Abstract
Objective: To explore the molecular mechanism by which hippocampal neural stem cell (HNSC) exosome (exo)-derived MIAT improves cognitive disorders in rats with vascular dementia (VD). Methods: Rat hippocampal tissues were collected, and HNSCs and hippocampal neuronal cells (HNCs) were isolated, purified, and identified. Then the exosomes (exo) of the HNSCs were extracted and identified. A VD rat model was constructed. HE staining was used to evaluate the hippocampal pathology in each group. The expressions of the RNAs in the HNSCs were intervened, and the cells were then grouped. ELISA was used to measure the of TNF-α, IL-1, and Aβ1-42 expression levels. The kits were used to determine the oxidative stress factor levels. The targeting relationships among MIAT, miR-34b-5p, and CALB1 were measured using dual-luciferase assays. The MIAT expressions in exo were measured using qRT-PCR. The proliferation and apoptosis of the HNCs were determined using CCK-8 and Annexin V-FITC/PI staining, respectively. The CALB1, TH, and Bcl-2 protein expressions were determined using Western blot. The Morris water maze test was used for the spatial learning and memory testing. Results: The hippocampal tissues in the model group were clearly damaged, but the pathological characteristics were significantly improved in the exo group. The exo group also showed an increased SOD level, decreased MDA and ROS levels, and down-regulated TNF-α, IL-1, and Aβ1-42 expressions (all P<0.05). MiR-34b-5p had a targeting relationship with both MIAT and CALB1, and MIAT was found to be expressed in exo. The oe-MIAT-exo group and the miR-34b-5p inhibitor group showed significantly up-regulated CALB1, TH, and Bcl-2 protein expressions in the HNCs, increased cell viability, as well as reduced apoptosis, but the si-MIAT-exo group showed the opposite results (all P<0.05). The MiR-34b-5p inhibitor partially reversed the effect on the si-MIAT-exo group. The miR-34b-5p mimic group showed significantly down-regulated CALB1, TH, and Bcl-2 protein expressions in the HNCs, inhibited cell viability, as well as increased apoptosis, but the oe-CALB1 group showed the opposite results (all P<0.05). Oe-CALB1 partially reversed the effect on the miR-34b-5p mimic group. The memory and learning abilities of the rats in the oe-MIAT-exo group and the model + exo group were significantly improved but not as much as they were in the normal rats. Conclusion: MIAT-containing exo from HNSCs can improve cognitive disorders in VD rats via the miR-34b-5p/CALB1 axis.
Keywords: MIAT, miR-34b-5p, CALB1, vascular dementia, exosome, hippocampal neural stem cells
Introduction
Vascular dementia (VD) the second most-common type of dementia, is caused by cardiovascular and cerebrovascular diseases, results in cognitive disorders, and is closely related to a decrease in and damage to hippocampal neuronal cells (HNCs) [1]. Hippocampal neural stem cells (HNSCs) are metrocytes with a strong differentiation ability. They can differentiate into HNCs, astrocytes, and oligodendrocytes, etc. HNSCs can also promote the renewal of brain tissue cells. With the continuous development of technology, stem cell therapy has been showing great repercussions in the market and has been used in treatments for many diseases [2,3]. Initially, the therapeutic mechanisms of stem cells were thought to have a strong differentiation ability, immune response, and cytokine secretions from the paracrine effect. Further discoveries showed that vesicles secreted by the paracrine effect also play a role in the treatments [4]. Exo is one of these vesicles. It has now been discovered that exo can interact with cells through secretions and the delivery of proteins and nucleotides. Exo can be used as diagnostic markers for many diseases, but its specific mechanism remains to be determined [5-7].
Studies have proved that long non-coding RNAs (LncRNAs) are involved in a variety of diseases, such as cancer, and metabolic, neurological, and skin diseases [8-11]. Gao et al. demonstrated that knocking down LncRNA SNHG1 could reduce amyloid-induced neuronal damage in Alzheimer’s dementia [12]. Zhuang et al. found decreased levels of LncRNA MALAT1 in the cerebrospinal fluid and plasma in patients with Alzheimer’s dementia [13]. The exo-derived LncRNA myocardial infraction association transcript (MIAT) is a gene located at chr22:27,042,392-27072441 (GRCh37/hg19). Moreover, a study showed that the loss of MIAT can lead to cerebral microvascular diseases, neurodegeneration, neuron loss, and Alzheimer’s dementia, suggesting that MIAT may be a key factor in neurovascular diseases [14]. The absence of MIAT may also promote the development of VD. Therefore, this study aimed to explore the biological effects of MIAT on the progression of VD.
LncRNAs affect physiological and pathological processes mainly through the competitive adsorption of microRNA to further regulate mRNAs, the mechanism of which has been studied in detail for many diseases [15]. In this study, we also further explored the downstream molecular pathways of MIAT and found that MIAT can regulate miR-34b-5p/CALB1. One study showed that treatment with miR-34 inhibitors can improve memory deficits in aged rats [16]. As a member of the superfamily of calcium-binding proteins, CALB1 widely presents in neurons. A study showed that the positive neurons of CALB1 are relatively reduced in Parkinson’s disease [17]. However, the role of MIAT in regulating the miR-34b-5p/CALB1 axis in VD remains to be investigated. Therefore, this study aimed to clarify the effect of HNSC exo-derived MIAT on improving cognitive disorders by regulating the miR-34b-5p/CALB1 axis in VD rats, in order to provide a new biomarker for the prevention and treatment of VD.
Materials and methods
Bioinformatics methods
We used the Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/geo/) to retrieve the microarray data of the VD-related expression profile by using “Vascular dementia” as the keyword, then GSE122063 was selected for the subsequent analysis. GSE122063 is the dataset of altered gene expressions in the aortas of 32-week ApoE-/- mice and wild-type mice. Meanwhile, the aorta expression profiles in ApoE-/- mice and wild-type mice were selected from GSE72248 to analyze the differentially expressed genes. The Affy package from the R programming language was used for the background corrections and normalization of the microarray data (http://www.bioconductor.org/packages/release/bioc/html/affy.html). The limma package (http://master.bioconductor.org/packages/release/bioc/html/limma.html) was used to standardize the microarray data and screen the differentially expressed genes (DEGs). The differential gene screening conditions were FDR<0.05 and |LogFoldChange|>1.5. The pheatmap package (https://cran.r-project.org/web/packages/pheatmap/index.html) was used to draw a heat map of the DEGs. “Calculate and draw custom Venn diagrams” (http://bioinformatics.psb.ugent.be/webtools/Venn/) was used to compare and analyze the different sets of data as well as to draw the Venn diagrams. The candidate miRNAs of the target genes were searched using miRDB (http://mirdb.org), miRWalk (http://mirwalk.umm.uni-heidelberg.de), and RNA22 (http://www.rna-society.org/raid/index.html). Then DisgeNET (http://www.disgenet.org/?1) was used to search for VD associated risk genes in the disease-related gene database. The interactions between the target gene and the protein were searched in the STRING database (https://string-db.org/). The visualization was implemented using the Cytoscape_v3.6.0 software.
Isolation and identification of the HNSCs and HNCs
This study was approved by the Medical Research Ethics Committee of Heilongjiang Provincial Hospital, and the animal experiments were carried out in strict accordance with the regulations of the Heilongjiang Provincial Hospital Committee on the Care and Use of Laboratory Animals, and we tried our best to alleviate the suffering of the animals. A total of 50 SD rats used in this experiment were provided by the Experimental Animal Center of Heilongjiang Provincial Hospital. The rats were sacrificed using intraperitoneal injections of 3% sodium pentobarbital (0.15 mL/100 g, Shanghai Sixin, China), then disinfected with 75% alcohol on the skin to harvest the brain tissues and the hippocampus using craniotomies. Part of each hippocampus was stored in liquid nitrogen for later use, and each remaining part was used to separate the HNSCs and HNCs. The meninges and blood vessels were removed in pre-cooled DMEM/F12 medium (Thermo Fisher, USA) and rinsed carefully. Then each hippocampus was cut into fragments of about 1 mm3. The HNSCs were separated using trypsin digestion. Trypsin (0.25%, Thermo Fisher, USA) was added for digestion to prepare the cell suspension which was filtered with a 200-mesh filter [18]. The HNCs were separated using trypsin-free mechanical pipetting. The hippocampus tissues were placed in a centrifuge tube, mixed with 4 mL of cell inoculum, pipetted with 1 mL and 200 μL pipettors. The pipetting was stopped when the clumpy tissues disappeared. The cells separated using the above two methods were centrifuged at 1000 r/min for 3 min, then the supernatant was removed, and a DMEF/F12 medium (containing 2% B27, 20 μg/L of epidermal growth factor and 20 μg/L of basic fibroblast growth factors) was added. Thereafter, the cells were cultivated in a 37°C, 5% CO2 incubator, then examined for the HNSC markers PE-CD133 (ab252128, ABCAM, UK), APC-Nestin (ab187846, ABCAM, UK), HNCs marker PE-CD24 (ab25494, ABCAM, UK, 0.2 µg for 106 cells), and FITC-Tyrosine hydroxylase (TH, 10009396-1, Cayman, USA) using flow cytometry [19].
Exo collection and identification in the HNSCs
The exo was extracted using ultracentrifugation. The HNSCs were cultured in a serum-free DMEF/F12 medium. After 24 hours, 225 mL of the HNSC conditioned medium was collected and centrifuged at 2000× g for 10 min at 4°C. The supernatant was centrifugated for another 10 min at 1000× g and 4°C, and another 3 h in a 60 Ti centrifuge (Beckman) at 126,000× g and 4°C. Then the sample was mixed with PBS containing a protease inhibitor and centrifuged for 1 h to obtain the exo-containing particles. The particles were resuspended in PBS containing protease inhibitors, identified and analyzed for the morphology of exo with a transmission electron microscope.
Construction of the animal model and the in vivo transfection
The VD rat model was established using bilateral common carotid artery occlusion [20]. A total of 50 SD rats weighing 200-230 g were used as the experimental animals, regardless of their sex. The rats were anesthetized using intraperitoneal injections of 250 mg/kg tribromoethanol, then placed supine on a constant temperature pad, shaved, had their skin disinfected, and a 3 cm incision was cut along the midline of the neck to expose the left and right common carotid arteries, which were ligated with 3-0 surgical sutures, then the incisions were sutured [21]. We applied the same operation in the sham group but without blocking the arteries. The survival rate of the model-operated rats was 73.5%, so 36 rats were used for the follow-up experiments. The experimental animals were divided into the sham group (n=9, with a tail vein injection of PBS equivalent to exo), the model group (n=9, with a tail vein injection of PBS equivalent to exo), the model + exo group (n=9, with a tail vein injection of 100 μg at 3*108 particles/μL in the modeled rats), and the oe-MIAT-exo group (n=9, with a tail vein injection of exo with overexpressed MIAT in the model rats) [22]. The sequence of the oe-MIAT was 5’-GACTTTATAGCGATCGAGCTCGATT-3’. The rats were anesthetized using pentobarbital sodium at a concentration of 40 mg/kg, and then the rats were sacrificed by having their necks broken. Their brain tissues were collected for the subsequent experiments. This study was conducted in accordance with the regulations for the management of experimental animals and was approved by the Animal Ethics Committee of our hospital (Approval No. (2021) 031).
HE staining
The brain tissues of the rats in each group were collected, routinely fixed, and embedded in paraffin. The embedded tissues were cut into 3 μm slices and prepared as paraffin sections. The sections were dewaxed using xylene, dehydrated using decreasing concentrations of ethanol, rinsed with distilled water for 2 min, and stained with a hematoxylin solution (Shanghai Gefan Biotechnology, China) for 10 min. The slices were immersed in tap water for 10 min, washed with distilled water, dehydrated with 95% ethanol, and then stained with an eosin solution (Shanghai Gefan Biotechnology, China) for 1 min. After being washed again with 70% ethanol, the slices were observed to determine the staining results under a microscope. Five fields of view were randomly selected to count the number of necrotic pyramidal nerve cells within 1 mm2 [23].
ELISA
In this experiment, ELISA was used to measure the tumor necrosis factor-α (TNF-α), interleukin-1 (IL-1), and amyloid (Aβ1-42) secretion levels in the rat serum. The blood samples from the abdominal aortae were collected, placed at 4°C for 30 min, then centrifugated at 4000 r for 10 min to obtain the serum samples. The measurements were conducted using TNF-α detection kits (LE-B0126), IL-1 detection kits, and Aβ1-42 detection kits from Hefei Laier, China. The experimental procedures were carried out in strict accordance with the kits’ instructions. A microplate reader (Thermo Fisher, USA) was used to measure the optical density (OD) at a wavelength of 450 nm. A curve of the OD values was plotted, and the concentration values were calculated.
Measurement of the oxidative stress levels
The oxidative stress levels were measured in the rat serum which was obtained using the same method we used with the ELISA. The superoxide dismutase (SOD), malondialdehyde (MDA), and reactive oxygen species (ROS) were used as evaluation indicators. The content of SOD was measured using WST-8 according to the instructions of the total SOD activity detection kit (Shanghai Beyotime, China), and the OD was measured at 450 nm. The MDA was measured using colorimetry according to the instructions of the MDA detection kit (Shanghai Beyotime, China), and the OD was determined with a microplate reader (Thermo Fisher, US) at 532 nm. The ROS content was determined using fluorescent probe DCFH-DA according to the instructions of the ROS detection kit (Shanghai Beyotime, China).
Western blot
The stored hippocampal tissues were ground and then mixed with a RIPA lysate (Beijing Soleibao, China) to extract the protein. The protein concentrations were determined using a BCA protein quantification kit (Shanghai Yisheng, China). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis was used to separate the total proteins at 45°C, and the separated proteins were then transferred to polyvinylidene fluoride membranes (Sigma-Aldrich, USA). The membranes were blocked in 5% skim milk and mixed with the primary antibodies CALB1 (1:3000, ab108404, Abcam, UK), TH (1:1000, ab137869, Abcam, UK), Bcl-2 (1:1000, Ab32124, Abcam, UK), and GAPDH (1:1000, ab181602, Abcam, UK) to incubate overnight at 4°C. The membranes were then washed with PBST, and we added horseradish peroxidase-labeled secondary antibody IgG (1:5000, ab190492, Abcam, UK) and incubate the membranes at 37°C for 2 h. Thereafter, an enhanced chemiluminescence reagent (Shanghai Yisheng, China) was used to visualize the protein signals. Image J software was used to analyze the gray values (the ratio of the gray values of the target proteins to the internal reference proteins).
FISH
FISH test kits (1002-30, Guangzhou Boxiny, China) were used to measure the MIAT localizations and expressions. The cells were routinely fixed and embedded. Then the pre-hybridization reactions, the hybridization reactions, and the staining of the cell nuclei with the fluorochromes DAPI were performed according to the kit instructions. The staining results were observed under a fluorescence microscope.
Dual-luciferase reporter assays
The online biological prediction tools predicted the downstream target miR-34b-5p for MIAT and the downstream target gene CALB1 for miR-34b-5p. A dual-luciferase reporter assay was designed according to the complementary base-pairing to verify the targeting relationships. The 3’UTR end of MIAT/CALB1 was inserted into the pGL3-basic vector to construct wild-type and mutant luciferase reporter plasmids. The reporter plasmids were then co-transfected with miR-34b-5p NC and miR-34b-5p mimics, and the transfection steps were carried out in accordance with the requirements of Lipofectamine 3000 (Thermo Fisher, USA). The medium was refreshed after 6 hours. The cells were washed with PBS after 48 hours and tested for luciferase activity according to the gene detection system of the dual-luciferase reporter assay (Shanghai, China). The Renilla luciferase activity was used as an internal reference.
qRT-PCR
Total RNA was extracted from the HNCs and exo using TRlzol reagent (Thermo Fisher, USA), and the purity of the RNA was measured using a spectrophotometer (Mettler-Toledo, Switzerland). Then cDNA was synthesized using reverse transcription kits (Shanghai Yisheng, China). Real-time PCR was conducted using 2× HieffCanace PCR Master Mix (Shanghai Yisheng, China). GAPDH was the internal reference for MIAT and CALB1, and U6 was the internal reference for miR-34b-5p. The relative expression levels were calculated using the 2-ΔΔCt formula. The primers were designed and synthesized by Shanghai Gema Gene, China. The sequences are shown in Table 1.
Table 1.
Primer sequence of qRT-PCR
| Gene | Primer sequence (5’-3’) | |
|---|---|---|
|
| ||
| Forward primer | Reverse primer | |
| MIAT | GGACGTTCACAACCACACTG | TCCCACTTTGGCATTCTAGG |
| miR-34b-5p | GGGTAGGCAGTGTCATTAGC | AACAACCAACACAACCCAAC |
| CALB1 | CTCCGACGGCAATGGGTAC | GGTGTTAAGTCCAAGCCTGCC |
| GAPDH | CACCCACTCCTCCACCTTTG | CCACCACCCTGTTGCTGTAG |
| U6 | GCCCTCTGTGCTACTTACTC | GCTGGTTGTGGGTTACTCTC |
Note: MIAT: myocardial infraction association transcript.
Collecting exo from the silenced and overexpressed genes
The exo were collected after the HNSCs were transfected with the plasmids of the silenced or overexpressed genes. The expression of RNAs was intervened in the HNCs. The cells were then divided into the following groups, the blank group (HNCs + empty-vector-exo), the NC group (HNCs + NC-vector-exo), the oe-MIAT-exo group (HNCs + oe-MIAT-exo), the si-MIAT-exo group (HNCs + si-MIAT-exo), the miR-34b-5p inhibitor group (HNCs + miR-34b-5p inhibitor), the miR-34b-5p mimic group (HNCs + miR-34b-5p mimic), the miR-34b-5p inhibitor + oe-MIAT-exo group (HNCs + miR-34b-5p inhibitor + oe-MIAT-exo), the oe-CALB1 group (HNCs + oe-CALB1), and the miR-34b-5p mimic + oe-CALB1 group (HNCs + miR-34b-5p mimic + oe-CALB1). The miR-34b-5p mimic and the miR-34b-5p inhibitor were purchased from Thermo Fisher, US. The recombinant plasmids were designed by Shanghai Gema Gene, China. Lipofectamine 3000 (Thermo Fisher, USA) was used for the plasmid transfection, and the operations were carried out in accordance with the kit instructions. The transfection sequences in this study are shown in Table 2.
Table 2.
Sequence of vectors
| NC | 5’-TCTATGCGAGCAGCGCGTC-3’ |
| oe-MIAT | 5’-GACTTTATAGCGATCGAGCTCGATT-3’ |
| si-MIAT | 5’-AACTGCTCGAATATCGCAGC-3’ |
| miR-34b-5p inhibitor | 5’-TAGCGAGCTCGCGAGCGAGCCT-3’ |
| miR-34b-5p mimic | 5’-GGCTATTAGCGCTTCGGCTTAT-3’ |
| oe-CALB1 | 5’-CATATCGAGGCACTTCTCGTC-3’ |
Note: MIAT: myocardial infraction association transcript.
CCK-8
The transfected HNCs were spread onto a 96-well plate for incubation. To each well, 10 μL of LCCK8 solution (Beijing Soleibao, China) was added at 24 h, 48 h, and 72 h. The plate was placed in a 5% CO2 incubator at 37°C for 4 h. Then, the OD value at 450 nm was measured using a microplate reader (Thermo Fisher, USA).
Annexin V-FITC/PI double labeling
Annexin V-FITC/PI cell apoptosis detection kits (Shanghai Qianchen, China) were used for this experiment. The cells were centrifuged at 300 g for 5 min at 4°C, washed with pre-cooled PBS, centrifuged again at 300 g for 5 min at 4°C, collected and resuspended in a 1× binding buffer. Then, the cells were mixed with 5 μL of Annexin V-FITC and 10 μL of PI staining solution, mixed and incubated in the dark for 15 min. 1× binding buffer was then added, and the cells were placed on ice, and examined for apoptosis with the use of a flow cytometer (Beckman Coulter, USA).
Water maze test
The Morris water maze test was used to evaluate the rats’ cognitive function [21]. The experiment was performed in a quiet and spacious room with a water maze basin (31 cm in depth, 94 cm in diameter) in the center of the room, and a FLOOR lamp with a red safety bulb in a corner of the room. A camera was fixed on the ceiling above the water maze basin and connected to a computer equipped with an automatic tracking system. A heater was used to keep the temperature of the water at around 23°C.
A circular platform was put in one of the quadrants, 15 cm from the edge. Then the safety light was turned on for testing. The rat was put in the water maze and observed to determine the time it took to find the platform within 90 s. If a rat could find the platform within 90 s, it was allowed to stay on the platform for 10 s and then moved out of the water maze. If a rat failed to find the platform within 90 s, it was manually guided to the platform and also stayed on the platform for 10 s. The experiments were performed 4 times a day (with four different quadrants as starting points) on each rat for 4 consecutive days. On the 5th day, the platform was removed, and a probe test was carried out by putting the rat at a random site in the basin, and we counted the number of times the rat crossed the original platform site within 90 seconds.
Statistical analyses
SPSS 23.0 software (IBM Corporation, USA) was used to conduct the data analyses in this study. The measurement data were expressed as the mean ± standard deviation. The comparisons between two groups were carried out using t tests, and the comparisons among multiple groups were done using one-way analyses of variance plus Tukey’s analysis. P<0.05 is considered to be statistically significant.
Results
Identification of the HNCs, HNSCs, and exo
The specific HNC and HNSC markers were identified using flow cytometry. The isolated HNCs were almost double positive for CD24 and TH (98.61%). See Figure 1A. The HNSCs were double positive for CD133 and Nestin (98.22%). See Figure 1B. The results indicated the high purity of the separated HNCs and HNSCs. The separated exo were observed in a transmission electron microscope and found to have elliptical shapes with bright outer edges, and they were mostly around 100-120 nm in size. See Figure 1C.
Figure 1.
Identification of the HNCs, HNSCs and exo. A: The identification of the HNCs using PE-CD24 and FITC-TH (double positive); B: The identification of the HNSCs sing PE-CD133 and APC-Nestin (double positive); C: The identification of exo (10000×, 40000×). N=3, and the experiment was repeated 3 times. HNSCs: hippocampal neural stem cells; HNCs: hippocampal neuronal cells; exo: exosome.
The exo treatment reduced the hippocampal pathological features
The HNSCs-derived exo were injected into the brains of the VD rats, and the pathological conditions in area CA1 of the hippocampi were observed using HE staining. It was found that the molecular layer (pentagonal), the pyramidal cell layer (rectangular), and the polymorphic layer (circular) showed clear boundaries and complete nuclei in the sham group. In the model group, the molecular layer, the pyramidal cell layer, and the polymorphic cell layer of the hippocampal tissues were disordered. The cell nuclei of the pyramidal layers were pykeptic, and the cells were dehydrated and necrotic (see the arrow) (P<0.05). However, after the exo (naïve) treatment, the boundaries between the cell layers became clear, and the pyramidal cell layer necrosis was reduced (P<0.05). See Figure 2.
Figure 2.
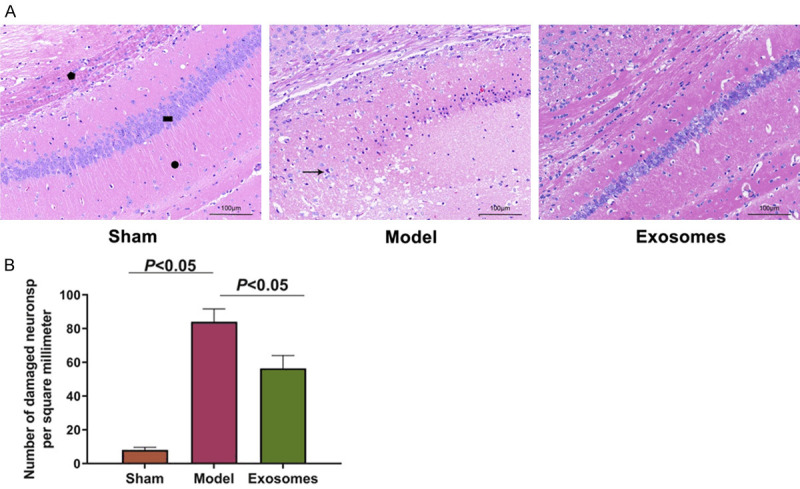
The pathological characteristics of the hippocampus (HE staining). A: The pathological results of the HE staining (100×); B: The number of necrotic pyramidal neurons per square millimeter. Pentagon: molecular layer; rectangle: pyramidal cell layer; circle: polymorphic cell layer; arrow: dehydration and pyknosis of pyramidal cell nucleus. N=9, and the experiment was repeated 3 times.
The exo treatment improved the oxidative stress and the inflammatory factor secretions
The role of the HNSCs-derived exo was clarified by measuring the oxidative stress and the secretions of the inflammatory factors (Figure 3). Compared with the sham group, the model group showed decreased SOD levels, increased MDA and ROS levels, and up-regulated TNF-α, IL-1, and Aβ1-42 expressions (all P<0.05). After the exo (naïve) treatment, the MDA, ROS, TNF-α, IL-1, and Aβ1-42 levels were all decreased, and the SOD level was increased (all P<0.05). The above results showed that exo can improve the various pathological indicators of VD rats.
Figure 3.
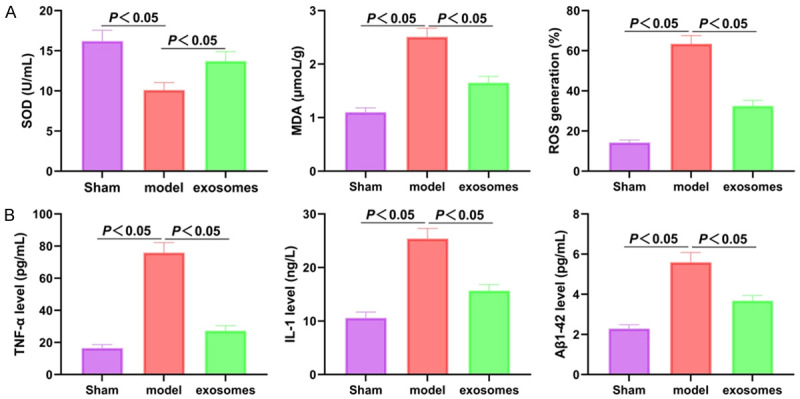
Exo improved the oxidative stress and secretion levels of the inflammatory factors in the VD rats. A: Comparison of SOD, MDA, and ROS in each group; B: Comparison of TNF-α, IL-1 and Aβ1-42 in each group. N=9, and the experiment was repeated 3 times. Exo: exosome; VD: vascular dementia; SOD: superoxide dismutase; MDA: malondialdehyde; ROS: reactive oxygen species; TNF-α: tumor necrosis factor-α; IL-1: interleukin-1; Aβ1-42: amyloid.
Exo improved brain inflammation and oxidative damage possibly via MIAT/miR-34b-5p/CALB1
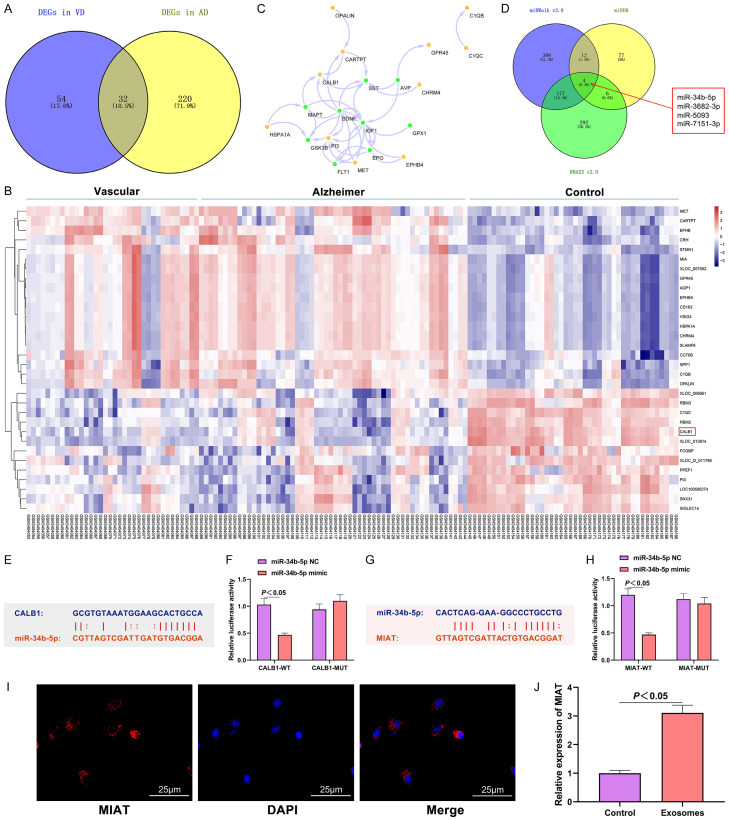
The possible mechanism of the HNSCs-derived exo regulating VD was analyzed with the use of molecular biology. The Limma package was used to analyze the differential genes (compared to the control) of Alzheimer’s dementia and VD in GSE122063 (|LogFC|>1.5, FDR<0.05). The mutual phenotypes of the two were analyzed to determine the differential genes that exist in both Alzheimer’s dementia and VD. See Figure 4A. A heat map representing the expression profile of the intersected genes was plotted to explore the protein interaction of the VD-associated risk genes. See Figure 4B and 4C. Among the above-mentioned intersected genes, CALB1, a member of the superfamily of calcium-binding proteins, was down-regulated in Alzheimer’s disease and VD compared with the controls, and CALB1 interacted more closely with the proteins of the VD associated genes MAPT, BDNF, and SST (Figure 4C). One study found that the superfamily of calcium-binding proteins is involved in the development of the hippocampus, and the low expressions of its members are related to neurodegenerative diseases such as Alzheimer’s disease [24].
Figure 4.
MIAT regulated the expression of CALB1 by adsorbing the miR-34b-5p. A: The intersection of the differential genes between Alzheimer’s disease and VD; B: Heat map of the expression profiles of the differential genes (horizontal axis: sample number; vertical axis: gene symbol); the red in the upper right corner indicates up-regulation, and the blue indicates down-regulation; C: The interaction network between the intersected genes and the VD risk gene set; the yellow circles represent the intersected genes; the green circles represent the VD risk genes; the arrows indicate the regulatory relationship; D: The miRNAs targeting CALB1 predicted by miRDB, miRWalk and RNA22; E: Complementary base-pairing of CALB1 and miR-34b-5p; F: Verification of the targeting relationship between CALB1 and miR-34b-5p; G: Complementary base-pairing of MIAT and miR-34b-5p; H: Verification of the targeting relationship between CALB1 and miR-34b-5p; I: Subcellular localization of MIAT in neural stem cells (400×); J: Expression of MIAT in exo. N=6, and the experiment was repeated 3 times. VD: vascular dementia; exo: exosome; MIAT: myocardial infraction association transcript.
We searched for the upstream targets based on the CALB1 protein using the prediction websites miRDB (http://mirdb.org), miRWalk (http://mirwalk.umm.uni-heidelberg.de), and RNA22 (http://www.rna-society. org/raid/index.html). We found miR-34b-5p, miR-3682-3p, miR-5093 and miR-7151-3p in all three websites (Figure 4D). Among them, the expressions of miR-34b were up-regulated in patients with Alzheimer’s disease, suggesting that the differential expression of miR-34b-5p might be related to the progression of VD [25]. Specific binding sites were found between miR-34b-5p and CALB1 (Figure 4E), and we verified their targeting relationship using a dual-luciferase reporter assay (Figure 4F).
During the search for the upstream target of miR-34b-5p and the exploration of the molecular pathway mechanism, we found complementary base-pairing between miR-34b-5p and MIAT on the biological information website (Figure 4G). The targeting relationship between the two was further verified using a dual-luciferase reporter assay (Figure 4H). The FISH results showed a co-localization of MIAT and the cytoplasms (Figure 4I). A high expression of MIAT was found in the exo of the HNSCs using qRT-PCR (Figure 4J). Through the above analyses, we speculated that the therapeutic effect of exo might come from the increased expression of MIAT which regulated VD through the molecular mechanism pathway miR-34b-5p/CALB1.
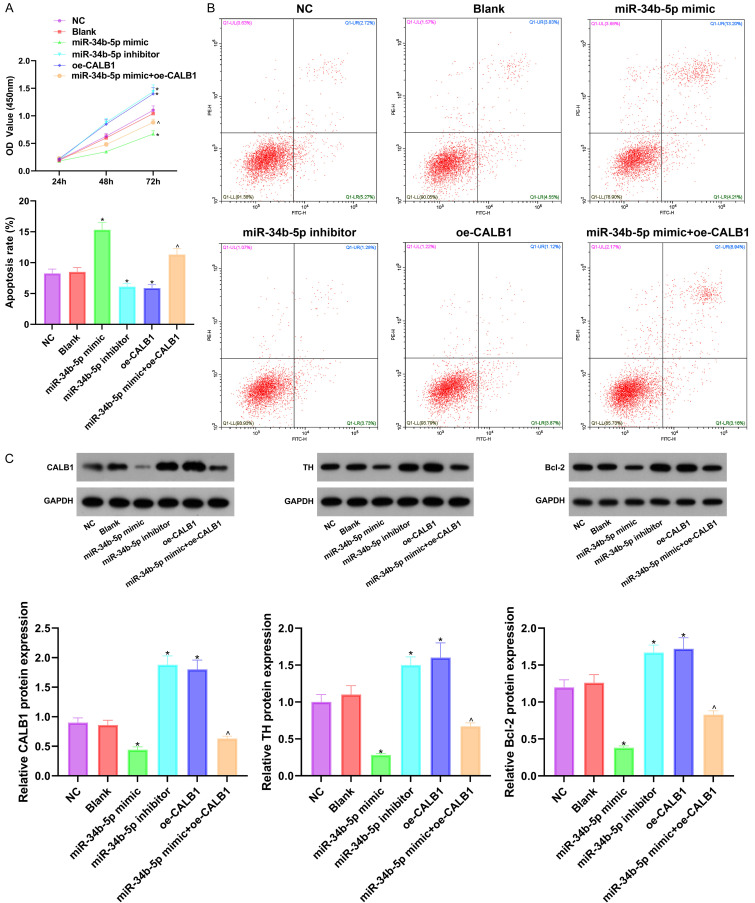
Exo from the HNSCs with MIAT knockdown damaged the HNCs, but the damage could be partially reversed by the miR-34b-5p inhibitor
After analyzing the possible mechanism of the HNSCs-derived exo regulating VD using bioinformatics, we further verified the biological effects of MIAT on neurological function (determined by CCK8, Western blot and Annexin V-FITC/PI double labeling). After overexpression or silencing MIAT in the HNSCs, the exo were collected and co-cultured with HNCs transfected with a miR-34b-5p inhibitor. Compared with the NC group, the oe-MIAT-exo group and the miR-34b-5p inhibitor group showed increased cell viability, up-regulated protein expressions of CALB1, TH, and Bcl-2, as well as reduced apoptosis, while the si-MIAT-exo group showed the opposite results (all P<0.05). After adding the miR-34b-5p inhibitor on the basis of si-MIAT-exo, the cell viability was increased; the CALB1, TH, and Bcl-2 protein levels were up-regulated, and the apoptosis was induced (Figure 5, all P<0.05), indicating that the effects of the si-MIAT-exo could be partially reversed by the miR-34b-5p inhibitor. The above experiments showed that the overexpression of MIAT can reduce the damage to the HNCs.
Figure 5.
The overexpression of MIAT can protect HNCs. A: Measuring the cell viability using CCK8; B: Measuring the apoptosis using Annexin V-FITC/PI double labeling; C: Measuring the protein expressions of CALB1, TH and Bcl-2 using Western blot. Compared with the NC group, *P<0.05; compared with the si-MIAT-exo group, #P<0.05. N=18, and the experiment was repeated 3 times. HNCs: hippocampal neuronal cells; MIAT: myocardial infraction association transcript.
miR-34b-5p damaged the HNCs, but the damage could be partially reversed by CALB1
The HNCs were injected with a miR-34b-5p mimic or inhibitor, transfected with CALB1 overexpressing plasmids and grouped. The results from the CCK8, Western blot and Annexin V-FITC/PI double labeling are shown in Figure 6. Compared with the NC group, the miR-34b-5p inhibitor group and the oe-CALB1 group showed increased HNC activity, and relatively increased CALB1, TH, and Bcl-2 protein levels, as well as decreased apoptosis, while the miR-34b-5p mimic group showed inhibited cell viability, decreased CALB1, TH, and Bcl-2 protein levels, as well as increased apoptosis. Compared with the miR-34b-5p mimic group, the miR-34b-5p mimic + oe-CALB1 group showed increased HNC activity, relatively increased CALB1, TH, and Bcl-2 protein levels, as well as decreased apoptosis (all P<0.05). The above experiments showed that the overexpression of miR-34b-5p can damage cells by inhibiting the viability of the HNCs and promoting apoptosis, but the damage can be partially reversed by CALB1.
Figure 6.
The overexpression of miR-34b-5p can inhibit the proliferation of HNCs and induce apoptosis, and the effects cab be partially reversed by CALB1. A: Measuring the cell viability using CCK8; B: Measuring the apoptosis using Annexin V-FITC/PI double labeling; C: Measuring the protein expressions of CALB1, TH, and Bcl-2 using Western blot. Compared with the NC group, *P<0.05; compared with the miR-34b-5p mimic group, ^P<0.05. N=18, and the experiment was repeated 3 times. HNCs: hippocampal neuronal cells.
Exo-derived MIAT significantly improves the learning abilities and the memories of VD rats
A VD rat model was established using bilateral common carotid artery occlusion. The MIAT-overexpressed exo or normal exo were co-cultured with HNSCs and injected into the modelled rats. A water maze experiment was used to observe the effect of the MIAT-overexpressed exo and normal exo on the memory and learning abilities of the VD rats. After the 5-day experiment, it was found that, compared with the sham group, the rats’ incubation periods were longer, and the number of times crossing the platform were reduced in the other three groups (all P<0.05). Compared with the model group, the incubation period was shorter, and the number of times crossing the platform were increased in the model + exo group and the oe-MIAT-exo group (all P<0.05). See Figure 7A and 7B. Then the expression of MIAT in the hippocampus was measured using qRT-PCR. The results showed that, compared with the sham group, the expression of MIAT decreased in the other three groups. Compared with the model group, the expression of MIAT was increased in both the model + exo group and the oe-MIAT-exo group (all P<0.05). See Figure 7C. The above results showed that the overexpression of both MIAT and exo can improve cognitive dysfunction and enhance learning abilities in VD rats. The mechanism is shown in Figure 8.
Figure 7.
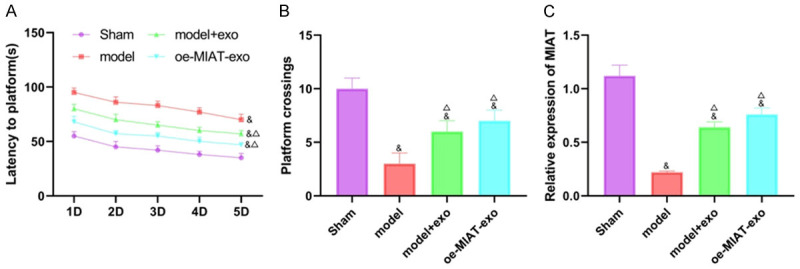
The effects of MIAT and exo on the VD rats measured with water maze tests. A: Comparison of the incubation periods; B: Comparison of the number of times crossing the platform; C: The mRNA levels of MIAT in the hippocampi. Compared with the Sham group, &P<0.05; compared with the model group, ΔP<0.05. N=12, and the experiment was repeated 3 times. VD: vascular dementia; exo: exosome.
Figure 8.
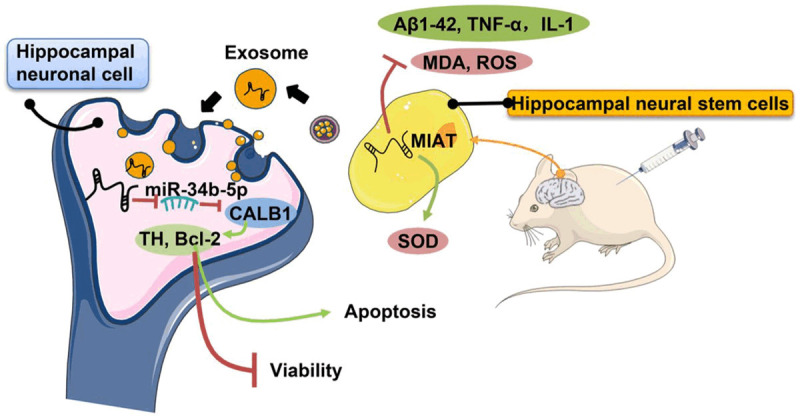
Exosome-derived MIAT from the hippocampal stem cells improves cognitive disorders in rats with vascular dementia through the miR-34b-5p/CALB1 axis. MIAT: myocardial infraction association transcript; SOD: superoxide dismutase; MDA: malondialdehyde; ROS: reactive oxygen species; TNF-α: tumor necrosis factor-α; IL-1: interleukin-1; Aβ1-42: amyloid.
Discussion
VD is a type of dementia second only to Alzheimer’s disease. People with cerebrovascular risks are likely to develop VD which threatens the quality of life of the elderly, especially among people over 65 years old [26]. LncRNAs regulate many diseases, including VD [27]. A previous study showed that the down-regulation of MIAT is associated with the pathogenesis of Alzheimer’s disease [14]. In this study, we have confirmed that MIAT exists in exo. We established a VD rat model using bilateral common carotid artery occlusion. After treatment with exo, the pathological characteristics of the rats were significantly improved. A previous study showed that the combination of receptors for advanced glycation end products and Aβ1-42 can mediate the inflammatory response, increase the expression of the pro-inflammatory factors (such as TNF-α), and accelerate the degeneration of nerve tissues [28]. This study also showed elevated expressions of these factors in the VD rats, and the secretions of these factors were controlled using exo treatment. Oxidative stress refers to excessive oxidation in the body which causes inflammatory stress, produces a large number of oxidative intermediates, and leads to various diseases and aging [29]. Our research also confirmed that exo treatment can improve the oxidative stress levels in VD rats. In addition, our in vivo experiments further proved that the overexpression of exo-derived MIAT can improve the memory and learning abilities of VD rats. Therefore, we believe that exo-derived MIAT is likely to be a new target for the treatment of VD.
In order to prove the mechanism of exo treatment for VD, we used online bioinformatics prediction tools to conduct research. The Gene Expression Omnibus database combined with miRDB, miRWalk and RNA22 predicted that MIAT can regulate miR-34b-5p and CALB1 in VD. The interaction of this molecular pathway was then verified using a dual-luciferase reporter assay. A type of miRNA, miR-34b-5p can be infiltrated by LncRNA, which inhibits the expression of miRNA to weaken its function. This mechanism has been confirmed in various studies [30,31]. The up-regulation of miR-34b is found in Parkinson’s disease and in multiple system atrophy [32]. The differential expression of miR-34b also plays a role in Alzheimer’s disease and VD [25]. In our study, it was proved that the overexpression of MIAT has a protective effect on neurological function, but this effect can be partially resisted by miR-34b-5p.
CALB1, a member of the superfamily of calcium-binding proteins, is a downstream target gene of miR-34b-5p. One study found that CHI3L2 might be involved in the progression of Alzheimer’s disease, but the expressions of CALB1 and CHI3L2 were negatively correlated, and the expression of CALB1 was correlated with age in patients with Alzheimer’s disease [33]. Studies have also shown that calcium binding proteins are involved in important developmental processes of the striatum, the hippocampal cortex, and the cerebellar cortex [34,35]. Our research found that miR-34b-5p can inhibit the growth of neuronal cells, but this effect is partially reversed by CALB1.
This study also has some limitations. The sample size was limited. Our In vivo experiments only verified the role of the exo-derived MIAT in improving VD through behavioral studies but did not measure the apoptosis or the oxidative stress levels. We will conduct future studies with improvements in the above aspects.
In summary, our experiment proves that exo can improve the pathological damage caused by VD, and exo treatment is achieved by MIAT regulating the miR-34b-5p/CALB1 axis, which suggests that MIAT has a protective effect on nerve function. Our study fills the vacancy of the effects of MIAT on the biological study of VD and provides a reference for the targeted therapy of VD.
Acknowledgements
This work was supported by the Heilongjiang Province Natural Joint Guidance Project (LH2021H064).
Disclosure of conflict of interest
None.
References
- 1.Zhou L, Yang R, Wu F. Efficacy and safety of butylphthalide as adjunctive therapy for vascular dementia: a protocol for systematic review and meta-analysis. Medicine (Baltimore) 2020;99:e23236. doi: 10.1097/MD.0000000000023236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li J, Shang Y, Wang L, Zhao B, Sun C, Li J, Liu S, Li C, Tang M, Meng FL, Zheng P. Genome integrity and neurogenesis of postnatal hippocampal neural stem/progenitor cells require a unique regulator filia. Sci Adv. 2020;6:eaba0682. doi: 10.1126/sciadv.aba0682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amin LE, Montaser M. Comparative evaluation of pulpal repair after direct pulp capping using stem cell therapy and biodentine: an animal study. Aust Endod J. 2021;47:11–19. doi: 10.1111/aej.12463. [DOI] [PubMed] [Google Scholar]
- 4.Li L, Wang YK, Yu XB, Bao YM, An LJ, Wei XW, Yu WT, Liu BY, Li JL, Yang JH, Xia Y, Liu G, Cao F, Zhang XZ, Zhao DW. Bone marrow mesenchymal stem cell-derived exosomes promote plasminogen activator inhibitor 1 expression in vascular cells in the local microenvironment during rabbit osteonecrosis of the femoral head. Stem Cell Res Ther. 2020;11:480. doi: 10.1186/s13287-020-01991-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Domenis R, Marino M, Cifù A, Scardino G, Curcio F, Fabris M. Circulating exosomes express α4β7 integrin and compete with CD4+ T cells for the binding to Vedolizumab. PLoS One. 2020;15:e0242342. doi: 10.1371/journal.pone.0242342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Y, Yin ZR, Fan JS, Zhang SY, Yang WB. The roles of exosomal miRNAs and lncRNAs in lung diseases. Signal Transduct Target Ther. 2019;4:47. doi: 10.1038/s41392-019-0080-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vlachakis D, Mitsis Τ, Nicolaides N, Efthimiadou A, Giannakakis A, Bacopoulou F, Chrousos GP. Functions, pathophysiology and current insights of exosomal endocrinology (review) Mol Med Rep. 2021;23:26. doi: 10.3892/mmr.2020.11664. [DOI] [PubMed] [Google Scholar]
- 8.Liu H, Zhang L, Ding X, Sui X. LINC00861 inhibits the progression of cervical cancer cells by functioning as a ceRNA for miR-513b-5p and regulating the PTEN/AKT/mTOR signaling pathway. Mol Med Rep. 2021;23:24. doi: 10.3892/mmr.2020.11662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhai HY, Yan RH, Yang TZ, Zhou ZH, Gao L, Li J. LncRNA BCAR4 up-regulates EGFR and thus promotes human thyrocyte proliferation. Neoplasma. 2019;66 doi: 10.4149/neo_2018_180105N12. 180105N12. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Y, Xia QM, Lin J. LncRNA H19 attenuates apoptosis in MPTP-induced Parkinson’s disease through regulating miR-585-3p/PIK3R3. Neurochem Res. 2020;45:1700–1710. doi: 10.1007/s11064-020-03035-w. [DOI] [PubMed] [Google Scholar]
- 11.Li HT, Yang C, Zhang J, Zhong W, Zhu L, Chen YF. Identification of potential key mRNAs and LncRNAs for psoriasis by bioinformatic analysis using weighted gene co-expression network analysis. Mol Genet Genomics. 2020;295:741–749. doi: 10.1007/s00438-020-01654-0. [DOI] [PubMed] [Google Scholar]
- 12.Gao Y, Zhang N, Lv C, Li N, Li X, Li W. lncRNA SNHG1 knockdown alleviates amyloid-β-induced neuronal injury by regulating ZNF217 via sponging miR-361-3p in Alzheimer’s disease. J Alzheimers Dis. 2020;77:85–98. doi: 10.3233/JAD-191303. [DOI] [PubMed] [Google Scholar]
- 13.Zhuang J, Cai P, Chen Z, Yang Q, Chen X, Wang X, Zhuang X. Long noncoding RNA MALAT1 and its target microRNA-125b are potential biomarkers for Alzheimer’s disease management via interactions with FOXQ1, PTGS2 and CDK5. Am J Transl Res. 2020;12:5940–5954. [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang Q, Shan K, Qun-Wang X, Zhou RM, Yang H, Liu C, Li YJ, Yao J, Li XM, Shen Y, Cheng H, Yuan J, Zhang YY, Yan B. Long non-coding RNA-MIAT promotes neurovascular remodeling in the eye and brain. Oncotarget. 2016;7:49688–49698. doi: 10.18632/oncotarget.10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y, Fu J, Yang L, Liang Z. Long non-coding RNA SNHG20 promotes colorectal cancer cell proliferation, migration and invasion via miR-495/STAT3 axis. Mol Med Rep. 2021;23:31. doi: 10.3892/mmr.2020.11669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hernandez-Rapp J, Rainone S, Hébert SS. MicroRNAs underlying memory deficits in neurodegenerative disorders. Prog Neuropsychopharmacol Biol Psychiatry. 2017;73:79–86. doi: 10.1016/j.pnpbp.2016.04.011. [DOI] [PubMed] [Google Scholar]
- 17.Mizuta I, Tsunoda T, Satake W, Nakabayashi Y, Watanabe M, Takeda A, Hasegawa K, Nakashima K, Yamamoto M, Hattori N, Murata M, Toda T. Calbindin 1, fibroblast growth factor 20, and alpha-synuclein in sporadic Parkinson’s disease. Hum Genet. 2008;124:89–94. doi: 10.1007/s00439-008-0525-5. [DOI] [PubMed] [Google Scholar]
- 18.Tian JY, Chen WW, Cui J, Wang H, Chao C, Lu ZY, Bi YY. Effect of lycium bararum polysaccharides on methylmercury-induced abnormal differentiation of hippocampal stem cells. Exp Ther Med. 2016;12:683–689. doi: 10.3892/etm.2016.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Monaco S, Baur K, Hellwig A, Hölzl-Wenig G, Mandl C, Ciccolini F. A flow cytometry-based approach for the isolation and characterization of neural stem cell primary cilia. Front Cell Neurosci. 2018;12:519. doi: 10.3389/fncel.2018.00519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Venkat P, Chopp M, Chen J. Models and mechanisms of vascular dementia. Exp Neurol. 2015;272:97–108. doi: 10.1016/j.expneurol.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim Y, Kim YJ. Effect of obesity on cognitive impairment in vascular dementia rat model via BDNF-ERK-CREB pathway. Biol Res Nurs. 2021;23:248–257. doi: 10.1177/1099800420951633. [DOI] [PubMed] [Google Scholar]
- 22.Gao BY, Zhou ST, Sun CC, Cheng DD, Zhang Y, Li XT, Zhang L, Zhao J, Xu DS, Bai YL. Brain endothelial cell-derived exosomes induce neuroplasticity in rats with ischemia/reperfusion injury. ACS Chem Neurosci. 2020;11:2201–2213. doi: 10.1021/acschemneuro.0c00089. [DOI] [PubMed] [Google Scholar]
- 23.Liu DH, Agbo E, Zhang SH, Zhu JL. Anticonvulsant and neuroprotective effects of paeonol in epileptic rats. Neurochem Res. 2019;44:2556–2565. doi: 10.1007/s11064-019-02874-6. [DOI] [PubMed] [Google Scholar]
- 24.Emmanuele V, Garcia-Cazorla A, Huang HB, Coku J, Dorado B, Cortes EP, Engelstad K, De Vivo DC, Dimauro S, Bonilla E, Tanji K. Decreased hippocampal expression of calbindin D28K and cognitive impairment in MELAS. J Neurol Sci. 2012;317:29–34. doi: 10.1016/j.jns.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barbagallo C, Mostile G, Baglieri G, Giunta F, Luca A, Raciti L, Zappia M, Purrello M, Ragusa M, Nicoletti A. Specific signatures of serum miRNAs as potential biomarkers to discriminate clinically similar neurodegenerative and vascular-related diseases. Cell Mol Neurobiol. 2020;40:531–546. doi: 10.1007/s10571-019-00751-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perera G, Rijnbeek PR, Alexander M, Ansell D, Avillach P, Duarte-Salles T, Gordon MF, Lapi F, Mayer MA, Pasqua A, Pedersen L, van Der Lei J, Visser PJ, Stewart R. Vascular and metabolic risk factor differences prior to dementia diagnosis: a multidatabase case-control study using European electronic health records. BMJ Open. 2020;10:e038753. doi: 10.1136/bmjopen-2020-038753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nuthikattu S, Milenkovic D, Rutledge J, Villablanca A. The western diet regulates hippocampal microvascular gene expression: an integrated genomic analyses in female mice. Sci Rep. 2019;9:19058. doi: 10.1038/s41598-019-55533-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.C RC, Lukose B, Rani P. G82S RAGE polymorphism influences amyloid-RAGE interactions relevant in Alzheimer’s disease pathology. PLoS One. 2020;15:e0225487. doi: 10.1371/journal.pone.0225487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gamage R, Wagnon I, Rossetti I, Childs R, Niedermayer G, Chesworth R, Gyengesi E. Cholinergic modulation of glial function during aging and chronic neuroinflammation. Front Cell Neurosci. 2020;14:577912. doi: 10.3389/fncel.2020.577912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen WJ, Zhai LL, Liu HM, Li YT, Zhang Q, Xu DD, Fan WY. Downregulation of lncRNA ZFAS1 inhibits the hallmarks of thyroid carcinoma via the regulation of miR-302-3p on cyclin D1. Mol Med Rep. 2021;23:2. doi: 10.3892/mmr.2020.11640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li XQ, Mo JX, Li J, Chen YL. lncRNA CASC2 inhibits lipopolysaccharide-induced acute lung injury via miR-27b/TAB2 axis. Mol Med Rep. 2020;22:5181–5190. doi: 10.3892/mmr.2020.11606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vallelunga A, Ragusa M, Di Mauro S, Iannitti T, Pilleri M, Biundo R, Weis L, Di Pietro C, De Iuliis A, Nicoletti A, Zappia M, Purrello M, Antonini A. Identification of circulating microRNAs for the differential diagnosis of Parkinson’s disease and multiple system atrophy. Front Cell Neurosci. 2014;8:156. doi: 10.3389/fncel.2014.00156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanfilippo C, Castrogiovanni P, Imbesi R, Di Rosa M. CHI3L2 expression levels are correlated with AIF1, PECAM1, and CALB1 in the brains of Alzheimer’s disease patients. J Mol Neurosci. 2020;70:1598–1610. doi: 10.1007/s12031-020-01667-9. [DOI] [PubMed] [Google Scholar]
- 34.Bernácer J, Prensa L, Giménez-Amaya JM. Distribution of GABAergic interneurons and dopaminergic cells in the functional territories of the human striatum. PLoS One. 2012;7:e30504. doi: 10.1371/journal.pone.0030504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Flace P, Lorusso L, Laiso G, Rizzi A, Cagiano R, Nico B, Ribatti D, Ambrosi G, Benagiano V. Calbindin-D28K immunoreactivity in the human cerebellar cortex. Anat Rec (Hoboken) 2014;297:1306–1315. doi: 10.1002/ar.22921. [DOI] [PubMed] [Google Scholar]