ABSTRACT
Clostridioides difficile spores, like the spores from most endospore-forming organisms, are a metabolically dormant stage of development with a complex structure that conveys considerable resistance to environmental conditions, e.g., wet heat. This resistance is due to the large amount of dipicolinic acid (DPA) that is taken up by the spore core, preventing rotational motion of the core proteins. DPA is synthesized by the mother cell, and its packaging into the spore core is mediated by the products of the spoVA operon, which has a variable number of genes, depending on the organism. C. difficile encodes 3 spoVA orthologues, spoVAC, spoVAD, and spoVAE. Prior work has shown that C. difficile SpoVAC is a mechanosensing protein responsible for DPA release from the spore core upon the initiation of germination. However, the roles of SpoVAD and SpoVAE remain unclear in C. difficile. In this study, we analyzed the roles of SpoVAD and SpoVAE and found that they are essential for DPA uptake into the spore, similar to SpoVAC. Using split luciferase protein interaction assays, we found that these proteins interact, and we propose a model where SpoVAC/SpoVAD/SpoVAE proteins interact at or near the inner spore membrane, and each member of the complex is essential for DPA uptake into the spore core.
IMPORTANCE C. difficile spore heat resistance provides an avenue for it to survive the disinfection protocols in hospital and community settings. The spore heat resistance is mainly the consequence of the high DPA content within the spore core. By elucidating the mechanism by which DPA is taken up by the spore core, this study may provide insight into how to disrupt the spore heat resistance with the aim of making the current disinfection protocols more efficient at preventing the spread of C. difficile in the environment.
KEYWORDS: Clostridium difficile, DPA, SpoVAC, SpoVAD, SpoVAE, spores
INTRODUCTION
Clostridioides difficile is a Gram-positive, spore-forming, strictly anaerobic bacterium that is opportunistically pathogenic in humans. According to the most recent report by the Centers for Disease Control and Prevention, issued in 2019, it is estimated that 223,900 cases of C. difficile occurred in 2017 (1, 2). Of these, 12,800 deaths can be directly attributed to C. difficile, with the majority of deaths occurring among people aged 65 and older. In recent years, there has been an emergence of antibiotic-resistant strains, as well as strains with increased virulence, making C. difficile a leading cause of hospital-associated infections with an estimated $5 billion in annual treatment-associated cost for C. difficile infections (CDI) in U.S. hospitals alone (3, 4).
C. difficile infections are initiated upon disruption to the normally protective microbiota, commonly due to broad-spectrum antibiotic use (5–8). Antibiotics are prescribed to treat CDI (i.e., vancomycin or fidaxomicin), but patients frequently relapse with recurring CDI due to the continued disruption to the colonic microbiome and the presence of antibiotic-resistant spores that remain in the gastrointestinal tract or in the surrounding environment (9). Though the C. difficile vegetative form is the disease-causing agent, it is the spore form that is the infective agent due to its ability to survive outside the host in the aerobic environment. C. difficile spores are structurally complex, composed of several distinct layers that are broadly similar to spores produced by other endospore-forming organisms, e.g., Bacillus subtilis (10, 11). In the spore core, pyridine-2,6-dicarboxylic acid (dipicolinic acid [DPA]), chelated with calcium (Ca-DPA), provides extreme heat-resistant properties (12, 13). The core is surrounded by an inner membrane that has low permeability, even to water, and potentially DNA-damaging molecules, from entering the core (14, 15). Surrounding the inner spore membrane is a germ cell wall that will become the cell wall of the vegetative cell postgermination. Surrounding the germ cell wall is a thick layer of cortex peptidoglycan. In the cortex, a proportion of muramic acid residues is converted to muramic-δ-lactam (16, 17). The muramic-δ-lactam residues are the targets of cortex lytic enzymes during spore germination (18, 19). The cortex is surrounded by the outer spore membrane and by the coat, which provides the spore with protection from environmental insults and decontaminants (20). In some endospore-forming bacteria, the outermost layer of spores is an exosporium layer, which may have a role in adherence to host intestinal epithelial cells and other surfaces (21, 22).
Spores are metabolically dormant until the detection of germinants by germinant receptors. Germinant recognition initiates a cascade of events that irreversibly commits the spore to the germination pathway (23–25). This event is followed by the release of DPA from the core and degradation of the cortex by spore cortex lytic enzymes (SCLE), either simultaneously or sequentially, depending on the organism (19, 26, 27). In the model spore-forming bacterium, B. subtilis, SCLEs can be activated by exogenously added Ca-DPA or the release of Ca-DPA from the core, meaning that DPA release precedes cortex hydrolysis (19). However, in C. difficile, these two early germination steps are inverted, and cortex hydrolysis precedes DPA release (12, 28).
In C. difficile, activation of the SleC cortex lytic enzyme leads to the release of DPA stores from the spore core in exchange for water. In B. subtilis, the proteins encoded by the spoVA operon (SpoVAA-AB-AC-AD-AEa-AEb-AF) play a role in DPA uptake and release, but C. difficile encodes only 3 orthologues, spoVAC, spoVAD, and spoVAE. B. subtilis SpoVAC is a mechanosensing protein, similar to its role in C. difficile (28, 29). In B. subtilis, SpoVAD binds to DPA and is likely important in the uptake of DPA into the developing spore, and a spoVAEa mutation causes only a slight germination defect, while the role of spoVAEb is unclear (30, 31). Herein, we show that C. difficile spoVAD and spoVAE are essential for DPA uptake into the spore. Because in B. subtilis, the proteins of the spoVA operon are hypothesized to form a membrane channel enabling the uptake of DPA into the forming spore during sporulation and release of DPA from the spore during germination, we speculated that this also may be the case in C. difficile (31, 32). Using a split-luciferase reporter system, we find that, indeed, some of the products of the spoVA operon interact. This led us to propose a model of a SpoVAC/SpoVAD/SpoVAE interaction that describes their roles in DPA uptake and release during C. difficile spore formation and germination, respectively.
RESULTS
spoVAD and spoVAE are required for the uptake of DPA into the spore.
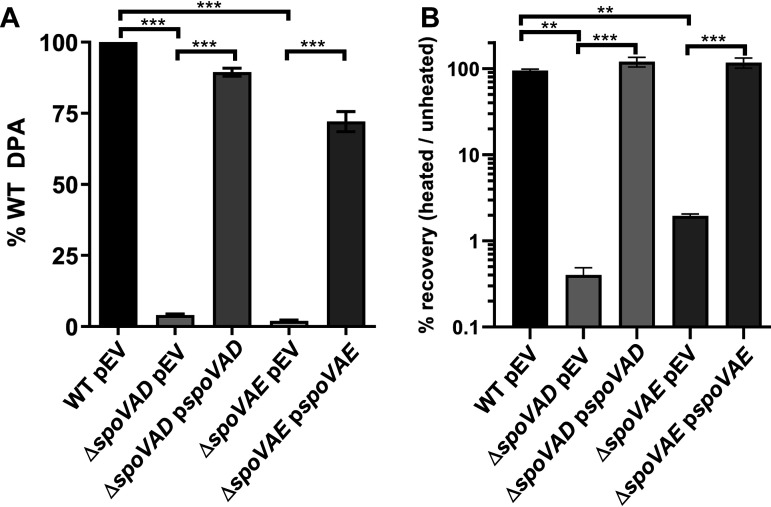
Prior research on C. difficile spoVAC found that a spoVAC deletion resulted in spores that contain only 1% of the wild-type level of DPA (28, 33, 34). To understand the contribution of spoVAD and spoVAE to sporulation and DPA uptake, we used an established pyrE-based allelic exchange system (35) to create spoVAD or spoVAE deletion strains in the C. difficile CRG2359 strain, restored the pyrE deletion, and then analyzed spores derived from the resulting strains for their DPA content using terbium fluorescence (36). Spores derived from C. difficile spoVAD and spoVAE mutant strains contained approximately 1% of the wild-type levels of DPA (Fig. 1A). The amount of spore DPA content could be restored to wild-type levels by expressing spoVAD or spoVAE, in trans, from a plasmid. These results suggest that spoVAD and spoVAE, like spoVAC, are required for DPA uptake into the C. difficile spore, similar to their roles in B. subtilis (30, 31).
FIG 1.
spoVAD and spoVAE are required for DPA uptake into the spore. (A) Equal amounts of spores purified from C. difficile strains RS19 (wild type [WT]), MB03 (ΔspoVAD), and MB04 (ΔspoVAE), containing an empty vector (pEV) or a complementing plasmid, were boiled for 20 min and DPA amounts quantified by Tb3+ fluorescence. Values are reported as percentage of the WT DPA content. (B) We serially diluted and plated 1 × 108 spores on BHIS plates supplemented with TA before and after heating at 65°C for 30 min. The values are reported as percentage of heated spores that formed colonies compared to unheated spores. The data represent results from 3 independent assays, and the error bars represent the standard errors of the mean. **, P < 0.001; ***, P < 0.0001 as determined by one-way ANOVA using Tukey’s multiple-comparison test.
Next, we tested if the absence of DPA led to a loss of heat resistance to spores derived from the mutant strains. When spores derived from the C. difficile ΔspoVAD and ΔspoVAE strains were heated at 65°C for 30 min and plated on brain heart infusion agar supplemented with 5 g/liter yeast extract and 0.1% l-cysteine (BHIS) medium supplemented with taurocholic acid (a potent spore germinant), we observed a >90% decrease in the number of CFU compared to the unheated samples or to the samples containing complementation plasmids (Fig. 1B). These results show that spores derived from the C. difficile ΔspoVAD and ΔspoVAE mutant strains are heat sensitive due to the lack of DPA or that heat treatment blocks an early germination event.
spoVAE may contribute to C. difficile spore germination.
C. difficile spore germination is activated upon binding of certain bile acid and certain amino acid germinants to receptors (24, 36–39). This results in the irreversible initiation of germination and the release of the majority of DPA from the spore core. Another indicator of germination is the change of optical density of the spore solution. This assay takes advantage of the transition of dormant spores from a phase-bright, dormant state to a phase-dark, germinated state. In both assays, equal numbers of spores derived from C. difficile RS19 (C. difficile CRG2359 with a restored pyrE [38]), C. difficile ΔspoVAD, or C. difficile ΔspoVAE strains containing empty vectors or the complementation constructs were suspended in buffer alone or buffer supplemented with the germinants taurocholate and glycine. Subsequently, the release of DPA and the change in optical density at 600 nm (OD600) values were measured over a period of 1 h.
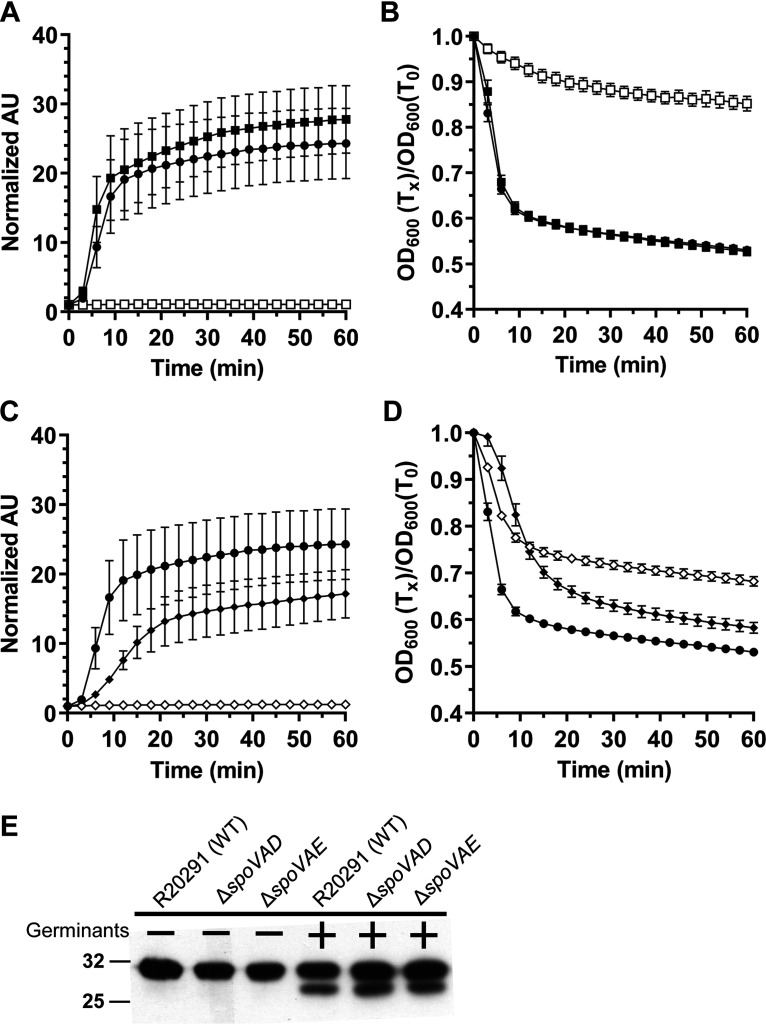
Due to the reduced abundance of DPA in the spores, C. difficile ΔspoVAD spores release very little DPA (Fig. 2A). This phenotype can be complemented by the expression of the wild-type copy of the gene in trans. In the optical density-based germination assay, we observed only a small, ∼10% reduction in OD600 values for C. difficile ΔspoVAD-derived spores. Moreover, due to the lack of DPA in the spores, these mutants are difficult to purify, as they do not migrate through density gradients well. To circumvent this issue, nearly 100 plates were required to purify spores from the mutant strains. The spores derived from the wild-type and the complemented strains germinated normally (Fig. 2B). Spores derived from the C. difficile ΔspoVAE strain also released very little DPA, and this phenotype was only partially complemented to wild-type levels (Fig. 2C). In the optical density germination assay, C. difficile ΔspoVAE-derived spores showed an intermediate phenotype, with an ∼30% reduction in OD600 values, which can again only be partially complemented to wild-type levels (Fig. 2D). Despite the lack of DPA, spores derived from the C. difficile ΔspoVAD and ΔspoVAE strains are capable of initiating germination. Western blot analysis showed that the cortex lytic enzyme SleC, required for the initiation of C. difficile spore germination, was activated to similar levels in the spores derived from the mutant and the wild-type strains (Fig. 2E). These results suggest that spores derived from C. difficile ΔspoVAD and ΔspoVAE strains activate SleC normally but that the C. difficile ΔspoVAE mutant strain has a small defect in germination.
FIG 2.
C. difficile spoVAD and spoVAE mutant spores initiate germination normally. Spores derived from C. difficile RS19 pEV (●), MB03 pEV (ΔspoVAD; □), MB03 pspoVAD (■), MB04 (ΔspoVAE; ◊), and MB04 pspoVAE (♦) were purified and germination was quantified by Tb3+ (A and C) or OD600 (B and D). For clarity, every fifth data point is plotted. The data represent results from 3 independent biological replicates, and the error bars represent standard error of the mean. (E) We incubated 108 spores derived from C. difficile CRG2359 (WT), MB03 (ΔspoVAD), and MB04 (ΔspoVAE) in buffer alone or in buffer supplemented with the germinants taurocholate and glycine, and they were then boiled in sample buffer and the proteins resolved by SDS-PAGE and probed with anti-SleC antibody. Inactive pro-SleC corresponds to the ∼32 kDa-band, while activated SleC is the ∼29-kDa band.
Testing the interaction of the C. difficile SpoVAC, SpoVAD, and SpoVAE proteins.
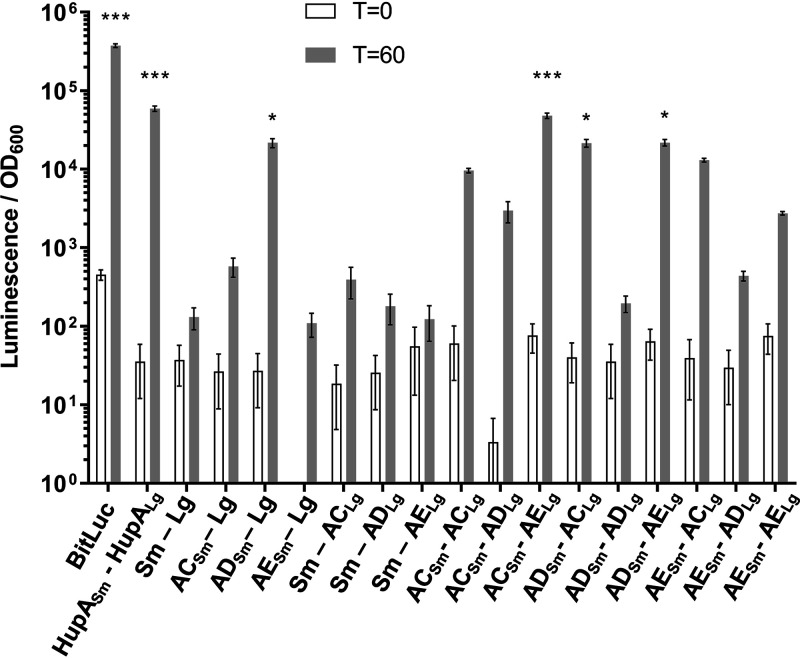
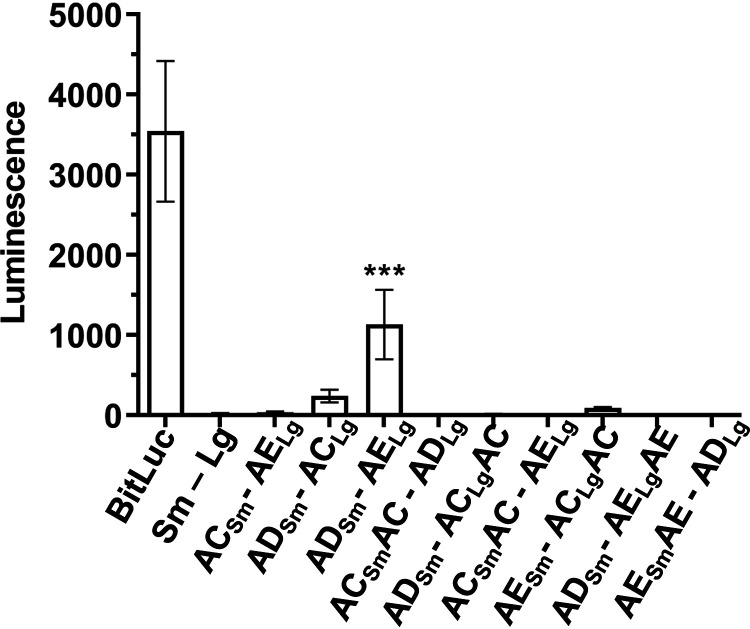
Our findings of C. difficile spoVAD and spoVAE mutant phenotypes indicate that deletion of either gene results in the absence of DPA in the spore core. Prior research has shown a similar phenotype in a C. difficile spoVAC mutant (28, 33). In both B. subtilis and C. difficile, the SpoVAC protein is a transmembrane protein, hypothesized to be embedded in the inner spore membrane, and acts in a mechanosensing fashion to release DPA from the spore core during germination (12, 29). While SpoVAD is not predicted to have transmembrane domains, we analyzed the C. difficile SpoVAE protein sequence with Constrained Consensus TOPology prediction server (CCTOPS) that predicts protein topology as a consensus of 10 different methods, and the results indicated that SpoVAE has several transmembrane domains (40). Thus, the locations of the C. difficile SpoVA proteins are consistent with those found in B. subtilis (29, 31, 32, 41, 42). The similar phenotype of single mutants and their predicted topology led us to hypothesize that SpoVAC, SpoVAD, and SpoVAE interact and form a complex at the inner spore membrane. To test the potential interaction of these proteins, we used the luciferase protein interaction assay, developed for use in C. difficile (43). Briefly, the codon-optimized luciferase reporter was split into two parts, SmBit and LgBit. Each reporter gene fragment was translationally fused to the 3′ end of C. difficile spoVAC, spoVAD, and spoVAE, and each fusion pair was expressed under the control of an anhydrotetracycline (aTc)-inducible promoter. The plasmids were introduced into the wild-type C. difficile R20291 strain, and luciferase activity was measured before induction and 1 h after induction with aTc. As positive controls, we used the full-length luciferase reporter BitLuc and the HupA-HupA fusions that were originally used to validate the assay (43). As negative controls, we used single spoVAC or spoVAD or spoVAE fusions (to one reporter fragment), while the second reporter fragment remained unfused. We detected a greater than 2-log10 increase in luminescence signal after 1 h of induction for the positive controls BitLuc and HupA-HupA. Interestingly, we observed an increase in luciferase signal for SpoVAC-SpoVAE, SpoVAD-SpoVAC, and SpoVAD-SpoVAE interaction pairs (Fig. 3). Though the signal for other interaction pairs after induction was increased, it did not reach statistical significance. In our negative controls, the signal did not increase significantly after induction except, notably, in SpoVAD-SmBit fusion where the signal significantly increased for unknown reasons. Nevertheless, the other negative control, SpoVAD-LgBit, does not exhibit a significant luciferase signal increase after induction.
FIG 3.
SpoVAC, SpoVAD, and SpoVAE interact in vivo. C. difficile vegetative cells transformed with the indicated plasmids were induced with 200 ng/ml of aTc for 60 min. Averages from 3 biological replicates are shown. Optical density-normalized luciferase activity (LU/OD) is shown before induction (white bars) and after 60 min (gray bars). Positive interaction was determined by comparison of LU/OD at time (T) of 60 min of transformed strains with the negative control (SmBit-LgBit). No significant difference was detected at T of 0. *, P < 0.05; ***, P < 0.0001 as determined by one-way ANOVA using Dunnett’s multiple-comparison test.
Because the uptake of DPA into the spore occurs in the late stages of sporulation (44, 45), we wanted to test for the potential SpoVA protein interactions in cultures that are actively undergoing sporulation. The full-length BitLuc control, the negative SmBit-LgBit control, and the interaction partners that showed a significant luciferase signal in induced vegetative cultures (SpoVAC-SpoVAE, SpoVAD-SpoVAC, and SpoVAD-SpoVAE) were cloned into a plasmid and placed under the control of a native C. difficile spoVAC promoter (a region of 500 bp upstream of spoVAC) and conjugated into the wild-type C. difficile R20291 strain. Additionally, we cloned either a SmBit or LgBit luciferase reporter fragment into a segment of the SpoVAC and SpoVAE that is predicted to face outside the core and, therefore, SpoVAD (based on the CCTOPs prediction) (32, 40, 42).
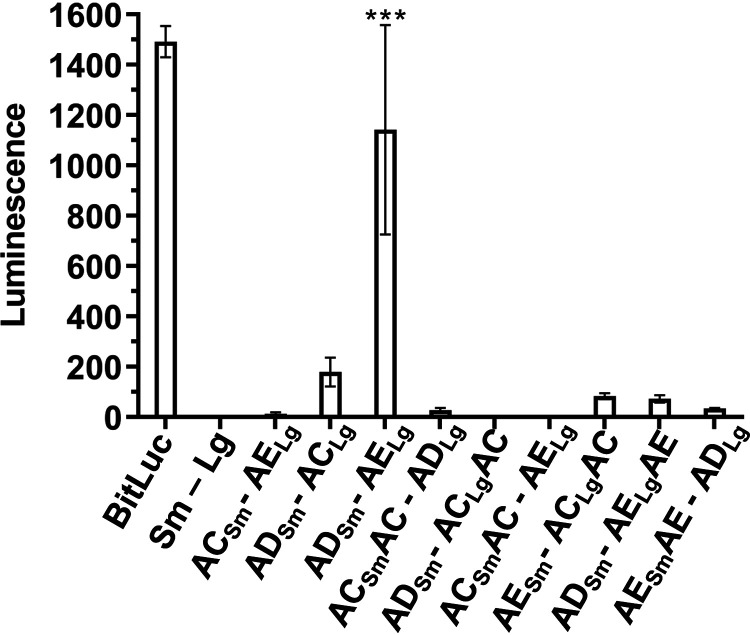
The strains were streaked onto sporulation medium and left to grow for 2 days, the period of time that would ensure that the sporulation has commenced, but not fully completed, for the majority of cells in the sample. The cell mass was scraped and resuspended in water, and an equal number of cells for each strain were incubated with luciferase substrate and luminescence signal measured (Fig. 4). Under these conditions, we observed that SpoVAD and SpoVAE yielded the greatest signal, indicating that, during sporulation, these two proteins come in close proximity. The other constructs showed a smaller luminescence signal but did not reach statistical significance.
FIG 4.
SpoVAD and SpoVAE interact in sporulating cultures. C. difficile cells transformed with the indicated plasmids were grown on sporulation medium. Luminescence of the sporulating culture was assayed after 2 days of growth. Averages from 4 biological replicates are shown. **, P < 0.001 as determined by one-way ANOVA using Dunnett’s multiple-comparison test.
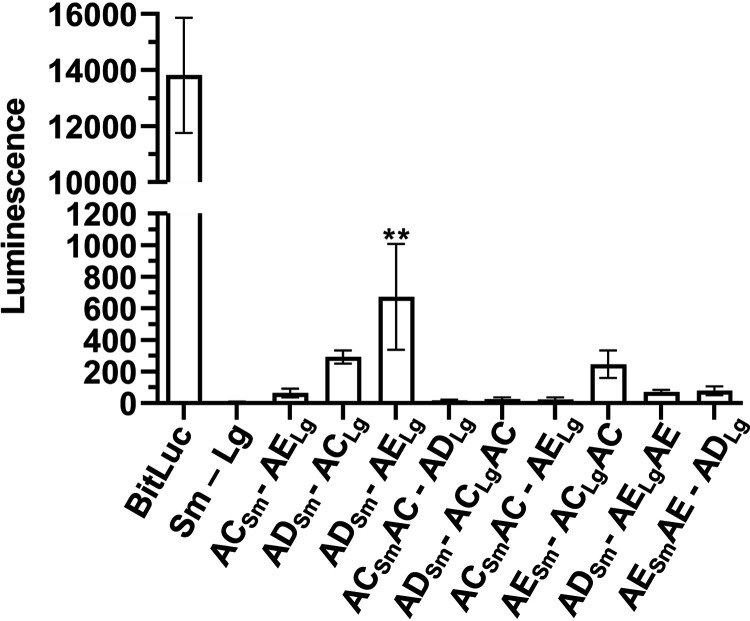
Because our data indicate that SpoVAD and SpoVAE interact during sporulation, it is plausible that these proteins would remain in close proximity in the metabolically dormant spore. We therefore tested the same protein interaction pairs in fully formed dormant spores. The strains were sporulated on sporulation medium, and spores purified as previously described. Next, 1 × 108 spores of each strain were assayed for luciferase activity (Fig. 5). Similar to what we observed for a sporulating culture, the SpoVAD-SpoVAE interacting pair showed the highest level of interaction, while SpoVAC-SpoVAD, SpoVAD-SpoVAC, and SpoVAE-SpoVAClgAC showed a small but nonsignificant signal. Taken together, our results suggest that SpoVAD-SpoVAE interact when expressed in vegetative cell or in sporulating/dormant spores, while the other SpoVA proteins may weakly and/or transiently interact.
FIG 5.
SpoVAD and SpoVAE demonstrate interaction in dormant spores. C. difficile cells transformed with the indicated plasmids were grown on sporulation medium, and the resulting spores were purified and assayed for luminescence. Averages from 3 biological triplicates are shown. ***, P < 0.0001 as determined by one-way ANOVA using Dunnett’s multiple-comparison test.
We hypothesized that the SpoVA proteins that interact during DPA uptake would still interact upon spore germination. To test this, we exposed 1 × 108 spores derived from the indicated strains to the germinants taurocholate and glycine in buffer and measured the luminescence signal over the period of 30 min. Similar to our observation in the sporulating culture and dormant spores, the SpoVAD-SpoVAE seems to interact the most strongly and gives the highest luminescence signal (Fig. 6). The other interacting pairs, the C-terminal fusions and midprotein fusions, also had an increase in signal. However, these signals did not reach statistical significance. But this and the similar signal increase in the previous assays using sporulating cultures and dormant spores suggest either a weak or transient interaction. We therefore conclude that SpoVAD and SpoVAE interact during all stages of C. difficile development cycle in vegetative cells, sporulating cells, spores, and germinating spores.
FIG 6.
SpoVAD and SpoVAE interact during spore germination. Spores derived from the strains containing the indicated plasmids were germinated in the presence of TA and glycine. Luminescence was measured over the period of 30 min, but only T of 15 min is shown for clarity. Averages from 3 biological triplicates are shown. ***, P < 0.0001 as determined by one-way ANOVA using Dunnett’s multiple-comparison test.
DISCUSSION
C. difficile spores, like most spores from endospore-forming organisms, are notable for their resilience to wet heat. Peak natural environment temperatures and even temperatures that are commonly recommended for heat treatment of foodstuffs are often insufficient to destroy the spores or render them incapable of germination (46). This heat resistance, along with other characteristics of spores that provide resistance to UV radiation, desiccation, and resistance to common disinfectants, makes C. difficile (or other spore-forming organisms) difficult to eradicate (47). The spore resistance to heat is largely the consequence of large amounts of DPA in the spore core that replace the majority of water and comprise 5 to 15% of dry weight of the spore (48). Since spores are metabolically dormant, in order for metabolic processes to initiate, this DPA must be released from the spore core in exchange for water during the initial stages of spore germination.
The initiation of germination is a nonreversible process, and the release of DPA, in exchange for water, is tightly regulated by the germinant receptors. To initiate C. difficile spore germination, the Csp-type germinant receptors, composed of a hypothesized CspB, CspA, and CspC protein complex, become activated by bile acid and cogerminants (i.e., certain amino acids or Ca2+). Germinant activation of the germinant receptors leads to the processing of the inhibitory propeptide from the cortex lytic enzyme SleC (38, 49). Activated SleC degrades the spore cortex layer, resulting in the activation of the SpoVAC mechanosensing protein and DPA release from the core (12, 28). Though the importance of spoVAC is known in both B. subtilis and C. difficile, the roles of spoVAD and spoVAE have not been tested in C. difficile. Here, we find that the mutations in C. difficile spoVAD and spoVAE prevent DPA uptake into the spore core. This is similar to prior observations with C. difficile spoVAC mutants and B. subtilis spoVAD mutants (28, 30, 33).
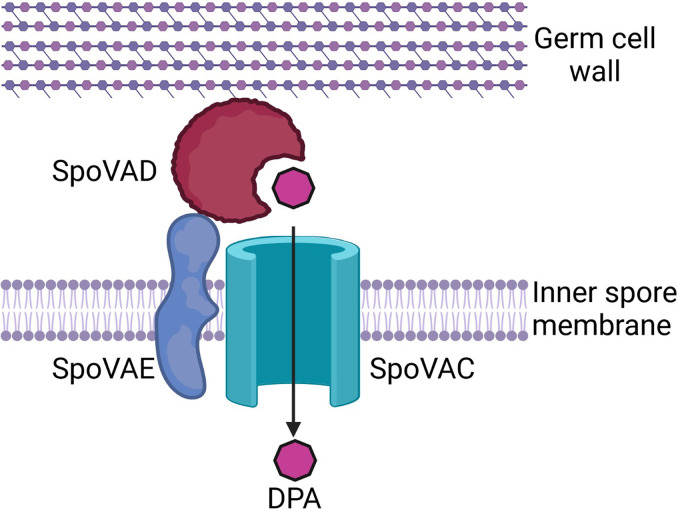
Because a mutation in any member of the C. difficile spoVA operon resulted in a similar phenotype, we hypothesized that the products of this operon form a complex and interact during DPA uptake. Because SpoVAC is a transmembrane protein that is embedded in the inner spore membrane and SpoVAE is predicted to have transmembrane domains, this complex should be located at, or near, the inner spore membrane. Even though SpoVAD is not predicted to have a transmembrane domain, consistent with its hypothesized role as a DPA binding protein (30), it should be located near the inner spore membrane during DPA uptake in order to transfer DPA via SpoVAC into the spore core. Using the recently developed split luciferase system for studying protein-protein interactions in C. difficile (43), we detected a large increase in luminescence for SpoVAD and SpoVAE and smaller but nonsignificant increases in other interaction pairs in vegetative cells, sporulating cells, dormant spores, and germinating spores. We therefore propose a model in which the DPA, synthesized in the mother cell in the late stage of sporulation, is taken up into the spore by the interaction between all 3 SpoVA proteins. In this model, SpoVAD acts as a DPA-binding protein, SpoVAC as a channel through which DPA passes into the spore core, and SpoVAE as an accessory protein (Fig. 7). Consistent with the predictions arising from this model, we first observed interaction between all 3 protein pairs in vegetative cells (i.e., SpoVAC-SpoVAE, SpoVAD-SpoVAC, and SpoVAD-SpoVAE). In our assay, the protein pair expression was driven by an aTc-induced pTet promoter, which likely induces protein expression above the level of baseline expression in vegetative C. difficile cells, but this served to show that the interaction partners could interact and to discover any interactions that may be transient, temporary, or at low incidence rate in the uninduced culture. However, this was dependent upon the fusions being located on the region of the proteins predicted to be on the outer surface of the inner membrane, where the proteins are likely to interact. We also discovered such interactions in sporulating cultures, in the dormant spores, and in the germinating spores. We found that, in all 3 stages of spore development, the strongest interacting partners were SpoVAD and SpoVAE. This has given us confidence in our model because SpoVAE has predicted transmembrane domains and is found in the inner spore membrane of B. subtilis spores (30, 31). Though SpoVAD has no predicted transmembrane domains, our data suggest it is situated on the inner spore membrane with SpoVAE, similar to its location in B. subtilis (41).
FIG 7.
Proposed model SpoVA protein interaction at the inner spore membrane by which DPA produced by the mother cell is taken up into the spore during sporulation and released in the initial stages of germination. Created with http://BioRender.com.
Because the expression of the fusions was driven by an inducible promoter, the data likely represent the signal at the maximum expression levels, which may be why the SpoVAC-SpoVAD interaction signal is quite high. Because the mother cell produces large amounts of DPA during the late stages of sporulation, the trafficking of DPA by SpoVAD to SpoVAC, and their interaction, must be a high-incidence event, if perhaps temporary. After sporulation is completed, the assays showed the highest interaction signal for SpoVAE-SpoVAD, suggesting that this pair forms a more stable interaction.
In the model spore-forming organism B. subtilis, the spoVA operon is composed of 8 genes, while the C. difficile spoVA operon is considerably less complex and encodes only 3 orthologues, making it a convenient system for study. Since our data suggest that all of the C. difficile SpoVA proteins may interact, we hypothesize that the protein products of a more complex B. subtilis spoVA operon form a complex as well.
MATERIALS AND METHODS
Bacteria and strains.
C. difficile CRG2359 and its derivative strains were grown at 37°C in an anaerobic chamber (Coy Laboratories; model B; >4% H2, 5% CO2, 85% N2) on brain heart infusion agar supplemented with 5 g/liter yeast extract and 0.1% l-cysteine (BHIS). The C. difficile CRG2359 strain is derived from the C. difficile R20291 strain from the Anaerobe Reference Laboratory (Cardiff, Whales, UK) (35, 50). Escherichia coli DH5α (51) was grown on LB medium. Chloramphenicol (20 μg/ml), thiamphenicol (10 μg/ml), kanamycin (50 μg/ml), and ampicillin (100 μg/ml) were added where indicated. Deletion mutants were selected on C. difficile minimal medium (CDMM) supplemented with 5 μg/ml 5-fluoroorotic acid (FOA) and 20 μg/ml uracil.
Construction of spoVAD and spoVAE mutants.
Deletion mutations were introduced using the established pyrE-mediated allelic exchange technique (35). Briefly, 1-kb upstream and downstream DNA regions that surround spoVAD and spoVAE (including the first 30 bp of the 5′ and 3′ ends of the gene) were amplified using primers spoVAD_ndeI_L, spoVAD_ndeI_R, spoVAD_KO_RHA_For, and spoVAD_xhoI_R for spoVAD and spoVAE_ndeI_L, 250_Downstream_VAE, 250_Upstream_VAE, and spoVAE_xhoI_R for spoVAE, assembled using PCR stitching, and inserted by Gibson assembly (52) into pMTL-YN4 plasmid digested with NdeI and XhoI, yielding plasmids pMB02 and pMB04. The plasmids were then transformed into E. coli DH5α. These plasmids were subsequently transformed into E. coli HB101/pRK24 and grown on LB medium supplemented with chloramphenicol and ampicillin. The resulting strain was grown overnight and then mixed with the C. difficile CRG2359 strain grown in BHIS in an anaerobic chamber. The conjugation mixtures were spotted onto BHI plates and allowed to grow for 24 h. Subsequently, the cells were washed with BHIS, and the slurry was transferred onto BHIS medium supplemented with thiamphenicol (for plasmid maintenance) and kanamycin (to counterselect E. coli growth) (BHIS[TK]). Individual colonies were passaged several times onto BHIS(TK) supplemented with uracil (BHIS[TKU]) to encourage the single-crossover events. Growth was then transferred to CDMM medium supplemented with FOA and uracil to select for colonies that have excised the plasmid from the chromosome. Thiamphenicol-sensitive colonies were tested for desired mutation by PCR and confirmed by sequencing. The wild-type pyrE allele was restored using the same technique. The resulting C. difficile ΔspoVAD strain was renamed MB03 and the C. difficile ΔspoVAE strain MB04. The mutations were complemented by introduction of a wild-type gene under the control of a native promoter on a plasmid.
Construction of split luciferase interaction plasmids.
Plasmids for luciferase assays were purchased from Addgene (IDs 105494 to 105497), and plasmids for testing interaction in vegetative cells were constructed following previously established protocols (43, 53). For these plasmids, spoVAC, spoVAD, and spoVAE were amplified using C. difficile R20291 DNA as a template to create fragments with the appropriate overlap for SmBit reporter fragment fusions (5′ SpoVAC SmBit Gibson and 3′ SpoVAC SmBit Gibson for spoVAC, 5′ SpoVAD SmBit Gibson and 3′ SpoVAD SmBit Gibson for spoVAD, and 5′ SpoVAE SmBit Gibson and 3′ SpoVAE SmBit Gibson for spoVAE). These fragments were then introduced by Gibson assembly into plasmid pAP118 digested with SacI/XhoI. Using the same approach, fragments for LgBit reporter fusions were created (5′ SpoVAC LgBit Gibson and 3′ SpoVAC LgBit Gibson, 5′ SpoVAD LgBit Gibson and 3′ SpoVAD LgBit Gibson, and 5′ SpoVAE LgBit Gibson and 3′ SpoVAE LgBit Gibson). These fragments were then ligated into the above plasmids and digested with PvuI/NotI, yielding the interaction plasmids. To construct the negative-control interaction plasmids, plasmid pAF256 was digested with SacI/XhoI and plasmid pAF257 digested with PvuI/NotI, and the required single-fusion fragments were introduced into them by Gibson assembly. To construct the negative-control interaction plasmid with only SmBit and LgBit reporters without the protein fusion, the LgBit reporter fragment was amplified from pAF256 using primers 5′ LgBit Gibson and 3′ LgBit Gibson and inserted by Gibson assembly (52) into plasmid pAF257 digested with PvuI/BamHI. This yielded plasmids pMB45 through pMB62 with all combinations of interacting partners and negative controls needed for vegetative cell expression. Next, we created plasmids with a native spoVAC promoter to drive the expression of the requisite constructs from the native promoter. For SpoVAC-SpoVAE, we amplified the promoter region fragment from a wild-type template using primers 5′ SpoVAC prm and 3′ SpoVAC prm-SpoVAC and the fusion partner sequence from pMB45 using primers 5′ SpoVAC RBS and 3′ LgBit-pJS116. For SpoVAD-SpoVAC, we amplified the promoter region from a wild-type template using primers 5′ SpoVAC prm and 3′ spoVAC pr.-spoVAD Gibson and the fusion partner sequence from pMB46 using primers 5′ SpoVAC prm-SpoVAD and 3′ LgBit-pJS116. For SpoVAD-SpoVAE, we amplified the promoter region from a wild-type template using primers 5′ SpoVAC prm/spoVAC pr.-spoVAD Gibson and the fusion partner sequence from pMB48 using primers 5′ SpoVAC prm-SpoVAD and 3′ LgBit-pJS116. Fragments were introduced by Gibson assembly into pMTL84151 digested with NotI/HinDIII. This yielded plasmids pMB72 (SpoVAC-SpoVAE), pMB73 (SpoVAD-SpoVAC), and pMB75 (SpoVAD-SpoVAE). We also created the plasmids where the SmBit or LgBit reporter fragment is inserted into an extra cytoplasmic-facing segment of SpoVAC or SpoVAE. For ACSmAC-ADLg, we amplified fragments from wild-type template using primers 5′ SpoVAC downstream (linker-SmBit) and 3′ SpoVAC downstream (linker-SmBit) and primers 5′ SpoVAC prm and 3′ SpoVAC upstream (linker-SmBit), from pMB54 template using primers 5′ SpoVAC-SpoVAD and 3′ LgBit-pJS116, and from pMB51 template using primers 5′ linker-SmBit (SpoVAC) and 3′ linker-SmBit (SpoVAC). For ADLg-ACSmAC, we amplified fragments from the wild-type template using primers 5′ SpoVAC prm and 3′ spoVAC pr.-spoVAD Gibson, primers 5′ SpoVAC LgBit Gibson and 3′ SpoVAC upstream (linker-SmBit), and primers 5′ linker-LgBit (SpoVAC) and 3′ Linker-LgBit (SpoVAC); from pMB51 template using primers 5′ SpoVAC prm-SpoVAD and 3′ SmBit-SpoVAC RBS; and from pMB54 template using primers 5′ linker-SmBit (SpoVAC) and 3′ Linker-LgBit. For ACSmAC-AELg, we amplified fragments from the wild-type template using primers 5′ SpoVAC prm and 3′ SpoVAC upstream (linker-SmBit) and primers 5′ SpoVAC downstream (linker-SmBit) and 3′ SpoVAC downstream (linker-SmBit)-SpoVAE; from pMB51 template using primers 5′ linker-SmBit (SpoVAC) and 3′ linker-SmBit (SpoVAC); and from pMB54 template using primers 5′ SpoVAC-SpoVAE and 3′ Linker-LgBit. For AESm-ACLgAC, we amplified fragments from the wild-type template using primers 5′ SpoVAC prm and 3′ spoVAC pr.-spoVAE Gibson, primers 5′ SpoVAC LgBit Gibson and 3′ SpoVAC upstream (linker-SmBit), and primers 5′ linker-LgBit (SpoVAC) and 3′ Linker-LgBit (SpoVAC); from pMB52 template using primers 5′ spoVAC pr.-spoVAE Gibson and 3′ SmBit-SpoVAC RBS; and from pMB54 template using primers 5′ linker-SmBit (SpoVAC) and 3′ Linker-LgBit (SpoVAC). For ADSm-AELgAE, we amplified fragments from the wild-type template using primers 5′ SpoVAC prm and 3′ spoVAC pr.-spoVAD Gibson, primers 5′ SpoVAE LgBit Gibson and 3′ spoVAE-SpoVADSm, and primers 5′ SpoVAE-plasmid LgBit Mid and 3′ SpoVAE-plasmid LgBit Mid; from pMB75 template using primers 5′ SpoVAC prm-SpoVAD and 3′ spoVAD-SmBit; and from pAP118 template using primers 5′ SpoVAE LgBit Mid and 3′ SpoVAE LgBit Mid. For AES/mAE- ADLg, we amplified fragments from the wild-type template using primers 5′ SpoVAC prm and 3′ spoVAC pr.-spoVAE Gibson, primers 5′ spoVAC pr.-spoVAE Gibson and 3′ SpoVAESm-SpoVADLg, and primers 5′ SpoVAE Smbit Mid and 3′ SpoVAE Smbit Mid; from pMB50 template using primers 5′ SpoVAE-SmBi and 3′ SpoVAE-SmBit; and from pMB56 template using primers 5′ SpoVAE SmBit mid-SpoVAD LgBit and 3′ LgBit-pJS116. The fragments were introduced by Gibson assembly (52) into the pMTL84151 plasmid digested with NotI/HinDII, yielding plasmids pMB82 to pMB85 and pMB88 to pMB89. All strains and plasmids in this study are listed in Table 1. Primers used to construct the strains and plasmids are listed in Table S1 in the supplemental material.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Reference no. or source |
|---|---|---|
| C. difficile strains | ||
| R20291 | Wild type, ribotype 027 | 55 |
| CRG2359 | R20291 ΔpyrE | 35 |
| RS19 | CRG2359-restored pyrE | 38 |
| MB03 | ΔspoVAD strain | This study |
| MB04 | ΔspoVAE strain | This study |
| Other strains | ||
| E. coli DH5α | Cloning strain | 51 |
| E. coli HB101 pRK24 | Conjugal donor strain for C. difficile | 56 |
| Plasmids | ||
| pMTL-YN4 | Backbone used to make deletions in R20291 by allelic exchange | 35 |
| pMTL84151 | Backbone used to make complementing plasmids and spore luciferase interaction plasmids | 57 |
| pMB02 | To create ΔspoVAD deletion | This study |
| pMB04 | To create ΔspoVAE deletion | This study |
| pMB34 | spoVAD-complementing plasmid | This study |
| pMB35 | spoVAE-complementing plasmid | This study |
| pAP118 | Plasmid encoding aTC-inducible HupA-SmBit and HupA-LgBit fusions | 43 |
| pAF256 | Plasmid encoding an aTC-inducible HupA-SmBit and LgBit | 43 |
| pAF257 | Plasmid encoding an aTC-inducible SmBit and HupA-LgBit | 43 |
| pAF259 | Plasmid encoding an aTC-inducible BitlucOpt | 43 |
| pMB50 | Luciferase SpoVAC/SmBit-LgBit | This study |
| pMB51 | Luciferase SpoVAD/SmBit-LgBit | This study |
| pMB52 | Luciferase SpoVAE/SmBit-LgBit | This study |
| pMB53 | Luciferase SmBit-SpoVAC/LgBit | This study |
| pMB54 | Luciferase SmBit-SpoVAD/LgBit | This study |
| pMB55 | Luciferase SmBit-SpoVAE/LgBit | This study |
| pMB62 | Luciferase SpoVAC/SmBit-SpoVAC/LgBit | This study |
| pMB45 | Luciferase SpoVAC/SmBit-SpoVAE/LgBit | This study |
| pMB46 | Luciferase SpoVAD/SmBit-SpoVAC/LgBit | This study |
| pMB47 | Luciferase SpoVAD/SmBit-SpoVAD/LgBit | This study |
| pMB48 | Luciferase SpoVAD/SmBit-SpoVAE/LgBit | This study |
| pMB49 | Luciferase SpoVAE/SmBit-SpoVAC/LgBit | This study |
| pMB57 | Luciferase SpoVAE/SmBit-SpoVAE/LgBit | This study |
| pMB60 | Luciferase SpoVAC/SmBit-SpoVAD/LgBit | This study |
| pMB61 | Luciferase SmBit-LgBit | This study |
| pMB72 | SpoVAC-SmBit/SpoVAE-LgBit luciferase spore complementation | This study |
| pMB73 | SpoVAD-SmBit/SpoVAC-LgBit luciferase spore complementation | This study |
| pMB75 | SpoVAD-SmBit/SpoVAE-LgBit luciferase spore complementation | This study |
| pMB79 | HupA-SmBit/HupA-LgBit luciferase spore complementation | This study |
| pMB81 | BitLuc luciferase spore complementation | This study |
| pMB82 | SpoVAC-SmBit-SpoVAC/SpoVAD-LgBit; small luciferase reporter inserted mid-AC gene, ADLg partner | This study |
| pMB83 | SpoVAD-SmBit/SpoVAC-LgBit-SpoVAC; large luciferase reporter inserted mid-AC gene, ADSm partner | This study |
| pMB84 | SpoVAC-SmBit-SpoVAC/SpoVAE-LgBit; small luciferase reporter inserted mid-AC gene, AELg partner | This study |
| pMB85 | SpoVE-SmBit/SpoVAC-LgBit-SpoVAC; large luciferase reporter inserted mid-AC gene, AESm partner | This study |
| pMB87 | Sm-Lg luciferase-spore interaction plasmid, negative control | This study |
| pMB88 | SpoVAD-Sm/SpoVAE-Lg-SpoVAE; large luciferase reporter inserted mid-AE gene, ADSm partner | This study |
| pMB89 | SpoVAE-Sm-SpoVAE/SpoVAD-Lg; small luciferase reporter inserted mid-AE gene, ADLg partner | This study |
Spore purification.
Spores were purified as previously described (12, 24, 28, 36). Briefly, strains were grown on 70:30 sporulation medium. After 5 days, growth from 2 plates each was scraped into 1 ml distilled water (dH2O) in microcentrifuge tubes and left overnight at 4°C. The cultures were then resuspended in the dH2O in the same microcentrifuge tubes and centrifuged at >14,000 × g for 10 min, the top layer containing vegetative cells and cell debris was removed by pipetting, and the rest of the sediment resuspended in fresh dH2O. The tubes, again, were centrifuged for 1 min at >14,000 × g, the top layer removed, and the sediment resuspended. This was repeated 5 more times, combining the sediment from 2 tubes into one. The spores were then separated from the cell debris by centrifugation through a 50% sucrose gradient for 20 min at 4°C and 3,500 × g. The resulting spore pellet was then washed 5 times with dH2O, resuspended in 1 ml dH2O, and stored at 4°C until use.
DPA and OD germination assays.
Ca-DPA release was measured using a SpectraMax M3 plate reader for 1 h at 37°C with excitation at 270 nM and emission at 545 nM and with a 420-nM cutoff. Spores were heat activated at 65°C for 30 min and suspended in water at an OD600 of 50. The spores were then added to final OD of 0.5 in 100 μl final volume of HEPES buffer, pH 7.5, containing 100 mM NaCl, 10 mM taurocholic acid (TA), 30 mM glycine, and 250 μM Tb3+ in a 96-well plate. OD600 was monitored using the same plate reader at 37°C for 1 h. The heat-activated spores were added to a final OD of 0.5 in in the same HEPES buffer composition as in Ca-DPA release assay, omitting Tb3+ (38, 54).
Luciferase assays.
Assays involving vegetative cells were performed using the previously published protocols (43). Briefly, strains were grown overnight in liquid BHIS supplemented with thiamphenicol. The following day, the liquid cultures were diluted to an OD600 of 0.05 and allowed to grow to an OD600 of 0.3 to 0.4. Next, 100 μl was removed and placed into wells of a transparent 96-well plate, suitable for OD600, and another 100 μl placed into white flat-bottom 96-well plate for the luminescence assay. The rest of the cultures were induced with 200 ng/ml of aTc for 1 h. We added 20 μl of NanoGlo luciferase (Promega; part no. N1110) to each sample well to measure luciferase activity using SpectraMax M3 plate reader in the luminescence mode, using all channels and 0.1 s sampling time. After 1 h, the induced cultures were assayed for OD and luciferase activity in the same way. The luciferase activity was normalized to culture optical density at OD600.
Western blot analysis.
Solutions of 1 × 108 spores of C. difficile R20291, C. difficile ΔspoVAD, and C. difficile ΔspoVAE were prepared, and 100 μl of each was incubated in HEPES buffer, pH 7.5, containing 100 mM NaCl, 10 mM TA, and 30 mM glycine for 15 min to induce germination. These samples and equal amounts of nongerminated control spore solutions were boiled for 20 min in 2× NuPage buffer at 95°C. Then, 10 μl of each sample was separated on 10% SDS-PAGE gel. The protein was transferred to polyvinylidene difluoride (PVDF) membrane and then blocked overnight at 4°C with 5% milk powder dissolved in Tris-buffered saline with Tween 20 (TBST). The membrane was washed thrice for 20 min at room temperature with TBST and then probed for 1 h in 5% milk dissolved in TBST with anti-SleC antibodies at room temperature. The membrane was then washed thrice for 20 min at room temperature with TBST before labeling with anti-rabbit IgG secondary antibody. The membrane was again washed and then incubated for 5 min with Pierce ECL Western blotting substrate (Thermo Scientific), overlaid with X-ray film, exposed, and developed.
Statistical analysis.
Data represent results from at least 3 independent experiments, and the error bars represent standard errors of the means. One-way analysis followed by Tukey’s or Dunnett’s multiple-comparison test, as indicated, was performed using GraphPad Prism version 9.0.2 (161) for Windows (GraphPad Software, San Diego, California USA).
ACKNOWLEDGMENTS
This project was supported by awards 5R01AI116895 and 1U01AI124290 to J.A.S. from the National Institute of Allergy and Infectious Diseases.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIAID. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material is available online only.
Contributor Information
Joseph A. Sorg, Email: jsorg@bio.tamu.edu.
Tina M. Henkin, Ohio State University
REFERENCES
- 1.Lessa FC, Mu Y, Bamberg WM, Beldavs ZG, Dumyati GK, Dunn JR, Farley MM, Holzbauer SM, Meek JI, Phipps EC, Wilson LE, Winston LG, Cohen JA, Limbago BM, Fridkin SK, Gerding DN, McDonald LC. 2015. Burden of Clostridium difficile infection in the United States. N Engl J Med 372:825–834. 10.1056/NEJMoa1408913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention. 2019. Antibiotic resistance threats in the United States, 2019. U.S. Department of Health and Human Services, CDC, Atlanta, GA. [Google Scholar]
- 3.McDonald LC, Killgore GE, Thompson A, Owens RC, Jr., Kazakova SV, Sambol SP, Johnson S, Gerding DN. 2005. An epidemic, toxin gene-variant strain of Clostridium difficile. N Engl J Med 353:2433–2441. 10.1056/NEJMoa051590. [DOI] [PubMed] [Google Scholar]
- 4.Abdrabou AMM, Ul Habib Bajwa Z, Halfmann A, Mellmann A, Nimmesgern A, Margardt L, Bischoff M, von Muller L, Gartner B, Berger FK. 2021. Molecular epidemiology and antimicrobial resistance of Clostridioides difficile in Germany, 2014-2019. Int J Med Microbiol 311:151507. 10.1016/j.ijmm.2021.151507. [DOI] [PubMed] [Google Scholar]
- 5.Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, No D, Liu H, Kinnebrew M, Viale A, Littmann E, van den Brink MR, Jenq RR, Taur Y, Sander C, Cross JR, Toussaint NC, Xavier JB, Pamer EG. 2015. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517:205–208. 10.1038/nature13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Theriot CM, Bowman AA, Young VB. 2016. Antibiotic-induced alterations of the gut microbiota alter secondary bile acid production and allow for Clostridium difficile spore germination and outgrowth in the large intestine. mSphere 1:e00045-15. 10.1128/mSphere.00045-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Theriot CM, Koenigsknecht MJ, Carlson PE, Jr., Hatton GE, Nelson AM, Li B, Huffnagle GB, J ZL, Young VB. 2014. Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat Commun 5:3114. 10.1038/ncomms4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilson KH, Perini F. 1988. Role of competition for nutrients in suppression of Clostridium difficile by the colonic microflora. Infect Immun 56:2610–2614. 10.1128/iai.56.10.2610-2614.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smits WK, Lyras D, Lacy DB, Wilcox MH, Kuijper EJ. 2016. Clostridium difficile infection. Nat Rev Dis Primers 2:16020. 10.1038/nrdp.2016.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Permpoonpattana P, Tolls EH, Nadem R, Tan S, Brisson A, Cutting SM. 2011. Surface layers of Clostridium difficile endospores. J Bacteriol 193:6461–6470. 10.1128/JB.05182-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paredes-Sabja D, Shen A, Sorg JA. 2014. Clostridium difficile spore biology: sporulation, germination, and spore structural proteins. Trends Microbiol 22:406–416. 10.1016/j.tim.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Francis MB, Allen CA, Sorg JA. 2015. Spore cortex hydrolysis precedes dipicolinic acid release during Clostridium difficile spore germination. J Bacteriol 197:2276–2283. 10.1128/JB.02575-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cowan AE, Koppel DE, Setlow B, Setlow P. 2003. A soluble protein is immobile in dormant spores of Bacillus subtilis but is mobile in germinated spores: implications for spore dormancy. Proc Natl Acad Sci USA 100:4209–4214. 10.1073/pnas.0636762100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cortezzo DE, Setlow P. 2005. Analysis of factors that influence the sensitivity of spores of Bacillus subtilis to DNA damaging chemicals. J Appl Microbiol 98:606–617. 10.1111/j.1365-2672.2004.02495.x. [DOI] [PubMed] [Google Scholar]
- 15.Setlow P. 2007. I will survive: DNA protection in bacterial spores. Trends Microbiol 15:172–180. 10.1016/j.tim.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 16.Coullon H, Rifflet A, Wheeler R, Janoir C, Boneca IG, Candela T. 2018. N-Deacetylases required for muramic-delta-lactam production are involved in Clostridium difficile sporulation, germination, and heat resistance. J Biol Chem 293:18040–18054. 10.1074/jbc.RA118.004273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Diaz OR, Sayer CV, Popham DL, Shen A. 2018. Clostridium difficile lipoprotein GerS is required for cortex modification and thus spore germination. mSphere 3:e00205-18. 10.1128/mSphere.00205-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Makino S, Moriyama R. 2002. Hydrolysis of cortex peptidoglycan during bacterial spore germination. Med Sci Monit 8:RA119-27. [PubMed] [Google Scholar]
- 19.Paredes-Sabja D, Setlow P, Sarker MR. 2011. Germination of spores of Bacillales and Clostridiales species: mechanisms and proteins involved. Trends Microbiol 19:85–94. 10.1016/j.tim.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 20.Driks A, Eichenberger P. 2016. The spore coat. In Driks A, Eichenberger P (ed), The bacterial spore: from molecules to systems. ASM Press, Washington, DC. [Google Scholar]
- 21.Paredes-Sabja D, Sarker MR. 2012. Adherence of Clostridium difficile spores to Caco-2 cells in culture. J Med Microbiol 61:1208–1218. 10.1099/jmm.0.043687-0. [DOI] [PubMed] [Google Scholar]
- 22.Castro-Cordova P, Mora-Uribe P, Reyes-Ramirez R, Cofre-Araneda G, Orozco-Aguilar J, Brito-Silva C, Mendoza-Leon MJ, Kuehne SA, Minton NP, Pizarro-Guajardo M, Paredes-Sabja D. 2021. Entry of spores into intestinal epithelial cells contributes to recurrence of Clostridioides difficile infection. Nat Commun 12:1140. 10.1038/s41467-021-21355-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gibbs PA. 1967. The activation of spores of Clostridium bifermentans. J Gen Microbiol 46:285–291. 10.1099/00221287-46-2-285. [DOI] [PubMed] [Google Scholar]
- 24.Sorg JA, Sonenshein AL. 2008. Bile salts and glycine as cogerminants for Clostridium difficile spores. J Bacteriol 190:2505–2512. 10.1128/JB.01765-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paredes-Sabja D, Udompijitkul P, Sarker MR. 2009. Inorganic phosphate and sodium ions are cogerminants for spores of Clostridium perfringens type A food poisoning-related isolates. Appl Environ Microbiol 75:6299–6305. 10.1128/AEM.00822-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moir A. 2006. How do spores germinate? J Appl Microbiol 101:526–530. 10.1111/j.1365-2672.2006.02885.x. [DOI] [PubMed] [Google Scholar]
- 27.Shen A. 2020. Clostridioides difficile spore formation and germination: new insights and opportunities for intervention. Annu Rev Microbiol 74:545–566. 10.1146/annurev-micro-011320-011321. [DOI] [PubMed] [Google Scholar]
- 28.Francis MB, Sorg JA. 2016. Dipicolinic acid release by germinating Clostridium difficile spores occurs through a mechanosensing mechanism. mSphere 1:e00306-16. 10.1128/mSphere.00306-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Velasquez J, Schuurman-Wolters G, Birkner JP, Abee T, Poolman B. 2014. Bacillus subtilis spore protein SpoVAC functions as a mechanosensitive channel. Mol Microbiol 92:813–823. 10.1111/mmi.12591. [DOI] [PubMed] [Google Scholar]
- 30.Li Y, Davis A, Korza G, Zhang P, Li YQ, Setlow B, Setlow P, Hao B. 2012. Role of a SpoVA protein in dipicolinic acid uptake into developing spores of Bacillus subtilis. J Bacteriol 194:1875–1884. 10.1128/JB.00062-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perez-Valdespino A, Li Y, Setlow B, Ghosh S, Pan D, Korza G, Feeherry FE, Doona CJ, Li YQ, Hao B, Setlow P. 2014. Function of the SpoVAEa and SpoVAF proteins of Bacillus subtilis spores. J Bacteriol 196:2077–2088. 10.1128/JB.01546-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Korza G, Setlow P. 2013. Topology and accessibility of germination proteins in the Bacillus subtilis spore inner membrane. J Bacteriol 195:1484–1491. 10.1128/JB.02262-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Donnelly ML, Fimlaid KA, Shen A. 2016. Characterization of Clostridium difficile spores lacking either SpoVAC or dipicolinic acid synthetase. J Bacteriol 198:1694–1707. 10.1128/JB.00986-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vepachedu VR, Setlow P. 2004. Analysis of the germination of spores of Bacillus subtilis with temperature sensitive spo mutations in the spoVA operon. FEMS Microbiol Lett 239:71–77. 10.1016/j.femsle.2004.08.022. [DOI] [PubMed] [Google Scholar]
- 35.Ng YK, Ehsaan M, Philip S, Collery MM, Janoir C, Collignon A, Cartman ST, Minton NP. 2013. Expanding the repertoire of gene tools for precise manipulation of the Clostridium difficile genome: allelic exchange using pyrE alleles. PLoS One 8:e56051. 10.1371/journal.pone.0056051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Francis MB, Allen CA, Shrestha R, Sorg JA. 2013. Bile acid recognition by the Clostridium difficile germinant receptor, CspC, is important for establishing infection. PLoS Pathog 9:e1003356. 10.1371/journal.ppat.1003356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Howerton A, Ramirez N, Abel-Santos E. 2011. Mapping interactions between germinants and Clostridium difficile spores. J Bacteriol 193:274–282. 10.1128/JB.00980-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shrestha R, Cochran AM, Sorg JA. 2019. The requirement for co-germinants during Clostridium difficile spore germination is influenced by mutations in yabG and cspA. PLoS Pathog 15:e1007681. 10.1371/journal.ppat.1007681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shrestha R, Sorg JA. 2018. Hierarchical recognition of amino acid co-germinants during Clostridioides difficile spore germination. Anaerobe 49:41–47. 10.1016/j.anaerobe.2017.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dobson L, Remenyi I, Tusnady GE. 2015. CCTOP: a Consensus Constrained TOPology prediction web server. Nucleic Acids Res 43:W408–412. 10.1093/nar/gkv451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Griffiths KK, Zhang J, Cowan AE, Yu J, Setlow P. 2011. Germination proteins in the inner membrane of dormant Bacillus subtilis spores colocalize in a discrete cluster. Mol Microbiol 81:1061–1077. 10.1111/j.1365-2958.2011.07753.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vepachedu VR, Setlow P. 2005. Localization of SpoVAD to the inner membrane of spores of Bacillus subtilis. J Bacteriol 187:5677–5682. 10.1128/JB.187.16.5677-5682.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paiva AMO, Friggen AH, Qin L, Douwes R, Dame RT, Smits WK. 2019. The bacterial chromatin protein HupA can remodel DNA and associates with the nucleoid in Clostridium difficile. J Mol Biol 431:653–672. 10.1016/j.jmb.2019.01.001. [DOI] [PubMed] [Google Scholar]
- 44.Daniel RA, Errington J. 1993. Cloning, DNA sequence, functional analysis and transcriptional regulation of the genes encoding dipicolinic acid synthetase required for sporulation in Bacillus subtilis. J Mol Biol 232:468–483. 10.1006/jmbi.1993.1403. [DOI] [PubMed] [Google Scholar]
- 45.Fimlaid KA, Bond JP, Schutz KC, Putnam EE, Leung JM, Lawley TD, Shen A. 2013. Global analysis of the sporulation pathway of Clostridium difficile. PLoS Genet 9:e1003660. 10.1371/journal.pgen.1003660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rodriguez-Palacios A, Lejeune JT. 2011. Moist-heat resistance, spore aging, and superdormancy in Clostridium difficile. Appl Environ Microbiol 77:3085–3091. 10.1128/AEM.01589-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lawley TD, Clare S, Deakin LJ, Goulding D, Yen JL, Raisen C, Brandt C, Lovell J, Cooke F, Clark TG, Dougan G. 2010. Use of purified Clostridium difficile spores to facilitate evaluation of health care disinfection regimens. Appl Environ Microbiol 76:6895–6900. 10.1128/AEM.00718-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Setlow P, Wang S, Li YQ. 2017. Germination of spores of the orders Bacillales and Clostridiales. Annu Rev Microbiol 71:459–477. 10.1146/annurev-micro-090816-093558. [DOI] [PubMed] [Google Scholar]
- 49.Kevorkian Y, Shirley DJ, Shen A. 2016. Regulation of Clostridium difficile spore germination by the CspA pseudoprotease domain. Biochimie 122:243–254. 10.1016/j.biochi.2015.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Monteford J, Bilverstone TW, Ingle P, Philip S, Kuehne SA, Minton NP. 2021. What's a SNP between friends: the lineage of Clostridioides difficile R20291 can effect research outcomes. Anaerobe 71:102422. 10.1016/j.anaerobe.2021.102422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166:557–580. 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 52.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, III, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345. 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 53.Paiva AMO, Friggen AH, Hossein-Javaheri S, Smits WK. 2016. The signal sequence of the abundant extracellular metalloprotease PPEP-1 can be used to secrete synthetic reporter proteins in Clostridium difficile. Acs Synth Biol 5:1376–1382. 10.1021/acssynbio.6b00104. [DOI] [PubMed] [Google Scholar]
- 54.Shrestha R, Sorg JA. 2019. Terbium chloride influences Clostridium difficile spore germination. Anaerobe 58:80–88. 10.1016/j.anaerobe.2019.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sorg JA, Sonenshein AL. 2010. Inhibiting the initiation of Clostridium difficile spore germination using analogs of chenodeoxycholic acid, a bile acid. J Bacteriol 192:4983–4990. 10.1128/JB.00610-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ma NJ, Moonan DW, Isaacs FJ. 2014. Precise manipulation of bacterial chromosomes by conjugative assembly genome engineering. Nat Protoc 9:2285–2300. 10.1038/nprot.2014.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Heap JT, Pennington OJ, Cartman ST, Minton NP. 2009. A modular system for Clostridium shuttle plasmids. J Microbiol Methods 78:79–85. 10.1016/j.mimet.2009.05.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S2. Download JB.00394-21-s0001.pdf, PDF file, 117 KB (116.1KB, pdf)