Abstract
Simple Summary
Immunoproteasome plays a key role in the generation of antigenic peptides. Immune checkpoints therapy is a front-line treatment of advanced/metastatic tumors, and to improve its efficacy, a broader knowledge of the dynamics of antigen repertoire processing by cancer cells is mandatory. The scope of this review is to offer a picture of the role of immunoproteasome in antigen presentation to fuel the hypothesis of novel therapeutic interventions based on the modulation of this proteolytic complex and immune checkpoints.
Abstract
Immunoproteasome is a noncanonical form of proteasome with enzymological properties optimized for the generation of antigenic peptides presented in complex with class I MHC molecules. This enzymatic property makes the modulation of its activity a promising area of research. Nevertheless, immunotherapy has emerged as a front-line treatment of advanced/metastatic tumors providing outstanding improvement of life expectancy, even though not all patients achieve a long-lasting clinical benefit. To enhance the efficacy of the currently available immunotherapies and enable the development of new strategies, a broader knowledge of the dynamics of antigen repertoire processing by cancer cells is needed. Therefore, a better understanding of the role of immunoproteasome in antigen processing and of the therapeutic implication of its modulation is mandatory. Studies on the potential crosstalk between proteasome modulators and immune checkpoint inhibitors could provide novel perspectives and an unexplored treatment option for a variety of cancers.
Keywords: immunoproteasome, ubiquitin–proteasome system, immune checkpoints, proteasome inhibitors, immunotherapy
1. Introduction
Cancer immunotherapy is conceptually based on therapeutic stimulation of cancer immunosurveillance, a theory that states that tumor cells, mostly through the phenotypic alteration and the repertoire of tumor-associated neoantigens they often present, can be recognized and targeted by the immune system in the attempt to prevent disease progression. Nowadays, the dynamics of cancer and immune system crosstalk have been unequivocally unveiled to be extremely complex and has been renamed as cancer immunoediting. According to the three Es theory, this process comprises three defined phases: elimination of cancer cells by the immune system; equilibrium between tumor growth and control by the immune system; escape of neoplastic cells from immunosurveillance. The first two phases in some way coincide with the concepts described by the former theory, but the second phase can last long, running asymptomatically, until it is overrun by the third one, which results in tumor growth and dissemination. In an immunocompetent host, this breakthrough results from a positive selection of tumor clones that are able to evade the inhibitory control of the immune system by downregulating/masking antigen epitopes or by increasing the immunosuppressive properties of the tumor microenvironment [1,2].
Modern approaches of cancer immunotherapy, designed to restore a robust degree of immune activity against tumor cells, encompass immune checkpoint blockade, adoptive cellular therapies, and cancer vaccines [1,2,3,4,5]. Among these therapeutic interventions, immune checkpoint inhibitors (ICKi) have substantially revolutionized the oncology field by prolonging the survival of patients affected by highly aggressive/advanced stage cancers, such as metastatic melanoma and non-small-cell lung cancer (NSCLC). This approach is currently based on the use of monoclonal antibodies targeting inhibitory ICKs, such as CTLA4 (cytotoxic T lymphocyte antigen 4) and PD-1 (programmed cell death 1) or PD-L1 (programmed cell death 1 ligand) that regulate activated T lymphocytes function, switching off the immune response. A number of preclinical and clinical studies have revealed that antibodies raised against the immune checkpoint molecules enhance the antitumor immunity through not completely identified mechanisms of action, which differ depending on the target and specificity of the monoclonal antibody used (see Appendix A) [1,3,4,5]. Despite the undoubtful clinical success of ICK blockade, the on-field experimentation has opened several tasks that are worth being addressed: organ involvement and cohorts of signs and symptoms; second, the clinical efficacy is limited to subgroups of responder patients or, in many other subjects, after an initial response, drug resistance takes over. The latter event is generally due to the genetic and phenotypic heterogeneity of cancer cells and/or to tumor microenvironment remodeling during disease progression and dissemination [5,6,7]. Nonetheless, the overall efficacy of ICKi is affected by multiple factors, spanning from the expression and distribution of the target within the tumor microenvironment, the tumor mutational rate, and alterations of antigen presentation [8,9,10]. In this context, a crucial role is played by components of the host microenvironment that infiltrate the tumor and exert immunosuppressive effects, thus counteracting ICKi activity (i.e., infiltration by T regulatory cells (Tregs), dendritic cells, immunosuppressive myeloid cells, cancer-associated fibroblasts) [11].
Cancer cell immunogenicity is a key determinant for ICKi efficacy: malignancies which do not express tumor-specific antigens are not potentially susceptible to this approach. This is somewhat strengthened by the evidence that an immunotherapy-non-responsive cancer can turn into an immunotherapy-responsive one upon enhancement of tumor immunogenicity [8,9,10,12].
Hence, a better knowledge of the “immunopeptidome” (the repertoire of peptides bound to and presented by MHC molecules) and how tumor-specific antigen repertoire change during tumor progression is expected to improve the current therapeutic strategies; thus, a challenging issue is the identification and characterization of the MHC-I-presented peptides that modulate T cell-based tumor response [12,13,14].
At the molecular level, the generation of effective T cells that fight cancer requires a functional and efficient machinery for the multistep and multicellular mediated process, including antigen processing and presentation. The intracellular antigen processing pathway almost exclusively deals with the ubiquitin proteasome system (UPS) activity by which proteins are first ubiquitinated and then degraded. Indeed, the UPS carries out multiple functions in cells and in the onset and progression of different human pathologies spanning from neurodegeneration to cancer [15]. A major role in antigen processing is covered by a specialized and inducible form of proteasome (i.e., the multi-catalytic machine which degrades ubiquitinated proteins) named “immunoproteasome” [16]. This review focuses on immunoproteasome, its involvement in antigen generation, and on the therapeutic implications of its modulation to halt cancer progression. Finally, the potential crosstalk between proteasome modulators and immune checkpoint inhibitors is discussed.
2. Ubiquitin–Proteasome System: Cellular Biocomputing Machinery
In living cells, the proteome is constantly tuned through a complex, entwined, and multi-subcellular compartments network which coordinates the synthesis, folding, conformational upkeep and degradation of individual proteins [1]. The removal of undesired proteins is carried out by two main intracellular proteolytic systems, namely the ubiquitin–proteasome system (UPS) and autophagy [17,18,19,20]. The UPS is the major actor in the turnover of more than half of intracellular proteins that play fundamental roles in several facets of cell life, such as cell cycle, apoptosis, DNA repair, antigen presentation, inflammation, cellular response to environmental stress, and morphogenesis of neuronal networks [21,22,23]. The UPS’s hierarchical organization includes two intertwined and consecutive steps, i.e., the covalent ATP-dependent attachment of ubiquitin (Ub) polymers to a given substrate (target protein conjugation cascade), which is catalyzed by three classes of ubiquitin ligases, E1 (Ub-activating enzyme), E2 (Ub-conjugating enzyme), and E3 ligase, and its degradation by the 26S proteasome, followed by recycling of ubiquitin moieties along with the release of cleared protein oligopeptides (Figure 1) [15,24,25,26,27,28]. In the final step of ubiquitination, the E3 ligase, which is committed with substrate specificity [25,26,27,29,30] (Figure 1), mediates the formation of an isopeptide bond between the carboxyl C-terminal group of Ub and the ε-amino group of the lysine residue of the target protein. Thereafter, the reaction can be repeated multiple times, allowing the polyubiquitin chain to increase by 6 Ub moieties in average, in which each subsequent Ub monomer is connected to the previous one through an isopeptide covalent bond similar to that of the first Ub–substrate bond [30,31,32,33,34,35]. The process of ubiquitylation is a highly dynamic and reversible equilibrium; in fact, deubiquitinases or deubiquitinating enzymes (DUBs) can reverse the effect of E3 ligases by removing ubiquitin from target proteins. Furthermore, they mediate the polyubiquitin chain release during the hydrolysis of substrates by the proteasome [36,37].
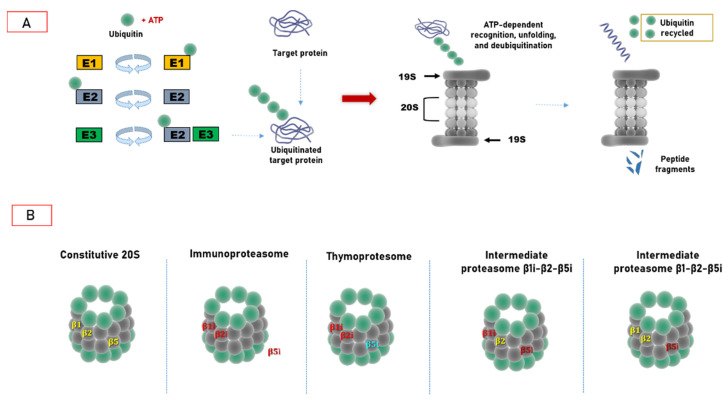
Figure 1.
Ubiquitin–proteasome system. (A) General UPS organization. The Ub target protein conjugation cascade consists of three classes of ubiquitin ligases: E1, E2, and E3. Ub is activated by E1, forming a high-energy thioester bond; then, activated Ub is transferred to E2, and then E3 attaches Ub to a specific polypeptide substrate. At the end stage, the substrates are recognized and subjected to ATP-dependent degradation by the 26S proteasome. (B) Structural heterogeneity of the proteasome core particle; 20S constitutive proteasome contains the standard catalytic subunits β1 (caspase-like activity), β2 (trypsin-like activity), and β5 (chymotrypsin-like activity). The immunoproteasome incorporates the inducible catalytic subunits β1i, β2i, and β5i, which have different catalytic specificities. The thymoproteasome is expressed only by cortical thymic epithelial cells and is characterized by the presence of β1i-β2i, and β5t subunits. Finally, intermediate proteasomes contain a mix of constitutive and inducible subunits: β1i,-β2-β5i and β1-β2, and β5i, respectively.
Canonical 26S proteasome holoenzyme is a multifunctional proteolytic machine composed by the 20S proteasome core particle (CP), which features the proteolytic activity, capped by the 19S regulatory subunit (RP, also known as PA700), which carries out the ATP-dependent recognition, unfolding, and translocation into the 20S of the polyubiquitinated substrate [4,6,20]. The 20S core particle is a cylinder-like packed particle which contains four axial stacking heptameric rings arranged into two outer α rings and two inner β rings (i.e., α1–7β1–7α1–7β1–7) [38,39,40]. Eukaryotic 20S has a central channel, which houses six catalytically active β subunits, three for each β ring: the chymotryptic-like (β5 subunit, which hydrolyses at the C-terminus of hydrophobic residues), the trypsin-like (β2, which hydrolyses at the C-terminus of basic residues), and caspase-like (β1, which hydrolyses at the C-terminus of acidic residues) sites [40,41]. In the free 20S not engaged with a regulatory particle, the N-terminal tails of the α subunits point inwards to the center of the ring and the neighboring tails form an intricate network of inter-subunit interactions, constituting “the gate” of 20S, which regulates the substrate access through a 13 Å entry pore: the insertion of the substrate through this “N-terminal gate” is the rate-limiting step of proteasome activity [39,42,43]. Since “gate opening” is a key step in inducing the 20S function during evolution, cells have evolved different regulators (see also Section 2) which control this process and adapt proteasome functionality to the metabolic condition [44]. In fact, within the proteasome architecture, the outer α rings of the 20S make a nearly flat surface that binds the 19S and other alternative regulatory particles (i.e., PA28, see Section 3.2). The best known 20S activator is indeed the 19S which interacts, in the presence of ATP, with one or both ends of the 20S to form proteasome holocomplexes: the 26S, referred to as “single-capped”, and the 30S, named “doubly-capped” [44,45]. Once bound over the axial 20S pores, the 19S RP binding induces the displacement of the 20S α subunits’ N-terminal tails, thus opening the gate and promoting substrate access and translocation into the catalytic chamber [42,46,47]. From the structural point of view, the 19S is composed by two main elements, the lower “base” which directly binds to the 20S and the upper “lid”, thus forming a conformational dynamic complex [42,48]. The base consists of six structurally different subunits with ATPase activity (Rpt1–6), the structural subunits Rpn1 and Rpn2, and two ubiquitin-binding subunits, Rpn10 and Rpn13. The energy of ATP hydrolysis is spent to unfold the protein substrate and pull it down into the catalytic chamber of the 20S. Five ATPases (Rpt1–3; Rpt5 and Rpt6) present a hydrophobic tyrosine HbYX motif at the C-termini that insert into the α subunits pocket to induce gate opening.
Conversely, the peripheral lid consists of nine non-ATPase subunits: Rpn 3, 5–9, 11, 12, and 15, whose main functions are the strengthening of the 20S–19S interaction and deubiquitination of substrates before their processing by the ATPases [49,50,51]. Therefore, the 19S carries out different and fundamental functions: the recognition and unfolding of ubiquitinated substrates, the opening of the 20S pore, the entry of substrates into the catalytic chamber, and the release of ubiquitin moieties during substrate degradation [15,44,52]. Recently, the structure, function, and biogenesis of the 20S and 19S as well as the structural conformation of the proteasome holoenzyme have been extensively reviewed [6,20]. Emerging evidence shows that a protein tagged with at least four ubiquitin molecules is not the unique signal for proteasome degradation by 26S: in fact, multiple or single monoubiquitination appears to be sufficient to label a substrate for proteasomal degradation [53,54]. Additionally, a series of proteins has been reported to be degraded by the 26S regardless of ubiquitination [20,55,56], implying the existence of alternative molecular signals, such as a specific amino acidic sequence or structural elements (also called “degrons”), that mediate substrate recognition and degradation [19,56,57,58]. An additional topic that underlines the complexity of proteasome degradation pathway is the ubiquitin-independent degradation of macromolecular substrates by the uncapped 20S: several studies demonstrate that the 20S degrades natively unfolded and oxidative stress-damaged proteins [15,59,60,61]. The exact biological meaning and the molecular basis of these different degradation pathways are unclear and deserve additional investigations.
3. Hats off to Proteasome Variability
Since the UPS catalyzes the degradation of the majority of intracellular proteins and its organization is tuned on the cellular metabolic demands, the maintenance of an adequate activity is essential for cell homeostasis as much as an appropriate plasticity, in terms of structural and functional organization, is required for cells to adapt to the stimuli they experience [15]. Thus, the proteasome machine is a highly dynamic complex whose structural and conformational composition, substrate specificity is regulated at multiple steps encompassing transcriptional regulation, kinetics of assembly, post-synthetic modifications, and the interaction with a number of proteasome-interacting proteins (PIPs) which act as regulatory factors [40,62,63,64,65]. Focusing on the proteasome structural composition, two main elements of proteasome plasticity and variability are represented by (1) the interchangeability between constitutive and inducible catalytic subunits of the 20S; (2) the presence of different regulatory particles which can associate to just one or both free ends of the 20S. This allows generating different subtypes of proteasome that can coexist in a single cell and whose ratios may change among tissues. The metabolic and pathological stimuli that allow these canonical and noncanonical particles to form have been partially described, but to unequivocally address the interconnected, sometimes overlapping, or specific biological functions they carry out in vivo is a challenging task [62,66,67]. Noteworthily, in vertebrates, proteasome has gained considerable tissue specificity, as indicated by the existence of alternative forms of proteasome: immunoproteasome, also known as inducible 20S (i20S), thymoproteasome (Appendix B), and spermatoproteasome, in which the constitutive catalytic subunits of the 20S are replaced by inducible/tissue-specific homologs [62,68,69,70]. Immunoproteasome and thymoproteasome serve critical roles in the immunity, whereas spermatoproteasome is a testis-specific and chronologically-defined form of proteasome, exclusively identified in spermatocytes, spermatids, and sperm. It is characterized by the presence of a specific α4 subunit (α4s) (PMSA8 gene) that replaces the constitutive α4: the incorporation of this subunit into a newly formed 20S is mutually exclusive with wild-type α4 and seems not to alter the constitutive catalytic specificity of the 20S [69,70]. Nevertheless, α4s incorporation seems to promote the association of the 20S with an alternative regulatory particle named PA200, a nuclear-specific proteasome activator expressed in all mammalian tissues, but particularly abundant in the testis, where it plays a crucial role in spermatogenesis [71,72,73,74,75]. Remarkably, an increase of PSMA8 expression has been reported in different tumors, such as large B cell lymphoma, thymomas, and testicular germ cell tumors, even though its pathophysiological meaning and relevance as a novel therapeutic target have not been investigated yet [69,76,77]. As mentioned above, the reversible binding of activators (i.e., 19S and PA200) to either one or both α subunit rings of the 20S is another important level of proteasome organization that contributes to the overall heterogeneity of proteasomes (Figure 1) [15]. Besides the 19S, the best-characterized regulatory particle is PA28 (i.e., 11S regulatory particle), which preferentially binds the immunoproteasome, forming the PA28/i20S complex as discussed further (see Section 3.2) [16,78]. The binding of activators increases the proteolytic activity of the catalytic core, promoting the α-gate opening, influencing the substrate specificity of the complex, and, more importantly, affecting the repertoire of cleavage products [15]. Hybrid proteasomes (i.e., 19S–20S 11S, 19S–20S-PA200) have also been identified. Their biological function remains obscure, but their identification underlies how the cells edit the proteasome repertoire in relation to their specific needs [79,80,81].
3.1. Immunoproteasome as a Specialized Apparatus of Self-Target Designation
In the early nineties, proteasome was discovered as the crucial player for the class I MHC-restricted antigen processing pathway and two proteasome genes, namely PSMB9 (LMP2) and PSMB8 (LMP7), which encode two alternative subunits of the 20S, β1 and β5, respectively, were identified in close proximity of the transporter associated with the antigen-processing (TAP) gene in the MHC class II genomic region [82,83]. Concurrently, it was shown that synthesis and incorporation of these subunits into the 20S was driven by interferon γ (IFNγ) [84,85,86,87] (Figure 2). Thus, immunoproteasome, also known as inducible proteasome, is a specialized form of the 20S with a prominent role in immunity. Immunoproteasome preferentially and cooperatively incorporates three immune subunits, β1i, β2i (MECL-1), and β5i, to replace the constitutive catalytic subunits into the β ring of the 20S within its biogenesis pathway. The preferential assembly of inducible subunits is likely due to the higher affinity of β5i than of β5c for the proteasome maturation protein (POMP) which mediates the β ring formation [15,88,89]. The i20S assembly is four times faster than c20S, clearly reflecting the need for a rapid and transient response upon exposure to a proinflammatory stimulus. In fact, IFNγ induces, via the JAK/STAT signaling, the transcription of immune catalytic subunits, the MHC-I and TAP genes, thus enhancing the entire class I antigen presentation machinery [62,82,84,90]. Immunoproteasome is constitutively expressed at the basal level in hematopoietic cells and has a shorter half-life than c20S (average 27 h for immunoproteasome and 133 h for constitutive proteasome). Such a rapid turnover has the purpose of efficiently adapting to the environmental changes [91,92]. It has been shown that during the course of viral, bacterial, and fungal infections, immunoproteasome replaces up to 90% of the c20S pool [93,94]. As a matter of fact, besides the pioneering contribution of IFNγ, immunoproteasome was shown to be further transcriptionally induced by a plethora of inflammatory stimuli, such as IFNα and IFNβ, tumor necrosis factor α (TNF-α), lipopolysaccharides (LPS), as well as by redox unbalance [95,96,97,98]. It is important to recall that the different forms of proteasome particles can coexist inside the cells as well as the different peptide antigens generated. Anyway, in dependence to the different stimuli the cells are exposed to, the relative abundance of antigens generated by each specific subpopulation can be adapted [99].
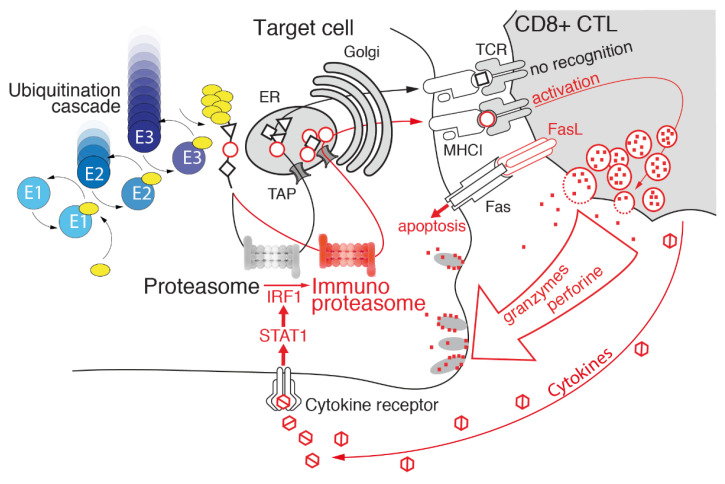
Figure 2.
Immunoproteasome mediates cell cytotoxicity. The development of an inflammatory reaction, particularly the production of IFNγ and other inflammatory cytokines, induces the expression of the proteasome immune subunits in target cells and leads to efficient production of peptides beneficial in terms of presentation on the MHC class I and recognizable by CD8+ CTLs. Further elevation of the immunoproteasome (in red) mediated by increased production of inflammatory cytokines and subsequent generation of immunodominant peptides may function as a positive feedback loop, enhancing T cell-mediated cytotoxicity.
A side-by-side comparison of the three different substrate-binding pockets of the c20S and i20S points out the enzymological differences of the two complexes [100]. In general, the i20S is characterized by increased chymotrypsin-like and trypsin-like activities, but a lower caspase activity [101]. In detail, the caspase-like subunit β1 accommodates peptides with an acidic residue in the P1 position, whereas β1i binds to peptides with a hydrophobic residue in the same position, exerting a branched-chain amino acid-preferring activity.
The folded trimeric complex formed by a given peptide and the MHC-I ligand cleft is exposed to the cell surface for presentation to the immune cells. The requisites for tight peptide MHC class I binding are essentially two: (1) the length of 8–9 amino acids and (2) an anchor of basic or hydrophobic residues located at the C-terminus or within the peptide sequence [68]. MHC-I does not accept C-termini with acidic anchor residues; thus, the substitution of β1c with β1i produces antigenic peptides with hydrophobic C-termini that can efficiently bind to MHC-1 molecules. Additionally, the structural properties of β5i also contribute to generating peptides with preferred C-terminal anchor amino acids for MHC-I molecules. In fact, the β5i S1 pocket accommodates larger hydrophobic amino acids chains than β5c (which presents, instead, a “small neutral amino acids-preferring activity”) and is characterized by a more hydrophilic environment around the catalytic threonine favoring the chymotryptic-like catalytic properties of the inducible subunit. Despite the β5 and β1 subunits, the active sites of β2c and β2i seem to be structurally identical, rendering this substitution more enigmatic, even though several studies reported an increase in trypsin-like activity of i20S with respect to c20S [68,100].
Additional forms of proteasome bearing a mix of standard and inducible subunits were identified. These intermediate proteasomes, which represent from one third to one half of the overall proteasome content in different tissues, such as liver, colon, and kidney, contain these triads of subunits, β1/β2/β5i or β1i/β2/β5i. Remarkably, a recently discovered mechanism of antigen generation through which proteasome increases the repertoire of antigens for presentation to the immune system is the “proteasome-catalyzed peptide splicing”: spliced peptides, which are made by two not contiguous fragments of parental proteins, are produced efficiently both by immunoproteasome and constitutive particles [102,103,104,105]. The existence of these mechanisms along with the copresence of different proteasome populations beyond the constitutive proteasome involved in antigen–peptide generation broadens the repertoire of antigens produced by a cell [92,106,107] (Figure 3). However, whether the incorporation of immune subunits triggers qualitative or quantitative effects on peptide repertoire generation is still not resolved since different studies report somewhat controversial results. In fact, a number of studies highlighted the positive role of immunoproteasome mainly against viral and bacterial antigens, whereas some studies reported that immunoproteasome expression can abrogate the presentation of some tumor epitopes [106,107,108,109,110,111,112,113].
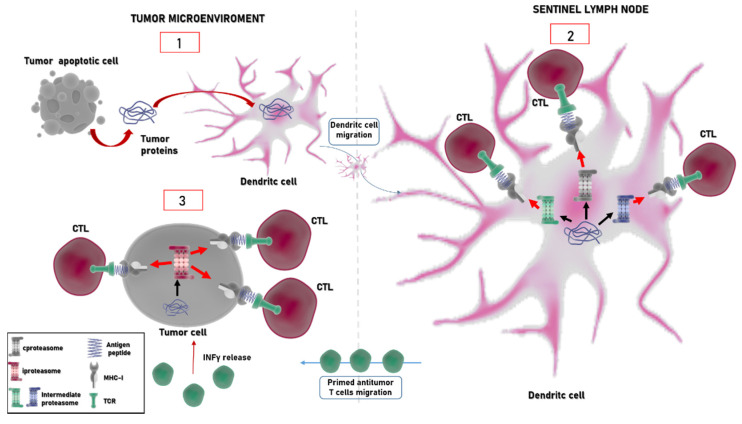
Figure 3.
Activation of antitumor T cells mediated by proteasomes. At the tumor site, apoptotic cells release tumor proteins which are internalized by dendritic cells (1). These cells migrate to the sentinel lymph node, where peptides are derived by proteasome (mainly constitutive (in grey) and intermediate proteasome (in green and in blue) particles). Different populations of proteasome are represented with different colors in the figure, as indicated also in the legend. Degradation of tumor-associated proteins is presented to naïve T cells in complex with MHC-I molecules (2). Primed T cells migrate to the tumor site (3). The in loco production of IFNγ induces the expression of i26S (in red) which becomes the prevalent form of proteasome in the “hot tumor site”. Thus, tumor cells with a high content of immunoproteasome change their antigenic peptide repertoire, which can be targeted by other circulating T cells.
Anyway, a defect in antigen presentation was found in the triple-inducible-subunit KO mice, and this alteration is now reported to be much broader qualitatively and quantitatively than that previously described in any of the β1i, β2i, or β5i single-subunit KO mice and still far greater than the sum of the defects these single-subunit KO animals were reported to bear [92,114,115]. Moreover, analysis of MHC class I-bound peptides shows that the antigen repertoire of KO mice differs from that of WT mice, reinforcing the hypothesis that immunoproteasomes generate peptides that, apparently, cannot be produced by constitutive proteasomes [108,116,117]. On the other hand, other studies suggest that immunoproteasomes affect the quantity rather than the quality of the given generated peptides, influencing also in this case the immune response [112,118,119]. Thus, some antigens are exclusively produced by the immunoproteasome or the constitutive proteasome, while others can be processed by both, and some others can be preferentially processed by intermediate-type proteasomes [120,121]. Of note, this distinction between the quantitative and qualitative effects of the antigen repertoire depending on the expression rate of immune–constitutive or mixed proteasome is not simply semantic. In fact, it is of basic significance not only to better understand the enzymatic properties of the different proteasome populations, but also to better define the MHC class I-dependent CD8+ T cell response in the context of specific physiopathological conditions [122].
As mentioned above, there is now extensive evidence that the UPS generates MHC class I-presented peptides; however, the source of the presented peptides is far less clear [123,124,125]. In this framework, the DRIP (defective ribosomal product) hypothesis is a novel concept about the source of peptides targeted by proteasome for antigen presentation. A considerable amount of newly synthesized polypeptides does not reach their native state after leaving the ribosome. It has been suggested that the defective proteins (indeed, DRiPs) are ubiquitinated and rapidly degraded by the proteasome and DRIPs-derived peptides may be preferentially loaded on class I MHC molecules through an unknown mechanism (possibly relying on the preferential uptake into the ER and/or exclusion of peptides derived from mature proteins) [126,127]. This proposed model challenges the older one in which MHC I-presented peptides are generated from the turnover of all cellular proteins. Therefore, further research is required to quantify the relative bulk of the newly synthesized and mature proteins in generating MHC class I-presented peptides by proteasome, and it will be important to unveil which proteins immunoproteasome shows cleavage preference for and how DRIPs-derived peptides produced by different proteasome populations are involved in mediating the immune response [124,127].
3.2. Regulatory Particles Which Associate with Immunoproteasome
As mentioned in the previous sections, besides the 19S, the second most common proteasome regulator is the ATP and ubiquitin-independent PA28 activator family that includes three highly homologous, ~28 kDa subunits, α, β, and γ, which form a ring-shaped 200 kDa multimeric complex (i.e., PA28αβ and PA28γ). This complex binds, in an ATP-dependent manner, to the two free ends of the 20S or associates with a single-capped 26S (19S:20S), forming the hybrid proteasome 19S–20S–PA28 [16,128,129,130,131]. This hybrid assembly hydrolyses tri- and tetra-peptides at a higher rate than constitutive 26S. The structure, function, and mechanism through which it induces the 20S gate opening was recently extensively and more competently reviewed [16]. PA28α and PA28β, which show a prevalent cytosolic localization, are upregulated by IFNγ and other proinflammatory cytokines. Consistently with this, their biological function is related to MHC-I antigen processing. Conversely, PA28γ is a primary nuclear homoheptamer, and its expression is not significantly upregulated by inflammatory stimuli. Thus, the role of PA28γ in antigenic processing is controversial, even though some studies have reported that alteration of PA28γ expression influences the bioavailability of epitopes derived from the pioneer translation products (PTPs)—polypeptides produced in the nucleus by a nonstandard translation process which occurs prior to mRNA splicing [78,120,132]. Interestingly, these epitopes show increased abundance in different cancers and are of key relevance in eliciting effective antitumor responses [133]. Remarkably, PA28γ binding to the i20S seems to mediate the ubiquitin-independent degradation of the key regulatory proteins and, thus, it is involved in different biological processes (besides the antigenic processing), spanning from cell growth and angiogenesis to apoptosis [79]. PA28αβ, a heteroheptamer assembled from four α (PSME1) and three β (PSME2) subunits, is constitutively expressed in lymphoid organs, but it can also be detected in tissues that lack immunoproteasome, where it likely exerts its biological functions also in association with the c20S. PA28αβ levels dramatically increase virtually in any other tissues in response to inflammatory stimuli, concomitant with the increase in other components of the MHC-I antigen processing pathway [120,134]. The i20S in association with PA28αβ mediates Ub-independent hydrolysis of the myelin basic protein during autoimmune neurodegeneration, accomplished through a novel class of charge-dependent proteasomal degrons [19,59].
Although several lines of evidence indicate the role of PA28αβ in MHC-I processing, its precise function and molecular mechanism of action are still poorly understood [78]. In 59fact, it has been reported that PA28αβ reduces the size and increases the hydrophilicity of peptides generated by i20S catalysis, thus producing peptides potentially suitable for MHC-I binding [62,129]. However, its genetic ablation does not lead to severe abnormalities in immune response against infections and causes the loss of only certain antigens. Nevertheless, this activator seems to stimulate the antigen production for some MHC-I alleles, but does not alter or downregulate the generation of other ones [78,110,135].
3.3. Immunoproteasome: Beyond Antigen Processing
In addition to the role in MHC-I antigen processing discussed above, immunoproteasome carries out other important functions in the dynamics of the immune system. In fact, its involvement in the molecular onset of autoimmune diseases has been proposed [136,137].
T cells knocked out for β2i, β5i, and, to a lesser extent, β1i show impaired proliferation and survival when transferred into virus-infected wild-type mice, suggesting a role in T cell expansion [138,139]. Moreover, ablation of the β5i activity (by either enzymatic inhibition or genetic depletion) suppresses the differentiation of Th-1 and Th-17 lineages and promotes the development of T regulatory cells (Tregs). The molecular basis of this intriguing β5i role remains to be determined: a major working hypothesis is that it may be involved in the turnover of factors that promotes Th1 and Th17 differentiation. This evidence about immunoproteasome involvement in T cells maturation and differentiation has fueled the hypothesis that the selective inhibition of β5i could be a suitable strategy in autoimmune disease therapy [114,116,140,141]. In this regard, a β5i selective inhibitor, ONX0914 (also called PR-957) has been shown to prevent the progression of rheumatoid arthritis in preclinical mouse models [141]. Moreover, β5i inhibition blocks the induction of experimental colitis, the development of lupus erythematosus-like disease, Hashimoto’s thyroiditis, and other autoimmune diseases [141,142,143,144,145,146]. Furthermore, the first selective inhibitor that targets all three proteolytically active immunoproteasome subunits (LU-005) has recently shown therapeutic efficacy against autoimmunity [124]. The UPS is well-known to cover a broad range of functions in immune cells of myeloid origin, being involved in the regulation of key crucial cell signaling pathways, such as NF-kB and IFN regulatory factors (IRFs), that mediate the production of inflammatory cytokines [15,113,147,148]. Interestingly, it has been reported that the natural inhibitor of NF-kB, IkBα, which is a prototypical proteasome substrate, is degraded by immunoproteasome faster than canonical proteasome and immunoproteasome asymmetrically capped with the 19S and PA28 (see also Section 3.2) [149,150,151,152,153]. Since immunoproteasome is constitutively expressed in myeloid cells, these results provide an intriguing hypothesis to explain the exceedingly rapid production of proinflammatory cytokines in response to various stimuli these cells are known to trigger [113,147]. However, the role of immunoproteasome in the myeloid maturation process is not completely understood. Despite this, the question whether immunoproteasome subunits affect NF-kB signal transduction differently from constitutive particles is still debated. In fact, studies concerning the effect of immunoproteasome subunits on NF-kB activation yielded contradictory results [154,155,156]. As a matter of fact, it has been recently reported that neither the kinetics of nuclear translocation nor the DNA-binding activity of NF-kB as well as the production of NF-kB-dependent proinflammatory cytokines differed between immunoproteasome-deficient (LMP2 KO and LMP7/MECL-1 double KO) and proficient cells [157].
Besides the role in the immune system, immunoproteasome seems to cover key functions in the regulation of protein homeostasis in the presence of redox unbalance, maintenance of stem cell pluripotency, neurodegenerative insults, muscle differentiation, and visual transmission [149,158,159,160,161]. Interestingly, concerning this last role, immunoproteasome is constitutively highly expressed in photoreceptors and synaptic regions of the immune-privileged retina, suggesting a role in normal neuronal functions of this highly differentiated nervous tissue [158,162]. Moreover, immunoproteasome is expressed in the lens and the cornea [163,164]. The precise role of immunoproteasome in eye functionality is still debated, even though it has been reported that its deficiency causes defects in bipolar response and abnormalities in retinal development [165,166]. Moreover, immunoproteasome expression is upregulated in stressed and injured retina, sustaining the investigation of the therapeutic potential of specific immunoproteasome inhibitors beyond the constitutive proteasome ones for the treatment of eye diseases [167].
4. Immunoproteasome: An Emerging Target in Cancer
Alterations of different genes belonging to the UPS are a hallmark of cancer. UPS dysregulation may occur at multiple levels, spanning from genetic modifications (i.e., mutations, amplifications, deletions), transcriptional network alterations (i.e., p53; NRF-1 and NRF-2) to epigenetic and post-translational modifications [15,168].
A number of studies underline the role of the i20S in cancer progression, strengthening its therapeutic potential. The tumor microenvironment is profoundly different from that of healthy tissues; this is also due to the presence of tumor-infiltrating lymphocytes (TILs) that release IFNγ and other inflammatory cytokines. Interestingly, immunoproteasome seems to possess both pro- and antitumorigenic properties, which are associated with the modulation of cytokine expression and tumor-associated peptide presentation, respectively [121].
Tumor cells can evade recognition by cytotoxic T lymphocytes (CTLs or CD8+ T cells) through downregulation of MHC-I at the cell surface or, additionally, by reducing immunoproteasome subunits expression [121,169] (Figure 4). In fact, non-small-cell lung cancer that undergoes the epithelial–mesenchymal transition shows a reduced immunoproteasome subunits expression: this leads to a dramatic drop of heterogeneity of the antigen/peptide repertoire produced by tumor cells and to poor clinical outcomes [170]. Thus, it has been proposed that a decrease in immunoproteasome expression might represent a mechanism of immune escape in tumor cells which present with a mesenchymal phenotype since this downregulation is associated with a decline in the amount and diversity of MHC-I-presented peptides [170]. In accordance with these data, transforming growth factor β (TGF-β)-induced epithelial–mesenchymal transition leads to a decrease in the immunoproteasome content [170]. Moreover, in the early stage of NSCLC, low expression of the i20S is linked to an increased risk of recurrence and metastasis onset [170]. At the molecular level, one proposed mechanism which links carcinogenesis with immunoproteasome deficiency is the differential expression of the β5i subunit. Two main β5i variants have been described, which are both induced by IFNγ: LMP7E2 and LMP7E1. LMP7E2, usually expressed in normal cells and in certain cancer types, is regularly incorporated into the mature i20S. However, many cancer cell lines express only the LMP7E1 isoform that does not interact with the 20S assembly chaperone POMP and thus cannot be integrated into the mature i20S, leading to a deficiency of functional immunoproteasome [171]. Moreover, a polymorphism at amino acid 49 of LMP7 (K49 instead of Q49), localized at the pre-sequence of β5i, reduces the rate of proteasome assembly and is associated with a higher risk of developing colon carcinoma [154]. On the other hand, overexpression of immunoproteasome subunits due to an increase in IFNγ production by TILs correlates with a better prognosis in different tumors, such as melanoma and breast cancer [121,168].
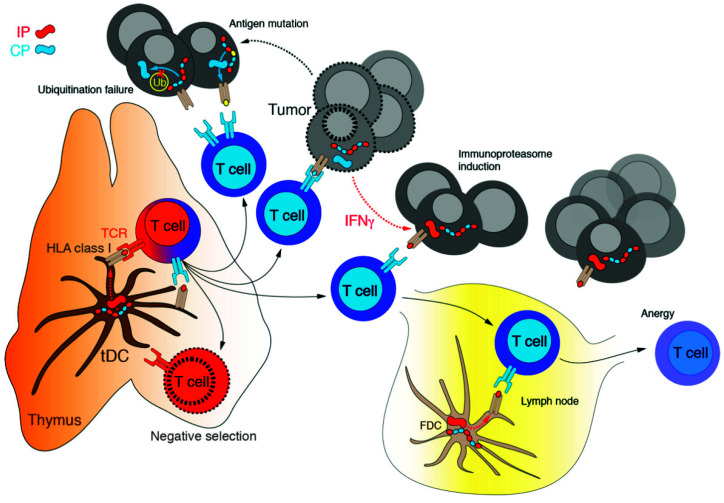
Figure 4.
Mechanisms of tumor immunoescape. T cell clones that passed negative selection recognize a limited number of tumor self-antigens. Moreover, cancer cells may escape from cytotoxic CD8+ T cells by (i) IFNγ-mediated induction of immunoproteasome with altered pattern of antigens processing, (ii) mutation of the antigen leading to the failure of the recognition by TCR, and (iii) inactivation of intracellular ubiquitination of tumor antigens caused by either suppression of expression or functional mutation of the E2/E3 ligases specific to tumor antigens.
Immunoproteasome expression is not only triggered by paracrine production of proinflammatory cytokines (such as IFNγ) by immune cells, but it is constitutively elevated in hematological malignancies [155,172]. In myeloid leukemia cells, the i20S increase was associated with a higher survival rate [156]. Interestingly, the upregulation of immunoproteasome by IFNγ overcomes resistance to the proteasome inhibitor bortezomib and sensitizes hematological malignant cells (such as multiple myeloma and leukemia) to a selective immunoproteasome inhibitor ONX0914 [173]. This opens up the perspective of developing therapeutic approaches based on selective inhibition of immunoproteasome subunits different from those targeting the constitutive ones. Moreover, some evidence indicates that immunoproteasome alterations can have an impact on the onset of inflammation-driven carcinogenesis: indeed, β5i inhibition prevents colitis associated with colon carcinoma [144]. Thus, the emerging complex picture indicates that the altered expression of immunoproteasome subunits (mainly LMP2 and LMP7) is common in various tumors, but the extent of the expression and its biological significance vary depending on cancer type and grading [94,144]. As a matter of fact, the immunopeptidome changes in the context of tumor microenvironment and depending on the relative abundance of constitutive or inducible proteasome. For example, a number of cancer antigens derived from members of the melanoma antigen gene protein family (MAGE), whose expression is restricted to reproductive tissues but which are also aberrantly expressed in a wide variety of cancer types, such as MAGA3(114–122), MAGEC(42–50), and MAGEA2(338–344), are produced by immunoproteasome but not by the constitutive proteasome [103,174]. Since the identification and characterization of neoantigens is of clinical relevance, modern strategies which combine genomic, proteomic, and immunopeptidomic approaches are a powerful way of discovering novel presented antigens and tumor-associated antigens, paving the way to the novel therapeutic potential [175,176]. As mentioned in the previous section, intermediate proteasomes broaden the repertoire of MHC-I antigenic peptides and, intriguingly, are involved in the production of unique tumor antigens (Figure 2 and Figure 3). In fact, it has been reported that some peptides derived from proteins belonging to the melanoma antigen gene (MAGE) family are generated by intermediate forms. Specifically, the β1i–β2–β5i intermediate produces the MAGE-A10(254–262) peptide, whereas the β1–β2–β5i intermediates generate the MAGE-C2(191–200) and MAGE-A3(271–279) peptides. On the other hand, other antigenic peptides, such as MAGE-A3(114–122) and MAGE-C2(42–50), are produced with equal efficiency by the i20S and intermediate proteasomes [92,177,178]. Moreover, intermediate proteasomes were detected in a number of tumor cells, including lung carcinoma, myeloma, osteosarcoma, and melanoma [66,79,92,177]. Despite this evidence, the role of these forms of proteasome in cancer onset and development is poorly known.
Immunoproteasome Inhibitors
The discovery and application in clinical practice of proteasome inhibitors have revolutionized the therapeutic approach of multiple myeloma, further improving the survival rate of patients with refractory or relapsed forms of this devastating tumor. Currently, three clinically approved proteasome inhibitors are available, namely (i) bortezomib (approved in 2003 and 2004 by the FDA and the EMA, respectively), (ii) carfilzomib (approved in 2012 and 2015 by the FDA and the EMA, respectively), and (iii) the first oral inhibitor, ixazomib (approved in 2015 and 2016 by the FDA and the EMA, respectively). As extensively reviewed elsewhere [15], proteasome inhibition results in multiple deleterious downstream effects in cancer cells, including downregulation of NF-κB signaling, alteration of cytokine secretion, stabilization of p53, and cell cycle arrest. A poorly explored issue that should be clarified concerns the contribution of proteasome inhibition to PD-L1 degradation and its impact on cancer immunotherapy [179]. PD-L1 undergoes ubiquitination and proteasome degradation by different E3 ligases (STUB1, Cullin3, and β-TrCP) [180,181,182]. Cancer cells exhibit the ability to inhibit this process with a consequent impairment of T cell activity. Remarkably, within the tumor microenvironment, TNF-α increase activates NF-kB in cancer cells, leading to the further increase of deubiquitinase CSN5 (COP9 signalosome 5) expression which inhibits PD-L1 degradation, facilitating immune escape of cancer cells. How proteasome inhibitors used in clinical practice impact PD-L1 degradation and, therefore, immune escape of cancer cells mediated by the PD-1/PD-L1 pathway is unclear and represents an important aspect to be investigated.
Despite the beneficial effects, the therapeutic potential of PI is limited by several drawbacks, including the low potency and specificity, the onset of adverse effects, and development of drug resistance [15,183]. Moreover, the main therapeutic benefits are limited to hematological malignancies since proteasome inhibitors are largely ineffective against solid tumors [184,185]. Therefore, there is a growing demand for novel inhibitors with different mechanisms of action and more favorable pharmacological profiles. To achieve this goal, one of the currently explored strategies is the development of selective immunoproteasome inhibitors. In fact, by targeting immunoproteasome subunits, it is expected to achieve a more selective inhibition of proteasomal activity in cancer cells, thereby widening the therapeutic window. Since immunoproteasome plays pivotal roles in antigen presentation, participates in a variety of immune processes and, in general, regulates protein homeostasis, selective inhibitors are expected to bring new therapeutic opportunities for the treatment of various diseases (beyond cancer), spanning from neurodegeneration to inflammatory and autoimmune disorders [186,187]. Therefore, several covalent and noncovalent peptidyl inhibitors selective for immunoproteasome subunits with different structural and biochemical properties have been developed and are currently studied. A number of detailed reviews on immunoproteasome inhibitors are already available [186,188,189,190,191]. Of note, ONX0914 was the first developed selective epoxy ketone peptide which showed an improved activity towards the β5i subunit (IC50 = 5.7 nM) with respect to β5c (IC50 = 54 nM). Starting from the success of ONX0914, a series of β5i-selective inhibitors with comparable activities and increased selectivity have been synthesized, such as PR-924, LU-015i, UK-101, LU-002i, and YU-102 [186]. Importantly, on the ONX0914 backbone, the only immunoproteasome inhibitor synthesized so far is KZR-616 that has entered the stage of clinical trials. In fact, several phase 1/2 trials are ongoing with the aim of evaluating the safety, tolerability, efficacy, pharmacokinetics, and pharmacodynamics of KZR-616 treatment in patients with autoimmune diseases (such as active polymyositis, dermatomyositis, lupus erythematosus) (NCT03393013, NCT04628936, NCT04033926) [186,192]. As already mentioned, a number of preclinical studies evaluated the potential of selective immunoproteasome inhibitors as anti-cancer therapeutic agents. It is important to note that the currently approved proteasome inhibitors target both constitutive proteasome and immunoproteasome subunits and this lack of selectivity, and primarily the inhibition of the constitutive proteasome, is postulated to account for the onset of several severe adverse events [189,193,194]. In fact, the target tissues of proteasome inhibitor toxicities predominantly express constitutive proteasomes; thus, selective inhibition of immunoproteasome might reduce or eliminate most of these toxicities [195,196]. Among the plethora of proposed immunoproteasome inhibitors for cancer treatment, the most promising ones, at least in preclinical models, are PR-924, which specifically inhibits the β5i subunit, and UK-101, that targets β1i. Importantly, both inhibitors reduce the proliferation rate of different tumor types (e.g., leukemia, multiple myeloma, and prostate cancer), including those resistant to bortezomib. Moreover, in mouse models, they are characterized by lower toxicities as compared to other therapeutic approaches and are generally better tolerated [186,197]. Interestingly, it has been demonstrated that exposure of multiple myeloma cells to bortezomib before administration of the immunoproteasome inhibitor ONX914 renders cells more sensitive to i20S inhibition, providing the biological rationale for using a combination therapy that includes selective immunoproteasome inhibitors and pan-proteasome inhibitors [198,199]. More recently, a novel, highly selective i20S inhibitor, M3258, was developed, characterized by a 500-fold greater specificity for the β5i subunit as compared to other proteasome inhibitors and oral bioavailability. In a xenograft model of multiple myeloma, M3258 showed strong target inhibition and in vivo efficacy, with some animals revealing complete tumor eradication. It also demonstrated an attractive overall profile with regard to physicochemical properties, biotransformation, and pharmacokinetics. Daily M3258 administration was associated with a more durable in vivo efficacy in multiple myeloma models compared with intermittent schedule, supporting the hypothesis of the need of continuous LMP7 inhibition to sustain tumor cell apoptosis. Additionally, lower systemic toxicity was observed than for other inhibitors of constitutive proteasome or immunoproteasome [200,201,202]. The robust efficacy of the M3258 inhibitor in preclinical models supported the rationale for a phase 1 clinical trial (https://clinicaltrials.gov/ct2/show/NCT04075721, accessed date: 25 July 2021) to determine the safety, tolerability, pharmacokinetics, pharmacodynamics, and early efficacy signs of this drug as a single agent (dose escalation) and as coadministered with dexamethasone (dose expansion) in patients with relapsed/refractory multiple myeloma [202]. Unfortunately, this trial was discontinued due to the changed therapeutic landscape and the lack of recruitment (see the ClinicalTrials.gov identifier accessed date: 25 July 2021). The overall bulk of described data indicate that the development and introduction in clinical practice of an immunoproteasome-specific inhibitor is largely awaited [189,203,204]. However, the onset of side effects and the extent of off-targets as well as the development of resistance following administration of selective immunoproteasome inhibitors remain unknown [200].
5. Immunoproteasome and Immune Checkpoint Inhibitors: A Glance to the Future?
As mentioned in the previous sections, the production of tumor-associated antigenic peptides recognized by CTLs is a process that starts in the cytoplasm with the degradation of cellular proteins mainly by immunoproteasome (see also Appendix B) [205,206]. Peptide antigens produced by cancer cells are commonly classified into two main groups, namely with high and low specificity. Antigens with high tumor specificity are encoded by viral genes (expressed only in infected cells), mutated genes (generated by the intrinsic instability of cancer cells and hereafter referred to as neoantigens), and cancer germinal genes (expressed as a result of genome-wide demethylation occurring in germinal cells) [99,207]. Moreover, it is known that tumorigenesis is strictly related to genetic diversity and high mutational burden of cancer cells, which increase the possibility of production of neoantigens [168,176,208,209]. This high mutational heterogeneity and neoantigens frequency positively correlate with the response to ICKi therapy. In fact, ICKi are particularly effective against cancers that present with a high burden of mutations and are characterized by DNA mismatch repair deficiency, such as colorectal cancer and NSCLC [210,211,212]. Thus, neoantigens have been proposed to be a prognostic marker for positive clinical outcomes [168,176,208,209]. As a matter of fact, one of the main reasons of acquired resistance to the ICKi therapy seems to be the loss of neoantigens recognized by circulating T cells, suggesting that tumors are “able” to eliminate some mutations during the acquisition of a resistant phenotype [212]. Remarkably, despite the approval of the ICK therapy for cancers characterized by a high mutational burden, a very recent study failed to support the concept that a high mutational burden is a positive biomarker for the ICKi treatment in all solid tumors. In fact, a high mutational burden seems to behave as a predictive marker of response to ICK-based therapies only when the CD8+ infiltration level correlates with the neoantigen load (such as melanoma, lung, bladder cancers, and colon cancer). On the other hand, for tumors in which no relationship between CD8+ levels and the neoantigens load is reported (such as glioma), the high mutational burden failed to predict a positive response to therapy [213]. Thus, it clearly emerges that additional tumor type-specific studies should be performed to unveil the role of this biomarker in the ICKi response. Anyway, the identification of an additional biomarker as well as of non-invasive techniques that monitor the microenvironment before and during the course of the treatment (e.g., imaging-based radiogenomics) are urgently needed for selecting patients who will benefit from immunotherapy [214,215].
In light of the plethora of antigenic peptides produced by the proteasome pathway, it is not surprising that alterations of the proteasome activity and composition could be linked to antigen processing and the ICKi response. Of note, the local production of IFNγ within the tumor microenvironment by infiltrating T lymphocytes positively correlates with clinical outcomes to the ICKi therapy and cancer vaccination in tumors like metastatic melanoma [216,217]. In fact, the upregulation and secretion of chemotactic cytokines (such as IFNγ and TNF-α) increase the recruitment of additional immune cells and alter the tumor microenvironment, stimulating the inhibition of immune exclusion of cancer signatures [218]. Interestingly, some studies show that the primary response to anti-CTLA4 antibodies required a high-level exposure of MHC-1 on the surface of cancer cells at baseline; on the other hand, the response to anti-PD-1 antibodies is linked to a pre-existing IFNγ transcriptome signature [219]. In a recent study, the transcriptome of baseline and on-therapy tumor biopsies from a cohort of 101 patients with advanced melanoma included within the CheckMate 038 study (https://clinicaltrials.gov/ct2/show/NCT01621490, accessed date: 25 July 2021) and treated with nivolumab alone or in combination with ipilimumab (see also Appendix A) has been analyzed [220]. These data, together with in vitro studies, suggest that the immune activation, which follows the administration of ICKi, is associated with the expression of IFNγ response genes, mediated by the increase of T cell infiltration. Among the different sets of genes induced by IFNγ, the most relevant in mediating the response to the therapy are those involved in antigen-presenting machinery [220]. Thus, a combination therapy of ICKi with agents that independently trigger the intratumoral production of IFNγ could become meaningful [220,221,222,223]. Consistently with the role of IFNγ signature in driving ICKi response, it has been proposed that upregulation of immunoproteasome subunits in tumor cells might be also involved in this process [216,220]. In fact, the local production of IFNγ induced by ICKi, which are routinely used in the treatment of advanced/metastatic melanoma, leads to the modulation of proteasome composition, thus inducing the generation of antigenic peptides [216,217]. Consistent with this observation, the expression of immunoproteasome subunits β1i and β5i was associated with a better prognosis in the case of tumors with a high mutational burden (i.e., melanoma and NSCLC) and positively correlated with the response to ICKi and the survival rate of patients [170,216,224]. Thus, it has been proposed that at least in some tumors the expression level of the β1i and β5i subunits might represent a predictive marker of response to ICKi [216,225]. As a matter of fact, the overexpression of these subunits is linked to longer survival and improved response to the ICKi therapy in melanoma patients and the proposed mechanism underlying this connection consists in enhanced reactivity of TILs toward melanoma cells as a consequence of an altered repertoire of the presented antigens [216]. Importantly, it has also been reported that immunoproteasome subunit overexpression sometimes occurs regardless of IFNγ and T cell infiltration, suggesting that these subunits should be independent prognostic biomarker with respect to the IFNγ level and the rate of T cell infiltration in the context of tumor microenvironment [216]. Thus, this last observation opens up a possible and yet poorly investigated scenario concerning the role of immunoproteasome in mediating the ICKi response independently of the IFNγ pathway. Therefore, even though many crucial points deserve to be clarified, it clearly emerges that immunoproteasome expression seems to be an important predictive marker in colorectal cancer, melanoma, and NSCLC, for which the ICKi therapy has proven to be effective. Thus, an intriguing therapeutic strategy that should be explored in the near future is the combination of ICKi and drugs that directly modulate immunoproteasome activity and/or induce immunoproteasome expression in order to increase its pool inside the cells.
6. Conclusions
Cancer cells are often more dependent on a proper integrity and functionality of UPS than nonmalignant cells due to the rapid proliferation rate, increased metabolic activity, and continuous exposure to a variety of extrinsic stress perturbations (such as nutrient deprivation, hypoxia, and acidosis) under which cancer cells live. All these conditions lead to a decrease in protein quality control and make UPS a suitable target for cancer therapy [218,219]. Accordingly, a number of proteasome based-strategies have been proposed, spanning from (i) inhibition of proteasome proteolytic activity, (ii) modulation of the abundance of proteasome regulatory particles (i.e., 19S or PA28) and of their interaction with the 20S to (iii) modulation of the activity of enzymes involved in proteasome subunit post-translational modifications and (iv) interference with transport of natural low-molecular-weight proteasome activators (e.g., spermine) [62,226,227,228,229,230,231,232]. Additionally, a series of strategies targeting the ubiquitination cascade have been studied. One of the most intriguing and novel therapeutic approaches involves the use of PROTAC (proteolysis-targeting chimeric molecules), which are hetero-bifunctional molecules that recruit specific target proteins to the E3 ligase, thus inducing the increase of target ubiquitination and degradation. This strategy has already been applied to the degradation of a number of selected targets [233,234,235]. However, even though promising, it is still in its infancy for application to immunotherapy. On the other hand, the most deeply investigated strategy consists in the use of broad-specificity inhibitors of the 20S activity, like bortezomib, carfilzomib, and ixazomib that inhibit proteolysis of all the proteasome forms present in different cells [15,62]. The second approach encompasses the identification of specific inhibitors targeting inducible tissue-specific forms of proteasome, mainly the i20S, which is involved in the production of antigenic peptides. Though this mechanism is well-known, how such inhibitors might either decrease or stimulate cancer cell recognition by T cells is debated (see Section 4) [62]. As a matter of fact, a challenging but poorly investigated issue is the precise in vivo impact of broad-specificity 20S proteasome inhibitors commonly used in clinical practice on antigen presentation by cancer cells [99], and it should deserve more attention. A key question for improving the efficacy and safety profile of immunotherapy includes the identification of the most appropriate strategy to optimize the antigenic peptide repertoire of the tumor required for an efficient immune response [99]. Notably, some recent data suggest that enhanced immunoproteasome activity might play an important role in the response of melanoma to ICKi [216]. It seems to indicate that, at least in some tumors, the more efficient strategy could be to “enhance” immunoproteasome expression and activity. Thus, the choice of the best strategy whether to inhibit or activate immunoproteasome should take into consideration the biological features of the specific tumor that has to be treated. Even though additional in vitro and in vivo investigation needs to be performed, the current evidence indicates that selective modulation of proteasome activity might have a role in improving the outcome of the ICKi or other immunotherapeutic approaches.
Acknowledgments
This work was supported by the Italian Ministry of University and Research (PRIN 2017SNRXH3), the Italian Association for Cancer Research (AIRC) (project IG 2017—ID. 20353—PI to Grazia Graziani), the Italian Ministry of Health (RC18-2638151; to P.M.L.), POR 2014-2020 “Progetto gruppi di ricercar” A0375-2020-36583, Department of Systems Medicine, University of Rome Tor Vergata. Special thanks are due to Omikron Italia for a liberal donation and to Fondazione Roma. The reported study was supported by the Russian Scientific Foundation, project No. 19-14-00262 (A.A.B.J. and A.A.K.). A.A.B.J. would like to acknowledge personal fellowship MD-5902.2021.1.4.
Appendix A. Spotlights on Immune Checkpoints
Several negative regulators of T cell activation act as “checkpoint molecules” to finely tune the immune response and regulate their hyperactivation [1]. CTLA4 and the PD-1–PD-L1 axis, which exert their biological function at distinct sites and with different timing during the T cell lifespan, are the most studied and potent examples of immune checkpoint molecules [1,4]. CTLA4 is operative during the priming phase of T cells activation. In resting naïve T cells, CTLA4, which is a costimulatory receptor CD28 homolog with a higher affinity for the common ligand B7, expressed by antigen-presenting cells (APC), localizes primarily in the intracellular compartment. Following antigen activation, CTLA4 is strongly induced, with a peak of expression within 2–3 days from the stimulatory signal, and translocated from vesicles to the T cell surface. The binding between CTLA4 and B7 transmits an inhibitory signal inside T cells that reduces the proliferation rate and survival of activated T cells as well as cytokines production. CTLA4 exerts its inhibitor effect predominantly into lymphoid organs [126,236,237,238]. On the other hand, PD-1 acts during the effector phase, turning off cell activation mainly in the peripheral tissues. It is absent in naïve T cells, whereas, after TCR stimulation, it is expressed on the T cell surface, where it binds the B7 homologues PD-L1 and PD-L2, which are constitutively expressed by APCs. After PD-1–PD-L1/PD-L2 engagement, a negative costimulatory signal is transmitted, tempering TCR cascade [239,240,241,242]. PD-L1 can also be expressed by tumor cells and its upregulation may represent a possible mechanism of immune evasion. The two described pathways (CTLA4 and PD-1–PD-L1 axis) cooperate in turning off T cell activation and antibodies directed against these molecules hamper the inhibitory signal, favoring T lymphocytes reactivation [1,5]. The recognition of CTLA4 and the PD-1–PD-L1 axis as a negative regulator of T cell activation gave rise to the idea that inhibition of their pathways could be useful for cancer treatment. In 2011, ipilimumab became the first clinically approved monoclonal antibody blocking an immune checkpoint molecule (CTLA4), followed, starting from 2014, by monoclonal antibodies targeting PD-1 (pembrolizumab, nivolumab, and cemiplimab) and PD-L1 (atezolizumab, avelumab, and durvalumab). ICKi are now regarded as the most commonly prescribed anticancer therapy, as single agents or in combination with chemotherapy, in the first or second line of treatment of about 50 cancer types [243,244]. Despite the undoubted therapeutic efficacy, clinical benefit is limited to subsets of patients for each cancer type, and, in responder patients, acquired resistance may develop [5]. Therefore, there is an increasing interest in the identification of predictive biomarkers of response and development of add-on therapies that may enhance ICKi efficacy (see also the main text) [5].
Appendix B. Thymoproteasome
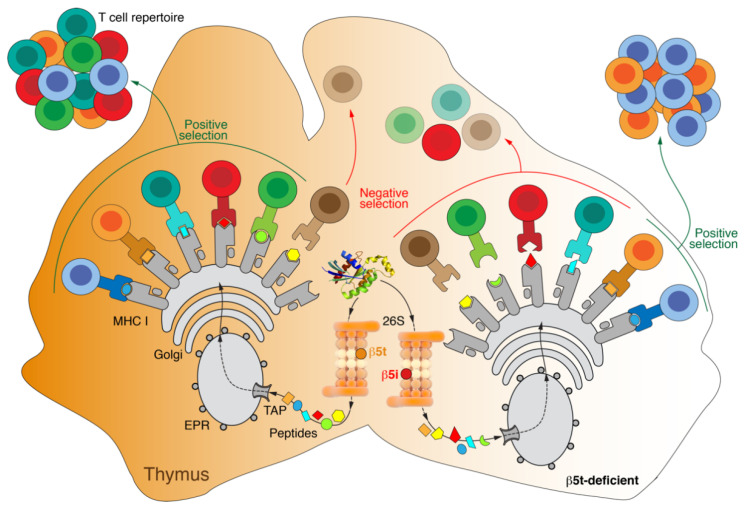
Thymoproteasome is a noncanonical form of proteasome whose expression is restricted to the cortex of the thymus, a primary lymphoid organ that screens out T cell receptor specificity. Within the mechanisms governing the central tolerance, thymoproteasome activity is supposed to generate antigenic peptides required for the positive selection of T cell clones. This process allows immature CD4 and CD8 double-positive T cells expressing T cell receptors (TCRs), which bind with low affinity to HLA molecules loaded with self-peptides, to differentiate into mature single-positive cells [245,246]. Conversely, the negative selection that occurs at the cortical–medullary junction of the gland and, conceptually, is the reverse process of positive selection, refers to the clearance of double-positive or single-positive T cells expressing TCRs displaying high affinity for self-peptide–MHC complexes. However the negative selection appears to be achieved through a repertoire of self-peptides generated by the proteolytic activity of immunoproteasome and constitutive proteasomes [203,247,248,249,250,251]. From the structural point of view, thymoproteasome is equipped with the inducible immune subunits β1i and β2i and a distinctive catalytic subunit β5 (designated β5t, where “t” stands for thymus) (PSMB11), which is the homolog of the constitutive β5 and the inducible β5i [62,252,253] (Figure 1); β5t is selectively expressed and incorporated together with β1i and β2i into the newly assembling 20S in cortical epithelial cells. In accordance with a less relevant role of thymoproteasome in negative selection, the epithelial cells populating the thymus medulla do not express appreciable levels of these specific subunits. The transcription of thymoproteasome subunit genes is carried out by the Foxn1 transcription factor which is generally involved in the regulation of the key target genes for T cell development [253,254,255]. Regarding the enzymological properties, compared with the c20S and i20S, thymoproteasome is characterized by a robust drop of chymotrypsin-like activity (60–70% decrease) but not of trypsin-like and caspase-like activities, at least on specific fluorogenic substrates. In fact, the catalytic properties of β5t seem to be quite different from those of β5i and β5t. The substrate-binding S1 pocket of β5i and β5t mainly accommodates hydrophobic amino acids, which are preferred cleavage sites for the chymotrypsin-like activity, whereas β5t shows preference for hydrophilic residues. This biochemical feature endows thymoproteasome with a specific pattern of endopeptidase proteolysis which releases a repertoire of peptide fragments that show weak binding to class I HLA molecules. Thus, the affinity model for positive selection of T cells assumes that thymoproteasome produces a unique class of MHC-binding peptides with low affinity for TCR which is the driving force of the whole process [256]. Accordingly, the repertoire of self-antigens differs between the cortex and the medulla, where class I-binding peptides are mainly released by immunoproteasome and constitutive proteasome, as anticipated before. The fact that different thymic compartments display distinct sets of peptides seems to increase the number of surviving CD8+ clones, avoiding overlaps in the positive and negative selection [203,257,258] (Figure A1). In this regard, transgenic mice which express thymoproteasome both in the cortex and the medulla show an increased rate of negative selection. Nonetheless, negative selection is almost blocked in the mice expressing only the c20S [258,259,260]. Furthermore, β5t−/− mice appear to develop normally and have a thymus of a typical size, with the cortico–medullary structure undisturbed. However, these animals express immunoproteasome subunits but not those of thymoproteasome in all the thymus layers and show aberrant TCR responsiveness, reduction in the CD8+ T cells number (approximately by 80%), and altered response to infection. Conversely, no defects in the CD4+ T cells selection have been observed [249,250,259,261]. Additional insights on thymoproteasome come from studies on β5i−/− mice, which express an aberrant level of thymoproteasome in antigen-presenting cells (APCs), including dendritic cells and medullar epithelial cells. These animals develop several metabolic abnormalities (obesity, hepatic steatosis, increase in the adipose differentiation-related protein). Thus, extracortical expression of β5t induces autoreactive responsiveness of CD8+ T cells, suggesting that specific cortical expression is fundamental not only for the correct positive selection, but also for the maintenance of peripheral tolerance [259,260,262]. Recent studies support the hypothesis that thymoproteasome alterations are involved in the onset and progression of human diseases, even though additional preclinical and clinical data are required to address its specific pathogenic role. Immunohistochemistry analyses show that β5t is overexpressed in B and AB thymomas, but not in type A thymomas and thymic carcinoma. Thus, it has been suggested that β5t could be used as a diagnostic biomarker to discriminate between the different types of thymomas and thymic carcinoma. Moreover, β5t has been proposed as a reliable marker for rare cervical ectopic thymomas. However, the precise diagnostic and prognostic value of this marker is still poorly understood [262,263,264,265]. Unlike immunoproteasome and constitutive proteasome, the involvement of thymoproteasome in extrathymic diseases not related with cancer is poorly exlpored. Recently, it has been shown that patients with Down syndrome have a decreased β5t expression, which seems to contribute to the elevated susceptibility to infections these subjects suffer from [251,263,266]. Furthermore, a single-nucleotide polymorphism that replaces glycine with serine at the 49th amino acid residue (G49S) is associated with Sjogren’s syndrome. These amino acid substitutions, which occur at a significantly high frequency in the Japanese population, partially prevent the carboxyl terminus cleavage that turns the β5t proproteins into the catalytic active form [267,268].
Figure A1.
Thymoproteasome in the positive selection of MHC-I-specific T cells. A unique catalytic subunit, β5t, which, together with β1i and β2i, is part of the 20S proteasome instead of β5 or β5i, forms a thymoproteasome (in orange) with reduced chymotrypsin-like activity. This type of proteasome plays an important role in the positive selection of MHC-I-specific T cells, as a result of which the final repertoire of cytotoxic lymphocytes is significantly expanded.
Author Contributions
Conceptualization, G.R.T., D.S. and G.G.; writing—original draft preparation, G.R.T., D.S., A.A.B.J., A.A.K., F.O., P.M.L. and G.G.; writing—review and editing G.R.T., D.S., A.A.B.J., A.A.K., F.O., P.M.L. and G.G.; supervision G.R.T., D.S., G.G. and S.M.; funding acquisition D.S. and F.O. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare that there are no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Waldman A.D., Fritz J.M., Lenardo M.J. A Guide to Cancer Immunotherapy: From T Cell Basic Science to Clinical Practice. Nat. Rev. Immunol. 2020;20:651–668. doi: 10.1038/s41577-020-0306-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Emens L.A., Ascierto P.A., Darcy P.K., Demaria S., Eggermont A.M.M., Redmond W.L., Seliger B., Marincola F.M. Cancer Immunotherapy: Opportunities and Challenges in the Rapidly Evolving Clinical Landscape. Eur. J. Cancer. 2017;81:116–129. doi: 10.1016/j.ejca.2017.01.035. [DOI] [PubMed] [Google Scholar]
- 3.Fife B.T., Bluestone J.A. Control of Peripheral T-Cell Tolerance and Autoimmunity via the CTLA-4 and PD-1 Pathways. Immunol. Rev. 2008;224:166–182. doi: 10.1111/j.1600-065X.2008.00662.x. [DOI] [PubMed] [Google Scholar]
- 4.Decker W.K., da Silva R.F., Sanabria M.H., Angelo L.S., Guimarães F., Burt B.M., Kheradmand F., Paust S. Cancer Immunotherapy: Historical Perspective of a Clinical Revolution and Emerging Preclinical Animal Models. Front. Immunol. 2017;8:829. doi: 10.3389/fimmu.2017.00829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tundo G.R., Sbardella D., Lacal P.M., Graziani G., Marini S. On the Horizon: Targeting Next-Generation Immune Checkpoints for Cancer Treatment. Chemotherapy. 2019;64:62–80. doi: 10.1159/000500902. [DOI] [PubMed] [Google Scholar]
- 6.Fritz J.M., Lenardo M.J. Development of Immune Checkpoint Therapy for Cancer. J. Exp. Med. 2019;216:1244–1254. doi: 10.1084/jem.20182395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kumar V., Chaudhary N., Garg M., Floudas C.S., Soni P., Chandra A.B. Current Diagnosis and Management of Immune Related Adverse Events (IrAEs) Induced by Immune Checkpoint Inhibitor Therapy. Front. Pharm. 2017;8:49. doi: 10.3389/fphar.2017.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang S., He Z., Wang X., Li H., Liu X.-S. Antigen Presentation and Tumor Immunogenicity in Cancer Immunotherapy Response Prediction. Elife. 2019;8:e49020. doi: 10.7554/eLife.49020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chowell D., Morris L.G.T., Grigg C.M., Weber J.K., Samstein R.M., Makarov V., Kuo F., Kendall S.M., Requena D., Riaz N., et al. Patient HLA Class I Genotype Influences Cancer Response to Checkpoint Blockade Immunotherapy. Science. 2018;359:582–587. doi: 10.1126/science.aao4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zaretsky J.M., Garcia-Diaz A., Shin D.S., Escuin-Ordinas H., Hugo W., Hu-Lieskovan S., Torrejon D.Y., Abril-Rodriguez G., Sandoval S., Barthly L., et al. Mutations Associated with Acquired Resistance to PD-1 Blockade in Melanoma. [(accessed on 21 June 2021)]. Available online: https://www.nejm.org/doi/10.1056/NEJMoa1604958. [DOI] [PMC free article] [PubMed]
- 11.Labani-Motlagh A., Ashja-Mahdavi M., Loskog A. The Tumor Microenvironment: A Milieu Hindering and Obstructing Antitumor Immune Responses. Front. Immunol. 2020;11:940. doi: 10.3389/fimmu.2020.00940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gubin M.M., Zhang X., Schuster H., Caron E., Ward J.P., Noguchi T., Ivanova Y., Hundal J., Arthur C.D., Krebber W.-J., et al. Checkpoint Blockade Cancer Immunotherapy Targets Tumour-Specific Mutant Antigens. Nature. 2014;515:577–581. doi: 10.1038/nature13988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seliger B., Maeurer M.J., Ferrone S. Antigen-Processing Machinery Breakdown and Tumor Growth. Immunol. Today. 2000;21:455–464. doi: 10.1016/S0167-5699(00)01692-3. [DOI] [PubMed] [Google Scholar]
- 14.Vizcaíno J.A., Kubiniok P., Kovalchik K.A., Ma Q., Duquette J.D., Mongrain I., Deutsch E.W., Peters B., Sette A., Sirois I., et al. The Human Immunopeptidome Project: A Roadmap to Predict and Treat Immune Diseases. Mol. Cell Proteom. 2020;19:31–49. doi: 10.1074/mcp.R119.001743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tundo G.R., Sbardella D., Santoro A.M., Coletta A., Oddone F., Grasso G., Milardi D., Lacal P.M., Marini S., Purrello R., et al. The Proteasome as a Druggable Target with Multiple Therapeutic Potentialities: Cutting and Non-Cutting Edges. Pharm. Ther. 2020;213:107579. doi: 10.1016/j.pharmthera.2020.107579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cascio P. PA28γ: New Insights on an Ancient Proteasome Activator. Biomolecules. 2021;11:228. doi: 10.3390/biom11020228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sala A.J., Bott L.C., Morimoto R.I. Shaping Proteostasis at the Cellular, Tissue, and Organismal Level. J. Cell Biol. 2017;216:1231–1241. doi: 10.1083/jcb.201612111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klaips C.L., Jayaraj G.G., Hartl F.U. Pathways of Cellular Proteostasis in Aging and Disease. J. Cell Biol. 2018;217:51–63. doi: 10.1083/jcb.201709072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kudriaeva A.A., Sokolov A.V., Belogurov A.A.J. Stochastics of Degradation: The Autophagic-Lysosomal System of the Cell. Acta Nat. 2020;12:18–32. doi: 10.32607/actanaturae.10936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kudriaeva A., Kuzina E.S., Zubenko O., Smirnov I.V., Belogurov A. Charge-Mediated Proteasome Targeting. FASEB J. 2019;33:6852–6866. doi: 10.1096/fj.201802237R. [DOI] [PubMed] [Google Scholar]
- 21.Glickman M.H., Ciechanover A. The Ubiquitin-Proteasome Proteolytic Pathway: Destruction for the Sake of Construction. Physiol Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- 22.Kunjappu M.J., Hochstrasser M. Assembly of the 20S Proteasome. Biochim. Biophys. Acta. 2014;1843:2–12. doi: 10.1016/j.bbamcr.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lander G.C., Estrin E., Matyskiela M.E., Bashore C., Nogales E., Martin A. Complete Subunit Architecture of the Proteasome Regulatory Particle. Nature. 2012;482:186–191. doi: 10.1038/nature10774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scheffner M., Nuber U., Huibregtse J.M. Protein Ubiquitination Involving an E1-E2-E3 Enzyme Ubiquitin Thioester Cascade. Nature. 1995;373:81–83. doi: 10.1038/373081a0. [DOI] [PubMed] [Google Scholar]
- 25.Leestemaker Y., Ovaa H. Tools to Investigate the Ubiquitin Proteasome System. Drug Discov. Today Technol. 2017;26:25–31. doi: 10.1016/j.ddtec.2017.11.006. [DOI] [PubMed] [Google Scholar]
- 26.Pickart C.M. Mechanisms Underlying Ubiquitination. Annu. Rev. Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 27.Windheim M., Peggie M., Cohen P. Two Different Classes of E2 Ubiquitin-Conjugating Enzymes Are Required for the Mono-Ubiquitination of Proteins and Elongation by Polyubiquitin Chains with a Specific Topology. Biochem. J. 2008;409:723–729. doi: 10.1042/BJ20071338. [DOI] [PubMed] [Google Scholar]
- 28.Bellia F., Lanza V., García-Viñuales S., Ahmed I.M.M., Pietropaolo A., Iacobucci C., Malgieri G., D’Abrosca G., Fattorusso R., Nicoletti V.G., et al. Ubiquitin Binds the Amyloid β Peptide and Interferes with Its Clearance Pathways. Chem. Sci. 2019;10:2732–2742. doi: 10.1039/C8SC03394C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ciechanover A. The Unravelling of the Ubiquitin System. Nat. Rev. Mol. Cell Biol. 2015;16:322–324. doi: 10.1038/nrm3982. [DOI] [PubMed] [Google Scholar]
- 30.Zheng N., Shabek N. Ubiquitin Ligases: Structure, Function, and Regulation. Annu. Rev. Biochem. 2017;86:129–157. doi: 10.1146/annurev-biochem-060815-014922. [DOI] [PubMed] [Google Scholar]
- 31.Berndsen C.E., Wolberger C. New Insights into Ubiquitin E3 Ligase Mechanism. Nat. Struct. Mol. Biol. 2014;21:301–307. doi: 10.1038/nsmb.2780. [DOI] [PubMed] [Google Scholar]
- 32.Komander D., Rape M. The Ubiquitin Code. Annu. Rev. Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- 33.Deshaies R.J., Joazeiro C.A.P. RING Domain E3 Ubiquitin Ligases. Annu. Rev. Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 34.Kudriaeva A.A., Livneh I., Baranov M.S., Ziganshin R.H., Tupikin A.E., Zaitseva S.O., Kabilov M.R., Ciechanover A., Belogurov A.A., Jr. In-depth characterization of ubiquitin turnover in mammalian cells by fluorescence tracking. Cell Chem. Biol. 2021;28:1192–1205.e9. doi: 10.1016/j.chembiol.2021.02.009. [DOI] [PubMed] [Google Scholar]
- 35.Yuan T., Yan F., Ying M., Cao J., He Q., Zhu H., Yang B. Inhibition of Ubiquitin-Specific Proteases as a Novel Anticancer Therapeutic Strategy. Front. Pharm. 2018;9:1080. doi: 10.3389/fphar.2018.01080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oikawa D., Sato Y., Ito H., Tokunaga F. Linear Ubiquitin Code: Its Writer, Erasers, Decoders, Inhibitors, and Implications in Disorders. Int. J. Mol. Sci. 2020;21:3381. doi: 10.3390/ijms21093381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pfoh R., Lacdao I.K., Saridakis V. Deubiquitinases and the New Therapeutic Opportunities Offered to Cancer. Endocr. Relat. Cancer. 2015;22:T35–T54. doi: 10.1530/ERC-14-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Groll M., Ditzel L., Löwe J., Stock D., Bochtler M., Bartunik H.D., Huber R. Structure of 20S Proteasome from Yeast at 2.4 A Resolution. Nature. 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- 39.Groll M., Bajorek M., Köhler A., Moroder L., Rubin D.M., Huber R., Glickman M.H., Finley D. A Gated Channel into the Proteasome Core Particle. Nat. Struct. Biol. 2000;7:1062–1067. doi: 10.1038/80992. [DOI] [PubMed] [Google Scholar]
- 40.Tanaka K. The Proteasome: Overview of Structure and Functions. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2009;85:12–36. doi: 10.2183/pjab.85.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Groll M., Huber R. Substrate Access and Processing by the 20S Proteasome Core Particle. Int. J. Biochem. Cell Biol. 2003;35:606–616. doi: 10.1016/S1357-2725(02)00390-4. [DOI] [PubMed] [Google Scholar]
- 42.Bajorek M., Glickman M.H. Keepers at the Final Gates: Regulatory Complexes and Gating of the Proteasome Channel. Cell Mol. Life Sci. 2004;61:1579–1588. doi: 10.1007/s00018-004-4131-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ruschak A.M., Religa T.L., Breuer S., Witt S., Kay L.E. The Proteasome Antechamber Maintains Substrates in an Unfolded State. Nature. 2010;467:868–871. doi: 10.1038/nature09444. [DOI] [PubMed] [Google Scholar]
- 44.Finley D., Prado M.A. The Proteasome and Its Network: Engineering for Adaptability. Cold Spring Harb. Perspect. Biol. 2020;12 doi: 10.1101/cshperspect.a033985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmidt M., Finley D. Regulation of Proteasome Activity in Health and Disease. Biochim. Biophys. Acta. 2014;1843:13–25. doi: 10.1016/j.bbamcr.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Choi W.H., de Poot S.A.H., Lee J.H., Kim J.H., Han D.H., Kim Y.K., Finley D., Lee M.J. Open-Gate Mutants of the Mammalian Proteasome Show Enhanced Ubiquitin-Conjugate Degradation. Nat. Commun. 2016;7:10963. doi: 10.1038/ncomms10963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Finley D., Chen X., Walters K.J. Gates, Channels, and Switches: Elements of the Proteasome Machine. Trends Biochem. Sci. 2016;41:77–93. doi: 10.1016/j.tibs.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Budenholzer L., Cheng C.L., Li Y., Hochstrasser M. Proteasome Structure and Assembly. J. Mol. Biol. 2017;429:3500–3524. doi: 10.1016/j.jmb.2017.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pathare G.R., Nagy I., Bohn S., Unverdorben P., Hubert A., Körner R., Nickell S., Lasker K., Sali A., Tamura T., et al. The Proteasomal Subunit Rpn6 Is a Molecular Clamp Holding the Core and Regulatory Subcomplexes Together. Proc. Natl. Acad. Sci. USA. 2012;109:149–154. doi: 10.1073/pnas.1117648108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pathare G.R., Nagy I., Śledź P., Anderson D.J., Zhou H.-J., Pardon E., Steyaert J., Förster F., Bracher A., Baumeister W. Crystal Structure of the Proteasomal Deubiquitylation Module Rpn8-Rpn11. Proc. Natl. Acad. Sci. USA. 2014;111:2984–2989. doi: 10.1073/pnas.1400546111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martinez-Fonts K., Davis C., Tomita T., Elsasser S., Nager A.R., Shi Y., Finley D., Matouschek A. The Proteasome 19S Cap and Its Ubiquitin Receptors Provide a Versatile Recognition Platform for Substrates. Nat. Commun. 2020;11:477. doi: 10.1038/s41467-019-13906-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marshall R.S., Vierstra R.D. Dynamic Regulation of the 26S Proteasome: From Synthesis to Degradation. Front. Mol. Biosci. 2019;6:40. doi: 10.3389/fmolb.2019.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kravtsova-Ivantsiv Y., Cohen S., Ciechanover A. Modification by Single Ubiquitin Moieties Rather than Polyubiquitination Is Sufficient for Proteasomal Processing of the P105 NF-KappaB Precursor. Mol. Cell. 2009;33:496–504. doi: 10.1016/j.molcel.2009.01.023. [DOI] [PubMed] [Google Scholar]
- 54.Shabek N., Herman-Bachinsky Y., Buchsbaum S., Lewinson O., Haj-Yahya M., Hejjaoui M., Lashuel H.A., Sommer T., Brik A., Ciechanover A. The Size of the Proteasomal Substrate Determines Whether Its Degradation Will Be Mediated by Mono- or Polyubiquitylation. Mol. Cell. 2012;48:87–97. doi: 10.1016/j.molcel.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 55.Yau R., Rape M. The Increasing Complexity of the Ubiquitin Code. Nat. Cell Biol. 2016;18:579–586. doi: 10.1038/ncb3358. [DOI] [PubMed] [Google Scholar]
- 56.Ju D., Xie Y. Proteasomal Degradation of RPN4 via Two Distinct Mechanisms, Ubiquitin-Dependent and -Independent. J. Biol. Chem. 2004;279:23851–23854. doi: 10.1074/jbc.C400111200. [DOI] [PubMed] [Google Scholar]
- 57.Baugh J.M., Viktorova E.G., Pilipenko E.V. Proteasomes Can Degrade a Significant Proportion of Cellular Proteins Independent of Ubiquitination. J. Mol. Biol. 2009;386:814–827. doi: 10.1016/j.jmb.2008.12.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Belogurov A., Kuzina E., Kudriaeva A., Kononikhin A., Kovalchuk S., Surina Y., Smirnov I., Lomakin Y., Bacheva A., Stepanov A., et al. Ubiquitin-Independent Proteosomal Degradation of Myelin Basic Protein Contributes to Development of Neurodegenerative Autoimmunity. FASEB J. 2015;29:1901–1913. doi: 10.1096/fj.14-259333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Belogurov A., Kudriaeva A., Kuzina E., Smirnov I., Bobik T., Ponomarenko N., Kravtsova-Ivantsiv Y., Ciechanover A., Gabibov A. Multiple Sclerosis Autoantigen Myelin Basic Protein Escapes Control by Ubiquitination during Proteasomal Degradation. J. Biol. Chem. 2014;289:17758–17766. doi: 10.1074/jbc.M113.544247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Raynes R., Pomatto L.C.D., Davies K.J.A. Degradation of Oxidized Proteins by the Proteasome: Distinguishing between the 20S, 26S, and Immunoproteasome Proteolytic Pathways. Mol. Asp. Med. 2016;50:41–55. doi: 10.1016/j.mam.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shringarpure R., Grune T., Mehlhase J., Davies K.J.A. Ubiquitin Conjugation Is Not Required for the Degradation of Oxidized Proteins by Proteasome. J. Biol. Chem. 2003;278:311–318. doi: 10.1074/jbc.M206279200. [DOI] [PubMed] [Google Scholar]
- 62.Morozov A.V., Karpov V.L. Proteasomes and Several Aspects of Their Heterogeneity Relevant to Cancer. Front. Oncol. 2019;9:761. doi: 10.3389/fonc.2019.00761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sbardella D., Tundo G.R., Sciandra F., Bozzi M., Gioia M., Ciaccio C., Tarantino U., Brancaccio A., Coletta M., Marini S. Proteasome Activity Is Affected by Fluctuations in Insulin-Degrading Enzyme Distribution. PLoS ONE. 2015;10:e0132455. doi: 10.1371/journal.pone.0132455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tundo G.R., Sbardella D., Ciaccio C., Grasso G., Gioia M., Coletta A., Polticelli F., Di Pierro D., Milardi D., Van Endert P., et al. Multiple Functions of Insulin-Degrading Enzyme: A Metabolic Crosslight? Crit. Rev. Biochem. Mol. Biol. 2017;52:554–582. doi: 10.1080/10409238.2017.1337707. [DOI] [PubMed] [Google Scholar]
- 65.Sbardella D., Tundo G.R., Coletta A., Marcoux J., Koufogeorgou E.I., Ciaccio C., Santoro A.M., Milardi D., Grasso G., Cozza P., et al. The Insulin-Degrading Enzyme Is an Allosteric Modulator of the 20S Proteasome and a Potential Competitor of the 19S. Cell Mol. Life Sci. 2018;75:3441–3456. doi: 10.1007/s00018-018-2807-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fabre B., Lambour T., Garrigues L., Ducoux-Petit M., Amalric F., Monsarrat B., Burlet-Schiltz O., Bousquet-Dubouch M.-P. Label-Free Quantitative Proteomics Reveals the Dynamics of Proteasome Complexes Composition and Stoichiometry in a Wide Range of Human Cell Lines. J. Proteome Res. 2014;13:3027–3037. doi: 10.1021/pr500193k. [DOI] [PubMed] [Google Scholar]
- 67.Dahlmann B. Mammalian Proteasome Subtypes: Their Diversity in Structure and Function. Arch. Biochem. Biophys. 2016;591:132–140. doi: 10.1016/j.abb.2015.12.012. [DOI] [PubMed] [Google Scholar]
- 68.Kniepert A., Groettrup M. The Unique Functions of Tissue-Specific Proteasomes. Trends. Biochem. Sci. 2014;39:17–24. doi: 10.1016/j.tibs.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 69.Qian M.-X., Pang Y., Liu C.H., Haratake K., Du B.-Y., Ji D.-Y., Wang G.-F., Zhu Q.-Q., Song W., Yu Y., et al. Acetylation-Mediated Proteasomal Degradation of Core Histones during DNA Repair and Spermatogenesis. Cell. 2013;153:1012–1024. doi: 10.1016/j.cell.2013.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Uechi H., Hamazaki J., Murata S. Characterization of the Testis-Specific Proteasome Subunit A4s in Mammals. J. Biol. Chem. 2014;289:12365–12374. doi: 10.1074/jbc.M114.558866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Khor B., Bredemeyer A.L., Huang C.-Y., Turnbull I.R., Evans R., Maggi L.B., White J.M., Walker L.M., Carnes K., Hess R.A., et al. Proteasome Activator PA200 Is Required for Normal Spermatogenesis. Mol. Cell Biol. 2006;26:2999–3007. doi: 10.1128/MCB.26.8.2999-3007.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ustrell V., Pratt G., Gorbea C., Rechsteiner M. Purification and Assay of Proteasome Activator PA200. Methods Enzym. 2005;398:321–329. doi: 10.1016/S0076-6879(05)98026-9. [DOI] [PubMed] [Google Scholar]
- 73.Toste Rêgo A., da Fonseca P.C.A. Characterization of Fully Recombinant Human 20S and 20S-PA200 Proteasome Complexes. Mol. Cell. 2019;76:138–147.e5. doi: 10.1016/j.molcel.2019.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Guan H., Wang Y., Yu T., Huang Y., Li M., Saeed A.F.U.H., Perčulija V., Li D., Xiao J., Wang D., et al. Cryo-EM Structures of the Human PA200 and PA200–20S Complex Reveal Regulation of Proteasome Gate Opening and Two PA200 Apertures. PLoS Biol. 2020;18:e3000654. doi: 10.1371/journal.pbio.3000654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang X., Meul T., Meiners S. Exploring the Proteasome System: A Novel Concept of Proteasome Inhibition and Regulation. Pharm. Ther. 2020;211:107526. doi: 10.1016/j.pharmthera.2020.107526. [DOI] [PubMed] [Google Scholar]
- 76.Bruggeman J.W., Koster J., Lodder P., Repping S., Hamer G. Massive Expression of Germ Cell-Specific Genes Is a Hallmark of Cancer and a Potential Target for Novel Treatment Development. Oncogene. 2018;37:5694–5700. doi: 10.1038/s41388-018-0357-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gómez-H L., Felipe-Medina N., Condezo Y.B., Garcia-Valiente R., Ramos I., Suja J.A., Barbero J.L., Roig I., Sánchez-Martín M., de Rooij D.G., et al. The PSMA8 Subunit of the Spermatoproteasome Is Essential for Proper Meiotic Exit and Mouse Fertility. PLoS Genet. 2019;15:e1008316. doi: 10.1371/journal.pgen.1008316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cascio P. PA28αβ: The Enigmatic Magic Ring of the Proteasome? Biomolecules. 2014;4:566–584. doi: 10.3390/biom4020566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Morozov A.V., Karpov V.L. Biological Consequences of Structural and Functional Proteasome Diversity. Heliyon. 2018;4:e00894. doi: 10.1016/j.heliyon.2018.e00894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cascio P., Call M., Petre B.M., Walz T., Goldberg A.L. Properties of the Hybrid Form of the 26S Proteasome Containing Both 19S and PA28 Complexes. EMBO J. 2002;21:2636–2645. doi: 10.1093/emboj/21.11.2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Thibaudeau T.A., Smith D.M. A Practical Review of Proteasome Pharmacology. Pharm. Rev. 2019;71:170–197. doi: 10.1124/pr.117.015370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Martinez C.K., Monaco J.J. Homology of Proteasome Subunits to a Major Histocompatibility Complex-Linked LMP Gene. Nature. 1991;353:664–667. doi: 10.1038/353664a0. [DOI] [PubMed] [Google Scholar]
- 83.Rock K.L., Gramm C., Rothstein L., Clark K., Stein R., Dick L., Hwang D., Goldberg A.L. Inhibitors of the Proteasome Block the Degradation of Most Cell Proteins and the Generation of Peptides Presented on MHC Class I Molecules. Cell. 1994;78:761–771. doi: 10.1016/S0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- 84.Aki M., Shimbara N., Takashina M., Akiyama K., Kagawa S., Tamura T., Tanahashi N., Yoshimura T., Tanaka K., Ichihara A. Interferon-Gamma Induces Different Subunit Organizations and Functional Diversity of Proteasomes. J. Biochem. 1994;115:257–269. doi: 10.1093/oxfordjournals.jbchem.a124327. [DOI] [PubMed] [Google Scholar]
- 85.Boes B., Hengel H., Ruppert T., Multhaup G., Koszinowski U.H., Kloetzel P.M. Interferon Gamma Stimulation Modulates the Proteolytic Activity and Cleavage Site Preference of 20S Mouse Proteasomes. J. Exp. Med. 1994;179:901–909. doi: 10.1084/jem.179.3.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Driscoll J., Brown M.G., Finley D., Monaco J.J. MHC-Linked LMP Gene Products Specifically Alter Peptidase Activities of the Proteasome. Nature. 1993;365:262–264. doi: 10.1038/365262a0. [DOI] [PubMed] [Google Scholar]
- 87.Gaczynska M., Rock K.L., Goldberg A.L. Role of Proteasomes in Antigen Presentation. Enzym. Protein. 1993;47:354–369. doi: 10.1159/000468693. [DOI] [PubMed] [Google Scholar]
- 88.Griffin T.A., Nandi D., Cruz M., Fehling H.J., Kaer L.V., Monaco J.J., Colbert R.A. Immunoproteasome Assembly: Cooperative Incorporation of Interferon Gamma (IFN-Gamma)-Inducible Subunits. J. Exp. Med. 1998;187:97–104. doi: 10.1084/jem.187.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Groettrup M., Standera S., Stohwasser R., Kloetzel P.M. The Subunits MECL-1 and LMP2 Are Mutually Required for Incorporation into the 20S Proteasome. Proc. Natl. Acad. Sci. USA. 1997;94:8970–8975. doi: 10.1073/pnas.94.17.8970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tanaka K. Role of Proteasomes Modified by Interferon-Gamma in Antigen Processing. J. Leukoc Biol. 1994;56:571–575. doi: 10.1002/jlb.56.5.571. [DOI] [PubMed] [Google Scholar]
- 91.Heink S., Ludwig D., Kloetzel P.-M., Krüger E. IFN-Gamma-Induced Immune Adaptation of the Proteasome System Is an Accelerated and Transient Response. Proc. Natl. Acad. Sci. USA. 2005;102:9241–9246. doi: 10.1073/pnas.0501711102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Guillaume B., Chapiro J., Stroobant V., Colau D., Van Holle B., Parvizi G., Bousquet-Dubouch M.-P., Théate I., Parmentier N., Van den Eynde B.J. Two Abundant Proteasome Subtypes That Uniquely Process Some Antigens Presented by HLA Class I Molecules. Proc. Natl. Acad. Sci. USA. 2010;107:18599–18604. doi: 10.1073/pnas.1009778107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Khan S., van den Broek M., Schwarz K., de Giuli R., Diener P.A., Groettrup M. Immunoproteasomes Largely Replace Constitutive Proteasomes during an Antiviral and Antibacterial Immune Response in the Liver. J. Immunol. 2001;167:6859–6868. doi: 10.4049/jimmunol.167.12.6859. [DOI] [PubMed] [Google Scholar]
- 94.Groettrup M., Kirk C.J., Basler M. Proteasomes in Immune Cells: More than Peptide Producers? Nat. Rev. Immunol. 2010;10:73–78. doi: 10.1038/nri2687. [DOI] [PubMed] [Google Scholar]
- 95.Reis J., Guan X.Q., Kisselev A.F., Papasian C.J., Qureshi A.A., Morrison D.C., Van Way C.W., Vogel S.N., Qureshi N. LPS-Induced Formation of Immunoproteasomes: TNF-α and Nitric Oxide Production Are Regulated by Altered Composition of Proteasome-Active Sites. Cell Biochem. Biophys. 2011;60:77–88. doi: 10.1007/s12013-011-9182-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shin E.-C., Seifert U., Kato T., Rice C.M., Feinstone S.M., Kloetzel P.-M., Rehermann B. Virus-Induced Type I IFN Stimulates Generation of Immunoproteasomes at the Site of Infection. J. Clin. Investig. 2006;116:3006–3014. doi: 10.1172/JCI29832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Johnston-Carey H.K., Pomatto L.C.D., Davies K.J.A. The Immunoproteasome in Oxidative Stress, Aging, and Disease. Crit Rev. Biochem Mol. Biol. 2015;51:268–281. doi: 10.3109/10409238.2016.1172554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Höhn T.J.A., Grune T. The Proteasome and the Degradation of Oxidized Proteins: Part III-Redox Regulation of the Proteasomal System. Redox Biol. 2014;2:388–394. doi: 10.1016/j.redox.2013.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Vigneron N., Abi Habib J., Van den Eynde B.J. Learning from the Proteasome How to Fine-Tune Cancer Immunotherapy. Trends Cancer. 2017;3:726–741. doi: 10.1016/j.trecan.2017.07.007. [DOI] [PubMed] [Google Scholar]
- 100.Huber E.M., Basler M., Schwab R., Heinemeyer W., Kirk C.J., Groettrup M., Groll M. Immuno- and Constitutive Proteasome Crystal Structures Reveal Differences in Substrate and Inhibitor Specificity. Cell. 2012;148:727–738. doi: 10.1016/j.cell.2011.12.030. [DOI] [PubMed] [Google Scholar]
- 101.Akiyama K., Kagawa S., Tamura T., Shimbara N., Takashina M., Kristensen P., Hendil K.B., Tanaka K., Ichihara A. Replacement of Proteasome Subunits X and Y by LMP7 and LMP2 Induced by Interferon-Gamma for Acquirement of the Functional Diversity Responsible for Antigen Processing. FEBS Lett. 1994;343:85–88. doi: 10.1016/0014-5793(94)80612-8. [DOI] [PubMed] [Google Scholar]
- 102.Platteel A.C.M., Liepe J., van Eden W., Mishto M., Sijts A.J.A.M. An Unexpected Major Role for Proteasome-Catalyzed Peptide Splicing in Generation of T Cell Epitopes: Is There Relevance for Vaccine Development? Front. Immunol. 2017;8:1441. doi: 10.3389/fimmu.2017.01441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Vigneron N., Ferrari V., Stroobant V., Abi Habib J., Van den Eynde B.J. Peptide Splicing by the Proteasome. J. Biol. Chem. 2017;292:21170–21179. doi: 10.1074/jbc.R117.807560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Vigneron N., Stroobant V., Chapiro J., Ooms A., Degiovanni G., Morel S., van der Bruggen P., Boon T., Van den Eynde B.J. An Antigenic Peptide Produced by Peptide Splicing in the Proteasome. Science. 2004;304:587–590. doi: 10.1126/science.1095522. [DOI] [PubMed] [Google Scholar]
- 105.Liepe J., Marino F., Sidney J., Jeko A., Bunting D.E., Sette A., Kloetzel P.M., Stumpf M.P.H., Heck A.J.R., Mishto M. A Large Fraction of HLA Class I Ligands Are Proteasome-Generated Spliced Peptides. Science. 2016;354:354–358. doi: 10.1126/science.aaf4384. [DOI] [PubMed] [Google Scholar]
- 106.Morel S., Lévy F., Burlet-Schiltz O., Brasseur F., Probst-Kepper M., Peitrequin A.L., Monsarrat B., Van Velthoven R., Cerottini J.C., Boon T., et al. Processing of Some Antigens by the Standard Proteasome but Not by the Immunoproteasome Results in Poor Presentation by Dendritic Cells. Immunity. 2000;12:107–117. doi: 10.1016/S1074-7613(00)80163-6. [DOI] [PubMed] [Google Scholar]
- 107.Chapiro J., Claverol S., Piette F., Ma W., Stroobant V., Guillaume B., Gairin J.-E., Morel S., Burlet-Schiltz O., Monsarrat B., et al. Destructive Cleavage of Antigenic Peptides Either by the Immunoproteasome or by the Standard Proteasome Results in Differential Antigen Presentation. J. Immunol. 2006;176:1053–1061. doi: 10.4049/jimmunol.176.2.1053. [DOI] [PubMed] [Google Scholar]
- 108.Kincaid E.Z., Che J.W., York I., Escobar H., Reyes-Vargas E., Delgado J.C., Welsh R.M., Karow M.L., Murphy A.J., Valenzuela D.M., et al. Mice Completely Lacking Immunoproteasomes Show Major Changes in Antigen Presentation. Nat. Immunol. 2011;13:129–135. doi: 10.1038/ni.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Van Kaer L., Ashton-Rickardt P.G., Eichelberger M., Gaczynska M., Nagashima K., Rock K.L., Goldberg A.L., Doherty P.C., Tonegawa S. Altered Peptidase and Viral-Specific T Cell Response in LMP2 Mutant Mice. Immunity. 1994;1:533–541. doi: 10.1016/1074-7613(94)90043-4. [DOI] [PubMed] [Google Scholar]
- 110.de Graaf N., van Helden M.J.G., Textoris-Taube K., Chiba T., Topham D.J., Kloetzel P.-M., Zaiss D.M.W., Sijts A.J.A.M. PA28 and the Proteasome Immunosubunits Play a Central and Independent Role in the Production of MHC Class I-Binding Peptides in Vivo. Eur. J. Immunol. 2011;41:926–935. doi: 10.1002/eji.201041040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chapatte L., Ayyoub M., Morel S., Peitrequin A.-L., Lévy N., Servis C., Van den Eynde B.J., Valmori D., Lévy F. Processing of Tumor-Associated Antigen by the Proteasomes of Dendritic Cells Controls in Vivo T-Cell Responses. Cancer Res. 2006;66:5461–5468. doi: 10.1158/0008-5472.CAN-05-4310. [DOI] [PubMed] [Google Scholar]
- 112.Borissenko L., Groll M. Diversity of proteasomal missions: Fine tuning of the immune response. Biol. Chem. 2007;388:947–955. doi: 10.1515/BC.2007.109. [DOI] [PubMed] [Google Scholar]
- 113.Çetin G., Klafack S., Studencka-Turski M., Krüger E., Ebstein F. The Ubiquitin-Proteasome System in Immune Cells. Biomolecules. 2021;11:60. doi: 10.3390/biom11010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Basler M., Kirk C.J., Groettrup M. The Immunoproteasome in Antigen Processing and Other Immunological Functions. Curr. Opin. Immunol. 2013;25:74–80. doi: 10.1016/j.coi.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 115.Fehling H.J., Swat W., Laplace C., Kühn R., Rajewsky K., Müller U., von Boehmer H. MHC Class I Expression in Mice Lacking the Proteasome Subunit LMP-7. Science. 1994;265:1234–1237. doi: 10.1126/science.8066463. [DOI] [PubMed] [Google Scholar]
- 116.McCarthy M.K., Weinberg J.B. The Immunoproteasome and Viral Infection: A Complex Regulator of Inflammation. Front. Microbiol. 2015;6:21. doi: 10.3389/fmicb.2015.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.de Verteuil D., Muratore-Schroeder T.L., Granados D.P., Fortier M.-H., Hardy M.-P., Bramoullé A., Caron E., Vincent K., Mader S., Lemieux S., et al. Deletion of Immunoproteasome Subunits Imprints on the Transcriptome and Has a Broad Impact on Peptides Presented by Major Histocompatibility Complex I Molecules. Mol. Cell Proteom. 2010;9:2034–2047. doi: 10.1074/mcp.M900566-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Basler M., Youhnovski N., Van Den Broek M., Przybylski M., Groettrup M. Immunoproteasomes Down-Regulate Presentation of a Subdominant T Cell Epitope from Lymphocytic Choriomeningitis Virus. J. Immunol. 2004;173:3925–3934. doi: 10.4049/jimmunol.173.6.3925. [DOI] [PubMed] [Google Scholar]
- 119.Zanker D., Waithman J., Yewdell J.W., Chen W. Mixed Proteasomes Function to Increase Viral Peptide Diversity and Broaden Antiviral CD8+ T Cell Responses. J. Immunol. 2013;191:52–59. doi: 10.4049/jimmunol.1300802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kloetzel P.M. Antigen processing by the proteasome. Nat. Rev. Mol. Cell Biol. 2016;2:179–188. doi: 10.1038/35056572. [DOI] [PubMed] [Google Scholar]
- 121.Tubío-Santamaría N., Ebstein F., Heidel F.H., Krüger E. Immunoproteasome Function in Normal and Malignant Hematopoiesis. Cells. 2021;10:1577. doi: 10.3390/cells10071577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mishto M., Liepe J., Textoris-Taube K., Keller C., Henklein P., Weberruß M., Dahlmann B., Enenkel C., Voigt A., Kuckelkorn U., et al. Proteasome Isoforms Exhibit Only Quantitative Differences in Cleavage and Epitope Generation. Eur. J. Immunol. 2014;44:3508–3521. doi: 10.1002/eji.201444902. [DOI] [PubMed] [Google Scholar]
- 123.Trentini D.B., Pecoraro M., Tiwary S., Cox J., Mann M., Hipp M.S., Hartl F.U. Role for Ribosome-Associated Quality Control in Sampling Proteins for MHC Class I-Mediated Antigen Presentation. Proc. Natl. Acad. Sci. USA. 2020;117:4099–4108. doi: 10.1073/pnas.1914401117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Murata S., Takahama Y., Kasahara M., Tanaka K. The Immunoproteasome and Thymoproteasome: Functions, Evolution and Human Disease. Nat. Immunol. 2018;19:923–931. doi: 10.1038/s41590-018-0186-z. [DOI] [PubMed] [Google Scholar]
- 125.Yewdell J.W. DRiPs Solidify: Progress in Understanding Endogenous MHC Class I Antigen Processing. Trends Immunol. 2011;32:548–558. doi: 10.1016/j.it.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wei S.C., Duffy C.R., Allison J.P. Fundamental Mechanisms of Immune Checkpoint Blockade Therapy. Cancer Discov. 2018;8:1069–1086. doi: 10.1158/2159-8290.CD-18-0367. [DOI] [PubMed] [Google Scholar]
- 127.Rock K.L., Farfán-Arribas D.J., Colbert J.D., Goldberg A.L. Re-Examining Class-I Presentation and the DRiP Hypothesis. Trends Immunol. 2014;35:144–152. doi: 10.1016/j.it.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Cascio P., Goldberg A.L. Preparation of Hybrid (19S-20S-PA28) Proteasome Complexes and Analysis of Peptides Generated during Protein Degradation. Methods Enzym. 2005;398:336–352. doi: 10.1016/S0076-6879(05)98028-2. [DOI] [PubMed] [Google Scholar]
- 129.Raule M., Cerruti F., Benaroudj N., Migotti R., Kikuchi J., Bachi A., Navon A., Dittmar G., Cascio P. PA28αβ Reduces Size and Increases Hydrophilicity of 20S Immunoproteasome Peptide Products. Chem. Biol. 2014;21:470–480. doi: 10.1016/j.chembiol.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 130.Rechsteiner M., Realini C., Ustrell V. The Proteasome Activator 11 S REG (PA28) and Class I Antigen Presentation. Pt 1Biochem. J. 2000;345:1–15. doi: 10.1042/bj3450001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kopp F., Dahlmann B., Kuehn L. Reconstitution of Hybrid Proteasomes from Purified PA700–20 S Complexes and PA28alphabeta Activator: Ultrastructure and Peptidase Activities. J. Mol. Biol. 2001;313:465–471. doi: 10.1006/jmbi.2001.5063. [DOI] [PubMed] [Google Scholar]
- 132.Fort P., Kajava A.V., Delsuc F., Coux O. Evolution of Proteasome Regulators in Eukaryotes. Genome Biol. Evol. 2015;7:1363–1379. doi: 10.1093/gbe/evv068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Apcher S., Daskalogianni C., Fåhraeus R. Pioneer Translation Products as an Alternative Source for MHC-I Antigenic Peptides. Mol. Immunol. 2015;68:68–71. doi: 10.1016/j.molimm.2015.04.019. [DOI] [PubMed] [Google Scholar]
- 134.Goldberg A.L., Cascio P., Saric T., Rock K.L. The Importance of the Proteasome and Subsequent Proteolytic Steps in the Generation of Antigenic Peptides. Mol. Immunol. 2002;39:147–164. doi: 10.1016/S0161-5890(02)00098-6. [DOI] [PubMed] [Google Scholar]
- 135.Yamano T., Sugahara H., Mizukami S., Murata S., Chiba T., Tanaka K., Yui K., Udono H. Allele-Selective Effect of PA28 in MHC Class I Antigen Processing. J. Immunol. 2008;181:1655–1664. doi: 10.4049/jimmunol.181.3.1655. [DOI] [PubMed] [Google Scholar]
- 136.Eskandari S.K., Seelen M.a.J., Lin G., Azzi J.R. The Immunoproteasome: An Old Player with a Novel and Emerging Role in Alloimmunity. Am. J. Transpl. 2017;17:3033–3039. doi: 10.1111/ajt.14435. [DOI] [PubMed] [Google Scholar]
- 137.Kimura H., Caturegli P., Takahashi M., Suzuki K. New Insights into the Function of the Immunoproteasome in Immune and Nonimmune Cells. J. Immunol. Res. 2015;2015:541984. doi: 10.1155/2015/541984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Moebius J., van den Broek M., Groettrup M., Basler M. Immunoproteasomes Are Essential for Survival and Expansion of T Cells in Virus-Infected Mice. Eur. J. Immunol. 2010;40:3439–3449. doi: 10.1002/eji.201040620. [DOI] [PubMed] [Google Scholar]
- 139.Zaiss D.M.W., de Graaf N., Sijts A.J.A.M. The Proteasome Immunosubunit Multicatalytic Endopeptidase Complex-like 1 Is a T-Cell-Intrinsic Factor Influencing Homeostatic Expansion. Infect. Immun. 2008;76:1207–1213. doi: 10.1128/IAI.01134-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kalim K.W., Basler M., Kirk C.J., Groettrup M. Immunoproteasome Subunit LMP7 Deficiency and Inhibition Suppresses Th1 and Th17 but Enhances Regulatory T Cell Differentiation. J. Immunol. 2012;189:4182–4193. doi: 10.4049/jimmunol.1201183. [DOI] [PubMed] [Google Scholar]
- 141.Ichikawa H.T., Conley T., Muchamuel T., Jiang J., Lee S., Owen T., Barnard J., Nevarez S., Goldman B.I., Kirk C.J., et al. Beneficial Effect of Novel Proteasome Inhibitors in Murine Lupus via Dual Inhibition of Type I Interferon and Autoantibody-Secreting Cells. Arthritis Rheum. 2012;64:493–503. doi: 10.1002/art.33333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Li J., Basler M., Alvarez G., Brunner T., Kirk C.J., Groettrup M. Immunoproteasome Inhibition Prevents Chronic Antibody-Mediated Allograft Rejection in Renal Transplantation. Kidney Int. 2018;93:670–680. doi: 10.1016/j.kint.2017.09.023. [DOI] [PubMed] [Google Scholar]
- 143.Moritz K.E., McCormack N.M., Abera M.B., Viollet C., Yauger Y.J., Sukumar G., Dalgard C.L., Burnett B.G. The Role of the Immunoproteasome in Interferon-γ-Mediated Microglial Activation. Sci. Rep. 2017;7:9365. doi: 10.1038/s41598-017-09715-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Vachharajani N., Joeris T., Luu M., Hartmann S., Pautz S., Jenike E., Pantazis G., Prinz I., Hofer M.J., Steinhoff U., et al. Prevention of Colitis-Associated Cancer by Selective Targeting of Immunoproteasome Subunit LMP7. Oncotarget. 2017;8:50447–50459. doi: 10.18632/oncotarget.14579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Althof N., Goetzke C.C., Kespohl M., Voss K., Heuser A., Pinkert S., Kaya Z., Klingel K., Beling A. The Immunoproteasome-Specific Inhibitor ONX 0914 Reverses Susceptibility to Acute Viral Myocarditis. EMBO Mol. Med. 2018;10:200–218. doi: 10.15252/emmm.201708089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Basler M., Li J., Groettrup M. On the Role of the Immunoproteasome in Transplant Rejection. Immunogenetics. 2019;71:263–271. doi: 10.1007/s00251-018-1084-0. [DOI] [PubMed] [Google Scholar]
- 147.Ebstein F., Lange N., Urban S., Seifert U., Krüger E., Kloetzel P.-M. Maturation of Human Dendritic Cells Is Accompanied by Functional Remodelling of the Ubiquitin-Proteasome System. Int. J. Biochem. Cell Biol. 2009;41:1205–1215. doi: 10.1016/j.biocel.2008.10.023. [DOI] [PubMed] [Google Scholar]
- 148.Studencka-Turski M., Çetin G., Junker H., Ebstein F., Krüger E. Molecular Insight into the IRE1α-Mediated Type I Interferon Response Induced by Proteasome Impairment in Myeloid Cells of the Brain. Front. Immunol. 2019;10:2900. doi: 10.3389/fimmu.2019.02900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Seifert U., Bialy L.P., Ebstein F., Bech-Otschir D., Voigt A., Schröter F., Prozorovski T., Lange N., Steffen J., Rieger M., et al. Immunoproteasomes Preserve Protein Homeostasis upon Interferon-Induced Oxidative Stress. Cell. 2010;142:613–624. doi: 10.1016/j.cell.2010.07.036. [DOI] [PubMed] [Google Scholar]
- 150.Visekruna A., Joeris T., Seidel D., Kroesen A., Loddenkemper C., Zeitz M., Kaufmann S.H.E., Schmidt-Ullrich R., Steinhoff U. Proteasome-Mediated Degradation of IkappaBalpha and Processing of P105 in Crohn Disease and Ulcerative Colitis. J. Clin. Investig. 2006;116:3195–3203. doi: 10.1172/JCI28804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Mitchell S., Mercado E.L., Adelaja A., Ho J.Q., Cheng Q.J., Ghosh G., Hoffmann A. An NFκB Activity Calculator to Delineate Signaling Crosstalk: Type I and II Interferons Enhance NFκB via Distinct Mechanisms. Front. Immunol. 2019;10:1425. doi: 10.3389/fimmu.2019.01425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Maldonado M., Kapphahn R.J., Terluk M.R., Heuss N.D., Yuan C., Gregerson D.S., Ferrington D.A. Immunoproteasome Deficiency Modifies the Alternative Pathway of NFκB Signaling. PLoS ONE. 2013;8:e56187. doi: 10.1371/journal.pone.0056187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Hayashi T., Faustman D. Essential Role of Human Leukocyte Antigen-Encoded Proteasome Subunits in NF-ΚB Activation and Prevention of Tumor Necrosis Factor-α-Induced Apoptosis*. J. Biol. Chem. 2000;275:5238–5247. doi: 10.1074/jbc.275.7.5238. [DOI] [PubMed] [Google Scholar]
- 154.Fellerhoff B., Gu S., Laumbacher B., Nerlich A.G., Weiss E.H., Glas J., Kopp R., Johnson J.P., Wank R. The LMP7-K Allele of the Immunoproteasome Exhibits Reduced Transcript Stability and Predicts High Risk of Colon Cancer. Cancer Res. 2011;71:7145–7154. doi: 10.1158/0008-5472.CAN-10-1883. [DOI] [PubMed] [Google Scholar]
- 155.Yang X.-W., Wang P., Liu J.-Q., Zhang H., Xi W.-D., Jia X.-H., Wang K.-K. Coordinated Regulation of the Immunoproteasome Subunits by PML/RARα and PU.1 in Acute Promyelocytic Leukemia. Oncogene. 2014;33:2700–2708. doi: 10.1038/onc.2013.224. [DOI] [PubMed] [Google Scholar]
- 156.Rouette A., Trofimov A., Haberl D., Boucher G., Lavallée V.-P., D’Angelo G., Hébert J., Sauvageau G., Lemieux S., Perreault C. Expression of Immunoproteasome Genes Is Regulated by Cell-Intrinsic and -Extrinsic Factors in Human Cancers. Sci. Rep. 2016;6:34019. doi: 10.1038/srep34019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Bitzer A., Basler M., Krappmann D., Groettrup M. Immunoproteasome Subunit Deficiency Has No Influence on the Canonical Pathway of NF-ΚB Activation. Mol. Immunol. 2017;83:147–153. doi: 10.1016/j.molimm.2017.01.019. [DOI] [PubMed] [Google Scholar]
- 158.Hussong S.A., Roehrich H., Kapphahn R.J., Maldonado M., Pardue M.T., Ferrington D.A. A Novel Role for the Immunoproteasome in Retinal Function. Investig. Ophthalmol. Vis. Sci. 2011;52:714–723. doi: 10.1167/iovs.10-6032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Atkinson S.P., Collin J., Irina N., Anyfantis G., Kyung B.K., Lako M., Armstrong L. A Putative Role for the Immunoproteasome in the Maintenance of Pluripotency in Human Embryonic Stem Cells. Stem Cells. 2012;30:1373–1384. doi: 10.1002/stem.1113. [DOI] [PubMed] [Google Scholar]
- 160.Cui Z., Hwang S.M., Gomes A.V. Identification of the Immunoproteasome as a Novel Regulator of Skeletal Muscle Differentiation. Mol. Cell Biol. 2014;34:96–109. doi: 10.1128/MCB.00622-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Orre M., Kamphuis W., Dooves S., Kooijman L., Chan E.T., Kirk C.J., Dimayuga Smith V., Koot S., Mamber C., Jansen A.H., et al. Reactive Glia Show Increased Immunoproteasome Activity in Alzheimer’s Disease. Brain. 2013;136:1415–1431. doi: 10.1093/brain/awt083. [DOI] [PubMed] [Google Scholar]
- 162.Campello L., Esteve-Rudd J., Cuenca N., Martín-Nieto J. The Ubiquitin-Proteasome System in Retinal Health and Disease. Mol. Neurobiol. 2013;47:790–810. doi: 10.1007/s12035-012-8391-5. [DOI] [PubMed] [Google Scholar]
- 163.Ferrington D.A., Roehrich H., Chang A.A., Huang C.W., Maldonado M., Bratten W., Rageh A.A., Heuss N.D., Gregerson D.S., Nelson E.F., et al. Corneal Wound Healing Is Compromised by Immunoproteasome Deficiency. PLoS ONE. 2013;8:e54347. doi: 10.1371/journal.pone.0054347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Singh S., Awasthi N., Egwuagu C.E., Wagner B.J. Immunoproteasome Expression in a Nonimmune Tissue, the Ocular Lens. Arch. Biochem. Biophys. 2002;405:147–153. doi: 10.1016/S0003-9861(02)00341-7. [DOI] [PubMed] [Google Scholar]
- 165.Choi J.-H., Jo H.S., Lim S., Kim H.-T., Lee K.W., Moon K.H., Ha T., Kwak S.S., Kim Y., Lee E.J., et al. MTORC1 Accelerates Retinal Development via the Immunoproteasome. Nat. Commun. 2018;9:2502. doi: 10.1038/s41467-018-04774-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Schuld N.J., Hussong S.A., Kapphahn R.J., Lehmann U., Roehrich H., Rageh A.A., Heuss N.D., Bratten W., Gregerson D.S., Ferrington D.A. Immunoproteasome Deficiency Protects in the Retina after Optic Nerve Crush. PLoS ONE. 2015;10:e0126768. doi: 10.1371/journal.pone.0126768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Sbardella D., Coletta A., Tundo G.R., Ahmed I.M.M., Bellia F., Oddone F., Manni G., Coletta M. Structural and Functional Evidence for Citicoline Binding and Modulation of 20S Proteasome Activity: Novel Insights into Its pro-Proteostatic Effect. Biochem. Pharm. 2020;177:113977. doi: 10.1016/j.bcp.2020.113977. [DOI] [PubMed] [Google Scholar]
- 168.Chen Y., Zhang Y., Guo X. Proteasome Dysregulation in Human Cancer: Implications for Clinical Therapies. Cancer Metastasis Rev. 2017;36:703–716. doi: 10.1007/s10555-017-9704-y. [DOI] [PubMed] [Google Scholar]
- 169.Algarra I., Collado A., Garrido F. Altered MHC Class I Antigens in Tumors. Int. J. Clin. Lab. Res. 1997;27:95–102. doi: 10.1007/BF02912442. [DOI] [PubMed] [Google Scholar]
- 170.Tripathi S.C., Peters H.L., Taguchi A., Katayama H., Wang H., Momin A., Jolly M.K., Celiktas M., Rodriguez-Canales J., Liu H., et al. Immunoproteasome Deficiency Is a Feature of Non-Small Cell Lung Cancer with a Mesenchymal Phenotype and Is Associated with a Poor Outcome. Proc. Natl. Acad. Sci. USA. 2016;113:E1555–E1564. doi: 10.1073/pnas.1521812113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Heink S., Fricke B., Ludwig D., Kloetzel P.-M., Krüger E. Tumor Cell Lines Expressing the Proteasome Subunit Isoform LMP7E1 Exhibit Immunoproteasome Deficiency. Cancer Res. 2006;66:649–652. doi: 10.1158/0008-5472.CAN-05-2872. [DOI] [PubMed] [Google Scholar]
- 172.Yang C., Schmidt M. Cutting through Complexity: The Proteolytic Properties of Alternate Immunoproteasome Complexes. Chem Biol. 2014;21:435–436. doi: 10.1016/j.chembiol.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 173.Niewerth D., Jansen G., Assaraf Y.G., Zweegman S., Kaspers G.J.L., Cloos J. Molecular Basis of Resistance to Proteasome Inhibitors in Hematological Malignancies. Drug Resist. Updat. 2015;18:18–35. doi: 10.1016/j.drup.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 174.Weon J.L., Potts P.R. The MAGE Protein Family and Cancer. Curr. Opin. Cell Biol. 2015;37:1–8. doi: 10.1016/j.ceb.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Olsson N., Heberling M.L., Zhang L., Jhunjhunwala S., Phung Q.T., Lin S., Anania V.G., Lill J.R., Elias J.E. An Integrated Genomic, Proteomic, and Immunopeptidomic Approach to Discover Treatment-Induced Neoantigens. Front. Immunol. 2021;12:662443. doi: 10.3389/fimmu.2021.662443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Schumacher T.N., Schreiber R.D. Neoantigens in Cancer Immunotherapy. Science. 2015;348:69–74. doi: 10.1126/science.aaa4971. [DOI] [PubMed] [Google Scholar]
- 177.Guillaume B., Stroobant V., Bousquet-Dubouch M.-P., Colau D., Chapiro J., Parmentier N., Dalet A., Van den Eynde B.J. Analysis of the Processing of Seven Human Tumor Antigens by Intermediate Proteasomes. J. Immunol. 2012;189:3538–3547. doi: 10.4049/jimmunol.1103213. [DOI] [PubMed] [Google Scholar]
- 178.Schooten E., Di Maggio A., van Bergen En Henegouwen P.M.P., Kijanka M.M. MAGE-A Antigens as Targets for Cancer Immunotherapy. Cancer Treat. Rev. 2018;67:54–62. doi: 10.1016/j.ctrv.2018.04.009. [DOI] [PubMed] [Google Scholar]
- 179.Gou Q., Dong C., Xu H., Khan B., Jin J., Liu Q., Shi J., Hou Y. PD-L1 Degradation Pathway and Immunotherapy for Cancer. Cell Death Dis. 2020;11:955. doi: 10.1038/s41419-020-03140-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Mezzadra R., Sun C., Jae L.T., Gomez-Eerland R., de Vries E., Wu W., Logtenberg M.E.W., Slagter M., Rozeman E.A., Hofland I., et al. Identification of CMTM6 and CMTM4 as PD-L1 Protein Regulators. Nature. 2017;549:106–110. doi: 10.1038/nature23669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Zhang J., Bu X., Wang H., Zhu Y., Geng Y., Nihira N.T., Tan Y., Ci Y., Wu F., Dai X., et al. Cyclin D-CDK4 Kinase Destabilizes PD-L1 via Cullin 3-SPOP to Control Cancer Immune Surveillance. Nature. 2018;553:91–95. doi: 10.1038/nature25015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Deng L., Qian G., Zhang S., Zheng H., Fan S., Lesinski G.B., Owonikoko T.K., Ramalingam S.S., Sun S.-Y. Inhibition of MTOR Complex 1/P70 S6 Kinase Signaling Elevates PD-L1 Levels in Human Cancer Cells through Enhancing Protein Stabilization Accompanied with Enhanced β-TrCP Degradation. Oncogene. 2019;38:6270–6282. doi: 10.1038/s41388-019-0877-4. [DOI] [PubMed] [Google Scholar]
- 183.Narayanan S., Cai C.-Y., Assaraf Y.G., Guo H.-Q., Cui Q., Wei L., Huang J.-J., Ashby C.R., Chen Z.-S. Targeting the Ubiquitin-Proteasome Pathway to Overcome Anti-Cancer Drug Resistance. Drug Resist. Updat. 2020;48:100663. doi: 10.1016/j.drup.2019.100663. [DOI] [PubMed] [Google Scholar]
- 184.Grigoreva T.A., Tribulovich V.G., Garabadzhiu A.V., Melino G., Barlev N.A. The 26S Proteasome Is a Multifaceted Target for Anti-Cancer Therapies. Oncotarget. 2015;6:24733–24749. doi: 10.18632/oncotarget.4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Huang Z., Wu Y., Zhou X., Xu J., Zhu W., Shu Y., Liu P. Efficacy of Therapy with Bortezomib in Solid Tumors: A Review Based on 32 Clinical Trials. Future Oncol. 2014;10:1795–1807. doi: 10.2217/fon.14.30. [DOI] [PubMed] [Google Scholar]
- 186.Xi J., Zhuang R., Kong L., He R., Zhu H., Zhang J. Immunoproteasome-Selective Inhibitors: An Overview of Recent Developments as Potential Drugs for Hematologic Malignancies and Autoimmune Diseases. Eur. J. Med. Chem. 2019;182:111646. doi: 10.1016/j.ejmech.2019.111646. [DOI] [PubMed] [Google Scholar]
- 187.Ebstein F., Kloetzel P.-M., Krüger E., Seifert U. Emerging Roles of Immunoproteasomes beyond MHC Class I Antigen Processing. Cell Mol. Life Sci. 2012;69:2543–2558. doi: 10.1007/s00018-012-0938-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Kisselev A.F., Groettrup M. Subunit Specific Inhibitors of Proteasomes and Their Potential for Immunomodulation. Curr. Opin. Chem. Biol. 2014;23:16–22. doi: 10.1016/j.cbpa.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Ettari R., Zappalà M., Grasso S., Musolino C., Innao V., Allegra A. Immunoproteasome-Selective and Non-Selective Inhibitors: A Promising Approach for the Treatment of Multiple Myeloma. Pharm. Ther. 2018;182:176–192. doi: 10.1016/j.pharmthera.2017.09.001. [DOI] [PubMed] [Google Scholar]
- 190.Basler M., Mundt S., Muchamuel T., Moll C., Jiang J., Groettrup M., Kirk C.J. Inhibition of the Immunoproteasome Ameliorates Experimental Autoimmune Encephalomyelitis. EMBO Mol. Med. 2014;6:226–238. doi: 10.1002/emmm.201303543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Miller Z., Lee W., Kim K.B. The Immunoproteasome as a Therapeutic Target for Hematological Malignancies. Curr. Cancer Drug Targets. 2014;14:537–548. doi: 10.2174/1568009614666140723113139. [DOI] [PubMed] [Google Scholar]
- 192.Johnson H.W.B., Lowe E., Anderl J.L., Fan A., Muchamuel T., Bowers S., Moebius D.C., Kirk C., McMinn D.L. Required Immunoproteasome Subunit Inhibition Profile for Anti-Inflammatory Efficacy and Clinical Candidate KZR-616 ((2 S,3 R)- N-(( S)-3-(Cyclopent-1-En-1-Yl)-1-((R)-2-Methyloxiran-2-Yl)-1-Oxopropan-2-Yl)-3-Hydroxy-3-(4-Methoxyphenyl)-2-(( S)-2-(2-Morpholinoacetamido)Propanamido)Propenamide) J. Med. Chem. 2018;61:11127–11143. doi: 10.1021/acs.jmedchem.8b01201. [DOI] [PubMed] [Google Scholar]
- 193.Kubiczkova L., Pour L., Sedlarikova L., Hajek R., Sevcikova S. Proteasome Inhibitors—Molecular Basis and Current Perspectives in Multiple Myeloma. J. Cell Mol. Med. 2014;18:947–961. doi: 10.1111/jcmm.12279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Cengiz Seval G., Beksac M. The Safety of Bortezomib for the Treatment of Multiple Myeloma. Expert. Opin. Drug. Saf. 2018;17:953–962. doi: 10.1080/14740338.2018.1513487. [DOI] [PubMed] [Google Scholar]
- 195.Jenkins T.W., Downey-Kopyscinski S.L., Fields J.L., Rahme G.J., Colley W.C., Israel M.A., Maksimenko A.V., Fiering S.N., Kisselev A.F. Activity of Immunoproteasome Inhibitor ONX-0914 in Acute Lymphoblastic Leukemia Expressing MLL–AF4 Fusion Protein. Sci. Rep. 2021;11:10883. doi: 10.1038/s41598-021-90451-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Downey-Kopyscinski S., Daily E.W., Gautier M., Bhatt A., Florea B.I., Mitsiades C.S., Richardson P.G., Driessen C., Overkleeft H.S., Kisselev A.F. An Inhibitor of Proteasome Β2 Sites Sensitizes Myeloma Cells to Immunoproteasome Inhibitors. Blood Adv. 2018;2:2443–2451. doi: 10.1182/bloodadvances.2018016360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Zerfas B.L., Maresh M.E., Trader D.J. The Immunoproteasome: An Emerging Target in Cancer and Autoimmune and Neurological Disorders. J. Med. Chem. 2020;63:1841–1858. doi: 10.1021/acs.jmedchem.9b01226. [DOI] [PubMed] [Google Scholar]
- 198.Fang Y., Johnson H., Anderl J.L., Muchamuel T., McMinn D., Morisseau C., Hammock B.D., Kirk C., Wang J. Role of Epoxide Hydrolases and Cytochrome P450s on Metabolism of KZR-616, a First-in-Class Selective Inhibitor of the Immunoproteasome. Drug. Metab. Dispos. 2021;49:810–821. doi: 10.1124/dmd.120.000307. [DOI] [PubMed] [Google Scholar]
- 199.Ogorevc E., Schiffrer E.S., Sosič I., Gobec S. A Patent Review of Immunoproteasome Inhibitors. Expert Opin. Pat. 2018;28:517–540. doi: 10.1080/13543776.2018.1484904. [DOI] [PubMed] [Google Scholar]
- 200.Zhang C., Zhu H., Shao J., He R., Xi J., Zhuang R., Zhang J. Immunoproteasome-selective inhibitors: The future of autoimmune diseases? Future Med. Chem. 2020;12:269–272. doi: 10.4155/fmc-2019-0299. [DOI] [PubMed] [Google Scholar]
- 201.Klein M., Busch M., Friese-Hamim M., Crosignani S., Fuchss T., Musil D., Rohdich F., Sanderson M.P., Seenisamy J., Walter-Bausch G., et al. Structure-Based Optimization and Discovery of M3258, a Specific Inhibitor of the Immunoproteasome Subunit LMP7 (Β5i) J. Med. Chem. 2021;64:10230–10245. doi: 10.1021/acs.jmedchem.1c00604. [DOI] [PubMed] [Google Scholar]
- 202.Sanderson M.P., Friese-Hamim M., Walter-Bausch G., Busch M., Gaus S., Musil D., Rohdich F., Zanelli U., Downey-Kopyscinski S.L., Mitsiades C.S., et al. M3258 Is a Selective Inhibitor of the Immunoproteasome Subunit LMP7 (Β5i) Delivering Efficacy in Multiple Myeloma Models. Mol. Cancer. 2021;20:1378–1387. doi: 10.1158/1535-7163.MCT-21-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Kasahara M. Role of Immunoproteasomes and Thymoproteasomes in Health and Disease. Pathol. Int. 2021;71:371–382. doi: 10.1111/pin.13088. [DOI] [PubMed] [Google Scholar]
- 204.Kuhn D.J., Orlowski R.Z. The Immunoproteasome as a Target in Hematologic Malignancies. Semin. Hematol. 2012;49:258–262. doi: 10.1053/j.seminhematol.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Goldberg A.L., Gaczynska M., Grant E., Michalek M., Rock K.L. Functions of the Proteasome in Antigen Presentation. Cold Spring Harb. Symp. Quant. Biol. 1995;60:479–490. doi: 10.1101/SQB.1995.060.01.052. [DOI] [PubMed] [Google Scholar]
- 206.Fiebiger B.M., Pfister H., Behrends U., Mautner J. Polyubiquitination of Lysine-48 Is an Essential but Indirect Signal for MHC Class I Antigen Processing. Eur. J. Immunol. 2015;45:716–727. doi: 10.1002/eji.201444830. [DOI] [PubMed] [Google Scholar]
- 207.Vigneron N., Van den Eynde B.J. Insights into the Processing of MHC Class I Ligands Gained from the Study of Human Tumor Epitopes. Cell Mol. Life Sci. 2011;68:1503–1520. doi: 10.1007/s00018-011-0658-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Mpakali A., Stratikos E. The Role of Antigen Processing and Presentation in Cancer and the Efficacy of Immune Checkpoint Inhibitor Immunotherapy. Cancers. 2021;13:134. doi: 10.3390/cancers13010134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Turajlic S., Litchfield K., Xu H., Rosenthal R., McGranahan N., Reading J.L., Wong Y.N.S., Rowan A., Kanu N., Al Bakir M., et al. Insertion-and-Deletion-Derived Tumour-Specific Neoantigens and the Immunogenic Phenotype: A Pan-Cancer Analysis. Lancet Oncol. 2017;18:1009–1021. doi: 10.1016/S1470-2045(17)30516-8. [DOI] [PubMed] [Google Scholar]
- 210.Le D.T., Uram J.N., Wang H., Bartlett B.R., Kemberling H., Eyring A.D., Skora A.D., Luber B.S., Azad N.S., Laheru D., et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N. Engl. J. Med. 2015;372:2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Rizvi N.A., Hellmann M.D., Snyder A., Kvistborg P., Makarov V., Havel J.J., Lee W., Yuan J., Wong P., Ho T.S., et al. Mutational Landscape Determines Sensitivity to PD-1 Blockade in Non–Small Cell Lung Cancer. Science. 2015;348:124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Anagnostou V., Smith K.N., Forde P.M., Niknafs N., Bhattacharya R., White J., Zhang T., Adleff V., Phallen J., Wali N., et al. Evolution of Neoantigen Landscape during Immune Checkpoint Blockade in Non-Small Cell Lung Cancer. Cancer Discov. 2017;7:264–276. doi: 10.1158/2159-8290.CD-16-0828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.McGrail D.J., Pilié P.G., Rashid N.U., Voorwerk L., Slagter M., Kok M., Jonasch E., Khasraw M., Heimberger A.B., Lim B., et al. High Tumor Mutation Burden Fails to Predict Immune Checkpoint Blockade Response across All Cancer Types. Ann. Oncol. 2021;32:661–672. doi: 10.1016/j.annonc.2021.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Wu J., Mayer A.T., Li R. Integrated Imaging and Molecular Analysis to Decipher Tumor Microenvironment in the Era of Immunotherapy. Semin Cancer Biol. 2020:S1044–579X(20)30264–9. doi: 10.1016/j.semcancer.2020.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Wu J., Li C., Gensheimer M., Padda S., Kato F., Shirato H., Wei Y., Schönlieb C.-B., Price S.J., Jaffray D., et al. Radiological Tumour Classification across Imaging Modality and Histology. Nat. Mach. Intell. 2021;3:787–798. doi: 10.1038/s42256-021-00377-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Kalaora S., Lee J.S., Barnea E., Levy R., Greenberg P., Alon M., Yagel G., Bar Eli G., Oren R., Peri A., et al. Immunoproteasome Expression Is Associated with Better Prognosis and Response to Checkpoint Therapies in Melanoma. Nat. Commun. 2020;11:896. doi: 10.1038/s41467-020-14639-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Hugo W., Zaretsky J.M., Sun L., Song C., Moreno B.H., Hu-Lieskovan S., Berent-Maoz B., Pang J., Chmielowski B., Cherry G., et al. Genomic and Transcriptomic Features of Response to Anti-PD-1 Therapy in Metastatic Melanoma. Cell. 2016;165:35–44. doi: 10.1016/j.cell.2016.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Neubert N.J., Tillé L., Barras D., Soneson C., Baumgaertner P., Rimoldi D., Gfeller D., Delorenzi M., Fuertes Marraco S.A., Speiser D.E. Broad and Conserved Immune Regulation by Genetically Heterogeneous Melanoma Cells. Cancer Res. 2017;77:1623–1636. doi: 10.1158/0008-5472.CAN-16-2680. [DOI] [PubMed] [Google Scholar]
- 219.Rodig S.J., Gusenleitner D., Jackson D.G., Gjini E., Giobbie-Hurder A., Jin C., Chang H., Lovitch S.B., Horak C., Weber J.S., et al. MHC Proteins Confer Differential Sensitivity to CTLA-4 and PD-1 Blockade in Untreated Metastatic Melanoma. Sci. Transl. Med. 2018;10:eaar3342. doi: 10.1126/scitranslmed.aar3342. [DOI] [PubMed] [Google Scholar]
- 220.Grasso C.S., Tsoi J., Onyshchenko M., Abril-Rodriguez G., Ross-Macdonald P., Wind-Rotolo M., Champhekar A., Medina E., Torrejon D.Y., Shin D.S., et al. Conserved Interferon-γ Signaling Drives Clinical Response to Immune Checkpoint Blockade Therapy in Melanoma. Cancer Cell. 2020;38:500–515.e3. doi: 10.1016/j.ccell.2020.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Karachaliou N., Gonzalez-Cao M., Crespo G., Drozdowskyj A., Aldeguer E., Gimenez-Capitan A., Teixido C., Molina-Vila M.A., Viteri S., Gil M.d., et al. Interferon gamma, an important marker of response to immune checkpoint blockade in non-small cell lung cancer and melanoma patients. Ther. Adv. Med. Oncol. 2018;18 doi: 10.1177/1758834017749748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Ribas A., Medina T., Kummar S., Amin A., Kalbasi A., Drabick J.J., Barve M., Daniels G.A., Wong D.J., Schmidt E.V., et al. SD-101 in Combination with Pembrolizumab in Advanced Melanoma: Results of a Phase Ib, Multicenter Study. Cancer Discov. 2018;8:1250–1257. doi: 10.1158/2159-8290.CD-18-0280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Torrejon D.Y., Abril-Rodriguez G., Champhekar A.S., Tsoi J., Campbell K.M., Kalbasi A., Parisi G., Zaretsky J.M., Garcia-Diaz A., Puig-Saus C., et al. Overcoming Genetically Based Resistance Mechanisms to PD-1 Blockade. Cancer Discov. 2020;10:1140–1157. doi: 10.1158/2159-8290.CD-19-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Woods K., Knights A.J., Anaka M., Schittenhelm R.B., Purcell A.W., Behren A., Cebon J. Mismatch in Epitope Specificities between IFNγ Inflamed and Uninflamed Conditions Leads to Escape from T Lymphocyte Killing in Melanoma. J. Immunother. Cancer. 2016;4:10. doi: 10.1186/s40425-016-0111-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Peng W., Lizée G., Hwu P. Blockade of the PD-1 Pathway Enhances the Efficacy of Adoptive Cell Therapy against Cancer. Onco. Immunol. 2013;2:e22691. doi: 10.4161/onci.22691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Carvalho A.S., Rodríguez M.S., Matthiesen R. Review and Literature Mining on Proteostasis Factors and Cancer. Methods Mol. Biol. 2016;1449:71–84. doi: 10.1007/978-1-4939-3756-1_2. [DOI] [PubMed] [Google Scholar]
- 227.Vahid S., Thaper D., Zoubeidi A. Chaperoning the Cancer: The Proteostatic Functions of the Heat Shock Proteins in Cancer. Recent Pat. Anticancer Drug Discov. 2017;12:35–47. doi: 10.2174/1574892811666161102125252. [DOI] [PubMed] [Google Scholar]
- 228.Santoro A.M., D’Urso A., Cunsolo A., Milardi D., Purrello R., Sbardella D., Tundo G.R., Diana D., Fattorusso R., Dato A.D., et al. Cooperative Binding of the Cationic Porphyrin Tris-T4 Enhances Catalytic Activity of 20S Proteasome Unveiling a Complex Distribution of Functional States. Int. J. Mol. Sci. 2020;21:7190. doi: 10.3390/ijms21197190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Santoro A.M., Cunsolo A., D’Urso A., Sbardella D., Tundo G.R., Ciaccio C., Coletta M., Diana D., Fattorusso R., Persico M., et al. Cationic Porphyrins Are Tunable Gatekeepers of the 20S Proteasome. Chem. Sci. 2016;7:1286–1297. doi: 10.1039/C5SC03312H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Dato A.D., Cunsolo A., Persico M., Santoro A.M., D’Urso A., Milardi D., Purrello R., Stefanelli M., Paolesse R., Tundo G.R., et al. Electrostatic Map Of Proteasome α-Rings Encodes The Design of Allosteric Porphyrin-Based Inhibitors Able To Affect 20S Conformation By Cooperative Binding. Sci. Rep. 2017;7:17098. doi: 10.1038/s41598-017-17008-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Goldberg A.L., Kim H.T., Lee D., Collins G.A. Mechanisms That Activate 26S Proteasomes and Enhance Protein Degradation. Biomolecules. 2021;11:779. doi: 10.3390/biom11060779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Kudriaeva A.A., Saratov G.A., Kaminskaya A.N., Vladimirov V.I., Barzilovich P.Y., Belogurov A.A. Polyamines Counteract Carbonate-Driven Proteasome Stalling in Alkaline Conditions. Biomolecules. 2020;10:1597. doi: 10.3390/biom10121597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Cotton A.D., Nguyen D.P., Gramespacher J.A., Seiple I.A., Wells J.A. Development of Antibody-Based PROTACs for the Degradation of the Cell-Surface Immune Checkpoint Protein PD-L1. J. Am. Chem. Soc. 2021;143:593–598. doi: 10.1021/jacs.0c10008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Wang Y., Deng S., Xu J. Proteasomal and Lysosomal Degradation for Specific and Durable Suppression of Immunotherapeutic Targets. Cancer Biol. Med. 2020;17:583–598. doi: 10.20892/j.issn.2095-3941.2020.0066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Hu X., Wang J., Chu M., Liu Y., Wang Z., Zhu X. Emerging Role of Ubiquitination in the Regulation of PD-1/PD-L1 in Cancer Immunotherapy. Mol. Ther. 2021;29:908–919. doi: 10.1016/j.ymthe.2020.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Zhao Y., Yang W., Huang Y., Cui R., Li X., Li B. Evolving Roles for Targeting CTLA-4 in Cancer Immunotherapy. Cell Physiol. Biochem. 2018;47:721–734. doi: 10.1159/000490025. [DOI] [PubMed] [Google Scholar]
- 237.Wolchok J.D., Saenger Y. The Mechanism of Anti-CTLA-4 Activity and the Negative Regulation of T-Cell Activation. Oncologist. 2008;13:2–9. doi: 10.1634/theoncologist.13-S4-2. [DOI] [PubMed] [Google Scholar]
- 238.Krummel M.F., Allison J.P. CD28 and CTLA-4 Have Opposing Effects on the Response of T Cells to Stimulation. J. Exp. Med. 1995;182:459–465. doi: 10.1084/jem.182.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Buchbinder E.I., Desai A. CTLA-4 and PD-1 Pathways: Similarities, Differences, and Implications of Their Inhibition. Am. J. Clin. Oncol. 2016;39:98–106. doi: 10.1097/COC.0000000000000239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Boussiotis V.A. Molecular and Biochemical Aspects of the PD-1 Checkpoint Pathway. N. Engl. J. Med. 2016;375:1767–1778. doi: 10.1056/NEJMra1514296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Robainas M., Otano R., Bueno S., Ait-Oudhia S. Understanding the Role of PD-L1/PD1 Pathway Blockade and Autophagy in Cancer Therapy. Onco Targets. 2017;10:1803–1807. doi: 10.2147/OTT.S132508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Ribas A., Wolchok J.D. Cancer Immunotherapy Using Checkpoint Blockade. Science. 2018;359:1350–1355. doi: 10.1126/science.aar4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Xin Yu J., Hubbard-Lucey V.M., Tang J. Immuno-Oncology Drug Development Goes Global. Nat. Rev. Drug Discov. 2019;18:899–900. doi: 10.1038/d41573-019-00167-9. [DOI] [PubMed] [Google Scholar]
- 244.Robert C. A Decade of Immune-Checkpoint Inhibitors in Cancer Therapy. Nat. Commun. 2020;11:3801. doi: 10.1038/s41467-020-17670-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Murata S., Takahama Y., Tanaka K. Thymoproteasome: Probable Role in Generating Positively Selecting Peptides. Curr. Opin. Immunol. 2008;20:192–196. doi: 10.1016/j.coi.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 246.Klein L., Hinterberger M., Wirnsberger G., Kyewski B. Antigen Presentation in the Thymus for Positive Selection and Central Tolerance Induction. Nat. Rev. Immunol. 2009;9:833–844. doi: 10.1038/nri2669. [DOI] [PubMed] [Google Scholar]
- 247.Nil A., Firat E., Sobek V., Eichmann K., Niedermann G. Expression of Housekeeping and Immunoproteasome Subunit Genes Is Differentially Regulated in Positively and Negatively Selecting Thymic Stroma Subsets. Eur. J. Immunol. 2004;34:2681–2689. doi: 10.1002/eji.200425032. [DOI] [PubMed] [Google Scholar]
- 248.Macagno A., Gilliet M., Sallusto F., Lanzavecchia A., Nestle F.O., Groettrup M. Dendritic Cells Up-Regulate Immunoproteasomes and the Proteasome Regulator PA28 during Maturation. Eur. J. Immunol. 1999;29:4037–4042. doi: 10.1002/(SICI)1521-4141(199912)29:12<4037::AID-IMMU4037>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 249.Nitta T., Murata S., Sasaki K., Fujii H., Ripen A.M., Ishimaru N., Koyasu S., Tanaka K., Takahama Y. Thymoproteasome Shapes Immunocompetent Repertoire of CD8+ T Cells. Immunity. 2010;32:29–40. doi: 10.1016/j.immuni.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 250.Xing Y., Jameson S.C., Hogquist K.A. Thymoproteasome Subunit-Β5T Generates Peptide-MHC Complexes Specialized for Positive Selection. Proc. Natl. Acad. Sci. USA. 2013;110:6979–6984. doi: 10.1073/pnas.1222244110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Frantzeskakis M., Takahama Y., Ohigashi I. The Role of Proteasomes in the Thymus. Front. Immunol. 2021;12:646209. doi: 10.3389/fimmu.2021.646209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Murata S., Sasaki K., Kishimoto T., Niwa S.-I., Hayashi H., Takahama Y., Tanaka K. Regulation of CD8+ T Cell Development by Thymus-Specific Proteasomes. Science. 2007;316:1349–1353. doi: 10.1126/science.1141915. [DOI] [PubMed] [Google Scholar]
- 253.Uddin M.M., Ohigashi I., Motosugi R., Nakayama T., Sakata M., Hamazaki J., Nishito Y., Rode I., Tanaka K., Takemoto T., et al. Foxn1-Β5t Transcriptional Axis Controls CD8+ T-Cell Production in the Thymus. Nat. Commun. 2017;8:14419. doi: 10.1038/ncomms14419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Tomaru U., Ishizu A., Murata S., Miyatake Y., Suzuki S., Takahashi S., Kazamaki T., Ohara J., Baba T., Iwasaki S., et al. Exclusive Expression of Proteasome Subunit {beta}5t in the Human Thymic Cortex. Blood. 2009;113:5186–5191. doi: 10.1182/blood-2008-11-187633. [DOI] [PubMed] [Google Scholar]
- 255.Mayer C.E., Žuklys S., Zhanybekova S., Ohigashi I., Teh H.-Y., Sansom S.N., Shikama-Dorn N., Hafen K., Macaulay I.C., Deadman M.E., et al. Dynamic Spatio-Temporal Contribution of Single Β5t+ Cortical Epithelial Precursors to the Thymus Medulla. Eur J. Immunol. 2016;46:846–856. doi: 10.1002/eji.201545995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Sasaki K., Takada K., Ohte Y., Kondo H., Sorimachi H., Tanaka K., Takahama Y., Murata S. Thymoproteasomes Produce Unique Peptide Motifs for Positive Selection of CD8(+) T Cells. Nat. Commun. 2015;6:7484. doi: 10.1038/ncomms8484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Kasahara M., Flajnik M.F. Origin and Evolution of the Specialized Forms of Proteasomes Involved in Antigen Presentation. Immunogenetics. 2019;71:251–261. doi: 10.1007/s00251-019-01105-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Hogquist K.A., Xing Y. Why CD8+ T Cells Need Diversity When Growing Up. Immunity. 2010;32:5–6. doi: 10.1016/j.immuni.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 259.Kincaid E.Z., Murata S., Tanaka K., Rock K.L. Specialized Proteasome Subunits Have an Essential Role in the Thymic Selection of CD8(+) T Cells. Nat. Immunol. 2016;17:938–945. doi: 10.1038/ni.3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Tomaru U., Konno S., Miyajima S., Kimoto R., Onodera M., Kiuchi S., Murata S., Ishizu A., Kasahara M. Restricted Expression of the Thymoproteasome Is Required for Thymic Selection and Peripheral Homeostasis of CD8+ T Cells. Cell Rep. 2019;26:639–651.e2. doi: 10.1016/j.celrep.2018.12.078. [DOI] [PubMed] [Google Scholar]
- 261.Takada K., Van Laethem F., Xing Y., Akane K., Suzuki H., Murata S., Tanaka K., Jameson S.C., Singer A., Takahama Y. TCR Affinity for Thymoproteasome-Dependent Positively Selecting Peptides Conditions Antigen Responsiveness in CD8(+) T Cells. Nat. Immunol. 2015;16:1069–1076. doi: 10.1038/ni.3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Tomaru U., Takahashi S., Ishizu A., Miyatake Y., Gohda A., Suzuki S., Ono A., Ohara J., Baba T., Murata S., et al. Decreased Proteasomal Activity Causes Age-Related Phenotypes and Promotes the Development of Metabolic Abnormalities. Am. J. Pathol. 2012;180:963–972. doi: 10.1016/j.ajpath.2011.11.012. [DOI] [PubMed] [Google Scholar]
- 263.Tomaru U., Kasahara M. Thymoproteasome: Role in Thymic Selection and Clinical Significance as a Diagnostic Marker for Thymic Epithelial Tumors. Arch. Immunol. Exp. 2013;61:357–365. doi: 10.1007/s00005-013-0234-1. [DOI] [PubMed] [Google Scholar]
- 264.Yamada Y., Tomaru U., Ishizu A., Kiuchi T., Marukawa K., Matsuno Y., Kasahara M. Expression of Proteasome Subunit Β5t in Thymic Epithelial Tumors. Am. J. Surg. Pathol. 2011;35:1296–1304. doi: 10.1097/PAS.0b013e3182237f5d. [DOI] [PubMed] [Google Scholar]
- 265.Yamada Y., Tomaru U., Ishizu A., Kiuchi T., Kasahara M., Matsuno Y. Expression of Thymoproteasome Subunit Β5t in Type AB Thymoma. J. Clin. Pathol. 2014;67:276–278. doi: 10.1136/jclinpath-2013-201930. [DOI] [PubMed] [Google Scholar]
- 266.Tomaru U., Tsuji T., Kiuchi S., Ishizu A., Suzuki A., Otsuka N., Ito T., Ikeda H., Fukasawa Y., Kasahara M. Decreased Expression of Thymus-Specific Proteasome Subunit Β5t in Down Syndrome Patients. Histopathology. 2015;67:235–244. doi: 10.1111/his.12642. [DOI] [PubMed] [Google Scholar]
- 267.Nitta T., Kochi Y., Muro R., Tomofuji Y., Okamura T., Murata S., Suzuki H., Sumida T., Yamamoto K., Takayanagi H. Human Thymoproteasome Variations Influence CD8 T Cell Selection. Sci. Immunol. 2017;2:eaan5165. doi: 10.1126/sciimmunol.aan5165. [DOI] [PubMed] [Google Scholar]
- 268.Ohigashi I., Ohte Y., Setoh K., Nakase H., Maekawa A., Kiyonari H., Hamazaki Y., Sekai M., Sudo T., Tabara Y., et al. A Human PSMB11 Variant Affects Thymoproteasome Processing and CD8+ T Cell Production. JCI Insight. 2017;2:93664. doi: 10.1172/jci.insight.93664. [DOI] [PMC free article] [PubMed] [Google Scholar]