Abstract
Chemotherapeutic genotoxins induce apoptosis in epithelial-cell-derived cancer cells. The death receptor ligand TRAIL also induces apoptosis in epithelial-cell-derived cancer cells but generally fails to induce apoptosis in nontransformed cells. We show here that the treatment of four different epithelial cell lines with the topoisomerase II inhibitor etoposide in combination with TRAIL (tumor necrosis factor [TNF]-related apoptosis-inducing ligand) induces a synergistic apoptotic response. The mechanism of the synergistic effect results from the etoposide-mediated increase in the expression of the death receptors 4 (DR4) and 5 (DR5). Inhibition of NF-κB activation by expression of kinase-inactive MEK kinase 1(MEKK1) or dominant-negative IκB (ΔIκB) blocked the increase in DR4 and DR5 expression following etoposide treatment. Addition of a soluble decoy DR4 fusion protein (DR4:Fc) to cell cultures reduced the amount of etoposide-induced apoptosis in a dose-dependent manner. The addition of a soluble TNF decoy receptor (TNFR:Fc) was without effect, demonstrating the specificity of DR4 binding ligands in the etoposide-induced apoptosis response. Thus, genotoxin treatment in combination with TRAIL is an effective inducer of epithelial-cell-derived tumor cell apoptosis relative to either treatment alone.
The tumor necrosis factor (TNF) receptor superfamily consists of proteins involved in proliferation, differentiation, and apoptosis. A subgroup of these receptors collectively called death receptors, including the TNF receptor, FAS (also Apo1 or CD95), death receptor 3 (DR3; also Apo3, WSL-1, TRAMP, or LARD), death receptor 4 (DR4; also TRAIL-R1) and death receptor 5 (DR5; also Apo2, TRAIL-R2, TRICK2, or KILLER), is defined by their ability to induce apoptosis in a variety of cell types (27). The ligands for these receptors belong to a complementary family of structurally related molecules consisting of TNF-α, FAS ligand (FASL), APO3 ligand, and TRAIL (TNF-related apoptosis-inducing ligand) (27). Most of these ligands are primarily expressed as biologically active type II membrane proteins that are cleaved into soluble forms. Death receptor cytoplasmic sequences contain a shared 80-amino-acid domain called the death domain that, upon ligand binding, associates with a similar domain found in adapter proteins such as FAS-associating protein with death domain (FADD) and TNF-related associated death domain (TRADD) (10, 11, 27). The adapter proteins also contain an effector domain that constitutively binds to cysteine proteases called caspases that cleave specific proteins. The two caspases that are predominantly bound to these adapter molecules are caspase 8 and caspase 10 (2, 27). Once these caspases are recruited by the association of adapter proteins with death receptors, the caspases are trans-proteolyzed, resulting in their activation. Caspase 8 or caspase 10 activation initiates a cascade where other caspases, including caspase 3, are activated, ultimately resulting in an irreversible commitment of cells to undergo apoptosis (2, 27).
DR4 and DR5 have been recently identified and induce apoptosis after binding to their ligand TRAIL (10, 23, 25, 28, 30). Binding of TRAIL to DR4 and DR5 leads to the recruitment of an adaptor protein bound to a caspase. Complexes of FADD and caspase 8 have been implicated in DR4- and DR5-induced apoptosis (26); however, there is also evidence suggesting they might not be required for DR4- and DR5-induced apoptosis. In FADD−/− mice, cells are still capable of undergoing apoptosis upon ligation of DR4 and DR5, indicating that another FADD-like adapter protein might be involved (35). In contrast, FAS signaling is well defined and involves recruitment of the FADD-caspase 8 complex to the activated receptor (27).
Epithelial-cell-derived cancers are often treated with chemotherapeutic drugs that induce apoptosis. TRAIL also has been demonstrated to effectively induce apoptosis in transformed epithelial cells but is less effective at inducing apoptosis in nontransformed epithelial cells (7, 12, 31). In mice, administration of TRAIL was effective at reducing mammary adenocarcinoma growth without any of the toxic effects shown with administration of FASL or TNF (31). Thus, TRAIL might be an effective treatment of epithelial-cell-derived cancers.
We demonstrate here that the genotoxic agent etoposide induced the expression of DR4 and DR5, resulting in a synergistic apoptosis response to TRAIL in the epithelial cell lines. This increase requires the activation of the NF-κB signaling pathway. Furthermore, blocking DR4 and DR5 binding of ligand effectively reduced etoposide-induced apoptosis. Our studies suggest that combined chemotherapy and TRAIL administration could be an effective treatment of epithelial-cell-derived cancers.
MATERIALS AND METHODS
Materials.
Etoposide was purchased from Sigma and dissolved in dimethyl sulfoxide. Anti-DR4, anti-DR5, anti-FAS, anti-caspase 9, and anti-caspase 10 antisera were purchased from Santa Cruz, and anti-caspase 8 antiserum was purchased from Upstate Biotechnology. DR4:Fc and TNFR:Fc were purchased from Alexis Biochemical and soluble TRAIL (sTRAIL) was purchased from Upstate Biotechnology.
Cell culture.
Human embryonic kidney 293 (HEK293) cells and T47D breast cancer cells were maintained in a humidified 7.0% CO2 environment in Dulbecco's modified Eagle medium supplemented with 100 U of penicillin per ml, 100 μg of streptomycin (Gibco Laboratories, Grand Island, N.Y.) per ml, and 10% bovine calf serum. MDA-MD-468 and ZR-75-1 breast cancer cell lines were maintained in minimum essential medium (Gibco) supplemented with 5% fetal calf serum, 100 U of penicillin per ml, 100 μg of streptomycin per ml, 1% nonessential amino acids, and 40 ng of insulin per ml. HEK293 cells stably expressing vector alone (pcDNA3) and kinase-inactive MEK kinase 1 (MEKK1 KM), dominant-negative IκB (ΔIκB), Bcl2, and FADD dominant-negative (DN) proteins were under selection with 1.5 mg of G418 (Gibco) per ml. Each cell line was demonstrated to be positive for expression of the plasmid-encoded protein by Western blotting.
Treatment of cancer cells.
HEK293, T47D, MDA-MD-468, and ZR-75-1 cells (1 × 106 to 2 × 106) were incubated with or without 100 μM etoposide, 200 ng of sTRAIL per ml, or in combination where indicated. HEK293 cells were also incubated with 10 to 100 ng of DR4:Fc per ml or 100 ng of TNFR:Fc per ml, in addition to 100 μM etoposide, for 24 h. Lower concentrations of etoposide and TRAIL showed reduced apoptosis in a dose-response curve (data not shown). DR4:Fc is a fusion protein of the extracellular domain of DR4 and the immunoglobulin Fc region, while TNFR:Fc is a fusion protein of the tumor necrosis factor receptor (TNFR) and the immunoglobulin Fc region.
RNase protection assay.
A RiboQuant Multi-Probe RNase Protection Assay System (PharMingen) was used per the manufacturer's instructions. An hAPO3c probe set containing DNA templates for caspase 8, FASL, FAS, DR3, decoy receptor 1 (DcR1), DR4, DR5, TRAIL, TNFR p55, TRADD, receptor interacting protein (RIP), L32, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) or an hSTRESS probe set containing DNA templates for Bcl-x, Bcl2, Bax, p21, GADD45, p53, Mcl1, L32, and GAPDH (PharMingen) was used for T7 RNA-polymerase direct synthesis of [α32-P]UTP-labeled antisense RNA probes. The probes were hybridized with 20 μg of RNA isolated from HEK293, T47D, MDA-MD-468, and ZR-75-1 cells by using RNAzol B (Tel-Test, Inc.). Samples were then digested with RNase to remove single-stranded (nonhybridized) RNA. Remaining probes were resolved on denaturing 5% polyacrylamide gels. Quantitation was done by using a PhosphorImager (Molecular Dynamics).
Immunoblots.
HEK293 cells were lysed in NP-40 lysis buffer (50 mM HEPES, pH 7.25; 150 mM NaCl; 50 μM ZnCl2; 50 μM NaF; 2 mM EDTA; 1 mM sodium vanadate; 1.0% NP-40; 2 mM phenylmethylsulfonyl fluoride). Cell debris was removed by centrifugation at 8,000 × g for 5 min, and the protein concentration was determined by the Bradford assay. Four hundred micrograms of cell lysate protein was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membranes. The membranes were blocked in Tris-buffered saline–Tween 20 solution containing 5% milk. Blots were performed as described previously (6). The amount of cell lysate in Western blotting was used to visualize specific proteins because of the low expression of the proteins of interest.
Measurement of apoptosis.
Cells (1 × 106 to 2 × 106) were resuspended in 100 μl of medium by gentle vortexing, and 2 μl of acridine orange (100 μg/ml) and ethidium bromide (100 μg/ml) in phosphate-buffered saline was added. Then, 10 μl was removed and placed on a microscope slide, and a coverslip was applied over the 10 μl. The slide was viewed on a fluorescence microscope by using a fluorescein filter set for the detection of condensed DNA in apoptotic cells. The condensed DNA was determined by intense local staining of DNA in the nucleus compared to the diffuse staining of the DNA in normal cells. The percentage of apoptotic cells was determined from cells containing normal DNA staining compared to cells with condensed DNA. Apoptosis was verified by propidium iodide staining for DNA fragmentation and morphological changes consistent with apoptotic cells.
MEKK1 in vitro kinase assay.
MEK kinase 1 (MEKK1) was immunoprecipitated from cell lysates (500 μg) with antibodies raised against specific sequences of MEKK1. The immunoprecipitates were used in an in vitro kinase assay with recombinant kinase-inactive SEK1 (SEK1 KM) as previously described (6). The samples were analyzed by SDS-PAGE, after which the gel was fixed in methanol. The extent of MEKK1 autophosphorylation and SEK1 KM phosphorylation was determined on a PhosphorImager (Molecular Dynamics).
NF-κB luciferase assay.
HEK293 cells (5 × 105) were transfected with the reporter plasmids NF-κB–luciferase (prLUC) and CMV–β-GAL (CH110) by using Lipofectamine (Gibco) according to the manufacturer's instructions. The cells were then treated with 100 μM etoposide and lysed according to the Beta-Galactosidase Enzyme Assay System with lysis buffer protocol (Promega, Madison, Wis.). The lysate was measured for 10 s as relative light units by a luminometer (Monolight 2010; Analytical Luminescence Laboratory, San Diego, Calif.). Luciferase activity was normalized to β-galactosidase activity measured at 420 nm and presented as luciferase units.
RESULTS
Etoposide and TRAIL act synergistically in inducing apoptosis.
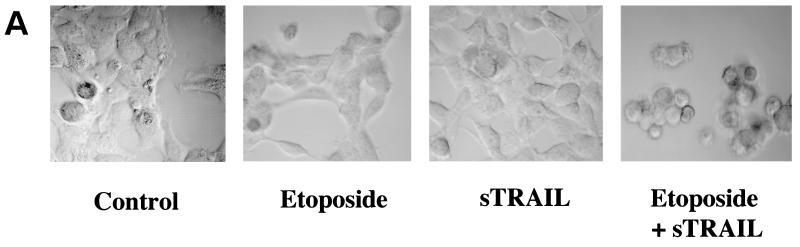
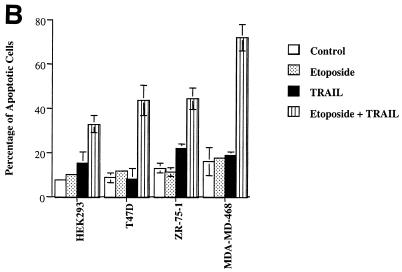
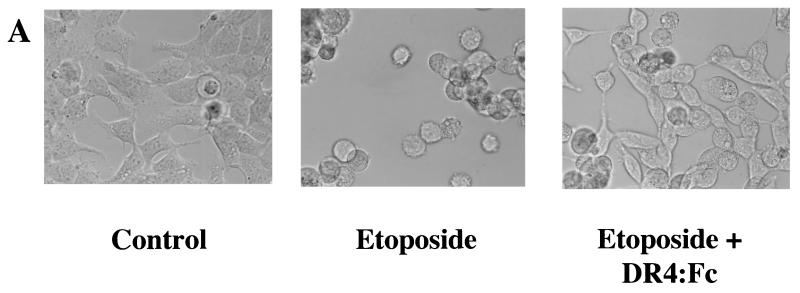
Exposure of HEK293 epithelial cells to etoposide or TRAIL failed to induce significant apoptosis as determined by both morphological changes and DNA condensation (Fig. 1). In addition to HEK293 cells, the T47D, MDA-MD-468, and ZR-75-1 breast epithelial cancer cell lines were exposed to etoposide, TRAIL, or a combination of etoposide and TRAIL for 16 h. The apoptotic index for treatment with etoposide or TRAIL was not significantly greater than that for untreated control cells (7 to 15%). The combination of etoposide and TRAIL increased the number of apoptotic cells in cultures of the four cell lines to 32 to 71% (Fig. 1B). TRAIL in combination with etoposide, therefore, induced apoptosis to a much greater extent than either did alone. This result indicates that etoposide and TRAIL acted synergistically to induce apoptosis.
FIG. 1.
Synergistic apoptotic response following combined treatment with etoposide and TRAIL. (A) HEK293 cells were treated with 100 μM etoposide, 200 ng of sTRAIL per ml, or both for 16 h. Untreated cells were designated the control. Cells were visualized by digital confocal microscopy by using a ×40 water objective for each treatment tested. (B) HEK293, T47D, ZR-75-1, and MDA-MD-468 cells were treated with 100 μM etoposide, 200 ng of sTRAIL per ml, or both for 16 h, and the percentage of apoptotic cells was determined. The cells were stained with acridine orange and counted by using a fluorescence microscope to quantitate condensed DNA. Cells having condensed DNA were scored as apoptotic (a total of 250 cells for each treatment were scored). All experiments were repeated three times. Error bars represent the standard deviations for three independent experiments.
DR4 and DR5 are increased after etoposide treatment in an NF-κB-dependent manner.
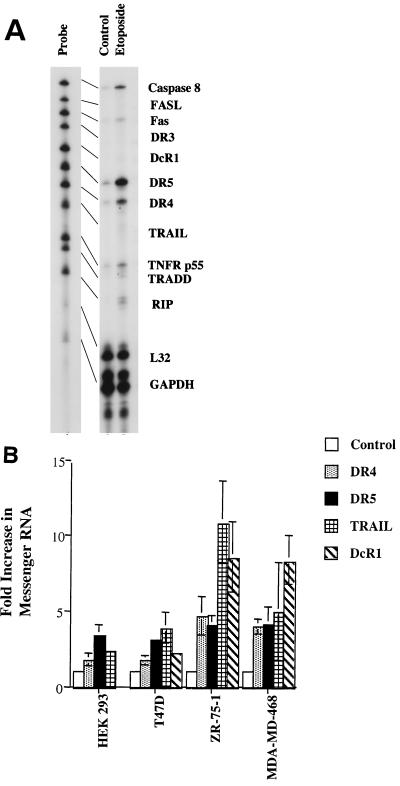
RNase protection assays were used to monitor death receptor mRNA expression in HEK293 cells treated with etoposide. DR4 and DR5 mRNA levels were increased in response to etoposide, while caspase 8, FAS, TNFR p55, TRADD, and RIP mRNA levels increased to a lesser extent. FASL, TRAIL, DcR1, and DR3 mRNA expression was weak or was not detected after 8 h (Fig. 2A). In addition to HEK293 cells, T47D, MDA-MD-468, and ZR-75-1 cells treated with etoposide also showed two- to threefold increases in DR4 and DR5 mRNA expression at 8 h (data not shown) and at 24 h (Fig. 2B). At 24 h, TRAIL expression was detectable in HEK293, T47D, MDA-MD-468, and ZR-75-1 cells. The breast cancer cell lines ZR-75-1 and MDA-MD-468 showed a 5- to 10-fold increase in TRAIL mRNA expression, whereas T47D and HEK293 cells showed 2- to 3-fold-increased expression of TRAIL. This higher expression of TRAIL in ZR-75-1 and MDA-MD-468 cells was accompanied by an eightfold increase in DcR1 mRNA expression (Fig. 2B). This increase in DcR1 expression likely results in the binding of endogenously expressed TRAIL, preventing significant induction of apoptosis and allowing for higher expression levels of TRAIL.
FIG. 2.
RNase protection assay for death receptor family members. (A) HEK293 cells were treated with 100 μM etoposide for 8 h. Cells were lysed, and RNA was extracted. RNase protection assay was performed by using the hAPO3 probe set (PharMingen) as described in Materials and Methods. The probe lane represents antisense RNA probes before hybridization with RNA. Housekeeping genes, L32, and GAPDH serve as normalized controls. (B) HEK293, T47D, ZR-75-1, and MDA-MD-468 cells were treated with 100 μM etoposide for 24 h and then lysed. An RNase protection assay was performed as described in Fig. 2A. The control represents untreated cells normalized to a value of 1 for mRNA expression of DR4, DR5, TRAIL, and DcR1. Error bars represent the standard deviation for three independent experiments.
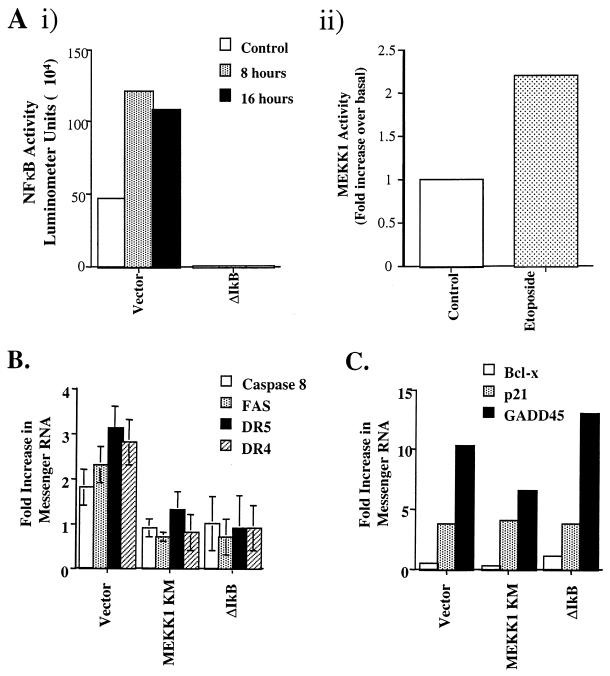
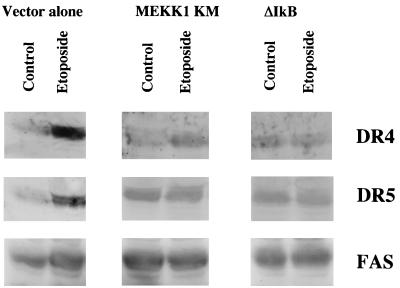
MEKK1 can activate IκB kinase (IKK), leading to the phosphorylation of IκB and subsequent activation of NF-κB transcriptional activity (21). Exposure of HEK293 cells to etoposide activates both NF-κB and MEKK1 (Fig. 3A) (4, 9). To test the role of MEKK1 and NF-κB in the apoptotic response of death receptor upregulation, cells stably overexpressing MEKK1 KM or ΔIκB in which the amino-terminal end of IκB is deleted were treated with etoposide. Overexpression of either ΔIκB (Fig. 3A) or MEKK1 KM (data not shown) effectively inhibited etoposide-induced activation of NF-κB. DR4 and DR5 mRNA expression was increased threefold, on average, over basal levels following 8 h of etoposide treatment in control cells expressing vector alone (Fig. 3B). Caspase 8 and FAS mRNA levels were measurably increased, on average, twofold in these same cells. In cells expressing MEKK1 KM, DR4 and DR5 mRNA levels were not measurably changed in response to etoposide (Fig. 3B). This corresponds with MEKK1 KM expression blocking etoposide-induced apoptosis. As in cells expressing MEKK1 KM, DR4 and DR5 mRNA expression was not significantly induced in etoposide-treated cells that overexpressed ΔIκB after 8 h. In addition to DR4 and DR5, upregulation of both caspase 8 and FAS was blocked in cells expressing either MEKK1 KM or ΔIκB (Fig. 3B). Since ΔIκB-expressing cells fail to upregulate DR4 and DR5 following etoposide treatment, the synergistic apoptotic response to combined treatment with etoposide and TRAIL should be diminished. Importantly, the synergy between etoposide and TRAIL in inducing apoptosis was lost in ΔIκB cells during a 16-h treatment with the two agents (data not shown). This result confirms the observation that etoposide and TRAIL synergy is regulated by an NF-κB-dependent increase in DR4 and DR5 expression. Cumulatively, the data demonstrate that an increase in DR4 and DR5 expression was significantly inhibited in MEKK1 KM- and ΔIκB-expressing cells (Fig. 4). FAS protein levels were increased in cells with vector alone but were not increased in MEKK1 KM and ΔIκB cells in response to etoposide (Fig. 4). In vector-alone cells, both TRAIL and FASL protein levels were increased after etoposide treatment, but to a much lower extent than DR4 and DR5. However, in MEKK1 KM and ΔIκB cells the TRAIL and FASL protein levels were not increased following etoposide treatment (data not shown). It should be noted that the Western blots are representative of several independent experiments. The quality of the antibodies and the level of expression of the proapoptotic proteins made analysis difficult. Nonetheless, the protein expression results confirm and substantiate the RNase protection assay results.
FIG. 3.
Activation of NF-κB and MEKK1 in response to etoposide. (Ai) NF-κB transcriptional activity was analyzed in HEK293 cells following etoposide treatment. Cells were transfected with NF-κB luciferase reporter plasmid and a β-GAL reporter plasmid as a control. The cells were then stimulated with 100 μM etoposide for the times indicated. The cells were lysed, and a luciferase assay was performed. NF-κB activity is represented by luciferase units. (Aii) MEKK1 kinase activity was determined in HEK293 cells following etoposide (100 μM) treatment. The increase in MEKK1 kinase activity is presented as a fold increase in SEK1 KM phosphorylation over basal levels. (B) HEK293 cells expressing vector alone, MEKK1 KM, or ΔIκB were treated with 100 μM etoposide for 8 h. An RNase protection assay was performed, and the mRNA levels for DR4, DR5, FAS, and caspase 8 were determined by phosphorimager analysis. The mRNA levels were then normalized to GAPDH mRNA expression. Differences following exposure to etoposide are represented as the fold increase over basal mRNA levels. (C) mRNA expression was also determined by RNase protection assay for p53-regulated genes p21 and GADD45 and the antiapoptotic Bcl-x gene by using the hSTRESS probe set as described in Materials and Methods. The fold increase was determined by the increase in normalized mRNA levels over basal levels. Standard deviations were determined based on the data from three separate experiments.
FIG. 4.
Western blot analysis of the expression of DR4, DR5, and FAS following etoposide treatment. HEK293 cells expressing vector alone, MEKK1 KM, or ΔIκB were treated or not treated (control) with 100 μM etoposide for 24 h. Cells were lysed in NP-40 lysis buffer and Western blotted by using αDR4, αDR5, and αFAS antibodies (1:100) as described in Materials and Methods. Proteins were detected by enhanced chemiluminescence. Western blot analysis was performed with anti-MKP-1 antibodies to determine and normalize for equal amounts of protein loaded under each condition tested. The apparent higher basal expression levels of FAS compared to DR4 and DR5 were due to differences in the length of exposure of the X-ray films. Experiments were repeated three times.
Ligation of DR4 and DR5 is involved in etoposide-induced apoptosis.
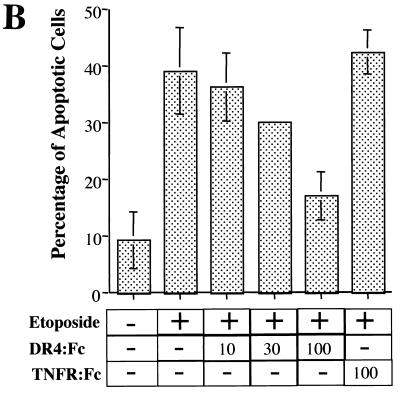
To determine if ligation of DR4 and DR5 plays a role in etoposide-induced apoptosis, a fusion protein having the DR4 extracellular domain fused to the immunoglobulin Fc region (DR4:Fc) was added to the cell culture. DR4:Fc binds ligand(s), including TRAIL for DR4 and DR5, and is thus capable of inhibiting the activation of endogenous DR4 and DR5. DR4:Fc reduced, in a concentration-dependent manner, the amount of etoposide-induced apoptosis from 38% to near- control cell levels (19%) and blocked morphological changes associated with apoptosis (Fig. 5). As a negative control, 100 ng of TNFR:Fc per ml, which binds TNF but not TRAIL, was added to the cells. This fusion protein failed to block etoposide-induced apoptosis (42% with TNF:Fc compared to 38% without TNF:Fc) (Fig. 5B). This demonstrates that etoposide induces apoptosis largely through DR4 and DR5 by a mechanism that involves a ligand that binds to DR4. This ligand is most likely TRAIL.
FIG. 5.
Blockage of TRAIL ligation to DR4 and DR5 decreased etoposide-induced apoptosis. Cells were treated with or without (control) 100 μM etoposide in the presence or absence of extracellular domain of DR4 fused to the immunoglobulin Fc region (200 ng/ml) for 24 h. This fusion protein binds to TRAIL, blocking its interaction with cellular DR4 and DR5. (A) To observe morphological changes associated with apoptosis, the cells were visualized on a digital confocal microscope by using a ×40 water objective for each treatment condition. (B) HEK293 cells were treated with 100 μM etoposide and increasing concentrations (10 to 100 ng/ml) of DR4:Fc protein. Cells treated with etoposide and TNFR:Fc (100 ng/ml) were used as a negative control. The percentage of apoptotic cells was determined by acridine orange staining as described in Fig. 1B. Standard deviations were determined based on three separate experiments.
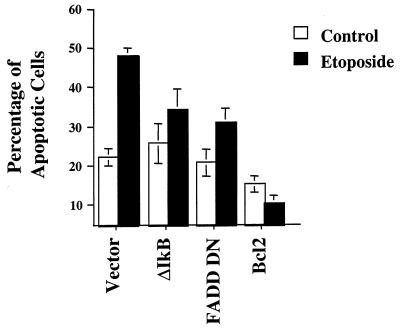
ΔIκB-, FADD DN-, and Bcl2-expressing cell lines show diminished etoposide-induced apoptosis.
We have demonstrated that MEKK1 KM blocks etoposide-induced apoptosis (9, 34). Since MEKK1 KM- and ΔIκB-expressing cells failed to upregulate DR4 and DR5 expression, we investigated the apoptotic response of ΔIκB cells following etoposide treatment. ΔIκB-expressing cells showed no change in the basal apoptotic index (23%) relative to that of control cells but showed significantly reduced apoptosis in response to etoposide (35% compared to 48%) (Fig. 6). Expression of a dominant-negative form of the adapter protein FADD (FADD DN) that lacks the ability to bind to death receptors but retains the ability to bind to caspases, including caspase 8, also effectively reduced etoposide-induced apoptosis (34% compared to 48% in vector-alone cells) (Fig. 6). When the antiapoptotic Bcl2 protein was expressed in HEK293 cells, the amount of basal and etoposide-induced apoptosis was markedly decreased (by 15 and 10%, respectively) reconfirming Bcl2's ability to block not only etoposide-induced apoptosis but also the other inputs contributing to apoptosis in rapidly growing cells in culture (Fig. 6). These results clearly show that NF-κB and FADD-like molecules contribute to the regulatory pathways involved in etoposide-induced apoptosis. NF-κB is required for upregulation of DR4 and DR5 expression following etoposide treatment. Even though the FADD-like proteins associated with DR4 and DR5 signaling are not defined, it is obvious that disruption of death receptor signaling in response to etoposide is interfered with by expression of a FADD dominant-negative mutant.
FIG. 6.
Percentage of apoptotic cells in HEK293 cells expressing ΔIκB, FADD DN, and Bcl2. Cells expressing ΔIκB, FADD DN, or Bcl2 were left untreated (control) or were exposed to 100 μM etoposide for 48 h. The percentage of apoptotic cells was determined by staining the cells with acridine orange. Apoptotic cells were scored for a total of 250 cells counted for each condition. Standard deviations were determined based on three separate experiments.
Both Bcl2 and FADD act downstream of DR4 and DR5 upregulation.
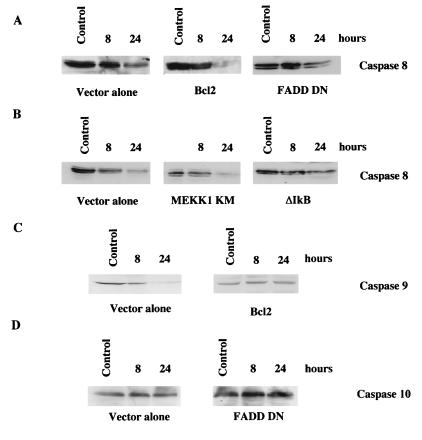
Bcl2- and FADD DN-expressing cells have a reduced level of etoposide-induced apoptosis compared to control cells (Fig. 6). By using RNase protection assays, treatment with etoposide was shown to increase DR4 and DR5 mRNA expression in both Bcl2 and FADD DN cells (data not shown). This indicates that Bcl2 and FADD-like proteins act downstream of the changes in DR4 and DR5 expression. Expression of FADD DN blocked the activation of caspase 8 following treatment with etoposide (Fig. 7A). Indeed, 24 h after the exposure to etoposide, FADD DN cells failed to cleave caspase 8, whereas vector-alone cells cleaved caspase 8 (Fig. 7A). In contrast, Bcl2 cells showed cleavage of full-length caspase 8 (Fig. 7A). Cleavage of caspase 8 following etoposide treatment was not detected at 8 h in cells expressing ΔIκB or MEKK1 KM; however, cleavage was detected in cells expressing MEKK1 KM at 24 h (Fig. 7B). This result correlates with increased expression of DR4 and DR5 24 h after etoposide treatment in MEKK1 KM cells (data not shown). In contrast, vector-alone cells show cleavage of caspase 8 at 8 h after etoposide treatment (Fig. 7B). These results indicate that caspase 8 cleavage is functionally upstream of Bcl2 but downstream of MEKK1 and NF-κB signaling. Several reports have suggested that caspase 10 is also involved in DR4 and DR5 activation (2, 27). Western blotting for caspase 10 failed to show cleavage in vector alone and FADD DN cells (Fig. 7D) or in MEKK1 KM and ΔIκB cells (data not shown). Since Bcl2 fails to block caspase 8 cleavage but effectively blocked caspase activity and apoptosis induced by etoposide, Bcl2 must block the activation of other caspases. By Western blotting for caspase 9 in which its activity is regulated by Bcl2 family members it was demonstrated that overexpression of Bcl2 prevented the cleavage of caspase 9 compared to that seen in vector control cells treated with etoposide (Fig. 7C). Taken together, these findings illustrate that FADD-like adapter proteins lead to the activation of caspase 8 following etoposide treatment and that Bcl2 inhibits etoposide-induced apoptosis by blocking the activation of caspase 9 downstream of DR4 and DR5 upregulation.
FIG. 7.
Western blot analysis of caspase 8, caspase 9, and caspase 10 following etoposide treatment. (A) Cells expressing either vector alone, Bcl2, or FADD DN were left untreated (control) or were treated with 100 μM etoposide for 8 or 24 h. The cells were then lysed and Western blotted by using anti-caspase 8 antibodies (1:1,000). (B) Cells expressing vector alone, MEKK1 KM or ΔIκB were left untreated (control) or were treated with 100 μM etoposide for 8 or 24 h. The cells were lysed and then Western blotted with anti-caspase 8 antibodies (1:1,000). (C) Cells expressing vector alone and Bcl2 were left untreated (control) or were treated with 100 μM etoposide for 8 or 24 h and then Western blotted with anti-caspase 9 antibodies (1:100). (D) Cells expressing vector alone or FADD DN were left untreated (control) or were treated with 100 μM etoposide for 8 or 24 h and then Western blotted with anti-caspase 10 antibodies (1:100). Proteins were visualized by enhanced chemiluminescence. The data shown are representative of three separate experiments.
DISCUSSION
Chemotherapeutic drugs are used in the treatment of most cancers; however, these drugs are only partially effective in the treatment of epithelial-cell-derived cancer. Thus, new ways of treating cancer must be developed (24). Ligation of death receptors to induce apoptosis in tumor cells has been proposed as an effective way of treating epithelial-cell-derived cancers; unfortunately, the administration of death receptor ligands FASL and TNF has toxic effects (7, 31). The discovery of DR4 and DR5, along with their ligand TRAIL, provided new death receptor family members that may offer less-toxic effects on noncancerous cells.
We have demonstrated that treatment of different epithelial cells with the genotoxic agent etoposide in combination with TRAIL synergistically induced apoptosis. The mechanism of this synergy involves the etoposide-induced expression of DR4 and DR5 that is mediated in part by activation of the NF-κB signaling pathway. The expression of ΔIκB has been demonstrated to inhibit apoptosis in response to a variety of extracellular stimuli (3, 4). For example, virus-induced apoptosis with Sindbis virus is blocked by inhibiting NF-κB activation (18). NF-κB activation has been shown to be required for the upregulation of the proapoptotic genes FAS and FASL (1, 14, 20). In contrast, NF-κB can also contribute to antiapoptotic survival responses. For example, mice deficient in NF-κB showed an increased apoptotic response after TNF-α and T-cell antigen receptor activation (3, 13). The expression of the dominant-negative ΔIκB mutant in tumor cells also resulted in an enhanced apoptosis in response to TNF-α treatment (32). TNF-α has been shown to stimulate the expression of inhibitor of apoptosis protein 1 (cIAP1) and cIAP2 in an NF-κB-dependent manner (33). These findings demonstrate that NF-κB is involved in the regulation of several proteins that contribute to the apoptotic potential of different stimuli. However, NF-κB activation is only one signaling component of many that are activated by different cytokines and cellular stresses. It seems to be the integration of these additional signals in combination with NF-κB activation that ultimately determines the role of NF-κB in pro- or antiapoptotic responses to extracellular stimuli.
Our data indicate that neither JNK nor p53 signaling pathways are involved in the upregulation of DR4 and DR5 expression. The JNK pathway, however, mediates regulation of FASL in Jurkat T cells and neuroblastoma cells following chemotherapeutic drug treatment (14, 16). This suggests that JNK is important in FASL expression but not essential for DR4 and DR5 expression. In some cell types, DR5 expression is regulated in a p53-dependent manner, while in other cells increased DR5 expression seems p53 independent (29). This is reminiscent of p53 involvement in the apoptosis of some cells but not of others (5).
Ligation of FAS can also play a role in chemotherapeutic drug-induced apoptosis in epithelial cells, since both FAS and FASL are upregulated following exposure of cells to etoposide (14). Both FADD and caspase 8 are involved in FAS-induced apoptosis (27). The reduction in etoposide-induced apoptosis after expression of dominant-negative FADD correlates with an inhibition of FAS-mediated caspase 8 cleavage. However, expression of DR4 and DR5 is more highly regulated than FAS. Blockage of DR4 and DR5 activation by DR4:Fc strongly inhibited etoposide-induced apoptosis. Furthermore, combined treatment with etoposide and TRAIL gave a synergistic apoptotic response. Thus, activation of DR4 and DR5 plays a prominent role in etoposide-induced apoptosis in epithelial cells.
Activation of DR4 and DR5 is also regulated by the level of expression of decoy receptors such as DcR1 that bind the ligand(s) of DR4 and DR5 but do not induce apoptosis (19, 22). In HEK293 cells, DcR1 mRNA and protein levels (data not shown) were not increased following etoposide treatment. In the T47D, MDA-MD-468, and ZR-75-1 breast cancer cell lines, DcR1 expression was increased. Most importantly, however, the addition of TRAIL to the cultures of etoposide-treated cells caused a synergistic apoptotic response. Thus, an increase in DR4 and DR5 levels is capable of enhancing apoptosis in response to added TRAIL even with increased DcR1 expression in these cells.
Etoposide treatment of cells activates caspases (9, 14). Caspase 8 is cleaved after ligation of DR4 and DR5. Upon etoposide treatment, caspase 8 is also cleaved and FADD dominant-negative expression blocks etoposide-mediated caspase 8 activation. In contrast, caspase 10 is not cleaved by etoposide treatment, suggesting that caspase 10 is not involved in DR4 and DR5 signaling in HEK293 cells. Overexpression of Bcl2 effectively blocked etoposide-induced apoptosis but failed to block DR4 and DR5 upregulation and cleavage of caspase 8 in response to etoposide. Bcl2 overexpression blocked caspase 9 cleavage to its active form in etoposide-treated cells. It is probable that overexpression of Bcl2 is sequestering Apaf1, a regulator of caspase 9 activation, preventing the activation of caspase 9 (17, 36). Taken together, ligation of DR4 and DR5 activates pathways leading to caspase 8 and caspase 9 that commits the cell to apoptosis. The blocking of either caspase 8 or caspase 9 inhibits etoposide-induced apoptosis.
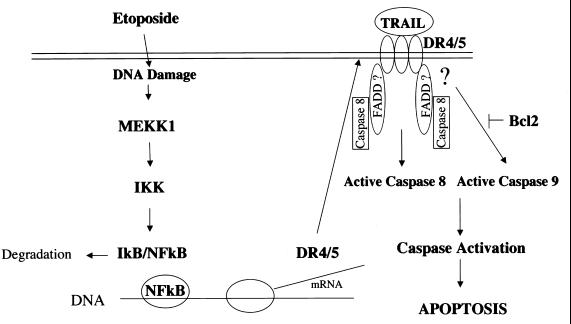
We propose a model (Fig. 8) in which etoposide treatment of cells leads to DNA damage and activation of MEKK1. MEKK1 activation could lead to phosphorylation and activation of IKKα/β. Subsequent phosphorylation of IκB by IKKα/β leads to the degradation of IκB. This allows NF-κB to migrate to the nucleus, where it is required for the transcription of DR4 and DR5 genes. Upregulation of DR4 and DR5 enhances the responsiveness of cells to TRAIL. TRAIL-activated DR4 and/or DR5 requires FADD-like adapter proteins that regulate the activation of caspase 8. Ultimately, DR4 and DR5 activation of caspases results in the commitment to apoptosis. The sensitivity of cells to TRAIL or related ligands is regulated by etoposide-induced upregulation of DR4 and DR5.
FIG. 8.
Proposed model for increased expression and activation of DR4 and DR5 following etoposide treatment. Exposure of cells to etoposide causes DNA damage, leading to activation of MEKK1. This activation leads to the phosphorylation and activation of IKKα/β that subsequently phosphorylates and degrades IκB. The degradation of IκB leads to NF-κB-dependent transcription of DR4 and DR5. Increased DR4 and DR5 expression increases TRAIL binding to its receptors. This leads to activation of caspase 8 by its recruitment to the receptor via FADD-like proteins and the subsequent downstream activation of caspase 9. Caspase 8 and caspase 9 activation causes further caspase activation, leading to apoptosis. The antiapoptotic Bcl2 protein inhibits the activation of caspase 9 and prevents etoposide-induced apoptosis.
By defining the mechanism of genotoxin-induced upregulation of DR4 and DR5 in different tumors, strategies could be developed for effective combined treatment regimes that would enhance the apoptotic response to TRAIL. Indeed, the combination of the antimetabolite doxorubicin and death receptor ligands can synergistically induce apoptosis in certain breast cancer lines (15). Optimizing the drug-induced expression of DR4 and DR5 in tumors should allow TRAIL to be an effective tumorigenic agent in treating cancer.
ACKNOWLEDGMENTS
We thank Kenneth Tyler and Penny Clarke for their helpful advice during the development of this project.
This work was supported by NIH grants DK48845, DK37871, GM303024, and CA58157. S.G. is a Leukemia Society Fellow.
REFERENCES
- 1.Ashkenazi A, Dixit V M. Apoptosis control by death and decoy receptors. Curr Opin Cell Biol. 1999;11:255–260. doi: 10.1016/s0955-0674(99)80034-9. [DOI] [PubMed] [Google Scholar]
- 2.Ashkenazi A, Dixit V M. Death receptors: signaling and modulation. Science. 1998;281:1305–1308. doi: 10.1126/science.281.5381.1305. [DOI] [PubMed] [Google Scholar]
- 3.Baeuerle P A, Baltimore D. NF-kappa B: ten years after. Cell. 1996;87:13–20. doi: 10.1016/s0092-8674(00)81318-5. [DOI] [PubMed] [Google Scholar]
- 4.Basu S, Rosenzweig K R, Youmell M, Price B D. The DNA-dependent protein kinase participates in the activation of NF kappa B following DNA damage. Biochem Biophys Res Commun. 1998;247:79–83. doi: 10.1006/bbrc.1998.8741. [DOI] [PubMed] [Google Scholar]
- 5.Bates S, Vousden K H. Mechanisms of p53-mediated apoptosis. Cell Mol Life Sci. 1999;55:28–37. doi: 10.1007/s000180050267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fanger G R, Johnson N L, Johnson G L. MEK kinases are regulated by EGF and selectively interact with Rac/Cdc42. EMBO J. 1997;16:4961–4972. doi: 10.1093/emboj/16.16.4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.French L E, Tschopp J. The TRAIL to selective tumor death. Nat Med. 1999;5:146–147. doi: 10.1038/5505. [DOI] [PubMed] [Google Scholar]
- 8.Fuchs S Y, Adler V, Pincus M R, Ronai Z. MEKK1/JNK signaling stabilizes and activates p53. Proc Natl Acad Sci USA. 1998;95:10541–10546. doi: 10.1073/pnas.95.18.10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gibson S, Widmann C, Johnson G L. Differential involvement of MEK kinase 1 (MEKK1) in the induction of apoptosis in response to microtubule-targeted drugs versus DNA damaging agents. J Biol Chem. 1999;274:10916–10922. doi: 10.1074/jbc.274.16.10916. [DOI] [PubMed] [Google Scholar]
- 10.Golstein P. Cell death: TRAIL and its receptors. Curr Biol. 1997;7:R750–R753. doi: 10.1016/s0960-9822(06)90000-1. [DOI] [PubMed] [Google Scholar]
- 11.Griffith T S, Lynch D H. TRAIL: a molecule with multiple receptors and control mechanisms. Curr Opin Immunol. 1998;10:559–563. doi: 10.1016/s0952-7915(98)80224-0. [DOI] [PubMed] [Google Scholar]
- 12.Gura T. How TRAIL kills cancer cells, but not normal cells. Science. 1997;277:768. doi: 10.1126/science.277.5327.768. [DOI] [PubMed] [Google Scholar]
- 13.Jarpe M B, Widmann C, Knall C, Schlesinger T K, Gibson S, Yujiri T, Fanger G R, Gelfand E W, Johnson G L. Anti-apoptotic versus pro-apoptotic signal transduction: checkpoints and stop signs along the road to death. Oncogene. 1998;17:1475–1482. doi: 10.1038/sj.onc.1202183. [DOI] [PubMed] [Google Scholar]
- 14.Kasibhatla S, Brunner T, Genestier L, Echeverri F, Mahboubi A, Green D R. DNA damaging agents induce expression of Fas ligand and subsequent apoptosis in T lymphocytes via the activation of NF-kappa B and AP-1. Mol Cell. 1998;1:543–551. doi: 10.1016/s1097-2765(00)80054-4. [DOI] [PubMed] [Google Scholar]
- 15.Keane M M, Ettenberg S A, Nau M M, Russell E K, Lipkowitz S. Chemotherapy augments TRAIL-induced apoptosis in breast cell lines. Cancer Res. 1999;59:734–741. [PubMed] [Google Scholar]
- 16.Le-Niculescu H, Bonfoco E, Kasuya Y, Claret F X, Green D R, Karin M. Withdrawal of survival factors results in activation of the JNK pathway in neuronal cells leading to Fas ligand induction and cell death. Mol Cell Biol. 1999;19:751–763. doi: 10.1128/mcb.19.1.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li P, Nijhawan D, Budihardjo I, Srinivasula S M, Ahmad M, Alnemri E S, Wang X. Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell. 1997;91:479–489. doi: 10.1016/s0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- 18.Lin K I, DiDonato J A, Hoffmann A, Hardwick J M, Ratan R R. Suppression of steady-state, but not stimulus-induced NF-kappaB activity inhibits alphavirus-induced apoptosis. J Cell Biol. 1998;141:1479–1487. doi: 10.1083/jcb.141.7.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marsters S A, Sheridan J P, Pitti R M, Huang A, Skubatch M, Baldwin D, Yuan J, Gurney A, Goddard A D, Godowski P, Ashkenazi A. A novel receptor for Apo2L/TRAIL contains a truncated death domain. Curr Biol. 1997;7:1003–1006. doi: 10.1016/s0960-9822(06)00422-2. [DOI] [PubMed] [Google Scholar]
- 20.Mo Y Y, Beck W T. DNA damage signals induction of fas ligand in tumor cells. Mol Pharmacol. 1999;55:216–222. doi: 10.1124/mol.55.2.216. [DOI] [PubMed] [Google Scholar]
- 21.Nemoto S, DiDonato J A, Lin A. Coordinate regulation of I-κB kinases by mitogen-activated protein kinase kinase kinase 1 and NF-κB-inducing kinase. Mol Cell Biol. 1998;18:7336–7343. doi: 10.1128/mcb.18.12.7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pan G, Ni J, Wei Y F, Yu G, Gentz R, Dixit V M. An antagonist decoy receptor and a death domain-containing receptor for TRAIL. Science. 1997;277:815–818. doi: 10.1126/science.277.5327.815. [DOI] [PubMed] [Google Scholar]
- 23.Pan G, O'Rourke K, Chinnaiyan A M, Gentz R, Ebner R, Ni J, Dixit V M. The receptor for the cytotoxic ligand TRAIL. Science. 1997;276:111–113. doi: 10.1126/science.276.5309.111. [DOI] [PubMed] [Google Scholar]
- 24.Potmesil M, Hsiang Y H, Liu L F, Bank B, Grossberg H, Kirschenbaum S, Forlenza T J, Penziner A, Kanganis D, Forlenzar T J, et al. Resistance of human leukemic and normal lymphocytes to drug-induced DNA cleavage and low levels of DNA topoisomerase II. Cancer Res. 1988;48:3537–3543. [PubMed] [Google Scholar]
- 25.Schneider P, Bodmer J L, Thome M, Hofmann K, Holler N, Tschopp J. Characterization of two receptors for TRAIL. FEBS Lett. 1997;416:329–334. doi: 10.1016/s0014-5793(97)01231-3. [DOI] [PubMed] [Google Scholar]
- 26.Schneider P, Thome M, Burns K, Bodmer J L, Hofmann K, Kataoka T, Holler N, Tschopp J. TRAIL receptors 1 (DR4) and 2 (DR5) signal FADD-dependent apoptosis and activate NF-kappaB. Immunity. 1997;7:831–836. doi: 10.1016/s1074-7613(00)80401-x. [DOI] [PubMed] [Google Scholar]
- 27.Schulze-Osthoff K, Ferrari D, Los M, Wesselborg S, Peter M E. Apoptosis signaling by death receptors. Eur J Biochem. 1998;254:439–459. doi: 10.1046/j.1432-1327.1998.2540439.x. [DOI] [PubMed] [Google Scholar]
- 28.Screaton G R, Mongkolsapaya J, Xu X N, Cowper A E, McMichael A J, Bell J I. TRICK2, a new alternatively spliced receptor that transduces the cytotoxic signal from TRAIL. Curr Biol. 1997;7:693–696. doi: 10.1016/s0960-9822(06)00297-1. [DOI] [PubMed] [Google Scholar]
- 29.Sheikh M S, Burns T F, Huang Y, Wu G S, Amundson S, Brooks K S, Fornace A J, Jr, el-Deiry W S. p53-dependent and -independent regulation of the death receptor KILLER/DR5 gene expression in response to genotoxic stress and tumor necrosis factor alpha. Cancer Res. 1998;58:1593–1598. [PubMed] [Google Scholar]
- 30.Walczak H, Degli-Esposti M A, Johnson R S, Smolak P J, Waugh J Y, Boiani N, Timour M S, Gerhart M J, Schooley K A, Smith C A, Goodwin R G, Rauch C T. TRAIL-R2: a novel apoptosis-mediating receptor for TRAIL. EMBO J. 1997;16:5386–5397. doi: 10.1093/emboj/16.17.5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walczak H, Miller R E, Ariail K, Gliniak B, Griffith T S, Kubin M, Chin W, Jones J, Woodward A, Le T, Smith C, Smolak P, Goodwin R G, Rauch C T, Schuh J C, Lynch D H. Tumoricidal activity of tumor necrosis factor-related apoptosis-inducing ligand in vivo. Nat Med. 1999;5:157–163. doi: 10.1038/5517. [DOI] [PubMed] [Google Scholar]
- 32.Wang C Y, Cusack J C, Jr, Liu R, Baldwin A S., Jr Control of inducible chemoresistance: enhanced anti-tumor therapy through increased apoptosis by inhibition of NF-kappaB. Nat Med. 1999;5:412–417. doi: 10.1038/7410. [DOI] [PubMed] [Google Scholar]
- 33.Wang C Y, Mayo M W, Korneluk R G, Goeddel D V, Baldwin A S., Jr NF-kappaB antiapoptosis: induction of TRAF1 and TRAF2 and c-IAP1 and c-IAP2 to suppress caspase-8 activation. Science. 1998;281:1680–1683. doi: 10.1126/science.281.5383.1680. [DOI] [PubMed] [Google Scholar]
- 34.Widmann C, Gerwins P, Johnson N L, Jarpe M B, Johnson G L. MEK kinase 1, a substrate for DEVD-directed caspases, is involved in genotoxin-induced apoptosis. Mol Cell Biol. 1998;18:2416–2429. doi: 10.1128/mcb.18.4.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yeh W C, Pompa J L, McCurrach M E, Shu H B, Elia A J, Shahinian A, Ng M, Wakeham A, Khoo W, Mitchell K, El-Deiry W S, Lowe S W, Goeddel D V, Mak T W. FADD: essential for embryo development and signaling from some, but not all, inducers of apoptosis. Science. 1998;279:1954–1958. doi: 10.1126/science.279.5358.1954. [DOI] [PubMed] [Google Scholar]
- 36.Zou H, Li Y, Liu X, Wang X. An APAF-1.Cytochrome c multimeric complex is a functional apoptosome that activates procaspase-9. J Biol Chem. 1999;274:11549–11556. doi: 10.1074/jbc.274.17.11549. [DOI] [PubMed] [Google Scholar]