Abstract
Purpose: Inflammation out of control may induce many diseases. Baicalin has certain anti-inflammatory effects, but its mechanism of action is not clear. Therefore, this study was designed to explore a potential mechanism of anti-inflammation. Methods: In this study, RAW264.7 cells were induced by 1.0 g/mL lipopolysaccharide (LPS) and then exposed to baicalin at various concentrations (0.1-1.0 μmol/L). Then, we investigated the effect of baicalin in RAW264.7 inflammation models. Results: In this study, 0.1-1.0 μmol/L baicalin, especially baicalin at 1.0 μmol/L, effectively inhibited the expression of inflammatory factors (TNF-α, IL-1β, IL-6, Cox, and iNOS), decreased the activity of High Mobility Group Box 1 (HMGB1)/Toll-like Receptor 4 (TLR4)/nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) pathway, and stimulated miR-181b expression. HMGB1 was proved to be negatively regulated by miR-181b. Here, up-regulation of miR-181b or down-regulation of HMGB1 exerted similar effects as baicalin and down-regulated miR-181b reversed the anti-inflammatory effect of baicalin in RAW264.7 inflammation models. Conclusion: Baicalin can inhibit LPS-induced inflammation in RAW264.7 cells via the miR-181b/HMGB1/TRL4/NF-κB pathway.
Keywords: Murine macrophage RAW264.7, inflammation, baicalin, miR-181b/HMGB1/TRL4/NF-κB pathway
Introduction
Inflammation refers to physiological responses to external stimuli such as infection, injury, or chemical stimulus [1]. It is classified into acute inflammation and chronic inflammation according to its duration. Chronic inflammation lasts longer than acute inflammation and involves immune cells, leading to angiogenesis, tissue fibrosis, and necrosis [2]. The inflammatory response induced by lipopolysaccharide (LPS) is chronic and is closely related to the Toll-like receptor (TLR) family. For example, LPS induces macrophages to secrete factors from the TLRs, thereby activating the expression or abnormal activity of mediators including tumor necrosis factor-α (TNF-α), and ultimately initiating inflammatory responses in endothelial cells [3]. Many studies [4-8] have revealed that macrophages are crucial during inflammation, so regulation of macrophages may relieve inflammation. High-mobility group box 1 (HMGB1) is a protein encoded by the HMGB1 gene, containing the HMG-box domain, which is secreted by immune cells and can bind to TLR4 protein to affect the inflammatory response [9-11].
MiR-181b is a microRNA located on human chromosome 2, which can regulate gene expression via its target binding to mRNA at the post-transcriptional level. A previous study [12] suggests that the up-regulation of miR-181b is related to the endotoxin resistance of RAW264.7 and can suppress excessive immune response induced by LPS. Therefore, miR-181b is essential in the inflammatory response network.
Baicalin is a flavonoid compound extracted from the roots of Scutellaria baicalensis [13]. A growing number of studies have confirmed the anti-inflammatory role of baicalin. The research by Wu et al. [14] found that baicalin alleviated cellular inflammation caused by mycoplasma infection through TLR2/NF-κB (Nuclear factor kappa light chain enhancer of activated B cells). In the study by Ishfaq et al. [15], they also noted that baicalin inhibited inflammation and apoptosis caused by mycoplasma infection through regulating energy metabolism. Ishfaq et al. [16] suggested that baicalin regulated oxidative stress and apoptosis caused by mycoplasma infection through NF-κB-mediated signaling pathways. Another study [17] suggested that baicalin might relieve LPS-induced inflammation in macrophages via the TLR4/NF-κB pathway. But the mechanism underlying the regulation of baicalin on macrophage inflammation is not clear. MiRNAs are important cell regulators, and baicalin may modulate miRNAs to regulate inflammation [18].
Here, we established RAW264.7 cell models (murine macrophages) of inflammation induced by 1.0 g/ml lipopolysaccharide (LPS) and exposed cells to baicalin at various concentrations (0.1-1.0 μmol/L). We examined the effects of different doses of baicalin on inflammation and explored the mechanism of baicalin in RAW264.7 inflammation models, aiming to identify the potential value of baicalin in inflammation mitigation and treatment.
Methods
RAW264.7 cell models of inflammation induced by LPS
We purchased murine macrophages (RAW264.7) from the American Type Culture Collection (ATCC) cell bank and cultured them with RPMI1640 (Cell Biotechnology Saba Arna, Fars, Iran) basal medium supplemented with 10% fetal bovine serum (FBS; Invitrogen, Thermo Fisher Scientific, Inc., Waltham, Massachusetts, USA) and antibiotics (1%) until they grew well. Then, we seeded RAW264.7 cells into the 6-well plate and added 1 g/ml LPS (Sigma-Aldrich, Inc., St. Louis, MO, USA) into each well. After 24 hours, we examined whether the RAW264.7 inflammation models were successfully constructed. MiR-181b mimic (5’-AACAUUCAUUGCUGUCGGUGGGU-3’)/inhibitor (5’-ACCCACCGACAGCAAUGAAUGUU-3’) was used to up/down-regulate the expression of miR-181b, and HMGB1 siRNA to downregulate the expression of HMGB1. The miR-181b mimic/inhibitor and HMGB1 siRNA (5’-GGUUCUGUGUCCUAGGAUAAC-3’) were purchased from Shanghai Sangon Biotech.
Exposure of RAW264.7 inflammation models to baicalin
Successful RAW264.7 models of inflammation induced by LPS were exposed to baicalin (Sigma, St. Louis, Missouri, USA) at different concentrations (0.1-1.0 μmol/L). After 24 hours, we tested the inflammatory responses, cell activity, and HMGB1/TRL4/NF-κB pathway activity in RAW264.7 cells.
qPCR assay
We prepared cell suspension, isolated the total RNA with the Trizol reagent (Invitrogen, Carlsbad, CA, USA), and quantified its purity. Then, miR-181b and HMGB1 mRNAs were subjected to reverse transcription and amplification. The primer sequences were designed and synthesized by Sangon Biotech (Shanghai, China). Reverse transcription and qPCR amplification kits were purchased from Solarbio (Beijing, China). All procedures were carried out following the kit manual. After determining the Ct value of samples, we calculated the expression levels with the 2-ΔΔt method. The sequences of primers are shown in Table 1.
Table 1.
Primer sequences
| Gene | Upstream primer (5’-3’) | Downstream primer (5’-3’) |
|---|---|---|
| miR-181b | CCAGCTGGGCTCACTGAACAATGA | CAACTGGTGTCGTGGAGTCGGC |
| HMGB1 | TCAAAGGAGAACATCCTGGCCTGT | CTGCTTGTCATCTGCAGCAGTGTT |
| U6 | CTCGCTTCGGCAGCACA | TGGTGTCGTGGAGTCG |
| GAPDH | TCAACGACCACTTTGTCAAGCTCA | GCTGGTGGTCCAGGGGTCTTACT |
Western blot assay
We prepared cell suspension and lysed cells with the RIPA lysis buffer (Cell Signaling Technology, Inc., Danvers, MA, USA), followed by centrifugation for 20 minutes. We discarded the precipitate and collected the supernatant for testing the protein concentration using the BCA kit (Thermo Fisher, Waltham, MA, USA). The protein was subjected to electrophoresis separation with the sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (Solarbio, Beijing, China) and then transferred to a polyvinylidene fluoride membrane (Solarbio, Beijing, China). We mixed the protein to be tested with the primary antibodies and kept them still over 12 h at 4°C. After being washed 3 times with PBS buffer, the membrane was incubated with goat anti-rabbit secondary antibody for 1 hour. Finally, after PBS washing, the membrane was mixed with the ECL kit. β-actin and LaminB were chosen as internal reference protein controls. The proteins to be tested were acquired from Abcam (Cambridge, MA, USA). The proteins to be tested include HMGB1 (1:500), TLR4 (1:1000), phosphorylation of IκB (1:1000), phosphorylation of p65 (1:1000), nuclear p65 (1:1000), TNF-α (1:1000), IL-1β (1:1000), IL-6 (1:1000), COX (1:500) and iNOS (1:1000). All antibodies were purchased from Abcam Company (USA).
MTT assay
We seeded RAW264.7 cells with inflammation into 12 wells of a 96-well plate and equally divided the 12 wells into 4 groups: LPS group, LPS+0.1 μmol/L baicalin group, LPS+0.5 μmol/L baicalin group, and LPS+1.0 μmol/L baicalin group. After 24 hours, we added 10 μL MTT solutions (5 mg/ml) into each well and cultured at 37°C for 4 hours (5% CO2). Then we removed the medium, added 100 μL dimethyl sulfoxide (Beijing Solarbio Science & Technology Co., Ltd., Beijing, China) into each well and shook it well. Finally, we measured the OD value of each well at 570 nm with a microplate reader (Thermo Fisher Scientific).
Dual-luciferase reporter assay
The sequences of miR-181b and HMGB1 mRNAs were analyzed using Targetscan7.2. We constructed wild-type HMGB1 (with miR-181b binding site) and mutant-type HMGB1 (without HMGB1 binding site) using the pmirGLO vector. Then we transfected the vectors separately into cells and tested the luciferase intensity using the dual-luciferase reporter assay system (Promega, Madison, WI, USA). The 293T cell lines were used for dual-luciferase reporter assay. The mutant sequence of HMGB1 (5’-3’): UUAAAGAAGACCUGA-CGGUCUAU.
Apoptosis
The apoptosis of bladder cancer cells was detected with an Annexin V-FITC/PI fluorescence double staining kit (Procell, Wuhan, China) and flow cytometer (BD Biosciences, USA). First, cell suspension was prepared, followed by 5 min centrifugation, and then the supernatant was discarded. Subsequently, 500 μL 1× Annexin V binding buffer, 5 μL Annexin V-FITC and 5 μL propidium iodide were successively added into the solution, followed by 20 min incubation at room temperature in the dark. Finally, the cell apoptosis was analyzed with a flow cytometer and CellQuest software (BD Biosciences).
Statistical analysis
All tests were repeated 3 times. SPSS20.0 software (IBM, Armonk, NY, USA) was used for statistical analysis. The results were expressed as mean ± standard deviation (mean ± SD). The independent sample t-test was used for the comparison of data between any two groups, and one-way ANOVA followed with LSD t-test for the comparison among multiple groups. The confidence interval in this study was 95%. P<0.05 was regarded as statistically significant.
Results
LPS induced inflammation and oxidative stress in RAW264.7 cells
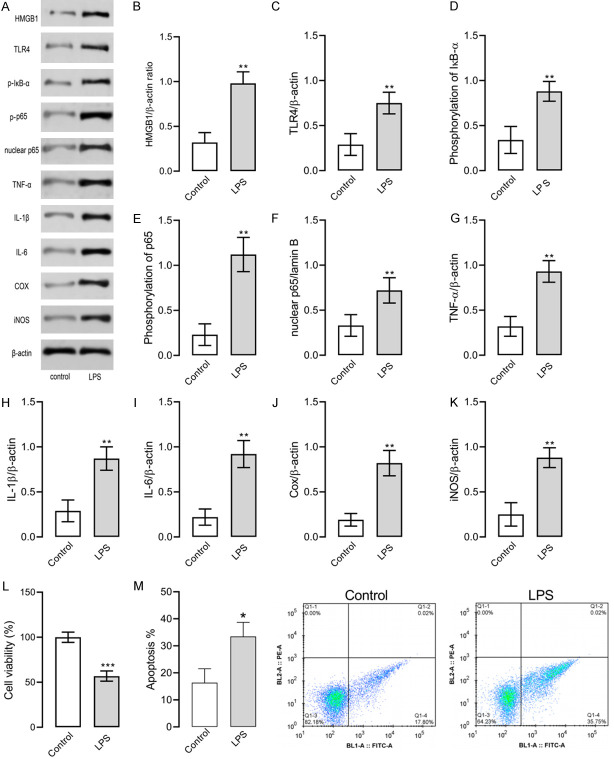
Here we established RAW264.7 cell models of inflammation induced by 1 g/ml LPS. Then we tested cell viability with the MTT assay, and the levels of TNF-α, IL-1β, IL-6, Cox, iNOS and proteins involved in the HMGB1/TLR4/NF-κB pathway were determined with the Western blot assay. Figure 1 illustrates that LPS treatment decreased cell viability, induced apoptosis, increased concentration of proteins involved in the HMGB1/TLR4/NF-κB pathway, and up-regulated the expression of TNF-α, IL-1β, IL-6, Cox, and iNOS in RAW264.7 cells. Such results indicate that the RAW264.7 cell model of inflammation was successfully constructed.
Figure 1.
RAW264.7 cell models of inflammation induced by LPS. A. Western blot results, and P-IκB means phosphorylation of IκB and P-p65 means phosphorylation of p65. B. HMGB1 protein level. C. TLR4 protein level. D. IκB-α phosphorylation level. E. NF-κB p65 phosphorylation level. F. Nuclear NF-κB p65 level. G. TNF-α level. H. IL-1β level. I. IL-6 level. J. Cox level. K. iNOS level. L. Cell viability. M. Apoptosis of each group. ** indicates P<0.01 and *** indicates P<0.001 when compared with the control group. The experiment was repeated 3 times (n=3).
Baicalin inhibited LPS-induced inflammation and oxidative stress in RAW264.7 cells
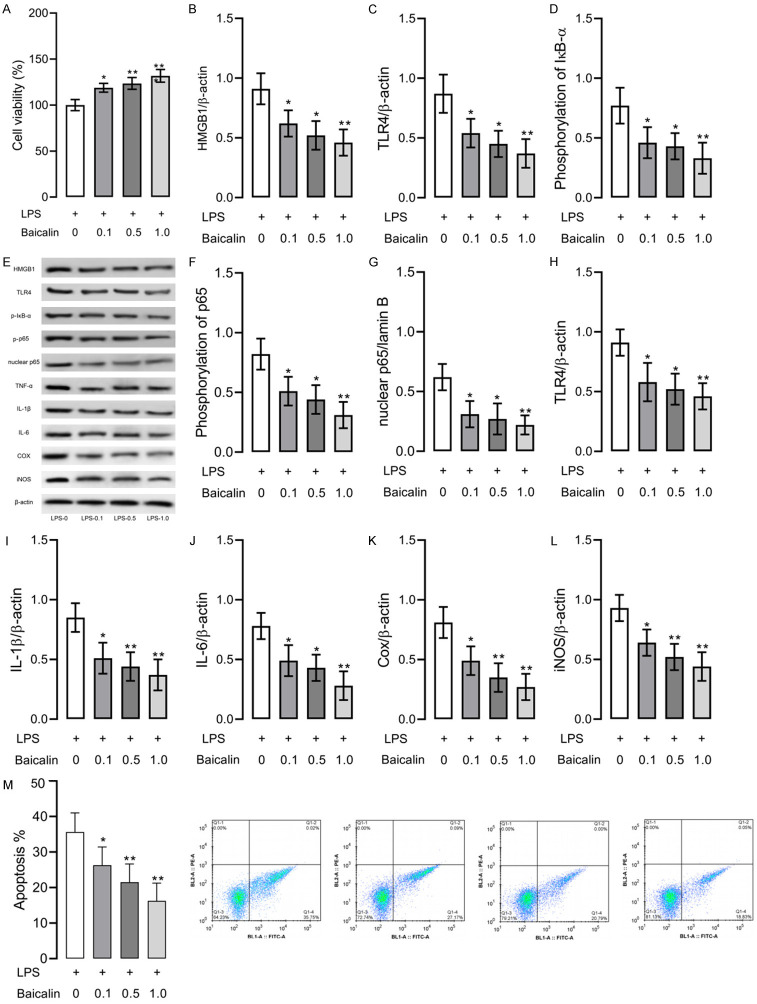
Successful RAW264.7 models of inflammation induced by LPS were exposed to baicalin at different concentrations (0.1-1.0 μmol/L). Figure 2 illustrates that RAW264.7 cells exposed to baicalin garnered higher cell activity, lower concentrations of proteins involved in the HMGB1/TLR4/NF-κB pathway, and lower expression of TNF-α, IL-1β, IL-6, Cox, and iNOS, as compared with cells not exposed to baicalin, with baicalin at 1.0 μmol/L showing the strongest inhibition of inflammation and apoptosis in cells. Such results indicate that baicalin can effectively control LPS-induced inflammation in RAW264.7 cells and suppress the activity of the HMGB1/TLR4/NF-κB pathway.
Figure 2.
RAW264.7 cell models of LPS-induced inflammation exposed to various baicalin concentrations (0.1-1.0 μmol/L). A. Cell viability. B. HMGB1 protein level. C. TLR4 protein level. D. IκB-α phosphorylation level. E. Western blot results, and P-IκB means phosphorylation of IκB and P-p65 means phosphorylation of p65. F. NF-κB p65 phosphorylation level. G. Nuclear NF-κB p65 level. H. TNF-α level. I. IL-1β level. J. IL-6 level. K. Cox level. L. iNOS level. M. Apoptosis of each group. * indicates P<0.05, ** indicates P<0.01 and *** indicates P<0.001 when compared with the LPS group. The experiment was repeated 3 times (n=3).
HMGB1 is a target gene of miR-181b
Figure 3A shows that different doses of baicalin can increase miR-181b expression in RAW264.7 cells. Figure 3B and 3C show that up-regulated miR-181b caused down-regulation of HMGB1 mRNA. Figure 3D indicates that there are binding sites of miR-181b in the 3’ non-transcribed region of HMGB1. Figure 3E illustrates that the co-transfection of HMGB1_wt and miR-181b mimics resulted in the decrease in relative luciferase activity, which imply that HMGB1 could bind to miR-181b via predictive site. The above results suggest that HMGB1 is a target gene of miR-181b.
Figure 3.
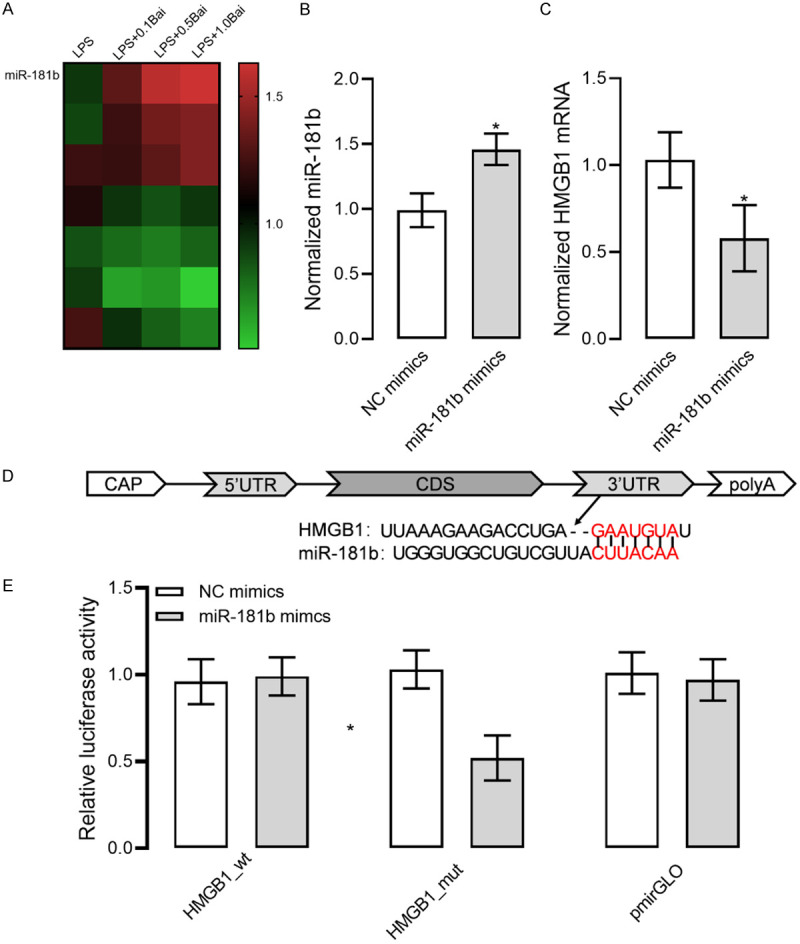
HMGB1 as a target gene of miR-181b. A. Increased miR-181b expression in RAW264.7 cell models of LPS-induced inflammation exposed to baicalin. B. Up-regulated miR-181b expression in RAW264.7 cells. C. Decreased HMGB1 mRNA expression in RAW264.7 cells with up-regulated miR-181b. D. Targetscan7.2 prediction of the targeting relationship between miR-181b and HMGB1 mRNA. E. Results of the dual-luciferase reporter assay. * indicates P<0.05 when compared with the NC mimics group. The experiment was repeated 3 times (n=3).
Upregulated miR-181b inhibited inflammation in RAW264.7 cells
Here we discovered that baicalin increased miR-181b expression in cells, so we up-regulated miR-181b expression to determine its effect on LPS-induced RAW264.7 cells and HMGB1/TLR4/NF-κB pathway. As shown in Figure 4, up-regulation of miR-181b led to decreased levels of HMGB1, TLR4, nuclear NF-κB p65, TNF-α, IL-1β, IL-6, Cox, and iNOS, as well as lowered levels of phosphorylated NF-κB p65 and IκB-α. Such results indicate that up-regulated miR-181b can inhibit LPS-induced inflammation and apoptosis in RAW264.7 cells and suppress the activity of the HMGB1/TLR4/NF-κB pathway.
Figure 4.
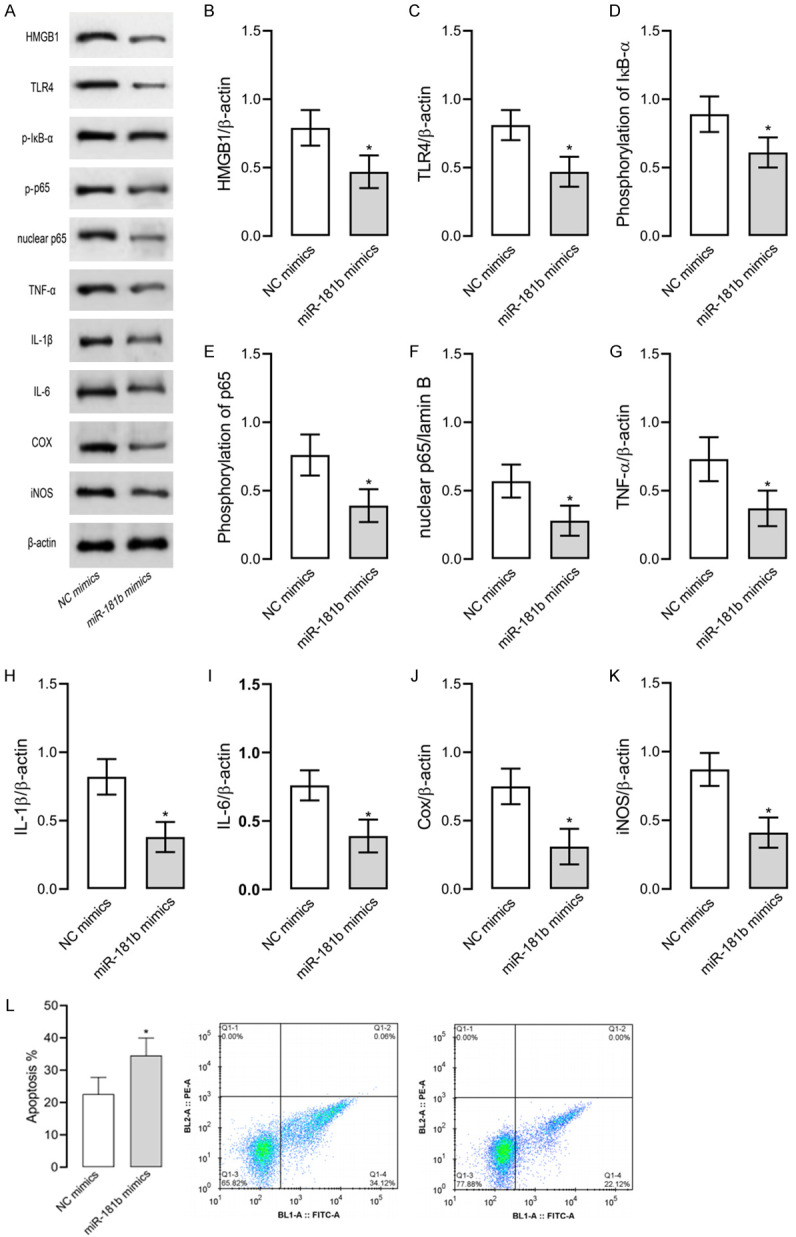
Up-regulated miR-181b in RAW264.7 models of inflammation. Up-regulation of miR-181b leads to decreased levels of HMGB1, TLR4, nuclear NF-κB p65, TNF-α, IL-1β, IL-6, Cox, and iNOS, as well as lowered NF-κB p65 and IκB-α phosphorylation levels. A. Western blot results. P-IκB means phosphorylation of IκB and P-p65 means phosphorylation of p65. B. HMGB1 protein level. C. TLR4 protein level. D. IκB-α phosphorylation level. E. NF-κB p65 phosphorylation level. F. Nuclear NF-κB p65 level. G. TNF-α level. H. IL-1β level. I. IL-6 level. J. Cox level. K. iNOS level. L. Apoptosis of each group. * indicates P<0.05 when compared with the NC mimics group. The experiment was repeated 3 times (n=3).
Downregulated HMGB1 inhibited inflammation in RAW264.7 cells
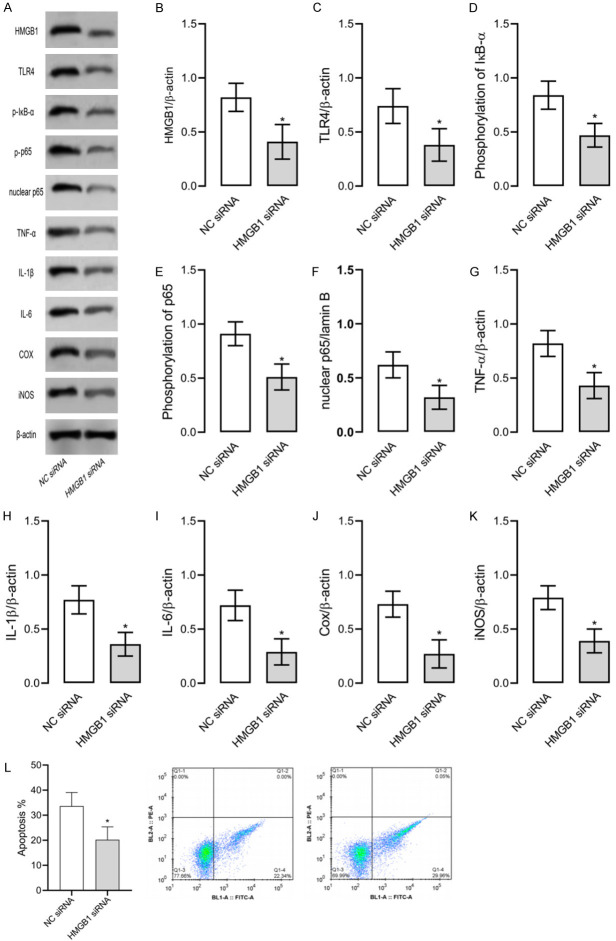
Here we discovered that baicalin decreased HMGB1 expression in cells, so we down-regulated HMGB1 expression to determine its effect on LPS-induced RAW264.7 cells and HMGB1/TLR4/NF-κB pathway. As shown in Figure 5, down-regulation of HMGB1 led to decreased levels of HMGB1, TLR4, nuclear NF-κB p65, TNF-α, IL-1β, IL-6, Cox, and iNOS, as well as phosphorylated NF-κB p65 and IκB-α. Such results indicate that down-regulation of HMGB1 can inhibit LPS-induced inflammation and apoptosis in RAW264.7 cells and suppress the activity of the HMGB1/TLR4/NF-κB pathway.
Figure 5.
Down-regulated HMGB1 expression in RAW264.7 models of inflammation. Down-regulation of HMGB1 leads to decreased levels of HMGB1, TLR4, nuclear NF-κB p65, TNF-α, IL-1β, IL-6, Cox, and iNOS, as well as lowered NF-κB p65 and IκB-α phosphorylation levels. A. Western blot results. P-IκB means phosphorylation of IκB and P-p65 means phosphorylation of p65. B. HMGB1 protein level. C. TLR4 protein level. D. IκB-α phosphorylation level. E. NF-κB p65 phosphorylation level. F. Nuclear NF-κB p65 level. G. TNF-α level. H. IL-1β level. I. IL-6 level. J. Cox level. K. iNOS level. L. Apoptosis of each group. * indicates P<0.05 when compared with the NC mimics group. The experiment was repeated 3 times (n=3).
Baicalin inhibited LPS-induced inflammation in RAW264.7 cells via miR-181b/HMGB1/TLR4/NF-κB pathway
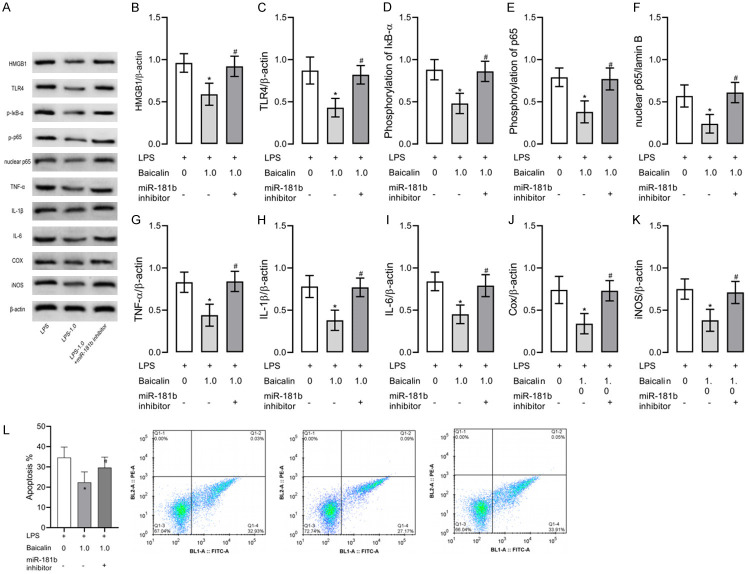
Here we exposed LPS-induced RAW264.7 cells to 1.0 μmol/L baicalin and down-regulated miR-181b expression in cells (Figure 6). Down-regulation of miR-181b reversed the inhibitory effect of 1.0 μmol/L baicalin on cell inflammation, apoptosis and HMGB1/TLR4/NF-κB pathway. These results indicate that baicalin can inhibit LPS-induced inflammation in RAW264.7 cells through the miR-181b/HMGB1/TLR4/NF-κB pathway (Figures 7, 8).
Figure 6.
RAW264.7 cell models of inflammation with down-regulated miR-181b expression and exposed to 1.0 µmol/L baicalin. Down-regulation of miR-181b reverses the inhibition on cell inflammation and HMGB1/TLR4/NF-κB pathway by 1.0 µmol/L baicalin. A. Western blot results. P-IκB means phosphorylation of IκB and P-p65 means phosphorylation of p65. B. HMGB1 protein level. C. TLR4 protein level. D. IκB-α phosphorylation level. E. NF-κB p65 phosphorylation level. F. Nuclear NF-κB p65 level. G. TNF-α level. H. IL-1β level. I. IL-6 level. J. Cox level. K. iNOS level. L. Apoptosis of each group. * indicates P<0.05 when compared with the LPS group. # indicates P<0.05 when compared with the LPS+1.0 µmol/L baicalin group. The experiment was repeated 3 times (n=3).
Figure 7.
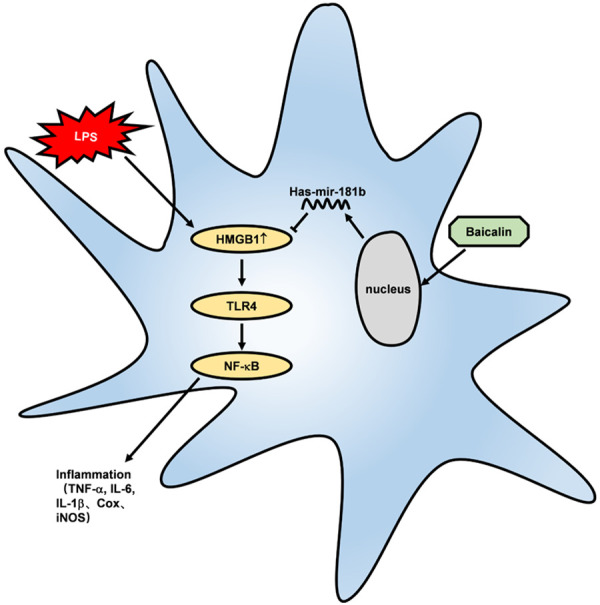
LPS activates the HMGB1/TLR4/NF-κB pathway in RAW264.7 cells and induces the abnormal secretion of inflammatory factors (TNF-α, IL-6, IL-1β, Cox, and iNOS), triggering Inflammation. Baicalin inhibits the expression of HMGB1 by promoting miR-181b, thus inhibiting the HMGB1/TLR4/NF-κB pathway mediated by HMGB1 and relieving inflammation.
Figure 8.
The morphology of cells in each group was observed under light microscope. A. Control group and LPS group. B. Lps-0, lps-0.1, lps-0.5 and lps-1 groups. The experiment was repeated 3 times (n=3).
Discussion
Inflammation may induce tumor growth [19], pain [20], endothelial dysfunction [21], metabolic disorders [22], and other subsequent diseases, endangering the health of the patients. A variety of diseases or external damages will cause inflammation in organisms, along with abnormally expressed genes or proteins. LncRNA NEAT1 can regulate sepsis-induced inflammation through miR-495, miR-211, and miR-125a-2p [23,24]. MiR-155 is abnormally up-regulated in patients with inflammation caused by kidney injury, and inhibiting miR-155 expression can relieve macrophage infiltration and reduce inflammatory toxicity [25]. Inflammation, if not controlled in time, may lead to life-threatening outcomes. Baicalin has potential anti-inflammatory effects. Here we explored the mechanism of baicalin in inhibiting inflammation, aiming to provide an accurate reference for its application.
In this study, we established RAW264.7 cell models of inflammation induced by LPS and exposed them to various baicalin concentrations (0.1-1.0 μmol/L). During the baicalin exposure, the activity of the HMGB1/TLR4/NF-κB pathway in RAW264.7 cells was inhibited, the levels of downstream factors including TNF-α, IL-6, IL-1β, Cox, and iNOS were decreased, and cell viability was increased. The exposure to baicalin also promoted miR-181b expression in cells. MiR-181b is involved in the occurrence and development of diseases such as prostate cancer bone metastasis [26], liver fibrosis [27], and breast cancer [28]. Hence, miR-181b, which is affected by baicalin, may also play a significant role in inflammation. In this study, HMGB1 was proved to be negatively regulated by miR-181b. Many studies [9-11,29-31] revealed that HMGB1 was closely related to inflammatory response. When miR-181b expression is affected by baicalin, HMGB1 may be negatively regulated by miR-181b, thereby altering the downstream inflammation pathways. These results suggest that the up-regulated miR-181b may be the effective target of baicalin to inhibit inflammation in RAW264.7 cells.
The results of this study showed that up-regulation of miR-181b or down-regulation of HMGB1 could inhibit LPS-induced inflammation in RAW264.7 cells. In this study, the exposure to baicalin along with the down-regulation of miR-181b at the same time enhanced the activity of HMGB1/HMGB1/TLR4/NF-κB pathway and increased the levels of TNF-α, IL-6, IL-1β, Cox, and iNOS in RAW264.7 cells.
We speculate that LPS activates the HMGB1/TLR4/NF-κB pathway in RAW264.7 cells and induces the abnormal secretion of inflammatory factors (TNF-α, IL-6, IL-1β, Cox, and iNOS), finally triggering inflammation. In this study, baicalin inhibited the expression of HMGB1 by promoting miR-181b, thus inhibiting the HMGB1/TLR4/NF-κB pathway mediated by HMGB1 and relieving inflammation.
This study selected RAW264.7 cells for construction of inflammation model and discovered the possible mechanism of baicalin in the treatment of inflammation. But there are still some limitations, for example, this study only investigated cell models of inflammation but did not explore the application of baicalin in animal models of inflammation. Besides, this study confirmed that baicalin at 1.0 μmol/L had a good anti-inflammation effect, but it did not identify the optimal concentration of baicalin in treating inflammation. Therefore, we should conduct high-throughput screening in the future to determine the optimal concentration of baicalin. Here, we note that baicalin regulates inflammation through the miR-181b/HMGB1/TLR4/NF-κB pathway, but whether baicalin can inhibit inflammation through other pathways needs further exploration.
Conclusion
In summary, baicalin inhibits LPS-induced inflammation in RAW264.7 cells through the miR-181b/HMGB1/TLR4/NF-κB pathway and its inhibition on inflammation is fairly effective at the concentration of 1.0 μmol/L. Therefore, the effect of baicalin is worthy of further study.
Acknowledgements
This study was financially supported by Project within Budget of Shanghai University of Traditional Chinese Medicine (2016YSN55).
Disclosure of conflict of interest
None.
References
- 1.Wang Y, Wang L, Wise JTF, Shi X, Chen Z. Verteporfin inhibits lipopolysaccharide-induced inflammation by multiple functions in RAW 264.7 cells. Toxicol Appl Pharmacol. 2020;387:114852. doi: 10.1016/j.taap.2019.114852. [DOI] [PubMed] [Google Scholar]
- 2.Fujiwara N, Kobayashi K. Macrophages in inflammation. Curr Drug Targets Inflamm Allergy. 2005;4:281–286. doi: 10.2174/1568010054022024. [DOI] [PubMed] [Google Scholar]
- 3.Medzhitov R. Origin and physiological roles of inflammation. Nature. 2008;454:428–435. doi: 10.1038/nature07201. [DOI] [PubMed] [Google Scholar]
- 4.Cao Y, Chen J, Ren G, Zhang Y, Tan X, Yang L. Punicalagin prevents inflammation in LPS-induced RAW264.7 macrophages by inhibiting FoxO3a/autophagy signaling pathway. Nutrients. 2019;11:2794. doi: 10.3390/nu11112794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang J, Yan S, Lu H, Wang S, Xu D. METTL3 attenuates LPS-induced inflammatory response in macrophages via NF-kappaB signaling pathway. Mediators Inflamm. 2019;2019:3120391. doi: 10.1155/2019/3120391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang WB, Yang F, Wang Y, Jiao FZ, Zhang HY, Wang LW, Gong ZJ. Inhibition of HDAC6 attenuates LPS-induced inflammation in macrophages by regulating oxidative stress and suppressing the TLR4-MAPK/NF-kappaB pathways. Biomed Pharmacother. 2019;117:109166. doi: 10.1016/j.biopha.2019.109166. [DOI] [PubMed] [Google Scholar]
- 7.Nagai J, Balestrieri B, Fanning LB, Kyin T, Cirka H, Lin J, Idzko M, Zech A, Kim EY, Brennan PJ, Boyce JA. P2Y6 signaling in alveolar macrophages prevents leukotriene-dependent type 2 allergic lung inflammation. J Clin Invest. 2019;129:5169–5186. doi: 10.1172/JCI129761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nepal S, Tiruppathi C, Tsukasaki Y, Farahany J, Mittal M, Rehman J, Prockop DJ, Malik AB. STAT6 induces expression of Gas6 in macrophages to clear apoptotic neutrophils and resolve inflammation. Proc Natl Acad Sci U S A. 2019;116:16513–16518. doi: 10.1073/pnas.1821601116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klune JR, Dhupar R, Cardinal J, Billiar TR, Tsung A. HMGB1: endogenous danger signaling. Mol Med. 2008;14:476–484. doi: 10.2119/2008-00034.Klune. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Michel HE, Menze ET. Tetramethylpyrazine guards against cisplatin-induced nephrotoxicity in rats through inhibiting HMGB1/TLR4/NF-kappaB and activating Nrf2 and PPAR-gamma signaling pathways. Eur J Pharmacol. 2019;857:172422. doi: 10.1016/j.ejphar.2019.172422. [DOI] [PubMed] [Google Scholar]
- 11.Shang J, Liu W, Yin C, Chu H, Zhang M. Cucurbitacin E ameliorates lipopolysaccharide-evoked injury, inflammation and MUC5AC expression in bronchial epithelial cells by restraining the HMGB1-TLR4-NF-kappaB signaling. Mol Immunol. 2019;114:571–577. doi: 10.1016/j.molimm.2019.09.008. [DOI] [PubMed] [Google Scholar]
- 12.Zhang W, Shen X, Xie L, Chu M, Ma Y. MicroRNA-181b regulates endotoxin tolerance by targeting IL-6 in macrophage RAW264.7 cells. J Inflamm (Lond) 2015;12:18. doi: 10.1186/s12950-015-0061-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang Y, Sun M, Yang X, Ma A, Ma Y, Zhao A. Baicalin relieves inflammation stimulated by lipopolysaccharide via upregulating TUG1 in liver cells. J Physiol Biochem. 2019;75:463–473. doi: 10.1007/s13105-019-00698-0. [DOI] [PubMed] [Google Scholar]
- 14.Wu Z, Chen C, Miao Y, Liu Y, Zhang Q, Li R, Ding L, Ishfaq M, Li J. Baicalin attenuates mycoplasma gallisepticum-induced inflammation via inhibition of the TLR2-NF-kappaB pathway in chicken and DF-1 cells. Infect Drug Resist. 2019;12:3911–3923. doi: 10.2147/IDR.S231908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ishfaq M, Zhang W, Hu W, Waqas Ali Shah S, Liu Y, Wang J, Wu Z, Ahmad I, Li J. Antagonistic effects of baicalin on mycoplasma gallisepticum-induced inflammation and apoptosis by restoring energy metabolism in the chicken lungs. Infect Drug Resist. 2019;12:3075–3089. doi: 10.2147/IDR.S223085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ishfaq M, Chen C, Bao J, Zhang W, Wu Z, Wang J, Liu Y, Tian E, Hamid S, Li R, Ding L, Li J. Baicalin ameliorates oxidative stress and apoptosis by restoring mitochondrial dynamics in the spleen of chickens via the opposite modulation of NF-kappaB and Nrf2/HO-1 signaling pathway during Mycoplasma gallisepticum infection. Poult Sci. 2019;98:6296–6310. doi: 10.3382/ps/pez406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fu YJ, Xu B, Huang SW, Luo X, Deng XL, Luo S, Liu C, Wang Q, Chen JY, Zhou L. Baicalin prevents LPS-induced activation of TLR4/NF-kappaB p65 pathway and inflammation in mice via inhibiting the expression of CD14. Acta Pharmacol Sin. 2021;42:88–96. doi: 10.1038/s41401-020-0411-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fu S, Liu J, Xu J, Zuo S, Zhang Y, Guo L, Qiu Y, Ye C, Liu Y, Wu Z, Hou Y, Hu CA. The effect of baicalin on microRNA expression profiles in porcine aortic vascular endothelial cells infected by Haemophilus parasuis. Mol Cell Biochem. 2020;472:45–56. doi: 10.1007/s11010-020-03782-y. [DOI] [PubMed] [Google Scholar]
- 19.Perfilyeva YV, Abdolla N, Ostapchuk YO, Tleulieva R, Krasnoshtanov VC, Perfilyeva AV, Belyaev NN. Chronic inflammation contributes to tumor growth: possible role of L-selectin-expressing Myeloid-Derived Suppressor Cells (MDSCs) Inflammation. 2019;42:276–289. doi: 10.1007/s10753-018-0892-6. [DOI] [PubMed] [Google Scholar]
- 20.Sommer C, Leinders M, Uceyler N. Inflammation in the pathophysiology of neuropathic pain. Pain. 2018;159:595–602. doi: 10.1097/j.pain.0000000000001122. [DOI] [PubMed] [Google Scholar]
- 21.Yang X, Chang Y, Wei W. Endothelial dysfunction and inflammation: immunity in rheumatoid arthritis. Mediators Inflamm. 2016;2016:6813016. doi: 10.1155/2016/6813016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Q, Liu D, Song P, Zou MH. Tryptophan-kynurenine pathway is dysregulated in inflammation, and immune activation. Front Biosci (Landmark Ed) 2015;20:1116–1143. doi: 10.2741/4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xia D, Yao R, Zhou P, Wang C, Xia Y, Xu S. LncRNA NEAT1 reversed the hindering effects of miR-495-3p/STAT3 axis and miR-211/PI3K/AKT axis on sepsis-relevant inflammation. Mol Immunol. 2020;117:168–179. doi: 10.1016/j.molimm.2019.10.009. [DOI] [PubMed] [Google Scholar]
- 24.Wang W, Guo ZH. Downregulation of lncRNA NEAT1 ameliorates LPS-induced inflammatory responses by promoting macrophage M2 polarization via miR-125a-5p/TRAF6/TAK1 axis. Inflammation. 2020;43:1548–1560. doi: 10.1007/s10753-020-01231-y. [DOI] [PubMed] [Google Scholar]
- 25.Zheng C, Zhang J, Chen X, Zhang J, Ding X, You X, Fan L, Chen C, Zhou Y. MicroRNA-155 mediates obesity-induced renal inflammation and dysfunction. Inflammation. 2019;42:994–1003. doi: 10.1007/s10753-019-00961-y. [DOI] [PubMed] [Google Scholar]
- 26.Han Z, Zhan R, Chen S, Deng J, Shi J, Wang W. miR-181b/Oncostatin m axis inhibits prostate cancer bone metastasis via modulating osteoclast differentiation. J Cell Biochem. 2020;121:1664–1674. doi: 10.1002/jcb.29401. [DOI] [PubMed] [Google Scholar]
- 27.Geng W, Zhou G, Zhao B, Xiao Q, Li C, Fan S, Dong P, Zheng J. Liquiritigenin suppresses the activation of hepatic stellate cells via targeting miR-181b/PTEN axis. Phytomedicine. 2020;66:153108. doi: 10.1016/j.phymed.2019.153108. [DOI] [PubMed] [Google Scholar]
- 28.Zeng C, Fan D, Xu Y, Li X, Yuan J, Yang Q, Zhou X, Lu J, Zhang C, Han J, Gu J, Gao Y, Sun L, Wang S. Curcumol enhances the sensitivity of doxorubicin in triple-negative breast cancer via regulating the miR-181b-2-3p-ABCC3 axis. Biochem Pharmacol. 2020;174:113795. doi: 10.1016/j.bcp.2020.113795. [DOI] [PubMed] [Google Scholar]
- 29.Kim YM, Park EJ, Kim HJ, Chang KC. Sirt1 S-nitrosylation induces acetylation of HMGB1 in LPS-activated RAW264.7 cells and endotoxemic mice. Biochem Biophys Res Commun. 2018;501:73–79. doi: 10.1016/j.bbrc.2018.04.155. [DOI] [PubMed] [Google Scholar]
- 30.Qi Z, Zhang Y, Qi S, Ling L, Gui L, Yan L, Lv J, Li Q. Salidroside inhibits HMGB1 acetylation and release through upregulation of SirT1 during inflammation. Oxid Med Cell Longev. 2017;2017:9821543. doi: 10.1155/2017/9821543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou S, Lu H, Chen R, Tian Y, Jiang Y, Zhang S, Ni D, Su Z, Shao X. Angiotensin II enhances the acetylation and release of HMGB1 in RAW264.7 macrophage. Cell Biol Int. 2018;42:1160–1169. doi: 10.1002/cbin.10984. [DOI] [PubMed] [Google Scholar]