Abstract
Basic helix-loop-helix (bHLH) proteins perform a wide variety of biological functions. Most bHLH proteins recognize the consensus DNA sequence CAN NTG (the E-box consensus sequence is underlined) but acquire further functional specificity by preferring distinct internal and flanking bases. In addition, induction of myogenesis by MyoD-related bHLH proteins depends on myogenic basic region (BR) and BR-HLH junction residues that are not essential for binding to a muscle-specific site, implying that their BRs may be involved in other critical interactions. We have investigated whether the myogenic residues influence DNA sequence recognition and how MyoD, Twist, and their E2A partner proteins prefer distinct CAN NTG sites. In MyoD, the myogenic BR residues establish specificity for particular CAN NTG sites indirectly, by influencing the conformation through which the BR helix binds DNA. An analysis of DNA binding by BR and junction mutants suggests that an appropriate BR-DNA conformation is necessary but not sufficient for myogenesis, supporting the model that additional interactions with this region are important. The sequence specificities of E2A and Twist proteins require the corresponding BR residues. In addition, mechanisms that position the BR allow E2A to prefer distinct half-sites as a heterodimer with MyoD or Twist, indicating that the E2A BR can be directed toward different targets by dimerization with different partners. Our findings indicate that E2A and its partner bHLH proteins bind to CAN NTG sites by adopting particular preferred BR-DNA conformations, from which they derive differences in sequence recognition that can be important for functional specificity.
A large family of transcriptional regulators is defined by the basic helix-loop-helix (bHLH) motif (40), in which a DNA-binding basic region (BR) lies immediately amino terminal to the HLH dimerization segment (17, 41, 55). In metazoans, bHLH proteins are involved in specification of multiple cell types (33, 43, 56). Some bHLH family members function as homodimers, but others appear to act together with a heterodimeric partner (56). For example, the closely related bHLH proteins that mediate myogenic differentiation, including MyoD, are thought to function as heterodimers with E proteins, a widely expressed bHLH protein subgroup that is exemplified by the E2A proteins (14, 17, 32, 42). Most bHLH protein dimers bind to the consensus CAN NTG (the E box; the consensus sequence is underlined throughout the text) with each respective BR binding to a half site (9, 19–21, 35, 45, 48). Given the many regulatory processes in which bHLH proteins are involved, the apparent simplicity of the CAN NTG consensus raises the important question of how different bHLH proteins act only on appropriate target genes (56).
In part, the specificity with which bHLH proteins function derives from preferential recognition of different classes of CAN NTG sites by different bHLH protein subgroups. The HLH segment consists of a parallel, left-handed, four-helix bundle (Fig. 1) (19–21, 35, 45, 48). The BR is unstructured in solution (2) but when bound to DNA, it extends N terminally from the HLH segment as an α helix that crosses the major groove (Fig. 1). Crystallographic analyses have revealed some differences in how these proteins bind DNA. For example, in Myc family and related bHLH proteins, an arginine (Arg) residue at BR position 13 (Fig. 2) specifies recognition of CACGTG sites (7, 16, 25, 54) by contacting bases in the center (20, 21, 48). However, it still is not understood how bHLH proteins which have a different amino acid at BR position 13 (Fig. 2) bind preferentially to distinct CAN NTG sites (9, 16) or how bHLH proteins establish differences in flanking sequence selectivity (9, 23, 24) that can be of biological importance (1, 30).
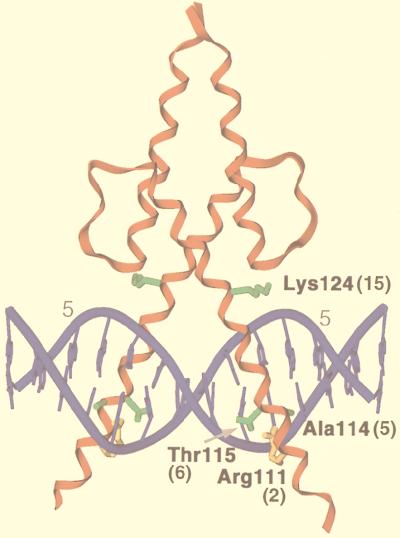
FIG. 1.
A MyoD-DNA complex. In this X-ray crystallographic structure (35), a MyoD homodimer is bound to the sequence AACAGCTGTT, which corresponds to its preferred recognition consensus (9). Residues are numbered as in full-length MyoD, and their positions as specified in Fig. 2 and the text are indicated in parentheses. Binding site positions ±5 (numbered as in Fig. 3A) are indicated by grey numerals. Side chains are shown only for the myogenic residues (green) (18) and Arg 111 (R2) (gold).
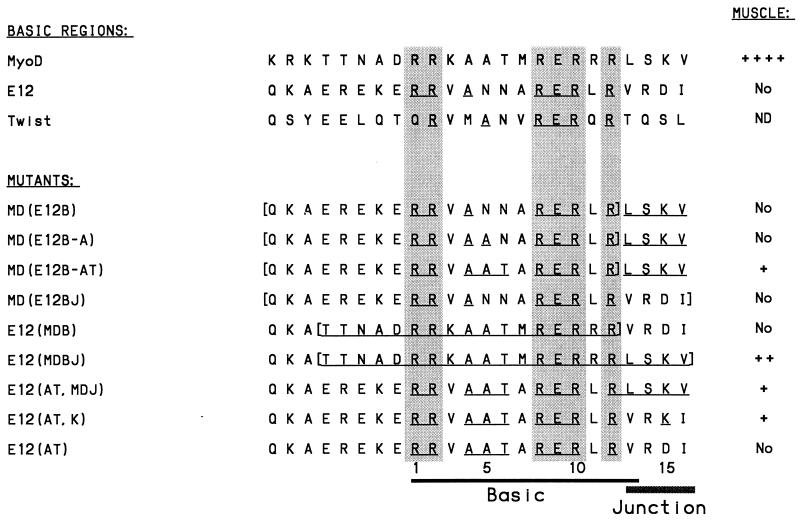
FIG. 2.
Myogenic activity of MyoD and E12 BR and junction mutants. Each of these mutants has been described previously (18, 57), and their sequences are compared with sequences from mouse MyoD, E12, and Twist. Amino acids that are identical to those of MyoD are underlined, positions that are conserved in most bHLH proteins are shaded, and entire BR and junction regions that have been swapped are bracketed. The column at the right indicates the relative activities of these proteins when assayed previously by transfection for conversion of cultured cells into muscle (18, 57); activity is denoted as ++++ (frequency of myogenic conversion obtained with wild-type MyoD), ++ (30 to 50% of that obtained with MyoD), + (5 to 30% of that obtained with wild-type MyoD), No (myogenic conversion was not detected), or ND (not done).
Many bHLH proteins that lack R13, including MyoD and other E2A partners (Fig. 2), can bind to similar DNA sequences in vitro but act on different tissue-specific genes (56). Cooperative or inhibitory relationships with other transcriptional regulators might contribute to this specificity (34, 39, 46, 58), but it is not likely to derive entirely from other lineage-specific factors, because MyoD can induce myogenesis in many different cell types (56). Initiation of myogenesis by MyoD and other myogenic bHLH proteins depends on three residues that are located within the BR and the BR-HLH junction (A5, T6, and K15 [Fig. 1 and 2]). These residues, which we refer to as myogenic are not essential for binding a muscle-specific site in vitro or in vivo, suggesting that they are involved in other critical interactions (11, 17, 18, 47, 57). These interactions have been proposed to involve distinct cofactors (11, 17, 57) and the unmasking of an activation domain in MyoD or the myogenic cofactor MEF2 (3, 5, 29, 57). In the MyoD-DNA structure, K15 is oriented away from the DNA, but A5 and T6 face the major groove and could not contact other proteins directly (35) (Fig. 1). However, the latter two residues could influence protein-protein interactions indirectly, by affecting how the BR helix is positioned on the DNA (35). Although substitutions at these positions might not substantially impair binding to particular CAN NTG sites, it is important to determine whether they might have more subtle influences on sequence specificity that could reflect conformational effects.
We have determined that the myogenic residues A5 and T6 establish the characteristic MyoD sequence preference, which includes a CAGCTG core. Individual substitutions at these BR positions simultaneously alter preferences for multiple bases that MyoD does not contact directly (35), indicating that these preferences are determined indirectly, by how the BR helix is positioned on the DNA. This mechanism is distinct from the standard model for sequence specificity, in which preferred bases are contacted directly (44, 50). The corresponding BR residues are also required for the sequence preferences of E2A proteins, which can recognize either of two distinct half-sites depending on their dimerization partner. E2A homodimers and E2A-MyoD heterodimers bind to asymmetric sites that include a CACCTG core. In contrast, as a heterodimer with the bHLH protein Twist, E2A binds preferentially to half of the symmetric sequence CATATG. The preference of E2A for the former asymmetric sites depends not only on the BR sequence but also on BR positioning that involves the junction region. An analysis of DNA binding by MyoD and E2A junction and BR mutants indicates that a MyoD-like sequence specificity is associated with, but not sufficient for, myogenesis. This supports the model that the BR-junction region is also involved in other critical interactions. The results suggest that E2A and its partner bHLH proteins bind DNA by adopting a limited number of preferred BR conformations, each of which is associated with a characteristic DNA sequence preference. They also indicate that binding of cofactors to the MyoD BR might be influenced by how it is positioned on the DNA and are consistent with the idea that relatively subtle differences in binding sequence recognition can modulate bHLH protein activity in vivo.
MATERIALS AND METHODS
Mutagenesis, protein expression, and DNA binding assays.
The various MyoD and E2A mutants used in this study have been described previously (17, 18, 57), with the exception of the MyoD mutants shown in Fig. 8A. For construction of those mutants, a SalI site that did not alter the encoded amino acid sequence was created at MyoD BR positions 10 and 11 (Fig. 2) by PCR. BR mutants were then generated by PCR using Pfu or Vent polymerase and introduced into this MyoD (SalI) construct as PmlI-SalI fragments. Junction region mutations were created similarly by PCR and inserted into MyoD (SalI) as SalI-NarI fragments. Constructs with both BR and junction mutations were produced by introduction of a mutant PmlI-SalI or SalI-NarI fragment into the appropriate BR or junction mutant construct. All of these mutations were confirmed by DNA sequencing.
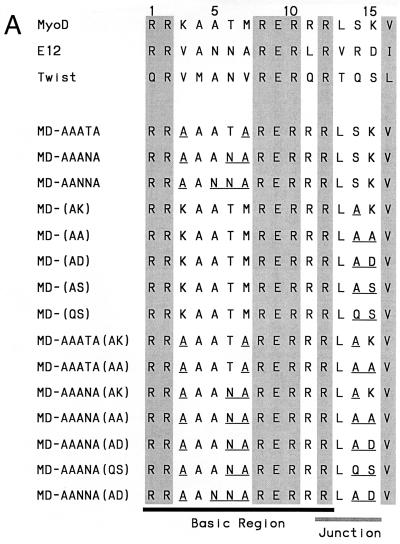
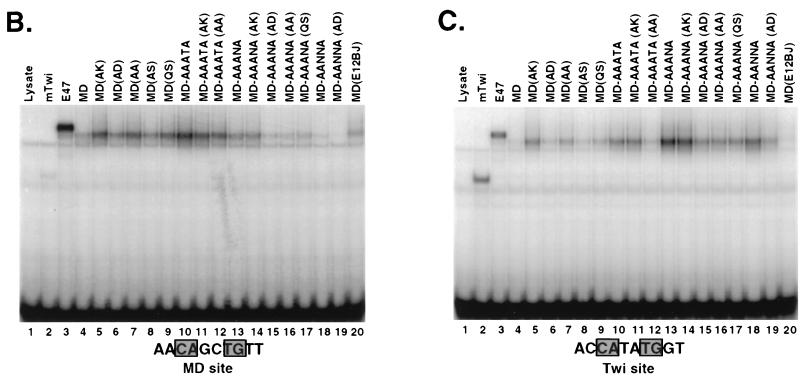
FIG. 8.
Effects of bHLH BR and BR-HLH junction residues on MyoD binding preferences. (A) Mutagenesis analysis of the MyoD BR and junction. MyoD BR mutant sequences are compared with the MyoD, E12, and Twist BR sequences (Fig. 2). Conserved bHLH residues are shaded, and residues that are altered within full-length MyoD are underlined. (B) Binding of MyoD mutants described in panel A to the MyoD preferred site. These mutants are compared with the indicated wild-type proteins, and binding is assayed as for Fig. 3D except that each protein is present at 40 pM and DNA labeled to the same specific activity is present at 400 pM. E47 is an alternatively spliced E2A protein that binds DNA well as a homodimer (40). Twi, Twist. (C) Binding of MyoD mutants to the Twist preferred site, assayed as for panel B.
For the in vitro selection experiment shown in Fig. 3, full-length MyoD was expressed in bacteria from a pRK171a-based construct (pT7-MyoD) described previously (53). The MD(E12B), MD(E12B-A), and MD(E12B-AT) mutations (57) were each introduced into this construct within a PmlI-MluI fragment. These proteins were expressed by isopropyl-β-d-thiogalactopyranoside induction in Escherichia coli BL21(DE3)/pLysS cells as described previously (51) and then purified to >90% homogeneity by precipitation in 0.6 M (NH4)2SO4. Precipitated protein was resuspended in a mixture containing 10% glycerol, 20 mM HEPES (pH 7.6), 100 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 1 μg of leupeptin per ml, and 1 μg of pepstatin per ml.
FIG. 3.
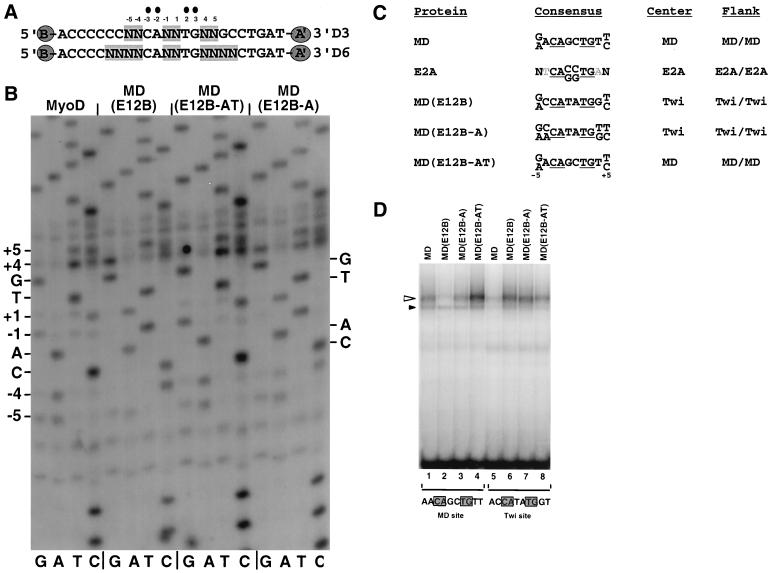
In vitro selection assay of binding site preferences. (A) Core sequences of the random sequence oligonucleotide libraries D3 and D6 (8, 9). In each library, the bases shown are flanked by sequences which correspond to primers (A and B) that allow selected sequences to be recovered by PCR. A′ indicates that primer A corresponds to the opposite strand. (B) Sequences of preferred binding sites. Starting with the D6 oligonucleotide random sequence library (A), three rounds of sequential selection and PCR amplification were performed for binding to the proteins indicated. A sample of the final selected population of binding sites was then sequenced directly as a pool and analyzed by autoradiography. The MyoD preferences at positions ±1 described previously (9) are more prominent after additional selection rounds (not shown). (C) Summary of sequence preferences identified by in vitro selection in panel B. MyoD and E2A homodimer preferences were described in reference 9. Binding site positions are numbered as in panel B, and grey letters indicate bases that were selected against. The CAN NTG consensus that was fixed in these experiments is underlined. Twi, Twist. (D) Binding of MyoD BR mutants to individual oligonucleotide sites, which differed only at the sequences shown. In this EMSA, which was analyzed by phosphorimaging, each sample contained the indicated in vitro-translated protein at a concentration of 40 pM and DNA that was labeled to the same specific activity at 550 pM. Specific and background species are indicated by open and closed triangles, respectively.
Other proteins were expressed by in vitro translation (Promega), with in vitro transcription and translation performed in separate steps. Protein expression was carefully quantitated by 35S-labeled translation and sodium dodecyl sulfate-polyacrylamide gel electrophoresis. These procedures and those for electrophoretic mobility shift assay (EMSA) have been described previously (31). Each EMSA was performed at room temperature and analyzed by autoradiography or phosphorimaging. Individual oligonucleotide sites were 21 bp in length and differed from the MyoD consensus oligonucleotide (7) only at the positions indicated. The MCK-R site corresponds to the right E-box site in the muscle creatine kinase enhancer (18).
In vitro selection experiments.
Populations of preferred binding sites were isolated by sequential in vitro selection and PCR amplification essentially as described previously (6, 7, 9). During each selection round, DNA that was bound by the protein complex of interest was isolated by EMSA and then amplified by PCR for the next round. In each EMSA selection, care was taken to ensure that sufficient quantities of labeled bound DNA were recovered to maintain a representative population of sequences. These experiments were initiated with 0.5 ng of 32P-end-labeled starting library. In each subsequent selection round, selections were performed with approximately 0.1 ng of amplified 32P-body-labeled DNA. In selections for binding to partially purified bacterially expressed MyoD mutant proteins (Fig. 3), these protein preparations were not quantitated, but instead sequential dilutions of these samples were tested for binding. Bound sequences were then recovered and amplified from a sample in which less than 10% of the input DNA was in the bound fraction. This strategy ensured selection of optimal binding sequences. The final selected binding site pool was sequenced directly, using a 32P-end-labeled primer as described previously (7). Mouse proteins were used in these selections with the exception of Twist, which was from Xenopus. Binding site competition analyses (not shown) demonstrated that its binding preferences were indistinguishable from those of mouse Twist, which was used in the EMSA analyses shown.
RESULTS
Myogenic BR residues and MyoD DNA binding preferences.
Identification of the myogenic BR residues stemmed originally from studies in which the MyoD BR was replaced with that of E12, a product of the alternatively spliced E2A gene (40). This MyoD mutant [MD(E12B) (Fig. 2)] binds to a muscle-specific regulatory site as a heterodimer with E2A proteins either in vitro or in vivo, but it cannot induce myogenesis in a cell culture assay or activate transcription through a muscle-specific enhancer (17, 57). Resubstitution of the myogenic residues A5 and T6 (Fig. 2) in MD(E12B) restores its activity in these functional assays (57). Similar results are obtained when A5 and T6 are mutated within MyoD (18, 29, 57) and when analogous substitutions are made in the context of the myogenic bHLH protein myogenin (11). These experiments implicate A5 and T6 in mechanisms that are of functional importance but not essential for binding to a particular muscle-specific DNA sequence.
We used an in vitro selection strategy (9) to test whether such mutations might have more subtle effects on how MyoD binds specifically to DNA. To identify sequences to which these mutants bind preferentially, we used sequence libraries in which only positions within and flanking the CAN NTG consensus are randomized (Fig. 3A), so that the position of bHLH protein binding along the DNA is fixed. This strategy makes it possible to sequence the selected sites as a pool and thereby to analyze a very large population of selected sites simultaneously (8, 9). It reveals the relative preferences for individual bases at each site position and can detect subtle differences that might not be identified through more conventional approaches.
This assay has previously shown that the preferred MyoD binding consensus is (G/A) ACAGCTG(T/C) (Fig. 3B and C) and that the E2A proteins E12 and E47 overlap considerably with MyoD in their binding properties but prefer sites that have an asymmetric CACCTG core sequence (Fig. 3C) (9). However, in contrast to either of these proteins, the MD(E12B) mutant prefers the sequence (G/A)CCATATGG(T/C), which differs from the MyoD preferred site over the eight central base pairs and contains the distinct core sequence CAT ATG (Fig. 3B and C). This sequence and related elements are normally targeted by the bHLH protein Twist, an E-protein partner that is involved in mesodermal cell fate specification (15, 27, 37, 52, 60) (Fig. 2). Back-substitution of A5 of MyoD into MD(E12B), which is not sufficient for myogenic activity in cell culture assays (57), results in preferences that are slightly more similar to those of MyoD at positions ±4 [MD(E12B-A) (Fig. 2, 3B, and 3C)]. However, introduction of both A5 and T6, which restores myogenesis (11, 57), results in preferences across the entire site that are indistinguishable from those of MyoD [MD(E12B-AT) (Fig. 2, 3B, and 3C)].
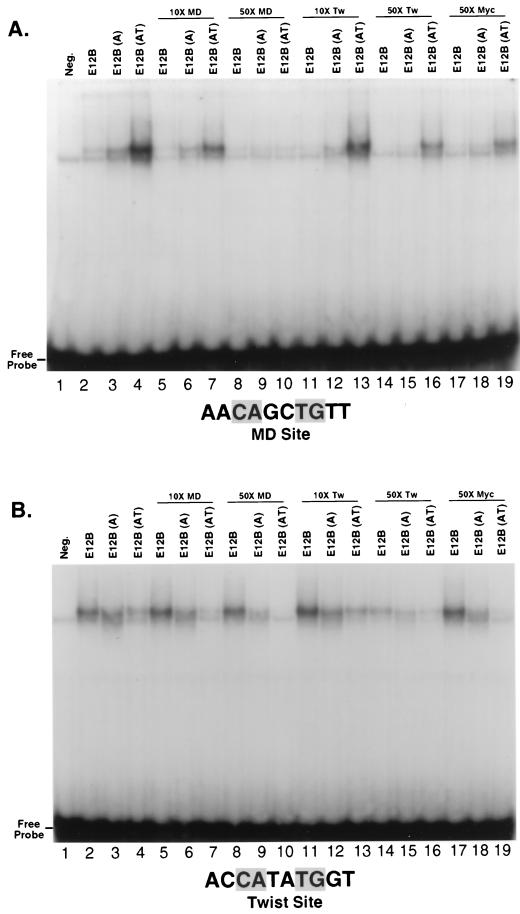
To determine whether these sequence preferences reflect significant differences in binding affinity and specificity, we compared levels of binding of these proteins to individual oligonucleotides that correspond to the MyoD and Twist preferences and differ only at positions within and adjacent to the CAN NTG consensus (Fig. 3D). Supporting the in vitro selection findings, both MyoD and MyoD(E12B-AT) homodimers bound with higher affinity to the preferred MyoD site than to the Twist site (Fig. 3D, lanes 1, 4, 5, and 8). In contrast, the Twist site was preferred by MD(E12B) and, to a lesser extent, MyoD(E12B-A) (Fig. 3D, lanes 2, 3, 6, and 7). In a binding competition assay, specific DNA binding by MD(E12B-AT) was competed much more effectively by the MyoD site (Fig. 4A, lanes 4, 7, 10, 13, and 16), and binding by either MD(E12B) or MD(E12B-A) was competed better by the Twist site (Fig. 4B, lanes 2, 3, 8, 9, 14, and 15). A c-Myc preferred site (CACGTG [not shown]) was a relatively poor competitor of binding by each of these proteins (Fig. 4A and B, lanes 17 to 19). The data show that introduction of A5 and T6 into MD(E12B) restores not only myogenic activity (Fig. 2) but also the MyoD DNA binding preference. This substitution affects sequence recognition across 4 bp within each half-site (Fig. 3A and B), indicating a global effect on how the BR helix is positioned on the DNA. The finding that MD(E12B) is distinct from either MyoD or E12 in its binding sequence preference also indicates that DNA recognition by an E2A BR can be profoundly influenced by its molecular context.
FIG. 4.
Specificity of MyoD BR mutant DNA binding. (A) Competition analysis of binding to the labeled MyoD preferred site, analyzed by EMSA and autoradiography. The indicated in vitro-translated proteins and DNA labeled to the same specific activity were present at concentrations of 50 and 900 pM, respectively. When the samples were mixed, unlabeled competitor DNA sites were added at the indicated ratios relative to the labeled probe. Tw, Twist. (B) Competition analysis of binding to the Twist preferred site, performed as for panel A.
Influence of BR positioning on MyoD-E2A and Twist-E2A heterodimer sequence preferences.
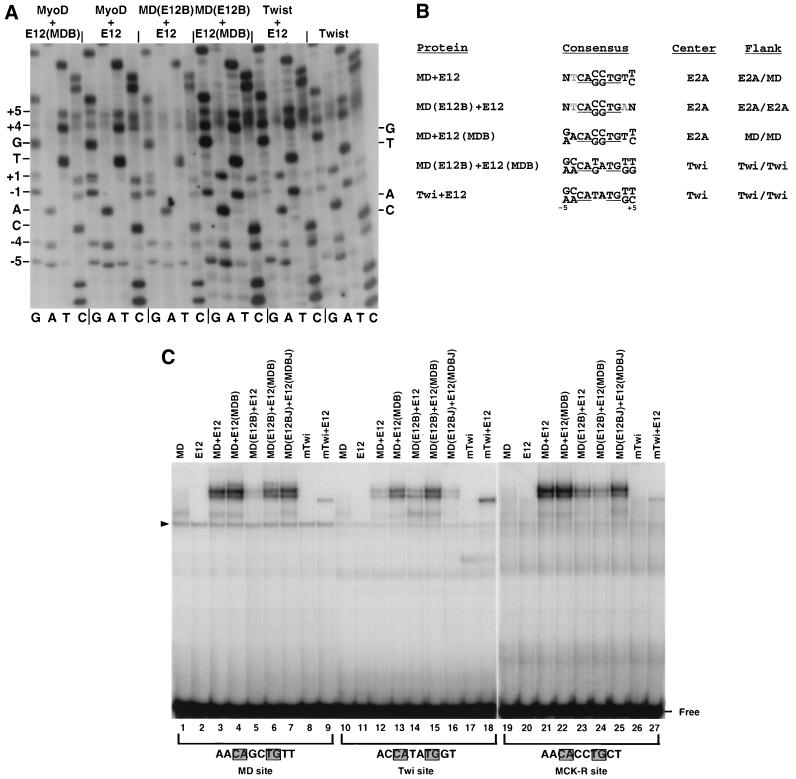
Twist and E2A proteins appear to cooperate in vivo to regulate transcription through CAT ATG sites (27), suggesting that the DNA sequence recognition properties of E2A might be altered by heterodimerization with Twist. However, an alternative possibility is that functional Twist-E2A recognition sites are distinct from their in vitro binding preference (28). To address this question, we performed in vitro selection on Twist-E12 complexes. Twist homodimers and Twist-E12 heterodimers both preferred sites that contain the core sequence CAT ATG (Fig. 5A and B). They were similar to MD(E12B) and especially to MD(E12B-A) in their preferences at ±4 but selected MyoD-like sequences at ±5 (Fig. 3B and 3C, 5A, and 5B). The symmetry of this preferred sequence suggests that in the Twist-E12 protein-DNA complex, the Twist and E12 BRs each prefer the same half-site sequence. In contrast, and as observed previously (9), MyoD-E12 heterodimers selected a MyoD-like half site at positions +4 and +5, an E2A-like half-site at −4 and −5, and CC or GG bases in the center of the site (Fig. 5A and B), indicating asymmetric binding. Apparently, an E2A BR normally prefers distinct half-sites in the context of these two bHLH dimerization partners, indicating an intermolecular effect on how it interacts specifically with DNA.
FIG. 5.
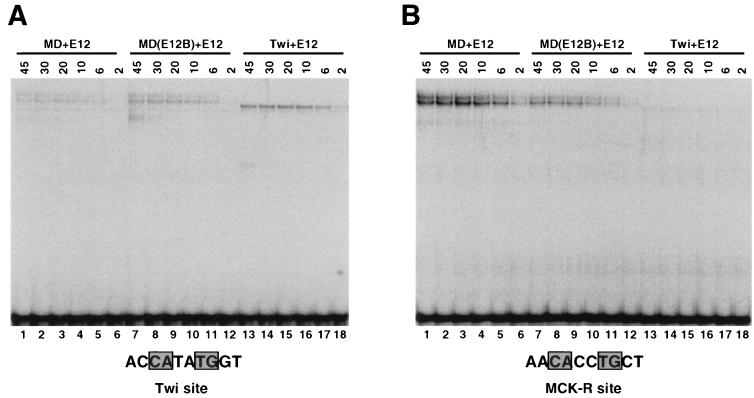
Binding site preferences of MyoD, E2A, and Twist heterodimer complexes. (A) In vitro selection analysis of binding site preferences. Four rounds of selection from the D3 library (Fig. 3A) were performed as for each in vitro-translated protein complex. In each case, the heterodimer complex could be easily identified in the EMSA on the basis of mobility (9), particularly because E12 homodimers bind DNA poorly (Fig. 9). In the Twist homodimer selection, binding to Twist-E12 heterodimers was selected for in the first round, because of the relatively low level of Twist homodimer binding. Subsequent rounds were performed with Twist homodimers. Each sample was analyzed by sequencing and autoradiography as for Fig. 3B. (B) Summary of sequence preferences identified in panel A, depicted as in Fig. 3C. MyoD-E2A heterodimer preferences were also described previously (9). Twi, Twist. (C) Binding of bHLH heterodimers to individual preferred sites, analyzed by EMSA and phosphorimaging. E2A-derived proteins were present at a concentration of 8 pM, and Twist and MyoD-derived proteins were present at 19 pM. The indicated DNA sites that had been labeled to the same specific activity were present at 550 pM. The MCK-R site differs from the others only at the positions shown. A background species is indicated by a triangle.
To investigate how heterodimer formation influences the binding preferences of the E12 and MyoD BRs, we performed in vitro selection on combinations of MyoD and E12 BR mutants. When the BR of one partner within a MyoD-E12 heterodimer was substituted with that of the other, the heterodimer binding preferences outside the CAN NTG consensus corresponded to those of the individual BRs. For example, unlike MD(E12B) homodimers (Fig. 3B and C), heterodimers of MD(E12B)+E12 preferred wild-type heterodimer sequences in the center of the site, and selected E2A-like sequences in both flanking regions, at ±4 and ±5 (Fig. 5A and B). A heterodimer of MyoD and an E12 protein containing the MyoD BR [E12(MDB) (Fig. 2A)] similarly selected a wild-type heterodimer preference within the CAN NTG motif but preferred a MyoD-like sequence at ±4 and ±5 (Fig. 5A and B). In contrast, MD(E12B)-E12(MDB) heterodimers had a binding preference more similar to that of Twist (Fig. 5A and B), indicating that placement of each BR in the protein context of the other partner affected binding over the entire site. A striking aspect of our findings is that each of the mutant homo- or heterodimer protein complexes that we have examined selected sequences that correspond to particular patterns preferred by MyoD, E2A, or Twist protein (Fig. 3C and 5B).
These in vitro selection findings were supported by assays of binding to individual sites, including a sequence from a muscle-specific regulatory region (MCK-R). This site, which corresponds to the MyoD-E12 heterodimer in vitro binding preference and responds to MyoD in vivo, was used in the original analysis of the myogenic residues (9, 17, 57). In an EMSA, MyoD-E12 heterodimers bound with higher affinity to either the MCK-R or MyoD site than to the Twist site (Fig. 5C, lanes 3, 12, and 21). MyoD(E12B)-E12 heterodimers only slightly preferred the MCK-R heterodimer site to the Twist site but appeared to prefer either of these sequences to the MyoD site (Fig. 5C, lanes 5, 14, and 23). As the preferences of MD(E12B-A) and MD(E12B-AT) homodimers would predict, introduction of both A5 and T6 into MD(E12B) altered its sequence preferences as a heterodimer with E12, so that they were more similar to those of MyoD (not shown). MyoD-E12(MDB) heterodimers only modestly preferred the MyoD or MCK-R site in comparison to the Twist site (Fig. 5C, lanes 4, 13, and 22). In contrast, the Twist site was preferred by MD(E12B)-E12(MDB), Twist, and Twist-E12 complexes (Fig. 5C, lanes 6, 8, 9, 15, 17, 18, 24, 26, and 27).
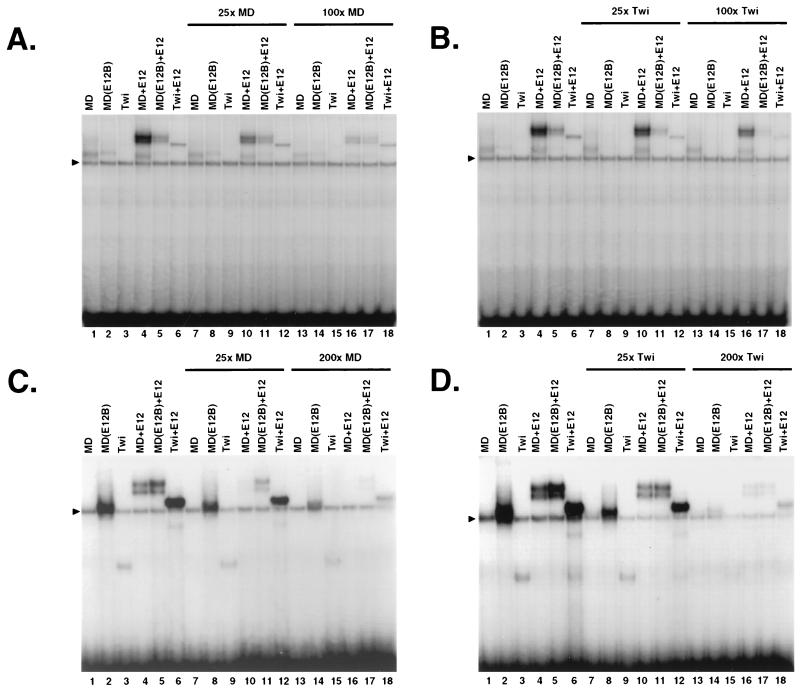
Binding site competition and protein titration assays also supported the in vitro selection data. The MyoD site competed more effectively than the Twist site for binding by either MyoD homodimers or MyoD-E12 heterodimers (Fig. 6A and B, lanes 1, 4, 7, 10, 13, and 16). In contrast, the Twist site competed more effectively for binding by MD(E12B), MD(E12B)-E12, Twist, and Twist-E12 complexes, although these latter complexes appeared to bind with less specificity than did MyoD-E12 complexes (Fig. 6C and D, lanes 2, 3, 5, 6, 8, 9, 11, 12, 14, 15, 17, and 18). However, the distinct binding specificities of MyoD-E12 and Twist-E12 heterodimers were apparent in a protein titration assay in which the amount of MyoD or Twist protein was varied under conditions of low DNA concentration (Fig. 7A and B, lanes 1 to 6 and 13 to 18) that more closely represent differences in binding affinity (13). Also in agreement with results described above (Fig. 5C, lanes 14 and 23), heterodimers of MD(E12B) plus E12 bind to the MCK-R site with decreased specificity and with slightly lower affinity than MyoD-E12 complexes (Fig. 7A and B, lanes 7 to 12).
FIG. 6.
Binding competition analysis of DNA binding by bHLH heterodimers. (A and B) Binding of the indicated protein complexes to the labeled MyoD site (Fig. 3D) was competed by addition of an unlabeled binding site at ratios indicated above the gel. These experiments were performed and analyzed as for Fig. 4 except that labeled DNA was present at 600 pM, E12 protein present at 8 pM, and all other proteins were present at 19 pM. Twi, Twist. (C and D) Binding of the indicated protein complexes to the labeled Twist site (Fig. 3D) was competed by addition of the indicated unlabeled sites. These experiments were performed described for panel A and B except that labeled DNA was present at 1.1 nM, and they were analyzed by autoradiography. Note that the gel shown in panel C was exposed longer than that shown in panel D, as indicated by comparison of lanes 1 to 6. A background species is indicated by a triangle.
FIG. 7.
Protein titration of DNA binding by bHLH heterodimers. (A) Binding to the Twist (Twi) site, analyzed by EMSA and phosphorimaging. In each experiment, E12 was present at 8 pM and DNA that had been labeled to the same specific activity was present at 5 pM. The indicated partner proteins were present at the concentrations (picomolar) shown above the gel. (B) Binding to the MCK-R site, analyzed as for panel A.
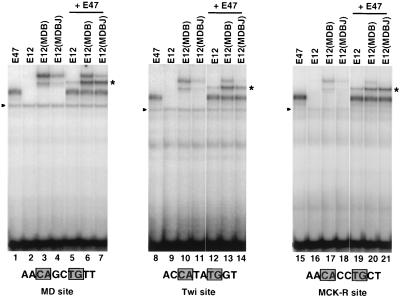
To investigate the role of the BR-HLH junction region in BR positioning, we examined the DNA binding preferences of the MD(E12BJ) and E12(MDBJ) mutants, each of which contains both the BR and junction of the other partner (Fig. 2). In contrast to MD(E12B)-E12(MDB) heterodimers (Fig. 5A and B; Fig. 5C, lanes 6, 15, and 24), MD(E12BJ)-E12(MDBJ) heterodimers (Fig. 2A) bound to the MyoD, Twist, and MCK-R sites with relative preferences that are comparable to those of MD-E12 heterodimers (Fig. 5C, lanes 3, 7, 12, 16, 21, and 25). Apparently, the Twist-like sequence preference resulting from simultaneous mispairing of both the MyoD and E12 BRs (Fig. 5A and B) can be corrected by matching each of these BRs with the corresponding junction region. Similarly, and in contrast to MD(E12B) homodimers, MD(E12BJ) homodimers bind to the MyoD, Twist, and MCK-R sites with preferences that are similar to those of E2A proteins (Fig. 8B and C, lane 20, and data not shown). These findings indicate that the BR-HLH junction can be critical for establishing the sequence specificity of an E2A BR, presumably because it influences how the BR is positioned on the DNA.
Contributions of the BR and junction to binding affinity and specificity.
It has been shown previously that introduction of A5, T6, and either the junction region or K15 of MyoD confers upon E12 the capacity to induce myogenesis (Fig. 2) (18). In the MyoD-DNA complex, A5 and T6 are not positioned to allow direct protein-protein contact (Fig. 1) (35), but we have shown that they are critical for the DNA sequence preferences of MyoD, apparently because they affect the conformation of the BR-DNA complex. We have also determined that the junction region can influence how the E2A BR binds DNA. These observations suggest the possibility that the capacity for myogenesis derives entirely from the conformation of the DNA-bound MyoD BR, a model which would predict that the sequence preference of each of these bHLH proteins is established by amino acids at BR positions 5, 6, and 15. We have investigated this model by determining how individual substitutions at these positions, which have been shown to be critical in vivo, influence the DNA binding preferences of MyoD.
To address the importance of the MyoD junction region for DNA binding, we altered MyoD positions 14 and 15 (Fig. 8A) and left position 13 intact because it is not required for the MyoD sequence preference in the MD(E12B-AT) mutant (Fig. 2 and 3C). Substitution of alanine for S14, which does not interact with DNA (35), increased binding affinity [MD(AK) (Fig. 8A; Fig. 8B and C, lanes 4 and 5)], perhaps by stabilizing the BR helix. The preference of MD(AK) for the MyoD site was not substantially altered by replacement of position 15 with alanine [MD(AA)] or with either glutamic acid [MD(AD)] or serine [MD(AS) and MD(QS)], which correspond to residues from E12 or Twist, respectively (Fig. 8; Fig. 8B and C, lanes 5 to 9). The relative preferences of these mutants for the MyoD site are comparable to the binding preferences of other proteins that were confirmed by binding competition analysis (Fig. 4 and 6). Apparently, appropriately specific DNA binding by MyoD homodimers is not impaired by a variety of BR-HLH junction substitutions, including nonconservative mutations of K15. This flexibility contrasts with the importance of the junction region for positioning the E12 BR and with the requirement for K15 for myogenesis.
To investigate the role of BR positions 5 and 6 in a neutral context, we first substituted alanine for two nonconserved BR residues (MD-AAATA [Fig. 8A]) that are not predicted to be required for DNA binding (22, 35). This substitution proportionally increased binding to both sites in the context of MyoD (MD-AAATA [Fig. 8B and C, lanes 10]) and enhanced specificity for the MyoD site in the context of MD(AA) (Fig. 8A; Fig. 8B and C, lanes 12). Replacement of T6 with asparagine conferred a preference for the Twist site (MD-AAANA [Fig. 8A; Fig. 8B and C, lanes 10 and 13]), a finding that parallels the preferences of MD(E12B-AT) and MD(E12B-A) (Fig. 3B and C). This effect was not diminished by various BR-HLH junction mutations or enhanced by presence of Twist junction residues (Fig. 8B and C, lanes 13 to 17), indicating that N6 is the most important of these residues for the Twist sequence preference. To test whether E2A amino acids that correspond to the three myogenic residues could specify an E2A-like DNA binding preference, we introduced an asparagine at BR position 7 into MD-AAANA and MD-AAANA(AD), the latter of which contains the D15 residue characteristic of E2A proteins (Fig. 8A). In contrast to MD(E12BJ), these mutants strongly preferred the Twist site to the MyoD or MCK-R sites (Fig. 8B and C, lanes 18 to 20, and data not shown), indicating that establishment of an E2A homodimer sequence preference requires additional E2A BR or junction residues and that the conformational mechanisms that dictate this asymmetric sequence preference might be complex.
In the examples that we have analyzed, MyoD mutants that lack myogenic activity bind preferentially to the Twist site (Fig. 2 and 3C), raising the question of whether changes in DNA binding preferences accompany conversion of E12 to a myogenic protein through introduction of MyoD BR and junction residues. E12 homodimers do not bind DNA as well as the E2A protein E47 (Fig. 9, lanes 1, 2, 8, 9, 15, and 16), which also cannot induce myogenesis (18). Introduction of the MyoD BR into E12 is not sufficient for myogenesis [E12(MDB) (Fig. 2)] but sharply increased binding of E12 to all three sites and was associated with a modest preference for the MyoD site (Fig. 9, lanes 3, 10, and 17). The E12(MDBJ) mutant, which can induce myogenesis (Fig. 2), bound to each of the three sites at a lower level than E12(MDB) and did not have a markedly increased preference for either the MyoD or MCK-R sites (Fig. 9, lanes 4, 11, and 18). Heterodimerization with E47 increased the relative levels with which E12(MDBJ) bound to the MyoD and MCK-R sites (Fig. 9, lanes 6, 7, 13, 14, 20, and 21) but also did not identify DNA binding effects that appear to be sufficient to account for the different functional properties of E12(MDB) and E12(MDBJ). These findings further support the idea that the MyoD junction region is not critical for DNA binding (Fig. 8B and C, lanes 4 to 9) and instead is important for myogenesis because it is involved in other interactions (18).
FIG. 9.
DNA binding by E12 mutants. DNA binding by the indicated protein complexes is assayed as for Fig. 5C except that all E12 derivatives are present at 8 pM and E47 is present at 19 pM. A protein-DNA complex of intermediate mobility that corresponds to E47-E12 heterodimers is indicated by an asterisk, and a background species is indicated by a closed triangle.
DISCUSSION
bHLH protein DNA binding specificity deriving from effects on BR-DNA conformation.
The myogenic MyoD BR residues A5 and T6 are essential for myogenesis but not for binding of MyoD-E2A heterodimers to a muscle-specific site in vitro or in vivo (18, 57). However, we have determined that these residues are required for MyoD to bind DNA with its characteristic specificity for particular CAN NTG sites. Substitution of asparagine for T6, and especially for both A5 and T6, results in MyoD binding preferentially to a Twist site (Fig. 8B and C, lanes 10, 13, and 18). The Twist-like MD(E12B) sequence preference is affected partially by substitution of A5 for the corresponding asparagine [MD(E12B-A) (Fig. 3C)] but is reconfigured by introduction of both A5 and T6 so that it is indistinguishable from that of wild-type MyoD [MD(E12B-AT), Fig. 3C)]. The data indicate that MyoD residues A5 and T6 are each critical for its DNA binding sequence preferences and that the N6 residue, which is common to the Twist and MD(E12B-A) BRs (Fig. 2), is important for the Twist-like preference. Mutations of these individual BR residues alter sequence preferences across each half-site (Fig. 3C), raising the question of how they might have such a global effect on how the BR helices and the DNA interact preferentially with each other.
A structure of MyoD obtained by X-ray crystallography suggests how A5 and T6 might influence binding sequence specificity. When bound to its preferred recognition site, MyoD does not directly contact base pairs that it specifies in the center of and flanking the CAN NTG consensus (35). However, A5 and T6 allow the MyoD BR helix to pack more tightly into the major groove than do the corresponding N5 and N6 residues of E2A proteins, in part because of their smaller sizes (Fig. 1 and 2) (35). As a result, the MyoD BR residues T6 and R2 directly contact CAN NTG bases at ±2 and ±3 respectively, and R1 binds a backbone phosphate at ±6 (Fig. 1) (35). In contrast, in E47 R2 swings out of the major groove and contacts the backbone, and the residue at position 1 does not interact directly with the DNA (12, 19). Supporting the idea that A5 and T6 influence the conformation of the DNA-bound BR, substitution of asparagine for A5 in MyoD increases its sensitivity to protease digestion (29). Our findings suggest that protein-DNA interactions that depend specifically on the MyoD A5 and T6 residues may directly influence how the BR helix interacts preferentially with the DNA and thereby indirectly specify its characteristic sequence preferences at positions within and flanking the CAN NTG consensus.
Such indirect conformational effects also appear to be critical for the E2A and Twist sequence preferences. When E47 homodimers bind DNA, a single subunit contacts a base in the center of the site through R10 (Fig. 2). This interaction could be important for the asymmetric E2A homodimer sequence preference (19). However, the Twist-like sequence preference that is characteristic of Twist-E2A heterodimers and MD(E12B) homodimers is different across each 5-bp half-site and symmetric (Fig. 3C and 5B), suggesting that it is likely to be established indirectly, through an intermolecular effect that involves a distinct positioning of the BR helix. Introduction of the E12 BR-HLH junction region into MD(E12B) corrects its binding preference so it is like that of E2A homodimers [MD(E12BJ) (Fig. 5C, lanes 7, 16, and 25; Fig. 8B and C, lanes 20)], implicating the BR-HLH junction in this effect. Presumably, the E2A junction acts in concert with the asparagines at BR positions 5 and 6 (Fig. 2), although the Twist-like preference of the MD-AANNA(AD) mutant (Fig. 8B and C, lane 19, and data not shown) suggests that the E2A junction residue D15 is not sufficient. The finding that E2A proteins can be targeted to different DNA sequences by different dimer partners may have important implications for their in vivo functions.
In contrast, the BR-HLH junction region does not have a strong influence on the MyoD DNA binding preference. Various MyoD junction mutations do not substantially diminish its preference for a MyoD site (Fig. 8B and C, lanes 5 to 9). In addition, the similar sequence preferences of E12(MDB) and E12(MDBJ) homodimers (Fig. 9, lanes 3, 4, 10, 11, 17, and 18) contrast sharply with the different specificities of MD(E12B) and MD(E12BJ) (Fig. 3D, lanes 2 and 6; Fig. 8B and C, lanes 20). This apparent difference between MyoD and E2A proteins might derive from the distinct arrangement of the BR helix on the DNA that results from presence of MyoD residues A5 and T6.
It is striking that as a group, these various bHLH mutants and dimer combinations bind DNA with a limited number of discrete sequence preferences (Figs. 3C and 5B). Presumably, each of these preferences reflects a preferred conformational state that is dictated by how each BR helix and the corresponding DNA sequence conform to each other in an induced fit (49). This mechanism for recognizing particular CAN NTG sites appears to be different from the direct recognition of central bases that is characteristic of bHLH proteins that contain R13 and bind to CACGTG or CATGTG sites (20, 21, 48). Consistent with this idea, BR residues 5 and 6 do not appear to be important for the function of the R13-containing bHLH protein c-Myc (10). In E2A and its tissue-specific dimerization partners, a more flexible conformation-based mechanism might have evolved to increase adaptability in both sequence recognition and function, so that different combinations of these proteins can result in distinct protein-DNA conformations that correspond to particular DNA sequence preferences. Such a model may be particularly plausible for bHLH proteins, because folding of the BR into an α helix is driven by its interaction with the DNA (2).
BR-DNA conformation, DNA binding specificity, and myogenesis.
The observation that the MyoD junction and K15 are not required for an appropriate DNA binding specificity (Fig. 8B and C, lanes 6 to 9; Fig. 9) supports the model that K15 is involved in other essential interactions (18). However, our experiments also pose the question of how the functional importance of A5 and T6 might be related to their effects on DNA recognition. Of the MyoD BR mutants that we have analyzed, those that do not induce myogenesis bind to DNA as homodimers with a Twist-like preference [MD(E12B) and MD(E12B-A) (Fig. 2 and 3C)]. Heterodimers of MD(E12B) with E12 prefer a heterodimer site (Fig. 5B), but with markedly diminished specificity compared to MyoD-E12 dimers (Fig. 5C, lanes 3, 5, 12, 14, 21, and 23; Fig. 6; Fig. 7A and B, lanes 1 to 12). This finding suggests that at least in part, A5 and T6 may be significant for myogenesis because they restrict the DNA binding specificity of MyoD and other myogenic bHLH proteins, so that they are less likely to bind inappropriate sites. However, other observations support a role for the A5 and T6 residues in protein-protein interactions. They have been implicated in binding to other proteins when they are not bound to DNA (26, 38), and evidence indicates that they are required for activation domain exposure (5, 29, 57) and cooperative DNA binding (3). Finally, unlike MyoD, MD(E12B) can activate transcription of a reporter only in particular cell lines, implicating the BR in protein-protein interactions (57).
In light of evidence that A5 and T6 establish the conformation of the DNA-bound BR, it is an attractive model that this effect might influence the function of myogenic bHLH proteins directly, by affecting their interactions with other proteins. Given that relatively subtle alterations of the MyoD BR and junction region can enhance MyoD DNA binding significantly [MD(AK) and MD(AAATA) (Fig. 8B and C, lanes 4, 5, and 10)], it appears likely that cooperative protein-protein interactions with the BR and junction could influence binding affinity. It has been demonstrated recently that MyoD binds cooperatively with other DNA binding proteins to a particular muscle-specific promoter (4). The E box sequences through which MyoD activates transcription in the context of this promoter can differ from those that it binds preferentially in vitro (28), suggesting that DNA sequence recognition may be influenced by interactions with cooperating proteins in vivo. In addition, interactions with cooperating proteins might be influenced in turn by the specificity of DNA sequence recognition, as suggested by evidence that for MyoD and E proteins, the choice between homo- or heterodimer formation may be dictated by the DNA binding affinities of the individual BRs (36, 59). Our findings are consistent with the idea that deceptively subtle aspects of sequence recognition could be important for the biological activity of MyoD, if they influence functionally critical interactions that might also involve K15 or other MyoD regions.
ACKNOWLEDGMENTS
For critically reading the manuscript we thank Robert Davis, Stephen Tapscott, members of the Blackwell laboratory, and Tom Ellenberger, whom we also thank for help with computer graphics. We thank Lauren Snider for advice on bacterial expression and partial purification of MyoD, and we thank Amy Chen and Jonathan Mitchell for contributing to early stages of this project. The Xenopus Twist cDNA was provided by Ralph Rupp, and the murine Twist cDNA was donated by Doug Spicer and Andrew Lassar. This work was initiated in the laboratory of Harold Weintraub, to whom T.K.B. is particularly grateful for support, advice, and inspiration.
This work was supported by grant DAMD17-94-J-4063 from the U.S. Army Breast Cancer Program to T.K.B.
REFERENCES
- 1.Aksan I, Goding C R. Targeting the microphthalmia basic helix-loop-helix-leucine zipper transcription factor to a subset of E-box elements in vitro and in vivo. Mol Cell Biol. 1998;18:6930–6938. doi: 10.1128/mcb.18.12.6930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anthony-Cahill S J, Benfield P A, Fairman R, Wasserman Z R, Brenner S L, Stafford W F, Altenbach C, Hubbell W L, DeGrado W F. Molecular characterization of helix-loop-helix peptides. Science. 1992;255:979–983. doi: 10.1126/science.1312255. [DOI] [PubMed] [Google Scholar]
- 3.Bengal E, Flores O, Rangarajan P N, Chen A, Weintraub H, Verma I M. Positive control mutations in the MyoD basic region fail to show cooperative DNA binding and transcriptional activation in vitro. Proc Natl Acad Sci USA. 1994;91:6221–6225. doi: 10.1073/pnas.91.13.6221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biesiada E, Hamamori Y, Kedes L, Sartorelli V. Myogenic basic helix-loop-helix proteins and Sp1 interact as components of a multiprotein transcriptional complex required for activity of the human cardiac alpha-actin promoter. Mol Cell Biol. 1999;19:2577–2584. doi: 10.1128/mcb.19.4.2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Black B L, Molkentin J D, Olson E N. Multiple roles for the MyoD basic region in transmission of transcriptional activation signals and interaction with MEF2. Mol Cell Biol. 1998;18:69–77. doi: 10.1128/mcb.18.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blackwell T K. Selection of protein binding sites from random nucleic acid sequences. Methods Enzymol. 1995;254:604–618. doi: 10.1016/0076-6879(95)54043-1. [DOI] [PubMed] [Google Scholar]
- 7.Blackwell T K, Huang J, Ma A, Kretzner L, Alt F W, Eisenman R N, Weintraub H. Binding of Myc proteins to canonical and noncanonical DNA sequences. Mol Cell Biol. 1993;13:5216–5224. doi: 10.1128/mcb.13.9.5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blackwell T K, Kretzner L, Blackwood E M, Eisenman R N, Weintraub H. Sequence-specific DNA binding by the c-Myc protein. Science. 1990;250:1149–1151. doi: 10.1126/science.2251503. [DOI] [PubMed] [Google Scholar]
- 9.Blackwell T K, Weintraub H. Differences and similarities in DNA-binding preferences of MyoD and E2A protein complexes revealed by binding site selection. Science. 1990;250:1104–1110. doi: 10.1126/science.2174572. [DOI] [PubMed] [Google Scholar]
- 10.Bodis S, Hemesath T, Fisher D E. Highly conserved asparagine in the basic domain of Myc is dispensable for DNA binding, transformation, and apoptosis. Biochem Mol Med. 1997;60:102–107. doi: 10.1006/bmme.1997.2575. [DOI] [PubMed] [Google Scholar]
- 11.Brennan T J, Chakraborty T, Olson E N. Mutagenesis of the myogenin basic region identifies an ancient protein motif critical for activation of myogenesis. Proc Natl Acad Sci USA. 1991;88:5675–5679. doi: 10.1073/pnas.88.13.5675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brownlie P, Ceska T, Lamers M, Romier C, Stier G, Teo H, Suck D. The crystal structure of an intact human Max-DNA complex: new insights into mechanisms of transcriptional control. Structure. 1997;5:509–520. doi: 10.1016/s0969-2126(97)00207-4. [DOI] [PubMed] [Google Scholar]
- 13.Carey J. Gel retardation. Methods Enzymol. 1991;208:103–117. doi: 10.1016/0076-6879(91)08010-f. [DOI] [PubMed] [Google Scholar]
- 14.Chakraborty T, Brennan T J, Li L, Edmondson D, Olson E N. Inefficient homooligomerization contributes to the dependence of myogenin on E2A products for efficient DNA binding. Mol Cell Biol. 1991;11:3633–3641. doi: 10.1128/mcb.11.7.3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cripps R M, Black B L, Zhao B, Lien C L, Schulz R A, Olson E N. The myogenic regulatory gene Mef2 is a direct target for transcriptional activation by Twist during Drosophila myogenesis. Genes Dev. 1998;12:422–434. doi: 10.1101/gad.12.3.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dang C V, Dolde C, Gillison M L, Kato G J. Discrimination between related DNA sites by a single amino acid residue of Myc-related basic-helix-loop-helix proteins. Proc Natl Acad Sci USA. 1992;89:599–602. doi: 10.1073/pnas.89.2.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davis R L, Cheng P F, Lassar A B, Weintraub H. The MyoD DNA binding domain contains a recognition code for muscle-specific gene activation. Cell. 1990;60:733–746. doi: 10.1016/0092-8674(90)90088-v. [DOI] [PubMed] [Google Scholar]
- 18.Davis R L, Weintraub H. Acquisition of myogenic specificity by replacement of three amino acid residues from MyoD into E12. Science. 1992;256:1027–1030. doi: 10.1126/science.1317057. [DOI] [PubMed] [Google Scholar]
- 19.Ellenberger T, Fass D, Arnaud M, Harrison S C. Crystal structure of transcription factor E47: E-box recognition by a basic region helix-loop-helix dimer. Genes Dev. 1994;8:970–980. doi: 10.1101/gad.8.8.970. [DOI] [PubMed] [Google Scholar]
- 20.Ferre-D'Amare A R, Prendergast G C, Ziff E B, Burley S K. Recognition by Max of its cognate DNA through a dimeric b/HLH/Z domain. Nature. 1993;363:38–45. doi: 10.1038/363038a0. [DOI] [PubMed] [Google Scholar]
- 21.Ferre-D'Amare A R, Pognonec P, Roeder R G, Burley S K. Structure and function of the b/HLH/Z domain of USF. EMBO J. 1994;13:180–189. doi: 10.1002/j.1460-2075.1994.tb06247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fisher D E, Parent L A, Sharp P A. High affinity DNA-binding Myc analogs: recognition by an α helix. Cell. 1993;72:467–476. doi: 10.1016/0092-8674(93)90122-7. [DOI] [PubMed] [Google Scholar]
- 23.Fisher F, Crouch D H, Jayaraman P-S, Clark W, Gillespie D A F, Goding C R. Transcription activation by Myc and Max: flanking sequences target activation to a subset of CACGTG motifs in vivo. EMBO J. 1993;12:5075–5082. doi: 10.1002/j.1460-2075.1993.tb06201.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gould K A, Bresnick E H. Sequence determinants of DNA binding by the hematopoietic helix-loop-helix transcription factor TAL1: importance of sequences flanking the E-box core. Gene Expr. 1998;7:87–101. [PMC free article] [PubMed] [Google Scholar]
- 25.Halazonetis T D, Kandil A N. Predicted structural similarities of the DNA binding domains of c-Myc and endonuclease Eco RI. Science. 1992;255:464–466. doi: 10.1126/science.1734524. [DOI] [PubMed] [Google Scholar]
- 26.Hamamori Y, Wu H Y, Sartorelli V, Kedes L. The basic domain of myogenic basic helix-loop-helix (bHLH) proteins is the novel target for direct inhibition by another bHLH protein, Twist. Mol Cell Biol. 1997;17:6563–6573. doi: 10.1128/mcb.17.11.6563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harfe B D, Branda C S, Krause M, Stern M J, Fire A. MyoD and the specification of muscle and non-muscle fates during postembryonic development of the C. elegans mesoderm. Development. 1998;125:2479–2488. doi: 10.1242/dev.125.13.2479. [DOI] [PubMed] [Google Scholar]
- 28.Huang J, Blackwell T K, Kedes L, Weintraub H. Differences between MyoD DNA binding and activation site requirements revealed by a functional random sequence selection. Mol Cell Biol. 1996;16:3893–3900. doi: 10.1128/mcb.16.7.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang J, Weintraub H, Kedes L. Intramolecular regulation of MyoD activation domain conformation and function. Mol Cell Biol. 1998;18:5478–5484. doi: 10.1128/mcb.18.9.5478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jennings B H, Tyler D M, Bray S J. Target specificities of Drosophila enhancer of split basic helix-loop-helix proteins. Mol Cell Biol. 1999;19:4600–4610. doi: 10.1128/mcb.19.7.4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kophengnavong T, Carroll A S, Blackwell T K. The SKN-1 amino-terminal arm is a DNA specificity segment. Mol Cell Biol. 1999;19:3039–3050. doi: 10.1128/mcb.19.4.3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lassar A B, Davis R L, Wright W E, Kadesch T, Murre C, Voronova A, Baltimore D, Weintraub H. Functional activity of myogenic HLH proteins requires hetero-oligomerization with E12/E47-like proteins in vivo. Cell. 1991;66:305–315. doi: 10.1016/0092-8674(91)90620-e. [DOI] [PubMed] [Google Scholar]
- 33.Lee J E. Basic helix-loop-helix genes in neural development. Curr Opin Neurobiol. 1997;7:13–20. doi: 10.1016/s0959-4388(97)80115-8. [DOI] [PubMed] [Google Scholar]
- 34.Lemercier C, To R Q, Carrasco R A, Konieczny S F. The basic helix-loop-helix transcription factor Mist1 functions as a transcriptional repressor of myoD. EMBO J. 1998;17:1412–1422. doi: 10.1093/emboj/17.5.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma P C, Rould M A, Weintraub H, Pabo C O. Crystal structure of MyoD bHLH domain-DNA complex: perspectives on DNA recognition and implications for transcriptional activation. Cell. 1994;77:451–459. doi: 10.1016/0092-8674(94)90159-7. [DOI] [PubMed] [Google Scholar]
- 36.Maleki S J, Royer C A, Hurlburt B K. MyoD-E12 heterodimers and MyoD-MyoD homodimers are equally stable. Biochemistry. 1997;36:6762–6767. doi: 10.1021/bi970262m. [DOI] [PubMed] [Google Scholar]
- 37.Michelson A. A new turn (or two) for Twist. Science. 1996;272:1449–1450. doi: 10.1126/science.272.5267.1449. [DOI] [PubMed] [Google Scholar]
- 38.Molkentin J D, Black B L, Martin J F, Olson E N. Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell. 1995;83:1125–1136. doi: 10.1016/0092-8674(95)90139-6. [DOI] [PubMed] [Google Scholar]
- 39.Molkentin J D, Olson E N. Combinatorial control of muscle development by basic helix-loop-helix and MADS-box transcription factors. Proc Natl Acad Sci USA. 1996;93:9366–9373. doi: 10.1073/pnas.93.18.9366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murre C, McCaw P S, Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989;56:777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- 41.Murre C, McCaw P S, Vaessin H, Caudy M, Jan L Y, Jan Y N, Cabrera C V, Buskin J N, Hauschka S D, Lassar A B, Weintraub H, Baltimore D. Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell. 1989;58:537–544. doi: 10.1016/0092-8674(89)90434-0. [DOI] [PubMed] [Google Scholar]
- 42.Neuhold L, Wold B. HLH forced dimers: tethering MyoD to E47 generates a dominant positive myogenic factor insulated from negative regulation by Id. Cell. 1993;74:1033–1042. doi: 10.1016/0092-8674(93)90725-6. [DOI] [PubMed] [Google Scholar]
- 43.Olson E N, Klein W H. bHLH factors in muscle development: dead lines and commitments, what to leave in and what to leave out. Genes Dev. 1994;8:1–8. doi: 10.1101/gad.8.1.1. [DOI] [PubMed] [Google Scholar]
- 44.Pabo C O, Sauer R T. Transcription factors: structural families and principles of DNA recognition. Annu Rev Biochem. 1992;61:1053–1095. doi: 10.1146/annurev.bi.61.070192.005201. [DOI] [PubMed] [Google Scholar]
- 45.Parraga A, Bellsolell L, Ferre-D'Amare A R, Burley S K. Cocrystal structure of sterol regulatory element binding protein 1a at 2.3 A resolution. Structure. 1998;6:661–672. doi: 10.1016/s0969-2126(98)00067-7. [DOI] [PubMed] [Google Scholar]
- 46.Postigo A A, Dean D C. ZEB represses transcription through interaction with the corepressor CtBP. Proc Natl Acad Sci USA. 1999;96:6683–6688. doi: 10.1073/pnas.96.12.6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schwarz J J, Chakraborty T, Martin J, Zhou J M, Olson E N. The basic region of myogenin cooperates with two transcription activation domains to induce muscle-specific transcription. Mol Cell Biol. 1992;12:266–275. doi: 10.1128/mcb.12.1.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shimizu T X, Toumoto A, Ihara K, Shimizu M, Kyogoku Y, Ogawa N, Oshima Y, Hakoshima T. Crystal structure of PHO4 bHLH domain-DNA complex: flanking base recognition. EMBO J. 1997;16:4689–4697. doi: 10.1093/emboj/16.15.4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spolar R S, Record M T., Jr Coupling of local folding to site-specific binding of proteins to DNA. Science. 1994;263:777–784. doi: 10.1126/science.8303294. [DOI] [PubMed] [Google Scholar]
- 50.Steitz T A. Structural studies of protein-nucleic acid interaction: the sources of sequence-specific binding. Q Rev Biophys. 1990;23:205–280. doi: 10.1017/s0033583500005552. [DOI] [PubMed] [Google Scholar]
- 51.Studier F W. Use of bacteriophage T7 lysozyme to improve an inducible T7 expression system. J Mol Biol. 1991;219:37–44. doi: 10.1016/0022-2836(91)90855-z. [DOI] [PubMed] [Google Scholar]
- 52.Szymanski P, Levine M. Multiple modes of dorsal-bHLH transcriptional synergy in the Drosophila embryo. EMBO J. 1995;14:2229–2238. doi: 10.1002/j.1460-2075.1995.tb07217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thayer M J, Weintraub H. A cellular factor stimulates the DNA-binding activity of MyoD and E47. Proc Natl Acad Sci USA. 1993;90:6483–6487. doi: 10.1073/pnas.90.14.6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Van Antwerp M E, Chen D G, Chang C, Prochownik E V. A point mutation in the MyoD basic domain imparts c-Myc-like properties. Proc Natl Acad Sci USA. 1992;89:9010–9014. doi: 10.1073/pnas.89.19.9010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Voronova A, Baltimore D. Mutations that disrupt DNA binding and dimer formation in the E47 helix-loop-helix protein map to distinct domains. Proc Natl Acad Sci USA. 1990;87:4722–4726. doi: 10.1073/pnas.87.12.4722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weintraub H, Davis R, Tapscott S, Thayer M, Krause M, Benezra R, Blackwell T K, Turner D, Rupp R, Hollenberg S, Zhuang Y, Lassar A. The MyoD gene family: nodal point during specification of the muscle cell lineage. Science. 1991;251:761–766. doi: 10.1126/science.1846704. [DOI] [PubMed] [Google Scholar]
- 57.Weintraub H, Dwarki V J, Verma I, Davis R, Hollenberg S, Snider L, Lassar A, Tapscott S J. Muscle-specific transcriptional activation by MyoD. Genes Dev. 1991;5:1377–1386. doi: 10.1101/gad.5.8.1377. [DOI] [PubMed] [Google Scholar]
- 58.Weintraub H, Genetta T, Kadesch T. Tissue-specific gene activation by MyoD: determination of specificity by cis-acting repression elements. Genes Dev. 1994;8:2203–2211. doi: 10.1101/gad.8.18.2203. [DOI] [PubMed] [Google Scholar]
- 59.Wendt H, Thomas R M, Ellenberger T. DNA-mediated folding and assembly of MyoD-E47 heterodimers. J Biol Chem. 1998;273:5735–5743. doi: 10.1074/jbc.273.10.5735. [DOI] [PubMed] [Google Scholar]
- 60.Yin Z, Xu X L, Frasch M. Regulation of the twist target gene tinman by modular cis-regulatory elements during early mesoderm development. Development. 1997;124:4971–4982. doi: 10.1242/dev.124.24.4971. [DOI] [PubMed] [Google Scholar]