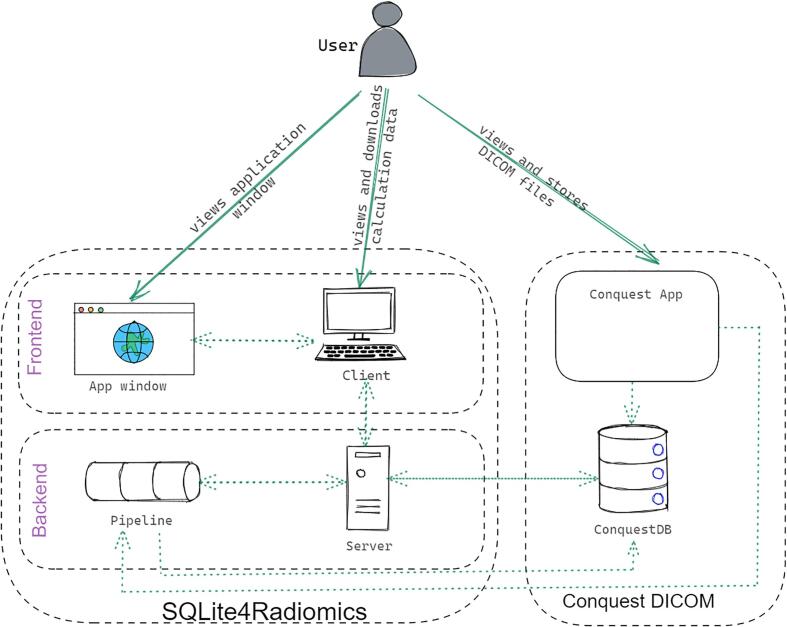
Graphical abstract

Keywords: Radiomics, Conquest DICOM, PACS
Abstract
Radiomics is referred to as quantitative imaging of biomarkers used for clinical outcome prognosis or tumor characterization. In order to bridge radiomics and its clinical application, we aimed to build an integrated solution of radiomics extraction with an open-source Picture Archiving and Communication System (PACS). The integrated SQLite4Radiomics software was tested in three different imaging modalities and its performance was benchmarked in lung cancer open datasets RIDER and MMD with median extraction time of 10.7 (percentiles 25–75: 8.9–18.7) seconds per ROI in three different configurations.
1. Introduction
Radiological images have been acquired routinely for decades in the process of radiation treatment. Commonly used imaging modalities are (cone beam) computed tomography (CT), magnetic resonance imaging (MRI), and positron emission tomography (PET). The aim of these imaging modalities is to give either anatomical (CT and MRI) or functional (MRI and PET) information. Generally, the interpretation of medical images is performed by visual inspection. With the advent of advanced computer vision, it was hypothesized that algorithms could add quantitative and objective measurements to visual interpretation, as for example, in Computer-aided diagnosis (CAD) systems [1].
Recently, radiomics, where properties of the image, such as local textures, can be used as imaging biomarkers was proposed. Radiomics has been extensively applied to quantify biological properties of the tumor for better prognostication or treatment response predictions [2]. However, many studies have shown that radiomic values can be influenced by specific acquisition settings [3], leading to a poor generalizability of radiomic signatures on multi-center data.
Radiomic feature computations need to be combined with the corresponding image acquisition settings (Digital Imaging and Communications in Medicine, DICOM, metadata) so that the feature values can be harmonized against these settings using appropriately calibrated algorithms [3], [4]. Conquest is an open-source Picture Archiving and Communication System (PACS) that stores DICOM metadata as database tables, opening the possibility of combining imaging data and metadata hosted on a PACS with radiomics. There are several radiomics extraction solutions available [5], [6], [7], [8], however, from the best of our knowledge, current existing radiomics extraction software solutions do not support direct PACS integration and database-centered radiomic feature storage.
So far, the calculation of radiomic features required some knowledge of programming. As all the imaging data is already stored in a PACS, we viewed radiomics-PACS integration as a logical step to facilitate the introduction of radiomics in a clinical environment. We believe that due to the DICOM connectivity of Conquest, SQLite4Radiomics can be used to form a pipeline within the hospital wide PACS system, enabling routine analysis of the data.
The aim of this study was to develop a solution for wider radiomics adoption in radiation oncology by integrating a publicly available library, pyradiomics, with the open-source Conquest DICOM PACS software. In this technical note, we described the architecture, workflow, and benchmarking of SQLite4Radiomics – an integration software for pyradiomics and Conquest DICOM. The code and the user manual are available as open-source.
2. Materials and methods
2.1. SQLite4Radiomics
Conquest (or Conquest DICOM) is open-source PACS software (https://ingenium.home.xs4all.nl/dicom.html) that stores imaging data as DICOM files and the DICOM metadata in a SQL database. It runs a DICOM server and has full DICOM functionality, which makes it suitable for integration in a radiation oncology department. Pyradiomics is an open-source Python package (https://github.com/Radiomics/pyradiomics) for radiomics extraction [5] from medical images as defined in the Image Biomarker Standardization Initiative (IBSI) [9]. SQLite4Radiomics further broadens Conquest’s functionality by integrating pyradiomics feature extraction into the PACS. The supplementary materials describe SQLite4Radiomics application customization, pipeline, graphical user interface (GUI) frontend and backend.
2.2. Case study: Datasets
Open data from the Maastro LUNG1 cohort [10], [11] was used to develop and test the software. These data included CT scans with manual delineations stored in the RTSTRUCT format. The LUNG1 dataset with the detailed cohort description is publicly available at the XNAT repository (https://xnat.bmia.nl/). Individual users collected their internal CT, MR, and PET data with RTSTRUCT delineations to test SQLite4Radiomics application in those modalities.
To benchmark SQLite4Radiomics performance, two open lung cancer cohorts were selected: Interobserver MMD PET-CT dataset and RIDER CT dataset, both available at the XNAT repository (https://xnat.bmia.nl/). The former consists of 22 unique PET-CT lung cancer images with multiple gross tumor volume delineations, and it was originally described and used in Aerts et al [10]. The latter, consists of 32 pairs of test–retest scans of lung cancer patients. The ROI delineations in both the datasets were provided as DICOM RTSTRUCT files suitable for SQLite4Radiomics. These data were previously used to benchmark the performance of the O-RAW software, which we compared SQLite4Radiomics performance to [6].
2.3. Case study: Benchmarking and performance evaluation
The MMD PET-CT and RIDER CT datasets were used to evaluate the performance of SQLite4Radiomics. The benchmarking pyradiomics parameter file corresponds to the default parameter file of SQLite4Radiomics stored on GitHub. With this file, 107 features including shape, first order statistics, and texture (GLCM, GLRLM, GLDM, GLSZM, NGTDM) categories were extracted from each of two ROIs per DICOMSeries. For instance, an MMD patient instance contains two series (PET and CT) – for both of those two, we extracted radiomics from two ROIs – the total of four extractions per patient.
Three system configurations were used to evaluate the performance of SQLite4Radiomics including SQLite query, plastimatch conversion, pyradiomics feature extraction, feature storage in the Conquest database. The first system configuration was represented with a HP EliteDesk 800 G2 TWR workstation (Windows 7 Enterprise, 16 GB RAM, Processor Intel(R) Core(TM) i7-6700). The second configuration was represented with HP EliteBook 840 G4 laptop (Windows 10 Enterprise, 8 GB RAM, Processor Intel(R) Core(TM) i5-7200U). The third configuration was represented by a Lenovo ThinkPad L480 laptop (Windows 10 Pro, 16 GB RAM, Processor Intel(R) Core(TM) i5-8250U). All the three system configurations were benchmarked using Novabench System Benchmarking Software (version 4.0.9 – January 2021).
A total of 107 features were extracted from two ROIs per DICOMSeries (either CT or PET) with the default parameter and configuration files listed on SQLite4Radiomics repository. The extraction time in seconds per ROI was chosen as a performance benchmarking metric in our study. There is, however, the possibility to customize the SQLite4Radiomics performance evaluation by, for instance, extraction time per voxel of ROI – this might be beneficial when there is a high ROI volume variation in a dataset.
2.4. User testing
In addition to the developers, two users independently tested the standalone pipeline application, while five users tested the GUI version. Two of the users are clinical physicists, one is a radiobiologist, and two are researchers. The user tests were performed by observation (a user performs a set of tasks, while a developer observes the process without interfering and takes notes) and in a remote fashion (a user freely tested and used SQLite4Radiomics tool in their own time). In both cases, feedback was given by the users at the end of each test.
3. Results
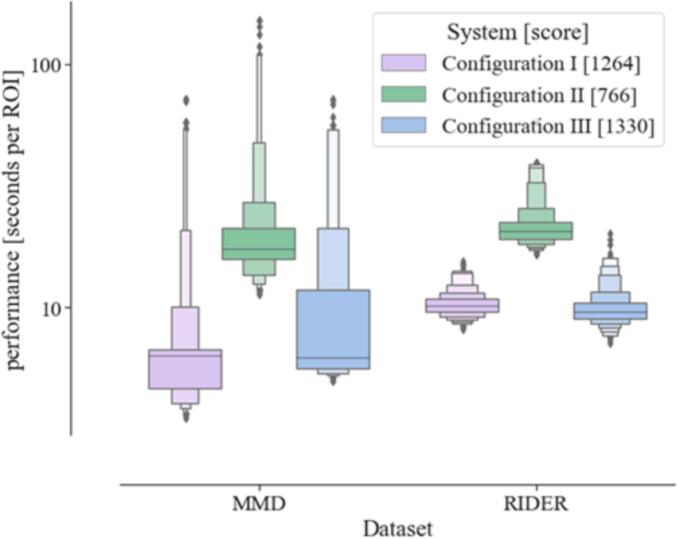
In system configuration I, it took 4057, 4074, and 4204 s to process the RIDER dataset with a median extraction time per ROI of 10.1 s and it took 1592, 1537, and 1547 s to run the MMD dataset with a median extraction time per ROI of 6.3 s. In system configuration II, it took 7492, 7527, and 8745 s to run the RIDER dataset with a median extraction time per ROI of 20.5 s and it took 3472, 3843, and 3727 s to run the MMD dataset with a median extraction time per ROI of 17.4 s. In system configuration III, it took 4687, 4817, and 4776 s to run the RIDER dataset with a median extraction time per ROI of 9.7 s and it took 1632, 1596, and 1696 s to run the MMD dataset with a median extraction time per ROI of 6.2 s. All three configurations resulted in the total of 18 successful runs of SQLite4Radiomics. The results are represented in Fig. 1.
Fig. 1.
Performance letter-value plot (generalization of the boxplot [15]). The graph shows the performance in seconds per region-of-interest in both MMD PET-CT and RIDER CT datasets for the three system configurations we used to benchmark SQLite4Radiomics’ performance in. The letter-value plot generalizes the classical boxplot by visualizing more percentiles (in %): [(3.125, 96.875), (6.25, 93.75), (12.5, 87.5), (25.0, 75.0)]. The box area is proportional to the percentage of covered data. Each system configuration contains its Novabench score for reference.
Each of the three configurations were benchmarked with the Novabench System Benchmarking Software. The configuration I received a total score of 1264 (CPU 920, RAM 257, Disk 87, GPU unavailable); the configuration II received 766 points (CPU 395, RAM 169, Disk 55, GPU 147), and the configuration III received 1330 (CPU 821, RAM 254, Disk 52, GPU 203). The GPU score was irrelevant for this specific study because the radiomics calculation and SQLite4Radiomics operation were CPU-based.
4. Discussion
Radiomics currently lacks integration in clinical pipelines. At the same time, we routinely generate, archive, and store images routinely in hospital PACS systems. In order to bridge radiomics and its clinical application, we presented SQLite4Radiomics – an integration software of pyradiomics and Conquest DICOM – two popular open-source tools of both worlds of radiomic analysis imaging and clinical imaging. SQLite4Radiomics can receive data through the standard DICOM frameworks present as a part of treatment planning workflow in radiation oncology.
We have built-in plastimatch (https://plastimatch.org/) conversion that removes the burden from the user regarding the conversion of the contour data of DICOM RTSTRUCT into binary mask files. The volumes (ROIs) selection is customizable, which allows for better match of local ROI labeling in a particular clinic. The calculated radiomic data is stored in a alongside the DICOM metadata and images. Therefore, the data can be easily coupled to statistical environments such as Python and R, which have database connection facilities. Due to the integration of the DICOM metadata and the radiomics output, the data can be combined to examine dependencies between the two [3], [4]. Although SQLite4Radiomics allows for simpler radiomics extraction and storage, it also gives the users opportunity to investigate the relationship between a radiomic feature and image acquisition settings. We would like to encourage the radiomics community to use this opportunity to further improve the reporting quality on radiomics reproducibility [12]. In addition, our approach will lead to simpler integration of radiomics and conventional clinical variables, such as performance and tumor stage, into cancer prognostic models [13].
We found that the computer performance benchmarking score (e.g. with Novabench) may be a relevant predictor in SQLite4Radiomics time efficiency approximation as the score is based on the processor and RAM performance. The time efficiency of SQLite4Radiomics is comparable to that of pyradiomics and it outperforms O-RAW. The main difference in the execution times between O-RAW and SQLite4Radiomics may be due to O-RAW’s RDF-conversion of radiomic data, which is not present in SQLite4Radiomics [6]. Yet the extraction can be performed without the user input if the RTSTRUCT trigger is enabled, which makes the access time to radiomic data comparable with executing an SQLite query only.
The radiomics extraction and storage process was automated using SQLIte4Radiomic tool and can be customized according to research or clinical requirements. The software was developed with the possibility to be extended upon by the research community and is open-source. With regards to the future functionality, we see the following possible developments. Currently, SQLite4Radiomics is hosted on a local machine, therefore, an extension may include scaling up with authentication and proper online hosting. SQLite4Radiomics can possibly be extended by converting the SQLite radiomics tables to SPARQL RDF triples to match Radiomics Ontology [6]. This integration with RO and machine learning toolkits will allow to perform rapid learning to automatically re-adjust prognostic models whenever new data comes in, as was proposed by Deist et al [14].
Interestingly, SQLite4Radiomics is based on pyradiomics, which was not originally intended to mimic IBSI’s image processing exactly. This causes a mismatch while executing feature extraction in the Lung Cancer CT phantom. This issue was addressed on pyradiomics github page (https://github.com/AIM-Harvard/pyradiomics/issues/498). For the purpose of matching the pre-processing of IBSI and pyradiomics, we listed IBSI-compliant pre-processing methods in the IBSI-pyradiomics_discrepancies branch.
SQLite4Radiomics reduces the entrance threshold for clinical researchers and makes radiomics extraction from organic: whenever new images arrive in a PACS, radiomic features can be extracted and stored as SQLite tables. Radiomics extraction is based on a popular open-source pyradiomics library and DICOM-RT conversion is performed by plastimatch – reliable open-source ITK software. SQLite4Radiomics makes radiomics data easy to store, query and combine with clinical data at any time with no need for additional wrappers.
Funding
This work is part of the research program STRaTeGy with project number 14930, which is (partly) financed by the Netherlands Organization for Scientific Research (NWO).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors thank Paul Rijken, Guy Warmerdam, and Theo Lam for helping with SQLite4Radiomics quality assurance and for giving their feedback.
References
- 1.Doi K. Computer-aided diagnosis in medical imaging: Historical review, current status and future potential. Comput. Med. Imaging Graph. 2007;31(4-5) doi: 10.1016/j.compmedimag.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gillies R.J., Kinahan P.E., Hricak H. Radiomics: Images are more than pictures, they are data. Radiology. 2016;278(2) doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhovannik I. Learning from scanners: Bias reduction and feature correction in radiomics. Clin. Transl. Radiat. Oncol. 2019;19:33–38. doi: 10.1016/j.ctro.2019.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Orlhac F. A postreconstruction harmonization method for multicenter radiomic studies in PET. J. Nucl. Med. 2018;59:8. doi: 10.2967/jnumed.117.199935. [DOI] [PubMed] [Google Scholar]
- 5.Van Griethuysen J.J.M. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017;77(21) doi: 10.1158/0008-5472.CAN-17-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi Z., Traverso A., van Soest J., Dekker A., Wee L. Technical Note: Ontology-guided radiomics analysis workflow (O-RAW) Med. Phys. 2019;46(12) doi: 10.1002/mp.13844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pfaehler E., Zwanenburg A., de Jong J.R., Boellaard R. RACAT: An open source and easy to use radiomics calculator tool. PLoS One. 2019;14:2. doi: 10.1371/journal.pone.0212223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Szczypiński P.M., Strzelecki M., Materka A., Klepaczko A. MaZda-A software package for image texture analysis. Comput. Methods Programs Biomed. 2009;94(1) doi: 10.1016/j.cmpb.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 9.Zwanenburg A. The image biomarker standardization initiative: Standardized quantitative radiomics for high-throughput image-based phenotyping. Radiology. 2020;295(2) doi: 10.1148/radiol.2020191145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aerts H.J.W.L., Velazquez E.R., Leijenaar R.T.H., Parmar C., Grossmann P., Carvalho S. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 2014;5(1) doi: 10.1038/ncomms5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kalendralis P. FAIR-compliant clinical, radiomics and DICOM metadata of RIDER, interobserver, Lung1 and head-Neck1 TCIA collections. Med. Phys. 2020;47(12) doi: 10.1002/mp.14805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Traverso A., Wee L., Dekker A., Gillies R. Repeatability and Reproducibility of Radiomic Features: A Systematic Review. Int. J. Radiat. Oncol. Biol. Phys. 2018;102(4) doi: 10.1016/j.ijrobp.2018.05.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foley K.G. External validation of a prognostic model incorporating quantitative PET image features in oesophageal cancer. Radiother. Oncol. 2019;133:205–212. doi: 10.1016/j.radonc.2018.10.033. [DOI] [PubMed] [Google Scholar]
- 14.Deist T.M., Jochems A., van Soest J., Nalbantov G., Oberije C., Walsh S. Infrastructure and distributed learning methodology for privacy-preserving multi-centric rapid learning health care: euroCAT. Clin. Transl. Radiat. Oncol. 2017;4:24–31. doi: 10.1016/j.ctro.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hofmann H., Wickham H., Kafadar K. Letter-Value Plots: Boxplots for Large Data. J. Comput. Graph. Stat. 2017;26 doi: 10.1080/10618600.2017.1305277. [DOI] [Google Scholar]