ABSTRACT
The ability of Enterococcus faecalis to use a variety of carbon sources enables colonization at various anatomic sites within a mammalian host. N-Acetylglucosamine (GlcNAc) is one of the most abundant natural sugars and provides bacteria with a source of carbon and nitrogen when metabolized. N-Acetylglucosamine is also a component of bacterial peptidoglycan, further highlighting the significance of N-acetylglucosamine utilization. In this study, we show that CcpA-regulated enzymes are required for growth on the poly-β1,4-linked GlcNAc substrate, chitopentaose (β1,4-linked GlcNAc5). We also show that EF0114 (EndoE) is required for growth on chitobiose (β1,4-linked GlcNAc2) and that the GH20 domain of EndoE is required for the conversion of GlcNAc2 to N-acetylglucosamine. GlcNAc is transported into the cell via two separate phosphotransferase system (PTS) complexes, either the PTS IICBA encoded by ef1516 (nagE) or the Mpt glucose/mannose permease complex (MptBACD). The Mpt PTS is also the primary glucosamine transporter. In order for N-acetylglucosamine to be utilized as a carbon source, phosphorylated N-acetylglucosamine (GlcNAc-6-P) must be deacetylated, and here, we show that this activity is mediated by EF1317 (an N-acetylglucosamine-6-phosphate deacetylase; NagA homolog), as a deletion of ef1317 is unable to grow on GlcNAc as the carbon source. Deamination of glucosamine to fructose-6-phosphate is required for entry into glycolysis, and we show that growth on glucosamine is dependent on EF0466 (a glucosamine-6-phosphate deaminase; NagB homolog). Collectively, our data highlight the chitinolytic machinery required for breaking down exogenous chitinous substrates, as well as the uptake and cytosolic enzymes needed for metabolizing N-acetylglucosamine.
IMPORTANCE Enterococcus faecalis causes life-threatening health care-associated infections in part due to its intrinsic and acquired antibiotic resistance, its ability to form biofilms, and its nutrient versatility. Alternative nutrient acquisition systems are key factors that contribute to enterococcal colonization at biologically unique host anatomic sites. Although E. faecalis can metabolize an array of carbon sources, little is known of how this bacterium acquires these secondary nutrient sources in mammalian hosts. Our research identifies the glycosidase machinery required for degrading exogenous chitinous substrates into N-acetylglucosamine monomers for transport and metabolism of one of the most abundant naturally occurring sugars, N-acetylglucosamine. Disrupting the function of this N-acetylglucosamine acquisition pathway may lead to new treatments against multidrug-resistant enterococcal infections.
KEYWORDS: N-acetylglucosamine, GlcNAc; CcpA; glycosyl hydrolase; GH18 family; GH20 family; metabolism; Enterococcus faecalis; chitin
INTRODUCTION
Enterococcus faecalis is a Gram-positive commensal that primarily inhabits the mucosa of human gastrointestinal tracts and the oral cavity (1). However, enterococci have emerged as a hospital-adapted, multidrug-resistant (MDR) pathogen that now ranks second on the list of causative agents of disease in health care settings in the United States (2–4). This opportunistic, nosocomial pathogen causes a variety of infections, including endocarditis, bacteremia associated with central-line catheters, surgical site infections, and catheter-associated urinary tract infections (CAUTI) (4). The ability of E. faecalis to colonize these host anatomic sites is, in part, governed by its ability to metabolize secondary carbon sources in nutrient-poor host environments (1, 5). Although E. faecalis can metabolize approximately 70 different carbon sources (6), little is known of the metabolic machinery required for the uptake and utilization of these substrates.
N-Acetylglucosamine (GlcNAc) is a monosaccharide derivative of glucose that is an important carbon and nitrogen source for several bacteria (7–12). Additionally, this amino sugar is a structural component of bacterial peptidoglycan (13, 14), the cell walls of fungi (15), and the exoskeleton of arthropods (16). N-Acetylglucosamine is the monomeric unit of the second most abundant natural polysaccharide, chitin, which is a linear polymer of β-1,4-linked GlcNAc (17, 18). Chitin also provides prebiotic and antioxidant properties when consumed as a source of insoluble fiber (19–21). Bacteria have evolved specific machinery to efficiently degrade chitin and GlcNAc-containing substrates, including chitinases and chitin-binding proteins (CBPs) with lytic polysaccharide monooxygenase activity (22). Recent evidence indicates that chitinases serve as virulence factors for bacterial pathogens during infection of mammalian hosts (23, 24). Although chitin is not naturally present in mammalian hosts, the biological role of bacterial chitinases during infection has been characterized from bacterial pathogens, such as Listeria monocytogenes (25, 26) and Legionella pneumophila (27), suggesting that additional substrates for these chitinases likely exist in mammalian hosts. Recent observations indicate that chitinases produced by Salmonella and Listeria also have specificity toward LacNac (Galβ1-4GlcNAc) and LacdiNAc (GalNAcβ1-4GlcNAc) sugar linkages present in mammalian N- and O-linked glycans, glycolipids, and glycosaminoglycans (24, 28). Although chitinolytic machinery has been identified in E. faecalis (29, 30), little is known of how it contributes to enterococcal metabolism and overall pathogenesis.
Based on observations from Vaaje-Kolstad et al. (30), all currently available enterococcal genomes contain an enzymatic system for cleaving chitin, chitobiose, and extracellular GlcNAc. Vaaje-Kolstad et al. show by analogy with other known microbial chitinolytic systems that EF0362 and EF0361 (EfCBM33A and EfChi18A, respectively) act synergistically to depolymerize poly-N-acetylglucosamine (poly-GlcNAc) substrates into GlcNAc2 dimers (30). EfCBM33A is a copper-dependent enzyme that cleaves chitin via an oxidative mechanism and belongs to a growing number of lytic polysaccharide monooxygenases (22). EfChi18A is a glycosyl family 18 hydrolase and possesses chitinolytic activity and is homologous to the related ChiA enzyme in L. monocytogenes (25).
It was of interest to determine the enzyme(s) responsible for converting GlcNAc2 to GlcNAc monomers as well as the transport system(s) required for GlcNAc and glucosamine (GlcN) importation. Entry of phosphorylated GlcNAc into glycolysis requires deacetylation and deamination into fructose-6-phosphate. We predicted the E. faecalis homologs responsible for these activities and provide further characterization of their function in the use of GlcNAc as a key carbon source.
RESULTS
The deletion of the enterococcal chitinase and chitin-binding protein, ef0362-61, impacts growth on a poly-β1,4-N-acetylglucosamine substrate.
Based on the observations that recombinant EF0362 (EfCBM33A) and EF0361 (EfChi18A) act synergistically to hydrolyze poly-β1,4-N-acetylglucosamine substrates into β1,4-GlcNAc2 oligomeric dimers (30), it was of interest to assess whether the deletion of ef0362-61 impacted growth with a poly-β1,4-GlcNAc substrate as the sole carbon source. Since the enterococcal chitinase and chitin binding protein are encoded in a single operon and both are involved in the degradation of chitinous substrates, we therefore constructed the ef0362-61 deletion mutant, where we removed the entire operon from the E. faecalis V583 genome. Since EF0361 is one of three glycosyl hydrolases encoded in the E. faecalis V583 genome that belong to the glycosyl hydrolase family 18 (GH18) (29, 31–33), we also assessed single deletion mutants of ef0114 and ef2863, as well as a triple glycosidase mutant (Δef0114 Δef0362-61 Δef2863) for their ability to utilize chitopentaose (β1,4-GlcNAc5) as the sole carbon source in a chemically defined medium (CDM) (34, 35). The deletion of ef2863 did not impact growth when chitopentaose was present as the sole carbon source. However, the deletion of ef0362-61 or ef0114 significantly impacted growth relative to the parental strain (Fig. 1). Theses phenotypes are complementable as the ef0362-61 genetic revertant, and the ef0114 complement grew equivalently to the parental strain. This indicates that the ability of E. faecalis to sustain growth on poly-β1,4-N-acetylglucosamine substrates is dependent on the activity of EF0362-61 and EF0114.
FIG 1.
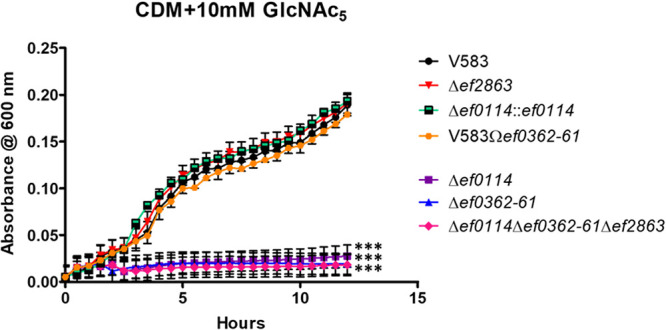
Growth of E. faecalis in CDM supplemented with 10 mM chitopentaose (GlcNAc5). Each graph shows the average from three biological replicates, with three technical replicates each time (n = 9). The statistical significance was calculated using a two-way analysis of variance (ANOVA) test. ***, P < 0.0001.
EF0114 is responsible for degrading β1,4-GlcNAc2 into GlcNAc prior to importation into the bacterial cell.
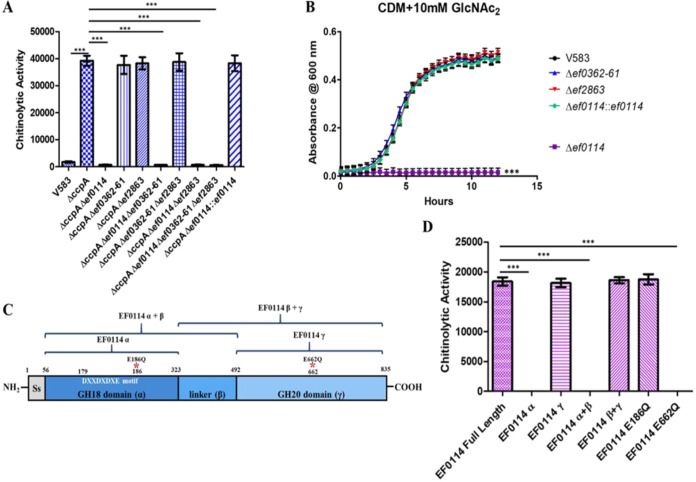
Vaaje-Kolstad et al. (30) proposed that an essential component of the chitinolytic machinery to metabolize chitinous substrates is a secreted glycosyl hydrolase that would be responsible for cleaving β1,4-GlcNAc2 into GlcNAc, prior to the transport of the amino sugar into the cell. Possible glycosyl hydrolase candidates for degrading β1,4-GlcNAc2 into GlcNAc are EF0114 and EF2863, as both are capable of cleaving β1,4-linked GlcNAc glycosidic bonds (31–33, 36). Since the expression of ef0114 and ef2863 are negatively regulated by CcpA (33), we hypothesized that culture supernatant from a ccpA mutant would hydrolyze the fluorogenic β1,4-GlcNAc2 analogue, 4MU-methylumbelliferyl-β-d-N-acetylglucosamine (4-MU-GlcNAc), resulting in an increase in fluorescence due to the release of the fluorogenic product after enzymatic cleavage. To assess this hypothesis, supernatants were collected from the E. faecalis V583 wild-type strain and the ccpA mutant strain grown overnight in CDM supplemented with 42 mM glucose to ensure glucose-replete conditions during growth (33). The filtered supernatants were subsequently incubated with 4-MU-GlcNAc to assess glycosidase activity. As shown in Fig. 2A, the ccpA mutant supernatant possesses significant chitinolytic activity against the fluorogenic β1,4-GlcNAc2 analogue relative to filtered supernatant collected from the parental strain.
FIG 2.
(A) Chitinolytic activity from culture supernatants were measured by determining the ability to hydrolyze the fluorogenic substrate 4MU-methylumbelliferyl-β-d-N-acetylglucosamine (GlcNAc2 analogue). Chitinolytic activity is expressed as relative fluorescence emitted at 485 nm by the absorbance of the culture at OD600. Each graph shows the average from three biological replicates, with three technical replicates each time (n = 9). The statistical significance was calculated using an unpaired t test; ***, P < 0.0001. (B) Growth of E. faecalis in CDM supplemented with 10 mM chitobiose (GlcNAc2). Each graph shows the average from three biological replicates, with three technical replicates each time (n = 9). The statistical significance was calculated using a two-way ANOVA test; ***, P < 0.0001. (C) Representation of the EF0114 domain structure. The putative signal peptide sequence is located between amino acid residues 1 and 56. The GH18 catalytic domain, described as α, is located between amino acid residues 56 and 323, with the GH18 DXXDXDXE catalytic motif located between amino acid residues 179 and 186. The uncharacterized linker region is described as the β domain (between amino acid residues 323 and 492). The GH20 domain (γ) is located between amino acid residues 492 and 835. Listed above the EF0114 domain schematic are representations of the various EF0114 recombinant domain variants used in panel D. (D) Chitinolytic activity of purified recombinant EF0114 variants against the fluorogenic substrate 4MU-methylumbelliferyl-β-d-N-acetylglucosamine (GlcNAc2 analogue). Chitinolytic activity is expressed as the fluorescence emitted at 485 nm. Each graph shows the average from three biological replicates, with three technical replicates each time (n = 9). The statistical significance was calculated using an unpaired t test; ***, P < 0.0001.
To determine which of the CcpA-regulated glycosyl hydrolases is responsible for degrading β1,4-GlcNAc2 to GlcNAc, supernatants were also collected from various glycosyl hydrolase deletion mutants in the ccpA mutant background grown overnight in CDM supplemented with glucose (42 mM). Figure 2A illustrates that only the deletion of ef0114 results in the elimination of chitinolytic activity against the fluorogenic β1,4-GlcNAc2 analogue relative to the ccpA mutant supernatant. The phenotype associated with the various ef0114 mutants was corroborated by assessing the ΔccpA Δef0114::ef0114 complement strain under the same 4MU-GlcNAc assay conditions (Fig. 2A). Incubating the fluorogenic 4MU-GlcNAc substrate with the ΔccpA Δef0114::ef0114 complement supernatant resulted in the restoration of glycosidase activity against this substrate. The data indicate that EF0114 is responsible for degrading extracellular β1,4-GlcNAc2 into GlcNAc.
Because EF0114 displays activity against the fluorogenic β1,4-GlcNAc2 analogue, it was of interest to assess the ability of the ef0114 genetic mutant to sustain growth with GlcNAc2 as the sole carbon source. We assessed the growth of single deletions of ef0114, ef0362-61, and ef2863 and the parental V583 strain in CDM supplemented with 10 mM GlcNAc2. The deletion of ef0114 significantly impacts growth on this substrate relative to the parental strain (Fig. 2B), whereas growth of ef0362-61 and ef2863 were unchanged relative to V583 (Fig. 2B), further highlighting that EF0114 is principally responsible for hydrolyzing dimers of GlcNAc into its monomeric form.
The GH20 domain of EF0114 is responsible for degrading β1,4-GlcNAc2 into GlcNAc monomers.
EF0114 possesses two catalytic domains, an N-terminal GH18 domain and a C-terminal GH20 domain (Fig. 2C). The GH18 domain of EF0114 has been previously characterized for deglycosylating complex glycoproteins, such as IgG and lactoferrin, by hydrolyzing the Asn-attached chitobiose (β-1,4-GlcNAc2) (33, 36). On the other hand, glycosyl hydrolases that possess a GH20 catalytic domain are predicted to cleave either nonreducing β-1,4- or β-1,6-glycosidic bonds between two adjacent N-acetylglucosamine moieties (37–39). Because EF0114 possess both a GH18 and a GH20 catalytic domain, we assessed individual recombinant domain constructs as well as two site-directed mutants for their ability to cleave the fluorogenic β1,4-GlcNAc2 analogue, 4MU-GlcNAc. The GH18 domain of EF0114 is referred to as EF0114 α, whereas the GH20 domain is referred to as EF0114 γ (33) (Fig. 2C). The central linker domain was termed EF0114 β due to its secondary structure and therefore was evaluated to determine if it contributed to overall enzymatic activity (33) (Fig. 2C). Enzymatic activity of recombinant EF0114 domains against the fluorogenic 4MU-GlcNAc substrate shows that protein constructs containing the GH20 catalytic domain (EF0114 γ or EF0114 β+γ) possess glycosidase activity against the β1,4-GlcNAc2 analogue (Fig. 2D). In contrast, EF0114 domain variants possessing the GH18 domain did not result in glycosidase activity against 4MU-GlcNAc (Fig. 2D). Additionally, the replacement of a key glutamate residue involved in the proton transfer during hydrolysis within the GH20 domain of EF0114 with a glutamine (EF0114 E662Q) results in the elimination of glycosidase activity against the 4MU-GlcNAc substrate relative to native EF0114 (Fig. 2D), highlighting the important role of the GH20 domain in the enzymatic activity to degrade GlcNAc2 into GlcNAc monomers. In contrast, the replacement of a key enzymatic residue in the GH18 domain (EF0114 E186Q) retained glycosidase activity toward the 4MU-GlcNAc substrate (Fig. 2D).
Importation of GlcNAc is mediated through the Mpt PTS complex and EF1516.
The uptake of N-acetylglucosamine has previously been studied in various Gram-positive bacteria, such as Bacillus subtilis, Streptococcus mutans, and Streptococcus pneumoniae (9, 10, 12, 40, 41). In S. mutans and S. pneumoniae, the glucose/mannose-specific EII permease (ManLMN) is capable of transporting a variety of carbohydrates, including glucose and mannose, but also has shown specificity for galactose, N-acetylglucosamine, and glucosamine (12, 40, 41); however, NagP (phosphotransferase system [PTS] EIICB) has been shown to be the main transporter of N-acetylglucosamine in B. subtilis (9, 10). It was of interest to identify the homologs of both the ManLMN PTS and NagP in E. faecalis V583. The MptBACD PTS in E. faecalis shares extensive sequence similarity with ManLMN in S. mutans (12, 40) and has been characterized as the primary glucose transporter and sole mannose transporter in E. faecalis (42, 43). In E. faecalis, ef1516 encodes a IICBA PTS and shares approximately 41% amino acid sequence identity and 55% sequence similarity with NagE (PTS IICBA) from Escherichia coli (44). EF1516 also shares 41% sequence identity and 60% sequence similarity with NagP (EIICB) in B. subtilis (9, 10). The major difference between EF1516 and NagP from B. subtilis is the presence of a PTS EIIA domain in EF1516 that is absent from NagP.
To address whether the enterococcal Mpt PTS and/or EF1516 are responsible for the importation of N-acetylglucosamine, single in-frame deletions of the mptBACD operon and ef1516, as well as the ΔmptBACD Δef1516 double deletion mutant, were grown in CDM supplemented with N-acetylglucosamine (10 mM) as the sole carbon source (Fig. 3A). As shown in Fig. 3A, the deletion of either mptBACD or ef1516 does not impede the overall growth relative to the parental strain in CDM with N-acetylglucosamine as the principal carbon source. However, a double deletion mutant of mptBACD and ef1516 results in a significant attenuation in growth relative to that of the parental or single transporter deletion mutants. Individual gene complements from the double deletion background (ΔmptBACD Δef1516::mptBACD and ΔmptBACD Δef1516::ef1516) fully restored growth, demonstrating that either PTS can functionally transport GlcNAc. To ascertain that the poor growth of the ΔmptBACD Δef1516 mutant cannot be rescued by increasing the concentration of N-acetylglucosamine, we also grew cells in CDM with 100 mM GlcNAc. Figure 3B illustrates that increasing the concentration of N-acetylglucosamine does not rescue the growth defect of the ΔmptBACD Δef1516 mutant. Collectively, this indicates that E. faecalis is capable of importing N-acetylglucosamine via two separate PTS complexes, the Mpt PTS complex and EF1516. Here, EF1516 is referred to as NagE, based on the full-length sequence similarity with the E. coli NagE homolog.
FIG 3.
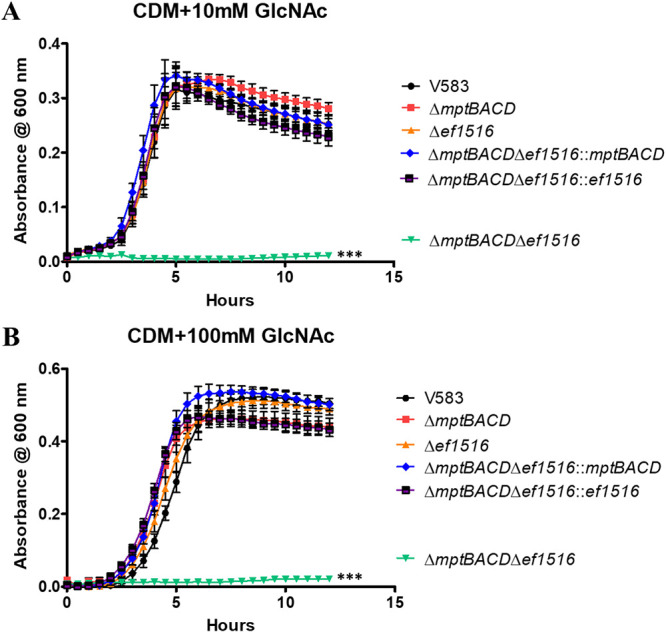
Growth of E. faecalis in CDM supplemented with 10 mM GlcNAc (A) or 100 mM GlcNAc (B). Each graph shows the average from three biological replicates, with three technical replicates each time (n = 9). The statistical significance was calculated using a two-way ANOVA test; ***, P < 0.0001.
The Mpt PTS is the primary glucosamine (GlcN) transporter in E. faecalis.
It was of interest to determine the transport machinery for the uptake of glucosamine (GlcN) in E. faecalis. Because the enterococcal NagE and Mpt PTS complexes are responsible for importing N-acetylglucosamine into the bacterial cell, we hypothesized that they also play a role in the uptake of glucosamine. To assess this hypothesis, the ΔmptBACD mutant, the Δef1516 mutant, and the ΔmptBACD Δef1516 double mutant were grown in CDM supplemented with 10 mM glucosamine. Figure 4A illustrates that the deletion of the mptBACD PTS operon eliminates growth with 10 mM glucosamine as the sole carbon source relative to the growth of the parental strain and the mptBACD genetic revertant strain. There was no significant difference in growth between the Δef1516 mutant relative to the parental strain in CDM with 10 mM glucosamine (Fig. 4A). In contrast to the growth assessment in 10 mM GlcN, the growth defect of the mptBACD mutant is partially rescued by increasing the concentration of glucosamine to 100 mM (Fig. 4B), suggesting that an additional transporter contributes to GlcN uptake at high concentrations. This partial rescue is lost in the double deletion mutant of mptBACD and ef1516 (nagE). Complementation of the double mutant with nagE partially restores growth to levels similar to that observed in the mptBACD mutant (Fig. 4B). The growth defect associated with the double mutant (ΔmptBACD Δef1516) in CDM with 100 mM GlcN is fully complementable with mptBACD, as the ΔmptBACD Δef1516::mptBACD strain grew similarly to that of the parental strain. Collectively, these results indicate that although NagE has some specificity for importing glucosamine at high concentrations, the Mpt PTS complex is the primary transport system for the uptake of glucosamine in E. faecalis.
FIG 4.
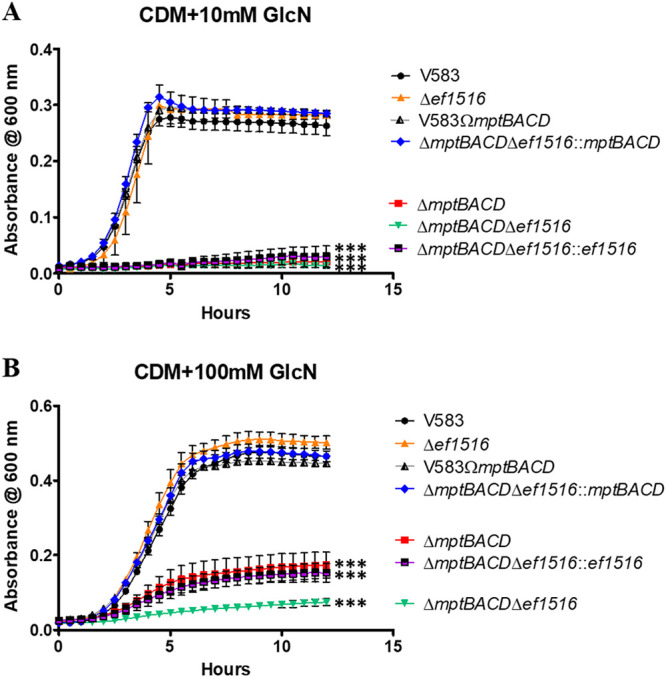
Growth of E. faecalis in CDM supplemented with 10 mM GlcN (A) or 100 mM GlcN (B). Each graph shows the average from three biological replicates, with three technical replicates each time (n = 9). The statistical significance was calculated using a two-way ANOVA test; ***, P < 0.0001.
Growth on N-acetylglucosamine is dependent on EF1317, an N-acetylglucosamine-6-phosphate deacetylase (NagA) homolog.
After PTS-mediated importation of GlcNAc into the cell via either the Mpt PTS complex or NagE, deacetylation of the phosphorylated sugar (GlcNAc-6-P) would be required prior to the conversion into fructose-6-phosphate (Fru-6-P). As observed in numerous bacteria, the enzyme responsible for deacetylating N-acetylglucosamine-6-phosphate is the N-acetylglucosamine-6-phosphate deacetylase (GlcNAc-6-P deacetylase), also referred to as NagA (7, 45–47). A predicted homolog of NagA in E. faecalis is encoded by ef1317 and shares approximately 63% amino acid sequence identity and 77% sequence similarity with NagA in S. pneumoniae (7). We therefore constructed a deletion mutant of ef1317 and assessed its growth in CDM supplemented with N-acetylglucosamine (10 mM). Figure 5A shows that the deletion of ef1317 eliminated the ability of E. faecalis to grow in CDM supplemented with 10 mM GlcNAc and that this phenotype is complementable, as the ef1317 complement strain restored growth equivalent to that of the parental strain. The growth of the ef1317 mutant can also be experimentally rescued by growing this mutant in CDM supplemented with glucosamine (Fig. 5B).
FIG 5.
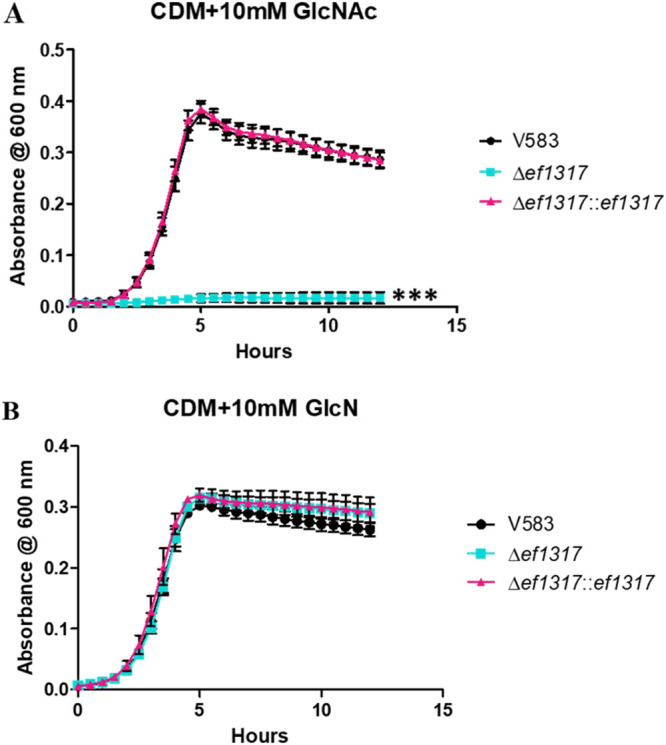
Growth of E. faecalis in CDM supplemented with 10 mM GlcNAc (A) or 10 mM GlcN (B). Each graph shows the average from three biological replicates, with six technical replicates each time (n = 18). The statistical significance was calculated using a two-way ANOVA test; ***, P < 0.0001.
Growth on glucosamine is dependent on the activity of EF0466, a glucosamine-6-phosphate deaminase (NagB) homolog.
Following deacetylation of GlcNAc-6-P into GlcN-6-P, an additional reaction is required before the amino sugar is funneled into the glycolysis pathway. This reaction entails the conversion of GlcN-6-P to fructose-6-phosphate (Fru-6-P) by the glucosamine-6-phosphate deaminase (GlcN-6-P deaminase), referred to as NagB (7, 45, 48). The NagB homolog in E. faecalis was identified as EF0466, as it shares 60% amino acid sequence identity and 78% sequence similarity with NagB of S. pneumoniae (7). When grown in CDM supplemented with glucosamine (10 mM), the deletion of ef0466 results in a significant decrease in growth relative to the parental and complement strains (Fig. 6A). The growth defect associated with the ef0466 mutant when grown in glucosamine can be restored by growing this mutant in CDM supplemented with fructose (Fig. 6B), highlighting that pathways downstream of the glucosamine to fructose-6-phosphate remain undisturbed.
FIG 6.
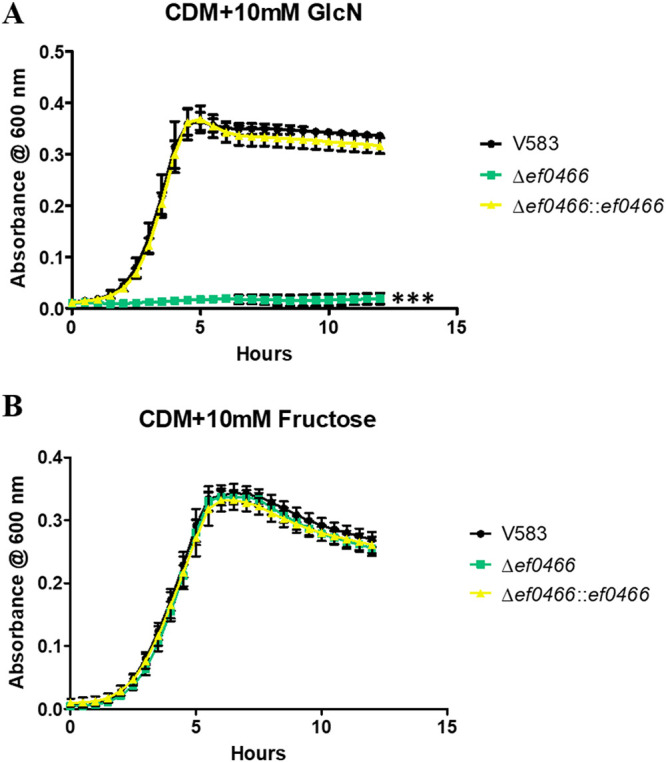
Growth of E. faecalis in CDM supplemented with 10 mM GlcN (A) or 10 mM fructose (B). Each graph shows the average from three biological replicates, with six technical replicates each time (n = 18). The statistical significance was calculated using a one-way ANOVA test; ***, P < 0.0001.
DISCUSSION
The plethora of functionally encoded sugar transport systems in the E. faecalis genome emphasizes the importance of carbohydrate utilization and confers an advantage to survive in nutrient-varying host environments. Although glucose is the preferred carbon source for bacteria, this carbohydrate is often present at concentrations that are growth limiting for most bacteria, including enterococci (49, 50). Extensive studies have been performed regarding the regulation of secondary nutrient acquisition systems that allow E. faecalis to survive in glucose-limiting environments (42, 51, 52). In Gram-positive bacteria, the transcriptional regulator CcpA plays a key role in regulating the expression of alternative nutrient acquisition systems via carbon catabolite repression (CCR) (42, 53, 54). Previously, the GH18 containing enterococcal glycosyl hydrolases (EF0114 [EndoE], EF0361 [EfChi18A], and EF2863 [EfEndo18A]) and the enterococcal chitin-binding protein/lytic polysaccharide monooxygenase (EF0362 [EfCBM33A]) were not only found to be significantly expressed in the absence of glucose and secreted (42), but were also found to be negatively regulated by CcpA (33). Their enzymatic activity was also previously assessed, and these studies revealed that they have enzymatic specificity toward the β1,4-linked N-acetylglucosamine glycosidic bond present in chitinous substrates (30) and glycoproteins (high-mannose and complex-type glycoproteins) (32, 33, 36), suggesting that they are involved in acquiring alternative carbon sources under glucose-limiting conditions. The genes encoding these enzymes have also been shown to be induced when E. faecalis is grown in blood or urine (51, 52), consistent with glucose-limited host environments.
Previous observations show that the enterococcal chitinase (EF0361; EfChi18A) is capable of degrading poly-β1,4-linked GlcNAc substrates (i.e., chitin [poly-β1,4-GlcNAc], chitohexaose [GlcNAc6], chitopentaose [GlcNAc5], chitotetraose [GlcNAc4], or chitotriose [GlcNAc3]) (30). However, in the presence of its cognate chitin binding protein/lytic polysaccharide monooxygenase (EF0362; EfCBM33A), EfChi18A degrades chitin five times more efficiently relative to chitin degradation by the chitinase alone (30). Therefore, EF0362-61 acts synergistically to catalyze the degradation of poly-β1,4-linked GlcNAc substrates. The previous analyses were conducted with recombinant protein and not in the context of other enterococcal glycosyl hydrolases that have specificity toward the β-1,4-linked glycosidic bond between two adjoining N-acetylglucosamine moieties. Using genetic mutants of ef0362-61 and the other GH18-containing glycosyl hydrolases present in the enterococcal genome (ef0114 and ef2863), our analysis suggests that EF0362-61 and EF0114 are required for growth on poly-β-1,4-linked GlcNAc substrates. It is noteworthy that Vaaje-Kolstad et al. found that EF0362-61 can only synergistically degrade poly-β-1,4-GlcNAc substrates into dimers of β-1,4-linked N-acetylglucosamine, indicating that an additional enzyme(s) is required for fully degrading these poly-β-1,4-GlcNAc substrates into monomers of N-acetylglucosamine prior to uptake into the bacterial cell. Since EF0114 and EF2863 target β1,4-linked GlcNAc (31–33), these glycosidases are good candidates for hydrolyzing extracellular GlcNAc2 into GlcNAc prior to importation into the bacterial cell. Based on our analysis using both genetic mutants and purified recombinant protein, this study revealed that the GH20 catalytic domain of EF0114 is responsible for cleaving the β1,4-linked glycosidic bond of extracellular chitobiose (β1,4-linked GlcNAc2). To our knowledge, this would be the first instance in which a precise enzymatic function has been determined for the GH20 catalytic domain of EF0114. Collectively, these observations indicate that the degradation of extracellular poly-β1,4-linked GlcNAc-containing substrates is dependent on the enzymatic activity of CcpA-regulated enzymes, EF0362-61 and EF0114.
Recent evidence indicates that chitinases and CBPs secreted by bacterial pathogens, such as Listeria monocytogenes (25, 26) and Legionella pneumophila (27), serve as virulence factors during infection of mammalian hosts (23, 24). Recent observations indicate that chitinases produced by Salmonella and Listeria also have specificity toward LacNac (Galβ1-4GlcNAc) and LacdiNAc (GalNAcβ1-4GlcNAc) sugar linkages present in mammalian N- and O-linked glycans, glycolipids and glycosaminoglycans (24, 28). These LacNAc- and LacdiNAc-containing substrates present in a mammalian host are likely an additional substrate for bacterial chitinases, and chitinolytic cleavage of these substrates may contribute to bacterial pathogenesis in a host. L. monocytogenes produces two chitinases, annotated as ChiA and ChiB. Chaudhuri et al. observed that the deletion of chiB resulted in an approximately 8-fold and 14-fold decrease in bacterial burden in the liver and spleen, respectively, of infected mice (25). However, the chiA deletion mutant exhibited a more dramatic virulence defect, as bacterial recovery from the liver and spleen of infected mice was reduced 19-fold and 45-fold, respectively (25). Using NCBI BLAST, the ChiA chitinase of L. monocytogenes was found to share approximately 65% sequence identity and 77% sequence similarity with EF0361 (EfChi18A) produced by E. faecalis. In contrast, there is no apparent enterococcal homolog of the listerial ChiB chitinase or the chitinase produced by L. pneumophila. Determining a functional role for EF0362-61 in the context of mammalian infection by E. faecalis will be a component of ongoing studies.
Although evidence indicates that E. faecalis can utilize N-acetylglucosamine and glucosamine as a carbon source (6), little is known of the uptake machinery and cytosolic enzymes required for the importation and metabolism of these amino sugars. The uptake of N-acetylglucosamine has previously been studied in various Gram-positive bacteria, such as Bacillus subtilis (9, 10), Streptococcus mutans (12, 40), and Streptococcus pneumoniae (41). In S. mutans and S. pneumoniae, the glucose/mannose-specific EII permease (ManLMN) is capable of transporting a variety of carbohydrates, including glucose and mannose, but also has shown specificity for galactose, N-acetylglucosamine, and glucosamine (12, 40, 41); however, NagP (PTS EIICB) has been shown to be the main transporter of N-acetylglucosamine in B. subtilis (9, 10). Our study indicates that E. faecalis encodes two PTS complexes that are involved in the uptake of N-acetylglucosamine, as single deletions of either the mpt PTS operon or ef1516 did not display a significant change in growth; however, deletion of ef1516 in the mptBACD mutant background resulted in complete attenuation of growth in CDM with either 10 mM or 100 mM N-acetylglucosamine. With respect to glucosamine uptake, the Mpt PTS is the primary transporter for importing glucosamine into the cell, as the deletion of mptBACD resulted in an attenuation in growth with 10 mM glucosamine as the sole carbon source. However, the PTS encoded by nagE (ef1516) does have a low affinity toward importing glucosamine into the cell, as increasing the concentration of glucosamine to 100 mM partially rescued the growth of the mptBACD mutant, and growth of the ΔmptBACD Δef1516 double mutant was significantly impaired relative to that of the ΔmptBACD mutant alone.
The functional redundancy of two GlcNAc/GlcN transporters in E. faecalis may be attributed to the fact that components of the Mpt PTS complex are known cellular receptors for class IIa and IIc bacteriocins (55). In complex human ecologies such as the oral cavity and gastrointestinal tract, E. faecalis is likely exposed to such bacteriocins that may inactivate sugar transport through the Mpt system (56) and therefore limit its metabolic capacity. The Mpt PTS is the principal glucose transporter and the sole mannose transporter in E. faecalis (42); additionally, here, we have shown it to be the primary transporter of glucosamine and a major contributor to GlcNAc uptake. The ability of E. faecalis to take advantage of a dysbiotic intestinal microflora may stem from the ability to efficiently transport multiple key host sugars via the Mpt PTS. The presence of a complex intestinal microflora is known to limit the growth of E. faecalis (57, 58) and highlights that maintaining this competitive microflora is a key aspect of disease prevention. Furthermore, we have recently shown that the alternative sigma factor, RpoN, is required for mptBACD expression and significantly contributes to infection outcome in both endocarditis and urinary tract infection models (42), highlighting an important role for sugar metabolism during infection.
For catabolic processing of GlcNAc, the enzymatic function of NagA (N-acetylglucosamine-6-phosphate deacetylase), followed by the activity of NagB (glucosamine-6-phosphate deaminase) is required to metabolize GlcNAc via glycolysis (7, 45–48). Our study shows that EF1317 (NagA) and EF0466 (NagB) are essential for E. faecalis to utilize N-acetylglucosamine as a carbon source. A review of enterococcal physiology and metabolism (59) identified two potential NagA homologs in the E. faecalis genome, encoded by ef1317 and ef3044, but our analysis provides direct evidence that EF1317 serves the primary activity of deacetylating GlcNAc.
N-Acetylglucosamine has a dual purpose in bacterial catabolism. When supplied exogenously, the catabolism of N-acetylglucosamine or glucosamine by NagB generates fructose-6-phosphate to provide a valuable source of energy to the cell, and ammonia is liberated to assist with cellular nitrogen needs (48). N-Acetylglucosamine is also an essential component of bacterial peptidoglycan (13, 60). Throughout the bacterial growth cycle, peptidoglycan is continuously made and degraded to accommodate cell growth and division into daughter cells (13, 60, 61). The involvement of GlcNAc for both metabolism and peptidoglycan synthesis requires a regulatory circuit(s) for this dual utilization, and in related Gram-positive bacteria, such as B. subtilis and S. mutans, this regulatory control is governed by NagR (62). NagR belongs to the GntR family of transcriptional regulators and directly regulates the expression of nagA, nagB, and glmS in S. mutans (63). However, in B. subtilis, the expression of colocalized nagAB is regulated by NagR, while the regulation of glmS expression is independent of NagR (45). In both S. mutans and B. subtilis, NagR is encoded distantly from nagA, nagB, or glmS. Currently, there are 11 annotated GntR regulators in the E. faecalis V583 genome with similar sequence identity and similarity to NagR in S. mutans, none of which are in close proximity to ef1317 (nagA) or ef0466 (nagB). Elucidating which GntR homolog is responsible for regulating the expression of nagA and nagB in E. faecalis will require further assessment and will be an area of future research interest.
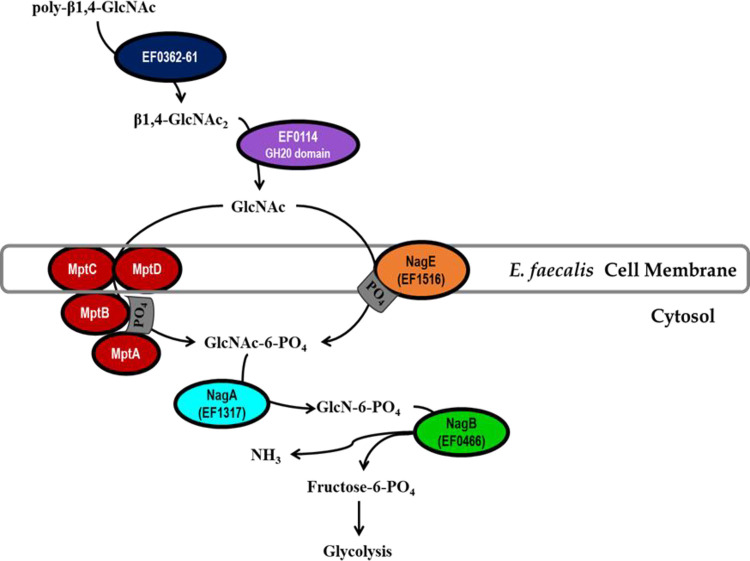
We present here a model of the metabolic pathway by which E. faecalis utilizes poly-β1,4-linked N-acetylglucosamine as a carbon source (Fig. 7). Extracellular polymers of β1,4-linked GlcNAc are hydrolyzed into β1,4-linked GlcNAc dimers by the combined activity of the chitin-binding protein, EF0362 (EfCBM33A), and the chitinase, EF0361 (EfChi18A). Subsequently, the GH20 catalytic domain of EF0114 (EndoE) is responsible for degrading β-1,4-linked GlcNAc2 into monomers of N-acetylglucosamine. The monomeric form of this amino sugar can be imported into the bacterial cell and simultaneously phosphorylated via either the Mpt PTS complex or NagE (EF1516). Once inside the bacterial cell, prior to downstream glycolytic processing, phosphorylated GlcNAc (GlcNAc-6-P) is deacetylated by NagA (EF1317), followed by deamination by NagB (EF0466), converting glucosamine-6-P into fructose-6-P and ammonia. It is noteworthy that as a result of catabolizing GlcNAc or GlcN, ammonia is generated and can be utilized in amino acid synthesis or other essential components (12). The accumulation of cytosolic ammonia can increase the difference in pH inside relative to outside the cell (i.e., ΔpH) and therefore contributes to acid tolerance (12). Although our study identified the chitinolytic machinery, the GlcNAc and GlcN-specific uptake systems, and cytosolic enzymes that are required for enterococcal metabolism of β-1,4-linked N-acetylglucosamine substrates, how they contribute to overall enterococcal pathogenesis will be an area of future research interest.
FIG 7.
Model for poly-β1,4-GlcNAc degradation and GlcNAc uptake and metabolism in Enterococcus faecalis. Extracellular poly-β1,4-N-acetylglucosamine substrates (β1,4-GlcNAc3-n) are hydrolyzed by EF0362-61 into dimers of β1,4-N-acetylglucosamine (β1,4-GlcNAc2), followed by hydrolysis into monomeric GlcNAc by the GH20 catalytic domain of EF0114. N-Acetylglucosamine is transported into the cell and simultaneously phosphorylated upon uptake by two PTS complexes, the Mpt PTS complex (MptBACD) and EF1516 (NagE). For the phosphorylated amino sugar to be metabolized via glycolysis, it first requires deacetylation by EF1317 (NagA), followed by conversion into fructose-6-PO4 by EF0466 (NagB) with ammonia generation (NH3).
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used in this study are listed in Table S1 in the supplemental material. For propagation of plasmids, Escherichia coli ElectroTen-Blue from Stratagene was cultivated in Luria-Bertani (LB) broth, supplemented with appropriate antibiotics when necessary. Unless mentioned otherwise, E. faecalis was cultured in Todd-Hewitt broth (THB) (BD Biosciences), supplemented with appropriate antibiotics when necessary. For antibiotic selection, chloramphenicol (Cm) at a concentration of 10 μg/ml and 15 μg/ml was used for E. coli and E. faecalis, respectively; ampicillin (Amp) at a concentration of 100 μg/ml was used for E. coli, and gentamicin (Gent) at a concentration of 250 μg/ml was used for E. faecalis.
Construction of in-frame markerless deletion strains.
Using the temperature-sensitive cloning vector pLT06 (64), isogenic in-frame deletion mutants were generated in E. faecalis V583. Upstream and downstream flanking DNA regions of the respective gene target were amplified using the primers listed in Table S2. For example, the primer pairs EF1317P1/EF1317P2 and EF1317P3/EF1317P4 were used to amplify flanking regions upstream and downstream of ef1317, respectively. To facilitate cloning, primers EF1317P1 and EF1317P2 were designed with EcoRI and BamHI (New England Biolabs [NEB]) restriction sites, respectively, whereas, EF1317P3 and EF1317P4 were designed with BamHI and PstI (NEB) restriction sites, respectively. For the construction of the insert, the amplified regions were digested with BamHI, ligated, and subsequently reamplified using primers EF1317P1 and EF1317P4. To generate pSP20 (ef1317 deletion construct), the amplified insert fragment was digested with EcoRI and PstI and ligated into the digested pLT06 cloning vector. pSP20 was electroporated into E. coli ElectroTen-Blue cells, and the plasmid was identified by colony PCR. The confirmed construct was screened by restriction digest analysis and subsequently electroporated into E. faecalis V583 cells. V583Δef1317 (SP27) was subsequently generated as previously described following plasmid integration and excision events (64) and confirmed by colony PCR using the primers EF1317Up and EF1317Down. In addition to the gene deletion mutants, paired genetic revertants (57, 65) from this PCR screen were also selected based on the restoration to the parental genotype following excision of the integrated plasmid and were designated V583Ωef1317. A similar approach was used to create all the gene deletion mutants and paired genetic revertants used in this and prior studies (33, 42) referenced in Table S1.
Construction of in-frame markerless complement strains.
Using the temperature-sensitive cloning vector pLT06 (64), the isogeneic in-frame EF1317 complement strain was generated in E. faecalis V583. The entirety of the ef1317 gene, including its upstream and downstream flanking DNA regions, was amplified using the primer pair EF1317P1/EF1317P4 (Table S2). For cloning purposes, EF1317P1 and EF1317P4 were designed with EcoRI and PstI restriction sites, respectively. The amplified region was digested with EcoRI and PstI and ligated into the digested pLT06 cloning vector, resulting in the creation of pEK60 (ef1317 complement construct). pEK60 was electroporated into E. coli ElectroTen-Blue cells, and the plasmid was identified by colony PCR. The confirmed construct was screened by restriction digest analysis and subsequently electroporated into E. faecalis V583Δef1317 (SP27) cells. V583Δef1317::ef1317 (EK47) was subsequently generated as previously described (64) and confirmed by colony PCR using the primers EF1317Up and EF1317Down.
Growth assessment in nutrient-limiting conditions.
Using a single colony of each strain, liquid cultures were started in THB and incubated at 37°C overnight. For growth analysis, overnight cultures were diluted 1:100 in a chemically defined medium (CDM) (34, 35), supplemented with either chitopentaose (GlcNAc5) (Seikagaku Kogyo Co.), chitobiose (GlcNAc2) (Seikagaku Kogyo Co.), N-acetylglucosamine (GlcNAc) (Acros Organics), glucosamine (GlcN) (Acros Organics), or fructose (Sigma-Aldrich). Growth was monitored for 12 h in an Infinite M200 Pro plate reader (Tecan Trading AG, Switzerland) at 37°C with orbital shaking at 250 rpm.
Expression and purification of EF0114 variants.
The plasmids bearing the protein overexpression constructs, listed in Table S3, are all derivatives of the pET21b vector (Novagen), and details of their construction are described in Keffeler et al. (33). For overexpression and purification, E. coli BL21(DE3)-RIPL cells (Agilent Technologies) harboring protein overexpression constructs were grown to an optical density at 600 nm (OD600) of 0.6 of 0.7 at 37°C, with shaking, and then induced using 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) and further incubated for 16 h at 16°C, after which the cells were harvested by centrifugation and resuspended in buffer A (100 mM Tris-HCl, 0.5 M NaCl, 10% glycerol, and 15 mM imidazole, pH 8.0). The bacteria were lysed using a French press, and the soluble fractions were loaded onto a cobalt-affinity column (GE Healthcare TALON Superflow) equilibrated with buffer A. Recombinant proteins were washed with buffer A and eluted with an imidazole gradient using buffers A and B (100 mM Tris-HCl, 0.5 M NaCl, 10% glycerol, and 500 mM imidazole, pH 8.0). Protein purity was analyzed by SDS-PAGE, and the protein concentration was determined using the Bradford microassay (Bio-Rad Laboratories) according to the supplier’s procedure.
Chitinolytic assays.
Glycosidase activity against the fluorogenic substrate 4MU-methylumbelliferyl-β-d-N-acetylglucosamine (GlcNAc2 analogue) (Sigma-Aldrich) was determined by incubating the fluorogenic substrate with filtered supernatants of bacterial cultures grown overnight (37°C with shaking) in a chemically defined medium (CDM) supplemented with 42 mM glucose. Briefly, 4MU-GlcNAc (50 μM) was incubated in prewarmed 25 mM citrate buffer (pH 6.0) for 15 min at 37°C, in a reaction volume of 150 μl. Filtered supernatant (50 μl) was subsequently added to the prewarmed chitinolytic reactions, briefly mixed, and incubated for 15 min at 37°C, resulting in a total reaction volume of 200 μl. The fluorescence intensity (excitation, 390 nm; emission, 485 nm) was measured using an Infinite M200 Pro plate reader (Tecan Trading AG, Switzerland). Chitinolytic activity is expressed as the relative fluorescence calculated by dividing the fluorescence emitted at 485 nm by the OD600 of the culture. For analysis of purified recombinant EF0114 variants, 1 μM recombinant enzyme was incubated with prewarmed chitinolytic reactions in a total reaction volume of 200 μl. Chitinolytic activity is expressed as the fluorescence emitted at 485 nm.
Statistical analyses.
The statistical analyses of the various experiments were conducted using GraphPad Prism 5 software (San Diego, CA).
ACKNOWLEDGMENTS
We gratefully acknowledge Subbaratnam Muthukrishna (Kansas State University, Department of Biochemistry & Molecular Biophysics) for providing the chitopentaose (GlcNAc5) and chitobiose (GlcNAc2) substrates.
Portions of this work were supported by Public Health Service grants AI77782 and AI117424 (L.E.H.) from the National Institutes of Health and a McNair Scholars Program grant (Z.H.A.) funded by the U.S. Department of Education TRIO program. The content of the sponsored research is solely the responsibility of the authors and does not necessarily represent the official views of the sponsoring agencies.
Footnotes
Supplemental material is available online only.
Contributor Information
Lynn E. Hancock, Email: lynnh@ku.edu.
Michael J. Federle, University of Illinois at Chicago
REFERENCES
- 1.Lebreton F, Willems RJL, Gilmore MS. 2014. Enterococcus diversity, origins in nature, and gut colonization. In Gilmore MS, Clewell DB, Ike Y, Shankar N (ed), Enterococci: from commensals to leading causes of drug resistant infection. Massachusetts Eye and Ear Infirmary, Boston, MA. [PubMed] [Google Scholar]
- 2.Garcia-Solache M, Rice LB. 2019. The Enterococcus: a model of adaptability to its environment. Clin Microbiol Rev 32:e00058-18. 10.1128/CMR.00058-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van Tyne D, Gilmore MS. 2014. Friend turned foe: evolution of enterococcal virulence and antibiotic resistance. Annu Rev Microbiol 68:337–356. 10.1146/annurev-micro-091213-113003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weiner-Lastinger LM, Abner S, Edwards JR, Kallen AJ, Karlsson M, Magill SS, Pollock D, See I, Soe MM, Walters MS, Dudeck MA. 2020. Antimicrobial-resistant pathogens associated with adult healthcare-associated infections: summary of data reported to the National Healthcare Safety Network, 2015-2017. Infect Control Hosp Epidemiol 41:1–18. 10.1017/ice.2019.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fiore E, Van Tyne D, Gilmore MS. 2019. Pathogenicity of enterococci. Microbiol Spectr 7:7.4.9. 10.1128/microbiolspec.GPP3-0053-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lebreton F, Manson AL, Saavedra JT, Straub TJ, Earl AM, Gilmore MS. 2017. Tracing the enterococci from Paleozoic origins to the hospital. Cell 169:849–861.e13. 10.1016/j.cell.2017.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Afzal M, Shafeeq S, Manzoor I, Henriques-Normark B, Kuipers OP. 2016. N-acetylglucosamine-mediated expression of nagA and nagB in Streptococcus pneumoniae. Front Cell Infect Microbiol 6:158. 10.3389/fcimb.2016.00158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dobrogosz WJ. 1968. Effect of amino sugars on catabolite repression in Escherichia coli. J Bacteriol 95:578–584. 10.1128/jb.95.2.578-584.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaugue I, Oberto J, Plumbridge J. 2014. Regulation of amino sugar utilization in Bacillus subtilis by the GntR family regulators, NagR and GamR. Mol Microbiol 92:100–115. 10.1111/mmi.12544. [DOI] [PubMed] [Google Scholar]
- 10.Gaugue I, Oberto J, Putzer H, Plumbridge J. 2013. The use of amino sugars by Bacillus subtilis: presence of a unique operon for the catabolism of glucosamine. PLoS One 8:e63025. 10.1371/journal.pone.0063025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mobley HL, Doyle RJ, Streips UN, Langemeier SO. 1982. Transport and incorporation of N-acetyl-D-glucosamine in Bacillus subtilis. J Bacteriol 150:8–15. 10.1128/jb.150.1.8-15.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moye ZD, Burne RA, Zeng L. 2014. Uptake and metabolism of N-acetylglucosamine and glucosamine by Streptococcus mutans. Appl Environ Microbiol 80:5053–5067. 10.1128/AEM.00820-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park JT, Uehara T. 2008. How bacteria consume their own exoskeletons (turnover and recycling of cell wall peptidoglycan). Microbiol Mol Biol Rev 72:211–227. 10.1128/MMBR.00027-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wheeler R, Chevalier G, Eberl G, Gomperts Boneca I. 2014. The biology of bacterial peptidoglycans and their impact on host immunity and physiology. Cell Microbiol 16:1014–1023. 10.1111/cmi.12304. [DOI] [PubMed] [Google Scholar]
- 15.Garcia-Rubio R, de Oliveira HC, Rivera J, Trevijano-Contador N. 2019. The fungal cell wall: Candida, Cryptococcus, and Aspergillus species. Front Microbiol 10:2993. 10.3389/fmicb.2019.02993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu X, Zhang J, Zhu KY. 2019. Chitin in arthropods: biosynthesis, modification, and metabolism. Adv Exp Med Biol 1142:169–207. 10.1007/978-981-13-7318-3_9. [DOI] [PubMed] [Google Scholar]
- 17.Chen JK, Shen CR, Liu CL. 2010. N-Acetylglucosamine: production and applications. Mar Drugs 8:2493–2516. 10.3390/md8092493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liao C, Rigali S, Cassani CL, Marcellin E, Nielsen LK, Ye BC. 2014. Control of chitin and N-acetylglucosamine utilization in Saccharopolyspora erythraea. Microbiology (Reading) 160:1914–1928. 10.1099/mic.0.078261-0. [DOI] [PubMed] [Google Scholar]
- 19.Ismail SA, El-Sayed HS, Fayed B. 2020. Production of prebiotic chitooligosaccharide and its nano/microencapsulation for the production of functional yoghurt. Carbohydr Polym 234:115941. 10.1016/j.carbpol.2020.115941. [DOI] [PubMed] [Google Scholar]
- 20.van Huis A. 2020. Nutrition and health of edible insects. Curr Opin Clin Nutr Metab Care 23:228–231. 10.1097/MCO.0000000000000641. [DOI] [PubMed] [Google Scholar]
- 21.Verkhnyatskaya S, Ferrari M, de Vos P, Walvoort MTC. 2019. Shaping the infant microbiome with non-digestible carbohydrates. Front Microbiol 10:343. 10.3389/fmicb.2019.00343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Forsberg Z, Sørlie M, Petrović D, Courtade G, Aachmann FL, Vaaje-Kolstad G, Bissaro B, Røhr ÅK, Eijsink VG. 2019. Polysaccharide degradation by lytic polysaccharide monooxygenases. Curr Opin Struct Biol 59:54–64. 10.1016/j.sbi.2019.02.015. [DOI] [PubMed] [Google Scholar]
- 23.Bhattacharya D, Nagpure A, Gupta RK. 2007. Bacterial chitinases: properties and potential. Crit Rev Biotechnol 27:21–28. 10.1080/07388550601168223. [DOI] [PubMed] [Google Scholar]
- 24.Frederiksen RF, Paspaliari DK, Larsen T, Storgaard BG, Larsen MH, Ingmer H, Palcic MM, Leisner JJ. 2013. Bacterial chitinases and chitin-binding proteins as virulence factors. Microbiology (Reading) 159:833–847. 10.1099/mic.0.051839-0. [DOI] [PubMed] [Google Scholar]
- 25.Chaudhuri S, Bruno JC, Alonzo F, 3rd, Xayarath B, Cianciotto NP, Freitag NE. 2010. Contribution of chitinases to Listeria monocytogenes pathogenesis. Appl Environ Microbiol 76:7302–7305. 10.1128/AEM.01338-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Larsen MH, Leisner JJ, Ingmer H. 2010. The chitinolytic activity of Listeria monocytogenes EGD is regulated by carbohydrates but also by the virulence regulator PrfA. Appl Environ Microbiol 76:6470–6476. 10.1128/AEM.00297-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DebRoy S, Dao J, Soderberg M, Rossier O, Cianciotto NP. 2006. Legionella pneumophila type II secretome reveals unique exoproteins and a chitinase that promotes bacterial persistence in the lung. Proc Natl Acad Sci USA 103:19146–19151. 10.1073/pnas.0608279103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frederiksen RF, Yoshimura Y, Storgaard BG, Paspaliari DK, Petersen BO, Chen K, Larsen T, Duus J, Ingmer H, Bovin NV, Westerlind U, Blixt O, Palcic MM, Leisner JJ. 2015. A diverse range of bacterial and eukaryotic chitinases hydrolyzes the LacNAc (Galβ1-4GlcNAc) and LacdiNAc (GalNAcβ1-4GlcNAc) motifs found on vertebrate and insect cells. J Biol Chem 290:5354–5366. 10.1074/jbc.M114.607291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leisner JJ, Larsen MH, Ingmer H, Petersen BO, Duus JO, Palcic MM. 2009. Cloning and comparison of phylogenetically related chitinases from Listeria monocytogenes EGD and Enterococcus faecalis V583. J Appl Microbiol 107:2080–2087. 10.1111/j.1365-2672.2009.04420.x. [DOI] [PubMed] [Google Scholar]
- 30.Vaaje-Kolstad G, Bohle LA, Gaseidnes S, Dalhus B, Bjoras M, Mathiesen G, Eijsink VG. 2012. Characterization of the chitinolytic machinery of Enterococcus faecalis V583 and high-resolution structure of its oxidative CBM33 enzyme. J Mol Biol 416:239–254. 10.1016/j.jmb.2011.12.033. [DOI] [PubMed] [Google Scholar]
- 31.Bohle LA, Mathiesen G, Vaaje-Kolstad G, Eijsink VG. 2011. An endo-beta-N-acetylglucosaminidase from Enterococcus faecalis V583 responsible for the hydrolysis of high-mannose and hybrid-type N-linked glycans. FEMS Microbiol Lett 325:123–129. 10.1111/j.1574-6968.2011.02419.x. [DOI] [PubMed] [Google Scholar]
- 32.Collin M, Fischetti VA. 2004. A novel secreted endoglycosidase from Enterococcus faecalis with activity on human immunoglobulin G and ribonuclease B. J Biol Chem 279:22558–22570. 10.1074/jbc.M402156200. [DOI] [PubMed] [Google Scholar]
- 33.Keffeler EC, Iyer VS, Henderson AJ, Huck IL, Schwarting N, Cortez A, Hancock LE. 2021. Activity of CcpA-regulated GH18 family glycosyl hydrolases that contribute to nutrient acquisition and fitness in Enterococcus faecalis. Infect Immum 10.1128/IAI.00343-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brown SA, Whiteley M. 2007. A novel exclusion mechanism for carbon resource partitioning in Aggregatibacter actinomycetemcomitans. J Bacteriol 189:6407–6414. 10.1128/JB.00554-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Socransky SS, Dzink JL, Smith CM. 1985. Chemically defined medium for oral microorganisms. J Clin Microbiol 22:303–305. 10.1128/jcm.22.2.303-305.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garbe J, Sjogren J, Cosgrave EF, Struwe WB, Bober M, Olin AI, Rudd PM, Collin M. 2014. EndoE from Enterococcus faecalis hydrolyzes the glycans of the biofilm inhibiting protein lactoferrin and mediates growth. PLoS One 9:e91035. 10.1371/journal.pone.0091035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang YL, Yu WL, Zhang JW, Frolet C, Di Guilmi AM, Zhou CZ, Vernet T, Chen Y. 2011. Structural basis for the substrate specificity of a novel beta-N-acetylhexosaminidase StrH protein from Streptococcus pneumoniae R6. J Biol Chem 286:43004–43012. 10.1074/jbc.M111.256578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ramasubbu N, Thomas LM, Ragunath C, Kaplan JB. 2005. Structural analysis of dispersin B, a biofilm-releasing glycoside hydrolase from the periodontopathogen Actinobacillus actinomycetemcomitans. J Mol Biol 349:475–486. 10.1016/j.jmb.2005.03.082. [DOI] [PubMed] [Google Scholar]
- 39.Val-Cid C, Biarnes X, Faijes M, Planas A. 2015. Structural-functional analysis reveals a specific domain organization in family GH20 hexosaminidases. PLoS One 10:e0128075. 10.1371/journal.pone.0128075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abranches J, Chen YY, Burne RA. 2003. Characterization of Streptococcus mutans strains deficient in EIIAB Man of the sugar phosphotransferase system. Appl Environ Microbiol 69:4760–4769. 10.1128/AEM.69.8.4760-4769.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bidossi A, Mulas L, Decorosi F, Colomba L, Ricci S, Pozzi G, Deutscher J, Viti C, Oggioni MR. 2012. A functional genomics approach to establish the complement of carbohydrate transporters in Streptococcus pneumoniae. PLoS One 7:e33320. 10.1371/journal.pone.0033320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Keffeler EC, Iyer VS, Parthasarathy S, Ramsey MM, Gorman MJ, Barke TL, Varahan S, Olson S, Gilmore MS, Abdullahi ZH, Hancock EN, Hancock LE. 2021. Influence of the alternative sigma factor RpoN on global gene expression and carbon catabolism in Enterococcus faecalis V583. mBio 12:e00380-21. 10.1128/mBio.00380-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Opsata M, Nes IF, Holo H. 2010. Class IIa bacteriocin resistance in Enterococcus faecalis V583: the mannose PTS operon mediates global transcriptional responses. BMC Microbiol 10:224. 10.1186/1471-2180-10-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rogers MJ, Ohgi T, Plumbridge J, Söll D. 1988. Nucleotide sequences of the Escherichia coli nagE and nagB genes: the structural genes for the N-acetylglucosamine transport protein of the bacterial phosphoenolpyruvate: sugar phosphotransferase system and for glucosamine-6-phosphate deaminase. Gene 62:197–207. 10.1016/0378-1119(88)90558-6. [DOI] [PubMed] [Google Scholar]
- 45.Bertram R, Rigali S, Wood N, Lulko AT, Kuipers OP, Titgemeyer F. 2011. Regulon of the N-acetylglucosamine utilization regulator NagR in Bacillus subtilis. J Bacteriol 193:3525–3536. 10.1128/JB.00264-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M. 2014. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res 42:D199–205. 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vincent F, Yates D, Garman E, Davies GJ, Brannigan JA. 2004. The three-dimensional structure of the N-acetylglucosamine-6-phosphate deacetylase, NagA, from Bacillus subtilis: a member of the urease superfamily. J Biol Chem 279:2809–2816. 10.1074/jbc.M310165200. [DOI] [PubMed] [Google Scholar]
- 48.Vincent F, Davies GJ, Brannigan JA. 2005. Structure and kinetics of a monomeric glucosamine 6-phosphate deaminase: missing link of the NagB superfamily? J Biol Chem 280:19649–19655. 10.1074/jbc.M502131200. [DOI] [PubMed] [Google Scholar]
- 49.Shepard BD, Gilmore MS. 2002. Differential expression of virulence-related genes in Enterococcus faecalis in response to biological cues in serum and urine. Infect Immun 70:4344–4352. 10.1128/IAI.70.8.4344-4352.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wright EM, Hirayama BA, Loo DF. 2007. Active sugar transport in health and disease. J Intern Med 261:32–43. 10.1111/j.1365-2796.2006.01746.x. [DOI] [PubMed] [Google Scholar]
- 51.Vebo HC, Snipen L, Nes IF, Brede DA. 2009. The transcriptome of the nosocomial pathogen Enterococcus faecalis V583 reveals adaptive responses to growth in blood. PLoS One 4:e7660. 10.1371/journal.pone.0007660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vebo HC, Solheim M, Snipen L, Nes IF, Brede DA. 2010. Comparative genomic analysis of pathogenic and probiotic Enterococcus faecalis isolates, and their transcriptional responses to growth in human urine. PLoS One 5:e12489. 10.1371/journal.pone.0012489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fujita Y. 2009. Carbon catabolite control of the metabolic network in Bacillus subtilis. Biosci Biotechnol Biochem 73:245–259. 10.1271/bbb.80479. [DOI] [PubMed] [Google Scholar]
- 54.Warner JB, Lolkema JS. 2003. CcpA-dependent carbon catabolite repression in bacteria. Microbiol Mol Biol Rev 67:475–490. 10.1128/MMBR.67.4.475-490.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miwa Y, Nakata A, Ogiwara A, Yamamoto M, Fujita Y. 2000. Evaluation and characterization of catabolite-responsive elements (cre) of Bacillus subtilis. Nucleic Acids Res 28:1206–1210. 10.1093/nar/28.5.1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sedgley CM, Clewell DB, Flannagan SE. 2009. Plasmid pAMS1-encoded, bacteriocin-related “Siblicide” in Enterococcus faecalis. J Bacteriol 191:3183–3188. 10.1128/JB.00147-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gilmore MS, Rauch M, Ramsey MM, Himes PR, Varahan S, Manson JM, Lebreton F, Hancock LE. 2015. Pheromone killing of multidrug-resistant Enterococcus faecalis V583 by native commensal strains. Proc Natl Acad Sci USA 112:7273–7278. 10.1073/pnas.1500553112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ubeda C, Taur Y, Jenq RR, Equinda MJ, Son T, Samstein M, Viale A, Socci ND, van den Brink MR, Kamboj M, Pamer EG. 2010. Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J Clin Invest 120:4332–4341. 10.1172/JCI43918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ramsey M, Hartke A, Huycke M. 2014. The physiology and metabolism of enterococci. In Gilmore MS, Clewell DB, Ike Y, Shankar N (ed), Enterococci: from commensals to leading causes of drug resistant infection. Massachusetts Eye and Ear Infirmary, Boston, MA. [PubMed] [Google Scholar]
- 60.Plumbridge J. 2015. Regulation of the utilization of amino sugars by Escherichia coli and Bacillus subtilis: same genes, different control. J Mol Microbiol Biotechnol 25:154–167. 10.1159/000369583. [DOI] [PubMed] [Google Scholar]
- 61.Reith J, Mayer C. 2011. Peptidoglycan turnover and recycling in Gram-positive bacteria. Appl Microbiol Biotechnol 92:1–11. 10.1007/s00253-011-3486-x. [DOI] [PubMed] [Google Scholar]
- 62.Kawada-Matsuo M, Oogai Y, Komatsuzawa H. 2016. Sugar allocation to metabolic pathways is tightly regulated and affects the virulence of Streptococcus mutans. Genes (Basel) 8:1–11. 10.3390/genes8010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zeng L, Burne RA. 2015. NagR differentially regulates the expression of the glmS and nagAB genes required for amino sugar metabolism by Streptococcus mutans. J Bacteriol 197:3533–3544. 10.1128/JB.00606-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thurlow LR, Thomas VC, Fleming SD, Hancock LE. 2009. Enterococcus faecalis capsular polysaccharide serotypes C and D and their contributions to host innate immune evasion. Infect Immun 77:5551–5557. 10.1128/IAI.00576-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Flannagan SE, Clewell DB. 2002. Identification and characterization of genes encoding sex pheromone cAM373 activity in Enterococcus faecalis and Staphylococcus aureus. Mol Microbiol 44:803–817. 10.1046/j.1365-2958.2002.02922.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 to S3. Download JB.00371-21-s0001.pdf, PDF file, 0.4 MB (389.4KB, pdf)