Abstract
In most trinucleotide repeat (TNR) diseases, the primary factor determining the likelihood of expansions is the length of the TNR. In some diseases, however, stable alleles contain one to three base pair substitutions that interrupt the TNR tract. The unexpected stability of these alleles compared to the frequent expansions of perfect TNRs suggested that interruptions somehow block expansions and that expansions occur only upon loss of at least one interruption. The work in this study uses a yeast genetic assay to examine the mechanism of stabilization conferred by two interruptions of a 25-repeat tract. Expansion rates are reduced up to 90-fold compared to an uninterrupted allele. Stabilization is greatest when the interruption is replicated early on the lagging strand, relative to the rest of the TNR. Although expansions are infrequent, they are often polar, gaining new DNA within the largest available stretch of perfect repeats. Surprisingly, interruptions are always retained and sometimes even duplicated, suggesting that expansion in yeast cells can proceed without loss of the interruption. These findings support a stabilization model in which interruptions contribute in cis to reduce hairpin formation during TNR replication and thus inhibit expansion rates.
Trinucleotide repeat (TNR) instability has been found in more than 12 human neurological diseases that have many common genetic traits (1, 7, 18). One of the most important predictors of TNR expansions is the length of the triplet tract. In normal populations, the TNR is highly polymorphic but relatively short. In affected individuals the TNR is expanded anywhere from 5 to 2,000 repeats, depending on the disease locus (1, 7, 18). Not only does expansion often lead to disease but longer repeats are also further destabilized, expanding with even higher frequency upon subsequent transmissions. The cutoff between short, stable alleles and long, unstable alleles has been termed the threshold (1, 7, 18). Once a threshold of about 35 repeats is reached, the likelihood of expansion in the next generation is greatly increased.
The purity of the TNR tract also influences its mutability. There are three examples of TNR diseases in which most normal, stable alleles contain one to three point mutations, or interruptions, interspersed within the perfect repeat tract. SCA1 (spinocerebellar ataxia type 1 gene) contains CAT interruptions in a CAG tract (2), the CGG tract of the fragile X syndrome gene FMR1 is punctuated with AGGs (3, 9, 13, 25), and CAAs are dispersed through the CAG tract of SCA2 (10, 21, 23). About 1% of normal Friedreich's ataxia alleles have five to eight GAGGAA hexanucleotide interruptions within a GAA tract (17). In contrast, expanded alleles of these genes have fewer or no interruptions. This correlation has led to the suggestion that one or more interruptions must be lost before expansion can occur (2, 3, 9, 10, 13, 22, 25). The most direct test of this hypothesis would be to look at expansions that arise directly from interrupted alleles. However, it has not been possible to address this question directly owing to the lack of such events in humans.
Molecular models for the stabilizing influence of interruptions include the idea that they provide an anchoring sequence which helps keep the two strands of the duplex properly aligned to prevent replicational slippage (19, 26). Alternatively, interruptions may reduce the thermodynamic stability of DNA hairpins that are thought to be important intermediates in the expansion process (5, 19, 21). Another possibility is that interruptions stabilize TNRs by breaking up the perfect repeat stretches into smaller, subthreshold lengths. Expansions would occur only if the perfect repeat regions can lengthen to achieve the threshold, as has been suggested for fragile X (3, 9, 22, 25). In vitro, interruptions have been shown to decrease formation of slipped-strand DNA (S-DNA) structures and also to limit the number of different S-DNA isomers (19), which is consistent with the idea that interruptions somehow physically demarcate the positions where perfect repeats start and stop.
Using a yeast genetic assay, we were able to test these models. Our results support the idea that interruptions in yeast can help prevent the formation of hairpin DNA intermediates thought to be important for the expansion process.
MATERIALS AND METHODS
Strains.
Escherichia coli DH5α [endA1 hsdR17 (rK− mK+)] supE44 thi-1 recA1 gyrA (Nalr) relA1 Δ(lacI ZYA-argF) U169 deoR)] was used for plasmid construction of the interrupted TNR repeats and large-scale plasmid preparations. The Saccharomyces cerevisiae strain used was MW 3317-21A (MATα Δtrp1 ura3-52 ade2Δ ade8 hom3-10 his3-KpnI met4 met13) (11). TNR-containing plasmids were directed to integrate at LYS2 by Bsu36I digestion of the appropriate plasmids. The linearized plasmids were then integrated via lithium acetate transformation (24). Single integration of the TNR sequence at LYS2 was confirmed by Southern hybridization by using a 1.1-kb fragment of LYS2 as the probe sequence.
Plasmid constructs.
All plasmids were constructed using the pBL94 vector (16), which contains the URA3 gene driven by the Schizosaccharomyces pombe adh1 promoter, with a unique SphI site separating the two elements. Variants of pBL94 containing interrupted TNR sequences were generated by annealing 5′-phosphorylated complementary oligonucleotides as follows: oligonucleotides were melted 10 min at 90°C, allowed to anneal by cooling to 37°C for 40 min, and then held at 25°C for 10 min. Annealed oligonucleotides were cloned into the SphI site of pBL94. To generate the 5′ interrupted sequence, oBL 199 [5′-(CTG)6 ATGATG(CTG)17 CATG 3′] was annealed to oBL 198 [5′-(CAG)17 CATCAT(CAG)6 CATG-3′]. The 3′ interruption was created by pairing oBL 197 [5′ (CTG)17 ATGATG(CTG)6 CATG-3′] with oBL 196 [5′-(CAG)6 CATCAT(CAG)17 CATG-3′]. The centrally interrupted sequence was derived from oBL 220 [5′-(CTG)11 ATGATG(CTG)12 CATG-3′] and oBL 219 [5′ (CAG)12 CATCAT(CAG)11 CATG-3′]. Plasmids were transformed into DH5α by using the Hanahan procedure (8) or by electroporation at 2.5 mV with a Bio-Rad E. coli pulser. Plasmids were recovered from DH5α by using a QIAspin miniprep kit (Qiagen) according to the manufacturer's protocol. Plasmids were sequenced with Sequenase 2.0 by using the U.S. Biochemicals sequencing kit and protocol to confirm the accuracy of the cloned sequence prior to integration into yeast.
Fluctuation analysis.
Fluctuation analysis was performed as previously described (16). The rates of TNR instability were determined by the method of the median (14). Briefly, single yeast colonies harboring interrupted repeats were resuspended in water and appropriate dilutions were plated onto nonselective media (YPD). After 24 to 36 h of growth at 30°C, 7 to 10 colonies were resuspended in water, and an appropriate dilution was plated on YPD for total cell counts, while the remaining suspension was plated on selective complete medium lacking histidine but containing 1 mg of 5-fluoro-orotic acid (5FOA) per ml. To ensure reproducibility a minimum of three repetitions of the fluctuation assay were performed per strain, and at least three independently isolated clones were tested.
Molecular analysis of independent expansion events.
The colonies grown on the selective media were subjected to colony PCR by using published procedures (16). Briefly, template DNA was released from single colonies by heating in a solution containing dithiothreitol and Triton X-100. PCR, usually in the presence of 0.25 μCi of [α-32P]dCTP, was performed with primers that flank the triplet repeat tract. The products of the PCRs were analyzed on a denaturing 6% polyacrylamide gel, and the product sizes (± one to two repeats) were determined by comparison of the reaction products with a M13 DNA sequence ladder.
Two safeguards were included to minimize microheterogeneity of the starting tract size. First, prior to fluctuation analysis, a portion of each colony was examined by PCR to ensure that the starting tract contained 25 repeats. Second, after the fluctuation test, individual colonies from the YPD plate were tested by PCR for high-resolution analysis of the tract size. All 88 colonies tested gave PCR sizes corresponding to 25 ± 2 repeats. Therefore, the total tract size of 25 repeats in unselected cells was maintained throughout the experiment.
For determination of the retention and position of interruptions in the expanded alleles, PCR products were purified by using a QIAquick PCR purification kit (Qiagen) according to the manufacturer's protocol. Purified PCR products were subjected to restriction analysis with SfaNI (New England Biolabs). A typical reaction used 10,000 to 30,000 cpm of PCR product and 1.25 U of enzyme in a total volume of 9 μl. Digestion products were displayed on a 6% sequencing gel. In some cases, PCR products were sequenced to determine if either interruption was lost during an expansion event. PCR products for sequencing were purified by electrophoresis on a nondenaturing 7.5% polyacrylamide gel buffered with 45 mM Tris, 45 mM boric acid, and 1 mM EDTA. Products were visualized by ethidium bromide staining. Products were then excised from the gel and placed in 0.3 ml of 10 mM Tris-Cl (pH 7.6)–1 mM EDTA. After maceration, the gel fragments were soaked overnight at room temperature. Eluted PCR products were lyophilized to 100 μl and further purified via a QIAquick PCR purification kit (Qiagen) according to the manufacturer's protocol. The PCR products were sequenced using an ABI automated sequencer following the manufacturer's protocol for cycle sequencing to incorporate a fluorescently labeled nucleotide.
RESULTS
Model under investigation.
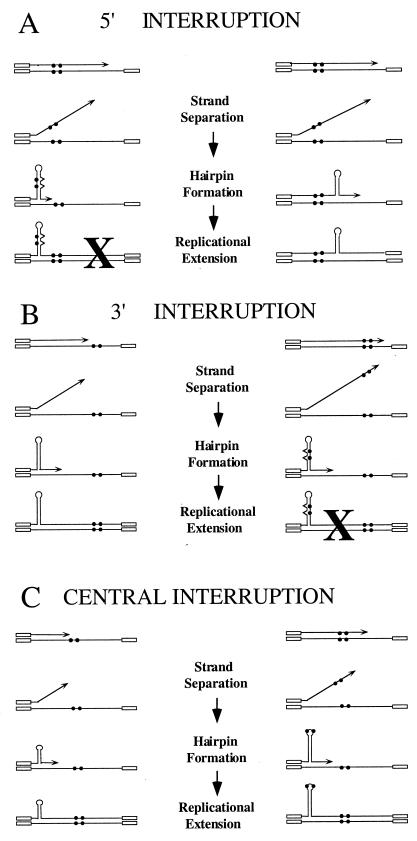
We tested predictions of the model that interruptions inhibit expansions by reducing hairpin stability (5, 19, 21). If this is so, placement of interruptions at different positions within the TNR tract, relative to the direction of replication, may lead to different outcomes (Fig. 1). Interruptions near the 5′ end of the TNR will likely be synthesized prior to hairpin formation (Fig. 1A). Interrupting bases in the newly synthesized strand might help prevent expansions in two ways, as shown in the left side of Fig. 1A. The interruptions might provide a reference point for proper reassociation of the separated strands. Alternatively, folding of the daughter strand might incorporate the interruptions into the hairpin stem, thus weakening it. Both factors would disfavor expansions, as indicated by the X in the left panel of Fig. 1A and as suggested previously (5, 19, 21, 26). The right panel depicts hairpin formation without including the interruptions. Under these circumstances, expansions would proceed unhindered. This model predicts that 5′ interruptions should have a large stabilizing effect because many expansions would be precluded. Duplication of the interruption should be rare since only the left panel generates duplications. Events occurring via the scheme presented in the right panel would lead to polar expansions, with new repeats added after the interruptions. The size of these expansions would most frequently be limited by the size of the perfect tract.
FIG. 1.
Model of interrupt-mediated stabilization. In this schematic the open boxes represent nonrepetitive flanking sequence, the lines denote the TNR, the filled circles symbolize the interruptions, arrowheads mark the 3′ end of the Okazaki fragment, and carats represent mismatches. The illustration uses two interruptions, since this is a frequently observed allele in the relevant human disease genes. (A) Two possible outcomes are shown for the 5′ interruption. The left panel represents hairpin folding that incorporates the newly synthesized interruptions into the stem of the hairpin. Expansions in the left panel are disfavored, as denoted by the “X” and as described in the text. The right panel shows a possible mechanism for expansion in which the hairpin does not include the interruptions. Replicational extension leads to the precursor for an expanded allele. Note that the expansion in the right panel is predicted to occur downstream (3′) of the interruption. (B) Two outcomes are displayed for the 3′ interruption. The left panel would be expected to produce expansions more often than the right panel (denoted by “X”). Expansions arising from the left panel should occur upstream (5′) of the interruption. (C) Two pathways leading to expansion of the central interruption. In the left panel, expansions are predicted to be ≤11 repeats and to occur upstream of the interruption. In the right panel, interruptions are incorporated in or near the loop of the hairpin and thus have a limited negative impact on hairpin formation. Expansions arising from the right panel are expected to duplicate the interruption. Although not shown, disfavored structures like those in panels A and B are also possible when the interruptions are incorporated into the stem of the hairpin.
Figure 1B indicates that 3′ interruptions should lead to a lesser degree of stabilization. In the left panel, hairpin formation frequently occurs prior to synthesis of the interruptions. Since the Okazaki fragment contains a perfect repeat, hairpin formation can proceed unhindered. The right panel shows the less-frequent case where synthesis includes the interruptions and thus hairpin formation would be disfavored. Based on the model, expansions from 3′ interruptions should be polar, with new repeats inserted before the interruptions, and duplications should be rare.
Centrally located interruptions (Fig. 1C) provide two scenarios where expansions might proceed unhindered. The left panel shows small hairpins forming prior to synthesis of the interruptions. These events are relatively uncommon, based on the fact that small expansions from perfect repeats make up only a minor portion of events in our system (16). Expansions that arise from this mechanism should be limited to the number of repeats preceding the interruptions and will not show duplication of the interruption. As shown, the polarity is 5′ to the interruption. Although expansions with 3′ polarity are also possible, no events with 3′ polarity were observed experimentally. The right panel of Fig. 1C indicates hairpin folding with the interruptions in or near the loop. This folding is expected to have less of an effect on hairpin stability than if the interruptions are in the stem. If so, expansions might proceed more readily and should yield relatively large expansions due to the requirement for added base pairing in the stem to overcome the slightly unfavorable effect of the interruptions in the loop. Duplication of the interruption will result by the scheme presented in the right panel of Fig. 1C. Our experimental results are consistent with expansions of this type. Disfavored events, where interruptions are included in the stem of the hairpin, have been omitted from Fig. 1C for reasons of clarity. These disfavored events are predicted to account for a relatively large portion of possible hairpins. Thus, expansions arising from central interruptions should show an intermediate level of stabilization, less than for 5′ alleles but greater than for 3′ interruptions.
All events in Fig. 1 are drawn as 3′ slippage events. However, Gordenin et al. (6) suggested that expansions can also occur by polymerase-mediated displacement of the 5′ end of an existing Okazaki fragment, followed by hairpin folding and ligation to the daughter strand. Our experiments with interrupted TNR tracts were not designed to distinguish between hairpins arising from 3′ slippage versus 5′ flaps, since the events in Fig. 1 can be drawn with 5′ flaps and yield virtually identical predictions.
Choice of interrupted TNRs.
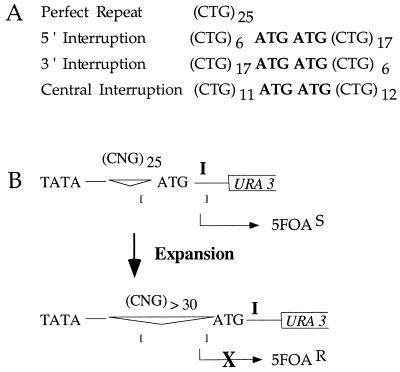
To address models for TNR stabilization by interruptions, we examined interrupted repeats in yeast, where rare events can be easily detected and characterized. We generated test sequences similar to interrupted alleles of SCA1, many of which contain one to three CAT interruptions of CAG repeats, resulting in total tract lengths equal to 23 to 36 repeats (2). The complementary strand therefore harbors ATG interruptions in a CTG tract. We have shown previously (16) that perfect (CTG)25 tracts are unstable in yeast. These tracts expand frequently (∼10−5 per cell generation) when present on the lagging daughter strand during DNA replication. Therefore, if ATG interruptions stabilize CTG tracts in yeasts as in humans, the rate of expansion should be reduced relative to an uninterrupted control. In this work, TNR sequences were constructed that contained two adjacently placed interruptions to maximize the stabilizing effect (Fig. 2A). Because the interrupted alleles were integrated at the LYS2 locus where the direction of DNA replication is known (4), the assignment of 5′ and 3′ interruptions is accurately reflected in Fig. 2A.
FIG. 2.
Selective identification of expanded TNR. (A) Sequences represent the TNR sequences used for this study. Nomenclature is that of the lagging daughter strand. (B) Genetic assay to select for TNR expansions in yeast. The region controlling expression of the reporter gene URA3 is shown, including the TATA box, the TNR region, an out-of-frame initiation codon, the preferred transcription initiation site (I), and the start of the URA3 gene. The upper diagram demonstrates the starting construct. The brackets represent a window of potential transcription initiation sites located 55 to 125 bp from the TATA box. With a 25-TNR tract, the transcription window includes the preferred site I, thus leading to expression of URA3 and making the yeast 5FOA sensitive. The lower diagram demonstrates what happens when the TNR tract expands to ≥30 repeats. The bracketed window does not include site I. Transcription initiating 5′ of the preferred initiation site will include the out-of-frame ATG, resulting in translational incompetence and therefore resistance to 5FOA.
Genetic assay.
Stabilizing effects of interruptions were assayed by using a selective, sensitive, and quantitative genetic assay in S. cerevisiae (16). This assay (Fig. 2B) allows the identification of expanded TNR alleles based on phenotypic changes. Briefly, a starting tract of 25 repeats, either uninterrupted or interrupted, allows expression of the URA3 reporter gene with concomitant sensitivity of the cells to the cytotoxic effects of 5-fluoro-orotic acid (5FOA). Expansions of the tract to lengths of ≥30 repeats inactivate the URA3 reporter, and the cells are accordingly resistant to 5FOA. The rate of expansion is therefore proportional to the number of 5FOA-resistant colonies. This assay was specifically designed to reveal expansions of ≥5 repeats because this size class is among the most frequent in human TNR diseases (1, 7, 18). The 5FOA assay avoids interference by small expansions of one to four repeats or contractions. The yeast assay also allows use of single-colony PCR to characterize the expanded alleles (16).
Early replicated interruptions provide the greatest TNR stabilization.
Interrupted TNRs were stabilized relative to an uninterrupted control, as judged by lower expansion rates (Table 1). The perfect (CTG)25 tract yielded a rate of 1.0 × 10−5 per cell generation, a result consistent with our previous work (16). In contrast, there was a stabilization of 90-fold when the interruption was placed 5′ in the TNR tract. Stabilization was also observed for the 3′ allele, but the extent was much smaller, only 2.4-fold relative to the control. When the interruption was placed in the center of the tract, the expansion rate was reduced 16-fold compared to the uninterrupted tract. In addition to these stabilizing effects with the reporter integrated at LYS2, similar results were observed for integration at URA3. The direction of replication through the repeat tract is the same for integration at both loci (4, 16). Compared to the uninterrupted repeat, the rate of 5FOA resistance was reduced >20-fold by the 5′ interruption and 2.9-fold by the 3′ interruption, indicating that the stabilizing influence is general rather than LYS2 specific. We conclude that interruptions reduce the rate of TNR expansions in yeast as they do in humans. Clearly, the extent of stabilization depends on the placement of the interruption with respect to the direction of DNA replication. The rate results shown in Table 1 are in agreement with predictions from the model in Fig. 1.
TABLE 1.
Expansion rates of perfect and interrupted CTG tracts
| TNRa | Rateb/cell generation | Stability increasec |
|---|---|---|
| Perfect repeat | 1.0 (±0.3) × 10−5 | 1 |
| 5′ interrupt | 1.1 (±0.5) × 10−7 | 90 |
| 3′ interrupt | 4.2 (±1.7) × 10−6 | 2.4 |
| Central interrupt | 6.1 (±4.3) × 10−7 | 16 |
Repeat sequences are specified in Fig. 2A.
Rates are given as the mean (± standard deviation).
Stability increase is calculated as the expansion rate of perfect repeats divided by the expansion rate of an interrupted repeat.
The interruptions are always retained in the expanded alleles.
The ATG interruptions within the CTG tract provide a recognition site for the restriction enzyme SfaNI. If expanded alleles retain the interruptions, then PCR products from these tracts should be sensitive to the enzyme. As described in detail in the next section, we tested a total of 60 genetically independent expansions by this method (17 from the 5′ interruption, 22 from the 3′ interruption, and 21 from the central interruption). Cleavage was observed in every case. In contrast, a control strain with a perfect CTG repeat did not give a cleavable PCR product. Therefore, at least one interruption was maintained in 60 of 60 expansions from the interrupted tracts. Since SfaNI cleavage requires that only one of the two ATGs be retained, it was possible that the expansions had lost one interruption either prior to or during expansion. To address this question, we sequenced five expansions (three from the 5′ interruption and two from the 3′ interruption). Sequencing determined that all five alleles retained both ATGs.
Polarity of expansions.
Mapping of the SfaNI site in the expanded alleles provided a means for assessing whether expansions from interrupted tracts are polar. In other words, are the extra triplet repeats generated preferentially on one side of the interruption? The model in Fig. 1 predicts polarity in certain cases. For the 5′ interruptions, expansions should occur primarily downstream of the interruption. The situation is reversed for the 3′ alleles, where expansions are expected upstream of the substitution. For the centrally located interruptions, some events will show polarity on the 5′ side (left panel of Fig. 1C). Expansions resulting from the scheme in the right panel of Fig. 1C will not yield polar products because the expansion will occur between the original interruption and a new, duplicated copy of the interruption. If events of the latter class exist, the SfaNI site will be duplicated and therefore an extra fragment should be observed after digestion.
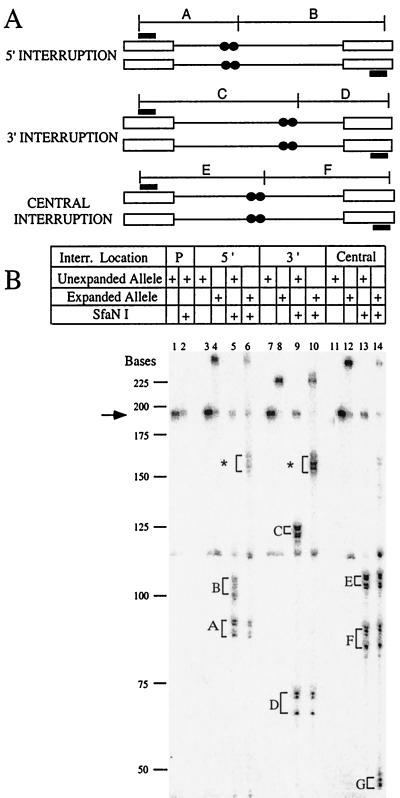
By comparing the sizes of PCR products that are uncut with those cleaved with SfaNI, we can deduce the location of extra triplet repeats in the expanded alleles. The starting tracts are shown in Fig. 3A, including the positions of the interrupting base pairs. Examples of this analysis are shown in Fig. 3B. The control sample in lanes 1 and 2 was amplified from a strain with a perfect tract of 25 repeats. Treatment of the sample with SfaNI did not cleave the DNA, as expected (compare lane 2 to lane 1). Lanes 3 and 4 were uncut samples from the 5′ interrupt; lane 3 was an unexpanded control, and lane 4 was one of the expanded alleles. When these samples were treated with SfaNI (lanes 5 and 6), cleavage occurred. The unexpanded sample in lane 5 gave the expected band sizes, corresponding to fragments A and B (Fig. 3A). The expanded sample in lane 6 showed that fragment A was unaltered but that fragment B was increased in size to the position marked with an asterisk and that this increase matched the overall expansion size. Therefore, this sample showed a polar expansion downstream of the interruption.
FIG. 3.
Example of SfaNI digests to identify interruptions and to determine polarity of expansions. (A) Schematic diagram of the PCR products of the unexpanded TNRs tested. The thin line represents the TNR, the open boxes represent the nonrepetitive flanking sequence, filled boxes show the location of the PCR primers, and the filled circles represent the location of the interruptions within the TNR tract. The expected fragments after digestion of these PCR products with SfaNI are shown (labeled A to F). The cut site for SfaNI is displaced by five nucleotides from the recognition sequence, and restriction fragments A to F are adjusted in size accordingly. Due to the four-nucleotide overhang generated by SfaNI and the fact that the PCR fragments are uniformly labeled in the presence of [α-32P]dCTP, cleaved samples should show two bands differing in size by four bases. The expected sizes (in nucleotides) are: fragment A, 90 and 94; fragment B, 101 and 97; fragment C, 123 and 127; fragment D, 68 and 64; fragment E, 105 and 109; and fragment F, 86 and 82. (B) Autoradiograph of representative digests of each of the TNRs tested. Markers were determined from a M13 sequence ladder, and the arrow indicates the size of the uncleaved starting tract. The table above the autoradiograph identifies which lanes have the starting or expanded products and which of these products were subjected to SfaNI digestion. The designation “P” for lanes 1 and 2 indicates a perfect repeat. Labeled fragments A to F refer to the expected fragments listed in panel A. The asterisks in lanes 6 and 10 designate the expanded alleles of fragments B and C, respectively. Fragment G is the result of a duplicated interruption. The brackets indicate the products of both strands of the digestion products, separated by four nucleotides as explained above. Shadow bands are due to polymerase “chatter” which is commonly seen during PCR of TNRs.
A parallel analysis of the 3′ interrupted allele also yielded polar expansions, but in this case the addition was before the interruption. Lanes 7 and 8 show the uncleaved PCR fragment of a control, unexpanded colony (lane 7) and an expansion (lane 8). Treatment with SfaNI yielded the expected fragments labeled C and D (lane 9). In the expanded sample of lane 10, the D fragment size was unchanged, whereas fragment C was increased to the position marked by the asterisk and this increase accounted for the overall expansion size. Comparison with the schematic diagram in Fig. 3A shows that polar expansions located 5′ to the interruption would lead to increased size of fragment C. A different pattern was frequently observed when the interruption was centrally located. Uncleaved samples (lanes 11 and 12) again demonstrated the expansion for the 5FOA-resistant colony. Cleavage of both samples yielded the same size fragments E and F (lanes 13 and 14), indicating that expansion did not affect these regions of DNA. Instead, a new fragment labeled G was observed (lane 14), a finding consistent with duplication of the interruption and therefore the creation of an extra SfaNI site. In the example shown in Fig. 3B, the duplication created a fragment of 18 repeats, one presumed to consist of 16 new CTG repeats and 2 extra ATG triplets.
Since the interruption was retained in all 60 independent expansions that were analyzed, polarity could be determined. As shown in Fig. 4A, expansions of the 5′ interrupted tract were exclusively polar, with extra DNA added after the interruptions. Similarly, expansions of the 3′ interrupted tract (Fig. 4B) were also polar but in this case addition was exclusively before the interruption. In all cases examined (39 of 39 events) for the 5′ and 3′ alleles, the repeats were added to the larger perfect repeat tract as predicted by the model in Fig. 1. For the central interruption, two short expansions showed a 5′ addition (Fig. 4C). One might expect an equivalent distribution of 5′ and 3′ additions to the centrally interrupted tract due to the nearly identical perfect repeat tract lengths on either side. However, no 3′ addition events were detected. This may be due to the few expansions of this size class that were observed. As seen in Fig. 4C, the great majority (19 of 21) of larger expansion events duplicated the interruption. Since duplications were defined by the generation of a new SfaNI fragment, there must be at least one ATG repeat at each end of the expansion. We presume that both ATG repeats were duplicated, although this point was not tested. It is also noteworthy that the size of the duplication frequently exceeded the largest perfect repeat stretch, 12 triplets, present in the starting tract. Therefore, any single expansion event must have bridged the site of the interruption.
FIG. 4.
Polarity of expansions for the interrupted TNRs. The open circles represent a CTG repeat, and the filled circles represent an ATG interruption. In panels A through C the starting allele is designated as Start. On the side of each represented TNR is the number of triplet repeats added (+13, etc.) and the number of times the expanded TNR was observed. (A) Expanded products of the 5′ interrupted TNR. (B) Expansions arising from the 3′ interrupted TNR. (C) Expansions of the centrally interrupted TNR. In panel C, expansions that led to duplication of the interruption are labeled as “dup.” The size values indicate the total number of repeats added, including the duplication. For example, “+14 dup.” means that 12 extra CTGs and 2 extra ATGs were present. The expansion polarity of the duplicated alleles could not be determined, and those events are diagrammed symmetrically below the starting allele.
Effects of the interruption location on the expansion product size.
The predictions from Fig. 1 suggest that expansions from tracts containing either 5′ or 3′ interruptions will be limited to increases of ≤17 repeats, since this limit is set by the longest uninterrupted repeat stretch for these alleles. For interruptions in the center of the TNR tract, a mixture of sizes was predicted. In addition to the events illustrated in Fig. 4B, we examined the sizes of more expansions arising from each type of tract to generate a mutational spectrum. Expansions from the perfect TNR tract provide a useful comparison. They range in size from 10 to 22 repeats with a median of 15 (Fig. 5A), a result consistent with a previous report (16). The expansion products seen in the 5′ interrupted TNR (Fig. 5B) were of similar size (13 to 23, with a median of 15). Of the 19 independent events examined for the 5′ interrupted tract, all but three fell within the predicted size limit of the model. Among the expansions from the 3′ interrupted tract (Fig. 5C), all 36 were of the predicted size class. However, all but one of the expanded alleles stemming from the 3′ interrupted TNR fell into an apparently smaller grouping (range, 5 to 11; median, 8) than for the perfect tract or for the 5′ interruption. The products of expansion seen for the 3′ interruption were significantly different than the products recovered from expansion of the perfect repeat tract or the 5′ interrupted tract, as judged by the Student's t test (P < 0.001 in each case). The major consequence of the 3′ interruption was a change in the spectrum of mutations and not a reduction in the rate of expansions.
FIG. 5.
Distribution of sizes of the expansion events. On the x axis is the number of repeats added with respect to the starting tract length (e.g., +11 repeats indicates a final length of 36 TNR). On the y axis is the observed number of expansion events of that size. (A) Twenty genetically independent expansions of the uninterrupted sequence (CTG)25. (B) Nineteen expansions of the 5′ interruption. (C) Thirty-six expansions of the 3′ interruption. (D) Thirty-three expansions of the centrally located interruption.
Expansions from the centrally interrupted TNR tract (Fig. 5D) showed a wide distribution ranging in size from 7 to 21 repeats. However, this distribution appeared bimodal, with a few small expansions of 7 to 9 repeats and the rest large expansions of 14 to 21 repeats. The smaller class of expansions appears to overlap the small size class observed with 3′ interrupted alleles (compare panels C and D of Fig. 5). From the model (left panel of Fig. 1C), we expected that some events might be limited to expansions of ≤11 repeats. We observed about 10% (3 of 33) fell into this grouping. In contrast, 90% of the expanded alleles were of a larger class that more closely resembled alleles arising from the perfect repeat or from the 5′ interruption (compare panel D with panels A and B of Fig. 5). We know from Fig. 4C that most of the larger class of expansions is due to events like those shown in the right panel of Fig. 1C, in which the expansion duplicated the interruption.
DISCUSSION
The results of this study indicate that interrupted trinucleotide repeats are stabilized in yeast, as has been suspected for humans. The yeast experiments support a model where interruptions contribute in cis to reduce the likelihood of secondary structure during TNR replication. By varying the position of the interruption, we provided supporting evidence that verifies several key predictions. First, the degree of stabilization depends on the location of the interruption. Replication early in the tract provides the greatest stabilization, late replicating interruptions are least stabilizing, and central interruptions give an intermediate amount of stabilization. Second, the expansion sizes for the asymmetrically placed interruptions were restricted to the larger available stretch of perfect repeats, closely mirroring the predictions from the model. Third, expansions from the asymmetric alleles were polar but did not lead to duplication of the interruption. Fourth, 90% of the expansions from a centrally placed interruption appear to span the interruption, leading to its duplication. We believe that the duplications provide a particularly compelling point in favor of the hairpin destabilization model. Although an anchoring effect by the interruptions may contribute to stability by reducing replicational slippage (19, 26), anchoring does not easily explain the duplications. Taken together, these results indicate that interruptions act in cis to help stabilize TNR tracts.
An alternative model for interruption-mediated stabilization is that the interruptions break up a long, unstable TNR into two smaller, genetically stable subthreshold lengths. For fragile X CGG repeats, one model is that subthreshold lengths of perfect repeats might lengthen by the accumulation of short slippage mutations (3, 9, 22, 25). Given the caveat that thresholds have not yet been demonstrated in yeast, our evidence suggests that interruptions do not act merely to generate a subthreshold length repeat. Comparison of the 5′ and 3′ interrupted tracts shows that both have 17 perfect contiguous repeats but that their expansion rates vary by 40-fold. Furthermore, the central interruption contains the shortest perfect repeat tract (12 repeats), yet it expands at a fivefold-higher rate than the 5′ interruption which has a longer perfect repeat tract. A second noteworthy point is that we found no evidence for gradual accumulation of short slippage events. PCR analysis of expanded and unexpanded siblings showed no tracts of intermediate lengths. Thus, in our yeast system, the stabilizing influence of interruptions on CTG tracts does not conform to the subthreshold model.
The work of Pearson et al. (19) examined SCA1 and FMR1 perfect and interrupted alleles from the human population. Their study demonstrated that interruptions decreased the total amount of S-DNA compared to perfect repeats. Their work also showed that interrupted tracts show less heterogeneity in the number of S-DNA isomers formed, suggesting that the interrupting base pairs limit S-DNA formation to regions of perfect repeats. Our data demonstrate that a smaller range of expansions is seen for asymmetrically located interruptions compared to a perfect repeat of the same length (Fig. 5A to C). Our model (Fig. 1) suggests that the interruptions reduce the likelihood of hairpin formation if the stem contains mispairs due to the interrupting bases. Another possible interpretation is that interruptions delimit boundaries for hairpins, like S-DNA. However, when we placed the interruption in the center of the TNR we observed frequent duplications, a result consistent with incorporation of the interruption in or near the stem of the hairpin. The latter result more closely supports the hairpin destabilization model compared to the boundary hypothesis.
One unexpected result from our work was that a few expansions of the 5′ interruptions were slightly larger (+18 to +23 repeats; Fig. 5) than that predicted by the hairpin destabilization model (+17 repeat maximum). These exceptional mutations accounted for 21% (4 of 19) of the expansions examined for the 5′ interrupted tract. If they occurred by a complex event, such as multiple slippage events in a single round of replication, the larger expansion size could be explained. One would expect multiple slippages to occur with low frequencies. Consistent with this prediction, we note that these alleles were only observed for the repeat tract with the lowest overall expansion rate and even then they comprised only a fraction of the total expansions. A second unanticipated result was that expansions from the 3′ interrupted allele tend to be shorter than for a perfect repeat or for the 5′ interruption (Fig. 5). Either small hairpins are somehow stabilized for the 3′ interrupted tract or large hairpins are selected against, even if the interrupting base pairs have not been replicated. Since the overall rate of expansion for the 3′ interruption was reduced only 2.4-fold relative to the perfect tract, it seems unlikely that there could be much selection against the large hairpins or else the rate would be more severely reduced. Thus, small expansions from the 3′ interrupted allele are not readily predicted either by the hairpin destabilization model or by the boundary model. The subthreshold model suggests that expansions can accumulate from several short slippage events. We note, however, that the accumulation hypothesis is an unlikely explanation for our result because examination of the unexpanded sibling colonies showed no evidence for lengthening of the starting tract. The observation of short expansions associated with 3′ interruptions will require additional experimentation for a satisfactory explanation.
In another yeast study, Maurer et al. (15) used tracts of 90 to 97 CAG repeats containing a single CAT interruption to investigate the polarity of repeat mutations. They observed only contractions in unselected populations of wild-type cells. Since no expansions were observed in their wild-type background, it is difficult to compare their results with our work.
The model presented in Fig. 1 depicts hairpin formation by 3′ slippage of Okazaki fragments. The 5′ flap model (6) can explain many of our results equally well. In the flap model, a DNA polymerase sometimes displaces the 5′ end of an existing Okazaki fragment within the TNR. Since the flap contains TNR sequences, it can fold into a hairpin. One of our results suggests that expansions may result more often from 3′ slippage than 5′ flaps. The observation is that interruptions that are replicated early are more stabilizing than late-replicated alleles. By the 3′ slippage model, slippage is more likely (based on probability) to occur after synthesis of the early-replicated interruptions, leading to hairpins that contain the substitutions. In contrast, slippage first is more likely for the late-replicated interruptions. One would then expect the 5′ interrupted TNR to provide greater stability than the 3′ interrupted TNR. Indeed, this was the observation. If, on the other hand, one envisions the flap model, it would seem more probable that the 5′ end of the displaced fragment would lie downstream of the 5′ interruption. Therefore, the interruption would not reside within the flap and would not affect hairpin formation. In contrast, late-replicated interruptions would more often be included in the flap and have a greater chance of inhibiting hairpin formation. By this view, the 3′ interruption would stabilize more than the 5′ interruption but the experimental observation is the opposite. From this analysis, TNR hairpin formation would seem more likely from 3′ slippage events than from 5′ flaps. However, this point is based on a number of assumptions, and therefore it provides only an indirect test of the 3′ slippage versus 5′ flap models.
Based on findings from human TNR diseases, we were surprised to find that the interruption was retained in all 60 independent events that were analyzed. Sequence analysis of five samples showed that both interrupting ATGs were still present. Thus, in our yeast system loss of the interruption is not required for expansions. Analysis of human pedigrees for disease loci such as SCA1 and FMR1 shows a majority of stable repeats have one to three interruptions, while the disease alleles have zero to one interruption (2, 3, 9, 13, 25). This correlative information has led to the belief that loss of an interruption precedes expansion in the human population, although it has not been possible to show this unambiguously. Perhaps there are multiple mechanisms by which interrupted alleles can expand and our yeast system exemplifies one of those mechanisms. Although the human diseases reported to date are suggestive of loss of interruptions upon expansion, it is possible that new TNR disease genes may exhibit genetic characteristics more like the yeast system.
The results of this study have been interpreted as the interruptions acting in cis to inhibit expansions. We note that trans-acting factors may also contribute to the stability provided by interruptions in TNRs, as suggested by others (12, 20). Candidate trans-acting factors are currently being investigated. It is also possible that the direction of transcription through the repeat vis-á-vis the direction of replication could affect stabilization. Evaluation of this possibility will require further experimentation.
ACKNOWLEDGMENTS
This work was supported by funds from the Eppley Institute (to R.S.L.), by National Cancer Institute (NCI) training grant T32 CA09476 (to M.L.R.), and by NCI Cancer Center support grant P30 CA36727 (to the Eppley Institute).
REFERENCES
- 1.Ashley C T, Jr, Warren S T. Trinucleotide repeat expansion and human disease. Annu Rev Genet. 1995;29:703–728. doi: 10.1146/annurev.ge.29.120195.003415. [DOI] [PubMed] [Google Scholar]
- 2.Chung M-Y, Ranum L P W, Duvick L A, Servadio A, Zoghbi H Y, Orr H T. Evidence for a mechanism predisposing to intergenerational CAG repeat instability in spinocerebellar ataxia type I. Nat Genet. 1993;5:254–258. doi: 10.1038/ng1193-254. [DOI] [PubMed] [Google Scholar]
- 3.Eichler E E, Holden J J A, Popovich B A, Reiss A L, Snow K, Thibodeau S N, Richards C S, Ward P A, Nelson D L. Length of uninterrupted CGG repeats determines instability of the FMR1 gene. Nat Genet. 1994;8:88–94. doi: 10.1038/ng0994-88. [DOI] [PubMed] [Google Scholar]
- 4.Freudenreich C H, Stavenhagen J B, Zakian V A. Stability of a CTG/CAG trinucleotide repeat in yeast is dependent on its orientation in the genome. Mol Cell Biol. 1997;17:2090–2098. doi: 10.1128/mcb.17.4.2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gacy A M, Goellner G, Juranic N, Macura S, McMurray C T. Trinucleotide repeats that expand in human disease form hairpin structure in vitro. Cell. 1995;81:533–540. doi: 10.1016/0092-8674(95)90074-8. [DOI] [PubMed] [Google Scholar]
- 6.Gordenin D A, Kunkel T A, Resnick M A. Repeat expansion—all in a flap? Nat Genet. 1997;16:116–118. doi: 10.1038/ng0697-116. [DOI] [PubMed] [Google Scholar]
- 7.Gusella J F, MacDonald M E. Trinucleotide instability: a repeating theme in human inherited disorders. Annu Rev Med. 1996;47:201–209. doi: 10.1146/annurev.med.47.1.201. [DOI] [PubMed] [Google Scholar]
- 8.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 9.Hirst M C, Grewal P K, Davies K E. Precursor arrays for triplet repeat expansion at the fragile X locus. Hum Mol Genet. 1994;3:1553–1560. doi: 10.1093/hmg/3.9.1553. [DOI] [PubMed] [Google Scholar]
- 10.Imbert G, Saudou F, Yvert G, Devys D, Trottier Y, Garnier J-M, Weber C, Mandel J-L, Cancel G, Abbas N, Durr A, Didierjean O, Stevanin G, Agid Y, Brice A. Cloning of the gene for spinocerebellar ataxia 2 reveals a locus with high sensitivity to expanded CAG/glutamine repeats. Nat Genet. 1996;14:285–291. doi: 10.1038/ng1196-285. [DOI] [PubMed] [Google Scholar]
- 11.Kramer B, Kramer W, Williamson M S, Fogel S. Heteroduplex DNA correction in Saccharomyces cerevisiae is mismatch specific and requires functional PMS genes. Mol Cell Biol. 1989;9:4432–4440. doi: 10.1128/mcb.9.10.4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kramer P R, Pearson C E, Sinden R R. Stability of triplet repeats of myotonic dystrophy and fragile X loci in human mismatch repair cell lines. Hum Genet. 1996;98:151–157. doi: 10.1007/s004390050179. [DOI] [PubMed] [Google Scholar]
- 13.Kunst C B, Warren S T. Cryptic and polar variation of the fragile X repeat could result in predisposing normal alleles. Cell. 1994;77:853–861. doi: 10.1016/0092-8674(94)90134-1. [DOI] [PubMed] [Google Scholar]
- 14.Lea D E, Coulson C A. The distribution of the number of mutants in bacterial populations. J Genet. 1948;49:264–284. doi: 10.1007/BF02986080. [DOI] [PubMed] [Google Scholar]
- 15.Maurer D J, O'Callaghan B L, Livingston D M. Mapping the polarity of changes that occur in interrupted CAG repeat tracts in yeast. Mol Cell Biol. 1998;18:4597–4604. doi: 10.1128/mcb.18.8.4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miret J J, Pessoa-Brandao L, Lahue R S. Orientation-dependent and sequence-specific expansions of CTG/CAG trinucleotide repeats in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1998;95:12438–12443. doi: 10.1073/pnas.95.21.12438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Montermini L, Andermann E, Labuda M, Richter A, Pandolfo M, Cavalcanti F, Pianese L, Iodice L, Farina G, Monticeli A, Turano M, Filla A, De Michele G, Cocozza S. The Friedreich ataxia GAA triplet repeat: premutation and normal alleles. Hum Mol Genet. 1997;6:1261–1266. doi: 10.1093/hmg/6.8.1261. [DOI] [PubMed] [Google Scholar]
- 18.Paulson H L, Fischbeck K H. Trinucleotide repeats in neurogenetic disorders. Annu Rev Neurosci. 1996;19:79–107. doi: 10.1146/annurev.ne.19.030196.000455. [DOI] [PubMed] [Google Scholar]
- 19.Pearson C E, Eichler E E, Lorenzetti D, Kramer S F, Zoghbi H Y, Nelson D L, Sinden R R. Interruptions in the triplet repeats of SCAI and FRAXA reduce the propensity and complexity of slipped strand DNA (S-DNA) formation. Biochemistry. 1998;37:2701–2708. doi: 10.1021/bi972546c. [DOI] [PubMed] [Google Scholar]
- 20.Pearson C E, Ewel A, Acharya S, Fishel R A, Sinden R R. Human MSH2 binds to trinucleotide repeat DNA structures associated with neurodegenerative diseases. Hum Mol Genet. 1997;6:1117–1123. doi: 10.1093/hmg/6.7.1117. [DOI] [PubMed] [Google Scholar]
- 21.Pulst S-M, Nechiporuk A, Nechiporuk T, Gispert S, Chen X-N, Lopes-Cendes I, Pearlman S, Starkman S, Orozco-Diaz G, Lunkes A, DeJong P, Rouleau G A, Auburger G, Korenberg J R, Figueroa C, Sahba S. Moderate expansion of a normally biallelic trinucleotide repeat in spinocerebellar ataxia type 2. Nat Genet. 1996;14:269–276. doi: 10.1038/ng1196-269. [DOI] [PubMed] [Google Scholar]
- 22.Richards R I, Sutherland G R. Dynamic mutations: a new class of mutations causing human disease. Cell. 1992;70:709–712. doi: 10.1016/0092-8674(92)90302-s. [DOI] [PubMed] [Google Scholar]
- 23.Sanpei K, Takano H, Igarashi S, Sato T, Oyake M, Sasaki H, Wakisaka A, Tashiro K, Ishida Y, Ikeuchi T, Koide R, Saito M, Sato A, Tanaka T, Hanyu S, Takiyama Y, Nishizawa M, Shimizu N, Nomura Y, Segawa M, Iwabuchi K, Eguchi I, Tanaka H, Takahashi H, Tsuji S. Identification of the spinocerebellar ataxia type 2 gene using a direct identification of repeat expansion and cloning technique, DIRECT. Nat Genet. 1996;14:277–284. doi: 10.1038/ng1196-277. [DOI] [PubMed] [Google Scholar]
- 24.Schiestl R H, Gietz D. High-efficiency transformation of intact yeast cells by single stranded nucleic acids as carrier. Curr Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- 25.Snow K, Tester D J, Kruckeberg K E, Schaid D J, Thibodeau S N. Sequence analysis of the fragile X trinucleotide repeat: implications for the origin of the fragile X mutation. Hum Mol Genet. 1994;3:1543–1551. doi: 10.1093/hmg/3.9.1543. [DOI] [PubMed] [Google Scholar]
- 26.Zhong N, Yang W, Dobkin C, Brown W T. Fragile X gene instability: anchoring AGGs and linked microsatellites. Am J Hum Genet. 1995;57:351–361. [PMC free article] [PubMed] [Google Scholar]