Abstract
The orphan nuclear hormone receptor SHP interacts with a number of other nuclear hormone receptors and inhibits their transcriptional activity. Several mechanisms have been suggested to account for this inhibition. Here we show that SHP inhibits transactivation by the orphan receptor hepatocyte nuclear factor 4 (HNF-4) and the retinoid X receptor (RXR) by at least two mechanisms. SHP interacts with the same HNF-4 surface recognized by transcriptional coactivators and competes with them for binding in vivo. The minimal SHP sequences previously found to be required for interaction with other receptors are sufficient for interaction with HNF-4, although deletion results indicate that additional C-terminal sequences are necessary for full binding and coactivator competition. These additional sequences include those associated with direct transcriptional repressor activity of SHP. SHP also competes with coactivators for binding to ligand-activated RXR, and based on the ligand-dependent interaction with other nuclear receptors, it is likely that coactivator competition is a general feature of SHP-mediated repression. The minimal receptor interaction domain of SHP is sufficient for full interaction with RXR, as previously described. This domain is also sufficient for full coactivator competition. Functionally, however, full inhibition of RXR transactivation requires the presence of the C-terminal repressor domain, with only weak inhibition associated with this receptor interaction domain. Overall, these results suggest that SHP represses nuclear hormone receptor-mediated transactivation via two separate steps: first by competition with coactivators and then by direct effects of its transcriptional repressor function.
Nuclear hormone receptors are transcription factors whose activities are regulated by direct binding of small lipophilic molecules such as steroids, thyroid hormone, retinoids, and vitamin D3 (24). These receptors have been studied extensively due to their important roles in the control of development and cellular homeostasis. The nuclear hormone receptor superfamily also includes numerous orphan receptors, which do not have identified ligands (10, 23). The structural similarities shared by superfamily members reside in two distinct functional domains. The first and most highly conserved is the DNA binding domain, also called the C domain, which targets receptors to specific DNA sequences known as hormone response elements. The second is the ligand binding domain (LBD), which is also called the E domain. It is located in the C-terminal region and is involved in several functions in addition to ligand binding, including dimerization and ligand-dependent transcriptional activation.
Crystallographic studies (e.g., references 30, 43, and 46) have revealed that ligand binding provokes a conformational change in the receptors that allows them to bind a diverse group of proteins termed coactivators (reviewed in references 14, 37, and 47). These coactivators bind to a common surface of the receptors formed by several alpha-helices (8, 27, 36), and it is thought that the appropriate positioning of the C-terminal helix 12 in response to ligand binding is of particular importance in promoting coactivator interaction. The ligand-dependent transcriptional activation function is referred to as AF-2, and a conserved sequence within helix 12 (7) that is required for coactivator binding is referred to as the AF-2 motif.
One important group of related coactivators is encoded by three genes, each of which has multiple names (37, 47). The first described was SRC-1 (27), which was followed by SRC-2, which is also known as TIF2 (39) or GRIP-1 (12), and by SRC-3, which is also known as ACTR (5), AIB1 (1), p/CIP (38), RAC3 (20), and TRAM-1 (40). These proteins, which are all approximately 160 kDa, show strong ligand-dependent interaction with the AF-2 surface of the receptors and are able to stimulate receptor transactivation when overexpressed. Another group of two related proteins, CBP and p300, also stimulate ligand-dependent transactivation by the receptors (e.g., reference 18). These two proteins, which can interact with members of the p160/SRC family, also function as coactivators for many other transcription factors (47). Finally, there are a number of other potentially important coactivators that also stimulate nuclear hormone-mediated transactivation and show either AF-2-dependent (e.g., reference 26) or AF-2-independent (e.g., reference 5) interaction with receptors.
At least two general mechanisms are thought to account for the effects of the coactivators. Several lines of evidence indicate that they can counteract inhibitory chromatin effects. At least for SRC-1 (39), ACTR/SRC-3 (6), p300/CBP (3, 28), and p/CAF (49), this is thought to be a consequence of their intrinsic histone acetyltransferase activity. It is also thought that these coactivators may function more directly to recruit RNA polymerase to promoters by interactions with components of the basal transcriptional apparatus (4).
Among the numerous proteins initially identified on the basis of interaction with receptor LBDs is an unusual orphan nuclear receptor, SHP. This orphan was originally isolated on the basis of its interaction with the new receptor CAR, using the Saccharomyces cerevisiae two-hybrid assay (32). Initial studies demonstrated that SHP lacks a DNA binding domain and that it exhibits functional interactions with several other nuclear hormone receptors in addition to CAR (32). In general, SHP acts to decrease transactivation by its partners in transient-transfection assays. This inhibitory effect was first attributed to an inhibition of DNA binding of the SHP targets. However, further studies revealed that SHP itself carries a novel autonomous repression domain in its C-terminal region (33). The existence of this domain suggested that SHP could also act to inhibit transcription if it could interact with DNA-bound receptor targets. Support for this alternative possibility was recently provided by results demonstrating that SHP is an effective inhibitor of estrogen receptor (ER) transactivation, even though it does not block binding of ER to estrogen response elements (34).
The inhibitory effect of SHP on ER raises two important questions. Because ER is in the group of receptors thought to function exclusively as homodimers, the first question is whether the range of SHP targets could include other superfamily members in this group. Of these superfamily members, the orphan HNF-4 is particularly interesting, since loss of function of a single HNF-4 allele causes the syndrome of mature-onset diabetes of the young (48). HNF-4 is expressed in liver and in the insulin-producing pancreatic beta cells, and the potential importance of any functional interaction between SHP and HNF-4 was recently extended by preliminary results indicating that SHP is expressed not only in liver, as previously described (32), but also in beta cells. The second question concerns the mechanism of the inhibitory effects of SHP on DNA-bound targets. In addition to the inherent SHP repressor function, a recent report has suggested yet another mechanism for repression, in which SHP interferes with coactivator binding to ER (17). The relative contributions of the direct repressor activity and the indirect interference with coactivator function to SHP-mediated inhibition of ER and other targets remain unclear.
We have examined both the potential interaction of SHP with HNF-4 and the molecular basis for its inhibitory effects. As with other superfamily members, SHP is an effective inhibitor of HNF-4 transactivation. SHP requires the conserved AF-2 motif for interaction with HNF-4 and also with retinoid X receptor (RXR) and can compete with coactivators for binding to both. At least in the case of RXR inhibition, loss of the SHP repressor function strongly weakens but does not completely block SHP-mediated repression. This supports the functional importance of this domain and suggests a two-step model for SHP inhibition. In this model, an initial inhibitory effect results from the loss of coactivator binding due to competition by SHP. Full inhibition depends on the second step, in which the SHP repressor domain acts directly to further decrease expression.
MATERIALS AND METHODS
Plasmids.
All of the HNF-4-related constructs, including mammalian expression vectors for wild-type HNF-4 and mutants, Gal4-HNF4 chimeras, and the reporter construct (BA1)5CAT were described previously (12). VP16-HNF4 was constructed by insertion of a PCR-amplified fragment containing the LBD of HNF-4 into the previously described mammalian VP16 fusion vector (11). Gal4 fusions of full-length and deletion mutants of murine SHP, CDM8SHP, and the Gal4Tkluc reporter construct were also described elsewhere (33). pCMX-SRC3 was a kind gift from J. D. Chen (University of Massachusetts, Worcester). Gal4SRC-3(RID) was constructed by insertion of PCR-amplified receptor interaction domain of SRC-3 (amino acids 601 to 761) into pCMXVP16. VP16RXRΔ19C was generated by insertion of human RXRα sequence from CDMhRXRΔ19C (51) into the previously described pCMXVP16 vector (11). For in vitro translation, HNF-4 inserts were reintroduced into pCMXGal4 plasmid. pT7lac-hRXRα and CDMhRXRΔ19C have been described previously (51).
Transient-transfection assays.
Plasmids were transfected into 50 to 60% confluent HepG2 cells by the calcium phosphate DNA coprecipitation method, as described previously (9, 20). For luciferase assays, cells were maintained in 24-well plates. For chloramphenicol acetyltransferase (CAT) assays, cell were maintained in 30-mm-diameter dishes. CAT, luciferase, and growth hormone (GH) activities were assayed approximately 48 h after addition of calcium phosphate DNA precipitates. For ligand treatment, cells were transfected in medium containing 10% charcoal-stripped fetal bovine serum and treated with the ligands for approximately 30 h. 9-cis-retinoic acid (9-cis-RA) was purchased from Biomol (Plymouth Meeting, Pa.) and LG1069 was from Glaxo Wellcome (Research Park Triangle, N.C.). For CAT assays, cells were collected in TEN solution (0.04 M Tris-HCl [pH 7.4], 1 mM EDTA, 0.15 M NaCl), and whole-cell extracts were prepared in 0.25 M Tris-HCl, pH 7.8, by three sequential freeze-thaw cycles. CAT activities were determined by using [14C]chloramphenicol and acetyl coenzyme A as previously described. Nonacetylated and acetylated chloramphenicol forms were separated on IB2 silica gel plates by using chloroform-methanol (95:5) for development, and radioactive spots, detected by autoradiography, were cut from the thin-layer plates and counted. CAT enzyme levels that exhibited more than 60% conversion of acetylated product were diluted and reassayed for CAT activity in the linear range.
In vitro binding assays.
In vitro binding assays were performed with glutathione S-transferase (GST) and GST fusion proteins expressed in Escherichia coli and bound to glutathione agarose, and appropriate [35S]methionine-labeled target proteins were produced by in vitro translation. Incubation and removal of nonspecifically bound proteins were performed by standard methods (2).
RESULTS
SHP represses HNF-4-mediated transactivation.
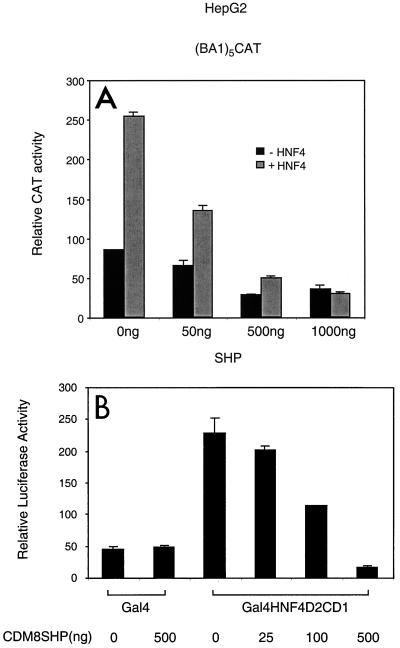
Based on both the association of SHP with a variety of nuclear hormone receptors and the expression of SHP in liver, we tested the capability of SHP to interact with the liver enriched orphan receptor HNF-4 (38) in the yeast two-hybrid system. In this assay, strong association was observed between a LexA-SHP fusion and a B42-HNF4 fusion (W. Seol, unpublished observation). In order to further test the effect of SHP on HNF-4-mediated transactivation, a reporter (12) containing a homopolymer of five HNF-4 binding sites from the apolipoprotein B promoter attached to a CAT reporter [(BA1)5CAT] was cotransfected into HepG2 cells with SHP and HNF-4 expression vectors. As shown in Fig. 1A, SHP inhibited HNF-4 transactivation in a dose-dependent manner. This functional interaction confirms the yeast results and indicates that HNF-4, like many other nuclear receptors, is an inhibitory target of SHP (17, 32–34).
FIG. 1.
Repression of HNF-4-mediated transactivation by SHP. (A) Repression of direct HNF-4 transactivation. HepG2 cells were cotransfected with 3 μg of (BA1)5CAT, 50 ng of HNF-4, 0.75 μg of CMVβ-Gal, and 50 ng to 1 μg of SHP plasmid. At 40 h posttransfection, the cells were harvested for CAT and β-galactosidase assay. The CAT values are the means of normalized three independent transfection experiments, each carried out in duplicate. (B) Repression of Gal4-HNF4 transactivation. HepG2 cells were cotransfected with 25 ng of Gal4-HNF4D2CD1 (Fig. 3A), 200 ng of TKGH, 200 ng of Gal4Tkluc reporter plasmid, and increasing amounts of SHP expression vector. CDM8 was used to maintain a constant amount of receptor expression vector, and total DNA was transfected. Cell extracts were prepared 48 h following transfection. Luciferase activities normalized with GH values are plotted as the mean ± standard deviation (n = 3).
Previous results have suggested that SHP inhibition may be a reflection of inhibition of DNA binding (32). However, in agreement with previous results with ER (34), no inhibitory effect of SHP on HNF-4 DNA binding was observed (data not shown). This result was not unexpected, since SHP targets the putative LBD of HNF-4 in yeast, and this domain is not essential for specific DNA binding by HNF-4 (16). To determine whether SHP could inhibit transactivation by an HNF-4 LBD tethered to DNA by a heterologous DNA binding domain, the effect of SHP on an appropriate Gal4-HNF4 fusion was determined. As previously described (12), this Gal4-HNF4 fusion is an apparently constitutive transactivator of a reporter containing Gal4 DNA binding sites. This constitutive activity was specifically inhibited by SHP in a dose-dependent manner (Fig. 1B), indicating that SHP can directly inhibit transactivation directed by the putative LBD of HNF-4. A direct role for the SHP repression function in this inhibition is suggested by the observation that, at the highest levels of SHP, expression was reduced below the basal level observed with Gal4 alone.
SHP interacts with the AF-2 domain of HNF-4 in mammalian cells.
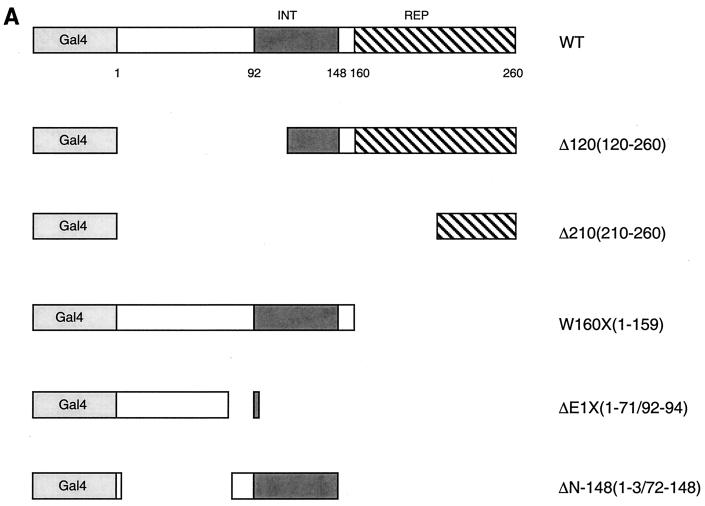
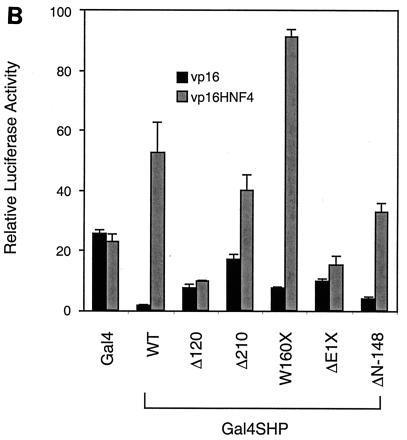
The association of SHP and HNF-4 previously indicated by yeast two-hybrid results was confirmed by a two-hybrid approach in mammalian cells, using either a Gal4-SHP fusion and a fusion of the VP16 transactivation domain to HNF-4 or the reciprocal combination. As indicated in Fig. 2B, coexpression of Gal4-SHP with the VP16-HNF4 fusion strongly stimulated reporter gene expression from the repressed state observed with Gal4-SHP alone. Coexpression of VP16-SHP also increased reporter gene expression beyond that observed with Gal4-HNF4 alone (Fig. 3B). To localize the SHP sequences required for HNF-4 association, a previously described series of SHP deletion mutants fused to Gal4 DNA binding domain (33) (Fig. 2A) were cotransfected into HepG2 cells with VP16-HNF4. As observed with other SHP targets, the constructs containing the SHP receptor association domain (W160X, ΔN-148) showed a significant stimulation of luciferase activity when coexpressed with VP16-HNF4, indicating that this region is also sufficient for HNF-4 interaction (Fig. 2B). Importantly, and in strong contrast to previous results with RXR (33), the loss of the SHP repression function in the W160X fusion did not result in a significant increase in apparent interaction. Previous results have demonstrated that the loss of such repressor function potently increases reporter gene expression in this system (33, 35). Thus, the lack of such an increase indicates that the truncated W160X product interacts more weakly with HNF-4 than the full-length SHP does. This is consistent with results with RAR, ER, and other receptor superfamily members (33, 34).
FIG. 2.
HNF-4 interacts with the receptor interaction domain of SHP. (A) Previously described SHP mutants (33) are diagrammed, fused to Gal4. INT and REP represent receptor interaction and direct repression domains, respectively. (B) SHP sequences required for SHP interaction in the mammalian two-hybrid assay. Fifty nanograms of each of the deletion versions of murine SHP fused to the Gal4 DNA binding domain was cotransfected into HepG2 cells with 50 ng of a vector expressing VP16 alone or VP16-HNF4D2. Normalized luciferase expression directed by the Gal4Tkluc reporter is indicated as the mean ± standard deviation (n = 3). WT, wild type.
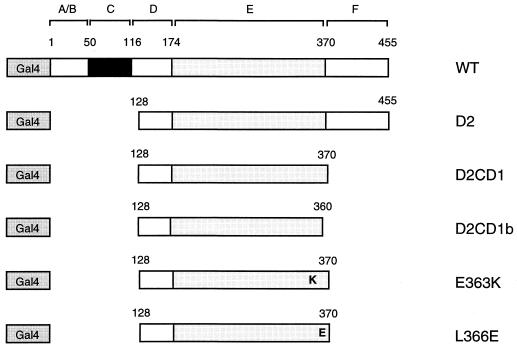
FIG. 3.
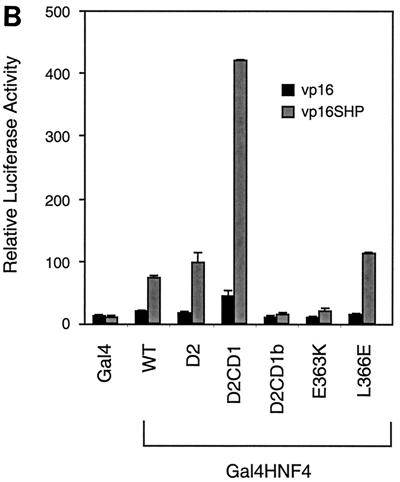
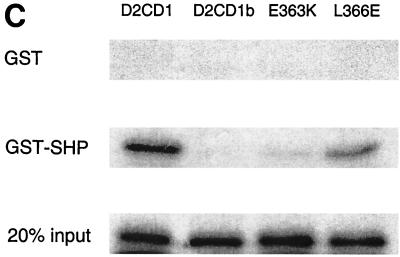
SHP interacts with the AF-2 surface of HNF-4. (A) Gal4-HNF4 constructs used in mammalian two-hybrid mapping are diagrammed. (B) HNF-4 sequences required for SHP interaction in the mammalian two-hybrid assay. Gal4 fusions (50 ng each) to the deletion or point mutant versions of HNF-4 were cotransfected with 50 ng of VP16 or VP16-SHP into HepG2 cells. Normalized luciferase expression from the Gal4Tkluc reporter is shown. (C) The AF-2 surface of HNF-4 is required for interaction with SHP in vitro. GST alone or a GST-SHP fusion protein were expressed in E. coli, bound to glutathione agarose, and incubated with the indicated Gal4-HNF4 fusion proteins, which were 35S labeled by in vitro translation. Specifically bound proteins were eluted by standard procedures (2) and are compared to 20% of the total input (bottom gel). WT, wild type.
To identify HNF-4 sequences required for SHP interaction in mammalian cells, a series of HNF-4 mutants fused to Gal4 (12) (Fig. 3A) was coexpressed with a VP16-SHP fusion in HepG2 cells. Interestingly, a deletion of 6 amino acids removing the conserved AF-2 motif (Gal4-D2CD1b) essentially abolished association with SHP, while an analogous construct with an intact AF-2 motif (Gal4-D2CD1) showed a significant association (Fig. 3B). Moreover, mutation of the conserved glutamic acid (Glu363) at the core of the AF-2 motif (Gal4-E363K) also blocked interaction with SHP. In contrast, mutation of a leucine residue (Leu366) located just outside of the conserved AF-2 motif (Gal4-L366E) did not impair SHP association. These mammalian two-hybrid results were confirmed and extended by biochemical results demonstrating specific binding of 35S-labeled HNF4D2CD1 to a GST-SHP fusion (Fig. 3C). As expected, binding was lost with the two HNF4 mutants affecting the AF-2 motif but retained with the L366E mutant.
Overall, these results demonstrate that SHP requires the HNF-4 AF-2 motif for interaction with HNF-4. This is in good agreement with recent, independently derived results indicating that SHP targets the AF-2 surface of ER (17).
SHP competes with the coactivator SRC-3 for binding to HNF-4.
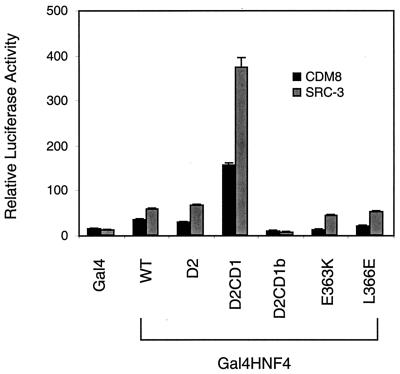
Two members of the p160 family, SRC-1 and SRC-2 (also known as TIF2 and GRIP-1) have been identified as coactivators for HNF-4 (44). The third member of this family, SRC-3 (also known as ACTR, AIB1, p/CIP, TRAM-1, and RAC3) was also tested for stimulation of HNF-4 transactivation. An SRC-3 expression vector was introduced into HepG2 cells along with an HNF-4 vector and the (BA1)5CAT reporter. As expected, SRC-3 coexpression further stimulated HNF-4 transactivation in a dose-dependent manner (data not shown). To confirm that the conserved AF-2 motif of HNF-4 is required for this SRC-3 stimulation, various deletion and point mutants of HNF-4 fused to Gal4 were cotransfected into HepG2 cells with the SRC-3 expression vector (Fig. 4). Gal4-HFN4 D2CD1, which contains an intact AF-2 motif, showed stimulation, but Gal4-HFN4 D2CD1b, which is missing this motif, did not. Mutation of the conserved glutamic acid in the AF-2 motif (E363K) decreased transactivation but, as previously observed with an analogous RXR mutant (42), did not prevent SRC-3 stimulation. Thus, these results indicate that the HNF-4 AF-2 motif is required for functional effects of SRC-3.
FIG. 4.
Stimulation of Gal4-HNF4 transactivation by SRC-3. Deletion or point mutant versions of HNF-4 (Fig. 3) fused to the Gal4 DNA binding domain (25 ng) were cotransfected into HepG2 cells with 500 ng of SRC-3. At 48 h after transfection, cells were harvested for luciferase and GH assays. Normalized luciferase expression directed by the Gal4Tkluc reporter is indicated as the mean ± standard deviation (n = 3). WT, wild type.
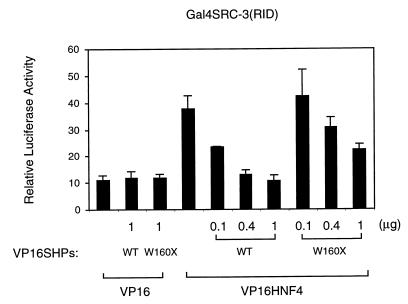
Together, the results with SHP and SRC-3 suggest that these two proteins should compete for the binding to the AF-2 surface of HNF-4. To test this, the effect of SHP on the interaction of SRC-3 and HNF-4 was examined in mammalian cells by a two-hybrid competition assay. A Gal4 fusion of the receptor-interacting domain of SRC-3 (amino acids 601 to 761) was generated and transfected into HepG2 cells with a VP16-HNF4 fusion. As expected, coexpression of these fusions resulted in increased luciferase expression, reflecting an effective SRC-3–HNF-4 interaction. As potential competitors, increasing amounts of either VP16-SHP or VP16-SHPW160X were added. (The VP16 activation domain was added to alleviate concerns related to secondary effects of recruiting an inhibitory SHP protein to an HNF-4–coactivator complex and to allow detection of potential interaction of SHP with SRC-3. However, similar results were observed with SHP and SHP W160X alone.) As indicated in Fig. 5, both competitors decreased the interaction of SRC-3 with HNF-4. Consistent with the apparently decreased affinity of the truncated mutant for HNF-4 as noted above, VP16-SHPW160X was an approximately 10-fold-weaker competitor than VP16-SHP. Since neither competitor showed any apparent interaction with Gal4-SRC3 and the total amount of VP16 expression vector was kept constant, we conclude that this decreased expression in this mammalian two-hybrid system is a result of competition between SRC-3 and SHP for binding to the AF-2 surface of HNF-4.
FIG. 5.
SHP competes specifically with SRC-3 for binding to HNF-4 in a mammalian two-hybrid assay. HepG2 cells were cotransfected with 50 ng (each) of Gal4SRC-3(RID), VP16 or VP16-HNF4, and the indicated amounts of VP16-SHP and VP16-SHPW160X. Normalized luciferase expression directed by the Gal4Tkluc reporter is indicated as the mean ± standard deviation (n = 3). The decreased luciferase expression in the presence of increasing amounts of the SHP proteins reflects decreased interaction between SRC-3 and HNF-4. Essentially identical results were observed with intact SHP.
SHP competes with coactivators to bind RXR.
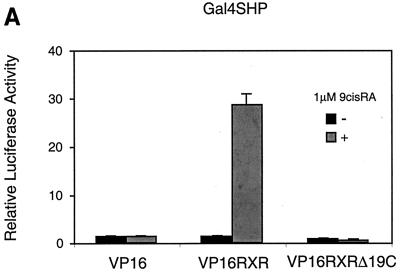
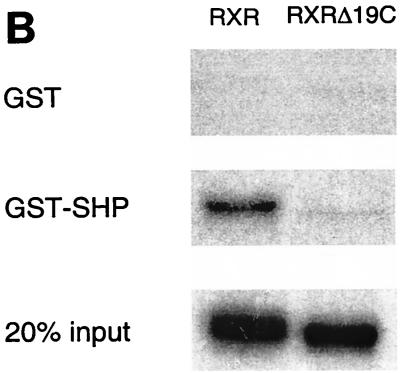
The ability of SHP to compete for coactivator binding to HNF-4 and the suggestion of a similar mechanism for ER (17) raise the question of the generality of this mechanism and also of its relationship to previously proposed direct activity of SHP as a transcriptional repressor. These issues were addressed by using RXR, which was previously shown to have a strong interaction with SHP (33). As with HNF-4, the potential involvement of the RXR AF-2 motif in the interaction with SHP was examined by the mammalian two-hybrid assay (Fig. 6A). Derivatives of the LBD of wild-type RXR and a mutant lacking the C-terminal AF-2 motif but capable of ligand binding (21, 52) were constructed and tested for interaction with GAL-SHP. As previously described, the full-length RXR exhibited a strong, ligand-dependent interaction with full-length SHP. However, SHP failed to bind the RXR lacking the AF-2 motif, indicating that SHP also targets the activation surface of RXR. This was confirmed by biochemical results demonstrating ligand-dependent interaction of 35S-labeled wild-type RXR, but not the RXR AF-2 mutant, with GST-SHP (Fig. 6B).
FIG. 6.
SHP interacts with the AF-2 surface of RXR. (A) Mammalian two-hybrid assay for interaction. Gal4SHP was transfected into HepG2 cells along with VP16RXR, VP16RXRΔ19C, or VP16 alone. Approximately 20 h after transfection, the cells were treated with 1 μM 9-cis-RA for 30 h. Normalized luciferase expression directed by the Gal4Tkluc reporter is indicated as the mean ± standard deviation (n = 3). (B) The AF-2 surface of RXR is required for interaction with SHP in vitro. GST alone or a GST-SHP fusion protein were expressed in E. coli, bound to glutathione agarose, and incubated with wild-type RXR or the C-terminal Δ19 deletion mutant, both of which were 35S labeled by in vitro translation. Specifically bound proteins were eluted by standard procedures (2) and are compared to 20% of the total input (bottom gel).
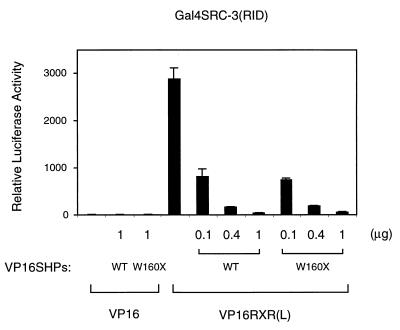
Since SRC-3 binds the same surface of RXR (6), the mammalian two-hybrid competition assay was used to determine whether SHP also competes with coactivators for binding to RXR in the presence of either 9-cis-RA or the synthetic RXR agonist LG1069. Increasing concentrations of SHP efficiently competed with SRC-3 for RXR binding (Fig. 7). In contrast to the results with HNF-4, however, the ability of the SHPW160X to compete was indistinguishable from that of full-length SHP. This is consistent with previous results demonstrating that the ability of this truncated SHP to interact with RXR is equivalent to that of wild-type SHP (33).
FIG. 7.
SHP competes specifically with SRC-3 for binding to activated RXR in a mammalian two-hybrid assay. HepG2 cells were cotransfected with 50 ng (each) of Gal4-SRC3, VP16 or a VP16 fusion to the RXR LBD [VP16-RXR(L)], and the indicated amounts of plasmids expressing VP16-SHP and VP16-SHPW160X. 9-cis-RA (1 μM) was added 20 h after transfection, and cells were further incubated for 30 h before harvest. Normalized luciferase expression is plotted as the mean ± standard deviation from three independent experiments. Essentially identical results were obtained in treatments with the specific RXR agonist LG1069. WT, wild type.
Two-step repression by SHP.
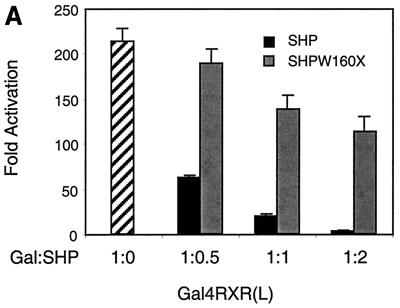
The efficient RXR interaction of this mutant, which lacks the direct repression activity of SHP, allowed a direct test of the relative importance of this activity and coactivator competition in RXR inhibition. As indicated in Fig. 8A, wild-type SHP efficiently repressed 9-cis-RA-dependent transactivation by Gal4-RXR, even at relatively low ratios of Gal4 and SHP expression vectors. However, the effect of W160X was significantly weaker. As expected from previous results with various Gal-SHP fusions (33), the wild-type and truncated SHP proteins were expressed at equivalent levels (data not shown). Thus, under circumstances where effects on DNA binding are excluded and the possible involvement of any other receptors are minimized, the SHP repression function is clearly required for full inhibition.
FIG. 8.
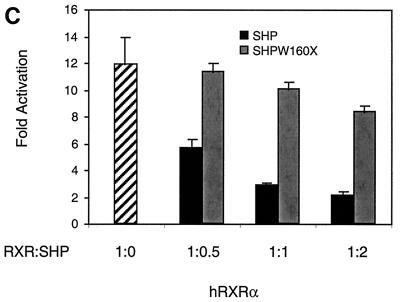
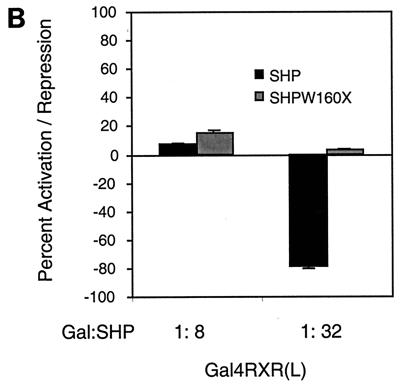
SHP requires the repression domain for full inhibition. (A) A Gal4-RXR(L) vector (50 ng) was cotransfected into HepG2 cells along with an SHP or SHPW160X vector in the indicated ratio (Gal:SHP). At 20 h posttransfection, cells were treated with 1 μM 9-cis-RA or vehicle alone and incubated for 30 h. Normalized luciferase expression is plotted as the mean fold activation by 9-cis-RA ± standard deviation from three independent experiments. (B) Gal4-RXR(L) vector (25 ng) was cotransfected into HepG2 cells with 200 and 800 ng of SHP or SHPW160X expression vector as indicated by concentration ratio (Gal:SHP). Normalized luciferase activities are plotted as percent activation or repression. Percent activation for each combination is relative to the activation observed with Gal4-RXR(L) in the presence of 9-cis-RA and the absence of SHP. Percent repression is relative to basal expression for each combination in the absence of 9-cis-RA. In this experiment, 800 ng of SHP or SHPW160X did not affect luciferase expression in the presence of Gal4 alone. (C) A thymidine kinase luciferase reporter containing the RXR response element from the CRBPII promoter was cotransfected into HepG2 cells with 50 ng of CDMhRXRα and either the wild-type SHP or the SHPW160X expression vector in the indicated ratio (RXR:SHP). Normalized luciferase expression was determined in three independent experiments, and the fold response to 1 μM LG1069 is shown.
As noted above, the potential importance of the SHP repression function was suggested by the effects of higher levels of SHP on Gal4-HNF4 (Fig. 1B). This was examined in more detail in the context of Gal4-RXR by a comparison of the effects of higher levels of wild-type SHP and SHP W160X. As demonstrated in Fig. 8B, the wild-type SHP decreased expression to levels well below the basal level observed with Gal4-RXR in the absence of ligand and SHP, but the repression-defective mutant failed to do so.
The importance of the SHP repression function was also examined in the context of native RXR. RXR transactivation of a reporter carrying the hormone response element from CRBPII, which is efficiently activated by RXR homodimers (25) (Fig. 8C), was decreased by both SHP and SHP W160X. As expected, however, the wild-type SHP was a much more effective inhibitor. In conclusion, while the SHP fragment capable of efficient coactivator competition in the previous results does show some inhibitory effects, they are much less than those observed with the full-length SHP. Full inhibition by SHP clearly requires its direct repressor activity, and at least for RXR, this activity appears to be responsible for the majority of the observed inhibition.
DISCUSSION
Previous results have demonstrated that the unusual orphan receptor SHP inhibits transactivation by many different members of the nuclear receptor superfamily. This was initially attributed to inhibition of DNA binding (32). However, SHP can also inhibit transactivation by targets that do not show such DNA binding effects, including the estrogen receptor (34), the Gal4 fusions described here, and at least a subset of the nuclear receptors that bind DNA as monomers (Y.-K. Lee, unpublished observations). Two additional mechanisms have been suggested to account for such inhibitory effects. The first is based on the actions of the previously described direct transcriptional repression function of SHP (33). In this case, recruitment of the active repressor function of SHP to DNA via interaction with another receptor would account for the decreased expression. The second is based on the competition between SHP and coactivators for binding to an activated receptor. This model, which was initially independently suggested based on biochemical results demonstrating the ability of SHP to inhibit coactivator binding to ER (17), is confirmed and significantly extended by the results described here.
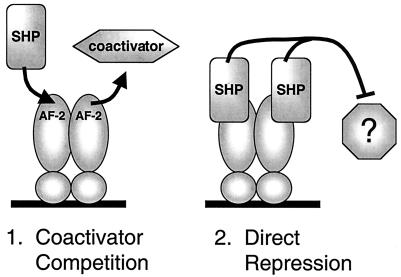
Importantly, these two mechanisms are not mutually exclusive. This suggests a two-step model for SHP repression, at least in cases where it does not affect DNA binding. In the first step, SHP binding reduces expression by decreasing coactivator binding to an activated receptor. In the second, the direct actions of SHP as a transcriptional repressor cause a further decrease in expression. This two-step model is diagrammed in Fig. 9.
FIG. 9.
Two-step repression by SHP. In the first step, SHP displaces coactivators by competing for binding to the receptor AF-2 surface. In the second step, the repressor function of the receptor-bound SHP further decreases expression via an unknown mechanism.
It is interesting that a very similar two-step repression model has also been suggested for SMAD-mediated transactivation. In this case, the factor TGIF was suggested to displace coactivators bound to SMADs and also to function directly as a corepressor by recruiting histone deacetylase activity (45). As previously described, SHP does not appear to interact with the nuclear receptor corepressor NCoR (33), and preliminary results indicate that SHP repression is not affected by trichostatin A, a potent inhibitor of histone deacetylase activity. Thus, the mechanism of the inhibitory effect of SHP remains unclear. Nonetheless, the parallels in the actions of SHP and TGIF emphasize the importance of mechanisms that function to rein in the effects of a variety of activating signals and also suggest that such two-step inhibition may be a general mechanism.
The existence of these two distinct steps for repression raises the question of their relative contribution to the observed inhibitory effects. The results described here demonstrate that the loss of the SHP repression function leads to a loss of the majority of the inhibition. Thus, at least for RXR, the direct activity of SHP as a transcriptional repressor is apparently responsible for the majority of the inhibitory effect. It is certainly possible that this activity could be less effective with other receptors or in other cell types. However, the apparent importance of this repressor function is consistent with genetic results with DAX-1, the only nuclear receptor superfamily member that is similar to SHP (50). These two orphans both lack a conventional DNA binding domain and also share a repression function that appears to be based on conserved C-terminal sequences (15, 19, 33). Surveys indicate that this C-terminal region is affected by all of the more than 40 mutations of the human DAX-1 gene identified in patients with congenital adrenal hypoplasia, including 7 single amino acid substitutions (19, 53). The striking prediction that all of these mutations affect repressor function has been supported by more recent results (e.g., reference 31). This clearly supports the importance of this function for DAX-1.
The results described here also add HNF-4 to the list of SHP targets. The potential importance of the functional interaction between these two orphans is supported by their coexpression in liver and pancreatic beta cells. This is reinforced by the fact that only a minor perturbation of HNF-4 activity results in the syndrome of mature-onset diabetes of the young (48), suggesting that the modulation of HNF-4 activity by SHP could have important metabolic effects. A determination of the consequences of the loss of SHP function will be required to assess the significance of this and other potential roles for SHP.
ACKNOWLEDGMENTS
We thank Wongi Seol for initial yeast results and Pavlos Pissios for helpful discussions. We also thank J. D. Chen for pCMX-SRC3 plasmid.
This work was supported by Public Health Service grants DK-46546 (to D.D.M.) and HL-56104 (to M.H.-C.).
REFERENCES
- 1.Anzick S L, Kononen J, Walker R L, Azorsa D O, Tanner M M, Guan X Y, Sauter G, Kallioniemi O P, Trent J M, Meltzer P S. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science. 1997;277:965–968. doi: 10.1126/science.277.5328.965. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. J. New York, N.Y: Wiley & Sons; 1999. [Google Scholar]
- 3.Bannister A J, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 4.Berk A J. Activation of RNA polymerase II transcription. Curr Opin Cell Biol. 1999;11:330–335. doi: 10.1016/S0955-0674(99)80045-3. [DOI] [PubMed] [Google Scholar]
- 5.Blanco J C, Minucci S, Lu J, Yang X J, Walker K K, Chen H, Evans R M, Nakatani Y, Ozato K. The histone acetylase PCAF is a nuclear receptor coactivator. Genes Dev. 1998;12:1638–1651. doi: 10.1101/gad.12.11.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen H, Lin R J, Schiltz R L, Chakravarti D, Nash A, Nagy L, Privalsky M L, Nakatani Y, Evans R M. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 7.Danielian P S, White R, Lee J A, Parker M G. Identification of a conserved region required for hormone dependent transcriptional activation by steroid hormone receptors. EMBO J. 1992;11:1025–1033. doi: 10.1002/j.1460-2075.1992.tb05141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Darimont B D, Wagner R L, Apriletti J W, Stallcup M R, Kushner P J, Baxter J D, Fletterick R J, Yamamoto K R. Structure and specificity of nuclear receptor-coactivator interactions. Genes Dev. 1998;12:3343–3356. doi: 10.1101/gad.12.21.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dell H, Hadzopoulou-Cladaras M. CREB-binding protein is a transcriptional coactivator for hepatocyte nuclear factor-4 and enhances apolipoprotein gene expression. J Biol Chem. 1999;274:9013–9021. doi: 10.1074/jbc.274.13.9013. [DOI] [PubMed] [Google Scholar]
- 10.Enmark E, Gustafsson J A. Orphan nuclear receptors—the first eight years. Mol Endocrinol. 1996;10:1293–1307. doi: 10.1210/mend.10.11.8923456. [DOI] [PubMed] [Google Scholar]
- 11.Forman B M, Umesono K, Chen J, Evans R M. Unique response pathways are established by allosteric interactions among nuclear hormone receptors. Cell. 1995;81:541–550. doi: 10.1016/0092-8674(95)90075-6. [DOI] [PubMed] [Google Scholar]
- 12.Hadzopoulou-Cladaras M, Kistanova E, Evagelopoulou C, Zeng S, Cladaras C, Ladias J A. Functional domains of the nuclear receptor hepatocyte nuclear factor 4. J Biol Chem. 1997;272:539–550. doi: 10.1074/jbc.272.1.539. [DOI] [PubMed] [Google Scholar]
- 13.Hong H, Kohli K, Garabedian M J, Stallcup M R. GRIP1, a transcriptional coactivator for the AF-2 transactivation domain of steroid, thyroid, retinoid, and vitamin D receptors. Mol Cell Biol. 1997;17:2735–2744. doi: 10.1128/mcb.17.5.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horwitz K B, Jackson T A, Bain D L, Richer J K, Takimoto G S, Tung L. Nuclear receptor coactivators and corepressors. Mol Endocrinol. 1996;10:1167–1177. doi: 10.1210/mend.10.10.9121485. [DOI] [PubMed] [Google Scholar]
- 15.Ito M, Yu R, Jameson J L. DAX-1 inhibits SF-1-mediated transactivation via a carboxy-terminal domain that is deleted in adrenal hypoplasia congenita. Mol Cell Biol. 1997;17:1476–1483. doi: 10.1128/mcb.17.3.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang G, Lee U, Sladek F M. Proposed mechanism for the stabilization of nuclear receptor DNA binding via protein dimerization. Mol Cell Biol. 1997;17:6546–6554. doi: 10.1128/mcb.17.11.6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johansson L, Thomsen J S, Damdimopoulos A E, Spyrou G, Gustafsson J, Treuter E. The orphan nuclear receptor SHP inhibits agonist-dependent transcriptional activity of estrogen receptors ERalpha and ERbeta. J Biol Chem. 1999;274:345–353. doi: 10.1074/jbc.274.1.345. [DOI] [PubMed] [Google Scholar]
- 18.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin S C, Heyman R A, Rose D W, Glass C K, Rosenfeld M G. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 19.Lalli E, Bardoni B, Zazopoulos E, Wurtz J-M, Strom T M, Moras D, Sassone-Corsi P. A transcriptional silencing domain in DAX-1 whose mutation causes adrenal hypoplasia congenita. Mol Endocrinol. 1997;11:1950–1960. doi: 10.1210/mend.11.13.0038. [DOI] [PubMed] [Google Scholar]
- 20.Lee Y K, Parker K L, Choi H S, Moore D D. Activation of the promoter of the orphan receptor SHP by orphan receptors that bind DNA as monomers. J Biol Chem. 1999;274:20869–20873. doi: 10.1074/jbc.274.30.20869. [DOI] [PubMed] [Google Scholar]
- 21.Leng X, Blanco J, Tsai S Y, Ozato K, O'Malley B W, Tsai M J. Mouse retinoid X receptor contains a separable ligand-binding and transactivation domain in its E region. Mol Cell Biol. 1995;15:255–263. doi: 10.1128/mcb.15.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H, Gomes P J, Chen J D. RAC3, a steroid/nuclear receptor-associated coactivator that is related to SRC-1 and TIF2. Proc Natl Acad Sci USA. 1997;94:8479–8484. doi: 10.1073/pnas.94.16.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mangelsdorf D J, Evans R M. The RXR heterodimers and orphan receptors. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- 24.Mangelsdorf D J, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans R M. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakshatri H, Chambon P. The directly repeated RG(G/T)TCA motifs of the rat and mouse cellular retinol-binding protein II genes are promiscuous binding sites for RAR, RXR, HNF-4, and ARP-1 homo- and heterodimers. J Biol Chem. 1994;269:890–902. [PubMed] [Google Scholar]
- 26.Nawaz Z, Lonard D M, Smith C L, Lev-Lehman E, Tsai S Y, Tsai M-J, O'Malley B W. The Angelman syndrome-associated protein, E6-AP, is a coactivator for the nuclear hormone receptor superfamily. Mol Cell Biol. 1999;19:1182–1189. doi: 10.1128/mcb.19.2.1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nolte R T, Wisely G B, Westin S, Cobb J E, Lambert M H, Kurokawa R, Rosenfeld M G, Willson T M, Glass C K, Milburn M V. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-gamma. Nature. 1998;395:137–143. doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]
- 28.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 29.Onate S A, Tsai S Y, Tsai M J, O'Malley B W. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 30.Renaud J P, Rochel N, Ruff M, Vivat V, Chambon P, Gronemeyer H, Moras D. Crystal structure of the RAR-gamma ligand-binding domain bound to all-trans retinoic acid. Nature. 1995;378:681–689. doi: 10.1038/378681a0. [DOI] [PubMed] [Google Scholar]
- 31.Reutens A T, Achermann J C, Ito M, Gu W X, Habiby R L, Donohoue P A, Pang S, Hindmarsh P C, Jameson J L. Clinical and functional effects of mutations in the DAX-1 gene in patients with adrenal hypoplasia congenita. J Clin Endocrinol Metab. 1999;84:504–511. doi: 10.1210/jcem.84.2.5468. [DOI] [PubMed] [Google Scholar]
- 32.Seol W, Choi H S, Moore D D. An orphan nuclear hormone receptor that lacks a DNA binding domain and heterodimerizes with other receptors. Science. 1996;272:1336–1339. doi: 10.1126/science.272.5266.1336. [DOI] [PubMed] [Google Scholar]
- 33.Seol W, Chung M, Moore D D. Novel receptor interaction and repression domains in the orphan receptor SHP. Mol Cell Biol. 1997;17:7126–7131. doi: 10.1128/mcb.17.12.7126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seol W, Hanstein B, Brown M, Moore D D. Inhibition of estrogen receptor action by the orphan receptor SHP (short heterodimer partner) Mol Endocrinol. 1998;12:1551–1557. doi: 10.1210/mend.12.10.0184. [DOI] [PubMed] [Google Scholar]
- 35.Seol W, Mahon M J, Lee Y K, Moore D D. Two receptor interacting domains in the nuclear hormone receptor corepressor RIP13/N-CoR. Mol Endocrinol. 1996;10:1646–1655. doi: 10.1210/mend.10.12.8961273. [DOI] [PubMed] [Google Scholar]
- 36.Shiau A K, Barstad D, Loria P M, Cheng L, Kushner P J, Agard D A, Greene G L. The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell. 1998;95:927–937. doi: 10.1016/s0092-8674(00)81717-1. [DOI] [PubMed] [Google Scholar]
- 37.Shibata H, Spencer T E, Onate S A, Jenster G, Tsai S Y, Tsai M J, O'Malley B W. Role of co-activators and co-repressors in the mechanism of steroid/thyroid receptor action. Recent Prog Horm Res. 1997;52:141–164. [PubMed] [Google Scholar]
- 38.Sladek F M, Zhong W, Lai E, Darnell J J E. Liver-enriched transcription factor HNF-4 is a novel member of the steroid hormone receptor superfamily. Genes Dev. 1990;4:2353–2365. doi: 10.1101/gad.4.12b.2353. [DOI] [PubMed] [Google Scholar]
- 39.Spencer T E, Jenster G, Burcin M M, Allis C D, Zhou J, Mizzen C A, McKenna N J, Onate S A, Tsai S Y, Tsai M J, O'Malley B W. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature. 1997;389:194–198. doi: 10.1038/38304. [DOI] [PubMed] [Google Scholar]
- 40.Takeshita A, Cardona G R, Koibuchi N, Suen C S, Chin W W. TRAM-1, a novel 160-kDa thyroid hormone receptor activator molecule, exhibits distinct properties from steroid receptor coactivator-1. J Biol Chem. 1997;272:27629–27634. doi: 10.1074/jbc.272.44.27629. [DOI] [PubMed] [Google Scholar]
- 41.Torchia J, Rose D W, Inostroza J, Kamei Y, Westin S, Glass C K, Rosenfeld M G. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature. 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- 42.Voegel J J, Heine M J, Zechel C, Chambon P, Gronemeyer H. TIF2, a 160 kDa transcriptional mediator for the ligand-dependent activation function AF-2 of nuclear receptors. EMBO J. 1996;15:3667–3675. [PMC free article] [PubMed] [Google Scholar]
- 43.Wagner R L, Apriletti J W, McGrath M E, West B L, Baxter J D, Fletterick R J. A structural role for hormone in the thyroid hormone receptor. Nature. 1995;378:690–697. doi: 10.1038/378690a0. [DOI] [PubMed] [Google Scholar]
- 44.Wang J C, Stafford J M, Granner D K. SRC-1 and GRIP1 coactivate transcription with hepatocyte nuclear factor 4. J Biol Chem. 1998;273:30847–30850. doi: 10.1074/jbc.273.47.30847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wotton D, Lo R S, Lee S, Massague J. A Smad transcriptional corepressor. Cell. 1999;97:29–39. doi: 10.1016/s0092-8674(00)80712-6. [DOI] [PubMed] [Google Scholar]
- 46.Wurtz J M, Bourguet W, Renaud J P, Vivat V, Chambon P, Moras D, Gronemeyer H. A canonical structure for the ligand-binding domain of nuclear receptors. Nature Struct Biol. 1996;3:87–94. doi: 10.1038/nsb0196-87. [DOI] [PubMed] [Google Scholar]
- 47.Xu L, Glass C K, Rosenfeld M G. Coactivator and corepressor complexes in nuclear receptor function. Curr Opin Genet Dev. 1999;9:140–147. doi: 10.1016/S0959-437X(99)80021-5. [DOI] [PubMed] [Google Scholar]
- 48.Yamagata K, Furuta H, Oda N, Kaisaki P J, Menzel S, Cox N J, Fajans S S, Signorini S, Stoffel M, Bell G I. Mutations in the hepatocyte nuclear factor-4alpha gene in maturity-onset diabetes of the young (MODY1) Nature. 1996;384:458–460. doi: 10.1038/384458a0. [DOI] [PubMed] [Google Scholar]
- 49.Yang X J, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 50.Zanaria E, Muscatelli F, Bardoni B, Strom T M, Guioli S, Guo W, Lalli E, Moser C, Walker A P, McCabe E R B, Meitinger T, Monaco A P, Sassone-Corsi P, Camerino G. An unusual member of the nuclear hormone receptor superfamily responsible for X-linked adrenal hypoplasia congenita. Nature. 1994;372:635–641. doi: 10.1038/372635a0. [DOI] [PubMed] [Google Scholar]
- 51.Zavacki A M, Lehmann J M, Seol W, Willson T M, Kliewer S A, Moore D D. Activation of the orphan receptor RIP14 by retinoids. Proc Natl Acad Sci USA. 1997;94:7909–7914. doi: 10.1073/pnas.94.15.7909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang X-K, Salbert G, Lee M-O, Pfahl M. Mutations that alter ligand-induced switches and dimerization activities in the retinoid X receptor. Mol Cell Biol. 1994;14:4311–4323. doi: 10.1128/mcb.14.6.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang Y H, Guo W, Wagner R L, Huang B L, McCabe L, Vilain E, Burris T P, Anyane-Yeboa K, Burghes A H, Chitayat D, Chudley A E, Genel M, Gertner J M, Klingensmith G J, Levine S N, Nakamoto J, New M I, Pagon R A, Pappas J G, Quigley C A, Rosenthal I M, Baxter J D, Fletterick R J, McCabe E R. DAX1 mutations map to putative structural domains in a deduced three-dimensional model. Am J Hum Genet. 1998;62:855–864. doi: 10.1086/301782. [DOI] [PMC free article] [PubMed] [Google Scholar]