Abstract
Background
A small but important proportion of patients (4–10 %) with AML have germline mutations. They can cause the development of AML at an earlier age, confer a higher risk of relapse or predispose to secondary leukemias, including therapy-related leukemias. The analysis of germline mutations in a patient and his/her family is also critical for the selection of suitable family donors if the patient is a candidate for hematopoietic stem cell transplantation (HSCT).
Methods
103 unrelated consecutive patients with de novo AML were enrolled in the study. Control group consisted of 103 persons from the general population. We performed NGS sequencing of bone marrow cells and buccal swabs DNA of six genes: CEBPA, DDX41, ETV6, TERT, GATA2, and IDH2 to detect germline pathogenic mutations.
Results
In the investigated group, 49 variants were detected in six genes. 26 of them were somatic and 23 germline. Germline variants were detected in all six tested genes. Eight pathogenic germline mutations were detected in 7 AML patients, in three genes: CEBPA, ETV6, and IDH2. One patient had two pathogenic germinal mutations, one in ETV6 and one in CEBPA gene. We identified one novel pathogenic germline mutation in CEBPA gene. The difference in frequency of all pathogenic germline mutations between the tested (7.77 %) and control groups (0.97 %) was statistically significant (p = 0.046). In the tested group, the median age at AML diagnosis was 11 years lower in patients with pathogenic germline mutations than in patients without them (p = 0.028).
Conclusions
We showed higher frequency of CEBPA, ETV6, and IDH2 germline mutations in AML patients than in control group, which confirms the role of these mutations in the development of AML. We also showed that the median age at the onset of AML in patients with pathogenic germline mutations is significantly lower than in patients without them.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13053-021-00200-2.
Keywords: Acute myeloid leukemia, AML, germline mutations, CEBPA, DDX41, ETV6, TERT, GATA2, IDH2
Background
AML (acute myeloid leukemia) is one of the most common types of leukemia in adults. AML and MDS (myelodysplastic syndromes) exist along a continuous disease spectrum starting with early-stage MDS, which may progress to advanced MDS, AML, resistant or cured AML. The disease is characterized by an overproduction of immature blood cells in the bone marrow (BM) and a lack of mature, healthy blood cells in the peripheral blood. This causes anemia and an increased risk for bleeding and infection. Moreover, there is a dysplasia in one to three main myeloblastic (erythro-, thrombo-, and granulopoietic) cell lines in BM. It was recognized that not only somatic mutations in neoplastic tissue but also germline mutations may influence disease development, progress, and prognosis. This is supported by the fact that the myeloid neoplasms with genetic predisposition represent a new category in the revised 2016 World Health Organization classification. The RUNX1 (Runt-related transcription factor 1), CEBPA (CCAAT enhancer binding protein), DDX41 (DEAD-box helicase 41), ETV6 (ETS translocation variant gene 6), GATA2 (GATA binding protein 2), TERC (Telomerase RNA component) and TERT (Telomerase reverse transcriptase) genes are important regulators of hematopoiesis and are frequently involved in the pathomechanism of leukemogenesis [1–3]. The importance of methylation disturbances is also emphasized during the development of AML, causing general hypomethylation of the genome and selective hypermethylation of suppressor genes. Methylation disorders are associated with translocations, rearrangements, and mutations of the IDH1 (Isocitrate dehydrogenase (NADP(+)1), IDH2 (Isocitrate dehydrogenase (NADP(+)2), TET2 (Tet methylcytosine dioxygenase 2) and DNMT3A (DNA (cytosine-5-)-methyltransferase 3 alpha) genes leading to changes in their expression. However, despite the growing knowledge of germline predisposition to myeloid malignancies, it is not fully explained yet [3, 4].
We performed NGS sequencing of six genes: CEBPA, DDX41, ETV6, TERT, GATA2, and IDH2 in the aim to detect germline pathogenic mutations in Polish patients with de novo AML.
AML associated with CEBPA or DDX41 mutations occurs without distinct clinical symptoms or antecedent hematological condition [2]. CEBPA gene encodes a transcription factor (TF) involved in the regulation of myelopoiesis. Two types of mutations may appear: an N-terminal frame-shift or overproduction mutation and a C-terminal mutation disrupting the DNA binding. Many AMLs with CEBPA mutations simultaneously carry 2 mutations (CEBPAdm) in trans position (on 2 different alleles), whereas single heterozygous mutations (CEBPAsm) occur less frequently. Clinically important CEBPAdm present in about 7 % of AML patients contain both, N-terminal and C-terminal mutations on the separate allele each. This mutational status is a favorable prognostic factor [5–8].
DDX41 encodes an RNA helicase, a protein involved in various processes of RNA metabolism, from the transcription to the degradation of RNA, including pre-mRNA splicing, mRNA export, ribosome biogenesis, translation initiation, and gene expression in organelles. However, DDX41 role in hematopoiesis and leukemogenesis remains unknown. Moreover, the prevalence and penetrance of DDX41 mutations are unclear [9, 10].
ETV6 gene encodes a main hematopoietic TF which is a part of a large, ETS family (E26 transformation-specific) composed of 28 TFs. The ETV6 protein plays a crucial role in embryonic development and hematopoietic regulation. The most frequent clinical feature associated with the germline mutations of ETV6 gene in the context of hematologic malignancy are younger age at the disease onset, platelet dysfunction, and bleeding disorders associated with an elevated risk of MDS/AML. [2, 11].
Mutations of TERT which appear frequently (2–19 %) in bone marrow failure syndromes are associated with an elevated risk of MDS/AML. TERT encodes the catalytic subunit of telomerase which catalyses the addition of TTAGGG telomeric repeat sequences at the ends of chromosomes in order to stabilize telomere length and achieve cell immortality. The abnormal reactivation of telomerase complex occurs in approximately 90 % of human tumors, and is considered a crucial element for cancer genesis and progression [12–15].
GATA2 mutations cause the development of familial MDS and often occur in the setting of cytopenias and rare immunological syndromes. The GATA2 gene encodes a TF which is expressed in hematopoietic stem cells. This protein contains two zinc finger domains that promote protein-protein and protein-DNA communication. GATA2 participates in the formation of early blood and lymphatic vessels [2, 16, 17].
The importance of methylation disturbances is also emphasized during the development of AML, causing general hypomethylation of the genome and selective hypermethylation of suppressor genes. IDH2 protein, encoded by IDH2 gene, plays a key role in the process of DNA methylation/demethylation. IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 gene function, and impair hematopoietic differentiation. Mutations in IDH2 have been first reported in glioblastoma multiforme, next in acute myeloid leukemia, and other malignancies such as breast invasive ductal carcinoma, colon adenocarcinoma, lung adenocarcinoma, and oligodendroglioma. Approximately 20 % of AML patients harbor a mutation in the isocitrate dehydrogenase (IDH) genes, IDH1 or IDH2 [18–21].
The importance of germline predisposition to myeloid malignancies is more and more emphasized due to its clinical significance; necessity of genetic counseling for family members and influence on hematopoietic stem cell donor selection. Assessment of the presence of germline mutations, particularly in younger patients or those with a positive family history, is important for the optimal care of patients [22].
Materials and methods
DNA samples stored at the Department of Clinical Genetics, Collegium Medicum in Bydgoszcz, Nicolaus Copernicus University in Torun, Poland (CM NCU) were used for the study. A total of 103 consecutive patients (54 men and 49 women) diagnosed with de novo AML, according to World Health Organization criteria, regardless of age at AML diagnosis and family history of cancer were involved in the study. None of the three most common in Polish population mutations in the BRCA1 gene were detected in the study group. The median age at diagnosis was 56 years (men – 53 years, women – 58 years, range 18–89). The pedigrees were made on the basis of the questionnaires. 39.80 % (41/103) of patients reported at least one first- or second-degree relative with cancer. In 33 families (32,04 %) lung, breast, ovary, stomach or colon cancer mainly occurred, in 3 families (2.91 %) Hodgkin’s lymphoma, in one family (0,97 %) 2 cases of AML and in 4 families (3.88 %) other leukemia types. Most investigated patients had M4 (31 patients), M1 (19), and M2 (18) AML cytomorphologic subtypes, according to the French-American-British (FAB) classification. 12 patients had M5, 5 - M3, 2 - M6, 2 - M7, and 1 - M0 subtypes. For thirteen patients, no FAB data were available.
The control group consisted of 103 volunteers out of the general population who on a questionnaire basis had no malignancies at the time of material collection, and originated from families without a history of cancer. Control group persons were matched by age and sex with patients from the investigated group.
Informed consent was obtained from all patients and control healthy persons. The study was approved by the Ethics Committee of the CM NCU.
DNA from 85 PB (peripheral blood) cell samples and 18 BM cell samples, collected at diagnosis of AML, was used for molecular testing. In the control group, the mutations were searched for in DNA from peripheral blood. Genomic DNA was extracted from leukocytes by QIAmp® DNA Mini Kit (QIAGEN) using standard procedures. In mutation-positive patients, the constitutional character of a mutation was verified by analysis of DNA from BS (buccal swabs), collected at AML diagnosis, extracted by Swab-Extract DNA Purification Kit (EURx) using standard procedures.
For the next-generation sequencing (NGS) reaction, the molecular inversion probes (MIPs) designed using the MIPGEN program were used. Procedures for the preparation of probes, hybridization reaction, complete ligation and amplification were based on the methods developed by Yoon et al. 2015 [23, 24]. Sequencing was performed on a MiSeq sequencer analyzer, in paired-end (PE) technology, 2 × 250nt, using Illumina’s v2 kit, according to the manufacturer’s protocol.
Mutation-positive cases were confirmed by sequencing analysis using ABI PRISM 3130 (Applied Biosystems). For Sanger sequencing, exons were amplified by PCR (PCR profiles, primers sequence available upon request). Primers were designed using the Primer3 tool (http://bioinfo.ut.ee/primer3-0.4.0). Sequencing reaction was conducted on PCR product with BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems), according to the manufacturer’s procedure, on the coding parts of the genes with parts of introns adjacent to 5’ and 3’ ends of all tested exons.
Pathogenicity of identified mutations was evaluated by the VarSome Clinical database. A variant was described as pathogenic if VarSome Clinical showed its pathogenicity or likely pathogenicity. Mutational analysis was performed on NCBI reference sequences: NM_004364.4, NM_032638.5, NM_001987.5, NM_002168.4, NM_016222.4, and NM_198253.3 for CEBPA, GATA2, ETV6, IDH2, DDX41, and TERT, respectively.
The karyotypes of heparinized BM cells of each patient were assessed at disease diagnosis, using classical GTG-banding and fluorescence in situ hybridization (FISH) techniques.
Statistical analysis included a comparison of the prevalence of variant alleles in the studied and control groups, calculation of odds ratios (ORs) from two-by-two tables, and calculation of statistical significance of differences between various tested groups using the Chi-square test. The Mann-Whitney U test was used to compare the median age in the groups of patients with and without pathogenic germline mutations. The normal distribution was verified using the Kolmogorov-Smirnov test. Statistical analysis was performed using the IBM SPSS 26 statistical package.
Results
We detected 49 types of variants in six genes (CEBPA, DDX41, ETV6, TERT, GATA2, IDH2). Among variants identified in the study were missense, frameshift, duplication, silent, and intronic variants. 26 types of somatic variants were detected: 5 pathogenic, 6 likely pathogenic, 5 of uncertain significance (VUS), and 10 benign. We detected also 23 types of germline variants: 2 pathogenic, 2 likely pathogenic, 1 VUS, and 18 benign (Tables 1 and 2).
Table 1.
Somatic mutations in CEBPA, GATA2, ETV6, DDX41, TERT and IDH2 found in patients with AML
| Gene | Mutation | Amino acid change | Exon/ Intron |
Prediction by VarSome | Variant ID | Carriers n = 103 |
Frequency in Europe (%) GnomAD |
|
|---|---|---|---|---|---|---|---|---|
| CEBPA | c.179dupC | p.Ser61ValfsTer47 | Ex1 | pathogenic | COSM18632 | 1 | – | |
| c.287_311del | p.Gly96AlafsTer56 | Ex1 | likely pathogenic | COSV57200088 | 1 | – | ||
| c.402G > A | p.Ala134= | Ex1 | benign |
rs752254340 COSV57196176 |
1 | 0.0 | ||
| c.536T > C | p.Phe179Ser | Ex1 | VUS | – | 1 | – | ||
| c.573 C > T | p.His191= | Ex1 | benign |
rs192240793 COSV57200682 |
1 | 0.9 | ||
| c.937_939dupAAG | p.Lys313dup | Ex1 | likely pathogenic | – | 1 | – | ||
| GATA2 | c.919 C > T | p.Arg307Trp | Ex5 | likely pathogenic | COSM306052 | 1 | – | |
| c.959G > A | p.Arg320His | Ex5 | likely pathogenic | rs121964985 COSM249854 | 1 | 0.1 | ||
| c.1085G > A | p.Arg362Gln | Ex6 | likely pathogenic |
rs867160952 COSM87004 |
1 | – | ||
| ETV6 | c.463 + 49G > A | NA | Int4 | benign | rs17210957 | 8 | – | |
| c.1090G > T | p.Glu364Ter | Ex6 | pathogenic | – | 1 | – | ||
| DDX41 | c.409 A > G | p.Thr137Ala | Ex5 | VUS | – | 16 | – | |
| TERT | c.835G > A | p.Ala279Thr | Ex2 | benign |
rs61748181 COSM3760906 |
2 | 3.6 | |
| c.915G > A | p.Ala305= | Ex2 | benign |
rs2736098 COSM3760904 |
15 | 23.5 | ||
| c.1573 + 39G > C | NA | Int2 | benign | rs79662648 | 2 | – | ||
| c.1574-16G > C | NA | Int2 | benign | rs79698601 | 1 | 0.6 | ||
| c.2007_2017del | p.Ala670ArgfsTer32 | Ex5 | pathogenic | – | 7 | – | ||
| c.2018_2019insTT | p.Gly674SerfsTer41 | Ex5 | pathogenic | – | 7 | – | ||
| c.2020G > A | p.Gly674Ser | Ex5 | VUS | rs773813809 COSM6919371 | 7 | 0.0 | ||
| c.2023 C > T | p.Leu675Phe | Ex5 | VUS | – | 7 | – | ||
| c.3184G > A | p.Ala1062Thr | Ex15 | benign |
rs35719940 COSM5019566 |
1 | 2.2 | ||
| c.3324G > A | p.Pro1108= | Ex16 | benign |
rs35033501 COSM5020092 |
2 | 2.9 | ||
| IDH2 | c.327G > A | p.Val109= | Ex3 | benign |
rs150943639 COSM6867404 |
1 | 0.4 | |
| c.404 C > A | p.Pro135His | Ex4 | likely pathogenic | – | 1 | – | ||
| c.515G > T | p.Arg172Met | Ex4 | pathogenic |
rs121913503 COSM33732 |
2 | – | ||
| c.709 C > A | p.Gln237Lys | Ex6 | VUS | – | 6 | – | ||
VUS - variant of uncertain significance, Variant ID - variant identifier, NA - no amino acid change, (–) - no data
Table 2.
Germline mutations in CEBPA, GATA2, ETV6, DDX41, TERT, and IDH2 found in patients with AML
| Gene | Mutation | Amino acid change | Exon/ Intron |
Prediction by VarSome | Variant ID | Carriers n = 103 |
Frequency in Europe (%) GnomAD |
|---|---|---|---|---|---|---|---|
| CEBPA | c.337_344del | p.Ala113ArgfsTer54 | Ex1 | likely pathogenic | – | 1 | – |
| c.590_591insACCCGC | p.Pro198_Ala199dup | Ex1 | likely pathogenic | COSM7448245 | 4 | – | |
| c.690G > T | p.Thr230= | Ex1 | benign |
rs34529039 COSV57196771 |
14 | 16.00 | |
| GATA2 | c.15 C > G | p.Pro5= | Ex3 | benign |
rs1573858 COSM3749812 |
21 | 66.40 |
| c.481 C > G | p.Pro161Ala | Ex4 | benign |
rs34799090 COSM5762885 |
1 | 0.30 | |
| c.490G > A | p.Ala164Thr | Ex4 | benign |
rs2335052 COSM445531 |
23 | 18.30 | |
| c.564G > C | p.Thr188= | Ex4 | benign | rs34870876 | 19 | 5.90 | |
| c.1018-19 C > T | NA | Int5 | benign | rs11708606 | 27 | 19.60 | |
| c.1233G > A | p.Ala411= | Ex7 | benign |
rs34172218 COSM5019736 |
1 | 2.40 | |
| ETV6 | c.258G > A | p.Thr86= | Ex3 | benign |
rs11611479 COSM3752958 |
12 | 10.90 |
| c.380G > A | p.Arg127Gln | Ex4 | benign | rs140357643 | 1 | – | |
| c.1075 C > T | p.Arg359Ter | Ex6 | pathogenic |
rs141938078 COSM3456975 |
1 | – | |
| DDX41 | c.936-23 C > G | NA | Int9 | benign | rs335439 | 56 | 99.90 |
| c.1200 C > T | p.Arg400= | Ex11 | benign |
rs335438 COSM3761138 |
39 | 47.10 | |
| c.1302 + 67_1303-67insAG | NA | Int12 | VUS | – | 1 | – | |
| c.1399 + 46T > C | NA | Int13 | benign | – | 64 | – | |
| c.1732 + 46 A > G | NA | Int16 | benign | – | 52 | – | |
| TERT | c.1770-24 C > T | NA | Int3 | benign | rs13167280 | 15 | 12.90 |
| c.1950 + 10 C > T | NA | Int4 | benign | rs33948291 | 1 | 1.90 | |
| c.3039 C > T | p.His1013= | Ex14 | benign |
rs33954691 COSM5019111 |
7 | 9.70 | |
| IDH2 | c.419G > A | p.Arg140Gln | Ex4 | pathogenic |
rs121913502 COSM41590 |
2 | 0.01 |
| c.535-40G > A | NA | Int4 | benign | rs142033117 | 2 | 0.80 | |
| c.996 C > T | p.Ser332= | Ex8 | benign |
rs61737003 COSM4128570 |
1 | 2.60 |
VUS - variant of uncertain significance, Variant ID - variant identifier, NA - no amino acid change, (–) - no data
Germline variants were detected in each of the six tested genes, but the pathogenic germline mutations were detected only in three genes: CEBPA, ETV6, and IDH2. We have detected 4 types of mutations in these genes: c.337_344del and c.590_591insACCCGC in CEBPA, c.1075 C > T in ETV6, and c.419G > A in IDH2. Overall, eight pathogenic germline mutations were detected in 7/103 of AML patients, one patient had two pathogenic germline mutations. In the CEBPA gene were detected two pathogenic mutations: c.337_344del in one patient and c.590_591insACCCGC in four patients. In one of four patients, c.590_591insACCCGC mutation coexisted with the pathogenic c.1075 C > T mutation in the ETV6 gene. In the IDH2 gene c.419G > A mutation occurred in two patients. These mutations were present in DNA not only from BM and/or PB, but also from BS of patients, which confirmed their constitutional character.
In the control group, we detected one pathogenic germline mutation (c.590_591insACCCGC) in the CEBPA gene. This mutation was detected in a 27-year-old healthy woman with no family history of cancer. No other variants (somatic or germline) were detected in the tested genes in this woman.
The difference in frequency of all germline mutations between the tested (7.77 %) and control groups (0.97 %) was statistically significant (p = 0.046), and the odds ratio was 8.59; 95 % CI: 1.054–69.975 (Table 3). Additionally, in one patient without pathogenic germline mutation, a germline intronic variant of uncertain significance (c.1302 + 67_1303-67insAG) in DDX41 gene was detected.
Table 3.
A correlation between pathogenic germline mutations and risk of AML in analyzed patients
| Gene | Mutations | Number of pathogenic germline mutations | OR | 95 % CI | p-value | |||
|---|---|---|---|---|---|---|---|---|
|
Tested group n = 103 [n/%] |
Control group n = 103 [n/%] |
|||||||
| CEBPA | c.337_344del | 1 | 0.97 % | 0 | 0.00 % | 3.03 | 0.122–75.235 | p = 0.499 |
| 590_591insACCCGC | 4 | 3,88 % | 1 | 0.97 % | 4.12 | 0.453–37.520 | p = 0.209 | |
| ETV6 | c.1075 C > T | 1 | 0.97 % | 0 | 0.00 % | 3.03 | 0.122–75.235 | p = 0.499 |
| IDH2 | c.419G > A | 2 | 1.94 % | 0 | 0.00 % | 5.10 | 0.241-107.521 | p = 0.295 |
| All mutations | 8 | 7.77 % | 1 | 0.97 % | 8.59 | 1.054–69.975 | p = 0.046 | |
We also analyzed a correlation between pathogenic germline mutations and karyotype of bone marrow cells at AML diagnosis. In the study group, 39 patients had chromosome aberrations, 38 had normal karyotypes, and in 26 the results of cytogenetic diagnostics were not available. 6 patients with germline mutations had normal karyotypes, and one had an aberrant karyotype (Table 4).
Table 4.
Clinical data of patients with pathogenic germline mutations and co-occurring mutations
| Patient laboratory number | Sex/age | AML classification (FAB) |
Karyotype | Family history of cancer | Nucleotide change | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| IDH2 | CEBPA | GATA2 | ETV6 | TERT | DDX41 | |||||
| 10 | M/44 | M5 | NK | ND |
c.419G > A* c.709 C > A |
– | c.1018-19 C > T | – |
c.1770-24 C > T c.3184G > A |
c.936-23 C > G c.1200 C > T |
| 13 | K/47 | M1 | NK | CLL60 | – | c.590_591insACCCGC* | – | c.1075 C > A* | – |
c.936-23 C > G c.1200 C > T |
| 33 | K/37 | M6 | NK | ND | – | c.337_344del* | – | c.258G > A | c.915G > A |
c.409 A > G c.936-23 C > G c.1200 C > T |
| 43 | M/20 | M1 | NK | ND | – | c.590_591insACCCGC* | c.490G > A | c.463 + 49G > A | – |
c.409 A > G c.936-23 C > G c.1200 C > T |
| 47 | M/51 | M4 | NK | Pr72, Ut70 | – | c.590_591insACCCGC* | – | c.258G > A | – |
c.936-23 C > G c.1200 C > T |
| 83 | K/49 | M4 | t(9;11) | Br50 | – | c.590_591insACCCGC* |
c.15 C > G c.490G > A |
– | – | c.936-23 C > G |
| 97 | M/48 | M1 | NK | Bn62 | c.419G > A* | c.690G > T | c.15 C > G | – | – | c.936-23 C > G |
CLL - chronic lymphocytic leukemia, Pr - prostate cancer, Ut - uterine cancer, Br - breast cancer, Bn - brain cancer, FAB - French-American-British classification, (*) - germline mutation, NK - normal karyotype, ND - no data, (–) - no mutation
We found that 21/103 patients (20.4 %) harbored a somatic and/or germline mutation in CEBPA gene. 18 patients had a single mutation, 2 patients two mutations (one somatic and one germline), and 1 patient had three mutations (two somatic and one germline). The location and combination pattern of all the detected CEBPA mutations are presented in Table 5.
Table 5.
Summary of CEBPA mutations detected in 21 acute myeloid leukemia patients
| Mutation status | Mutation 1 | Mutation 2 (Mutation 3) | No. of patients | ||||
|---|---|---|---|---|---|---|---|
| N-terminal AA 1-120 |
Middle AA 121–277 |
C-terminal AA 278–358 |
N-terminal AA 1-120 |
Middle AA 121–277 |
C-terminal AA 278–358 |
||
| CEBPA-single | p.Ala113ArgfsTer54* | 1 | |||||
| p.Ala134= | 1 | ||||||
| p.His191= | 1 | ||||||
| p.Pro198_Ala199dup* | 4 | ||||||
| p.Thr230=* | 11 | ||||||
| CEBPA-double | p.Gly96AlafsTer56 | p.Thr230=* | (p.Lys313dup) | 1 | |||
| p.Ser61ValfsTer47 | p.Thr230=* | 1 | |||||
| p.Phe179Ser | p.Thr230=* | 1 | |||||
(*) – germline mutation
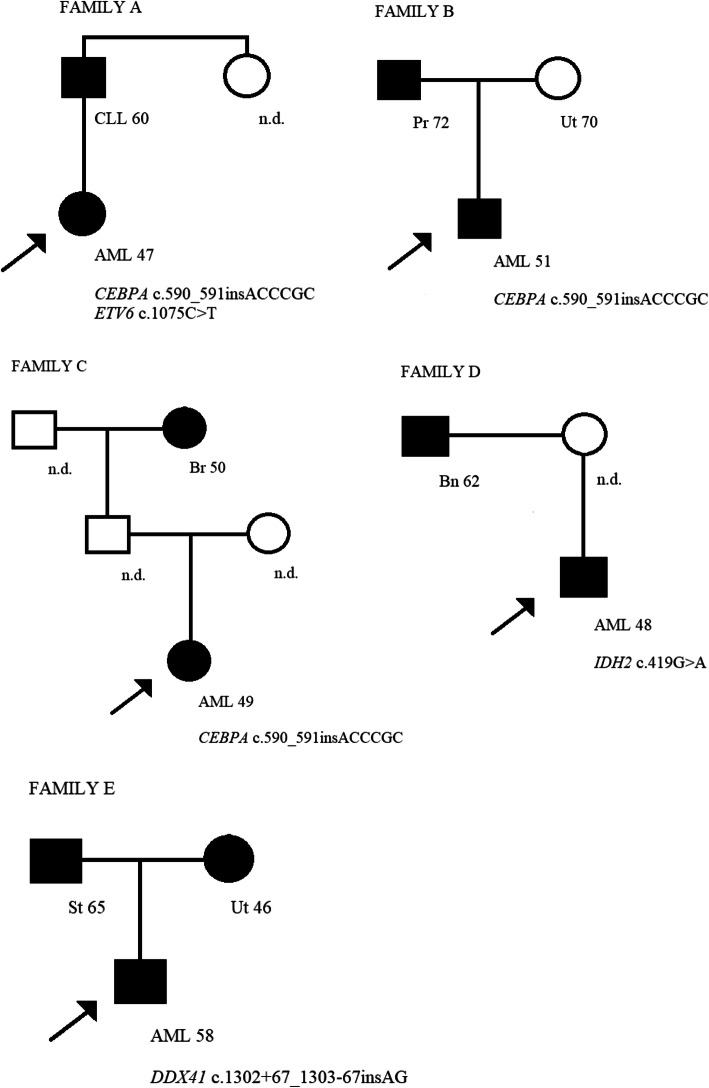
Single pathogenic germline mutation in CEBPA gene was found in 5 out of 103 AML patients (4.9 %), in two patients with M1-AML, two with M4-AML, and one with M6-AML. The c.590_591insACCCGC mutation in CEBPA occurred in four unrelated patients (age at diagnosis 20, 47, 49, and 51 years), while c.337_344del mutation occurred in one patient (age at diagnosis 37 years) (Tables 2 and 3). In the family of the patient with AML diagnosed at the age of 20, no other cancer was present. In one patient with M1-AML (age at diagnosis 47 years) two pathogenic germline mutations, c.590_591insACCCGC in CEBPA and c.1075 C > A in ETV6 (the only ETV6 germline mutation found in analyzed group), occurred. In his family, chronic lymphocytic leukemia (age at diagnosis 60 years) in second-degree relative appeared. In the family of the patient with AML diagnosed at the age of 49, breast cancer (age at diagnosis 50 years) in a second-degree relative occurred. In the family of the patient with AML diagnosed at the age 51, prostate cancer (age at diagnosis 72 years) and uterine cancer (age at diagnosis 70 years) in two first degree-relatives occurred (Fig. 1). In the family of the patient with AML diagnosed at the age of 37, and c.337_344del mutation, no other cancer was present.
Fig. 1.
The pedigrees of families with pathogenic germline mutations carriers (A-D) and germline variant of uncertain significance carrier (E). In all pedigrees, probands are indicated by arrows. The type of cancer and the age at disease diagnosis are described under the filled black square/circle. AML - acute myeloid leukemia, CLL - chronic lymphocytic leukemia, Pr - prostate cancer, Ut - uterine cancer, St - stomach cancer, Br -breast cancer, Bn - brain cancer, n.d. - no data
The c.419G > A mutation in IDH2 occurred in two unrelated patients (age at diagnosis 44 and 48 years) with M5-AML and M1-AML, respectively (Table 4). In the family of the patient with AML diagnosed at the age of 44, no other cancer was present. In the family of the patient with AML diagnosed at the age of 48, a brain tumor in the first-degree relative occurred (age at diagnosis 62 years) (Fig. 1).
The c.1302 + 67_1303-67insAG germline intronic variant of uncertain significance in DDX41 gene occurred in a patient (age at diagnosis 58 years) with M5-AML. Stomach cancer (age at diagnosis 65 years) and uterine cancer (age at diagnosis 46 years) in two first degree-relatives occurred in his family (Fig. 1).
The median age at AML diagnosis was 11 years lower in patients with germline pathogenic mutations (47 years, range 20–51) than in patients without them (58 years, range 18–89; p = 0.028).
Discussion
We tested CEBPA, GATA2, ETV6, IDH2, DDX41, and TERT genes, to detect germline pathogenic mutations in AML patients. We found germline variants in each of the tested genes, but only 8 of them were pathogenic – 5 in CEBPA, 1 in ETV6, and 2 in IDH2 genes. In the CEBPA gene were detected: c.337_344del mutation in one patient and c.590_591insACCCGC mutation in four patients, in the ETV6 gene c.1075 C > T mutation in one patient, and in the IDH2 gene c.419G > A mutation in two patients. The frequency of all pathogenic germline mutations was 7.77 % in tested group and 0.97 % in control group (p = 0.046).
We identified one novel pathogenic germline mutation, c.337_344del, in CEBPA gene. This mutation creates a stop codon within the TAD2 domain and results in a truncated protein. In our study, this mutation was disclosed with 0.97 % (1 patient) frequency in the study group, which was not statistically significantly different (NS) from its frequency in control group (no patient with this mutation). Moreover, in the CEBPA gene we identified a known pathogenic germline mutation, c.590_591insACCCGC, present with 3.88 % frequency in the study, and 0.97 % in the control group. The difference was not statistically significant, however the odds ratio was 4.12; 95 % CI: 0.453–37.520. According to the latest knowledge, the prevalence of this mutation in the European population has not yet been evaluated (https://varsome.com).
We also found a germline nonsense mutation c.1075 C > T in ETV6 gene, which creates a stop codon within the ETS domain and results in a truncated protein, without a DNA-binding function. This mutation was disclosed in one female patient (0.97 %) in tested group and in no person in control group (NS). Chronic lymphocytic leukemia (age at diagnosis 60 years) in second-degree relative occurred in patient’s family. ETV6 mutation coexisted with the c.590_591insACCCGC CEBPA mutation in one patient. The coexistence of two germline pathogenic mutations also was reported by Tsaousis et al. in hereditary breast cancer [25]. Moriyama et al. identified the same as in our patient ETV6 mutation in all investigated females from an acute lymphoblastic leukemia (ALL) family of European descent. The mother and 2 of her 3 daughters developed ALL in the childhood, at the ages of 9, 3, and 2 years, respectively. All 3 ALL cases were of B-lineage, although with various molecular subtypes. Mild congenital thrombocytopenia was noted in mother and one daughter, the second daughter was diagnosed with Turner syndrome and mild intellectual disability, and the third was diagnosed with a learning disability. The enlarged family history did not reveal other hematologic malignancies. Interestingly, the mutation was also present in the healthy daughter, suggesting incomplete penetrance. However, given her young age of 11 it is still possible that she has been at risk of ALL [26].
Mutations in the IDH1 and IDH2 genes are well described in lower-grade gliomas (grade II and III astrocytomas and oligodendrogliomas) and secondary glioblastomas, where they have an incidence of more than 70 %. Mutations and polymorphisms in these genes, predominantly somatic, are reported in 5–15 % of AML patients [27–29]. In AML, nearly all IDH2 mutations cause a single amino acid substitution, Arg172 to one of four different residues - Lys, Met, Gly, and Trp, and Arg140 to either Gln or Trp [29, 30]. Following the discoveries in glioma and AML, mutations in IDH2 gene were found in multiple types of human tumors, including thyroid carcinomas, cartilaginous tumors, and intrahepatic cholangiocarcinoma [31]. IDH2 mutations give rise to protein with newly acquired and distinct enzyme activity, which can catalyze NADPH-dependent reduction of α-KG to 2-hydroxyglutarate (2HG). Accumulation of this putative oncogenic metabolite has been observed in malignant gliomas and may be related to the pathogenesis of malignant brain tumors. Increased cellular 2HG levels contribute to epigenetic mechanisms of pathogenesis by inhibiting α-KG-dependent enzymes that are important for normal DNA methylation [32, 33].
A germline missense mutation, c.419G > A, in the IDH2 gene was detected in two patients in our study. According to GnomAD, this mutation occurs with a frequency of 0.01 % in the European population, as well as in the African and East Asian populations. The prevalence of this mutation was almost two-fold higher in relation to the control group, but this difference was not statistically significant (p = 0.295). In the family of one of two patients with c.419G > A mutation, a brain tumor occurred, at 48 years of age. Kranendijk et al. detected heterozygous germline mutations in IDH2 that alter enzyme residue Arg140 in patients with D-2-hydroxyglutaric aciduria (D-2-HGA). In 15 unrelated patients they detected a known heterozygous c.419G > A (p.Arg140Gln) mutation, the same as in our patients, and a novel heterozygous mutation c.418 C > G (p.Arg140Gly) [34, 35]. Molenaar et al. described a germline IDH2 mutation, c.782G > A (p.Arg261His), in a patient with AML, and another germline mutation, c.1304 C > T (p.Thr435Met), in two unrelated patients with MDS (RCMD - Refractory Cytopenia with Multilineage Dysplasia) [36].
In our study, one germline intronic variant (c.1302 + 67_1303-67insAG) in the DDX41 gene was detected, which was identified as VUS in VarSome. This variant occurred in a patient with AML diagnosed at 58 years. Stomach cancer and uterine cancer were diagnosed in two of the first degree-relatives of the patient. So far, this variant has not been described (https://varsome.com).
The results of our study suggest the need to increase the study group as well as to carry out the family studies to determine a heritability of a variant/mutation among cancer-positive family members.
In our study, we were interested only in germline mutations. They may be the best distinguished from somatic mutations by sequencing of a nonhematopoietic tissue, such as fibroblasts grown from a skin biopsy. However, a disadvantage of this approach is that a skin biopsy would be an additional invasive procedure in AML patients and that fibroblasts require several weeks of culture. Thus, we decided on testing buccal swabs. They were taken with due care and caution to avoid contamination with hematological material. Two mutations found in buccal swabs in our patients were confirmed as the germline ones in some other publications [24, 36]. The nature of other mutations found by us (c.337_344del, c.590_591insACCCGC, and c.1302 + 67_1303-67insAG) has not yet been confirmed elsewhere.
In the present study, we found no correlation between pathogenic germline mutations and karyotype of bone marrow cells at AML diagnosis.
Conclusions
Familial AML predisposition syndromes are rare inherited disorders characterized by significantly elevated risk of AML development. Although several disorders with germline predisposition have been included in the revised 2016 WHO classification of myeloid neoplasms and acute leukemia as “myeloid neoplasms with germline predisposition”, screening for known germline mutations in the background of these syndromes is not a part of the routine diagnostic algorithms. However, if a germline mutation is detected, this should be always taken into account in prophylactic and therapeutic decisions (WHO 2016) [1, 2]. Given the molecular heterogeneity of AML, a better understanding of mutational classes and their involvement in AML pathogenesis could improve risk stratification of patients for more effective and targeted therapy [37]. Identification of the affected families with inherited AML is of critical importance as they not only provide unique models to study the molecular pathogenesis of these diseases, but identification of a germline mutation may have immediate clinical implications as regards the pre-neoplastic monitoring of family members with this mutation. In addition, identification of germline mutations will make it possible to invent prophylactic and early diagnostic issues against germline mutations-driven AML. It is believed that familial AML is still underdiagnosed and its frequency is higher than currently reported. Further studies are necessary to assess the prevalence of germline mutations in the larger AML patient cohort and to establish their prognostic significance.
Supplementary information
Acknowledgements
The authors would like to thank all patients for participating in the study.
Abbreviations
- AML
Acute myeloid leukemia
- HSCT
Hematopoietic stem cells transplantation
- DNA
Deoxyribonucleic acid
- CEBPA
CCAAT/Enhancer-Binding Protein-A
- DDX41
DEAD/H-box helicase
- ETV6
EST Variant Transcription Factor 6
- TERT
Telomerase reverse transcriptase
- GATA2
GATA bining protein 2
- IDH2
Isocitrate dehydrogenase (NADP(+))2
- NGS
Next generation sequencing
- RUNX1
Runt-related transcription factor 1
- TERC
Telomerase RNA component
- IDH1
Isocitrate dehydrogenase (NADP(+))1
- DNMT3A
DNA methyltransferase 3 alpha
- TF
transcription factor
- RNA
Ribonucleic acid
- EST
E26 transformation specific
- MDS
Myelodysplastic syndromes
- TET2
Tet methylcytosine dioxygenase 2
- CM NCU
Nicolaus Copernicus University in Torun, Poland
- FAB
French-American-British classification
- PB
Peripheral bloom
- BM
Bone marrow
- BS
Buccal swabs
- MIP
Molecular inversion probe
- PE
Paired-end
- PCR
Polymerase chain reaction
- FISH
Fluorescence in situ hybridization
- OR
Odds ratio
- VUS
Uncertain significance variant
- NS
Not statistically significantly different
- ALL
Acute lymphoblastic leukemia
- RCMD
Refractory cytopenia with multilineage dysplasia
- WHO
World Health Organization
- ID
Identifier
- NA
No amino acid change
- CLL
Chronic lymphocytic leukemia
- Pr
Prostate cancer
- Ut
Uterine cancer
- Br
Breast cancer
- Bn
Brain cancer
- St
Stomach cancer
- NK
Normal karyotype
- ND
No data
Authors’ contributions
Aneta Bąk – study design, molecular investigation, data analysis, author of the manuscript (first and corresponding author), Katarzyna Skonieczka – cytogenetic investigation, Anna Jaśkowiec – cytogenetic investigation, Marta Heise – molecular investigation, Maria Pilarska-Deltow – molecular investigation, Anna Junkiert-Czarnecka – molecular investigation, Stanisław Potoczek – clinical evaluation of patients, sample collection, Maria Czyżewska – clinical evaluation of patients, sample collection, Olga Haus – clinical evaluation of patients, supervision of the research and manuscript (senior author). The author(s) read and approved the final manuscript.
Funding
This work was supported by the Nicolaus Copernicus University Grants number MN-SDL-2/WL/2018 and MN-SDL-2/WL/2019.
Availability of data and materials
All information about data and materials will be available on request after prior contact with me at my e-mail address: aneta.bak@cm.umk.pl.
Declarations
Ethics approval and consent to participate
The study was approved by the Ethics Committee of the CM NCU. Informed consent was obtained from all patients and control healthy persons for being included in the study.
Consent for publication
All authors gave their consent for the article publication.
Competing interests
Authors declare no competing of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Swerdlow SH, Campo E, Pileri SA, Harris LS, Stein H, Siebert R, et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. 2016;127(Suppl 20):2375–90. doi: 10.1182/blood-2016-01-643569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kiraly AP, Kallay K, Gango A, Kellner A, Egyed M, Szoke A, et al. Familial acute myeloid leukemia and myelodysplasia in Hungary. Pathol Oncol Res. 2018;24(Suppl 1):83–8. doi: 10.1007/s12253-017-0216-4. [DOI] [PubMed] [Google Scholar]
- 3.Rafei H, DiNardo CD. Hereditary myeloid malignancies. Best Pract Res Clin Haematol. 2019;32(Suppl 2):163–76. doi: 10.1016/j.beha.2019.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malcovati L, Papaemmanuil E, Ambaglio I, Elena C, Galli A, Della Porta MG, et al. Driver somatic mutations identify distinct disease entities within myeloid neoplasms with myelodysplasia. Blood. 2014;124(Suppl 9):1513–21. doi: 10.1182/blood-2014-03-560227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith ML, Cavenagh JD, Lister TA, Fitzgibbon J. Mutation of CEBPA in familial acute myeloid leukemia. N Engl J Med. 2004;351Suppl 23:2403–7. doi: 10.1056/NEJMoa041331. [DOI] [PubMed] [Google Scholar]
- 6.Taskesen E, Bullinger L, Corbacioglu A, Sanders MA, Erpelinck CAJ, Wouters BJ, et al. Prognostic impact, concurrent genetic mutations, and gene expression features of AML with CEBPA mutations in a cohort of 1182 cytogenetically normal AML patients: further evidence for CEBPA double mutant AML as a distinctive disease entity. Blood. 2011;117(Suppl 8):2469–75. doi: 10.1182/blood-2010-09-307280. [DOI] [PubMed] [Google Scholar]
- 7.Kim HS, Han E, Jang W, Kim M, Kim Y, Han K, et al. Germline CEBPA mutations in Korean patients with acute myeloid leukemia. Leuk Res. 2019;76(Suppl 2):84–6. doi: 10.1016/j.leukres.2018.12.003. [DOI] [PubMed] [Google Scholar]
- 8.van Waalwijk van Doorn-Khosrovani SB, Erpelinck C, Meijer J, van Oosterhoud S, van Putten WLJ, Valk PJM, et al. Biallelic mutations in the CEBPA gene and low CEBPA expression levels as prognostic markers in intermediate-risk AML. Hematol J. 2003;4(Suppl 1):31–40. [DOI] [PubMed]
- 9.Cheah JJ, Hahn CN, Hiwase DK, Scott HS, Brown AL. Myeloid neoplasms with germline DDX41 mutation. Int J Hematol. 2017;106(Suppl 2):163–74. doi: 10.1007/s12185-017-2260-y. [DOI] [PubMed] [Google Scholar]
- 10.Linder P. Dead-box proteins: a family affair-active and passive players in RNP-remodeling. Nucleic Acids Res. 2006;34(Suppl 15):4168–80. doi: 10.1093/nar/gkl468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feurstein S, Godley LA. Germline ETV6 mutations and predisposition to hematological malignancies. Int J Hematol. 2017;106(Suppl 2):189–95. doi: 10.1007/s12185-017-2259-4. [DOI] [PubMed] [Google Scholar]
- 12.Liu T, Yuan X, Xu D. Cancer-specific telomerase reverse transcriptase (TERT) promoter mutations: biological and clinical implications. Genes. 2016;7 Suppl 7:38. [DOI] [PMC free article] [PubMed]
- 13.Heidenreich B, Rachakonda PS, Hemminki K, Kumar R. TERT promoter mutations in cancer development. Curr Opin Genet Dev. 2014;24:30–7. doi: 10.1016/j.gde.2013.11.005. [DOI] [PubMed] [Google Scholar]
- 14.Du HY, Pumbo E, Ivanovich J, An P, Maziarz RT, Reiss UM, et al. TERC and TERT gene mutations in patients with bone marrow failure and the significance of telomere length measurements. Blood. 2009;113(Suppl 2):309–16. doi: 10.1182/blood-2008-07-166421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vulliamy T, Walne A, Baskaradas A, Marrone A, Dokal I. Mutations in the reverse transcriptase component of telomerase (TERT) in patients with bone marrow failure. Blood Cells Mol Dis. 2005;34(Suppl 3):257–63. doi: 10.1016/j.bcmd.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 16.McReynolds LJ, Calvo KR, Holland SM. Germline GATA2 mutation and bone marrow failure. Hematol Oncol Clin N Am. 2018;32(Suppl 4):713–28. doi: 10.1016/j.hoc.2018.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Collin M, Dickinson R, Bigley V. Haematopoietic and immune defects associated with GATA2 mutation. Brit J Haematol. 2015;169(Suppl 2):173–87. doi: 10.1111/bjh.13317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Issa GC, DiNardo CD. Acute myeloid leukemia with IDH1 and IDH2 mutations: 2021 treatment algorithm. Blood Cancer J. 2021;11:107. doi: 10.1038/s41408-021-00497-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Filipp FV, Scott DA, Ronai ZA, Smith JW. Reverse TCA cycle flux through isocitrate dehydrogenases 1 and 2 is required for lipogenesis in hypoxic melanoma cells. Pigment Cell Melanoma Res. 2012;25(Suppl 3):375–83. doi: 10.1111/j.1755-148X.2012.00989.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Green CL, Evans CM, Zhao L, Hills RK, Burnett AK, Linch DC, et al. The prognostic significance of IDH2 mutations in AML depends on the location of the mutation. Blood. 2011;118(Suppl 2):409–12. doi: 10.1182/blood-2010-12-322479. [DOI] [PubMed] [Google Scholar]
- 21.The AACR Project GENIE Consortium. AACR Project GENIE: powering precision medicine through an international consortium. Cancer Discov 2017;7 Suppl 8:818 – 31. [DOI] [PMC free article] [PubMed]
- 22.Jeong D, Lee DS, Kim N, Choi S, Kim K, Kim SM, et al. Prevalence of germline predisposition gene mutations in pediatric acute myeloid leukemia: Genetic background of pediatric AML. Leuk Res 2019;85. 10.1016/j.leukres.2019.106210. [DOI] [PubMed]
- 23.Boyle EA, O’Roak BJ, Martin BK. MIPgen: optimized modeling and design of molecular inversion probes for targeted resequencing. Bioinformatics. 2014;30(Suppl 18):2670–2. doi: 10.1093/bioinformatics/btu353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoon JK, Ahn J, Kim HS, Han SM, Jang H, Lee MG, et al. MicroDuMIP: target-enrichment technique for microarray-based duplex molecular inversion probes. Nucleic Acids Res. 2015;43 Suppl 5:28. [DOI] [PMC free article] [PubMed]
- 25.Tsaousis GN, Papadopoulou E, Apessos A, Agiannitopoulos K, Pepe G, Kampouri S, et al. Analysis of hereditary cancer syndromes by using a panel of genes: novel and multiple pathogenic mutations. BMC Cancer. 2019;19:535. doi: 10.1186/s12885-019-5756-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moriyama T, Metzger ML, Wu G, Nishii R, Qian M, Devidas M, et al. Germline genetic variation in ETV6 and risk of childhood acute lymphoblastic leukemia: a systematic genetic study. Lancet Oncol. 2016;16(Suppl 16):1659–66. doi: 10.1016/S1470-2045(15)00369-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patel KP, Barkoh BA, Chen Z, Ma D, Reddy N, Medeiros LJ, et al. Diagnostic testing for IDH1 and IDH2 variants in acute myeloid leukemia. An algorithmic approach using High-Resolution Melting curve analysis. J Mol Diagn. 2011;13(Suppl 6):678–86. doi: 10.1016/j.jmoldx.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 2010;18 Suppl 6:553 – 67. [DOI] [PMC free article] [PubMed]
- 29.Yan H, Parsons W, Jin G, McLendon R, Rasheed BA, Yuan W, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360(Suppl 8):765–73. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hamadou WS, Bourdon V, Letard S, Brenet F, Laarif S, Besbes S, et al. Familial hematological malignancies: new IDH2 mutation. Ann Hematol. 2016;95(Suppl 12):1943–7. doi: 10.1007/s00277-016-2813-9. [DOI] [PubMed] [Google Scholar]
- 31.Chou WC, Lei WC, Ko BS, Hou HA, Chen CY, Tang JL, et al. The prognostic impact and stability of isocitrate dehydrogenase 2 mutation in adult patients with acute myeloid leukemia. Leukemia. 2011;25(Suppl 2):246–53. doi: 10.1038/leu.2010.267. [DOI] [PubMed] [Google Scholar]
- 32.Yang H, Ye D, Guan KL, Xiong Y. IDH1 and IDH2 mutations in tumorigenesis: mechanistic insights and clinical perspectives. Clin Cancer Res. 2012;18(Suppl 20):5562–71. doi: 10.1158/1078-0432.CCR-12-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Im AP, Sehgal AR, Carroll MP, Smith BD, Tefferi A, Johnson DE, et al. DNMT3A and IDH mutations in acute myeloid leukemia and other myeloid malignancies: associations with prognosis and potential treatment strategies. Leukemia 2014;28 Suppl 9:1774–83. [DOI] [PMC free article] [PubMed]
- 34.Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17(Suppl 3):225–34. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kranendijk M, Struys EA, van Schaftingen E, Gibson KM, Kanhai WA, van der Knaap MS, et al. IDH2 mutations in patients with D-2-hydroxyglutaric aciduria. Science. 2010;330 Suppl 6002:336. [DOI] [PubMed]
- 36.Molenaar R, Sanikommu SR, Patel BJ, Tuominen J, Annala M, Hayrynen S, et al. Whole exome sequencing identifies germline IDH2 and IDH3 mutations that predispose to myeloid neoplasms. Blood 2015;126 Suppl 23:1405.
- 37.Yu J, Jiang PY, Sun H, Zhang X, Jiang Z, Li Y, et al. Advances in targeted therapy for acute myeloid leukemia. Biomark Res. 2020;8:17. doi: 10.1186/s40364-020-00196-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All information about data and materials will be available on request after prior contact with me at my e-mail address: aneta.bak@cm.umk.pl.