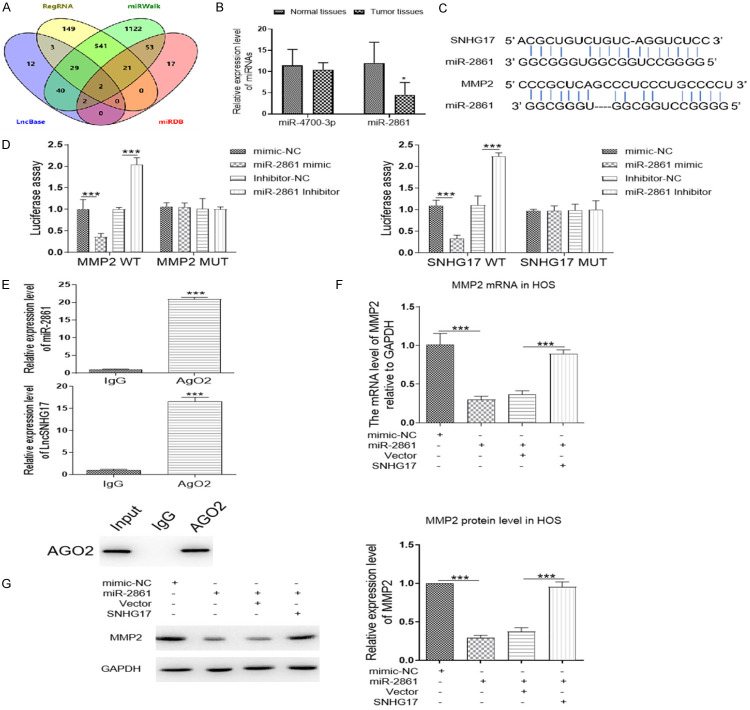
SNHG17 activates the MMP2 by acting as a competing endogenous RNA sponge for miR-2861
To date, abundant evidence pointed out that lncRNAs regulated target gene expression via functioned as a ceRNA for microRNAs [24]. To assess this hypothesis, LncBase and RegRNA databases were uses to predict the potential miRNAs targeting SNHG17. We prioritized the miRNAs following the prediction score and free-energy. There were 34 overlapping miRNAs between these databases. Moreover, miRDB and miWalk databases were performed to figure out the miRNAs targeting MMP2. Based on these data, two overlapping miRNAs of four databases, including miR-2861 and miR-4700-3p, were chosen for qRT-PCR analyses (Figure 8A). As shown in Figure 8B, the expression level of miR-2861 was dramatically downregulated in the tumor tissues as compared with the normal ones. Subsequently, to confirm the relationship between these three molecules, the dual-luciferase reporter assays were performed. The binding site among SNHG17, miR-2861 and MMP2 was demonstrated in Figure 8C. The HEK293T cells were co-transfected with a vector containing the 3’-UTR of MMP2 along with miR-2861 mimic, inhibitor, or related controls. Treatment with miR-2861 mimic suppressed MMP2-WT driven luciferase activity, and the inhibitor of this miRNA significantly increased the activity of wildtype MMP2 vector. Besides, the cells have also transfected the plasmid containing the sequence of SNHG17 prior to miRNA mimic, inhibitor, or controls treatment. The results showed that miR-2861 inhibitor upregulated the plasmid luciferase activity with wildtype SNHG17, while miR-2861 mimics downregulated the luciferase activity in the cells with SNHG17-WT transfection rather than the mutant ones (Figure 8D). Further, the data of RNA immunoprecipitation analysis demonstrated that miR-2861 and SNHG17 were both elevated in AgO2-immunoprecipitation compared with the IgG group in HOS cells (Figure 8E). In addition, we found that overexpression of miR-2861 significantly decreased the MMP2 expression at the protein level, with no effect in the mRNA level of MMP2. While co-transfection with SNHG17 reversed the decrease induced by miR-2861 (Figure 8F and 8G), these findings suggested that SNHG17 promoted OS proliferation and metastasis through interacting with miR-2861.
Figure 8.
SNHG17 activates the MMP2 by acting as a competing endogenous RNA sponge for miR-2861. A. The miRNAs targeting MMP2 and SNHG17 were predicted by four online databases. The overlapping miRNAs were performed by the Venn diagram. B. The expression level of miR-2861 and miR-4700-3p in normal tissues and tumor tissues were analyzed by qRT-PCR. C. The binding site between SNHG17 and miR-2861, or the binding sites between MMP2 and miR-2861, was predicted by RNAhybrid databases. D. Luciferase activity in HEK293T cells co-transfected with miR-2861 mimic, inhibitor or related controls (mimic-NC, inhibitor-NC), and a vector containing SNHG17 WT or SNHG17 MUT 3’-UTR, or vectors containing MMP2 WT or MMP2 MUT 3’-UTR. E. Immunoprecipitation using anti-AgO2 antibody or IgG followed by Western blot analysis. Immunoprecipitated RNA was isolated by TRIzol reagent, and the level of miR-2861 or SNHG17 was analyzed by qRT-PCR. ***P<0.001 vs. the IgG group. F and G. The mRNA and protein expression level of MMP2 in HOS cells after different transfections were determined by western blot or qRT-PCR. *P<0.05, **P<0.01, ***P<0.001.
Discussion
Abundant studies have explored that lncRNAs could involve in the initiation and development of various cancers, including osteosarcoma. For instance, Lu et al. revealed that LncRNA DANCR drove OS development via decoying miR-335-5p and miR-1972/ROCK2 axis [25]. Zhang et al. discussed that LncRNA MEG3/miR-200b-3p/AKT2 axis served as the novel mechanism of chemoresistance in patients with OS [26]. Another LncRNA loc285194 had also reported to suppress OS growth via sponging miR-211 and to regulate the p53 pathway [27]. In the present study, we identified LncRNA SNHG17 was highly expressed in the OS cells and associated with a poor overall survival rate. We also found SNHG17 promoted OS proliferation and metastasis in vivo and in vitro. Moreover, differential expression of SNHG17 in tumor cells and the stromal cell was investigated, suggesting that SNHG17 would participate in the communication between OS cells and stromal cells.
Within the stroma, CAFs have been identified to promote tumor progression and drug resistance via secreting various factors and extracellular matrix [28,29]. In this study, compared with the normal fibroblast, a higher level of SNHG17 was presented in the OS-related CAFs. Treatment with the conditioned medium of CAFs markedly enhanced tumor cell growth and migration in vitro. Our current study also revealed, for the first time, that CAF-derived exosomes played an essential role in the proliferation and migration of OS cells. This kind of small vesicles was released by almost all cell types and could be uptake by local or distant tissues [30]. Previous studies have confirmed that exosomes contained multiple non-coding RNAs, including lncRNAs, miRNAs, and circRNAs [31-33]. Using the qRT-PCR analysis, we identified significant higher enrichment of SNHG17 in CAFs-derived exosomes than those in the normal fibroblasts. However, the mechanism by which the SNHG17 affect tumor progression remains unclear. By silencing the expression of SNHG17 in OS cells, we found the MMP2 gene was dramatically downregulated. As expected, exosomes released by CAFs exhibited a similar effect on the MMP2 expression in vitro.
Additionally, in this study, we found SNHG17 located in the cytoplasm, which was consistent with the previous studies [34-36], suggesting that SNHG17 might function as ceRNA for sponging miRNA, thus regulating the post-transcription levels of target genes [37]. Through bioinformatics analysis, RIP assay, and luciferase assay, we confirmed that SNHG17 sponged miR-2861 in OS cells. The miR-2861 has been reported down-regulated in cervical cancer and suppressed tumor growth through regulating multiple genes, such as EGFR, AKT2, and CCND1 [38]. Here, our data demonstrated that miR-2861 level was downregulated in the OS tissues compared with the normal tissues, and this miRNA might regulate MMP2 mRNA expression via directly binding to its coding sequence. Collectively, based on the reverse assays, we found overexpression of miR-2861 could reverse the effect of SNHG17 on the expression of MMP2, indicating that SNHG17 controls the OS proliferation and metastasis through the miR-2861/MMP2 axis. Given that the function of SNHG17 needs to be confirmed by in vivo study and more complicated in the human body, there is a long way before clinical utilization. However, some questions remain unanswered at present. For example, Qiu et al. have also showed that elevated neoplastic MMP2 contents was observed in samples of osteosarcoma patients. Calycosin is identified to play the antitumor effect for suppressing osteosarcoma cell proliferation by reducing MMP2 concentration, which suggested that MMP2 may be a key factor during the progression of osteosarcoma [39]. Therefore, underlying mechanisms by which MMP2 affected the progression of osteosarcoma will be an important issue for future research.
In summary, through the integrating clinical data, we investigated the SNHG17 was upregulated in OS tissues and correlated with the poor prognosis. Based on the functional experiments, we explored SNHG17 played an oncogene role in vivo and in vitro through sponging miR-2861 to enhance MMP2 expression. Our study also illustrated CAFs-derived exosomes’ role in controlling tumor cell proliferation and migration by transferring the SNHG17. This study provided a novel idea for OS treatment.
Disclosure of conflict of interest
None.
References
- 1.Ritter J, Bielack SS. Osteosarcoma. Ann Oncol. 2010;21(Suppl 7):vii320–325. doi: 10.1093/annonc/mdq276. [DOI] [PubMed] [Google Scholar]
- 2.Whelan JS, Davis LE. Osteosarcoma, chondrosarcoma, and chordoma. J. Clin. Oncol. 2018;36:188–193. doi: 10.1200/JCO.2017.75.1743. [DOI] [PubMed] [Google Scholar]
- 3.Ottaviani G, Jaffe N. The epidemiology of osteosarcoma. Cancer Treat Res. 2009;152:3–13. doi: 10.1007/978-1-4419-0284-9_1. [DOI] [PubMed] [Google Scholar]
- 4.Isakoff MS, Bielack SS, Meltzer P, Gorlick R. Osteosarcoma: current treatment and a collaborative pathway to success. J. Clin. Oncol. 2015;33:3029–3035. doi: 10.1200/JCO.2014.59.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bielack SS, Rerin JS, Dickerhoff R, Dilloo D, Kremens B, von Stackelberg A, Vormoor J, Jürgens H Cooperative German-Austrian-Swiss Osteosarcoma Study Group (COSS) Primary metastatic osteosarcoma: presentation and outcome of patients treated on neoadjuvant cooperative osteosarcoma study group protocols. J. Clin. Oncol. 2003;21:2011–2018. [Google Scholar]
- 6.Brown HK, Tellez-Gabriel M, Heymann D. Cancer stem cells in osteosarcoma. Cancer Lett. 2017;386:189–195. doi: 10.1016/j.canlet.2016.11.019. [DOI] [PubMed] [Google Scholar]
- 7.Duan Z, Gao Y, Shen J, Choy E, Cote G, Harmon D, Bernstein K, Lozano-Calderon S, Mankin H, Hornicek FJ. miR-15b modulates multidrug resistance in human osteosarcoma in vitro and in vivo. Mol Oncol. 2017;11:151–166. doi: 10.1002/1878-0261.12015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang JW, Wu XF, Gu XJ, Jiang XH. Exosomal miR-1228 from cancer-associated fibroblasts promotes cell migration and invasion of osteosarcoma by directly targeting SCAI. Oncol Res. 2019;27:979–986. doi: 10.3727/096504018X15336368805108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang YM, Wang W, Qiu ED. Osteosarcoma cells induce differentiation of mesenchymal stem cells into cancer associated fibroblasts through Notch and Akt signaling pathway. Int J Clin Exp Pathol. 2017;10:8479–8486. [PMC free article] [PubMed] [Google Scholar]
- 10.Cortini M, Avnet S, Baldini N. Mesenchymal stroma: role in osteosarcoma progression. Cancer Lett. 2017;405:90–99. doi: 10.1016/j.canlet.2017.07.024. [DOI] [PubMed] [Google Scholar]
- 11.Kalluri R. The biology and function of exosomes in cancer. J Clin Invest. 2016;126:1208–1215. doi: 10.1172/JCI81135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ibrahim A, Marban E. Exosomes: fundamental biology and roles in cardiovascular physiology. Annu Rev Physiol. 2016;78:67–83. doi: 10.1146/annurev-physiol-021115-104929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang T, Deng CX. Current progresses of exosomes as cancer diagnostic and prognostic biomarkers. Int J Biol Sci. 2019;15:1–11. doi: 10.7150/ijbs.27796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Y, Zhao Z, Liu W, Li X. SNHG3 functions as miRNA sponge to promote breast cancer cells growth through the metabolic reprogramming. Appl Biochem Biotechnol. 2020;191:1084–1099. doi: 10.1007/s12010-020-03244-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ren J, Ding L, Zhang D, Shi G, Xu Q, Shen S, Wang Y, Wang T, Hou Y. Carcinoma-associated fibroblasts promote the stemness and chemoresistance of colorectal cancer by transferring exosomal lncRNA H19. Theranostics. 2018;8:3932–3948. doi: 10.7150/thno.25541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bai M, Lei Y, Wang M, Ma J, Yang P, Mou X, Dong Y, Han S. Long non-coding RNA SNHG17 promotes cell proliferation and invasion in castration-resistant prostate cancer by targeting the miR-144/CD51 axis. Front Genet. 2020;11:274. doi: 10.3389/fgene.2020.00274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu G, Hao C, Qi X, Nie J, Zhou W, Huang J, He Q. LncRNA SNHG17 aggravated prostate cancer progression through regulating its homolog SNORA71B via a positive feedback loop. Cell Death Dis. 2020;11:393. doi: 10.1038/s41419-020-2569-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu T, Yan S, Jiang L, Yu S, Lei T, Yang D, Lu B, Wei C, Zhang E, Wang Z. Gene amplification-driven long noncoding RNA SNHG17 regulates cell proliferation and migration in human non-small-cell lung cancer. Mol Ther Nucleic Acids. 2019;17:405–413. doi: 10.1016/j.omtn.2019.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma Z, Gu S, Song M, Yan C, Hui B, Ji H, Wang J, Zhang J, Wang K, Zhao Q. Long non-coding RNA SNHG17 is an unfavourable prognostic factor and promotes cell proliferation by epigenetically silencing P57 in colorectal cancer. Mol Biosyst. 2017;13:2350–2361. doi: 10.1039/c7mb00280g. [DOI] [PubMed] [Google Scholar]
- 20.Zhang H, Yue J, Jiang Z, Zhou R, Xie R, Xu Y, Wu S. CAF-secreted CXCL1 conferred radioresistance by regulating DNA damage response in a ROS-dependent manner in esophageal squamous cell carcinoma. Cell Death Dis. 2017;8:e2790. doi: 10.1038/cddis.2017.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang X, Chen Y, Zhao Z, Meng Q, Yu Y, Sun J, Yang Z, Chen Y, Li J, Ma T, Liu H, Li Z, Yang J, Shen Z. Engineered exosomes with ischemic myocardium-targeting peptide for targeted therapy in myocardial infarction. J Am Heart Assoc. 2018;7:e008737. doi: 10.1161/JAHA.118.008737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen J, Chen J, Cheng Y, Fu Y, Zhao H, Tang M, Zhao H, Lin N, Shi X, Lei Y, Wang S, Huang L, Wu W, Tan J. Mesenchymal stem cell-derived exosomes protect beta cells against hypoxia-induced apoptosis via miR-21 by alleviating ER stress and inhibiting p38 MAPK phosphorylation. Stem Cell Res Ther. 2020;11:97. doi: 10.1186/s13287-020-01610-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Faught E, Henrickson L, Vijayan MM. Plasma exosomes are enriched in Hsp70 and modulated by stress and cortisol in rainbow trout. J Endocrinol. 2017;232:237–246. doi: 10.1530/JOE-16-0427. [DOI] [PubMed] [Google Scholar]
- 24.Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17:272–283. doi: 10.1038/nrg.2016.20. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y, Zeng X, Wang N, Zhao W, Zhang X, Teng S, Zhang Y, Lu Z. Long noncoding RNA DANCR, working as a competitive endogenous RNA, promotes ROCK1-mediated proliferation and metastasis via decoying of miR-335-5p and miR-1972 in osteosarcoma. Mol Cancer. 2018;17:89. doi: 10.1186/s12943-018-0837-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu KP, Zhang CL, Ma XL, Hu JP, Cai T, Zhang L. Analyzing the interactions of mRNAs and ncRNAs to predict competing endogenous rna networks in osteosarcoma chemo-resistance. Mol Ther. 2019;27:518–530. doi: 10.1016/j.ymthe.2019.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Q, Huang J, Zhou N, Zhang Z, Zhang A, Lu Z, Wu F, Mo YY. LncRNA loc285194 is a p53-regulated tumor suppressor. Nucleic Acids Res. 2013;41:4976–4987. doi: 10.1093/nar/gkt182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Au Yeung CL, Co NN, Tsuruga T, Yeung TL, Kwan SY, Leung CS, Li Y, Lu ES, Kwan K, Wong KK, Schmandt R, Lu KH, Mok SC. Exosomal transfer of stroma-derived miR21 confers paclitaxel resistance in ovarian cancer cells through targeting APAF1. Nat Commun. 2016;7:11150. doi: 10.1038/ncomms11150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kitadai Y. Cancer-stromal cell interaction and tumor angiogenesis in gastric cancer. Cancer Microenviron. 2010;3:109–116. doi: 10.1007/s12307-009-0032-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simons M, Raposo G. Exosomes--vesicular carriers for intercellular communication. Curr Opin Cell Biol. 2009;21:575–581. doi: 10.1016/j.ceb.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 31.Pefanis E, Wang J, Rothschild G, Lim J, Kazadi D, Sun J, Federation A, Chao J, Elliott O, Liu ZP, Economides AN, Bradner JE, Rabadan R, Basu U. RNA exosome-regulated long non-coding RNA transcription controls super-enhancer activity. Cell. 2015;161:774–789. doi: 10.1016/j.cell.2015.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheng M, Yang J, Zhao X, Zhang E, Zeng Q, Yu Y, Yang L, Wu B, Yi G, Mao X, Huang K, Dong N, Xie M, Limdi NA, Prabhu SD, Zhang J, Qin G. Circulating myocardial microRNAs from infarcted hearts are carried in exosomes and mobilise bone marrow progenitor cells. Nat Commun. 2019;10:959. doi: 10.1038/s41467-019-08895-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, Chen D, Gu J, He X, Huang S. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25:981–984. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Du Y, Wei N, Hong J, Pan W. Long non-coding RNASNHG17 promotes the progression of breast cancer by sponging miR-124-3p. Cancer Cell Int. 2020;20:40. doi: 10.1186/s12935-020-1129-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H, Li T, Huang D, Zhang P. Long noncoding RNA SNHG17 induced by YY1 facilitates the glioma progression through targeting miR-506-3p/CTNNB1 axis to activate Wnt/beta-catenin signaling pathway. Cancer Cell Int. 2020;20:29. doi: 10.1186/s12935-019-1088-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu X, Zhang B, Jia Y, Fu M. SNHG17 enhances the malignant characteristics of tongue squamous cell carcinoma by acting as a competing endogenous RNA on microRNA-876 and thereby increasing specificity protein 1 expression. Cell Cycle. 2020;19:711–725. doi: 10.1080/15384101.2020.1727399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoon JH, Abdelmohsen K, Gorospe M. Posttranscriptional gene regulation by long noncoding RNA. J Mol Biol. 2013;425:3723–3730. doi: 10.1016/j.jmb.2012.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu J, Wan X, Chen X, Fang Y, Cheng X, Xie X, Lu W. miR-2861 acts as a tumor suppressor via targeting EGFR/AKT2/CCND1 pathway in cervical cancer induced by human papillomavirus virus 16 E6. Sci Rep. 2016;6:28968. doi: 10.1038/srep28968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qiu R, Li X, Qin K, Chen X, Wang R, Dai Y, Deng L, Ye Y. Antimetastatic effects of calycosin on osteosarcoma and the underlying mechanism. Biofactors. 2019;45:975–982. doi: 10.1002/biof.1545. [DOI] [PubMed] [Google Scholar]