Abstract
Objective: Osteosarcoma is a malignant bone tumor consisting of mesenchymal cells. This study aimed to investigate the inhibitory effects of human bone marrow mesenchymal stem cell (hBMSC)-derived miR-1913 on osteosarcoma. Methods: Cell viability was determined using CCK8 and colony formation assays. The cell migration and invasion abilities were assessed using wound healing and transwell assays. RT-qPCR and western blot were used to measure the miR-1913, Neurensin-2 (NRSN2), N-cadherin, and E-cadherin expression levels. Dual luciferase reporter assays were conducted to identify the target relationship between miR-1913 and NRSN2. The exosomes were extracted and identified using TEM and NTA assays. Results: In the osteosarcoma tumor tissues and cell lines, the NRSN2 expressions were up-regulated, which correlated with a poor osteosarcoma prognosis. MiR-1913 inhibited the cell viability, proliferation, migration, and invasion by negatively targeting NRSN2. Furthermore, the hBMSC-derived exosomes delivered miR-1913 to inhibit the NRSN2 expression in the osteosarcoma cells. Conclusion: The inhibitory role of hBMSC-derived miR-1913 on osteosarcoma progression was achieved by targeting NRSN2, indicating the potential therapeutic value of hBMSC-derived miR-1913.
Keywords: Osteosarcoma, exosomes, miR-1913, Neurensin-2, tumor progression
Introduction
Osteosarcoma is a subtype of bone cancer which most commonly occurs in children and adolescents. The current treatment for osteosarcoma involves the surgical removal of the tumors, combined with systemic chemotherapy to control the micrometastases. The 5-year event-free survival (EFS) rate is about 70% in patients with localized osteosarcoma, but the overall survival rate is lower than 20% in progressed patients with metastatic or recurrent disease [1]. Therefore, metastasis is the main challenge in osteosarcoma treatment, especially pulmonary osteosarcoma metastasis [2]. Neurensin-2 (NRSN2), a 24 kDa protein, promotes tumor cell proliferation, invasion and migration and plays an oncogenic role in osteosarcoma. However, the detailed mechanisms of NRSN2 responsible for the progression and metastasis of osteosarcoma need to be further explored.
Bone marrow mesenchymal stem cells (BMSCs) are pluripotent progenitors found in bone marrow. At present, the pivotal regulatory roles of BMSCs in the tumor microenvironment rely on various mechanisms, including secreting extracellular vesicles. Many studies have found that BMSC-derived exosomes, one of the external vesicles released by the cells, can regulate cell proliferation, migration, survival, and drug resistance [4]. However, it is not clear whether BMSC-derived exosomes are involved in osteosarcoma metastasis.
Exosomes can deliver a variety of nucleic acids, including microRNAs (miRNAs) [5]. MiRNAs are small non-coding RNAs that regulate gene expression at the post-transcriptional level by inhibiting mRNA translation or promoting mRNA degradation [6]. It has been shown that exosomes regulate cellular communication by transferring miRNAs [7]. An understanding of the rapid intercellular formation and the functional changes of miRNAs delivered by exosomes can lead to possible therapeutic applications [5,8,9]. MiR-1913 has been shown to be positively related to prostate cancer progression [10]. And our preliminary results determined that miR-1913 targets NRSN2. Since NRSN2 and hBMSC-derived exosomes are associated with tumor cell proliferation, invasion and migration, we innovatively speculated that there were regulatory effects of miR-1913 derived from hBMSCs exosomes in osteosarcoma metastasis, which is involved with NRSN2.
Materials and methods
Cell lines and clinical tissues
Human normal osteoblastic cells (hFOB1.19), as well as osteosarcoma cell lines, including 143B, U2OS, Saos-2, and HOS, were obtained from the American Type Culture Collection (ATCC) and incubated with DMEM medium (Gibco, Grand Island, USA) containing 12% fetal bovine serum (FBS) (Gibco). The cells were cultured under conditions of 37°C with 5% CO2. The clinical osteosarcoma tissue and adjacent normal tissue specimens, which were patient-matched, were collected from untreated, newly-diagnosed patients (n=58). This study was approved by the Ethics Committee of our hospital (Approval No. PJ2019MI-GS056-01). Written informed consent documents were obtained from the patients prior to their participation.
Microarray analysis
Five miRNA-mRNA predictors (miRDB, Targetscan, Starbase, and miRwalk) were used to predict the miRNAs regulating NRSN2. The osteosarcoma patients’ gene expression profiles and clinical data were obtained from the Cancer Genome Atlas (TCGA) database (https://cancergenome.nih.gov/). The Gene Expression Profiling Interactive Analysis (GEPIA) database (http://gepia.cancer-pku.cn/) was used to analyze the prognostic correlations between the patients with osteosarcoma and the related factors.
Cell transfection
For the NRSN2 knockdown, NRSN2 small interfering RNA (siRNA) was used. siRNA and miR-1913 miRNA mimics and miR-1913 inhibitor were synthesized by GenephPharma (Shanghai, China). The cells were cultured in 24 well dishes with a serum-free medium. Then, the transfection solution was diluted using a serum-free medium and mixed with Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA) carefully. After 48 h of incubation, the cells were used for the following experiments.
Dual-luciferase reporter assay
To examine whether miR-1913 targeted NRSN2 directly, NRSN2-WT and NRSN2-MUT reporter vectors were purchased from Promega (Madison, WI, USA). The assays were described previously [11]. The cells were cultured in 96-well plates until the cell confluence was 50-70%. Using Lipofectamine 2000 (Invitrogen), the cells were co-transfected with reporter vectors and an miR-1913-NC or an miR-1913 mimic. At 48 h post-transfection, the luciferase activity was estimated using the Dual-Luciferase Reporter Assay System (Promega).
Exosome extraction
The hBMSCs were cultured under 10% exosome-free FBS for 72 h. The cells, dead cells, and cell debris were removed using several low-speed centrifugations. Next, the exosomes were collected using high speed centrifugation.
Transmission electron microscope (TEM) detection and nanoparticle tracking analysis (NTA)
15 μL of the exosome solution was dropped onto a copper mesh and incubated at room temperature for 10 min, and then the copper net was washed with sterile distilled water. The excess liquid was absorbed with absorbent paper. Then, the copper mesh was negatively stained with 2% uranyl acetate for 1 min. After removing the excess liquid and drying the mesh for 2 min under an incandescent lamp, the copper mesh was observed under 80 kV TEM. As for NTA, the exosome solution was diluted to 5×107/mL~5×108/mL with PBS, and the sizes and trajectories of the particles were analyzed using NTA.
Wound healing assay
As previously described, the cells were cultured in 6-well plates [12]. A 10 μL pipette tip was used to scratch at the center of each well, and then the cells were incubated with serum-free DMEM and high glucose for 48 h. Images of each well in the 6-well plate were generated using an inverted microscope (Leica, Wetzlar, Germany), and the images were analyzed using ImageJ software (NIH, Bethesda, Maryland, USA) to calculate the relative distances according to the equation (W0-Wt)/W0×100%.
Transwell assays
The cells were cultured in the upper chamber with Matrigel-coated membrane (BD Biosciences, Bedford, MA, USA) under a serum-free culture medium for 48 h incubation. Using a cotton swab, the non-invading cells on the upper chamber were eliminated. After fixing the cells with paraformaldehyde (4%, Sigma-Aldrich, St Louis, MO, USA), the cells on the lower side of the chamber were incubated with crystal violet. The results were counted in five fields we randomly selected using a microscope (Leica, Wetzlar, Germany).
CCK8 assays
The cell viability was determined using CCK8 assays. The cells (50,000/mL) were seeded and incubated in a 96 well plate for the indicated time. 10 μL of CCK8 solution (Dojindo, Shanghai, China) were added and then reacted for 2 h. The optical density (OD) values were assessed using a microtitre plate reader (BioTek, Winooski, VT, USA) at 450 nm.
Colony formation
The cells were inoculated into a culture plate and incubated for 2 to 3 weeks. After the colony formation, paraformaldehyde (4%, Sigma-Aldrich) was used for fixation for 15 min. A crystalline violet solution (0.1%, Abcam) was then applied for 10 to 30 min. After washing the extra dye solution and drying them, the petri dishes were inverted and covered with a transparent grid. The results were visualized under general microscopy (Leica, Wetzlar, Germany).
Immunohistochemistry (IHC)
For the immunohistochemical staining, the tumor tissues were fixed with paraformaldehyde (10%, Sigma-Aldrich, St Louis, MS, USA). After being cut serially, slides (5 μm) were boiled in a citrate buffer solution (10 mM, pH 6.0) for 20 min of antigen retrieval and then blocked with 0.3% hydrogen peroxide to eliminate the endogenous peroxidase activity. After they were incubated with a primary antibody (anti-NRSN2, Abcam) at 4°C for 12 h, an HRP-labeled secondary antibody (1:1200, Abcam) was applied to the slides for 30 min and a DAB detection kit (Abcam, Cambridge, MA, USA) was applied for 10 min. After they were dehydrated and cleared, the slides were analyzed under a microscopy (Leica, Wetzlar, Germany).
Quantitative reverse transcription PCR (RT-qPCR)
The total RNA was extracted, reverse transcribed into cDNA, and amplified. This approach has been elaborated in past studies [13]. In brief, the total RNA of the transfected cells and liver tissues were first digested using TRIzol reagent (Invitrogen). After that, the reverse transcriptions of RNA into cDNA were conducted through a reverse transcriptase kit from Invitrogen Company. The used primers are shown in Table 1. The PCR processes were conducted on an ABI 7500 Real-Time PCR System. The relative expression levels of the RNA normalized to GAPDH or U6 were calculated using the 2-ΔΔCt method.
Table 1.
The primer sequences
| Forward | Reverse | |
|---|---|---|
| GAPDH | 5’-TCCGTGGTCCACGAGAACT-3’ | 5’-GAAGCATTTGCGGTGGACGAT-3’ |
| NRSN2 | 5’-CGGAGACGCAGGTCCAGAGGGAT-3’ | 5’-TATGCATCAACTGTTTATTGAAAGG-3’ |
| miR-1913 | 5’-ACCGTCGTCGCCTCCCCGTC-3’ | 5’-GACTGACATAAATGGGAGCAG-3’ |
| U6 | 5’-CTCGCTTCGGCAGCACA-3’ | 5’-AACGCTTCACGAATTTGCGT-3’ |
Western blot
The protein extraction (RIPA buffer, ThermoFisher Scientific, Waltham, MA, USA), concentration determination (BCA protein assay kit, ThermoFisher Scientific), separation and transfer to nitrocellulose filter membrane (Millipore, Boston, USA), incubation with BSA and antibodies were described previously [14]. The primary antibodies for NRSN2, E-cadherin, N-cadherin, CD63, calnexin, and Hsp70 (both of which were diluted to 1:1000; Cell Signaling Technology, USA) and GAPDH (Cell Signaling Technology, USA) were incubated at 4°C overnight. The secondary antibodies were obtained from Proteintech (USA). The protein expressions were measured using an ECL system (Amersham Pharmacia, Piscataway, NJ, USA).
Statistical analysis
SPSS 18.0 software was used for the statistical analyses. The data are presented as the means ± SD from at least three independent experiments. Spearman’s correlation was used to analyze the correlation of miR-1913 with NRNS2 in the clinical osteosarcoma tissues. The statistical analysis was performed using analyses of variance (ANOVA). The significance was determined using Newman-Keuls post hoc tests. Significant differences are indicated by P<0.05.
Results
NRSN2 was upregulated in the osteosarcoma tumor tissues and cell lines
According to the TCGA database, we found that the NRSN2 expression was significantly upregulated compared with its expression in normal tissues (P<0.001) (Figure 1A). A Kaplan-Meier curve was used to analyze the prognostic significance of NRSN2, and the results showed that the probability of patients with high expressions of NRSN2 was lower (P<0.05) (Figure 1B). Next, the NRSN2 expression in the clinical osteosarcoma samples was determined using IHC staining, western blot, and RT-qPCR. The IHC staining indicated that, compared with the adjacent normal tissues, the NRSN2 expression was upregulated in the osteosarcoma tissues (Figure 1C). The upregulated NRSN2 expression was also proved by western blot (P<0.001) (Figure 1D). Furthermore, the RT-qPCR results verified that the transcript level of NRSN2 was enhanced in the osteosarcoma tissues (P<0.001) (Figure 1E). Consistent with the TCGA database, a negative correlation between OS and the NRSN2 expression was observed (P<0.01). Figure 1F shows that the survival curves of the patients with high NRSN2 expressions (n=29) and low NRSN2 expressions, suggesting that the prognoses of the patients with high NRSN2 expression were poor. (Figure 1F). The NRSN2 expression was also determined in the different osteosarcoma cell lines. Compared with that of normal human osteoblast cells (hFOB 1.19), the NRSN2 expressions were upregulated in the osteosarcoma cell lines, including 143B, HOS, Saos-2, and U2OS (P<0.001) (Figure 1G), among which the 143B and U2OS cells had the highest NRSN2 expressions. These results indicate that NRSN2 is highly expressed in osteosarcoma and is associated with a poor prognosis.
Figure 1.
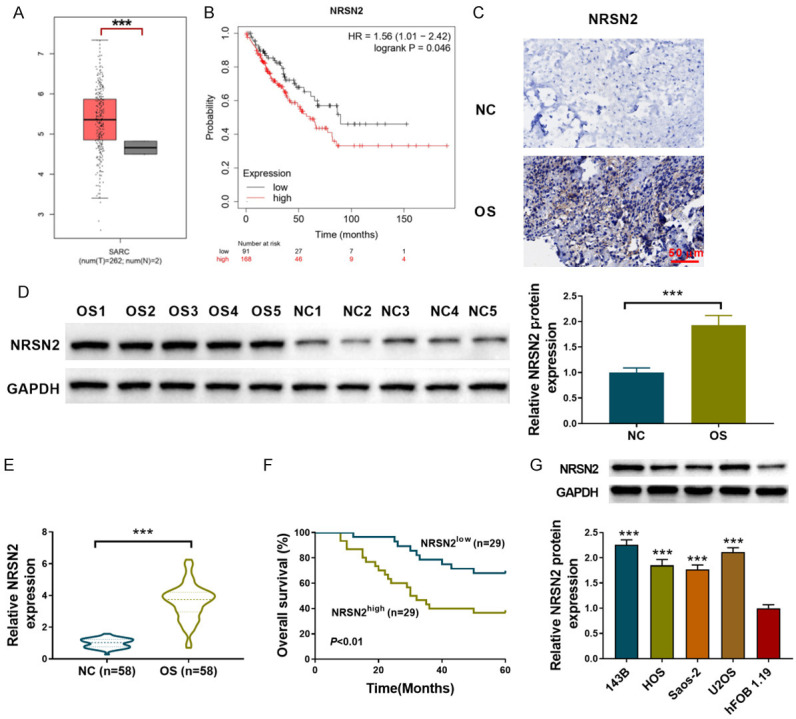
NRSN2 is up-regulated in osteosarcoma tumor tissues and cell lines. A. The TCGA database represents the NRSN2 expression in osteosarcoma tissues. B. The Kaplan-Meier curve of probability in osteosarcoma patients. C. IHC staining was used to determine the NRSN2 expression in the clinical osteosarcoma tissue and adjacent normal tissue specimens (n=2). Scale bar: 50 µM, 100×. D. Western blot was used to determine the protein NRSN2 expression in the clinical osteosarcoma tissue and adjacent normal tissue specimens (n=10). E. RT-qPCR was used to determine the NRSN2 expression in the clinical osteosarcoma tissue and adjacent normal tissue specimens (n=58). F. The Kaplan-Meier curve of OS in the clinical patients (n=58) with different NRSN2 expressions. G. The NRSN2 expressions in the different osteosarcoma cell lines (143B, HOS, Saos-2, and U2OS) and in the normal human osteoblast cells (hFOB 1.19) were determined using western blot. ***P<0.001.
NRSN2 was the direct target of miR-1913
MiRDB, Targetscan, Starbase and miRwalk were used to predict the miRNA targets of NRSN2, and miR-1913 was enriched in all four of the databases (Figure 2A). Therefore, the miR-1913 expression was measured in the osteosarcoma tumor tissues and cell lines. The results showed reduced expressions of miR-1913 in the osteosarcoma tissues compared to the adjacent normal tissues (P<0.001) (Figure 2B). Spearman’s correlation analysis indicated a negative correlation between miR-1913 and NRNS2 in the clinical osteosarcoma tissues (P<0.001) (Figure 2C). The miR-1913 expressions were also measured in the different osteosarcoma cell lines using RT-qPCR. Compared with the hFOB 1.19 cells, the miR-1913 expression was decreased in the 143B, HOS, Saos-2, and U2OS cells (P<0.001), among which miR-1913 had the lowest expression in the 143B and U2OS cells (Figure 2D). Figure 2E displays the predicted binding site between miR-1913 and NRSN2 directly as predicted (Figure 2E). The increased expression of miR-1913 was first verified in the 143B and U2OS cells transfected with the miR-1913 mimic (P<0.001) (Figure 2F), and then the dual-luciferase reporter gene assay demonstrated that the overexpression of miR-1913 inhibited the luciferase activity of NRSN2-WT, but the inhibitory effect was blocked by the mutated NRSN2 binding sites (P<0.001) (Figure 2G, 2H). The protein NRSN2 expression was decreased (P<0.001) in the 143B and U2OS cells overexpressing miR-1913 (Figure 2I). Therefore, miR-1913 regulated the NRSN2 expression negatively, and NRSN2 was the direct target of miR-1913.
Figure 2.
miR-1913 was down-regulated in the tumor tissues and cell lines of osteosarcoma and targeted NRSN2 directly. (A) A Venn diagram of the targeting miRNAs of NRSN2 after the prediction. (B) RT-qPCR was used to determine the miR-1913 expressions in the clinical osteosarcoma tissue and adjacent normal tissue specimens (n=58). (C) Spearman’s correlation was used to analyze the correlation between miR-1913 and NRNS2 in the clinical osteosarcoma tissues (n=58). (D) RT-qPCR was used to determine the miR-1913 expressions in the different osteosarcoma cell lines (143B, HOS, Saos-2, and U2OS) and in the hFOB 1.19 cells. (E) The predicted binding site between miR-1913 and NRSN2. (F) RT-qPCR was used to determine the miR-1913 expression in the 143B and U2OS cells transfected with the miR-1913 mimic. (G, H) A dual-luciferase reporter assay was used to verify the binding relationship between NRSN2 and miR-1913 in the 143B (G) and U2OS cells (H). (I) The protein NRSN2 expression was determined in the 143B and U2OS cells transfected with the miR-1913 mimic. ***P<0.001.
MiR-1913 inhibited osteosarcoma cell viability, proliferation and metastasis
The role of miR-1913 in osteosarcoma was then investigated in the 143B and U2OS cells. As shown in Figure 3A, 3B, the overexpression of miR-1913 reduced the cell viability and the colony formation ability (P<0.001). In addition, the results showed that the cell invasion and migration were inhibited significantly after the overexpression of miR-1913 (P<0.001) (Figure 3C, 3D). The western blot also showed that the overexpression of miR-1913 promoted the protein expression of E-cadherin, and it inhibited the N-cadherin expression (P<0.001) (Figure 3E). Therefore, miR-1913 exerted antitumor effects on osteosarcoma by inhibiting cell proliferation and metastasis.
Figure 3.
The overexpression of miR-1913 inhibited cell proliferation and metastasis. A. The cell viability was determined using CCK8 assays in the 143B and U2OS cells transfected with the miR-1913 mimic at 24 h, 48 h, and 72 h. B. The cell proliferation was determined using colony formation assays with the 143B and U2OS cells transfected with the miR-1913 mimic. C, D. The cell migration and invasion were determined using wound healing and transwell assays in the 143B and U2OS cells transfected with the miR-1913 mimic. Scale bar: 100 µM, 200×. E. The protein N-cadherin and E-cadherin expressions were determined using western blot. ***P<0.001.
MiR-1913 exerted antitumor effects on osteosarcoma via targeting NRSN2
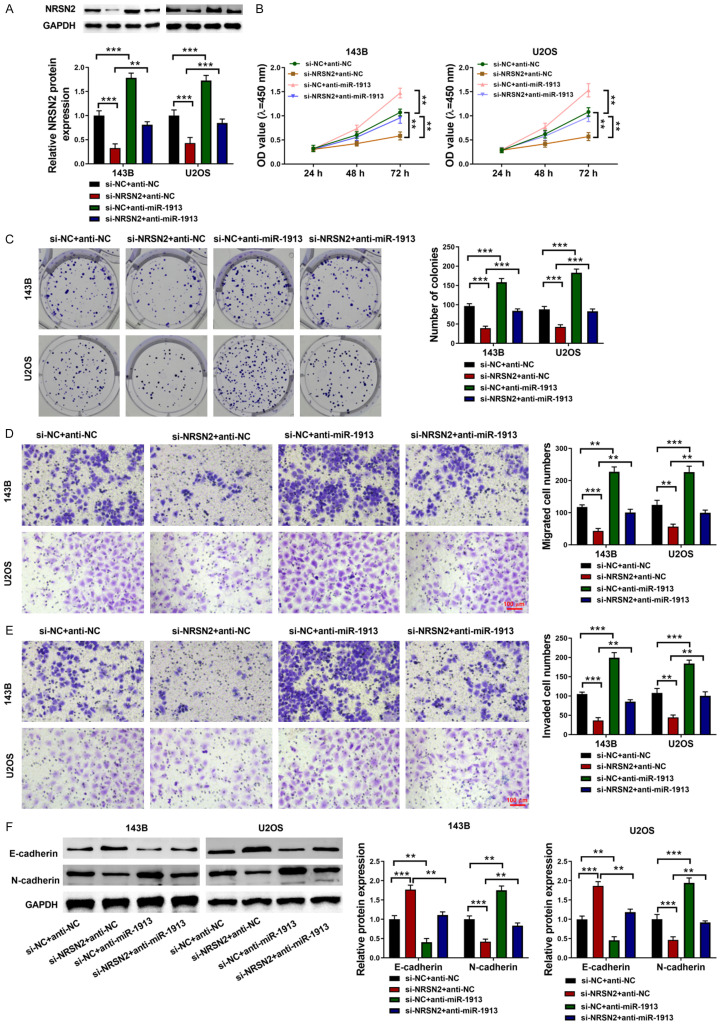
In order to further determine whether the antitumor effects of miR-191 were achieved by targeting NRSN2, 143B, and U2OS cells were transfected with NRSN2 siRNA and the miR-1913 inhibitor. After the NRSN2 siRNA transfection, the NRSN2 expression was decreased, and the single treatment of the miR-1913 inhibitor increased the NRSN2 expression. Without doubt, after co-transfecting with NRSN2 siRNA and the miR-1913 inhibitor, the effect of the NRSN2 siRNA was reversed by the miR-1913 inhibitor (P<0.001) (Figure 4A). The cell viability and proliferation were determined using CCK8 and colony formation assays. NRSN2 siRNA inhibited the cell viability and proliferation, which could be reversed by the miR-1913 inhibitor (P<0.01) (Figure 4B, 4C). Similarly, the cell migration and invasion abilities were inhibited by the NRSN2 siRNA and were increased significantly by the miR-1913 inhibitor alone. However, after co-transfecting with the NRSN2 siRNA transfection and the miR-1913 inhibitor, the inhibition of the cell migration and invasion induced by the NRSN2 siRNA could be rescued (P<0.01) (Figure 4D, 4E). The N-cadherin and E-cadherin expression levels also showed the same trend (P<0.001) (Figure 4F). All the results suggested that miR-1913 plays an antitumor role in osteosarcoma by targeting NRSN2.
Figure 4.
miR-1913 inhibited the cell viability, proliferation, and cell metastasis via targeting NRSN2. A. The NRSN2 expression was determined using western blot. B. The cell viability was determined using CCK8 assays in the 143B and U2OS cells at 24 h, 48 h and 72 h. C. The cell proliferation was determined using colony formation assays with the 143B and U2OS cells co-transfected with NRSN2 siRNA and the miR-1913 inhibitor. D, E. The cell migration and invasion were determined using wound healing and transwell assays in the 143B and U2OS cells co-transfected with NRSN2 siRNA and the miR-1913 inhibitor. Scale bar: 100 µM, 200×. F. The protein N-cadherin and E-cadherin expressions were measured using western blot in the 143B and U2OS cells co-transfected with NRSN2 siRNA and the miR-1913 inhibitor. **P<0.01, ***P<0.001.
hBMSC-derived exosomal miR-1913 reduced the NRSN2 expression
Exosomes derived from the hBMSCs were subjected to TEM and NTA assays to identify their morphology (Figure 5A, 5B). In addition, the expressions of the marker proteins including CD63 and Hsp70 were confirmed by western blot. The CD63 and Hsp70 expressions were significant in the isolated exosomes, which was barely detected in the hBMSCs (Figure 5C). To further investigated the role of exosomal miR-1913, the hBMSCs were transfected with an miR-1913 mimic, and the miR-1913 expression was enhanced both in the hBMSCs and in the hBMSC-derived exosomes (P<0.001) (Figure 5D). Then, 143B and U2OS cells were incubated with hBMSC-derived exosomes transfected with miR-1913 mimics or control mimics, respectively. The RT-qPCR results showed that miR-1913 was upregulated in the 143B and U2OS cells incubated with the hBMSC-derived exosomes (P<0.001) (Figure 5E), and the increased expression of miR-1913 was more prominent in the 143B and U2OS cells incubated with hBMSC-derived exosomes transfected with the miR-1913 mimics (P<0.001) (Figure 5E). Moreover, the NRSN2 expression was reduced in the 143B and U2OS cells incubated with hBMSC-derived exosomes, and it was further enhanced in the 143B and U2OS cells incubated with hBMSC-derived exosomes transfected the miR-1913 mimics (P<0.01) (Figure 5F).
Figure 5.

hBMSC-derived exosomal miR-1913 reduced the NRSN2 expression. A. Representative picture of the exosomes under TEM. Scale bar: 200 nM, 30000×. B. NTA was used to measure the size distribution of the exosomes. C. The marker proteins of the exosomes including Hsp70 and CD63 were measured using western blot. D. The miR-1913 expression was measured in the hBMSCs and in the hBMSC-derived exosomes both transfected with the miR-1913 mimic. E. The miR-1913 expressions were detected in the 143B and U2OS cells incubated with the hBMSC-derived exosomes transfected with the miR-1913 mimic. F. The NRSN2 expression was measured in the 143B and U2OS cells incubated with the hBMSC-derived exosomes transfected with the miR-1913 mimic. **P<0.01, ***P<0.001.
hBMSC-derived exosomal miR-1913 inhibited cell viability, proliferation, and metastasis
In order to determine the effects of hBMSC-derived exosomal miR-1913 on osteosarcoma, 143B and U2OS cells were incubated with hBMSC-derived exosomes transfected with miR-1913 mimics or control mimics, respectively. Compared with the NC group, the cell viability (Figure 6A), proliferation (Figure 6B), and metastasis (Figure 6C, 6D) were inhibited by the hBMSC-derived exosomes (P<0.01), and they were further inhibited by incubating the cells with hBMSC-derived exosomes transfected with miR-1913 mimics (P<0.001). The inhibitory effects of the hBMSC-derived exosomes were also proved by western blot. The phenomenon displaying the decreased expression of N-cadherin and the increased expression of E-cadherin in the exo-miR-NC group was more notable in the exo-miR-1913 group (P<0.01) (Figure 6E). In summary, hBMSC-derived exosomal miR-1913 inhibits cell viability, proliferation, and metastasis in osteosarcoma. The schematic diagram of our research summarized in Figure 7 shows that miR-1913 can negatively target NRSN2 to inhibit the progression of osteosarcoma by influencing the cell viability, proliferation, and migration and invasion, which is more significant if the miR-1913 is derived from hBMSC exosomes.
Figure 6.
The hBMSC-derived exosomal miR-1913 inhibits cell viability, proliferation, and metastasis. 143B and U2OS cells were incubated with hBMSC-derived exosomes transfected with miR-1913 mimics or control mimics, respectively. A. The cell viability was determined using CCK8 assays at 24 h, 48 h, and 72 h. B. The cell proliferation was determined using colony formation assays. C, D. The cell migration and invasion were determined using wound healing and transwell assays. Scale bar: 100 µM, 200×. E. The protein N-cadherin and E-cadherin expressions were determined using western blot. **P<0.01, ***P<0.001.
Figure 7.
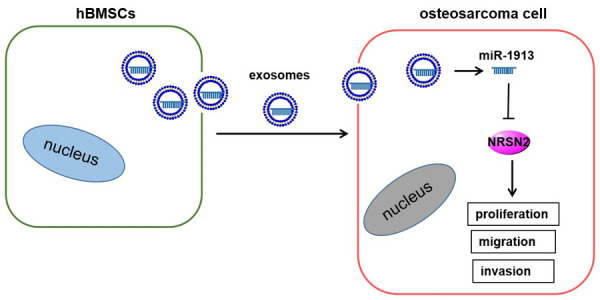
The schematic diagram of the hBMSC-derived exosomal miR-1913/NRSN2 axis.
Discussion
Osteosarcoma is a common bone cancer that tends to occur in teenagers and young adults [15]. In the present work, we demonstrated that hBMSC-derived exosomal miR-1913 alleviated cell viability, proliferation, and migration and invasion by targeting NRSN2, which thus blocks the progression of osteosarcoma.
The overall survival of osteosarcoma patients is influenced by the presence of pulmonary metastasis and by adherence to the treatment [16]. An increasing number of studies have shown the effects of exosomal miRNAs on tumor progression and metastasis. For example, hypoxic BMSC-derived exosomal miRNAs promote the metastasis of lung cancer cells via STAT3-induced EMT [17]. BMSC-derived exosomes are involved in the miRNA-dependent survival of retinal ganglion cells [18]. In terms of osteosarcoma, studies have also shown that BMSC-derived miR-208a and miR-122-5p are associated with osteosarcoma cell proliferation, migration, and invasion [19,20]. Herein, we showed that hBMSC-derived exosomal miR-1913 exerted an antitumor role in osteosarcoma by targeting NRSN2 negatively. Also, miR-1913 can effectively inhibit cell proliferation, migration and invasion, which is consistent with previous studies about the effects of BMSC-derived exosomal miRNAs.
NRSN2 is a 24 kDa protein located in the membrane. Evidence indicates that NRSN2 may regulate tumor progression. For example, NRSN2 promotes tumor cell proliferation, invasion and migration in colorectal cancer [21] and accelerates cell metastasis in breast cancer [22]. In ovarian cancer, its expression is associated with a malignant phenotype [23]. The NRSN2 mechanism on tumor progression uses the PI3K/AKT/mTOR pathway in non-small cell lung cancer [23] and the Wnt/β-catenin pathway in osteosarcoma [4]. NRSN2 also plays an oncogenic role by enhancing tumor progression via the PI3K/AKT/mTOR and Wnt/β-catenin pathways [3]. Similarly, our results show an aberrant increased NRSN2 expression in osteosarcoma tumor tissues and cell lines, and its expression is related to poor prognosis, indicating its oncogenic role in osteosarcoma. However, it is worth noting that NRSN2 also plays an antitumor role in hepatocellular carcinoma (HCC). Decreased NRSN2 expression correlates with poor HCC prognosis, and promotes tumor progression [24,25]. Unfortunately, we only proved the osteosarcoma-promoted effects of NRSN2 in vitro. Further in vivo studies should be performed to verify the role of NRSN2 in osteosarcoma.
In our study, four tools were used to predict the miRNAs targeting NRSN2, and miR-1913 was identified as an intersecting gene. Likewise, miR-1913 was predicted to be related to both acute lymphoblastic leukemia [26] and cerebral cavernous malformations [27]. And a previous study showed that miR-1913 is associated with poor overall survival in cervical cancer [28]. However, our research reported the antitumor role of miR-1913 in osteosarcoma. We found that miR-1913 is down-regulated in clinical osteosarcoma tissues and cell lines, and the overexpression of miR-1913 significantly inhibits cell viability, proliferation, and metastasis. The limitation of our study was the absence of in vivo evidence from animal experiments. And we will construct the animal experimental models of osteosarcoma to prove the mechanism involved in hBMSC-derived exosomal miR-1913.
In conclusion, our study shows that hBMSC-derived miR-1913 inhibits the progression of osteosarcoma by targeting NRSN2, which not only suggests a regulatory effect between hBMSC-derived exosomal miR-1913 and NRSN2, but it also provides a new therapeutic direction for osteosarcoma.
Acknowledgements
This work was supported by the High-level Hospital Construction Research Project of Maoming People’s Hospital.
Disclosure of conflict of interest
None.
References
- 1.Harrison DJ, Geller DS, Gill JD, Lewis VO, Gorlick R. Current and future therapeutic approaches for osteosarcoma. Expert Rev Anticancer Ther. 2018;18:39–50. doi: 10.1080/14737140.2018.1413939. [DOI] [PubMed] [Google Scholar]
- 2.Yao Z, Han L, Chen Y, He F, Sun B, Kamar S, Zhang Y, Yang Y, Wang C, Yang Z. Hedgehog signalling in the tumourigenesis and metastasis of osteosarcoma, and its potential value in the clinical therapy of osteosarcoma. Cell Death Dis. 2018;9:701. doi: 10.1038/s41419-018-0647-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Keremu A, Maimaiti X, Aimaiti A, Yushan M, Alike Y, Yilihamu Y, Yusufu A. NRSN2 promotes osteosarcoma cell proliferation and growth through PI3K/Akt/MTOR and Wnt/beta-catenin signaling. Am J Cancer Res. 2017;7:565–573. eCollection 2017. [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu W, Huang L, Li Y, Zhang X, Gu J, Yan Y, Xu X, Wang M, Qian H, Xu W. Exosomes derived from human bone marrow mesenchymal stem cells promote tumor growth in vivo. Cancer Lett. 2012;315:28–37. doi: 10.1016/j.canlet.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 5.Yu X, Odenthal M, Fries JW. Exosomes as miRNA carriers: formation-function-future. Int J Mol Sci. 2016;17:2028. doi: 10.3390/ijms17122028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Correia de Sousa M, Gjorgjieva M, Dolicka D, Sobolewski C, Foti M. Deciphering miRNAs’ action through miRNA editing. Int J Mol Sci. 2019;20:6249. doi: 10.3390/ijms20246249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun Z, Shi K, Yang S, Liu J, Zhou Q, Wang G, Song J, Li Z, Zhang Z, Yuan W. Effect of exosomal miRNA on cancer biology and clinical applications. Mol Cancer. 2018;17:147. doi: 10.1186/s12943-018-0897-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang J, Li S, Li L, Li M, Guo C, Yao J, Mi S. Exosome and exosomal microRNA: trafficking, sorting, and function. Genomics Proteomics Bioinformatics. 2015;13:17–24. doi: 10.1016/j.gpb.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manna I, Iaccino E, Dattilo V, Barone S, Vecchio E, Mimmi S, Filippelli E, Demonte G, Polidoro S, Granata A, Scannapieco S, Quinto I, Valentino P, Quattrone A. Exosome-associated miRNA profile as a prognostic tool for therapy response monitoring in multiple sclerosis patients. FASEB J. 2018;32:4241–4246. doi: 10.1096/fj.201701533R. [DOI] [PubMed] [Google Scholar]
- 10.Byun YJ, Piao XM, Jeong P, Kang HW, Seo SP, Moon SK, Lee JY, Choi YH, Lee HY, Kim WT, Lee SC, Cha EJ, Yun SJ, Kim WJ. Urinary microRNA-1913 to microRNA-3659 expression ratio as a non-invasive diagnostic biomarker for prostate cancer. Investig Clin Urol. 2021;62:340–348. doi: 10.4111/icu.20200488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xi Y, Jiang T, Wang W, Yu J, Wang Y, Wu X, He Y. Long non-coding HCG18 promotes intervertebral disc degeneration by sponging miR-146a-5p and regulating TRAF6 expression. Sci Rep. 2017;7:13234. doi: 10.1038/s41598-017-13364-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cory G. Scratch-wound assay. Methods Mol Biol. 2011;769:25–30. doi: 10.1007/978-1-61779-207-6_2. [DOI] [PubMed] [Google Scholar]
- 13.Sun L, Liu M, Luan S, Shi Y, Wang Q. MicroRNA-744 promotes carcinogenesis in osteosarcoma through targeting LATS2. Oncol Lett. 2019;18:2523–2529. doi: 10.3892/ol.2019.10530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo C, Liang C, Yang J, Hu H, Fan B, Liu X. LATS2 inhibits cell proliferation and metastasis through the hippo signaling pathway in glioma. Oncol Rep. 2019;41:2753–2761. doi: 10.3892/or.2019.7065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moore DD, Luu HH. Osteosarcoma. Cancer Treat Res. 2014;162:65–92. doi: 10.1007/978-3-319-07323-1_4. [DOI] [PubMed] [Google Scholar]
- 16.Faisham WI, Mat Saad AZ, Alsaigh LN, Nor Azman MZ, Kamarul Imran M, Biswal BM, Bhavaraju VM, Salzihan MS, Hasnan J, Ezane AM, Ariffin N, Norsarwany M, Ziyadi MG, Wan Azman WS, Halim AS, Zulmi W. Prognostic factors and survival rate of osteosarcoma: a single-institution study. Asia Pac J Clin Oncol. 2017;13:e104–e110. doi: 10.1111/ajco.12346. [DOI] [PubMed] [Google Scholar]
- 17.Xie Y, Lv Y, Zhang Y, Liang Z, Han L, Xie Y. LATS2 promotes apoptosis in non-small cell lung cancer A549 cells via triggering Mff-dependent mitochondrial fission and activating the JNK signaling pathway. Biomed Pharmacother. 2019;109:679–689. doi: 10.1016/j.biopha.2018.10.097. [DOI] [PubMed] [Google Scholar]
- 18.Mead B, Tomarev S. Bone marrow-derived mesenchymal stem cells-derived exosomes promote survival of retinal ganglion cells through mirna-dependent mechanisms. Stem Cells Transl Med. 2017;6:1273–1285. doi: 10.1002/sctm.16-0428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qin F, Tang H, Zhang Y, Zhang Z, Huang P, Zhu J. Bone marrow-derived mesenchymal stem cell-derived exosomal microRNA-208a promotes osteosarcoma cell proliferation, migration, and invasion. J Cell Physiol. 2020;235:4734–4745. doi: 10.1002/jcp.29351. [DOI] [PubMed] [Google Scholar]
- 20.Liao W, Ning Y, Xu HJ, Zou WZ, Hu J, Liu XZ, Yang Y, Li ZH. BMSC-derived exosomes carrying microRNA-122-5p promote proliferation of osteoblasts in osteonecrosis of the femoral head. Clin Sci (Lond) 2019;133:1955–1975. doi: 10.1042/CS20181064. [DOI] [PubMed] [Google Scholar]
- 21.Wang G, Yang K. Neurensin-2 promotes proliferation, invasion and migration of colorectal cancer cells via interaction with SOX12. Oncol Lett. 2020;20:389. doi: 10.3892/ol.2020.12252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ren F, Zhang W, Lu S, Ren H, Guo Y. NRSN2 promotes breast cancer metastasis by activating PI3K/AKT/mTOR and NF-kappaB signaling pathways. Oncol Lett. 2020;19:813–823. doi: 10.3892/ol.2019.11152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang W, Ren A, Xiao H, Sun H, Li B. Highly expressed NRSN2 is related to malignant phenotype in ovarian cancer. Biomed Pharmacother. 2017;85:248–255. doi: 10.1016/j.biopha.2016.11.012. [DOI] [PubMed] [Google Scholar]
- 24.Ma HQ, Liang XT, Zhao JJ, Wang H, Sun JC, Chen YB, Pan K, Xia JC. Decreased expression of neurensin-2 correlates with poor prognosis in hepatocellular carcinoma. World J Gastroenterol. 2009;15:4844–4848. doi: 10.3748/wjg.15.4844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X, Han L, Zhang J, Xia Q. Down-regulated NRSN2 promotes cell proliferation and survival through PI3K/Akt/mTOR pathway in hepatocellular carcinoma. Dig Dis Sci. 2015;60:3011–3018. doi: 10.1007/s10620-015-3736-3. [DOI] [PubMed] [Google Scholar]
- 26.Wang SM, Zeng WX, Wu WS, Sun LL, Yan D. Association between a microRNA binding site polymorphism in SLCO1A2 and the risk of delayed methotrexate elimination in Chinese children with acute lymphoblastic leukemia. Leuk Res. 2018;65:61–66. doi: 10.1016/j.leukres.2018.01.004. [DOI] [PubMed] [Google Scholar]
- 27.D’Angelo R, Alafaci C, Scimone C, Ruggeri A, Salpietro FM, Bramanti P, Tomasello F, Sidoti A. Sporadic cerebral cavernous malformations: report of further mutations of CCM genes in 40 Italian patients. BioMed Res Int. 2013;2013:459253. doi: 10.1155/2013/459253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bai M, Che Y, Lu K, Fu L. Analysis of deubiquitinase OTUD5 as a biomarker and therapeutic target for cervical cancer by bioinformatic analysis. PeerJ. 2020;8:e9146. doi: 10.7717/peerj.9146. [DOI] [PMC free article] [PubMed] [Google Scholar]