ABSTRACT
Enterococci are Gram-positive bacteria that have evolved to thrive as both commensals and pathogens, largely due to their accumulation of mobile genetic elements via horizontal gene transfer (HGT). Common agents of HGT include plasmids, transposable elements, and temperate bacteriophages. These vehicles of HGT have facilitated the evolution of the enterococci, specifically Enterococcus faecalis and Enterococcus faecium, into multidrug-resistant hospital-acquired pathogens. On the other hand, commensal strains of Enterococcus harbor CRISPR-Cas systems that prevent the acquisition of foreign DNA, restricting the accumulation of mobile genetic elements. In this review, we discuss enterococcal mobile genetic elements by highlighting their contributions to bacterial fitness, examine the impact of CRISPR-Cas on their acquisition, and identify key areas of research that can improve our understanding of enterococcal evolution and ecology.
KEYWORDS: enterococci, horizontal gene transfer, mobile genetic elements, pheromone-responsive plasmid, phage, transposon, virulence, antibiotic resistance, CRISPR
INTRODUCTION
Enterococci, commensal bacteria that make up less than 0.1% of the healthy intestinal microbiota (1), have continuously faced selective pressure for genes that provide beneficial adaptations, facilitating coevolution with eukaryotes since the early Paleozoic era (2). Around the 1960s, it was discovered that, in addition to being resident members of the human microbiota (3, 4), the enterococci colonize the gastrointestinal (GI) tracts and guts of other animals and insects (5, 6), and are members of food (7), plant (8), soil (9), and water ecosystems (10). Since the 1980s, enterococci have emerged as opportunistic pathogens, causing hospital-acquired urinary tract, wound, endocarditis, and bloodstream infections (11, 12). Antibiotic therapies can cause multidrug-resistant (MDR) enterococcal expansion in the intestine as other resident microbes are depleted, resulting in life-threatening infections due to enterococcal translocation across the intestinal barrier into the bloodstream (13, 14). MDR enterococci, including vancomycin-resistant enterococci (VRE), cause over 54,000 infections in the United States annually (15).
Studies on enterococcal epidemiology (discussed comprehensively in reference 16) reflect that enterococci emerged as a major cause of nosocomial infections in two distinct phases. During the late 1970s, Enterococcus faecalis strains dominated the clinical isolate pool (17). Since the early 2000s, however, vancomycin-resistant Enterococcus faecium isolates became more prevalent than E. faecalis (18, 19). Recently, health care systems of many countries, including those in North America and Europe, have witnessed increasing levels of VRE infections (20). According to the surveillance data from the National Healthcare Safety Network, VRE are responsible for 3% of all reported nosocomial infections (21). Several reports indicate that the risk of VRE infections is particularly high among intensive care unit (ICU) and immunocompromised patients (20). Further, mortality rates due to VRE-mediated bacteremia are notably higher than those caused by vancomycin-susceptible enterococci (20).
Excessive exposure to antimicrobials used to sterilize hospital surfaces and broad-spectrum antibiotics has resulted in the rise of MDR E. faecalis and E. faecium (22). These accessory resistance genes complement the enterococci’s intrinsic resistance to cephalosporins, penicillins, and clinically relevant concentrations of aminoglycosides (17, 23). Considering the clinical significance of vancomycin-resistant enterococcal infections, several studies have focused on the ecology, evolution, and dissemination of vancomycin resistance genes (reviewed in reference 24). Clinical E. faecalis and E. faecium isolates resistant to the “last-line-of-defense” antibiotics, linezolid and daptomycin, have also been identified (23, 25, 26). While exploring adaptive traits of vancomycin-resistant E. faecium in immunocompromised patients, researchers noted that the emergence of linezolid or daptomycin resistance is associated with prolonged antibiotic exposure in these patients (27).
E. faecalis and E. faecium have adapted to nosocomial settings due to their ability to acquire mobile genetic elements (MGEs) through horizontal gene transfer (HGT). HGT facilitates acquisition of novel traits, including genes that promote virulence and antibiotic resistance (28–32). Mobile genes that increase clinical resilience are genetically similar to genes found in other bacterial genera, suggesting their broad distribution among pathogens (29, 33, 34). Clinical E. faecium strains have twice as many genes associated with MGEs than nonclinical strains (35), while some clinical isolates of E. faecalis are comprised of up to 25% MGEs (36). Transferable plasmids (mobilizable and conjugative) and bacteriophages play central roles in HGT of bacterial genes and transposons (37), driving bacterial evolution. Many MGEs encode antimicrobial resistance (AMR) genes, which we will describe in this review. Antimicrobials used to combat enterococcal infections, such as tetracycline, aminoglycosides, and erythromycin, were nullified due to the emergence of resistance genes that were rapidly disseminated among the enterococci via MGEs (38). This has necessitated updated treatment guidelines for MDR enterococcal infections and development of the new antibiotic treatment regimens, such as daptomycin and linezolid (39–41).
Despite their benefits to the bacteria, MGEs can adversely affect their growth and survival (42, 43). Plasmid maintenance requires synthesis of additional cellular macromolecules and can be a metabolic burden on the host, while conjugation requires the production of pili, secretion systems, and additional DNA replication proteins following induction. It is important to note that, often, these fitness costs exist initially, but over time, these costs are mitigated via adaptive evolution, and the benefits of accessory plasmid genes can outweigh the costs (44, 45). Additionally, it is possible that conjugative structures may expose the bacteria to virulent phages that are otherwise unable to penetrate bacterial cellular barriers (46). This phenomenon has been studied for the F pili of Escherichia coli and filamentous phages but has not been thoroughly explored in enterococci (47). MGEs like prophages and transposons can integrate into bacterial chromosomes and disrupt gene function (48, 49). Stressors and nutrients, such as Casamino Acids, various temperatures, and DNA-damaging antimicrobials, drive prophage induction leading to bacterial lysis (50, 51). Hence, bacteria have acquired defense systems to protect themselves from these foreign MGEs. Clustered regularly interspaced short palindromic repeats (CRISPR) and corresponding Cas proteins represent an adaptive bacterial genetic system that can prevent MGE acquisition (52).
In this review, we (i) summarize current knowledge of enterococcal MGEs, with a specific emphasis on E. faecalis and E. faecium, (ii) highlight the contributions of MGEs to enterococcal fitness within the scope of intestinal colonization and infection, (iii) discuss the impact of CRISPR-Cas on enterococcal MGE acquisition, and (iv) identify key areas of MGE research that, if explored, should improve our understanding of enterococcal evolution and ecology.
CONTRIBUTIONS OF PHAGE-MEDIATED HGT TO ENTEROCOCCAL GENOME DIVERSITY AND NICHE ADAPTATION
Bacteriophages (phages) are viruses that infect and replicate in bacteria and are the most abundant organisms on earth (53). Since the initial discovery of enterococcal phages almost a century ago (54), these phages have been isolated and characterized from diverse environments, including wastewater, livestock runoff, and the mammalian intestine, with the most commonly studied enterococcal phages today being those that infect E. faecalis and E. faecium (55–57). Phages often lyse bacteria, but some phages (temperate phages) can integrate into the bacterial genome, an event termed lysogeny. Since phages can transfer DNA from cell to cell via transduction, phage-driven HGT can have a significant impact on host genetic composition and phenotypic characteristics as well as the surrounding microbial community (reviewed in references 58 and 59).
Phage-adapted replication and genome packaging mechanisms influence phage-mediated HGT. Upon infection, temperate phages integrate into the host genome and are maintained as prophages. Newly integrated prophages can supply genes which enhance host fitness, including antibiotic resistance, stress tolerance, and immunity against secondary infection by closely related phages (superinfection exclusion) (51, 60–62). In this scenario, the temperate phage is an MGE, whereupon integration, phage auxiliary genes bestow a benefit upon the host. HGT can also occur via specialized transduction when prophages excise specific bacterial DNA flanking the phage integration site and package it into progeny virions (59, 63). Additionally, generalized transduction occurs when nonphage, chromosomal DNA fragments are packaged into phage capsids (59, 63). The recently discovered process of lateral transduction occurs in situ prior to prophage excision, resulting in packing of up to several hundred kilobases of host DNA into infectious virions (64). The authors demonstrated that these Staphylococcus aureus prophages can integrate into a new host, carrying the DNA from the previous host. In each mode of transduction, host DNA-carrying phages can infect vulnerable hosts where the DNA can be integrated into the chromosome. Prophages can confer a competitive advantage to their host bacterium by switching to the lytic cycle and producing infective viruses which target phage-susceptible bacteria (65, 66). The shift from the lysogenic to lytic cycle can occur spontaneously or in response to UV radiation and antibiotics that induce DNA damage, triggering the bacterial SOS response (51, 67–71). Although the mobilization of phage-encoded virulence determinants and other factors are well studied in some Gram-positive pathogens such as S. aureus and group A Streptococcus (72–75), much remains unknown about the influence of prophages on enterococcal virulence and genetic plasticity.
The emergence of MDR enterococci makes it imperative to identify and characterize drivers of enterococcal evolution. With limited studies highlighting phage-mediated HGT in enterococci, we are just now appreciating potential contributions of phages to enterococcal evolution. Yasmin and colleagues demonstrated that temperate phages induced from clinical E. faecalis isolates are capable of intraspecies generalized transduction of antibiotic resistance genes and gelE encoding a metalloprotease implicated in virulence (76, 77). Enterococcal phages have also been shown to successfully transfer tetracycline and gentamicin resistance between enterococcal species via generalized transduction (78). Enterococci are not intrinsically resistant to tetracycline; enterococcal tetracycline resistance is common due to the distribution of the resistance genes tetM and tetL (23). While gentamicin monotherapy is not used to treat enterococcal infections, it is used synergistically with beta-lactams to achieve a 70% cure rate in susceptible isolates (23, 79). Other studies observed that the fitness and virulence of a probiotic E. faecalis strain were significantly enhanced when transduced by temperate phages from a pathogenic E. faecalis strain (80). Considering that enterococci regularly encounter prophage-inducing environmental stimuli in the GI tracts of animals, including bacterial signaling molecules (80), dietary components (50, 81), and sublethal antibiotic doses (51, 76), it is likely that temperate phage-mediated HGT can increase the distribution of pathogenic traits among enterococci.
Studies highlighting widespread prophage distribution among virulent enterococci underline the importance of prophage dynamics during enterococcal pathogenicity. The well-characterized clinical VRE isolate E. faecalis V583 harbors seven prophage-like elements. These elements, designated pp1 to pp6 and EfCIV583 (phage-related E. faecalis chromosomal island), constitute up to 10% of the V583 genome (36, 82). Upon excision of these prophage-like elements, host cell lysis is not detected (51). Polylysogeny, the presence of more than one prophage-like element in the genome, has been reported in multiple clinical isolates of E. faecalis besides V583 (36, 76, 82), suggesting that E. faecalis prophages are more widely distributed than previously thought. Contrastingly, there are still many strains of E. faecalis that do not harbor multiple prophages, such as the common lab strain OG1RF. Pp2, a cryptic prophage, is a part of the core E. faecalis genome (82). Pp2 has yet to be shown to excise from the bacterial chromosome and produce infectious virions; however, pp2 tail proteins have been identified in the supernatant of E. faecalis OG1RF cultures (83). Interestingly, it was recently observed that while the presence of clinically relevant antibiotics altered the excision of six of seven prophages via the E. faecalis V583 SOS response, including DNA-damaging antimicrobials and various temperatures, only four of these prophages were able to form infectious virions (51). Additionally, studies have elucidated interaction between coinhabiting prophages, ranging from cooperation to interference. EfCIV583 relies on pp1 for induction, genome packaging, and transmission but interferes with pp1 infectivity (50, 51, 84). On the other hand, the presence of pp1 impedes pp4 excision, while pp6 excises only when both pp3 and pp5 are deleted from the chromosome (51). Using an innovative DNA sequencing-based “transductomics” approach, wherein deep sequencing identifies DNA originating from ultrapurified virus-like particles, such as excised prophages, it was discovered that pp1, pp5, and EfCIV583 are capable of transducing at relatively high frequencies, potentially through lateral transduction, a recently discovered form of HGT (64, 85). The authors observed that these E. faecalis V583-transducing phages not only carried bacterial DNA surrounding either the region of the prophage integration site but also packaged several additional regions, including transposase-carrying insertion sequence (IS) elements and the three rRNA operons (85). The authors did not report mobilization of antibiotic resistance genes. Using this technology, future studies can reveal virus-like particle-host interactions in complex communities like the intestine.
Although prophages are pervasive among MDR enterococci, only a limited number of studies have addressed their impact on enterococcal fitness and evolution. E. faecalis V583 prophages pp1, pp4, and pp6, harboring homologs of the Streptococcus mitis platelet binding phage tail proteins PblA and PblB, have been shown to be crucial for adhesion to human platelets in vitro and are speculated to contribute to the development of infective endocarditis (51). In another study, investigators established that pp5 is required for efficient biofilm formation, and its induction facilitates biofilm dispersal (80). These findings emphasize the potential influence of enterococcal prophages in biofilm-associated infections, such as endocarditis, central-venous-catheter infections, or implant infections. Further, the therapeutic potential of virulent phages and their lysins is diminished against prophage-carrying clinical isolates of VRE (86, 87). Additionally, E. faecalis V583 can release EfCIV583 to target and lyse competitor E. faecalis strains in the GI tract of mice (50). Collectively, these studies provide evidence of the influence of prophages in enterococcal physiology, pathogenicity, and adaptation in competitive ecosystems.
While prophages constitute a large part of the E. faecalis mobilome, there are still major gaps in the field. To fully comprehend the influence of prophages on enterococcal physiology and adaptation, future research needs to explore the genetic content, composition, and molecular epidemiology of prophages. As many prophage genes are hypothetical or of unknown function, characterization of such prophage genes in relation to virulence or adaptation would divulge how maintaining prophages can augment enterococcal pathogenicity and fitness. The generation of a transposon library in E. faecalis MMH594, an MDR isolate that harbors prophage-like elements, could be a powerful tool for such future studies (88). Another noticeable knowledge gap is the lack of prophage-mediated HGT studies in other species of Enterococcus. E. faecium is an equal, if not greater, burden on the hospital system due to AMR gene acquisition. The study of temperate phages among E. faecium strains is limited, potentially due to the genetic intractability of the bacterium, although methodologies to overcome this are reported (30, 89–91). By expanding studies to include prophages of commensal enterococcal strains native to nonhuman hosts, we might elucidate mechanisms of temperate phage HGT relevant to hospital-adapted enterococci.
The fitness trade-offs of defending against phage infection versus allowing phage acquisition are complex, as lysogenic phages can act either as predators replicating within and lysing bacterial cells or as agents of HGT carrying novel genetic information. Investigation into the dynamics of temperate phage acquisition could provide insights into the complexity of microbial community interactions related to DNA transmission.
CONJUGATIVE AND BROAD-HOST-RANGE PLASMIDS OF ENTEROCOCCI
Plasmids are autonomously replicating, extrachromosomal genetic elements that are critical drivers of microbial adaptation and evolution via HGT. Although plasmids do not encode essential genes and can be a metabolic burden for the cell, they often provide beneficial genes that augment bacterial fitness in complex environments (92, 93). Plasmids are divided into narrow- or broad-host-range plasmids, referring to the specificity of the replication system encoded on the plasmid and the ability of the plasmid to be replicated within a single or multiple bacterial species.
PHEROMONE-RESPONSIVE PLASMIDS
Pheromone-responsive plasmids (PRPs) are narrow-host-range plasmids that are laterally transmitted via conjugation from plasmid-harboring donors to plasmid-free recipient cells (Fig. 1A). Briefly, recipient cells produce and secrete a chromosomally encoded, plasmid-specific conjugation peptide referred to as the pheromone, denoted with a “c” before the plasmid’s name. When the donor cell senses the pheromone, downstream transcriptional regulators are activated and mediate the expression of the PRP conjugation genes (94). Highly conserved, large cell wall-anchored surface proteins called aggregation substances (AS) encoded by PRPs facilitate physical contact between donor and recipient cells (95). The operon in which the AS is encoded collectively produces mating channels between the cells, resulting in horizontal plasmid transfer via type IV secretion system (T4SS) conjugation machinery (95). The T4SS is also involved with integrative and conjugative elements conjugation (discussed later) (96). Since pheromones are constitutively expressed, self-induction of plasmid conjugation is prevented by the production of an inhibitory protein encoded on the PRP, denoted “i.” PRP transfer has been thoroughly reviewed elsewhere (97–99).
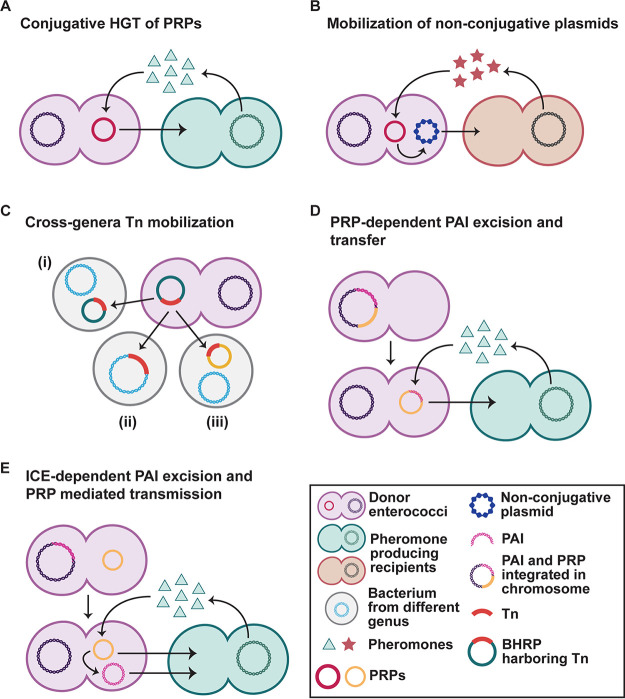
FIG 1.
Simplified models for intra- and interspecies dissemination of enterococcal plasmids. (A) PRP replication and conjugation is initiated by donor cells sensing pheromones produced by plasmid-less recipient cells. (B) Upon sensing noncognate peptide signals produced by bacteria from a different genus, PRPs can facilitate mobilization of nonconjugative plasmids. (C) Plasmid-mediated Tn mobilization across genera. A plasmid carrying the Tn can enter the new host; the Tn can replicate independently (i) or integrate into the chromosome (ii) or another plasmid (iii) of the recipient if the plasmid is unable to replicate in the new host. (D and E) Two mechanisms of PAI movement between enterococcal strains. (D) PRP integration and excision can mobilize the PAI by integrating near PAI on the chromosome and excise the PAI upon excision. This PAI-PRP hybrid circularizes and can be conjugated to pheromone producing E. faecalis cells. (E) Alternatively, the PAI can function as an integrative conjugative element (ICE) and relies on the direct repeats flanking the PAI and products of PAI-encoded enzymes to excise from and recombine into chromosomes. Mobilization to PAI-less cells is thought to rely on PRP presence.
Some PRPs encode antibiotic resistance genes and a wide variety of virulence factors (99). pHKK100, the first plasmid described to mediate vancomycin resistance, transfers via pheromone response (100). In addition to initiating PRP conjugation, AS also increases bacterial virulence by promoting adhesion to, internalization into, and survival within cultured human cells (101–105). PRPs also increase chromosomal diversity, resulting in transconjugants with hybrid donor-recipient genomes and the transfer of chromosomal virulence factors such as antibiotic resistance and the E. faecalis pathogenicity island (discussed later) (28, 31). To date, about 35 PRPs have been identified, but only in E. faecalis and E. faecium (28, 99), suggesting that PRPs are restricted to enterococci.
pCF10 is a well-studied PRP that encodes approximately 60 protein-coding genes that increase E. faecalis virulence. These genes include secreted proteins, transcription factors, and orthologs of UV and tetracycline resistance (98) that provide a selective advantage to pCF10-harboring cells. In the GI tract of germfree mice, rapid high-frequency pCF10 transfer was shown to be strongly dependent on the proximity of donor and recipient cells, but the transmission was not significantly impacted by the presence of a defined microbiota (106). Interestingly, the competitive advantage of pCF10 carriage in the intestine was distinct from antibiotic resistance or bacteriocin production and led to the speculation that the pCF10-encoded AS may influence bacterial fitness in the intestine (106). Although AS-mediated resistance to phagocytic killing has been suggested to benefit pCF10-carrying E. faecalis in an endocarditis model (105, 107), the contributions of AS in GI colonization are yet to be explored.
Understanding pheromone specificity is an expanding area of research. While some pheromones initiate replication and transfer of a specific plasmid, the PRP pMG2200 responds to synthetic cCF10 pheromone. This is because pMG2200 encodes genes identical to the replication and conjugation genes in pCF10, the cognate PRP for cCF10 (108). This phenomenon of pheromone cross talk has been documented for other pheromone-plasmid pairs (99). However, unlike pMG2200, it is unknown whether other PRPs that respond to cCF10, such as pHKK703 (109), pMB1 (110), and pAMS1 (111), encode identical PRP regulation genes and are therefore pCF10 chimeric derivatives. Future studies are necessary to determine in vivo cross talk between PRP pheromones and to determine genetic relatedness among plasmids that respond to the same pheromone.
The first identified PRP, pAD1 (112), follows typical PRP conjugation and encodes a plasmid addiction system to promote plasmid maintenance in the absence of selection (29, 113). The pAD1 toxin/antitoxin par locus encodes two convergently transcribed mRNAs, RNA I and RNA II. The stable RNA I encodes the membrane-active peptide toxin, Fst, that interferes with bacterial cell membrane integrity (114), whereas RNA II is an unstable sRNA antitoxin that represses Fst production by binding to RNA I. Upon loss of pAD1, the antitoxin is rapidly degraded, while the more stable toxin kills the E. faecalis cell (115, 116). E. faecalis V583 harbors the PRP pTEF1, a plasmid with a high level of nucleotide similarity to pAD1, and encodes a locus identical to par (36). Similar toxin/antitoxin systems have been identified on plasmids and chromosomes of species from Enterococcus, Lactobacillus, and Staphylococcus, indicating that par is the prototype of a family of type I toxin/antitoxin systems (117).
A recent study focusing on the correlation between antimicrobial use and resistance in livestock found that pAD1 is present in 38% and 30% of E. faecalis isolated from retail beef and human clinical samples, respectively, suggesting a clinical implication to ingesting pAD1-contaminated samples (118). Enhanced virulence of pAD1-carrying E. faecalis strains in mice is attributed to the plasmid-encoded hemolytic cytolysin (119). Epidemiological analysis of an MDR E. faecalis outbreak showed that patients infected with a cytolytic strain were at a 5-fold greater risk of death regardless of treatment, and of the patients that died, 71% were infected with a cytolytic strain (11). However, the cytolytic activity of this strain was not confirmed to be a result of pAD1 carriage. Despite this, other studies have shown that there is increased virulence associated with pAD1, specifically due to the cytolysin’s activity against multiple eukaryotic cells (120). It is hypothesized that lysing eukaryotic cells releases nutrients like cytochromes that are otherwise unavailable to noncytolytic bacteria, potentially allowing for increased survival through aerobic respiration (120, 121). Cytolytic E. faecalis strains also colonize the bloodstream significantly higher than noncytolytic strains in mice (11), supporting this theory. pAD1 also encodes UV resistance via uvrA. UV-resistant E. faecalis that was exposed to UV irradiation had a higher probability of developing spontaneous antibiotic resistance mutations than non-UV-resistant E. faecalis cells (122).
pPD1 is an E. faecalis PRP that encodes the bacteriocin Bac-21 (123). Bacteriocins are small cyclic peptides that are capable of killing or inhibiting growth of related bacteria by disrupting the proton motive force of the cytoplasmic membranes and energized membrane vesicles and are commonly produced by many lactic acid bacteria (124, 125). Bac-21 is a 70-amino-acid protein whose corresponding nucleotide sequence is identical to that of the PRP pMB2-encoded bacteriocin AS-48 (126). In the mammalian intestine, E. faecalis strains harboring pPD1 outcompete pPD1-deficient E. faecalis strains, suggesting that Bac-21 production confers a colonization benefit (127).
PRP conjugation has been documented within E. faecalis strains and between E. faecalis and E. faecium. The PRP pBRG1 is a vancomycin resistance (vanA)-carrying plasmid that can transfer from E. faecium to E. faecalis (128). There is no doubt that this intragenus transfer is concerning regarding the spread of antibiotic resistance genes. Although studies showing PRP transfer to nonenterococcal bacteria are limited, PRP transmission from E. faecalis to distant bacterial species, including S. aureus and Streptococcus gordonii, have been documented (129–131). Other than its cognate pheromone cAM373, the PRP pAM373 can also initiate conjugation in response to noncognate peptides produced by S. gordonii and S. aureus (132–134). Although cAM373 produced by S. gordonii and E. faecalis bear no structural resemblance, the S. gordonii-cAM373 could induce transfer of a lab-constructed, nonconjugative plasmid from E. faecalis into S. gordonii in vitro (133) (Fig. 1B). While PRPs do not appear to replicate efficiently in nonenterococcal hosts (28), the full breadth of PRP transmission and maintenance can only be realized if other bacterial genera that frequently coreside with enterococci in microbial communities are studied in this context.
While many enterococcal PRPs have been identified, not all are as well studied as the plasmids discussed above. Future studies focusing on PRP discovery, genetic diversity, and distribution will be critical to understanding their contributions to enterococcal fitness and pathogenicity. Over 900 E. faecalis and E. faecium genomes have been sequenced (135), but many of these genomes are deposited in public databases as draft genomes fragmented into multiple contigs. Draft genomes are useful for surveying genomic composition of isolates; however, it cannot be assumed that the entire genome is resolved in these assemblies. The combined use of overlapping short and long high-throughput sequencing reads will allow gap closures in these draft genomes, likely resulting in novel plasmid discovery. Determining if PRPs are truly enterococci specific will clarify how the PRP system arose and why it seems to be rare or nonexistent in other genera and could aid in restricting the potential transmission of these MGEs.
BROAD-HOST-RANGE PLASMIDS
Mobile broad-host-range plasmids (BHRPs) often harbor multiple origins of replication. Host range can be further expanded if plasmid replication initiation and essential proteins are also present on the BHRP and are not dependent on host factors (136, 137). These features allow BHRPs to replicate in distantly related bacteria and serve as mediators of HGT across genera (137). However, it is important to note that some narrow-host-range plasmids contain multiple replication origins (137). BHRPs can be divided into incompatibility groups (Inc-groups). Inc-groups are defined as the failure of two coresident plasmids to be inherited without external selection; if introducing a second plasmid interrupts the inheritance of the first, they are incompatible (138). Inc18-family plasmids are common among the enterococci, staphylococci, and streptococci (139, 140). Inc18-family plasmid biology is comprehensively reviewed by Kohler et al. (136).
Inc18 plasmids have been a focus of interest because they frequently carry antibiotic resistance genes, including resistance to chloramphenicol, vancomycin, and macrolide-lincosamide-streptogramin (MLS) antibiotics (136, 141). Chloramphenicol was used for the treatment of VRE (142, 143), while MLS antibiotics were used as a growth promoter in agriculture (144). pAM830, an Inc18-type plasmid encoding the vanA transposon Tn1546, is responsible for the transfer of vancomycin resistance from enterococci to methicillin-resistant S. aureus (MRSA) (28). Genomic analyses of vancomycin-resistant MRSA strains suggest that either the entire pAM830 was transferred or pAM830 facilitated conjugative dissemination of only the van gene carrying Tn1546, which ultimately recombined with the chromosome or another plasmid within MRSA (145–147) (Fig. 1C). pAMβ1 and pIP501 are two of the most well-studied Inc18-type plasmids originally isolated from E. faecalis (148) and Streptococcus agalactiae (149). pIP501 confers resistance to erythromycin and chloramphenicol, while pAMβ1 carries resistance markers for lincosamides, streptogramin B, and erythromycin. Both of these two BHPs are capable of antibiotic resistance transmission to other Gram-positive bacteria, including Listeria spp., Leuconostoc spp., Lactococcus spp., and Streptomyces lividans, while pIP501 can even disseminate antibiotic resistance to the Gram-negative bacterium E. coli (28). Another E. faecalis plasmid, pRE25, sharing 30.5 kb of sequence fragment with pIP501 and harboring 12 antibiotic resistance markers, is capable of conjugal transfer into Listeria innocua and Lactococcus lactis (28). In addition to disseminating antibiotic resistance genes, conjugative transfer of multiresistance plasmids from other Gram-positive bacteria into enterococci has also been reported. An L. monocytogenes plasmid, pIP811, which confers resistance to chloramphenicol, erythromycin, and streptomycin, can be transferred to enterococci via conjugation (150). Lateral transmission of pAMβ1 from Lactococcus to E. faecalis in vitro and in vivo has also been demonstrated (151–153). Collectively, these studies raise concerns regarding cross-genera horizontal transfer, not only of antibiotic resistance markers but also of other plasmid-encoded virulence and environmental adaptation traits. The high sequence identity among BHPs indicates that such interspecies plasmid transmission influences gene content of plasmids across species. Therefore, identifying factors that facilitate BHRP transmission and investigating the molecular mechanism(s) of BHRP dissemination are critical for understanding how these plasmids facilitate genome plasticity and bacterial evolution.
OTHER MOBILE ELEMENTS
Transposable elements (TEs) are widely distributed in prokaryotic genomes, including enterococci. TEs are DNA fragments that move autonomously to new locations within and between DNA molecules present in a single cell; however, they rely on conjugative plasmids and transducing phages for intercellular movement. All TEs encode a transposase, an enzyme that catalyzes excision and integration of TEs through transposition. Insertion of TEs can disrupt, activate, or silence gene functions (154). Broadly, TEs can be categorized into IS elements and composite transposons (Tns). IS elements are small MGEs composed of two inverted repeat sequences flanking the transposase open reading frame (ORF). In contrast, composite Tns consist of resistance genes or other adaptive traits bordered by a pair of IS elements.
The incorporation of TEs potentially facilitated the propagation of beneficial traits and genome plasticity in enterococci. TEs play a role in the shaping of the enterococcal genome through genomic rearrangements and recombination, thus facilitating E. faecium genome plasticity (155). Upon activation through cell stress, transposons can induce mutations through novel insertions that may allow for bacterial survival (141). Vancomycin resistance in E. faecalis V583 encoded by the vanB operon in Tn1549 found on a PRP was likely acquired via HGT (36, 156). Conjugative transposition of vanB Tn1549 from a plasmid results in transfer of antibiotic resistance between enterococci and human commensal bacteria in a germfree mouse model of GI colonization (157). Vancomycin resistance is associated with other transposons. Tn1547 encodes the vanB operon flanked by IS16- and IS25-like elements and can integrate into plasmids and chromosomes (158). E. faecium Tn5382 encodes the vanB2 operon and has been identified in multiple clinical isolates (159). Tn1546 encodes the vanA operon and has been identified in E. faecalis, E. faecium, as well as in other species, including Enterococcus gallinarum and Enterococcus casseliflavus (160). Apart from vancomycin, a gentamicin resistance-encoding transposon, Tn5281, is present on the PRP pBEM10 and is widely distributed among E. faecalis and E. faecium isolates from different geographical locations (161–164). The DNA sequence and genetic organization of Tn5281 are similar to those of other Tns such as Tn4001 and Tn4031, found in S. aureus and Staphylococcus epidermidis, respectively. This similarity suggests that these elements are closely related, possibly as a result of interspecies transfer (164, 165). Together, these studies indicate that transposon-encoded antibiotic resistance potentiates rapid and widespread dissemination and diversification among Enterococcus species and, more broadly, other bacterial species.
Pathogenicity islands (PAIs) are large chromosomal genetic elements that are horizontally transmitted among Gram-positive and Gram-negative bacteria (166). The E. faecalis PAI encodes nearly 150 genes, including virulence factors that vary between strains (82, 167–170). Examples include a cytolysin, a virulence-associated surface protein encoded by esp, and an AS (171, 172). The PAI is enriched in pathogenic E. faecalis strains and is excised at a rate of 1 in 1,000 cells (171). Evidence suggests that PAI intra- and interspecies mobilization is dependent on the PRPs pTEF1 and pTEF2 of E. faecalis V583 (173). These PRPs could facilitate PAI transmission in two ways. Manson and colleagues demonstrated that pTEFs integrate into the E. faecalis V583 chromosome in close proximity to the PAI, and subsequent excision of plasmid-PAI hybrids potentially allows for PAI movement between bacterial cells (31) (Fig. 2D). The authors also showed that the E. faecalis V583 pTEFs can mobilize random segments of chromosomal DNA, including vancomycin resistance, multilocus sequence type (MLST) markers, and capsule genes, indicating that plasmid-mediated HGT is a major driving force behind the evolution of MDR enterococci (31). In contrast, the E. faecalis UW3114 PAI is capable of precise excision and circularization and can undergo horizontal intra- and interspecies transfer with the help of a PRP (173) (Fig. 1E). It is suggested that the E. faecalis UW3114 PAI functions similarly to an integrative conjugative element (ICE), relying on the direct repeats flanking the PAI and phage-related excisionase and integrase genes to excise from and recombine into bacterial chromosomes (173). Nucleotide sequence identity exists at one end of the PAI with conjugation-related structural genes of enterococcal PRPs pAM373 and pAD1 (174), indicating that part of the PAI may have evolved from the integration of a PRP into the chromosome. A region present on the PAI has 87% nucleotide identity to a transfer origin in pAD1 (171), further supporting this theory. Although a number of PAI ORFs remain uncharacterized, the absence of these genes from certain human isolates suggests that the products of these genes could facilitate the commensal to pathogen transition of the enterococci. Additionally, tracing the evolutionary origins of enterococcal PAIs will provide further enlightenment as to how enterococci become successful pathogens.
FIG 2.
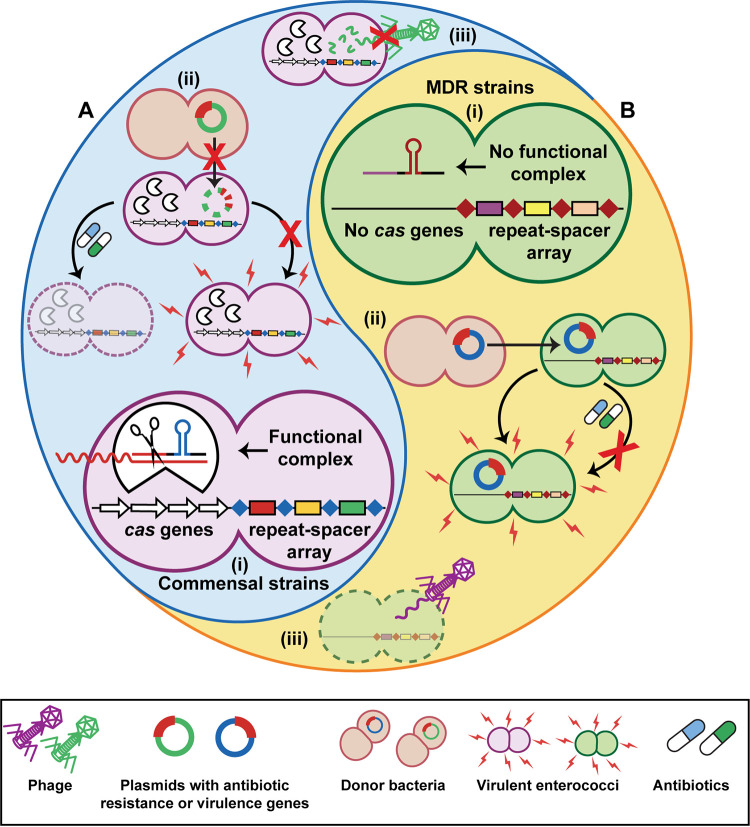
CRISPR-Cas is a fitness balance for enterococci. (A and B) Commensal strains typically harbor a functional CRISPR-Cas system (A), while MDR clinical isolates do not (B). Generally, commensal strains carry the cas genes and repeat spacer array, resulting in a fully functional system (A, i), while MDR strains contain only the orphan CRISPR2 repeat-spacer array (B, i). The presence of functional CRISPR-Cas results in reduced plasmid transfer, preventing transfer of virulence or antibiotic resistance genes (A, ii); however, CRISPR-Cas also blocks phage infection and killing of a host bacterium (A, iii). (B, ii) On the other hand, plasmid transfer is not blocked in MDR strains, allowing them to gain accessory genes. (B, iii) This trade-off makes MDR strains sensitive to phage infection.
CRISPR AND HGT
Many bacteria have established barriers to combat HGT (50, 51). One such defense system is CRISPR-Cas, an adaptive immune system that utilizes an RNA-guided nuclease to block acquisition of MGEs in a sequence-dependent manner (52). CRISPR-Cas systems are present throughout enterococcal species, most frequently in commensal isolates (175, 176). CRISPR loci consist of several direct repeat regions separated by variable spacers, which are sequences derived from foreign DNA which serve as adaptive memory units for identifying invading nucleic acid targets. Identified spacers match phage, plasmid, and nonkin chromosomal sequences (177, 178). CRISPR interference is adaptive, and new targeting spacers can be acquired as invading DNA threats are encountered (52, 179). However, spacer acquisition is tightly regulated and occurs at very low rates (180). Still, this may provide a benefit to cells over other phage protection mechanisms like surface receptor modification, as alteration of these receptors is known to increase antibiotic susceptibility (141, 181).
These repeat-spacer arrays are in close proximity to the cas genes (177). The cas genes encode a large and diverse family of proteins which interact with the transcribed spacers to form the CRISPR-Cas complex (182). Unique cas genes allow CRISPR systems to be divided into three major types, types I, II, and III, defined by the presence of cas3, cas9, and cas10, respectively (183). Type IV CRISPR has been identified but is much less common (184). CRISPR types are further divided into subtypes based on additional signature genes or gene variants (183). CRISPR classification and mechanisms of action have been thoroughly reviewed elsewhere (183, 184).
To date, type II-A and type I-C CRISPR loci have been found in Enterococcus (175). Type II-A CRISPR-Cas systems are significantly more common in commensal Enterococcus strains than in clinical isolates, and the lack of functional CRISPR-Cas is correlated with acquired antibiotic resistance in E. faecalis (176, 185, 186) (Fig. 2A and B). The type II-A subtype is broken down further into four systems within Enterococcus, CRISPR1-Cas, CRISPR2, CRISPR3-Cas, and CRISPR4. Of enterococcal genomes screened thus far, CRISPR1-Cas and CRISPR2 seem the most predominant (176, 187–189). CRISPR1-Cas is the most commonly identified functional CRISPR system in E. faecalis, with spacers varying by strain (176, 178, 187, 190, 191). CRISPR1-Cas has also been identified in E. faecium, primarily in isolates without antibiotic resistance genes, and in Enterococcus hirae (176, 187).
Many MDR E. faecalis strains lack functional CRISPR-Cas systems (176, 186). The CRISPR2 locus is nonfunctional, consisting only of the orphan repeat-spacer array and no cas genes (176, 192, 193). CRISPR2 is nearly ubiquitous within E. faecalis and common among E. faecium (176, 187, 189). It is theorized that CRISPR2 is maintained because of a self-preservation mechanism within the locus itself, using terminal repeats within the CRISPR array to prevent loss of the terminal spacer (182). Additionally, in the presence of cas9, the CRISPR2 locus becomes functional for defense (194). CRISPR systems are detected less frequently in other disease-associated species like E. gallinarum and E. casseliflavus (191).
Lack of CRISPR systems is associated with detergent and chlorohexidine resistance, increased biofilm production, and bacteriocin activity (186, 195). Conversely, the presence of a functional CRISPR-Cas locus correlates with a lack of virulence genes in both E. faecalis and E. faecium (176, 185, 191) (Fig. 2A). The E. faecalis oral commensal strain OG1RF contains both functional CRISPR1-Cas and an orphaned CRISPR2 locus, while the hospital-acquired strain V583 harbors only the CRISPR2 orphan locus (192, 193). This correlation between the absence of functional CRISPR-Cas in hospital-adapted strains indicates a fitness cost for harboring CRISPR-Cas; in order to protect against phage infection and/or plasmid acquisition, CRISPR-Cas is needed, which may not benefit enterococcal strains when adapting to environments outside the GI tract such as the hospital setting (Fig. 2B).
In addition to the above type II-A CRISPR systems, a type I-C CRISPR locus has been identified in three pathogenic strains of Enterococcus cecorum, a poultry pathogen, and predicted to be functional in two strains (175). This CRISPR type is rare in enterococci but commonly found in streptococci (175). Commensal strains of E. cecorum harbor the more typical type II-A CRISPR arrangement (175). The presence of a type 1-C locus in pathogenic strains is unusual, as it defies the established pattern of disease-associated strains being less likely to contain a functional CRISPR system (176, 185, 186, 191).
Although CRISPR-Cas was identified as an antiphage system, CRISPR also guards against invading MGEs. E. faecalis type II CRISPR-Cas blocks antibiotic resistance encoding plasmid conjugation in vivo (196, 197). Additional known targets of enterococcal CRISPR-Cas include the PRPs pTEF1 and pTEF2, phages, and prophages (176, 178). No spacers have been identified against conjugative transposons; the tet(M) tetracycline resistance gene, commonly carried by Tn916 and its relatives, is present in strains with functional CRISPR-Cas (176). Many spacers have been found in E. faecalis and E. hirae, which correspond to bacterial chromosomal sequences (178, 187). Specifically, spacers have been identified with homology to the E. faecalis V583 ref35B gene, potentially encoding an antisense ncRNA (178, 198). It is unknown if CRISPR-Cas systems target PAIs. Many spacers have yet to be matched with a target sequence, likely due to the lack of available MGE sequences in GenBank (178, 199).
Current research describes the ramifications of harboring a CRISPR-Cas system as it relates to the pathogenic potential of enterococci while also inspiring future research into the context of CRISPR-Cas in multispecies environments. Known spacers targeting the chromosome of non-CRISPR-harboring bacteria, in addition to the high abundance of CRISPR-positive strains in environments like endodontic biofilms, warrant investigations into how CRISPR might modulate polymicrobial interactions (186, 187). Experimentation on the role of CRISPR within a multispecies microbial community could provide insight into the complex nature of microbial interactions in various niches.
CONCLUSIONS
The recent accessibility of next-generation sequencing technologies has increased our knowledge of the role of MGEs in enterococcal biology. The discovery that MGEs can constitute 25% of the E. faecalis V583 genome opened the door to a multitude of studies elucidating the role of the mobilome in enterococcal evolution (36). By studying E. faecalis and E. faecium as model enterococci, it is becoming clear that the enterococcal mobilome drives environmental adaptation and niche colonization (Fig. 1). Similar studies in other species, including E. gallinarum and E. casseliflavus, are necessary to answer questions regarding the overall impact of MGEs on enterococcal evolution. Obtaining more complete genome sequences of these understudied species is a key first step. Closing genomes reveals plasmid carriage, prophage presence, and the distribution of transposable elements within the genome (200, 201). By comparing the mobilomes of E. faecalis and E. faecium to other species, we can resolve many of the questions regarding the role of MGEs in virulence and ecological competition in addition to better understanding the MGE host range.
Prophage contribution to enterococcal evolution is understudied. Even though E. faecalis V583 prophages are associated with virulence and interbacterial competition, the ecological and evolutionary impact remains underexplored. Studies focusing on single prophage-harboring strains of E. faecalis should reveal distinct prophage benefits. Homologs of the E. faecalis V583 prophages have been identified in other strains, but similarities and differences among these prophages are yet to be elucidated (76, 202). Prophage-mediated HGT and its contributions to virulence have been studied in other bacteria, such as S. aureus. Examples include pathogenicity and genomic island transfer between strains (203–205), gene regulation upon prophage excision and integration (206), and DNA transduction (207, 208). Similar studies in enterococci will further contribute to our understanding of the role of coevolution between phages and their hosts and the implications of undiscovered and uncharacterized prophages on enterococcal environmental adaptation.
Few studies have assessed the ability of the enterococci to share MGEs like plasmids with other genera (133). While PRPs have been extensively studied in E. faecalis, more work should focus on the ability of other bacterial species to induce the pheromone-responsive conjugation system through secreted peptides, especially since other bacteria such as staphylococci are known to produce a variety of small, secreted peptides (209). Alternatively, it is possible that PRPs will fail to replicate or will be subjected to CRISPR-Cas-targeted degradation in the new host, nullifying the hypothesis that PRPs can be maintained outside the enterococci (99, 130). Finally, chromosomal conformation capture (Hi-C) technology with deep sequencing that estimates physical proximity between DNA molecules (210, 211) could potentially be leveraged to track lateral dissemination of enterococcal prophages, plasmids, and PRPs within complex multispecies communities (212, 213).
Finally, the role of CRISPR-Cas systems in controlling enterococcal HGT has not yet been thoroughly characterized. While identified spacers indicate that CRISPR-Cas is able to target the genome of non-CRISPR-harboring strains (178), the circumstances upon which targeting of nonhost genomes may occur remains unclear. Further, contributions of CRISPR-Cas-mediated restriction of phage predation or MGE movement and subsequent impact on microbial community dynamics should be assessed in the context of polymicrobial environments, such as the GI tract. High-throughput sequencing could be leveraged to match enterococcal CRISPR spacers to their source in order to provide additional context on the role of CRISPR-Cas and identify common threats that enterococci face while also providing insight on spacer acquisition and retention mechanisms.
ACKNOWLEDGMENT
This work was supported by National Institutes of Health grant R01AI141479 (B.A.D.).
Contributor Information
Anushila Chatterjee, Email: anushila.chatterjee@cuanschutz.edu.
William Margolin, McGovern Medical School.
REFERENCES
- 1.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. 2005. Diversity of the human intestinal microbial flora. Science 308:1635–1638. 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lebreton F, Manson AL, Saavedra JT, Straub TJ, Earl AM, Gilmore MS. 2017. Tracing the enterococci from Paleozoic origins to the hospital. Cell 169:849–861.e13. 10.1016/j.cell.2017.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hayashi H, Takahashi R, Nishi T, Sakamoto M, Benno Y. 2005. Molecular analysis of jejunal, ileal, caecal and recto-sigmoidal human colonic microbiota using 16S rRNA gene libraries and terminal restriction fragment length polymorphism. J Med Microbiol 54:1093–1101. 10.1099/jmm.0.45935-0. [DOI] [PubMed] [Google Scholar]
- 4.Smyth CJ, Matthews H, Halpenny MK, Brandis H, Colman G. 1987. Biotyping, serotyping and phage typing of Streptococcus faecalis isolated from dental plaque in the human mouth. J Med Microbiol 23:45–54. 10.1099/00222615-23-1-45. [DOI] [PubMed] [Google Scholar]
- 5.Martin JD, Mundt JO. 1972. Enterococci in insects. Appl Microbiol 24:575–580. 10.1128/am.24.4.575-580.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mundt JO. 1963. Occurrence of enterococci in animals in a wild environment. Appl Microbiol 11:136–140. 10.1128/am.11.2.136-140.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giraffa G. 2002. Enterococci from foods. FEMS Microbiol Rev 26:163–171. 10.1111/j.1574-6976.2002.tb00608.x. [DOI] [PubMed] [Google Scholar]
- 8.Mundt JO. 1963. Occurrence of enterococci on plants in a wild environment. Appl Microbiol 11:141–144. 10.1128/am.11.2.141-144.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mundt JO. 1961. Occurrence of enterococci: bud, blossom, and soil studies. Appl Microbiol 9:541–544. 10.1128/am.9.6.541-544.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ator LL, Starzyk MJ. 1976. Distribution of group D streptococci in rivers and streams. Microbios 16:91–104. [PubMed] [Google Scholar]
- 11.Huycke MM, Sahm DF, Gilmore MS. 1998. Multiple-drug resistant enterococci: the nature of the problem and an agenda for the future. Emerg Infect Dis 4:239–249. 10.3201/eid0402.980211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weiner LM, Webb AK, Limbago B, Dudeck MA, Patel J, Kallen AJ, Edwards JR, Sievert DM. 2016. Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2011–2014. Infect Control Hosp Epidemiol 37:1288–1301. 10.1017/ice.2016.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilmore MS, Lebreton F, van Schaik W. 2013. Genomic transition of enterococci from gut commensals to leading causes of multidrug-resistant hospital infection in the antibiotic era. Curr Opin Microbiol 16:10–16. 10.1016/j.mib.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Donskey CJ, Chowdhry TK, Hecker MT, Hoyen CK, Hanrahan JA, Hujer AM, Hutton-Thomas RA, Whalen CC, Bonomo RA, Rice LB. 2000. Effect of antibiotic therapy on the density of vancomycin-resistant enterococci in the stool of colonized patients. N Engl J Med 343:1925–1932. 10.1056/NEJM200012283432604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Centers for Disease Control and Prevention. 2019. Antibiotic resistance threats in the United States, 2019. Centers for Disease Control and Prevention, Atlanta, GA. [Google Scholar]
- 16.Higuita NIA, Huycke MM. 2014. Enterococcal disease, epidemiology, and implications for treatment. In Enterococci: from commensals to leading causes of drug resistant infection [Internet]. Massachusetts Eye and Ear Infirmary, Boston, MA. [Google Scholar]
- 17.Murray BE. 1990. The life and times of the Enterococcus. Clin Microbiol Rev 3:46–65. 10.1128/CMR.3.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arias CA, Murray BE. 2012. The rise of the Enterococcus: beyond vancomycin resistance. Nat Rev Microbiol 10:266–278. 10.1038/nrmicro2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davis E, Hicks L, Ali I, Salzman E, Wang J, Snitkin E, Gibson K, Cassone M, Mody L, Foxman B. 2020. Epidemiology of vancomycin-resistant Enterococcus faecium and Enterococcus faecalis colonization in nursing facilities. Open Forum Infect Dis 7:ofz553. 10.1093/ofid/ofz553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Puchter L, Chaberny IF, Schwab F, Vonberg RP, Bange FC, Ebadi E. 2018. Economic burden of nosocomial infections caused by vancomycin-resistant enterococci. Antimicrob Resist Infect Control 7:1. 10.1186/s13756-017-0291-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sievert DM, Ricks P, Edwards JR, Schneider A, Patel J, Srinivasan A, Kallen A, Limbago B, Fridkin S, National Healthcare Safety Network T, Participating NF, National Healthcare Safety Network (NHSN) Team and Participating NHSN Facilities. 2013. Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2009–2010. Infect Control Hosp Epidemiol 34:1–14. 10.1086/668770. [DOI] [PubMed] [Google Scholar]
- 22.Van Tyne D, Gilmore MS. 2014. Friend turned foe: evolution of enterococcal virulence and antibiotic resistance. Annu Rev Microbiol 68:337–356. 10.1146/annurev-micro-091213-113003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.García-Solache M, Rice LB. 2019. The enterococcus: a model of adaptability to its environment. Clin Microbiol Rev 32:e00058-18. 10.1128/CMR.00058-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahmed MO, Baptiste KE. 2018. Vancomycin-resistant enterococci: a review of antimicrobial resistance mechanisms and perspectives of human and animal health. Microb Drug Resist 24:590–606. 10.1089/mdr.2017.0147. [DOI] [PubMed] [Google Scholar]
- 25.Rice LB, Desbonnet C, Tait-Kamradt A, Garcia-Solache M, Lonks J, Moon TM, D'Andréa ÉD, Page R, Peti W. 2018. Structural and regulatory changes in PBP4 trigger decreased β-lactam susceptibility in Enterococcus faecalis. mBio 9:e00361-18. 10.1128/mBio.00361-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tyson GH, Sabo JL, Hoffmann M, Hsu C-H, Mukherjee S, Hernandez J, Tillman G, Wasilenko JL, Haro J, Simmons M, Wilson Egbe W, White PL, Dessai U, Mcdermott PF. 2018. Novel linezolid resistance plasmids in Enterococcus from food animals in the USA. J Antimicrob Chemother 73:3254–3258. 10.1093/jac/dky369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chilambi GS, Nordstrom HR, Evans DR, Ferrolino JA, Hayden RT, Maron GM, Vo AN, Gilmore MS, Wolf J, Rosch JW, Van Tyne D. 2020. Evolution of vancomycin-resistant Enterococcus faecium during colonization and infection in immunocompromised pediatric patients. Proc Natl Acad Sci USA 117:11703–11714. 10.1073/pnas.1917130117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palmer KL, Kos VN, Gilmore MS. 2010. Horizontal gene transfer and the genomics of enterococcal antibiotic resistance. Curr Opin Microbiol 13:632–639. 10.1016/j.mib.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hegstad K, Mikalsen T, Coque TM, Werner G, Sundsfjord A. 2010. Mobile genetic elements and their contribution to the emergence of antimicrobial resistant Enterococcus faecalis and Enterococcus faecium. Clin Microbiol Infect 16:541–554. 10.1111/j.1469-0691.2010.03226.x. [DOI] [PubMed] [Google Scholar]
- 30.Mikalsen T, Pedersen T, Willems R, Coque TM, Werner G, Sadowy E, van Schaik W, Jensen LB, Sundsfjord A, Hegstad K. 2015. Investigating the mobilome in clinically important lineages of Enterococcus faecium and Enterococcus faecalis. BMC Genomics 16:282. 10.1186/s12864-015-1407-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manson JM, Hancock LE, Gilmore MS. 2010. Mechanism of chromosomal transfer of Enterococcus faecalis pathogenicity island, capsule, antimicrobial resistance, and other traits. Proc Natl Acad Sci USA 107:12269–12274. 10.1073/pnas.1000139107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lebreton F, van Schaik W, Manson McGuire A, Godfrey P, Griggs A, Mazumdar V, Corander J, Cheng L, Saif S, Young S, Zeng Q, Wortman J, Birren B, Willems RJL, Earl AM, Gilmore MS. 2013. Emergence of epidemic multidrug-resistant Enterococcus faecium from animal and commensal strains. mBio 4:e00534-13. 10.1128/mBio.00534-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roberts AP, Mullany P. 2009. A modular master on the move: the Tn916 family of mobile genetic elements. Trends Microbiol 17:251–258. 10.1016/j.tim.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 34.Jahan M, Holley RA. 2016. Transfer of antibiotic resistance from Enterococcus faecium of fermented meat origin to Listeria monocytogenes and Listeria innocua. Lett Appl Microbiol 62:304–310. 10.1111/lam.12553. [DOI] [PubMed] [Google Scholar]
- 35.Kim EB, Marco ML. 2014. Nonclinical and clinical Enterococcus faecium strains, but not Enterococcus faecalis strains, have distinct structural and functional genomic features. Appl Environ Microbiol 80:154–165. 10.1128/AEM.03108-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paulsen IT, Banerjei L, Myers GS, Nelson KE, Seshadri R, Read TD, Fouts DE, Eisen JA, Gill SR, Heidelberg JF, Tettelin H, Dodson RJ, Umayam L, Brinkac L, Beanan M, Daugherty S, DeBoy RT, Durkin S, Kolonay J, Madupu R, Nelson W, Vamathevan J, Tran B, Upton J, Hansen T, Shetty J, Khouri H, Utterback T, Radune D, Ketchum KA, Dougherty BA, Fraser CM. 2003. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 299:2071–2074. 10.1126/science.1080613. [DOI] [PubMed] [Google Scholar]
- 37.Frost LS, Leplae R, Summers AO, Toussaint A. 2005. Mobile genetic elements: the agents of open source evolution. Nat Rev Microbiol 3:722–732. 10.1038/nrmicro1235. [DOI] [PubMed] [Google Scholar]
- 38.Kristich CJ, Rice LB, Arias CA. 2014. Enterococcal infection—treatment and antibiotic resistance. In Gilmore MS, Clewell DB, Ike Y, Shankar N (ed), Enterococci: from commensals to leading causes of drug resistant infection. Massachusetts Eye and Ear Infirmary, Boston, MA. [PubMed] [Google Scholar]
- 39.Balli EP, Venetis CA, Miyakis S. 2014. Systematic review and meta-analysis of linezolid versus daptomycin for treatment of vancomycin-resistant enterococcal bacteremia. Antimicrob Agents Chemother 58:734–739. 10.1128/AAC.01289-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Narayanan N, Rai R, Vaidya P, Desai A, Bhowmick T, Weinstein MP. 2019. Comparison of linezolid and daptomycin for the treatment of vancomycin-resistant enterococcal bacteremia. Ther Adv Infect Dis 6:2049936119828964. 10.1177/2049936119828964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee T, Pang S, Abraham S, Coombs GW. 2019. Antimicrobial-resistant CC17 Enterococcus faecium: the past, the present and the future. J Glob Antimicrob Resist 16:36–47. 10.1016/j.jgar.2018.08.016. [DOI] [PubMed] [Google Scholar]
- 42.Starikova I, Al-Haroni M, Werner G, Roberts AP, Sorum V, Nielsen KM, Johnsen PJ. 2013. Fitness costs of various mobile genetic elements in Enterococcus faecium and Enterococcus faecalis. J Antimicrob Chemother 68:2755–2765. 10.1093/jac/dkt270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Diaz Ricci JC, Hernandez ME. 2000. Plasmid effects on Escherichia coli metabolism. Crit Rev Biotechnol 20:79–108. 10.1080/07388550008984167. [DOI] [PubMed] [Google Scholar]
- 44.Yano H, Wegrzyn K, Loftie-Eaton W, Johnson J, Deckert GE, Rogers LM, Konieczny I, Top EM. 2016. Evolved plasmid-host interactions reduce plasmid interference cost. Mol Microbiol 101:743–756. 10.1111/mmi.13407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stalder T, Top E. 2016. Plasmid transfer in biofilms: a perspective on limitations and opportunities. NPJ Biofilms Microbiomes 2:16022. 10.1038/npjbiofilms.2016.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jacobson A. 1972. Role of F pili in the penetration of bacteriophage fl. J Virol 10:835–843. 10.1128/JVI.10.4.835-843.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Caro LG, Schnös M. 1966. The attachment of the male-specific bacteriophage F1 to sensitive strains of Escherichia coli. Proc Natl Acad Sci USA 56:126–132. 10.1073/pnas.56.1.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brooks MR, Padilla-Vélez L, Khan TA, Qureshi AA, Pieper JB, Maddox CW, Alam MT. 2020. Prophage-mediated disruption of genetic competence in Staphylococcus pseudointermedius. mSystems 5:e00684-19. 10.1128/mSystems.00684-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Feiner R, Argov T, Rabinovich L, Sigal N, Borovok I, Herskovits AA. 2015. A new perspective on lysogeny: prophages as active regulatory switches of bacteria. Nat Rev Microbiol 13:641–650. 10.1038/nrmicro3527. [DOI] [PubMed] [Google Scholar]
- 50.Duerkop BA, Clements CV, Rollins D, Rodrigues JL, Hooper LV. 2012. A composite bacteriophage alters colonization by an intestinal commensal bacterium. Proc Natl Acad Sci USA 109:17621–17626. 10.1073/pnas.1206136109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Matos RC, Lapaque N, Rigottier-Gois L, Debarbieux L, Meylheuc T, Gonzalez-Zorn B, Repoila F, Lopes MdeF, Serror P. 2013. Enterococcus faecalis prophage dynamics and contributions to pathogenic traits. PLoS Genet 9:e1003539. 10.1371/journal.pgen.1003539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709–1712. 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 53.Mushegian AR. 2020. Are there 10(31) virus particles on earth, or more, or fewer? J Bacteriol 202:e00052-20. 10.1128/JB.00052-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Clark PF, Clark AS. 1927. A bacteriophage active against a virulent hemolytic Streptococcus. Proc Soc Exp Biol Med 24:635–639. 10.3181/00379727-24-3498. [DOI] [Google Scholar]
- 55.Bonilla N, Santiago T, Marcos P, Urdaneta M, Santo Domingo J, Toranzos GA. 2010. Enterophages, a group of phages infecting Enterococcus faecalis, and their potential as alternate indicators of human faecal contamination. Water Sci Technol 61:293–300. 10.2166/wst.2010.815. [DOI] [PubMed] [Google Scholar]
- 56.Santiago-Rodríguez TM, Dávila C, González J, Bonilla N, Marcos P, Urdaneta M, Cadete M, Monteiro S, Santos R, Domingo JS, Toranzos GA. 2010. Characterization of Enterococcus faecalis-infecting phages (enterophages) as markers of human fecal pollution in recreational waters. Water Res 44:4716–4725. 10.1016/j.watres.2010.07.078. [DOI] [PubMed] [Google Scholar]
- 57.Uchiyama J, Rashel M, Maeda Y, Takemura I, Sugihara S, Akechi K, Muraoka A, Wakiguchi H, Matsuzaki S. 2008. Isolation and characterization of a novel Enterococcus faecalis bacteriophage ΦEF24C as a therapeutic candidate. FEMS Microbiol Lett 278:200–206. 10.1111/j.1574-6968.2007.00996.x. [DOI] [PubMed] [Google Scholar]
- 58.Chatterjee A, Duerkop BA. 2018. Beyond bacteria: bacteriophage-eukaryotic host interactions reveal emerging paradigms of health and disease. Front Microbiol 9:1394. 10.3389/fmicb.2018.01394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Touchon M, Moura de Sousa JA, Rocha EP. 2017. Embracing the enemy: the diversification of microbial gene repertoires by phage-mediated horizontal gene transfer. Curr Opin Microbiol 38:66–73. 10.1016/j.mib.2017.04.010. [DOI] [PubMed] [Google Scholar]
- 60.Fortier LC, Sekulovic O. 2013. Importance of prophages to evolution and virulence of bacterial pathogens. Virulence 4:354–365. 10.4161/viru.24498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Howard-Varona C, Hargreaves KR, Abedon ST, Sullivan MB. 2017. Lysogeny in nature: mechanisms, impact and ecology of temperate phages. ISME J 11:1511–1520. 10.1038/ismej.2017.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Obeng N, Pratama AA, Elsas JDV. 2016. The significance of mutualistic phages for bacterial ecology and evolution. Trends Microbiol 24:440–449. 10.1016/j.tim.2015.12.009. [DOI] [PubMed] [Google Scholar]
- 63.Chiang YN, Penades JR, Chen J. 2019. Genetic transduction by phages and chromosomal islands: the new and noncanonical. PLoS Pathog 15:e1007878. 10.1371/journal.ppat.1007878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen J, Quiles-Puchalt N, Chiang YN, Bacigalupe R, Fillol-Salom A, Chee MSJ, Fitzgerald JR, Penades JR. 2018. Genome hypermobility by lateral transduction. Science 362:207–212. 10.1126/science.aat5867. [DOI] [PubMed] [Google Scholar]
- 65.Andersson DI, Hughes D. 2014. Microbiological effects of sublethal levels of antibiotics. Nat Rev Microbiol 12:465–478. 10.1038/nrmicro3270. [DOI] [PubMed] [Google Scholar]
- 66.Haaber J, Leisner JJ, Cohn MT, Catalan-Moreno A, Nielsen JB, Westh H, Penades JR, Ingmer H. 2016. Bacterial viruses enable their host to acquire antibiotic resistance genes from neighbouring cells. Nat Commun 7:13333. 10.1038/ncomms13333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Blanco M, Devoret R. 1973. Repair mechanisms involved in prophage reactivation and UV reactivation of UV-irradiated phage λ. Mutat Res 17:293–305. 10.1016/0027-5107(73)90001-8. [DOI] [PubMed] [Google Scholar]
- 68.DeMarini DM, Lawrence BK. 1992. Prophage induction by DNA topoisomerase II poisons and reactive-oxygen species: role of DNA breaks. Mutat Res 267:1–17. 10.1016/0027-5107(92)90106-c. [DOI] [PubMed] [Google Scholar]
- 69.Johnson TA, Looft T, Severin AJ, Bayles DO, Nasko DJ, Wommack KE, Howe A, Allen HK. 2017. The in-feed antibiotic carbadox induces phage gene transcription in the swine gut microbiome. mBio 8:e00709-17. 10.1128/mBio.00709-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Úbeda C, Maiques E, Knecht E, Lasa Í, Novick RP, Penadés JR. 2005. Antibiotic-induced SOS response promotes horizontal dissemination of pathogenicity island-encoded virulence factors in staphylococci. Mol Microbiol 56:836–844. 10.1111/j.1365-2958.2005.04584.x. [DOI] [PubMed] [Google Scholar]
- 71.Casjens SR, Hendrix RW. 2015. Bacteriophage lambda: early pioneer and still relevant. Virology 479–480:310–330. 10.1016/j.virol.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brouwer S, Barnett TC, Ly D, Kasper KJ, De Oliveira DMP, Rivera-Hernandez T, Cork AJ, McIntyre L, Jespersen MG, Richter J, Schulz BL, Dougan G, Nizet V, Yuen KY, You Y, McCormick JK, Sanderson-Smith ML, Davies MR, Walker MJ. 2020. Prophage exotoxins enhance colonization fitness in epidemic scarlet fever-causing Streptococcus pyogenes. Nat Commun 11:5018. 10.1038/s41467-020-18700-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Coombs GW, Baines SL, Howden BP, Swenson KM, O'Brien FG. 2020. Diversity of bacteriophages encoding Panton-Valentine leukocidin in temporally and geographically related Staphylococcus aureus. PLoS One 15:e0228676. 10.1371/journal.pone.0228676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rezaei Javan R, Ramos-Sevillano E, Akter A, Brown J, Brueggemann AB. 2019. Prophages and satellite prophages are widespread in Streptococcus and may play a role in pneumococcal pathogenesis. Nat Commun 10:4852. 10.1038/s41467-019-12825-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schroven K, Aertsen A, Lavigne R. 2021. Bacteriophages as drivers of bacterial virulence and their potential for biotechnological exploitation. FEMS Microbiol Rev 45:fuaa041. 10.1093/femsre/fuaa041. [DOI] [PubMed] [Google Scholar]
- 76.Yasmin A, Kenny JG, Shankar J, Darby AC, Hall N, Edwards C, Horsburgh MJ. 2010. Comparative genomics and transduction potential of Enterococcus faecalis temperate bacteriophages. J Bacteriol 192:1122–1130. 10.1128/JB.01293-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Waters CM, Antiporta MH, Murray BE, Dunny GM. 2003. Role of the Enterococcus faecalis GelE protease in determination of cellular chain length, supernatant pheromone levels, and degradation of fibrin and misfolded surface proteins. J Bacteriol 185:3613–3623. 10.1128/JB.185.12.3613-3623.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fard RMN, Barton MD, Heuzenroeder MW. 2011. Bacteriophage-mediated transduction of antibiotic resistance in enterococci. Lett Appl Microbiol 52:559–564. 10.1111/j.1472-765X.2011.03043.x. [DOI] [PubMed] [Google Scholar]
- 79.Miller WR, Munita JM, Arias CA. 2014. Mechanisms of antibiotic resistance in enterococci. Expert Rev Anti Infect Ther 12:1221–1236. 10.1586/14787210.2014.956092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rossmann FS, Racek T, Wobser D, Puchalka J, Rabener EM, Reiger M, Hendrickx AP, Diederich AK, Jung K, Klein C, Huebner J. 2015. Phage-mediated dispersal of biofilm and distribution of bacterial virulence genes is induced by quorum sensing. PLoS Pathog 11:e1004653. 10.1371/journal.ppat.1004653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Boling L, Cuevas DA, Grasis JA, Kang HS, Knowles B, Levi K, Maughan H, McNair K, Rojas MI, Sanchez SE, Smurthwaite C, Rohwer F. 2020. Dietary prophage inducers and antimicrobials: toward landscaping the human gut microbiome. Gut Microbes 11:721–734. 10.1080/19490976.2019.1701353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.McBride SM, Fischetti VA, Leblanc DJ, Moellering RC, Jr., Gilmore MS. 2007. Genetic diversity among Enterococcus faecalis. PLoS One 2:e582. 10.1371/journal.pone.0000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Afonina I, Tien B, Nair Z, Matysik A, Lam LN, Veleba M, Jie AKJ, Rashid R, Cazenave-Gassiot A, Wenk M, Wai SN, Kline KA. 2021. The composition and function of Enterococcus faecalis membrane vesicles. microLife 2:uqab002. 10.1093/femsml/uqab002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Martinez-Rubio R, Quiles-Puchalt N, Marti M, Humphrey S, Ram G, Smyth D, Chen J, Novick RP, Penades JR. 2017. Phage-inducible islands in the Gram-positive cocci. ISME J 11:1029–1042. 10.1038/ismej.2016.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kleiner M, Bushnell B, Sanderson KE, Hooper LV, Duerkop BA. 2020. Transductomics: sequencing-based detection and analysis of transduced DNA in pure cultures and microbial communities. Microbiome 8:158. 10.1186/s40168-020-00935-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lossouarn J, Briet A, Moncaut E, Furlan S, Bouteau A, Son O, Leroy M, DuBow MS, Lecointe F, Serror P, Petit MA. 2019. Enterococcus faecalis countermeasures defeat a virulent picovirinae bacteriophage. Viruses 11:48. 10.3390/v11010048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang H, Stevens R. 2021. Intrinsic resistance of Enterococcus faecalis strains to ΦEf11 phage endolysin is associated with the presence of ΦEf11 prophage. Arch Virol 166:249–258. 10.1007/s00705-020-04861-7. [DOI] [PubMed] [Google Scholar]
- 88.Gilmore MS, Salamzade R, Selleck E, Bryan N, Mello SS, Manson AL, Earl AM. 2020. Genes contributing to the unique biology and intrinsic antibiotic resistance of Enterococcus faecalis. mBio 11:e02962-20. 10.1128/mBio.02962-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Buultjens AH, Lam MMC, Ballard S, Monk IR, Mahony AA, Grabsch EA, Grayson ML, Pang S, Coombs GW, Robinson JO, Seemann T, Johnson PDR, Howden BP, Stinear TP. 2017. Evolutionary origins of the emergent ST796 clone of vancomycin resistant Enterococcus faecium. PeerJ 5:e2916. 10.7717/peerj.2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bayjanov JR, Baan J, Rogers MRC, Troelstra A, Willems RJL, van Schaik W. 2019. Enterococcus faecium genome dynamics during long-term asymptomatic patient gut colonization. Microb Genom 5:e000277. 10.1099/mgen.0.000277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.de Maat V, Stege PB, Dedden M, Hamer M, van Pijkeren J, Willems RJL, van Schaik W. 2019. CRISPR-Cas9-mediated genome editing in vancomycin-resistant Enterococcus faecium. FEMS Microbiol Lett 366:fnz256. 10.1093/femsle/fnz256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Vogwill T, MacLean RC. 2015. The genetic basis of the fitness costs of antimicrobial resistance: a meta-analysis approach. Evol Appl 8:284–295. 10.1111/eva.12202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.San Millan A, MacLean RC. 2017. Fitness costs of plasmids: a limit to plasmid transmission. Microbiol Spectr 5. 10.1128/microbiolspec.MTBP-0016-2017. [DOI] [PubMed] [Google Scholar]
- 94.Chen Y, Bandyopadhyay A, Kozlowicz BK, Haemig HAH, Tai A, Hu WS, Dunny GM. 2017. Mechanisms of peptide sex pheromone regulation of conjugation in Enterococcus faecalis. Microbiologyopen 6:e00492. 10.1002/mbo3.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dunny GM. 2013. Enterococcal sex pheromones: signaling, social behavior, and evolution. Annu Rev Genet 47:457–482. 10.1146/annurev-genet-111212-133449. [DOI] [PubMed] [Google Scholar]
- 96.Alvarez-Martinez CE, Christie PJ. 2009. Biological diversity of prokaryotic type IV secretion systems. Microbiol Mol Biol Rev 73:775–808. 10.1128/MMBR.00023-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Breuer RJ, Hirt H, Dunny GM. 2018. Mechanistic features of the enterococcal pCF10 sex pheromone response and the biology of Enterococcus faecalis in its natural habitat. J Bacteriol 200:e00733-17. 10.1128/JB.00733-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hirt H, Manias DA, Bryan EM, Klein JR, Marklund JK, Staddon JH, Paustian ML, Kapur V, Dunny GM. 2005. Characterization of the pheromone response of the Enterococcus faecalis conjugative plasmid pCF10: complete sequence and comparative analysis of the transcriptional and phenotypic responses of pCF10-containing cells to pheromone induction. J Bacteriol 187:1044–1054. 10.1128/JB.187.3.1044-1054.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sterling AJ, Snelling WJ, Naughton PJ, Ternan NG, Dooley JSG. 2020. Competent but complex communication: the phenomena of pheromone-responsive plasmids. PLoS Pathog 16:e1008310. 10.1371/journal.ppat.1008310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Handwerger S, Pucci MJ, Kolokathis A. 1990. Vancomycin resistance is encoded on a pheromone response plasmid in Enterococcus faecium 228. Antimicrob Agents Chemother 34:358–360. 10.1128/AAC.34.2.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chow JW, Thal LA, Perri MB, Vazquez JA, Donabedian SM, Clewell DB, Zervos MJ. 1993. Plasmid-associated hemolysin and aggregation substance production contribute to virulence in experimental enterococcal endocarditis. Antimicrob Agents Chemother 37:2474–2477. 10.1128/AAC.37.11.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Olmsted SB, Dunny GM, Erlandsen SL, Wells CL. 1994. A plasmid-encoded surface protein on Enterococcus faecalis augments its internalization by cultured intestinal epithelial cells. J Infect Dis 170:1549–1556. 10.1093/infdis/170.6.1549. [DOI] [PubMed] [Google Scholar]
- 103.Süssmuth SD, Muscholl-Silberhorn A, Wirth R, Susa M, Marre R, Rozdzinski E. 2000. Aggregation substance promotes adherence, phagocytosis, and intracellular survival of Enterococcus faecalis within human macrophages and suppresses respiratory burst. Infect Immun 68:4900–4906. 10.1128/IAI.68.9.4900-4906.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wells CL, Moore EA, Hoag JA, Hirt H, Dunny GM, Erlandsen SL. 2000. Inducible expression of Enterococcus faecalis aggregation substance surface protein facilitates bacterial internalization by cultured enterocytes. Infect Immun 68:7190–7194. 10.1128/IAI.68.12.7190-7194.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rakita RM, Vanek NN, Jacques-Palaz K, Mee M, Mariscalco MM, Dunny GM, Snuggs M, Van Winkle WB, Simon SI. 1999. Enterococcus faecalis bearing aggregation substance is resistant to killing by human neutrophils despite phagocytosis and neutrophil activation. Infect Immun 67:6067–6075. 10.1128/IAI.67.11.6067-6075.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hirt H, Greenwood-Quaintance KE, Karau MJ, Till LM, Kashyap PC, Patel R, Dunny GM. 2018. Enterococcus faecalis sex pheromone cCF10 enhances conjugative plasmid transfer in vivo. mBio 9:e00037-18. 10.1128/mBio.00037-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chuang ON, Schlievert PM, Wells CL, Manias DA, Tripp TJ, Dunny GM. 2009. Multiple functional domains of Enterococcus faecalis aggregation substance Asc10 contribute to endocarditis virulence. Infect Immun 77:539–548. 10.1128/IAI.01034-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zheng B, Tomita H, Inoue T, Ike Y. 2009. Isolation of VanB-type Enterococcus faecalis strains from nosocomial infections: first report of the isolation and identification of the pheromone-responsive plasmids pMG2200, encoding VanB-type vancomycin resistance and a Bac41-type bacteriocin, and pMG2201, encoding erythromycin resistance and cytolysin (Hly/Bac). Antimicrob Agents Chemother 53:735–747. 10.1128/AAC.00754-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Heaton MP, Discotto LF, Pucci MJ, Handwerger S. 1996. Mobilization of vancomycin resistance by transposon-mediated fusion of a VanA plasmid with an Enterococcus faecium sex pheromone-response plasmid. Gene 171:9–17. 10.1016/0378-1119(96)00022-4. [DOI] [PubMed] [Google Scholar]
- 110.Quirantes R, Valdivia E, Martín I, Martínez-Bueno M, Maqueda M, Gálvez A, Méndez E. 1995. Purification of sex pheromones specific for pMB1 and pMB2 plasmids of Enterococcus faecalis S-48. Can J Microbiol 41:629–632. 10.1139/m95-084. [DOI] [PubMed] [Google Scholar]
- 111.Flannagan SE, Clewell DB, Sedgley CM. 2008. A “retrocidal” plasmid in Enterococcus faecalis: passage and protection. Plasmid 59:217–230. 10.1016/j.plasmid.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 112.Weaver KE, Clewell DB. 1988. Regulation of the pAD1 sex pheromone response in Enterococcus faecalis: construction and characterization of lacZ transcriptional fusions in a key control region of the plasmid. J Bacteriol 170:4343–4352. 10.1128/jb.170.9.4343-4352.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Weaver K. 2020. The Fst/Ldr family of type I TA system toxins: potential roles in stress response, metabolism and pathogenesis. Toxins (Basel) 12:474. 10.3390/toxins12080474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Weaver KE, Weaver DM, Wells CL, Waters CM, Gardner ME, Ehli EA. 2003. Enterococcus faecalis plasmid pAD1-encoded Fst toxin affects membrane permeability and alters cellular responses to lantibiotics. J Bacteriol 185:2169–2177. 10.1128/JB.185.7.2169-2177.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Weaver KE, Walz KD, Heine MS. 1998. Isolation of a derivative of Escherichia coli-Enterococcus faecalis shuttle vector pAM401 temperature sensitive for maintenance in E. faecalis and its use in evaluating the mechanism of pAD1 par-dependent plasmid stabilization. Plasmid 40:225–232. 10.1006/plas.1998.1368. [DOI] [PubMed] [Google Scholar]
- 116.Greenfield TJ, Ehli E, Kirshenmann T, Franch T, Gerdes K, Weaver KE. 2000. The antisense RNA of the par locus of pAD1 regulates the expression of a 33-amino-acid toxic peptide by an unusual mechanism. Mol Microbiol 37:652–660. 10.1046/j.1365-2958.2000.02035.x. [DOI] [PubMed] [Google Scholar]
- 117.Weaver KE, Reddy SG, Brinkman CL, Patel S, Bayles KW, Endres JL. 2009. Identification and characterization of a family of toxin-antitoxin systems related to the Enterococcus faecalis plasmid pAD1 par addiction module. Microbiology (Reading) 155:2930–2940. 10.1099/mic.0.030932-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zaheer R, Cook SR, Barbieri R, Goji N, Cameron A, Petkau A, Polo RO, Tymensen L, Stamm C, Song J, Hannon S, Jones T, Church D, Booker CW, Amoako K, Van Domselaar G, Read RR, McAllister TA. 2020. Surveillance of Enterococcus spp. reveals distinct species and antimicrobial resistance diversity across a One-Health continuum. Sci Rep 10:3937. 10.1038/s41598-020-61002-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ike Y, Hashimoto H, Clewell DB. 1984. Hemolysin of Streptococcus faecalis subspecies zymogenes contributes to virulence in mice. Infect Immun 45:528–530. 10.1128/iai.45.2.528-530.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Coburn PS, Gilmore MS. 2003. The Enterococcus faecalis cytolysin: a novel toxin active against eukaryotic and prokaryotic cells. Cell Microbiol 5:661–669. 10.1046/j.1462-5822.2003.00310.x. [DOI] [PubMed] [Google Scholar]
- 121.Ritchey TW, Seeley HW. 1974. Cytochromes in Streptococcus faecalis var. zymogenes grown in a haematin-containing medium. J Gen Microbiol 85:220–228. 10.1099/00221287-85-2-220. [DOI] [PubMed] [Google Scholar]
- 122.Miehl R, Miller M, Yasbin RE. 1980. Plasmid mediated enhancement of UV resistance in Streptococcus faecalis. Plasmid 3:128–134. 10.1016/0147-619x(80)90104-3. [DOI] [PubMed] [Google Scholar]
- 123.Tomita H, Fujimoto S, Tanimoto K, Ike Y. 1997. Cloning and genetic and sequence analyses of the bacteriocin 21 determinant encoded on the Enterococcus faecalis pheromone-responsive conjugative plasmid pPD1. J Bacteriol 179:7843–7855. 10.1128/jb.179.24.7843-7855.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Alvarez-Sieiro P, Montalban-Lopez M, Mu D, Kuipers OP. 2016. Bacteriocins of lactic acid bacteria: extending the family. Appl Microbiol Biotechnol 100:2939–2951. 10.1007/s00253-016-7343-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Mokoena MP. 2017. Lactic acid bacteria and their bacteriocins: classification, biosynthesis and applications against uropathogens: a mini-review. Molecules 22:1255. 10.3390/molecules22081255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Maqueda M, Galvez A, Bueno MM, Sanchez-Barrena MJ, Gonzalez C, Albert A, Rico M, Valdivia E. 2004. Peptide AS-48: prototype of a new class of cyclic bacteriocins. Curr Protein Pept Sci 5:399–416. 10.2174/1389203043379567. [DOI] [PubMed] [Google Scholar]
- 127.Kommineni S, Bretl DJ, Lam V, Chakraborty R, Hayward M, Simpson P, Cao Y, Bousounis P, Kristich CJ, Salzman NH. 2015. Bacteriocin production augments niche competition by enterococci in the mammalian gastrointestinal tract. Nature 526:719–722. 10.1038/nature15524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Magi G, Capretti R, Paoletti C, Pietrella M, Ferrante L, Biavasco F, Varaldo PE, Facinelli B. 2003. Presence of a vanA-carrying pheromone response plasmid (pBRG1) in a clinical isolate of Enterococcus faecium. Antimicrob Agents Chemother 47:1571–1576. 10.1128/AAC.47.5.1571-1576.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Noble WC, Virani Z, Cree RG. 1992. Co-transfer of vancomycin and other resistance genes from Enterococcus faecalis NCTC 12201 to Staphylococcus aureus. FEMS Microbiol Lett 72:195–198. 10.1016/0378-1097(92)90528-v. [DOI] [PubMed] [Google Scholar]
- 130.Flannagan SE, Clewell DB. 2002. Identification and characterization of genes encoding sex pheromone cAM373 activity in Enterococcus faecalis and Staphylococcus aureus. Mol Microbiol 44:803–817. 10.1046/j.1365-2958.2002.02922.x. [DOI] [PubMed] [Google Scholar]
- 131.Showsh SA, De Boever EH, Clewell DB. 2001. Vancomycin resistance plasmid in Enterococcus faecalis that encodes sensitivity to a sex pheromone also produced by Staphylococcus aureus. Antimicrob Agents Chemother 45:2177–2178. 10.1128/AAC.45.7.2177-2178.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Clewell DB, An FY, White BA, Gawron-Burke C. 1985. Streptococcus faecalis sex pheromone (cAM373) also produced by Staphylococcus aureus and identification of a conjugative transposon (Tn918). J Bacteriol 162:1212–1220. 10.1128/jb.162.3.1212-1220.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Vickerman MM, Flannagan SE, Jesionowski AM, Brossard KA, Clewell DB, Sedgley CM. 2010. A genetic determinant in Streptococcus gordonii Challis encodes a peptide with activity similar to that of enterococcal sex pheromone cAM373, which facilitates intergeneric DNA transfer. J Bacteriol 192:2535–2545. 10.1128/JB.01689-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Nakayama J, Igarashi S, Nagasawa H, Clewell DB, An FY, Suzuki A. 1996. Isolation and structure of staph-cAM373 produced by Staphylococcus aureus that induces conjugal transfer of Enterococcus faecalis plasmid pAM373. Biosci Biotechnol Biochem 60:1038–1039. 10.1271/bbb.60.1038. [DOI] [PubMed] [Google Scholar]
- 135.Chen I-MA, Chu K, Palaniappan K, Ratner A, Huang J, Huntemann M, Hajek P, Ritter S, Varghese N, Seshadri R, Roux S, Woyke T, Eloe-Fadrosh EA, Ivanova NN, Kyrpides Nikos C. 2021. The IMG/M data management and analysis system v.6.0: new tools and advanced capabilities. Nucleic Acids Res 49:D751–D763. 10.1093/nar/gkaa939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kohler V, Vaishampayan A, Grohmann E. 2018. Broad-host-range Inc18 plasmids: occurrence, spread and transfer mechanisms. Plasmid 99:11–21. 10.1016/j.plasmid.2018.06.001. [DOI] [PubMed] [Google Scholar]
- 137.Jain A, Srivastava P. 2013. Broad host range plasmids. FEMS Microbiol Lett 348:87–96. 10.1111/1574-6968.12241. [DOI] [PubMed] [Google Scholar]
- 138.Novick RP. 1987. Plasmid incompatibility. Microbiol Rev 51:381–395. 10.1128/mr.51.4.381-395.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Brantl S, Behnke D, Alonso JC. 1990. Molecular analysis of the replication region of the conjugative Streptococcus agalactiae plasmid pIP501 in Bacillus subtilis. Comparison with plasmids pAMβ1 and pSM 19035. Nucleic Acids Res 18:4783–4790. 10.1093/nar/18.16.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zhu W, Murray PR, Huskins WC, Jernigan JA, McDonald LC, Clark NC, Anderson KF, McDougal LK, Hageman JC, Olsen-Rasmussen M, Frace M, Alangaden GJ, Chenoweth C, Zervos MJ, Robinson-Dunn B, Schreckenberger PC, Reller LB, Rudrik JT, Patel JB. 2010. Dissemination of an Enterococcus Inc18-like vanA plasmid associated with vancomycin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 54:4314–4320. 10.1128/AAC.00185-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Chatterjee A, Johnson CN, Luong P, Hullahalli K, McBride SW, Schubert AM, Palmer KL, Carlson PE, Duerkop BA. 2019. Bacteriophage resistance alters antibiotic-mediated intestinal expansion of enterococci. Infect Immun 87:e00085-19. 10.1128/IAI.00085-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lautenbach E, Schuster MG, Bilker WB, Brennan PJ. 1998. The role of chloramphenicol in the treatment of bloodstream infection due to vancomycin-resistant Enterococcus. Clin Infect Dis 27:1259–1265. 10.1086/515002. [DOI] [PubMed] [Google Scholar]
- 143.Ricaurte JC, Boucher HW, Turett GS, Moellering RC, Labombardi VJ, Kislak JW. 2001. Chloramphenicol treatment for vancomycin-resistant Enterococcus faecium bacteremia. Clin Microbiol Infect 7:17–21. 10.1046/j.1469-0691.2001.00189.x. [DOI] [PubMed] [Google Scholar]
- 144.De Graef EM, Decostere A, De Leener E, Goossens H, Baele M, Haesebrouck F. 2007. Prevalence and mechanism of resistance against macrolides, licosamides, and streptogramins among Enterococcus faecium isolates from food-producing animals and hospital patients in Belgium. Microb Drug Resist 13:135–141. 10.1089/mdr.2007.718. [DOI] [PubMed] [Google Scholar]
- 145.Zhu W, Clark NC, McDougal LK, Hageman J, McDonald LC, Patel JB. 2008. Vancomycin-resistant Staphylococcus aureus isolates associated with Inc18-like vanA plasmids in Michigan. Antimicrob Agents Chemother 52:452–457. 10.1128/AAC.00908-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zhu W, Clark N, Patel JB. 2013. pSK41-like plasmid is necessary for Inc18-like vanA plasmid transfer from Enterococcus faecalis to Staphylococcus aureus in vitro. Antimicrob Agents Chemother 57:212–219. 10.1128/AAC.01587-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Weigel LM, Clewell DB, Gill SR, Clark NC, McDougal LK, Flannagan SE, Kolonay JF, Shetty J, Killgore GE, Tenover FC. 2003. Genetic analysis of a high-level vancomycin-resistant isolate of Staphylococcus aureus. Science 302:1569–1571. 10.1126/science.1090956. [DOI] [PubMed] [Google Scholar]
- 148.Clewell DB, Yagi Y, Dunny GM, Schultz SK. 1974. Characterization of three plasmid deoxyribonucleic acid molecules in a strain of Streptococcus faecalis: identification of a plasmid determining erythromycin resistance. J Bacteriol 117:283–289. 10.1128/jb.117.1.283-289.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Horodniceanu T, Bouanchaud DH, Bieth G, Chabbert YA. 1976. R plasmids in Streptococcus agalactiae (group B). Antimicrob Agents Chemother 10:795–801. 10.1128/AAC.10.5.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Poyart-Salmeron C, Carlier C, Trieu-Cuot P, Courtieu AL, Courvalin P. 1990. Transferable plasmid-mediated antibiotic resistance in Listeria monocytogenes. Lancet 335:1422–1426. 10.1016/0140-6736(90)91447-i. [DOI] [PubMed] [Google Scholar]
- 151.Gasson MJ, Davies FL. 1980. Conjugal transfer of the drug resistance plasmid pAMβ in the lactic streptococci. FEMS Microbiol Lett 7:51–53. 10.1111/j.1574-6941.1980.tb01575.x. [DOI] [Google Scholar]
- 152.Gevers D, Huys G, Swings J. 2003. In vitro conjugal transfer of tetracycline resistance from Lactobacillus isolates to other Gram-positive bacteria. FEMS Microbiol Lett 225:125–130. 10.1016/S0378-1097(03)00505-6. [DOI] [PubMed] [Google Scholar]
- 153.Jacobsen L, Wilcks A, Hammer K, Huys G, Gevers D, Andersen SR. 2007. Horizontal transfer of tet(M) and erm(B) resistance plasmids from food strains of Lactobacillus plantarum to Enterococcus faecalis JH2-2 in the gastrointestinal tract of gnotobiotic rats. FEMS Microbiol Ecol 59:158–166. 10.1111/j.1574-6941.2006.00212.x. [DOI] [PubMed] [Google Scholar]
- 154.Durrant MG, Li MM, Siranosian BA, Montgomery SB, Bhatt AS. 2020. A bioinformatic analysis of integrative mobile genetic elements highlights their role in bacterial adaptation. Cell Host Microbe 27:140–153. 10.1016/j.chom.2019.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Top J, Arredondo-Alonso S, Schürch AC, Puranen S, Pesonen M, Pensar J, Willems RJL, Corander J. 2020. Genomic rearrangements uncovered by genome-wide co-evolution analysis of a major nosocomial pathogen, Enterococcus faecium. Microb Genom 6:mgen000488. 10.1099/mgen.0.000488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Sadowy E. 2021. Mobile genetic elements beyond the VanB-resistance dissemination among hospital-associated enterococci and other Gram-positive bacteria. Plasmid 114:102558. 10.1016/j.plasmid.2021.102558. [DOI] [PubMed] [Google Scholar]
- 157.Launay A, Ballard SA, Johnson PD, Grayson ML, Lambert T. 2006. Transfer of vancomycin resistance transposon Tn1549 from Clostridium symbiosum to Enterococcus spp. in the gut of gnotobiotic mice. Antimicrob Agents Chemother 50:1054–1062. 10.1128/AAC.50.3.1054-1062.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Quintiliani R, Jr., Courvalin P. 1996. Characterization of Tn1547, a composite transposon flanked by the IS16 and IS256-like elements, that confers vancomycin resistance in Enterococcus faecalis BM4281. Gene 172:1–8. 10.1016/0378-1119(96)00110-2. [DOI] [PubMed] [Google Scholar]
- 159.Lu JJ, Chang TY, Perng CL, Lee SY. 2005. The vanB2 gene cluster of the majority of vancomycin-resistant Enterococcus faecium isolates from Taiwan is associated with the pbp5 gene and is carried by Tn5382 containing a novel insertion sequence. Antimicrob Agents Chemother 49:3937–3939. 10.1128/AAC.49.9.3937-3939.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Simjee S, Gill MJ. 1997. Gene transfer, gentamicin resistance and enterococci. J Hosp Infect 36:249–259. 10.1016/s0195-6701(97)90051-7. [DOI] [PubMed] [Google Scholar]
- 161.Behnood A, Farajnia S, Moaddab SR, Ahdi-Khosroshahi S, Katayounzadeh A. 2013. Prevalence of aac(6')-Ie-aph(2″)-Ia resistance gene and its linkage to Tn5281 in Enterococcus faecalis and Enterococcus faecium isolates from Tabriz hospitals. Iran J Microbiol 5:203–208. [PMC free article] [PubMed] [Google Scholar]
- 162.Hodel-Christian SL, Murray BE. 1990. Mobilization of the gentamicin resistance gene in Enterococcus faecalis. Antimicrob Agents Chemother 34:1278–1280. 10.1128/AAC.34.6.1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Hodel-Christian SL, Murray BE. 1991. Characterization of the gentamicin resistance transposon Tn5281 from Enterococcus faecalis and comparison to staphylococcal transposons Tn4001 and Tn4031. Antimicrob Agents Chemother 35:1147–1152. 10.1128/AAC.35.6.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Hodel-Christian SL, Murray BE. 1992. Comparison of the gentamicin resistance transposon Tn5281 with regions encoding gentamicin resistance in Enterococcus faecalis isolates from diverse geographic locations. Antimicrob Agents Chemother 36:2259–2264. 10.1128/AAC.36.10.2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Lin Y-T, Tseng S-P, Hung W-W, Chang C-C, Chen Y-H, Jao Y-T, Chen Y-H, Teng L-J, Hung W-C. 2020. A possible role of insertion sequence IS 1216V in dissemination of multidrug-resistant elements MESPM1 and MES6272-2 between Enterococcus and ST50 Staphylococcus aureus. Microorganisms 8:1905. 10.3390/microorganisms8121905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Gal-Mor O, Finlay BB. 2006. Pathogenicity islands: a molecular toolbox for bacterial virulence. Cell Microbiol 8:1707–1719. 10.1111/j.1462-5822.2006.00794.x. [DOI] [PubMed] [Google Scholar]
- 167.Lepage E, Brinster S, Caron C, Ducroix-Crepy C, Rigottier-Gois L, Dunny G, Hennequet-Antier C, Serror P. 2006. Comparative genomic hybridization analysis of Enterococcus faecalis: identification of genes absent from food strains. J Bacteriol 188:6858–6868. 10.1128/JB.00421-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Nallapareddy SR, Wenxiang H, Weinstock GM, Murray BE. 2005. Molecular characterization of a widespread, pathogenic, and antibiotic resistance-receptive Enterococcus faecalis lineage and dissemination of its putative pathogenicity island. J Bacteriol 187:5709–5718. 10.1128/JB.187.16.5709-5718.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Shankar N, Baghdayan AS, Willems R, Hammerum AM, Jensen LB. 2006. Presence of pathogenicity island genes in Enterococcus faecalis isolates from pigs in Denmark. J Clin Microbiol 44:4200–4203. 10.1128/JCM.01218-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.McBride SM, Coburn PS, Baghdayan AS, Willems RJL, Grande MJ, Shankar N, Gilmore MS. 2009. Genetic variation and evolution of the pathogenicity island of Enterococcus faecalis. J Bacteriol 191:3392–3402. 10.1128/JB.00031-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Shankar N, Baghdayan AS, Gilmore MS. 2002. Modulation of virulence within a pathogenicity island in vancomycin-resistant Enterococcus faecalis. Nature 417:746–750. 10.1038/nature00802. [DOI] [PubMed] [Google Scholar]
- 172.Sahm DF, Kissinger J, Gilmore MS, Murray PR, Mulder R, Solliday J, Clarke B. 1989. In vitro susceptibility studies of vancomycin-resistant Enterococcus faecalis. Antimicrob Agents Chemother 33:1588–1591. 10.1128/AAC.33.9.1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Laverde Gomez JA, Hendrickx AP, Willems RJ, Top J, Sava I, Huebner J, Witte W, Werner G. 2011. Intra- and interspecies genomic transfer of the Enterococcus faecalis pathogenicity island. PLoS One 6:e16720. 10.1371/journal.pone.0016720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Francia MV, Haas W, Wirth R, Samberger E, Muscholl-Silberhorn A, Gilmore MS, Ike Y, Weaver KE, An FY, Clewell DB. 2001. Completion of the nucleotide sequence of the Enterococcus faecalis conjugative virulence plasmid pAD1 and identification of a second transfer origin. Plasmid 46:117–127. 10.1006/plas.2001.1533. [DOI] [PubMed] [Google Scholar]
- 175.Borst LB, Suyemoto MM, Scholl EH, Fuller FJ, Barnes HJ. 2015. Comparative genomic analysis identifies divergent genomic features of pathogenic Enterococcus cecorum including a type IC CRISPR-Cas system, a capsule locus, an epa-like locus, and putative host tissue binding proteins. PLoS One 10:e0121294. 10.1371/journal.pone.0121294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Palmer KL, Gilmore MS. 2010. Multidrug-resistant enterococci lack CRISPR-cas. mBio 1:e00227-10. 10.1128/mBio.00227-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Horvath P, Barrangou R. 2010. CRISPR/Cas, the immune system of bacteria and archaea. Science 327:167–170. 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- 178.Gawryszewska I, Malinowska K, Kuch A, Chrobak-Chmiel D, Łaniewska-Trokenheim Ł, Hryniewicz W, Sadowy E. 2017. Distribution of antimicrobial resistance determinants, virulence-associated factors and clustered regularly interspaced palindromic repeats loci in isolates of Enterococcus faecalis from various settings and genetic lineages. Pathog Dis 75:ftx021. 10.1093/femspd/ftx021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Sontheimer EJ, Marraffini LA. 2010. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat Rev Genet 11:181–190. 10.1038/nrg2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Shiriaeva A, Fedorov I, Vyhovskyi D, Severinov K. 2020. Detection of CRISPR adaptation. Biochem Soc Trans 48:257–269. 10.1042/BST20190662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Canfield GS, Chatterjee A, Espinosa J, Mangalea MR, Sheriff EK, Keidan M, McBride SW, McCollister BD, Hang HC, Duerkop BA. 2021. Lytic bacteriophages facilitate antibiotic sensitization of Enterococcus faecium. Antimicrob Agents Chemother 65:e00143-21. 10.1128/AAC.00143-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Hullahalli K, Rodrigues M, Palmer KL. 2017. Exploiting CRISPR-Cas to manipulate Enterococcus faecalis populations. Elife 6:e26664. 10.7554/eLife.26664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, van der Oost J, Koonin EV. 2011. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol 9:467–477. 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Makarova KS, Koonin EV. 2015. Annotation and classification of CRISPR-Cas systems. Methods Mol Biol 1311:47–75. 10.1007/978-1-4939-2687-9_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Lindenstrauss AG, Pavlovic M, Bringmann A, Behr J, Ehrmann MA, Vogel RF. 2011. Comparison of genotypic and phenotypic cluster analyses of virulence determinants and possible role of CRISPR elements towards their incidence in Enterococcus faecalis and Enterococcus faecium. Syst Appl Microbiol 34:553–560. 10.1016/j.syapm.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 186.Burley KM, Sedgley CM. 2012. CRISPR-Cas, a prokaryotic adaptive immune system, in endodontic, oral, and multidrug-resistant hospital-acquired Enterococcus faecalis. J Endod 38:1511–1515. 10.1016/j.joen.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 187.Katyal I, Chaban B, Ng B, Hill JE. 2013. CRISPRs of Enterococcus faecalis and E. hirae isolates from pig feces have species-specific repeats but share some common spacer sequences. Microb Ecol 66:182–188. 10.1007/s00248-013-0217-0. [DOI] [PubMed] [Google Scholar]
- 188.Lyons C, Raustad N, Bustos MA, Shiaris M. 2015. Incidence of type II CRISPR1-Cas systems in Enterococcus is species-dependent. PLoS One 10:e0143544. 10.1371/journal.pone.0143544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Hullahalli K, Rodrigues M, Schmidt BD, Li X, Bhardwaj P, Palmer KL. 2015. Comparative analysis of the orphan CRISPR2 locus in 242 Enterococcus faecalis strains. PLoS One 10:e0138890. 10.1371/journal.pone.0138890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Huescas CGY, Pereira RI, Prichula J, Azevedo PA, Frazzon J, Frazzon APG. 2019. Frequency of clustered regularly interspaced short palindromic repeats (CRISPRs) in non-clinical Enterococcus faecalis and Enterococcus faecium strains. Braz J Biol 79:460–465. 10.1590/1519-6984.183375. [DOI] [PubMed] [Google Scholar]
- 191.dos Santos BA, de Oliveira JS, Parmanhani-da-Silva BM, Ribeiro RL, Teixeira LM, Neves FPG. 2020. CRISPR elements and their association with antimicrobial resistance and virulence genes among vancomycin-resistant and vancomycin-susceptible enterococci recovered from human and food sources. Infect Genet Evol 80:104183. 10.1016/j.meegid.2020.104183. [DOI] [PubMed] [Google Scholar]
- 192.Bourgogne A, Garsin DA, Qin X, Singh KV, Sillanpaa J, Yerrapragada S, Ding Y, Dugan-Rocha S, Buhay C, Shen H, Chen G, Williams G, Muzny D, Maadani A, Fox KA, Gioia J, Chen L, Shang Y, Arias CA, Nallapareddy SR, Zhao M, Prakash VP, Chowdhury S, Jiang H, Gibbs RA, Murray BE, Highlander SK, Weinstock GM. 2008. Large scale variation in Enterococcus faecalis illustrated by the genome analysis of strain OG1RF. Genome Biol 9:R110. 10.1186/gb-2008-9-7-r110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Horvath P, Coute-Monvoisin AC, Romero DA, Boyaval P, Fremaux C, Barrangou R. 2009. Comparative analysis of CRISPR loci in lactic acid bacteria genomes. Int J Food Microbiol 131:62–70. 10.1016/j.ijfoodmicro.2008.05.030. [DOI] [PubMed] [Google Scholar]
- 194.Price VJ, Huo W, Sharifi A, Palmer KL. 2016. CRISPR-Cas and restriction-modification act additively against conjugative antibiotic resistance plasmid transfer in Enterococcus faecalis. mSphere 1:e00064-16. 10.1128/mSphere.00064-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Tong Z, Du Y, Ling J, Huang L, Ma J. 2017. Relevance of the clustered regularly interspaced short palindromic repeats of Enterococcus faecalis strains isolated from retreatment root canals on periapical lesions, resistance to irrigants and biofilms. Exp Ther Med 14:5491–5496. 10.3892/etm.2017.5205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Price VJ, McBride SW, Hullahalli K, Chatterjee A, Duerkop BA, Palmer KL. 2019. Enterococcus faecalis CRISPR-Cas is a robust barrier to conjugative antibiotic resistance dissemination in the murine intestine. mSphere 4:e00464-19. 10.1128/mSphere.00464-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Rodrigues M, McBride SW, Hullahalli K, Palmer KL, Duerkop BA. 2019. Conjugative delivery of CRISPR-Cas9 for the selective depletion of antibiotic-resistant enterococci. Antimicrob Agents Chemother 63:e01454-19. 10.1128/AAC.01454-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Fouquier d'Herouel A, Wessner F, Halpern D, Ly-Vu J, Kennedy SP, Serror P, Aurell E, Repoila F. 2011. A simple and efficient method to search for selected primary transcripts: non-coding and antisense RNAs in the human pathogen Enterococcus faecalis. Nucleic Acids Res 39:e46. 10.1093/nar/gkr012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Shmakov SA, Sitnik V, Makarova KS, Wolf YI, Severinov KV, Koonin EV. 2017. The CRISPR spacer space is dominated by sequences from species-specific mobilomes. mBio 8:e01397-17. 10.1128/mBio.01397-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Fadeev E, De Pascale F, Vezzi A, Hübner S, Aharonovich D, Sher D. 2016. Why close a bacterial genome? The plasmid of Alteromonas macleodii HOT1A3 is a vector for inter-specific transfer of a flexible genomic island. Front Microbiol 7:248. 10.3389/fmicb.2016.00248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Land M, Hauser L, Jun S-R, Nookaew I, Leuze MR, Ahn T-H, Karpinets T, Lund O, Kora G, Wassenaar T, Poudel S, Ussery DW. 2015. Insights from 20 years of bacterial genome sequencing. Funct Integr Genomics 15:141–161. 10.1007/s10142-015-0433-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Palmer KL, Godfrey P, Griggs A, Kos VN, Zucker J, Desjardins C, Cerqueira G, Gevers D, Walker S, Wortman J, Feldgarden M, Haas B, Birren B, Gilmore MS. 2012. Comparative genomics of enterococci: variation in Enterococcus faecalis, clade structure in E. faecium, and defining characteristics of E. gallinarum and E. casseliflavus. mBio 3:e00318-11. 10.1128/mBio.00318-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Ruzin A, Lindsay J, Novick RP. 2001. Molecular genetics of SaPI1 – a mobile pathogenicity island in Staphylococcus aureus. Mol Microbiol 41:365–377. 10.1046/j.1365-2958.2001.02488.x. [DOI] [PubMed] [Google Scholar]
- 204.Tormo MÁ, Ferrer MD, Maiques E, Úbeda C, Selva L, Lasa Í, Calvete JJ, Novick RP, Penadés JR. 2008. Staphylococcus aureus pathogenicity island DNA is packaged in particles composed of phage proteins. J Bacteriol 190:2434–2440. 10.1128/JB.01349-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Moon BY, Park JY, Hwang SY, Robinson DA, Thomas JC, Fitzgerald JR, Park YH, Seo KS. 2015. Phage-mediated horizontal transfer of a Staphylococcus aureus virulence-associated genomic island. Sci Rep 5:9784. 10.1038/srep09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Tran PM, Feiss M, Kinney KJ, Salgado-Pabón W. 2019. ϕSa3mw prophage as a molecular regulatory switch of Staphylococcus aureus β-toxin production. J Bacteriol 201:e00766-18. 10.1128/JB.00766-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Rudin L, Sjöström J-E, Lindberg M, Philipson L. 1974. Factors affecting competence for transformation in Staphylococcus aureus. J Bacteriol 118:155–164. 10.1128/jb.118.1.155-164.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.van der Mee-Marquet N, Corvaglia A-R, Valentin A-S, Hernandez D, Bertrand X, Girard M, Kluytmans J, Donnio P-Y, Quentin R, François P. 2013. Analysis of prophages harbored by the human-adapted subpopulation of Staphylococcus aureus CC398. Infect Genet Evol 18:299–308. 10.1016/j.meegid.2013.06.009. [DOI] [PubMed] [Google Scholar]
- 209.Cook LC, Federle MJ. 2014. Peptide pheromone signaling in Streptococcus and Enterococcus. FEMS Microbiol Rev 38:473–492. 10.1111/1574-6976.12046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, Sandstrom R, Bernstein B, Bender MA, Groudine M, Gnirke A, Stamatoyannopoulos J, Mirny LA, Lander ES, Dekker J. 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326:289–293. 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Marbouty M, Baudry L, Cournac A, Koszul R. 2017. Scaffolding bacterial genomes and probing host-virus interactions in gut microbiome by proximity ligation (chromosome capture) assay. Sci Adv 3:e1602105. 10.1126/sciadv.1602105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Zhang T, Zhang X-X, Ye L. 2011. Plasmid metagenome reveals high levels of antibiotic resistance genes and mobile genetic elements in activated sludge. PLoS One 6:e26041. 10.1371/journal.pone.0026041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Marbouty M, Thierry A, Millot GA, Koszul R. 2021. MetaHiC phage-bacteria infection network reveals active cycling phages of the healthy human gut. Elife 10:e60608. 10.7554/eLife.60608. [DOI] [PMC free article] [PubMed] [Google Scholar]