Abstract
In Drosophila, dosage compensation—the equalization of most X-linked gene products in males and females—is achieved by a twofold enhancement of the level of transcription of the X chromosome in males relative to each X chromosome in females. A complex consisting of at least five gene products preferentially binds the X chromosome at numerous sites in males and results in a significant increase in the presence of a specific histone isoform, histone 4 acetylated at lysine 16. Recently, RNA transcripts (roX1 and roX2) encoded by two different genes have also been found associated with the X chromosome in males. We have partially purified a complex containing MSL1, -2, and -3, MOF, MLE, and roX2 RNA and demonstrated that it exclusively acetylates H4 at lysine 16 on nucleosomal substrates. These results demonstrate that the MSL complex is responsible for the specific chromatin modification characteristic of the X chromosome in Drosophila males.
Dosage compensation is a regulatory mechanism to ensure that the level of expression of genes on the single X chromosome of Drosophila males equals the level attained from the two X chromosomes in females. This equalization, achieved by a twofold increase in the rate of X-linked gene transcription in males relative to females, has been observed for a wide variety of genes with promoters of different strengths, in many cell types, and at different developmental stages. For this reason, the study of dosage compensation may provide valuable insights into the mechanisms that regulate levels of transcription.
Five genes involved in dosage compensation have been identified based on the male-specific lethality of their loss-of-function alleles (26). The products of these genes, collectively referred to as MSL proteins, colocalize to the male X chromosome, a chromosome that is also highly enriched with histone H4 acetylated at lysine 16 (6, 41). Since in all eukaryotes acetylation of the histones has been correlated directly with the establishment and regulation of transcription (reviewed in reference 28), it is likely that the MSL complex mediates its effect, at least in part, through histone acetylation. Indeed, the most recent MSL to be discovered is MOF (for “males absent on the first”), a protein with homology to acetyltransferases of the MYST family (8, 18, 33).
Another protein component of the complex is MLE (for “maleless”), an ATP-dependent RNA or DNA helicase (25). Unlike the other members of the MSL complex, MLE can be dissociated from the X chromosome by treatment with RNase, suggesting that the complex may interact with either nascent or some other form of RNA (34). This speculation has been reinforced, if not validated, by the recent discovery of two genes, roX1 and roX2 (for “RNA on the X 1 and 2”) that encode RNAs with no apparent open reading frames (1, 27). These RNAs are found only in males, and their presence depends on the MSL complex, with which they are seen to colocalize on the X chromosome (15, 20).
In this paper, we report the initial functional characterization of the MSL complex. We demonstrate that all of the MSLs are associated in a complex that also contains a roX RNA. We also show that the complex requires MOF to acetylate H4 specifically on lysine 16, the isoform of H4 that colocalizes with the MSLs on the male X chromosome.
MATERIALS AND METHODS
Antisera.
12CA5 anti-HA monoclonal antibody was purchased from Boehringer Mannheim, and M2 Flag reagents were purchased from Sigma. Rabbit anti-H4Ac16 antibodies were generously provided by Brian Turner. Anti-MSL antibodies were raised against various fragments of MSL proteins fused to GST as follows: rabbit anti-MSL1 (amino acids [aa] 423 to 1029), guinea pig anti-MSL2 (aa 78 to 529), rabbit anti-MSL3 (full length), rabbit anti-MOF (aa 748 to 827), and rabbit anti-MLE (aa 1 to 359). Secondary antisera were purchased from Jackson ImmunoResearch. Indirect-immunofluorescence images were collected with a Bio-Rad confocal microscope and false colored. Horseradish peroxidase-conjugated secondaries were used for Western analysis and detected with enhanced chemiluminescence reagents (Amersham).
Transfection and cell culture.
Schneider 2 (S2) cells were grown in SFX serum-free medium (Hyclone). The calcium phosphate method of transfection was carried out as previously described (14). cDNAs were cloned into modified versions of the pMt/Hy vector (23) to allow for tagging with hemagglutinin (HA) or Flag epitopes. Stable transformants were selected with hygromycin in medium supplemented with 10% fetal calf serum. msl2-HA, mof-HA, and mof1-HA encode full-length proteins with an extra 2 aa at the N terminus (Met-Ser-Glu) in place of the start Met and the HA epitope at the C terminus. msl3-Flag encodes full-length MSL3 with no additional amino acids at the amino terminus and the Flag epitope at the C terminus. Further details of the cloning or construction of the HA and Flag vectors will be provided on request.
Preparation of nuclear extracts.
Cells were grown to a density of 3 × 106 cells/ml in 500-ml spinner flasks (Wheaton), pelleted at 750 × g for 5 min, washed in 200 ml of cell wash buffer (10 mM HEPES [pH 7.4], 140 mM NaCl), and resuspended in 80 ml of lysis buffer (20 mM HEPES [pH 7.4], 3 mM MgCl2, 0.1% Triton X-100, 1 mM dithiothreitol [DTT], 0.5 mM phenylmethylsulfonyl fluoride [PMSF]). Cells were homogenized with 30 to 40 strokes of a Dounce homogenizer (Wheaton; pestle clearance, 0.0035 to 0.0055 in.), and the nuclei were pelleted at 2,000 × g for 5 min. The nuclei were washed once in lysis buffer and used for one of two extraction protocols, salt extraction or sonication. For salt extraction, nuclei were resuspended in 2 ml of extraction buffer (20 mM HEPES [pH 7.4], 10% glycerol, 0.35 M NaCl, 1 mM MgCl2, 0.1% Triton X-100, 1 mM DTT, 0.5 mM PMSF) and rocked at 4°C for 1 h. Following centrifugation at 15,000 × g for 5 min, the supernatant was used for immunoprecipitations. For sonication, nuclei were resuspended in 2 ml of sonication buffer (20 mM HEPES [pH 7.4], 10% glycerol, 0.1 M NaCl, 1 mM MgCl2, 0.1% Triton X-100, 1 mM DTT, 0.5 mM PMSF, 5 U of RNasin [Promega] per ml) and sonicated on ice for three rounds of 10 s (output 4, 50% duty cycle [Branson Sonifier]) with 1-min rests between rounds. Nuclei were then pelleted for 5 min at 15,000 × g, and the supernatant was used for immunoprecipitations.
Immunoprecipitation.
Crude antiserum (2 μl) or monoclonal antibody (20 μg) was bound to 10 μl of protein A-agarose (BRL-Gibco) for 1 h. Antibody-labeled beads were washed and incubated with 250 μl of salt extracts for 1 h. The beads were washed 6 times in immunoprecipitation wash buffer (20 mM HEPES [pH 7.4], 10% glycerol, 0.4 M NaCl, 1 mM MgCl2, 0.1% Triton X-100, 1 mM DTT, 0.5 mM PMSF).
Two-step immunoprecipitation.
M2 Flag agarose (40 μl) was added to 1 ml of nuclear sonicate (from MSL3-Flag expressing cells) and rocked at 4°C for 1 h. The beads were then washed five times with 1 ml of sonication buffer without RNasin. Bound proteins were eluted by the addition of 200 μl of 0.2-mg/ml Flag peptide (Asp-Tyr-Lys-Asp-Asp-Asp-Asp-Lys) and rocking for 20 min at 4°C. The eluate was passed through microspin filters (Pierce), and the flowthrough was applied to 10 μl of PI- or MSL1-bound agarose beads (see above) that had been preincubated for 30 min with 5 U of RNasin per ml in sonication buffer. After a 1-h incubation, the beads were washed six times with 400 μl of sonication buffer.
Histone acetyltransferase assays. (i) Purification of substrates.
Drosophila histones (with H1) were extracted from S2 cell nuclei with 0.4 N H2SO4, precipitated with 20% (wt/vol) trichloroacetic acid, washed in acetone containing 0.1% (vol/vol) HCl, and then washed in acetone. Mononucleosomes were purified by micrococcal nuclease digestion and sucrose gradient purification as previously described (3). Briefly, nonhistone proteins were stripped from nuclei from 109 cells in 0.35 M NaCl–20 mM HEPES (pH 7.4)–1 mM MgCl2–1 mM PMSF for 30 min at 4°C, pelleted and resuspended in 2 ml of MNase solution (4 U of micrococcal nuclease [Sigma]/109 nuclei, 5 mM MgCl2, 1 mM CaCl2), and incubated at 37°C for 10 min, and the reaction was stopped with 5 mM EGTA. Digested nuclei were pelleted, and the supernatant was adjusted to 0.6 M NaCl and loaded on a 15 to 35% sucrose gradient (10 mM HEPES [pH 7.4], 0.6 M NaCl, 0.5 mM PMSF). Fractions were checked for the presence of core histones by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and were checked for DNA size by proteinase K digestion and agarose gel electrophoresis. Mononucleosome fractions were pooled, concentrated in Centricon 100s (Amicon), and dialyzed against 10 mM Tris-HCl–1 mM EDTA (pH 7.5). Aliquots were stored at −80°C.
(ii) Assays.
A recombinant MOF fragment (aa 518 to 827) was expressed in Escherichia coli as a His-tagged fusion protein in pET19c (Novagen). Recombinant yeast proteins Gcn5p and Esa1p were described previously (9, 37). A 5-μg portion of Drosophila histones was incubated with enzyme–0.2 μCi of 3H-labeled acetyl coenzyme A (acetyl-CoA) (7.2 Ci/mmol; Amersham)–1 mM DTT–1 mM PMSF–10% glycerol–50 mM Tris (pH 7.5) in a 50 μl-volume at room temperature (ca. 22°C) for 20 min. Aliquots were separated by SDS-PAGE, stained with Coomassie blue, and fluorographed with Amplify (Pharmacia) to identify which histones were labeled. Acetyltransferase assays with recombinant proteins were performed as above with the following changes: 2 μg of Drosophila nucleosomes was incubated with 10 μl of protein A beads-immune complexes and 0.2 μCi of 3H AcCoA in a volume of 30 μl for 45 min with occasional mixing. The beads were spun down, and the supernatant was used for SDS-PAGE and fluorography.
Acid-urea gels.
Aliquots of acetylation reaction mixtures were incubated with HCl (0.2 M final concentration) for 10 min on ice, trichloroacetic acid was added to 20% (wt/vol), and the mixture was incubated on ice overnight. After being washed with acetone, the pellets were resuspended in sample buffer and processed for discontinuous acid-urea gel electrophoresis as described previously (7). The gel was stained with Coomassie blue and processed for fluorography. Acid-extracted histones from S2 cells treated with butyrate were used as electrophoretic markers (16).
Determination of lysine specificity.
The acid-urea gel was blotted to a polyvinylidene difluoride (PVDF) membrane (Millipore P-SQ) in 0.7 M acetic acid at 1 A for 2 h. The blot was stained briefly with Coomassie blue. The monoacetylated band was cut out and treated for deblocking and microsequencing as previously described (38).
Northern analysis and RT-PCR.
RNA was extracted from adult male and female flies as previously described (2). RNA was extracted from S2 cells as specified by the manufacturer for suspension cells (RNeasy kit; Qiagen). Northern blotting was done according to standard procedures (36). roX1 (nucleotides 1 to 1945; GenBank accession no. U85980) and roX2 (nucleotides 158 to 1244; GenBank accession no. U85981) fragments were amplified by PCR from Samarkand genomic DNA. rp49 DNA was used as a control (29). The probes were generated by random priming (36).
Washed immunoprecipitation pellets (PI and MSL1) were first spiked with 5 pg of kanamycin kinase mRNA (Promega). RNA was then extracted with the RNA Easy Kit, treated with RQ1 DNase, and repurified. First-strand cDNA synthesis was primed with the roX2 specific primer (5′-CTTCAGTTTGCATTGCGACTTG-3′) and the kanamycin kinase primer (5′-CAGCCATTACGCTCGTCATC-3′) and was performed at 42°C for 1 h with Sensiscript reverse transcriptase (Qiagen). Parallel controls without reverse transcriptase were also performed. Reverse transcription (RT) products were amplified with the above primers and primer 5′-GCCATCGAAAGGGTAAATTGG-3′ for roX2 or 5′-GCAATCAGGTGCGACAATCTATC-3′ for kanamycin kinase.
Nucleotide sequence accession number.
A cDNA sequence corresponding to a 0.6-kb roX2 RNA has been submitted to GenBank (accession no. AF195878).
RESULTS
MOF is a histone acetyltransferase.
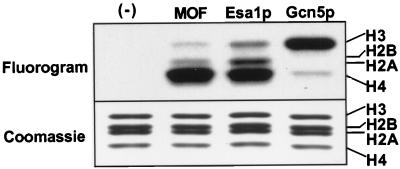
We expressed a cDNA fragment containing the putative MOF catalytic domain (aa 518 to 827) and determined that the recombinant peptide can acetylate Drosophila histones with a preference for histone H4 (Fig. 1). This pattern is similar to that for a related yeast protein, Esa1p (37), but different from that of the Gcn5-related enzymes (9). Since we were unsuccessful at expressing active full-length MOF, we proceeded to isolate MOF as a component of a partially purified MSL complex.
FIG. 1.
MOF acetylates histone H4. A recombinant fragment of MOF was expressed in E. coli and assayed for acetyltransferase activity on free histones. MOF acetylates histones predominantly on histone H4 and to a lesser extent on histones H3 and H2A. For comparison, the activities of yeast acetyltransferases Gcn5p and MOF-related Esa1p are shown.
Partial purification of the MSL complex.
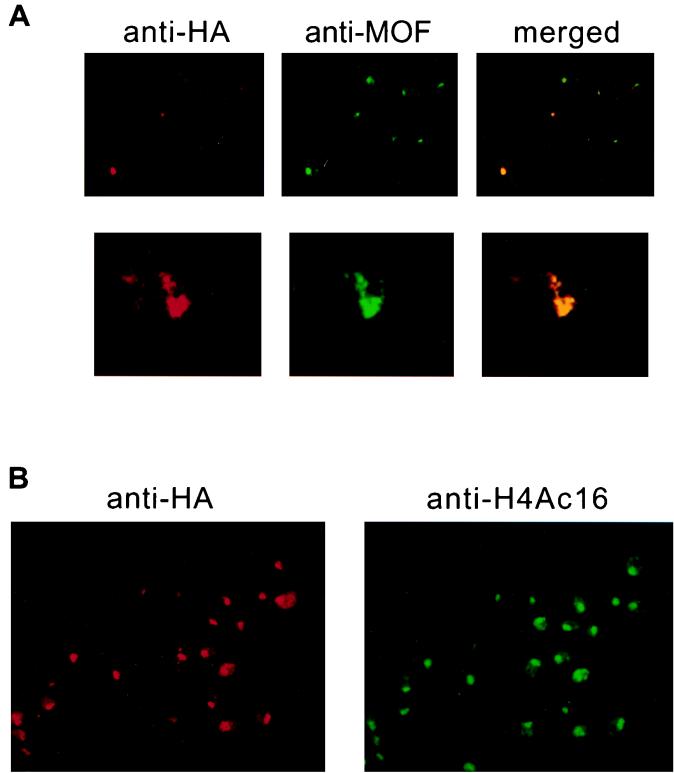
We have chosen to use tissue culture cells for the initial characterization of the MSL complex. S2 cells are male, based on the following criteria: they do not express the Sxl (Sex-lethal) gene product, which is necessary for female differentiation (35), and they express MSL2, a limiting component of the dosage compensation machinery whose synthesis is prevented by SXL (4, 5, 21, 22). S2 cells can be stably transfected, allowing the use of commercially available antibodies recognizing epitope tags. Transient transfection of S2 cells with MSL2 tagged at its carboxy terminus with the HA epitope revealed that the localization of the HA epitope is coincident with the location of endogenous MOF (Fig. 2A). After selection with hygromycin, most cells exhibit HA staining on the male X chromosome, the location of which is revealed by antibodies to H4Ac16 (Fig. 2B).
FIG. 2.
Epitope-tagged MSL2 colocalizes with MOF and histone H4 acetylated at lysine 16 (H4Ac16) on the X chromosome in S2 cells. (A) Cells were transfected with a construct expressing MSL2 fused to the HA epitope. Tagged MSL2 is detected with the 12CA5 monoclonal antibody, while endogenous MOF is detected with a rabbit polyclonal antibody to the C terminus of MOF. The top panel shows that a small fraction of the cells contain transfected MSL2-HA, which is seen colocalizing with endogenous MOF. The bottom panel shows an enlargement of a nucleus where both antibodies can be seen painting a chromosome that traverses the nucleus. (B) Stable cell lines were selected with hygromycin, and a line that expressed a low level of basal (uninduced) expression was chosen for further study. In this line, basal levels of MSL2-HA can be detected along the length of the X chromosome, coincident with H4Ac16 and the other MSLs (not shown). Occasional areas of nonoverlap may reflect the presence of partial or nonfunctional complexes.
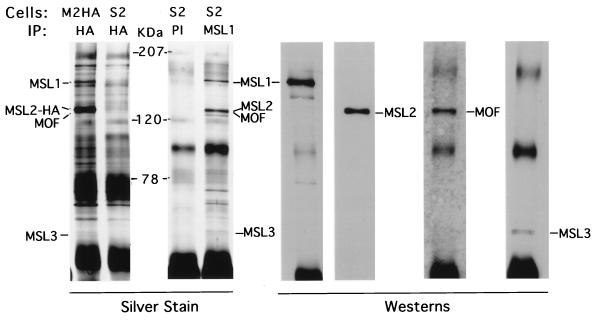
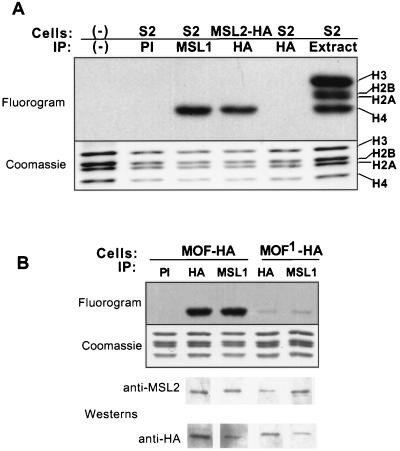
Immunoprecipitation of nuclear extracts from MSL2-HA cells with the 12CA5 (anti-HA) antiserum resulted in the same proteins as those obtained from S2 cells with an MSL1 antiserum (Fig. 3). As can be seen from comparisons of silver-stained gels and Western blots, major bands seen by silver staining correspond to known MSLs, with the exception of MLE, which is difficult to detect under these conditions (see below).
FIG. 3.
Major immunoprecipitated proteins detected by silver staining correspond to known MSLs. The 12CA5 antibody (HA) generates the same set of proteins from MSL2-HA (M2HA) nuclear extracts that the MSL1 antiserum generates from S2 cell extracts; differences in stochiometry are ascribable to the overexpression of HA-tagged MSL2. This set of proteins is absent from HA or control immunoglobulin G (PI) precipitates from (untransfected) S2 cells. Western analysis of the MSL1 immunoprecipitate shows that the major silver-stained bands correspond to known MSLs. Rabbit anti-MSL1 serum detects a 170-kDa band, guinea pig anti-MSL2 detects a 135-kDa band, rabbit anti-MOF detects a 135-kDa band, and rabbit anti-MSL3 detects a 58-kDa doublet of bands. Molecular masses are indicated in kilodaltons (KDa).
In salivary gland nuclei, MLE is released from the male X chromosome with RNase treatment (34). Furthermore, the roX1 and roX2 RNAs are found along the X chromosome with a distribution that mimics that of the MSL complex (15). Therefore, we wished to determine if we could obtain a partially purified complex containing MLE and a roX RNA and whether the presence of either of these components depended on the other.
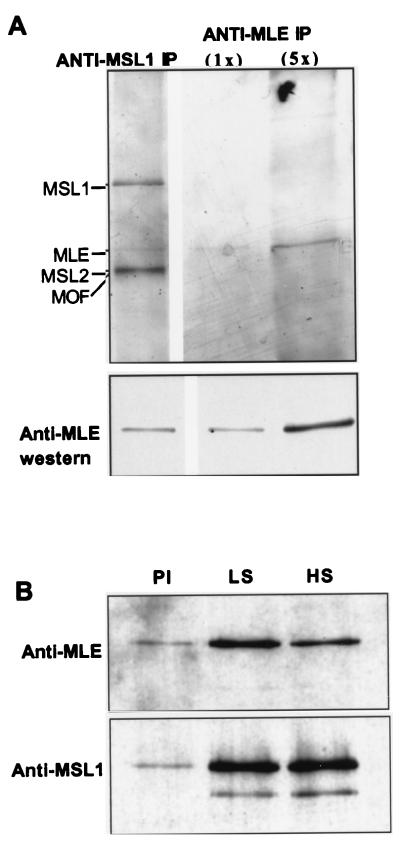
First, we developed “RNA-friendly” conditions to increase our chances of purifying MLE and roX RNA-containing complex. Our method involved a cell line expressing Flag-tagged MSL3 and sonication under low-salt conditions, immunoprecipitation with Flag antibodies followed by peptide elution, and a second immunoprecipitation with an MSL antibody or with the corresponding preimmune serum. By using this two-step procedure, we can detect a faint band by silver staining that corresponds to MLE protein (Fig. 4A). Clear enrichment of MLE was seen in the MSL1 immunoprecipitate relative to the preimmune serum (Fig. 4B). However, following a brief treatment with 0.4 M NaCl, the MLE levels were significantly reduced.
FIG. 4.
MLE association with the MSL complex. Significant levels of MLE are detected with other MSLs when immunoprecipitations are performed under low-salt conditions (see Materials and Methods). MSL complexes were eluted from M2-Flag agarose and subjected to a second immunoprecipitation with anti-MSL1. (A) A silver-stained protein is visible between MSL1 and MSL2/MOF. Two concentrations (1× and 5×) of a stringently washed anti-MLE immunoprecipitate (IP) were loaded on the same gel; correlation between silver staining and Western staining intensities as well as comigration by SDS-PAGE, confirms that this band is MLE. (B) Comparison of low-salt (LS) and high-salt (HS) washing conditions reveals a salt-sensitive association of MLE with the other MSLs. As seen by Western analysis, significant levels of MLE are released from immunoprecipitates when incubated with high-salt buffers but not low-salt buffers; MSL1 levels are unaffected. A preimmune control (PI) was washed under low-salt conditions and reveals a low level of contaminating MSLs in these preparations.
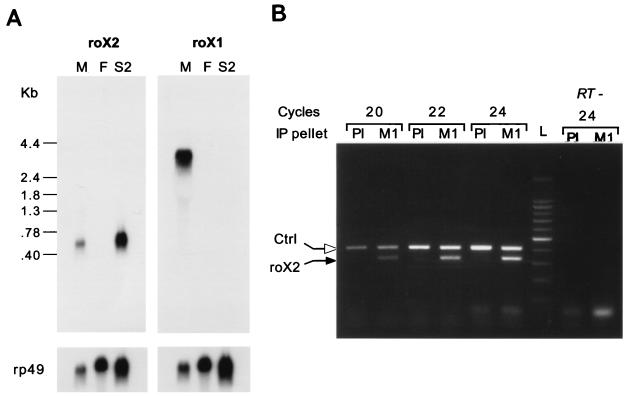
To determine if roX RNAs are expressed in S2 cells, we performed Northern blot analysis and observed that roX2, but not roX1, was expressed in these cells (Fig. 5A), consistent with the observation that roX1 is dispensable in flies (27). The size of the major roX2 transcript observed by Northern analysis was ca. 600 nucleotides. This size is consistent with the results reported by Amrein and Axel (1) in their Fig. 2, which demonstrate that roX2 transcripts migrate slightly faster than the 600-nucleotide rp49 mRNA (29). To test if roX2 RNA is present in the MLE-containing immunoprecipitates, RNA was extracted from the immunoprecipitation pellets and RT-PCR was performed with roX2-specific primers in the linear range. The results show a clear enrichment of roX2 RNA in the immune over the preimmune serum precipitates (Fig. 5B).
FIG. 5.
roX2 is expressed in S2 cells and associates with the MSL proteins. (A) Northern analysis shows that both roX RNAs are expressed in adult Drosophila males (M) but not females (F); only roX2 was detected in S2 cells. The filters were reprobed for rp49 RNA as a loading control. (B) RNA was extracted from anti-MSL1 immunoprecipitates (M1) or the corresponding preimmune serum (PI) and subjected to RT-PCR. Agarose gel electrophoresis of PCR products and staining with ethidium bromide detected significant levels of roX2 RNA in the anti-MSL1 immunoprecipitates, while a lower level of roX2 RNA (1 to 2%) was detected in the preimmune serum. This level of contamination is consistent with the amount of contaminating MSL proteins observed in these immunoprecipitates (Fig. 4B). A kanamycin kinase transcript was used to monitor variation in processing of samples; comparable levels of this control RNA (Ctrl) were detected by RT-PCR in the preimmune and anti-MSL1 immunoprecipitates.
The MSL complex specifically acetylates lysine 16 of histone H4.
When MSL-containing immunoprecipitates were incubated with nucleosomal substrates, significant acetyltransferase activity toward histone H4 was detected (Fig. 6A). MSL1 immunoprecipitates from S2 nuclear extracts and 12CA5 immunoprecipitates from MSL2-HA nuclear extracts contain H4-specific acetyltransferase activity, while control immunoglobulin G or 12CA5 immunoprecipitates from S2 cells do not.
FIG. 6.
The MSL complex acetylates nucleosomal H4 in a MOF-dependent manner. (A) Immunoprecipitates were assayed for acetyltransferase activity toward mononucleosomes and processed for SDS-PAGE and fluorography. Both MSL1 immunoprecipitates (IP) from S2 cells and 12CA5 immunoprecipitates (HA) from MSL2-HA-expressing cells acetylate nucleosomes specifically on histone H4. Preimmune serum (PI) or 12CA5 monoclonal antibody does not immunoprecipitate any histone acetyltransferase activity from S2 cells. (B) Acetyltransferase activity was also assayed from cells transfected with the HA-tagged mof+ or HA-tagged mof1 allele (Gly-to-Glu mutation at residue 691). Western analysis demonstrates that the extracts contain complexes with large amounts of HA-tagged MOF or MOF1. Histone acetyltransferase activity from MOF1-containing complexes is dramatically reduced.
To demonstrate that the acetyltransferase activity of the MSL complex is ascribable to MOF, we purified complexes containing either wild-type MOF or a protein produced by the mutant allele mof1 (18). This allele is a point mutation resulting in a glycine-to-glutamic acid replacement at the most highly conserved residue of the acetyl-CoA binding domain (G691E). We overexpressed wild-type MOF-HA or G691E MOF-HA in S2 cells and immunoprecipitated them with anti-HA antibodies to obtain complexes with only transfected MOF fusion proteins. Immunoprecipitates from G691E cells have markedly reduced acetylation, consistent with the conclusion that MOF is the sole acetyltransferase in the MSL complex (Fig. 6B).
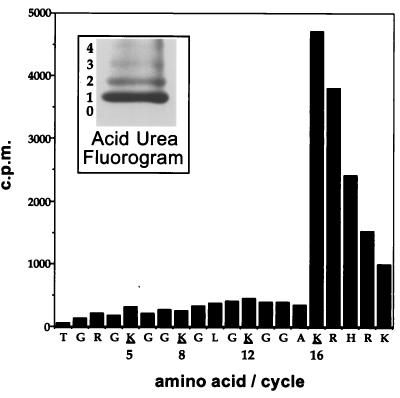
Given the specificity of the MSL complex toward H4, we wished to determine which particular lysines were acetylated. When acetylated histones were separated by acid-urea gel electrophoresis, predominantly monoacetylated H4 was created (Fig. 7, inset). A similar acid-urea gel was blotted to PVDF, and the mono-acetylated band was subjected to microsequencing. Counts were found at lysine 16, while other potential acetylation sites (at position 5, 8, or 12) were unlabeled (Fig. 7). This result provides a causative link between the presence of histone H4 acetylated at lysine 16 and the MSL complex on the X chromosome in Drosophila males.
FIG. 7.
The MSL complex specifically acetylates lysine 16 of H4 on Drosophila nucleosomes. The inset shows a fluorogram of an acid-urea gel-separated H4 that has been acetylated by the MSL complex, in the presence of 3H-labeled acetyl-CoA and mononucleosomes. Most of the [3H]acetate is found on the monoacetylated band. A similar acid-urea gel was blotted to PVDF, and the monoacetylated band was cut out and subjected to microsequencing. The counts per minute released at each cycle are plotted against the residue sequenced. Essentially all of the counts are found at lysine 16, except for the sequencing lag that results in the release of residual counts at positions 17 to 20. Additionally, phenylthiohydentoin analysis detected acetyl-lysine only at position 16, not at positions 5, 8, and 12.
DISCUSSION
The last few years have seen a remarkable increase in our knowledge of macromolecular complexes that modify chromatin and thereby modulate gene activity. These complexes interact with nucleosomal proteins, i.e., histones and in some cases with components of the preinitiation complex to regulate the level of transcription of a large number of different genes. At present, these multiprotein complexes can be placed into two broad categories: complexes that use the energy of ATP hydrolysis to alter nucleosomal conformation, and complexes that alter chromatin conformation via the modification of histone tails (reviewed in reference 45). A few examples of the former category are the RSC and SWI/SNF complexes in yeast (10, 11, 32), and the Brahma, NURF, CHRAC, and ACF complexes in Drosophila (19, 31, 40, 43). In these complexes the ATP requirement is ascribed to the activity of proteins which contain domains characteristic of helicases but which do not exhibit in vitro helicase activity. Complexes in the second category, including SAGA and NuA4 from yeast (17), are thought to function by targeting specific histone acetyltransferases to their site of action for the activation of transcription (24, 42). As described in this paper, the MSL complex includes both an ATP-dependent helicase (MLE) and a histone acetyltransferase (MOF); it also contains one or more noncoding RNAs of unknown function (roX).
It is notable that S2 cells express roX2 but not roX1 RNA. This, however, is consistent with the observation that these two RNAs are functionally redundant (15, 27). The roX RNAs differ vastly in size (3.5 and 0.6 kb for roX1 and roX2, respectively) and lack extensive similarities, although a small region of unknown function was identified by Franke and Baker (15).
One function of roX RNAs may be to help maintain the association of MLE with the other MSLs. MLE is distantly related to the SWI2 and ISWI ATPases, but it has been shown to exhibit RNA/DNA helicase, ATPase, and single-stranded RNA-DNA binding activities in vitro (25). As previously mentioned, MLE can be released from larval salivary gland chromosomes by RNase treatment (34) and appears to associate rather weakly with the other protein components of the complex (13). Our ability to detect significant amounts of MLE in the complex is probably due to shortened processing time, low-ionic-strength buffers, and RNase-free conditions.
MOF is a member of the MYST family of histone acetyltransferases. Two other members of this family, Esa1p and Tip60, acetylate histones H2A, H3, and especially H4 when tested in vitro as fusion proteins or as catalytic fragments (12, 37, 46). The specificity of the Esa1p-containing NuA4 complex, with respect to the sites of histone H4 acetylation, is identical to that of the recombinant Esa1p: lysines 5, 8, 12 and 16 (37). Recently, a protein (p80) has been identified in Tetrahymena that acetylates nucleosomal H4 in an identical pattern (30). In contrast, the MSL complex shows a clear preference for lysine 16.
One proposed role of histone acetylation in transcription is to help promote transcription factor access to DNA for the purposes of initiating transcription. Gcn5p activity is responsible for the acetylation of histone H3 at the HIS3 promoter, which results in gene activation (24). Additionally, histone acetylation may facilitate elongation by RNA polymerase. This is supported by in vitro experiments with hyperacetylated histones and RNA polymerase III (39) and by the recent discovery that an RNA polymerase II-associated elongation complex contains acetyltransferase activity (44).
In Drosophila, the preponderance of histone H4 acetylated at lysine 16 is found associated with the MSL complex on the X chromosome in males (6, 41). Since the other isoforms of histone H4 are found in similar amounts on both male and female X chromosomes (41), one might infer that MSL complexes are targeted to chromatin already acetylated at other lysines; the complex then adds an acetate on lysine 16 to further increase transcription. Whether the increase in transcription occurs at initiation or elongation is currently under study.
Finally, it should be noted that the MSL complex of Drosophila enhances transcription not by orders of magnitude (as appears to be the case with some of the other chromatin remodeling complexes) but, on average, only by a factor of 2. Therefore, it may be a very good model to study how chromatin-remodeling complexes do, in fact, achieve particular levels of gene activity.
ACKNOWLEDGMENTS
We thank Brian Turner (Birmingham, United Kingdom) for providing us with the anti-H4Ac16 sera and Michael Koelle for the pMK33/pMtHy vector. We thank Melissa Gilbert and Hisa Tajima for technical assistance and members of the Allis and Lucchesi laboratories for helpful discussions.
This work was supported by grants from the National Institutes of Health to J.C.L. (GM15961) and to C.D.A. (GM53512) and from the Human Frontiers of Science Program to C.D.A.
REFERENCES
- 1.Amrein H, Axel R. Genes expressed in neurons of adult male Drosophila. Cell. 1997;88:459–469. doi: 10.1016/s0092-8674(00)81886-3. [DOI] [PubMed] [Google Scholar]
- 2.Ashburner M. Drosophila: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 3.Ausio J, van Holde K E. Histone hyperacetylation: its effects on nucleosome conformation and stability. Biochemistry. 1986;25:1421–1428. doi: 10.1021/bi00354a035. [DOI] [PubMed] [Google Scholar]
- 4.Bashaw G J, Baker B S. The msl-2 dosage compensation gene of Drosophila encodes a putative DNA-binding protein whose expression is sex specifically regulated by Sex-lethal. Development. 1995;121:3245–3258. doi: 10.1242/dev.121.10.3245. [DOI] [PubMed] [Google Scholar]
- 5.Bashaw G J, Baker B S. The regulation of the Drosophila msl-2 gene reveals a function for Sex-lethal in translational control. Cell. 1997;89:789–798. doi: 10.1016/s0092-8674(00)80262-7. [DOI] [PubMed] [Google Scholar]
- 6.Bone J R, Lavender J, Richman R, Palmer M J, Turner B M, Kuroda M I. Acetylated histone H4 on the male X chromosome is associated with dosage compensation in Drosophila. Genes Dev. 1994;8:96–104. doi: 10.1101/gad.8.1.96. [DOI] [PubMed] [Google Scholar]
- 7.Bonner W M, West M H, Stedman J D. Two-dimensional gel analysis of histones in acid extracts of nuclei, cells, and tissues. Eur J Biochem. 1980;109:17–23. doi: 10.1111/j.1432-1033.1980.tb04762.x. [DOI] [PubMed] [Google Scholar]
- 8.Borrow J, Stanton V P, Jr, Andresen J M, Becher R, Behm F G, Chaganti R S, Civin C I, Disteche C, Dube I, Frischauf A M, Horsman D, Mitelman F, Volinia S, Watmore A E, Housman D E. The translocation t(8;16)(p11;p13) of acute myeloid leukaemia fuses a putative acetyltransferase to the CREB-binding protein. Nat Genet. 1996;14:33–41. doi: 10.1038/ng0996-33. [DOI] [PubMed] [Google Scholar]
- 9.Brownell J E, Zhou J, Ranalli T, Kobayashi R, Edmondson D G, Roth S Y, Allis C D. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 10.Cairns B R, Kim Y J, Sayre M H, Laurent B C, Kornberg R D. A multisubunit complex containing the SWI1/ADR6, SWI2/SNF2, SWI3, SNF5, and SNF6 gene products isolated from yeast. Proc Natl Acad Sci USA. 1994;91:1950–1954. doi: 10.1073/pnas.91.5.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cairns B R, Lorch Y, Li Y, Zhang M, Lacomis L, Erdjument-Bromage H, Tempst P, Du J, Laurent B, Kornberg R D. RSC, an essential, abundant chromatin-remodeling complex. Cell. 1996;87:1249–1260. doi: 10.1016/s0092-8674(00)81820-6. [DOI] [PubMed] [Google Scholar]
- 12.Clarke A S, Lowell J E, Jacobson S J, Pillus L. Esa1p is an essential histone acetyltransferase required for cell cycle progression. Mol Cell Biol. 1999;19:2515–2526. doi: 10.1128/mcb.19.4.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Copps K, Richman R, Lyman L M, Chang K A, Rampersad-Ammons J, Kuroda M I. Complex formation by the Drosophila MSL proteins: role of the MSL2 RING finger in protein complex assembly. EMBO J. 1998;17:5409–5417. doi: 10.1093/emboj/17.18.5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Di Nocera P P, Dawid I B. Transient expression of genes introduced into cultured cells of Drosophila. Proc Natl Acad Sci USA. 1983;80:7095–7098. doi: 10.1073/pnas.80.23.7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Franke A, Baker B S. The roX1 and roX2 RNAs are essential components of the compensasome, which mediates dosage compensation in Drosophila. Mol Cell. 1999;4:117–122. doi: 10.1016/s1097-2765(00)80193-8. [DOI] [PubMed] [Google Scholar]
- 16.Georgel P T, Tsukiyama T, Wu C. Role of histone tails in nucleosome remodeling by Drosophila NURF. EMBO J. 1997;16:4717–4726. doi: 10.1093/emboj/16.15.4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grant P A, Duggan L, Cote J, Roberts S M, Brownell J E, Candau R, Ohba R, Owen-Hughes T, Allis C D, Winston F, Berger S L, Workman J L. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- 18.Hilfiker A, Hilfiker-Kleiner D, Pannuti A, Lucchesi J C. mof, a putative acetyl transferase gene related to the Tip60 and MOZ human genes and to the SAS genes of yeast, is required for dosage compensation in Drosophila. EMBO J. 1997;16:2054–2060. doi: 10.1093/emboj/16.8.2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ito T, Bulger M, Pazin M J, Kobayashi R, Kadonaga J T. ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell. 1997;90:145–155. doi: 10.1016/s0092-8674(00)80321-9. [DOI] [PubMed] [Google Scholar]
- 20.Kelley R L, Meller V H, Gordadze P R, Roman G, Davis R L, Kuroda M I. Epigenetic spreading of the Drosophila dosage compensation complex from roX RNA genes into flanking chromatin. Cell. 1999;98:513–522. doi: 10.1016/s0092-8674(00)81979-0. [DOI] [PubMed] [Google Scholar]
- 21.Kelley R L, Solovyeva I, Lyman L M, Richman R, Solovyev V, Kuroda M I. Expression of msl-2 causes assembly of dosage compensation regulators on the X chromosomes and female lethality in Drosophila. Cell. 1995;81:867–877. doi: 10.1016/0092-8674(95)90007-1. [DOI] [PubMed] [Google Scholar]
- 22.Kelley R L, Wang J, Bell L, Kuroda M I. Sex lethal controls dosage compensation in Drosophila by a non-splicing mechanism. Nature. 1997;387:195–199. doi: 10.1038/387195a0. [DOI] [PubMed] [Google Scholar]
- 23.Koelle M R, Talbot W S, Segraves W A, Bender M T, Cherbas P, Hogness D S. The Drosophila EcR gene encodes an ecdysone receptor, a new member of the steroid receptor superfamily. Cell. 1991;67:59–77. doi: 10.1016/0092-8674(91)90572-g. [DOI] [PubMed] [Google Scholar]
- 24.Kuo M H, Zhou J, Jambeck P, Churchill M E, Allis C D. Histone acetyltransferase activity of yeast Gcn5p is required for the activation of target genes in vivo. Genes Dev. 1998;12:627–639. doi: 10.1101/gad.12.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee C G, Chang K A, Kuroda M I, Hurwitz J. The NTPase/helicase activities of Drosophila maleless, an essential factor in dosage compensation. EMBO J. 1997;16:2671–2681. doi: 10.1093/emboj/16.10.2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lucchesi J C. Dosage compensation in flies and worms: the ups and downs of X-chromosome regulation. Curr Opin Genet Dev. 1998;8:179–184. doi: 10.1016/s0959-437x(98)80139-1. [DOI] [PubMed] [Google Scholar]
- 27.Meller V H, Wu K H, Roman G, Kuroda M I, Davis R L. roX1 RNA paints the X chromosome of male Drosophila and is regulated by the dosage compensation system. Cell. 1997;88:445–457. doi: 10.1016/s0092-8674(00)81885-1. [DOI] [PubMed] [Google Scholar]
- 28.Mizzen C A, Allis C D. Linking histone acetylation to transcriptional regulation. Cell Mol Life Sci. 1998;54:6–20. doi: 10.1007/s000180050121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Connell P O, Rosbash M. Sequence, structure, and codon preference of the Drosophila ribosomal protein 49 gene. Nucleic Acids Res. 1984;12:5495–5513. doi: 10.1093/nar/12.13.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohba R, Steger D J, Brownell J E, Mizzen C A, Cook R G, Cote J, Workman J L, Allis C D. A novel H2A/H4 nucleosomal histone acetyltransferase in Tetrahymena thermophila. Mol Cell Biol. 1999;19:2061–2068. doi: 10.1128/mcb.19.3.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Papoulas O, Beek S J, Moseley S L, McCallum C M, Sarte M, Shearn A, Tamkun J W. The Drosophila trithorax group proteins BRM, ASH1 and ASH2 are subunits of distinct protein complexes. Development. 1998;125:3955–3966. doi: 10.1242/dev.125.20.3955. [DOI] [PubMed] [Google Scholar]
- 32.Peterson C L, Dingwall A, Scott M P. Five SWI/SNF gene products are components of a large multisubunit complex required for transcriptional enhancement. Proc Natl Acad Sci USA. 1994;91:2905–2908. doi: 10.1073/pnas.91.8.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reifsnyder C, Lowell J, Clarke A, Pillus L. Yeast SAS silencing genes and human genes associated with AML and HIV-1 Tat interactions are homologous with acetyltransferases. Nat Genet. 1996;14:42–49. doi: 10.1038/ng0996-42. [DOI] [PubMed] [Google Scholar]
- 34.Richter L, Bone J R, Kuroda M I. RNA-dependent association of the Drosophila maleless protein with the male X chromosome. Genes Cells. 1996;1:325–336. doi: 10.1046/j.1365-2443.1996.26027.x. [DOI] [PubMed] [Google Scholar]
- 35.Ryner L C, Baker B S. Regulation of doublesex pre-mRNA processing occurs by 3′-splice site activation. Genes Dev. 1991;5:2071–2085. doi: 10.1101/gad.5.11.2071. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 37.Smith E R, Eisen A, Gu W, Sattah M, Pannuti A, Zhou J, Cook R G, Lucchesi J C, Allis C D. ESA1 is a histone acetyltransferase that is essential for growth in yeast. Proc Natl Acad Sci USA. 1998;95:3561–3565. doi: 10.1073/pnas.95.7.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sobel R E, Cook R G, Allis C D. Non-random acetylation of histone H4 by a cytoplasmic histone acetyltransferase as determined by novel methodology. J Biol Chem. 1994;269:18576–18582. [PubMed] [Google Scholar]
- 39.Tse C, Sera T, Wolffe A P, Hansen J C. Disruption of higher-order folding by core histone acetylation dramatically enhances transcription of nucleosomal arrays by RNA polymerase III. Mol Cell Biol. 1998;18:4629–4638. doi: 10.1128/mcb.18.8.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsukiyama T, Daniel C, Tamkun J, Wu C. ISWI, a member of the SWI2/SNF2 ATPase family, encodes the 140 kDa subunit of the nucleosome remodeling factor. Cell. 1995;83:1021–1026. doi: 10.1016/0092-8674(95)90217-1. [DOI] [PubMed] [Google Scholar]
- 41.Turner B M, Birley A J, Lavender J. Histone H4 isoforms acetylated at specific lysine residues define individual chromosomes and chromatin domains in Drosophila polytene nuclei. Cell. 1992;69:375–384. doi: 10.1016/0092-8674(92)90417-b. [DOI] [PubMed] [Google Scholar]
- 42.Utley R T, Ikeda K, Grant P A, Cote J, Steger D J, Eberharter A, John S, Workman J L. Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature. 1998;394:498–502. doi: 10.1038/28886. [DOI] [PubMed] [Google Scholar]
- 43.Varga-Weisz P D, Wilm M, Bonte E, Dumas K, Mann M, Becker P B. Chromatin-remodelling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature. 1997;388:598–602. doi: 10.1038/41587. . (Erratum, 389:1003, 1997.) [DOI] [PubMed] [Google Scholar]
- 44.Wittschieben B O, Otero G, de Bizemont T, Fellows J, Erdjument-Bromage H, Ohba R, Li Y, Allis C D, Tempst P, Svejstrup J Q. A novel histone acetyltransferase is an integral subunit of elongating RNA polymerase II holoenzyme. Mol Cell. 1999;4:123–128. doi: 10.1016/s1097-2765(00)80194-x. [DOI] [PubMed] [Google Scholar]
- 45.Wolffe A P, Hayes J J. Chromatin disruption and modification. Nucleic Acids Res. 1999;27:711–720. doi: 10.1093/nar/27.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamamoto T, Horikoshi M. Novel substrate specificity of the histone acetyltransferase activity of HIV-1-Tat interactive protein Tip60. J Biol Chem. 1997;272:30595–30598. doi: 10.1074/jbc.272.49.30595. [DOI] [PubMed] [Google Scholar]