Abstract
Therapeutic bacteriophages, commonly called as phages, are a promising potential alternative to antibiotics in the management of bacterial infections of a wide range of organisms including cultured fish. Their natural immunogenicity often induces the modulation of a variated collection of immune responses within several types of immunocytes while promoting specific mechanisms of bacterial clearance. However, to achieve standardized treatments at the practical level and avoid possible side effects in cultivated fish, several improvements in the understanding of their biology and the associated genomes are required. Interestingly, a particular feature with therapeutic potential among all phages is the production of lytic enzymes. The use of such enzymes against human and livestock pathogens has already provided in vitro and in vivo promissory results. So far, the best-understood phages utilized to fight against either Gram-negative or Gram-positive bacterial species in fish culture are mainly restricted to the Myoviridae and Podoviridae, and the Siphoviridae, respectively. However, the current functional use of phages against bacterial pathogens of cultured fish is still in its infancy. Based on the available data, in this review, we summarize the current knowledge about phage, identify gaps, and provide insights into the possible bacterial control strategies they might represent for managing aquaculture-related bacterial diseases.
Keywords: aquaculture, bacteriophages, disease management, fish, immunology, lytic enzymes, pathogens
1. Phage Biology and Spatial Distribution
Bacteriophages or phages, in short, are an alternative to antimicrobials to fight against bacteria due to their unique host range that provides them with an excellent specificity. In addition, contrary to the antibiotic’s negative physiological effects on the host and the generation of bacterial resistance, the use of phages is eco-friendly and without major drawbacks [1,2]. Besides, phages produce lytic enzymes with the ability to act directly on the bacterial cell wall. An important associated advantage is that phages are ubiquitous to all fresh and saltwater environments representing a virtually unlimited source of virions and lytic enzymes. In seawater, the number and variety of phages have a direct and crucial impact on the variability of microbial communities which directly modulate the global biogeochemical cycles in the oceans [3,4]. Quantitative analyses of marine waters using transmission electron microscopy demonstrated that non-tailed viruses are the most abundant, followed by tailed viruses of the families Myoviridae and Podoviridae [5]. This example represents a huge gene reservoir across Earth’s ecosystems. Despite the great awakening interest in phage therapy and the discovery of a vast reservoir of new genes available in the phages of aquatic ecosystems, the composition the phage populations in the different fish species in aquaculture, either from freshwater or saltwater environments are not yet fully understood.
2. Phage’s Life Cycle
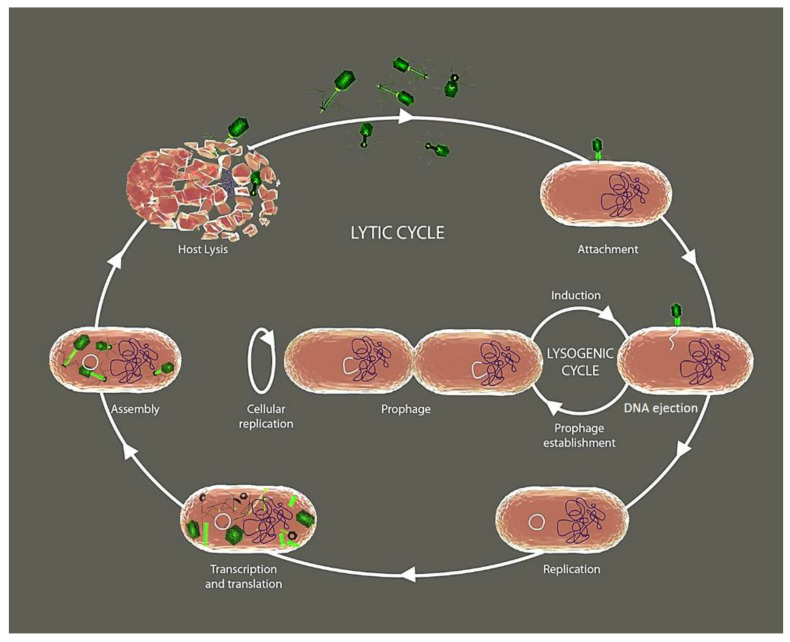
The phages like any other viruses depend on the metabolism of their bacterial host for reproduction. During the reproductive process, most phage types completely consume the resources of their host and kill them when releasing their progeny [6]. Initially, phages must infect their host bacteria through the binding of specific receptors that selectively sense specific components of the target bacterial cell wall such as the lipopolysaccharide in Gram-negative, or peptidoglycan in Gram-positive, capsular polysaccharides, and superficial appendages such as pili and flagella [7,8,9]. Following the classical viral reproductive strategies, once the phage inserts their nucleic acid into the bacterium’s cytoplasm, the host cellular machinery is highjacked to induce extensive replication through the lytic cycle (Figure 1). Alternatively, a phage also has the capacity to insert its genetic information into the genome of the host bacterium, thus becoming a prophage. The process of prophage incorporation into the host chromosome is called lysogenization, and the resulting bacterium with the prophage is called a lysogen. Therefore, the genetic material of the prophage is transferred to each daughter cell through cell division following the lysogenic cycle (Figure 1). A huge advantage associated with the lysogenic cycle is that daughter cells will not produce new virus particles until conditions are favorable for the virus or some external stimuli stress the cell and activate the highjacked genes. An additional less known phage reproductive cycle is the so-called pseudo-lysogenic. In the pseudo-lysogenic type, the information encoded by the genome of the phage is not translated immediately, perhaps due to the lack of nutrients and energy for the bacterium. However, it remains inactive inside the host, waiting until the optimal conditions recover for the bacterium to restart its metabolic processes. Then, the phage has the capacity to start again performing the lytic or lysogenic life cycles [10].
Figure 1.
The lytic and lysogenic cycle of bacteriophages. The lytic cycle comprises a series of events from attachment of the bacteriophage to the bacterial cell membrane, to the release of daughter phages by the destruction of its bacterial host. In the lysogenic cycle, phage DNA integrates into the bacterial genome without major consequences for the bacterial cell, and where the nucleic acid of the virus replicates along with that of its host.
3. Phage Lytic Enzymes and Depolymerases
With increasing development and with some of our hopes against antibiotic resistance placed on the use of complete phage particles of one or more types of phages (phage cocktails), the lytic enzymes of these viruses have begun to enter the race of clinical development [11,12]. Due to the possible break that legislation in human medicine may imply, researchers in various fields such as veterinary medicine or food science and technology have also begun to develop and produce these phage products [13,14,15]. Phage-encoded proteins with potential application in these different fields are divided into endolysins, exolysins, and depolymerases. Lysins derived from phages degrade bacterial peptidoglycans and are classified into five groups, depending on the bonds these enzymatic proteins cleave in the bacterial peptidoglycan [16]. Although their function is exclusively to degrade the cell wall of bacteria, the lytic enzymes of phages present a tremendous structural diversity and a significant number of different mechanisms of action [17,18,19,20].
A phage’s efficiency and the host’s specificity are determined by protein domains and their associated specific enzymatic activities. Their structure target and degrade the cell wall of Gram-positive or Gram-negative bacteria depending on whether they contain an enzymatic catalytic domain and a cell wall binding domain or a single catalytic domain, respectively. Most phage endolysins degrade the cell wall from inside the bacteria when the lytic cycle of the phages that grow in the bacterial cytoplasm mature [20]. However, there are also phage enzymes that degrade the bacterial cell wall from the outside; these proteins called—exolysins -or ectolysins—are of great interest for their application as antimicrobial agents because they can reach bacteria more quickly than if they must do so after a cycle of phage infection [21,22,23]. These proteins with enzymatic activity have also been used to destroy preformed biofilms by Gram-positive or Gram-negative bacteria, although most studies have been carried out in vitro [24,25,26]. Moreover, while some naturally occurring lysins have potent in vitro activity, they can be engineered to be more effective in binding or traversing the bacterial wall [27,28,29,30,31,32]. Although not well known as lysins, phage enzymes with depolymerase activity—or depolymerases—can be of great interest for fighting pathogens and the biofilms that they form since a critical component of the biofilm matrix is usually polymers of a different nature [33,34,35].
In general, lysins are more likely to lyse Gram-positive bacteria because their cell wall peptidoglycan is directly exposed on the cell surface unlike Gram-negative bacteria. However, the study of phages or their lysins has been limited to a few fish pathogens such as Streptococcus agalactiae, Lactococcus garvieae, Renibacterium salmoninarum, Streptococcus iniae, and S. dysgalactiae, which are highly associated with disease outbreaks in fish farms.
4. Interactions between Phage and the Fish Immune System
Contrary to the accomplishments garnered so far in mammals and cultured cells; limited studies have correlated the immune responses of cultured fish treatment with phages. Therefore, many knowledge gaps exist on the enhancement and link of phage activity and the fish immune system. However, the administration route of phages is perceived as crucial among the known interactions between phages and fish’s immune system [36]. In vitro studies with cultured cells obtained from mucosal surfaces and in vivo challenged experiments in fish by immersion have demonstrated that increased phage abundance in the mucus layer protects the underlying epithelium from bacterial infection [37,38]. Thus, the experimental results highlight that the immersion route enables phages to penetrate fish tissues [39]. Once inside the fish body, phages will first interact and overcome the cells of the mucosal innate immune system before searching for their bacterial prey.
Interestingly, it has been reported that phages lack an outer lipid layer, which is a typical target for the complement. Which leads to an intriguing question on how the phages evade adaptive and innate immunity elements like antibodies and phagocytes respectively to gain access to the eukaryotic cells. Transcytosis is a partial explanation which includes free uptake by endocytosis or crossing via a leaky gut allowing for passive transit through the epithelium. A less understood, but exciting mechanism exists that is regarded as the trojan horse. This mechanism explains the endocytosis of the bacterial host once colonized by the phage and is hidden inside to secure access free of recognition.
Caudovirales phages from the families Myoviridae, Siphoviridae, and Podoviridae have the ability to translocate across epithelial and endothelial cell lines [40]. Interestingly, the same three phage families reported are those commonly observed interacting with bacterial fish pathogens (See Table 1). Once phages have gained access to the fish mucosal tissue, the extra- and intra-cellular immune recognition mechanisms exert a systematic screening process [41]. However, the endocytic recognition starts only after lysosomal degradation occurs inside the cell, where pathogen recognition is mediated by classical conserved pattern recognition receptors (PRR). Some of the significant PRR responsible for sensing viral RNA identified so far include the retinoic acid inducible gene I (RIG-I) like receptors (RLRs) and several endosomal-associated TLRs, for example, TLR3, TLR7, TLR8, and TLR9 [42].
Table 1.
Phages used against Gram-negative bacterial fish and shellfish pathogens.
| Gram-Negative Targets |
Source | Enrichment ɸ | Characterization Method | Phage Strains Name | Family * | Genome Length | References |
|---|---|---|---|---|---|---|---|
| Aeromonas hydrophila | River water | No | TEM | ɸ2 and ɸ5 | Myoviridae | ~20 kb | [100] |
| Fishponds; Polluted rivers | Single | TEM | N21, W3, G65, Y71 and Y81 | Myoviridae; Podoviridae | n.d. | [101] | |
| Stream water | Single | TEM, dsDNA | pAh-1 | Myoviridae | ~64 kb | [102] | |
| Sea water | Single | TEM, DNA sequencing | Akh-2 | Siphoviridae | 114,901 bp | [103] | |
| Carp tissues | Single | TEM | AHP-1 | Myoviridae | n.d. | [104] | |
| Lake water | Single | TEM, dsDNA, DNA sequencing | AhyVDH1 | Myxoviridae | 39,175 bp | [105] | |
| River water | No | TEM, dsDNA, DNA sequencing | MJG | Podoviridae | 45,057 bp | [106] | |
| Sewage water | Single | TEM | AH1 | n.d. | n.d. | [107] | |
| Striped catfish pond water | Single | TEM, dsDNA, DNA sequencing | PVN02 | Myoviridae | 51,668 bp | [108,109] | |
| River water | TEM, dsDNA | pAh1-C pAh6-C |
Myoviridae | 55 kb 58 kb |
[110] | ||
| Wastewater | No | TEM, dsDNA, DNA sequencing | Ahp1 | Podoviridae | ~42 kb | [111] | |
| Aeromonas punctata | Stream water | Single | TEM, dsDNA | IHQ1 | Myoviridae | 25–28 kb | [112] |
| Aeromonas salmonicida | River waters, two passing through fish farms | Single | TEM, DNA sequencing | SW69-9 L9-6 Riv-10 |
Myoviridae | 173,097 bp, 173,578 bp and 174,311 bp | [113] |
| River water | Single | TEM, DNA sequencing | phiAS5 | Myoviridae | 225,268 bp | [114] | |
| Sediment of a Rainbow trout culture farm | Single | TEM, dsDNA, DNA sequencing | PAS-1 | Myoviridae | ~48 kb | [115] | |
| Wastewater from a seafood market | No | TEM, DNA sequencing | AsXd-1 | Siphoviridae | 39,014 bp | [116] | |
| Sewage network water from a lift station | Single | TEM | AS-A AS-D AS-E |
Myoviridae | n.d. | [117,118] | |
| River water | No | TEM | HER 110 | Myoviridae | n.d. | [119,120] | |
| Aeromonas spp. | Gastrointestinal content of variated fish species | No | TEM, DNA sequencing | phiA8-29 | Myoviridae | 144,974 bp | [66,121] |
| Citrobacter freundii | Sewage water | No | TEM, DNA sequencing | IME-JL8 | Siphoviridae | 49,838 bp | [122] |
| Edwardsiella ictaluri | Water from catfish ponds | Single | TEM, dsDNA, DNA sequencing | eiAU eiDWF eiMSLS |
Siphoviridae | 42.80 kbp 42.12 kbp 42.69 kbp |
[123,124] |
| River water | Multiple | DNA Sequencing | PEi21 | Myoviridae | 43,378 bp | [125,126] | |
| Striped catfish kidney and liver | Single | TEM, dsDNA | MK7 | Myoviridae | ~34 kb | [127] | |
| Edwardsiella tarda | Seawater | Single | TEM, dsDNA | ETP-1 | Podoviridae | ~40 kb | [54] |
| River water | No | TEM, DNA sequencing | pEt-SU | Myoviridae | 276,734 bp | [128] | |
| Wastewater | Single | DNA sequencing | PETp9 | Myoviridae | 89,762 bp | [129] | |
| Fish tissues and rearing seawater | No | TEM, DNA sequencing | GF-2 | Myoviridae | 43,129 bp | [130] | |
| Flavobacterium columnare | River water | Single | TEM, DNA sequencing | FCL-2 |
Myoviridae | 47,142 bp | [38,131,132] |
| Fishpond’s water and bottom sediments | No | TEM, dsDNA | FCP1-FCP9 | Podoviridae | n.d. | [133] | |
| Flavobacterium psychrophilum | Rainbow trout farm water | Single/double | TEM, dsDNA | ø (FpV-1 to FpV-22) |
Podoviridae
Siphoviridae Myoviridae |
(~8 to ~90 kb) | [134,135] |
| Ayu kidneys and pondwater collected from ayu farms | Multiple | TEM, dsDNA | PFpW-3, PFpC-Y PFpW-6, PFpW-7 PFpW-8 |
Myoviridae; Podoviridae; Siphoviridae | n.d. | [136] | |
| Photobacterium damselae subsp. damselae | Raw oysters | Single | TEM, dsDNA | Phda1 | Myoviridae | 35.2–39.5 kb | [137] |
| Gastrointestinal tract of lollipop catshark | Single | TEM, DNA sequencing | vB_Pd_PDCC-1 | Myoviridae | 237,509 bp | [138] | |
| Pseudomonas plecoglossicida | Ayu pond water and diseased fish | No | TEM, DNA sequencing | PPpW-3 PPpW-4 |
Myoviridae Podoviridae | 43,564 bp 41,386 bp |
[139,140] |
| Pseudomonas aeruginosa | Wastewater | No | TEM, DNA sequencing | MBL | n.d. | 42,519 bp | [141] |
| Shewanella spp. | Wastewater from a marketplace |
Single | TEM, DNA sequencing | SppYZU01 to SppYZU10 | Myoviridae; Siphoviridae. | SppYZU01 (43.567 bp) SppYZU5 (54.319 bp) |
[142] |
| Tenacibaculum maritimum | Seawater | Multiple | TEM, DNA sequencing | PTm1 PTm5 |
Myoviridae | 224,680 bp 226,876 bp |
[143] |
| Vibrio alginolyticus | Aquaculture tank water | Single | TEM, DNA sequencing | VEN | Podoviridae | 44,603 bp | [144] |
| Marine sediment | No | TEM, DNA sequencing | ValKK3 | Myoviridae | 248,088 bp | [145] | |
| Marine water | Single | TEM, dsDNA | St2 Grn1 |
Myoviridae | 250,485 bp 248,605 bp | [146] | |
| Vibrio anguillarum | Soft tissues from clams and mussels | No | TEM, dsDNA | 309 ALMED CHOED ALME CHOD CHOB |
Several shapes | ~47–48 kb | [147] |
| Sewage water | Double | dsDNA | VP-2 VA-1 |
n.d. | n.d. | [148] | |
| Water samples from fish farms | Multiple | TEM, DNA sequencing |
ø H1, H7, S4-7, H4, H5 H8, H20 S4-18, 2E-1, H2 |
Myoviridae Siphoviridae Podoviridae | ~194–195 kb ~50 kb ~45–51 kb |
[149] | |
| Vibrio campbellii | Host strain (V. campbellii) isolated form a dead shrimp | No | TEM, DNA sequencing | HY01 | Siphoviridae | 41.772 bp | [150] |
| Hepatopancreas of Pacific white shrimp |
Single | dsDNA, DNA sequencing | vB_Vc_SrVc9 | Autographiviridae | ~43.15 kb | [151] | |
| Vibrio harveyi | Shrimp farm, hatcheries and marine water | Multiple | TEM, dsDNA | A | Siphoviridae | n.d. | [152] |
| Vibrio harveyi | No | TEM, dsDNA | VHML | Myovirus-like | n.d. | [153] | |
| Shrimp pond water | Single | TEM, dsDNA | PW2 | Siphoviridae | ~46 kb | [154] | |
| Water and sediment samples | Single | TEM, dsDNA | VHM1, VHM2 VHS1 |
Myoviridae, Siphoviridae |
~55 kb, ~66 kb ~69 kb |
[155] | |
| Hatchery water and oyster tissues | Single | TEM, dsDNA | vB_VhaS-a vB_VhaS-tm |
Siphoviridae | ~82 kb ~59 kb |
[156] | |
| Commercial clam samples | Multiple | Genomic analysis, dsDNA |
ø VhCCS-01 VhCCS-02 VhCCS-04 VhCCS-06 VhCCS-17 VhCCS-20 VhCCS-19 VhCCS-21 |
Siphoviridae, Myoviridae |
n.d. | [157] | |
| Oyster, clam, shrimp, and seawater samples | No | TEM, DNA sequencing | VHP6b | Siphoviridae | 78,081 bp | [158] | |
| shrimp hatchery and farm water, oysters from estuaries, coastal sea water |
Multiple | TEM, dsDNA | Viha10 Viha8 Viha9 Viha11 Viha1 to Viha7 |
Siphoviridae - Siphoviridae Myoviridae (Viha4) |
n.d. ~44–94 kb ~85 kb (Viha4) |
[159,160] | |
| Seawater sample | Single | TEM | VhKM4 | Myoviridae | n.d. | [161] | |
| Vibrio ordalii | Macerated specimens of mussels | No | TEM, DNA sequencing | B_VorS-PVo5 | Siphoviridae | 80,578 bp | [162] |
| Vibrio parahaemolyticus | Sewage sample | No | TEM, dsDNA | VPp1 | Tectiviridae | ~15 kb | [163] |
| Polluted seawater | No | TEM, dsDNA | KVP40 KVP41 |
Myoviridae | n.d. | [164,165] | |
| Seawater or mussels | Single | dsDNA | SPA2 SPA3 |
n.d. | ~21 kb | [166] | |
| Coastal water | Single | TEM, DNA sequencing | pVP-1 | Siphoviridae | 111,506 bp | [167,168] | |
| V. parahaemolyticus isolated from sewage samples collected from an aquatic product market | No | TEM, DNA sequencing | vB_VpS_BA3 vB_VpS_CA8 | Siphoviridae | 58,648 bp 58,480 bp |
[169] | |
| Shrimp pond water | Single | TEM, DNA sequencing | VP-1 | Myoviridae | 150,764 bp | [170] | |
| Coastal sand sediment | double | TEM, DNA sequencing | VpKK5 | Siphoviridae | 56,637 bp | [171,172] | |
| Vibrio splendidus | Raw sewage obtained from local hatcheries | Single | TEM | PVS-1, PVS-2 PVS-3 |
Myoviridae; Siphoviridae | n.d. | [173] |
| Seawater near a fish farm cage | Single | TEM, DNA sequencing | vB_VspP_pVa5 | Podoviridae | 78,145 bp | [174] | |
| Vibrio coralliilyticus | sewage in oyster hatchery | Single | TEM | pVco-14 | Siphoviridae | n.d. | [175] |
| Vibrio vulnificus | Seawater sample | Single | TEM, DNA sequencing | SSP002 | Siphoviridae | 76,350 bp | [176,177] |
| Abalone samples | No | TEM, sequencing | VVPoo1 | Siphoviridae | 76,423 bp | [178] | |
| Initial host strain (V. vulnificus) | No | TEM | VV1 VV2 VV3 VV4 |
Tectiviridae | n.d. | [179] | |
| Vibrio sp. | Sewage draining exits | Single | TEM, DNA sequencing | VspDsh-1 VpaJT-1 ValLY-3 ValSw4-1 VspSw-1 |
Siphoviridae | 46,692 bp 60,177 bp 76,310 bp 79,545 bp 113,778 bp |
[180] |
| Yersinia ruckeri | Wastewater containing suspended trout feces from a settling pond at a trout farm | Single | TEM | NC10 | Podoviridae | n.d. | [181] |
| Sewage | No | TEM | YerA41 (several phages) | icosahedral head, contractile tail | n.d. | [182] | |
| Sewage | No | TEM, DNA sequencing, dsDNA | R1-37 | Myoviridae | ~270 kb | [183,184] |
ɸ Phage enrichment with “single” or “multiple” bacterial hosts; * Classification determined by the authors; TEM (Transmission Electron Microscopy); dsDNA (Double stranded DNA); n.d. (Not determined); ø Several phage strains were isolated but only selected strains were fully characterized.
After these initial encounters with the fish tissue, phages can stimulate and modulate the host’s innate immune response [43,44,45,46,47]. Several intracellular or facultative intracellular bacterial pathogens have been reported to trigger immune responses in cultured fish. Among them are Photobacterium damselae subsp. piscicida, Renibacterium salmoninarum, Piscirikettsia salmonis, Edwardsiella tarda, Edwardsiella ictaluri, Yersinia ruckeri, and Vibrio parahaemolyticus were identified. However, studies on fish immunity showing the detailed interaction of members in the genus Edwardsiella and Vibrio with phages have rarely been reported.
4.1. Phage-Mediated Activation of Inflammation
Bacteriophage treatment was associated with opposite shifts in the inflammatory response in several test models, both in vivo and in vitro [48,49,50,51]. However, the results seem to depend not only on the cellular or animal model used but also on the type of phage applied and the panel of cytokines analyzed. Phage therapy in humans can also modify the levels of some cytokines produced by blood cells in treated patients [39]. In fish, some researchers have analyzed the cytokines’ response to the presence of bacteriophages alone or the coinfection of phages with their target bacteria. For example, phage therapy reduced the expression of the proinflammatory cytokines tnfa and il1b in the inflammatory response generated by Pseudomonas aeruginosa infection in zebrafish embryos [52,53]. Besides, using the adult zebrafish (Danio rerio) and the E. tarda model of infection, other authors also showed that although a phage treatment induced the expression of cytokine genes at specific time points, a robust proinflammatory response was undetected in the host [54]. Furthermore, a recent study has shown that a phage lysate of A. hydrophila induced a more robust immune response in Cyprinus carpio when compared to a formalin killed vaccine [55]. As a proof-of-concept, a novel commercial preparation containing three bacterial phages (BAFADOR®) applied on European eel (Anguilla anguilla) caused the stimulation of cellular and humoral immune parameters in response to an experimental challenge with A. hydrophila and P. fluorecense [56].
4.2. Phage-Specific Adaptive Responses
Due to the protein structure of the phage envelope, these proteins are the target of the adaptive immune system, which response with the production of neutralizing antibodies against them. Early studies with mice and even amphibians showed that phage exposure of the animals induced primary and secondary antibody responses [57,58,59]. It is expected that some phage epitopes stimulate an antibody response in experimental models. However, antibody production depends on the route of phage administration, the application schedule and dose, and individual features of a phage. Consequently, the results of studies where an antibody response to phages has been verified are very heterogeneous. Phagocytosis by immune patrolling cells seems to be a significant process of bacteriophage neutralization within animal bodies [60]. Moreover, although blood in humans and animals, including fish, is deemed sterile, genomic analysis has shown a rich phage community, which inevitably comes into continuous contact with immune cells in this rich fluid [47]. Despite these mechanisms of phagocytosis, antigen presentation, and antibody production by the immune cells against phages, the number of antibodies produced does not affect phage therapy outcomes.
On the other hand, due to the numerous and constant presence of large numbers of phages in our microbiota, it is not surprising that a low but stable background of antibodies against them is produced. Therefore, in some human or animal tests, high antibody levels have not been found against the phages used. Phage-derived RNA and ssDNA could directly contribute to B cell activation and the synthesis of anti-bacteriophage antibodies [61,62]. Despite the production of antibodies by animals against phage core or tail proteins, the induction of antibodies seems irrelevant for treating infections because the antibacterial effects of phages are faster than antibody formation in acute infections [63]. Conversely, the production of antibodies against phages could interfere with the outcome of the infection in chronic infections [64]. However, no robust studies have demonstrated an antibody-mediated immune response after inoculation or experimental infection with phages in fish.
5. Phage Diversity in the Aquatic Environment
Bacteriophages or phages are relatively simple viruses that target a bacterial host. These viruses are extraordinarily abundant and diverse. Phages are common in soil and human and animal guts, so they are readily isolated from feces and sewage. Moreover, due to the exponential advancement of bioinformatics and massive sequencing techniques, the geospatial distribution of viruses in freshwater or saltwater ecosystems is an incipient area of research. Fish and shellfish that inhabit those environments also harbor copious amounts of phages in their digestive tracts [65,66]. Bacteriophages floating in waters are also very abundant, with an estimate of 10 million virus-like particles in 1 mL of seawater [67].
Virus classification is typically based on characteristics such as type of nucleic acid, virion structure or morphology, replication mode, host target and clinical and epidemiological features of disease they cause, and even geographical distribution [68]. Researchers working in aquaculture mainly use Transmission Electron Microscopy (TEM) and nucleic acid enzymatic sensitivity to determine the morphology and nucleic acid type of the phages used. In recent years, thanks to cryo-Electron Microscopy (cryo-EM), Nuclear Magnetic Resonance (NMR), and X-ray crystallography analysis, we have been able to study the structures of numerous bacteriophages. Genomic sequencing and analysis of the DNA isolated from phages can also provide reasonably accurate taxonomic information. The use of sequences alone sets a significant challenge for traditional phage taxonomy; historically it has been based primarily on descriptive definitions, but nothing prevents these techniques from being used simultaneously for the better characterization and study of phages.
There are three primary basic morphologies in bacteriophages: (1) filamentous form, (2) icosahedral form, and (3) icosahedral form with a tail through which it binds to its target bacteria. The latter is the best known and most studied morphology, with three components: a capsid where the dsDNA genome is packed, a tail—which can be rigid or flexible, short, or long—that serves as a syringe during infection to transfer the phage genome into the bacterial cell, and a unique recognition system at the end of the tail that specifically recognizes the host cell and penetrates its cell wall.
As temperate phages are expected to execute both types of infection modes, only lytic phages can be used to fight bacterial infections because their action destroys the host and causes new nascent phages to spread among adjacent bacterial populations. Phages showing the lytic mechanism of replication are also known as virulent phages, while phages exhibiting lytic and lysogenic cycles are temperate phages. The temperate phages exhibit horizontal gene transfer, where they can transfer antibiotic resistance genes to other bacteria, and therefore they are not preferred for phage therapy unless their genome is analyzed, and it is shown that they do not contain those resistance genes
In recent years, numerous bacteriophages that target the primary bacterial fish pathogens have been isolated and partially characterized (Table 1). Many of these studies with phages have proven their efficiency against pathogenic bacteria in vitro. In fact, some researchers have even carried out field trials over the last decade using different routes of administration (orally, intraperitoneally, or intramuscularly injected, administered in feed, or added to fish thank water). Most of these studies use the work of Ackermann or the criteria of the International Committee on Taxonomy of Viruses (ICTV, accessible online) as a basis to identify their phage isolates [69,70,71]. For further taxonomic classification and phage characterization, more detailed information, such as genomic data, has begun to be included in scientific publications [72,73,74].
As phages are ubiquitously present in nature, the sources of isolated phages against fish pathogens are quite diverse (Table 1). Fish can harbor phages easily as demonstrated long ago in an experiment in which phages were detected in fish tissue after bath exposure to a specified concentration of an E. coli phage [39]. Detection of bacteriophages can be carried out by classical infection assays such as single-host, double or multiple-host enrichment, followed by selection through phage plating with the target host strain, allowing the determination of phage infectivity. As observed in Table 1 and Table 2, most researchers working with pathogenic fish bacteria prefer to use enrichment with a single strain to isolate phages. Instead of enriching phage-containing samples, some authors use the phage isolation technique called the double-agar layer [75,76], choosing a single indicator strain.
Table 2.
Phages used against Gram-positive bacterial fish and shellfish pathogens.
| Gram-Positive Targets | Source | Enrichment ɸ | Characterization Method | Phage Strains Name | Family * | Genome Length | References |
|---|---|---|---|---|---|---|---|
| Lactococcus garvieae | L. garvieae isolated from diseased yellowtail | No | TEM, dsDNA | PLgY(16) | Siphoviridae | n.d. | [185] |
| Yellowtail (Y) Water (W) Sediments (S) |
Single | TEM, dsDNA | PLgW1-6 PLgY16 PLgY30 PLgY886 PLgS1 |
Siphoviridae | >20 kbp | [186,187,188] | |
| Domestic compost | Single | TEM, DNA sequencing | GE1 | Siphoviridae | 24,847 bp | [189] | |
| L. garvieae host | No | TEM, DNA sequencing | PLgT-1 | Siphoviridae | 29,284 bp | [190,191,192] | |
| Rainbow trout farm water | Single | TEM, DNA sequencing | WP-2 | Picovirinae | 18,899 bp | [193] | |
| Streptococcus agalactiae | Tilapia pond | No | TEM | HN48 | Caudoviridae | n.d. | [194] |
| S. iniae | S. iniae host | No | TEM, dsDNA | vB_SinS-44 vB_SinS-45 vB_SinS-46 vB_SinS-48 | Siphoviridae | ~51.7 kb ~28.4 kb ~66.3 kb ~27.5 kb |
[195] |
| Weissella ceti | W. ceti host strain | No | TEM | PWc | Siphoviridae | 38,783 bp | [196] |
ɸ Phage enrichment with “single” or “multiple” bacterial hosts; * Classification determined by the authors; TEM (Transmission Electron Microscopy); dsDNA (Double stranded DNA); n.d. (Not determined).
After phage amplification from their initial host strain, researchers use phage DNA extraction, DNA sensitivity to DNases or RNases, and virion microscopy visualization for quick taxonomic classification. Bacterial whole-genome sequencing (WGS), which focuses on the prophage state inside the bacterial chromosome, is also helpful for knowing the whole phage coding sequences and phage viability determination. Bacteria can carry more than half a dozen complete or incomplete prophages on their chromosomes. Therefore, researchers can use genomic characterization to choose lytic phages that do not contain enzymes that favor integration into the bacterial chromosome, such as recombinases, integrases, or repressor sequences of the lytic cycle—characteristics of lysogenic phages—and of course that do not contain resistance genes or bacterial virulence factors.
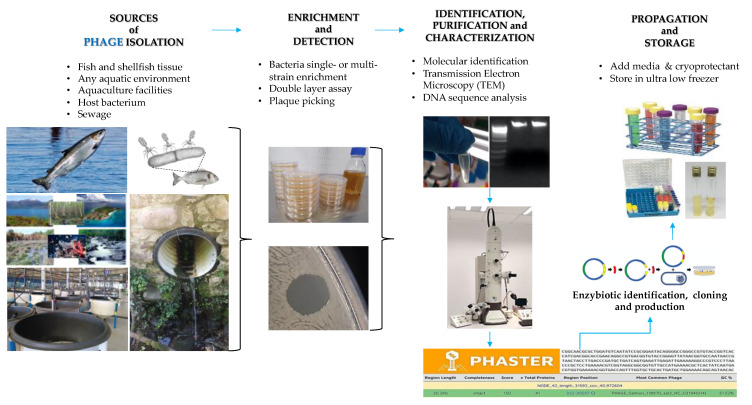
It is convenient to integrate as many techniques as possible in phage studies to offer more robust information regarding the phage of interest. Recently, new and more complex techniques such as Atomic Force Microscopy are able to provide information about phage integrity close to physiological conditions [77,78,79]. The preferred tool currently utilized for classifying phages is transmission electron microscopy, followed by phage DNA sequencing (Table 1). The steps followed to isolate and characterize phages used in in vitro assays or field trials against fish and shellfish bacterial pathogens are shown (Figure 2). The phages used against Gram-negative fish pathogens belong primarily to the Myoviridae family, followed by the Siphoviridae and Podoviridae families. Other minor families that display lytic activity against Gram-negative pathogens are Tectiviridae and Autographiviridae. Against Gram-positive pathogens, the Siphoviridae family predominates, although phages of the Picoviridae and Caudoviridae families with lytic activity against these pathogens have also been isolated.
Figure 2.
Schematic steps followed for the isolation and characterization of phages intended for use in aquaculture. From left to right: General steps for phage isolation, enrichment, detection, identification, characterization, propagation, and storage.
6. Bacteriophages against Biofilms in Aquaculture Facilities
All bacterial fish pathogens can form biofilms in vitro and in vivo given the proper environmental conditions [80,81,82]. Biofilm formation is an intrinsic characteristic of bacteria because living within these structures favors the uptake of nutrients and protection against the physical, chemical, or biological agents found in aquatic environments. This process always occurs sequentially, and the mechanisms have been understood for a couple of decades [83]. Firstly, the adhesion of the bacteria to a living or inert surface is required. This process used to be favored both by intrinsic factors of the bacteria such as the mobility or the expression of a polysaccharide capsule [80] and by physical—chemical and environmental factors, such as temperature [81], or the type of substrate to which the bacteria attach itself. After this union, the formation of microcolonies occurs, and these small groups of bacteria will evolve into big macro-colonies that make up the biofilm. An essential characteristic of mature biofilms is that they contain a heterogeneous matrix that maintains cohesion between the bacteria that form it and favors their adherence to the surface on which the biofilm is formed. Biofilms are extremely difficult to eradicate because the bacterial cells inside them are more protected from any chemical or biological agent, so they tend to easily resist conventional antibiotics and common disinfectants [84,85]. Biofilm formation is essential in veterinary medicine since they can represent the focus of recurrent infections [86,87]. From the fully formed and mature biofilms, bacteria are released and constitute a population of planktonic cells that will form new biofilms in other places, allowing recurrent infections or the contamination of new environments favorable for their proliferation.
Biofilms are also of particular interest in the food industry, where there are a series of ubiquitous microorganisms that are very difficult to eliminate from the food itself and food processing settings and environments. Phages alone or in phage cocktails were successfully used against both Gram-positive [88,89,90,91,92] and Gram-negative bacterial biofilms [93,94,95,96,97,98]. Despite these assays being performed in a wide variety of situations and against numerous pathogens in human medicine, veterinary medicine, and food production systems, these studies have been carried out mainly in the laboratory in vitro. Therefore, it will also be interesting to know the options that fish pathologists may have to eradicate the biofilms that bacteria produce in aquaculture. Phages were also tested against bacterial fish pathogens producing biofilms in vitro, but the number of studies conducted following these settings is very minimal [99]. Therefore, it is important that the action of the phages against fully formed biofilms is also evaluated when searching for lytic phages against pathogenic fish bacteria.
7. Potential of Phage Therapy in Aquaculture Settings
During the fish and shellfish production cycle, these animals are already in daily contact with billions of bacteriophages, which assures us that they are safe. However, in their use against bacterial infections where massive phage production is required, we must consider several factors.
As phage treatments constantly require isolating the bacterium causing the disease, once a helpful phage is characterized against this bacterial strain, a stable batch of technically challenging preparations must be produced for field use. Consequently, one of the most critical challenge for microbiologists working directly or indirectly with aquaculture is the standardization of stocks used to treat infections or combat biofilms in aquaculture facilities. These stocks require strict quality control for purity, viability, and stability, implying that the correct conservation of the stocks is necessary for preparations containing single or mixed phages (phage cocktail). Titer, dosage, and quality of phage preparations are crucial parameters in standardizing experiments in the laboratory and experimental infections in field trials. Since we know that while some phages can grow exponentially inside a bacterial population from a low initial concentration, other phages need to maintain a relationship between the number of bacteria and the number of phage particles to achieve an adequate performance. Therefore, we must empirically verify this critical parameter. Very recently, a phage cocktail containing seven bacteriophages (three against A. hydrophila and four against P. fluorescens) has been tested in the European eel (Anguilla anguilla) and rainbow trout (Oncorhynchus mykiss), reducing the mortality of fish challenged with strains of these two species of bacteria [56,197]. Cocktails have also been used successfully in laboratory tests or small field trials in food protection or veterinary and human medicine [198,199,200,201]. In these and other studies, many phages (cocktail) are used to carry out the experiments, but in most cases, only the phage that has presented better results in vitro is subsequently characterized [117,118,133,202]. Second, it would be desirable to know phage genetics with sufficient precision. After all, we must consider that when we intend to use bacteriophages in aquaculture, they may contain genes for resistance to antibiotics or bacterial virulence genes that can produce noticeable side effects because they replicate exponentially in contact with their target bacteria. We must also remember that many antibiotic residues end up in continental or oceanic waters due to anthropogenic activities. Therefore, we must be aware that even phages isolated from aquatic environments can carry antibiotic resistance genes or virulence factors [203,204]. At present, although each time their number increases, not all phages used in in vitro or in vivo assays against fish or shellfish bacterial pathogens have been entirely genetically analyzed or characterized (Table 1 and Table 2).
The list of species of fish bacterial pathogens in which lytic phages have been studied is not complete. It may be essential to conduct these studies in species of greater interest in aquaculture, such as Photobacterium damselae subsp. piscicida, bacterial anaerobes, mycobacteria, Nocardia, several Aeromonas species, Enterobacterales, pseudomonads, vibrios, and the Gram-positive bacteria mentioned above. Few studies with fish bacterial pathogens have characterized or evaluated the presence or evolution of phage-resistant strains. Some works have investigated this phenomenon in various fish pathogens such as Flavobacterium [205,206,207], Yersinia ruckeri [181], Aeromonas salmonicida [117,208], and Vibrio anguillarum [148]. The mechanisms by which bacteria become resistant to phages is also an area of intensive research, especially since the discovery and application of the clustered regularly interspaced short palindromic repeats (CRISPR) system.
Most of the studies with fish pathogens have used controlled laboratory conditions to verify the control exerted by these lytic phages to their pathogenic bacterial host. However, more studies on these interactions under natural conditions would be desirable. One of the critical parameters is the multiplicity of infection (MOI). The use of high or low multiplicities of infection seems to be a key parameter for achieving effective lysis of the bacterial population and the appearance of resistance to the phages used. Therefore, comparative studies are needed to relate MOIs used in vitro and in aquatic environments, where phages are exposed to environmental conditions and factors such as dilution or variability of the target bacteria in their natural environment. A better understanding of the biology of viruses and a greater capacity to standardize the settings related to preclinical or laboratory research can also help in the advancement of regulatory affairs. As bacteriophage research continues to grow, we believe that microbiologists and immunologists working in areas related to aquaculture can use phages or their lytic enzymes to offer many promising advances in the fight against pathogenic bacterial species affecting cultured fish and shellfish.
8. Future Perspectives
Innovative technologies to enhance fish health and decrease diseases are paramount to achieve the global perspectives on food sustainability required by the blue growth philosophy and fulfill the blue economy goals proposed by the new aquaculture 4.0 strategy. However, the growing food demands strongly stimulate the intensification of production, generating the need for economically viable and environmentally sustainable practices that may rely on improved health management strategies. The use of antibiotics and chemotherapies in aquaculture has been widely used to reduce infectious diseases and promote growth [209]. However, since antibiotics lead to bacterial resistance generation, their use is discouraged nowadays, and several countries with major fish-producing facilities have banned their inclusion in biosecurity plans. Therefore, to improve fish robustness in aquaculture settings, several strategies based on the use of alternative sources like phytogenics [210], live microbes [211,212], their metabolic products like short-chain fatty acids (SCFA) [213] and sphingolipids [214], or structural components like peptidoglycan [215] or B-glucan [216] have been extensively tested in the last century as possible therapeutics for fish. Nevertheless, despite the entire group being potentially effective, all the previous strategies lack any specificity against a particular fish disease.
In contrast, as discussed in the present review, phages are extremely specific in their ability to infect and destroy certain species or strains of bacteria without affecting the core commensal microbiota of the host. Therefore, phage therapy is well suited to be part of the multidimensional strategies focused on increasing fish health in culture and, at the same time, a promising tool in counteracting the rise of antibiotic-resistant bacteria. However, it is necessary to exercise caution since the potential evolution of phage resistance also exists [217]. Although resistance may occur, the concept of phage training for therapy through experimental coevolution has recently emerged and deserved future attention. Both Gram-negative and Gram-positive bacteria have been efficiently lysed by specific phages in vitro [218,219]. Therefore, the putative use of phage lytic enzymes and their interaction with the immune system of cultured aquatic organisms is an entirely new area demanding tremendous exploration. Although, the constant discovery of new phages with variated phenotypes and genotypes [220,221], hampers the integrative knowledge that requires stronger in vivo evidence to consolidate the specific phage-pathogenic bacteria as established biotechnology in aquaculture.
In the context of automation, recent researchers utilizing diverse mammalian models have demonstrated novel systems for single-virion identification of common pathogens using machine learning (ML) algorithm training. Consequently, easy access to big genomic data recently generated is fundamental to fuel the novel ML algorithms. The combination resulting from this may lead to the generation of a fast and accurate selection of bacteriophages with the specific characteristics and properties required to lysate the specific bacterial strains affecting fish culture grounds. Finally, creating biobanks of fish bacteriophages and their products (lysins) following established legal and regulatory frameworks for safe and stable use in countries with intensive aquaculture operations requires extensive maneuvers of the regulatory authorities and the pharmaceutical industry interested in their exploitation on a large-scale setting.
Author Contributions
Conceptualization, J.R.-V., J.G.-V. and F.A.; data curation, J.R.-V.; graphical material, J.R.-V.; Investigation and editing, J.S.; writing original draft, J.R.-V.; visualization, J.G.-V. and F.A.; supervision, F.A.; J.G.-V., writing, review, editing, and funding acquisition. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding, but “The APC was partially covered by Nord University Open Access Funds”.
Institutional Review Board Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ghosh C., Sarkar P., Issa R., Haldar J. Alternatives to Conventional Antibiotics in the Era of Antimicrobial Resistance. Trends Microbiol. 2019;27:323–338. doi: 10.1016/j.tim.2018.12.010. [DOI] [PubMed] [Google Scholar]
- 2.Golkar Z., Bagasra O., Pace D.G. Bacteriophage therapy: A potential solution for the antibiotic resistance crisis. J. Infect. Dev. Ctries. 2014;8:129–136. doi: 10.3855/jidc.3573. [DOI] [PubMed] [Google Scholar]
- 3.Wigington C.H., Sonderegger D., Brussaard C.P., Buchan A., Finke J.F., Fuhrman J.A., Lennon J.T., Middelboe M., Suttle C.A., Stock C., et al. Re-examination of the relationship between marine virus and microbial cell abundances. Nat. Microbiol. 2016;1:15024. doi: 10.1038/nmicrobiol.2015.24. [DOI] [PubMed] [Google Scholar]
- 4.Zhang R., Li Y., Yan W., Wang Y., Cai L., Luo T., Li H., Weinbauer M.G., Jiao N. Viral control of biomass and diversity of bacterioplankton in the deep sea. Commun. Biol. 2020;3:256. doi: 10.1038/s42003-020-0974-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brum J.R., Schenck R.O., Sullivan M.B. Global morphological analysis of marine viruses shows minimal regional variation and dominance of non-tailed viruses. ISME J. 2013;7:1738–1751. doi: 10.1038/ismej.2013.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dion M.B., Oechslin F., Moineau S. Phage diversity, genomics and phylogeny. Nat. Rev. Microbiol. 2020;18:125–138. doi: 10.1038/s41579-019-0311-5. [DOI] [PubMed] [Google Scholar]
- 7.Washizaki A., Yonesaki T., Otsuka Y. Characterization of the interactions between Escherichia coli receptors, LPS and OmpC, and bacteriophage T4 long tail fibers. Microbiologyopen. 2016;5:1003–1015. doi: 10.1002/mbo3.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunne M., Hupfeld M., Klumpp J., Loessner M.J. Molecular Basis of Bacterial Host Interactions by Gram-Positive Targeting Bacteriophages. Viruses. 2018;10:397. doi: 10.3390/v10080397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bertozzi Silva J., Storms Z., Sauvageau D. Host receptors for bacteriophage adsorption. FEMS Microbiol. Lett. 2016;363:fnw002. doi: 10.1093/femsle/fnw002. [DOI] [PubMed] [Google Scholar]
- 10.Los M., Wegrzyn G. Pseudolysogeny. Adv. Virus Res. 2012;82:339–349. doi: 10.1016/B978-0-12-394621-8.00019-4. [DOI] [PubMed] [Google Scholar]
- 11.Fischetti V.A. Development of Phage Lysins as Novel Therapeutics: A Historical Perspective. Viruses. 2018;10:310. doi: 10.3390/v10060310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trudil D. Phage lytic enzymes: A history. Virol. Sin. 2015;30:26–32. doi: 10.1007/s12250-014-3549-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coffey B., Mills S., Coffey A., McAuliffe O., Ross R.P. Phage and Their Lysins as Biocontrol Agents for Food Safety Applications. Annu. Rev. Food Sci. Technol. 2010;1:449–468. doi: 10.1146/annurev.food.102308.124046. [DOI] [PubMed] [Google Scholar]
- 14.Carvalho C., Costa A.R., Silva F., Oliveira A. Bacteriophages and their derivatives for the treatment and control of food-producing animal infections. Crit. Rev. Microbiol. 2017;43:583–601. doi: 10.1080/1040841X.2016.1271309. [DOI] [PubMed] [Google Scholar]
- 15.Rodriguez-Rubio L., Gutiérrez D., Donovan D.M., Martínez B., Rodríguez A., García P. Phage lytic proteins: Biotechnological applications beyond clinical antimicrobials. Crit. Rev. Biotechnol. 2016;36:542–552. doi: 10.3109/07388551.2014.993587. [DOI] [PubMed] [Google Scholar]
- 16.Vázquez R., García E., García P. Phage Lysins for Fighting Bacterial Respiratory Infections: A New Generation of Antimicrobials. Front. Immunol. 2018;9:2252. doi: 10.3389/fimmu.2018.02252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Broendum S.S., Buckle A.M., McGowan S. Catalytic diversity and cell wall binding repeats in the phage-encoded endolysins. Mol. Microbiol. 2018;110:879–896. doi: 10.1111/mmi.14134. [DOI] [PubMed] [Google Scholar]
- 18.Cahill J., Young R. Phage Lysis: Multiple Genes for Multiple Barriers. Adv. Virus Res. 2019;103:33–70. doi: 10.1016/bs.aivir.2018.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rodriguez-Rubio L., Martínez B., Donovan D.M., Rodriguez A., García P. Bacteriophage virion-associated peptidoglycan hydrolases: Potential new enzybiotics. Crit. Rev. Microbiol. 2013;39:427–434. doi: 10.3109/1040841X.2012.723675. [DOI] [PubMed] [Google Scholar]
- 20.Ghose C., Euler C.W. Gram-Negative Bacterial Lysins. Antibiotics. 2020;9:74. doi: 10.3390/antibiotics9020074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oliveira A., Leite M., Kluskens L.D., Santos S.B., Melo L.D., Azeredo J. The First Paenibacillus larvae Bacteriophage Endolysin (PlyPl23) with High Potential to Control American Foulbrood. PLoS ONE. 2015;10:e0132095. doi: 10.1371/journal.pone.0132095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fenton M., Ross P., Mcauliffe O., O’Mahony J., Coffey A. Recombinant bacteriophage lysins as antibacterials. Bioeng. Bugs. 2010;1:9–16. doi: 10.4161/bbug.1.1.9818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fischetti V.A. Bacteriophage endolysins: A novel anti-infective to control Gram-positive pathogens. Int. J. Med. Microbiol. 2010;300:357–362. doi: 10.1016/j.ijmm.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharma U., Vipra A., Channabasappa S. Phage-derived lysins as potential agents for eradicating biofilms and persisters. Drug Discov. Today. 2018;23:848–856. doi: 10.1016/j.drudis.2018.01.026. [DOI] [PubMed] [Google Scholar]
- 25.Chan B.K., Abedon S.T. Bacteriophages and Their Enzymes in Biofilm Control. Curr. Pharm. Des. 2015;21:85–99. doi: 10.2174/1381612820666140905112311. [DOI] [PubMed] [Google Scholar]
- 26.Meng X., Shi Y., Ji W., Zhang J., Wang H., Lu C., Sun J., Yan Y. Application of a bacteriophage lysin to disrupt biofilms formed by the animal pathogen Streptococcus suis. Appl. Environ. Microbiol. 2011;77:8272–8279. doi: 10.1128/AEM.05151-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gerstmans H., Grimon D., Gutierrez D., Lood C., Rodriguez A., van Noort V., Lammertyn J., Lavigne R., Briers Y. A VersaTile-driven platform for rapid hit-to-lead development of engineered lysins. Sci. Adv. 2020;6:eaaz1136. doi: 10.1126/sciadv.aaz1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gerstmans H., Rodriguez-Rubio L., Lavigne R., Briers Y. From endolysins to Artilysin(R)s: Novel enzyme-based approaches to kill drug-resistant bacteria. Biochem. Soc. Trans. 2016;44:123–128. doi: 10.1042/BST20150192. [DOI] [PubMed] [Google Scholar]
- 29.Wang Z., Kong L., Liu Y., Fu Q., Cui Z., Wang J., Ma J., Wang H., Yan Y., Sun J. A Phage Lysin Fused to a Cell-Penetrating Peptide Kills Intracellular Methicillin-Resistant Staphylococcus aureus in Keratinocytes and Has Potential as a Treatment for Skin Infections in Mice. Appl. Environ. Microbiol. 2018;84:e00380-18. doi: 10.1128/AEM.00380-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yan G., Liu J., Ma Q., Zhu R., Guo Z., Gao C., Wang S., Yu L., Gu J., Hu D., et al. The N-terminal and central domain of colicin A enables phage lysin to lyse Escherichia coli extracellularly. Antonie Van Leeuwenhoek. 2017;110:1627–1635. doi: 10.1007/s10482-017-0912-9. [DOI] [PubMed] [Google Scholar]
- 31.Cheleuitte-Nieves C., Heselpoth R.D., Westblade L.F., Lipman N.S., Fischetti V.A. Searching for a Bacteriophage Lysin to Treat Corynebacterium bovis in Immunocompromised Mice. Comp. Med. 2020;70:328–335. doi: 10.30802/AALAS-CM-19-000096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heselpoth R.D., Euler C.W., Schuch R., Fischetti V.A. Lysocins: Bioengineered Antimicrobials That Deliver Lysins across the Outer Membrane of Gram-Negative Bacteria. Antimicrob. Agents Chemother. 2019;63:e00342-19. doi: 10.1128/AAC.00342-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pires D.P., Oliveira H., Melo L.D., Sillankorva S., Azeredo J. Bacteriophage-encoded depolymerases: Their diversity and biotechnological applications. Appl. Microbiol. Biotechnol. 2016;100:2141–2151. doi: 10.1007/s00253-015-7247-0. [DOI] [PubMed] [Google Scholar]
- 34.Volozhantsev N.V., Shpirt A.M., Borzilov A.I., Komisarova E.V., Krasilnikova V.M., Shashkov A.S., Verevkin V.V., Knirel Y.A. Characterization and Therapeutic Potential of Bacteriophage-Encoded Polysaccharide Depolymerases with β Galactosidase Activity against Klebsiella pneumoniae K57 Capsular Type. Antibiotics. 2020;9:732. doi: 10.3390/antibiotics9110732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin H., Paff M.L., Molineux I.J., Bull J.J. Antibiotic Therapy Using Phage Depolymerases: Robustness across a Range of Conditions. Viruses. 2018;10:622. doi: 10.3390/v10110622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dabrowska K., Switała-Jelen K., Opolski A., Weber-Dabrowska B., Gorski A. Bacteriophage Penetration in Vertebrates. J. Appl. Microbiol. 2005;98:7–13. doi: 10.1111/j.1365-2672.2004.02422.x. [DOI] [PubMed] [Google Scholar]
- 37.Barr J.J., Auro R., Furlan M., Whiteson K.L., Erb M.L., Pogliano J., Stotland A., Wolkowicz R., Cutting A.S., Doran K.S. Bacteriophage Adhering to Mucus Provide a Non–Host-Derived Immunity. Proc. Natl. Acad. Sci. USA. 2013;110:10771–10776. doi: 10.1073/pnas.1305923110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Almeida G.M., Laanto E., Ashrafi R., Sundberg L.-R. Bacteriophage Adherence to Mucus Mediates Preventive Protection against Pathogenic Bacteria. MBio. 2019;10:e01984-19. doi: 10.1128/mBio.01984-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fattal B., Dotan A., Tchorsh Y., Parpari L., Shuval H. Penetration of E. coli and F2 Bacteriophage into Fish Tissues. Schr. Ver. Wasser- Boden-und Lufthyg. 1988;78:27–38. [PubMed] [Google Scholar]
- 40.Nguyen S., Baker K., Padman B.S., Patwa R., Dunstan R.A., Weston T.A., Schlosser K., Bailey B., Lithgow T., Lazarou M. Bacteriophage Transcytosis Provides a Mechanism to Cross Epithelial Cell Layers. MBio. 2017;8:e01874-17. doi: 10.1128/mBio.01874-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carroll-Portillo A., Lin H.C. Bacteriophage and the Innate Immune System: Access and Signaling. Microorganisms. 2019;7:625. doi: 10.3390/microorganisms7120625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Galindo-Villegas J., García-Moreno D., De Oliveira S., Meseguer J., Mulero V. Regulation of Immunity and Disease Resistance by Commensal Microbes and Chromatin Modifications during Zebrafish Development. Proc. Natl. Acad. Sci. USA. 2012;109:E2605–E2614. doi: 10.1073/pnas.1209920109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gorski A., Dabrowska K., Switala-Jeleń K., Nowaczyk M., Weber-Dabrowska B., Boratynski J., Wietrzyk J., Opolski A. New Insights into the Possible Role of Bacteriophages in Host Defense and Disease. Med. Immunol. 2003;2:1–5. doi: 10.1186/1476-9433-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.An T.-W., Kim S.-J., Lee Y.-D., Park J.-H., Chang H.-I. The Immune-Enhancing Effect of the Cronobacter sakazakii ES2 Phage Results in the Activation of Nuclear Factor-kB and Dendritic Cell Maturation via the Activation of IL-12p40 in the Mouse Bone Marrow. Immunol. Lett. 2014;157:1–8. doi: 10.1016/j.imlet.2013.10.007. [DOI] [PubMed] [Google Scholar]
- 45.Kurzępa A., Dąbrowska K., Skaradziński G., Górski A. Bacteriophage Interactions with Phagocytes and Their Potential Significance in Experimental Therapy. Clin. Exp. Med. 2009;9:93–100. doi: 10.1007/s10238-008-0027-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Górski A., Dąbrowska K., Międzybrodzki R., Weber-Dąbrowska B., Łusiak-Szelachowska M., Jończyk-Matysiak E., Borysowski J. Phages and Immunomodulation. Future Microbiol. 2017;12:905–914. doi: 10.2217/fmb-2017-0049. [DOI] [PubMed] [Google Scholar]
- 47.Van Belleghem J.D., Clement F., Merabishvili M., Lavigne R., Vaneechoutte M. Pro-and Anti-Inflammatory Responses of Peripheral Blood Mononuclear Cells Induced by Staphylococcus aureus and Pseudomonas aeruginosa Phages. Sci. Rep. 2017;7:1–13. doi: 10.1038/s41598-017-08336-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dufour N., Delattre R., Chevallereau A., Ricard J.-D., Debarbieux L. Phage Therapy of Pneumonia Is Not Associated with an Overstimulation of the Inflammatory Response Compared to Antibiotic Treatment in Mice. Antimicrob. Agents Chemother. 2019;63:e00379-19. doi: 10.1128/AAC.00379-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Khan Mirzaei M., Haileselassie Y., Navis M., Cooper C., Sverremark-Ekström E., Nilsson A.S. Morphologically Distinct Escherichia coli Bacteriophages Differ in Their Efficacy and Ability to Stimulate Cytokine Release in vitro. Front. Microbiol. 2016;7:437. doi: 10.3389/fmicb.2016.00437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Secor P.R., Michaels L.A., Smigiel K.S., Rohani M.G., Jennings L.K., Hisert K.B., Arrigoni A., Braun K.R., Birkland T.P., Lai Y. Filamentous Bacteriophage Produced by Pseudomonas aeruginosa Alters the Inflammatory Response and Promotes Noninvasive Infection in vivo. Infect. Immun. 2017;85:e00648-16. doi: 10.1128/IAI.00648-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trend S., Chang B.J., O’Dea M., Stick S.M., Kicic A., WAERP. AusREC. AREST CF Use of a Primary Epithelial Cell Screening Tool to Investigate Phage Therapy in Cystic Fibrosis. Front. Pharmacol. 2018;9:1330. doi: 10.3389/fphar.2018.01330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cafora M., Deflorian G., Forti F., Ferrari L., Binelli G., Briani F., Ghisotti D., Pistocchi A. Phage Therapy against Pseudomonas aeruginosa Infections in a Cystic Fibrosis Zebrafish Model. Sci. Rep. 2019;9:1–10. doi: 10.1038/s41598-018-37636-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cafora M., Forti F., Briani F., Ghisotti D., Pistocchi A. Phage Therapy Application to Counteract Pseudomonas aeruginosa Infection in Cystic Fibrosis Zebrafish Embryos. JoVE (J. Vis. Exp.) 2020:e61275. doi: 10.3791/61275. [DOI] [PubMed] [Google Scholar]
- 54.Nikapitiya C., Chandrarathna H., Dananjaya S., De Zoysa M., Lee J. Isolation and Characterization of Phage (ETP-1) Specific to Multidrug Resistant Pathogenic Edwardsiella tarda and Its in vivo Biocontrol Efficacy in Zebrafish (Danio rerio) Biologicals. 2020;63:14–23. doi: 10.1016/j.biologicals.2019.12.006. [DOI] [PubMed] [Google Scholar]
- 55.Yun S., Jun J.W., Giri S.S., Kim H.J., Chi C., Kim S.G., Kim S.W., Kang J.W., Han S.J., Kwon J. Immunostimulation of Cyprinus carpio Using Phage Lysate of Aeromonas hydrophila. Fish Shellfish. Immunol. 2019;86:680–687. doi: 10.1016/j.fsi.2018.11.076. [DOI] [PubMed] [Google Scholar]
- 56.Schulz P., Robak S., Dastych J., Siwicki A.K. Influence of Bacteriophages Cocktail on European Eel (Anguilla anguilla) Immunity and Survival after Experimental Challenge. Fish Shellfish. Immunol. 2019;84:28–37. doi: 10.1016/j.fsi.2018.09.056. [DOI] [PubMed] [Google Scholar]
- 57.Lin H., Caywood B.E., Rowlands D., Jr. Primary and Secondary Immune Responses of the Marine Toad (Bufo marinus) to Bacterophage F2. Immunology. 1971;20:373. [PMC free article] [PubMed] [Google Scholar]
- 58.Bradley S., Kim Y., Watson D. Immune Response by the Mouse to Orally Administered Actinophage. Proc. Soc. Exp. Biol. Med. 1963;113:686–688. doi: 10.3181/00379727-113-28462. [DOI] [PubMed] [Google Scholar]
- 59.Young R., Ruddle F.H. Inactivation of T-2 Bacteriophage by Sensitized Leucocytes in vitro. Nature. 1965;208:1105–1106. doi: 10.1038/2081105a0. [DOI] [PubMed] [Google Scholar]
- 60.Van Belleghem J.D., Dąbrowska K., Vaneechoutte M., Barr J.J., Bollyky P.L. Interactions between Bacteriophage, Bacteria, and the Mammalian Immune System. Viruses. 2019;11:10. doi: 10.3390/v11010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bekeredjian-Ding I.B., Wagner M., Hornung V., Giese T., Schnurr M., Endres S., Hartmann G. Plasmacytoid Dendritic Cells Control TLR7 Sensitivity of Naive B Cells via Type I IFN. J. Immunol. 2005;174:4043–4050. doi: 10.4049/jimmunol.174.7.4043. [DOI] [PubMed] [Google Scholar]
- 62.Hashiguchi S., Yamaguchi Y., Takeuchi O., Akira S., Sugimura K. Immunological Basis of M13 Phage Vaccine: Regulation under MyD88 and TLR9 Signaling. Biochem. Biophys. Res. Commun. 2010;402:19–22. doi: 10.1016/j.bbrc.2010.09.094. [DOI] [PubMed] [Google Scholar]
- 63.Hodyra-Stefaniak K., Miernikiewicz P., Drapała J., Drab M., Jończyk-Matysiak E., Lecion D., Kaźmierczak Z., Beta W., Majewska J., Harhala M. Mammalian Host-Versus-Phage Immune Response Determines Phage Fate in vivo. Sci. Rep. 2015;5:1–13. doi: 10.1038/srep14802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Krut O., Bekeredjian-Ding I. Contribution of the Immune Response to Phage Therapy. J. Immunol. 2018;200:3037–3044. doi: 10.4049/jimmunol.1701745. [DOI] [PubMed] [Google Scholar]
- 65.Bettarel Y., Combe M., Adingra A., Ndiaye A., Bouvier T., Panfili J., Durand J.-D. Hordes of Phages in the Gut of the Tilapia Sarotherodon melanotheron. Sci. Rep. 2018;8:1–6. doi: 10.1038/s41598-018-29643-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.He Y., Yang H. The Gastrointestinal Phage Communities of the Cultivated Freshwater Fishes. FEMS Microbiol. Lett. 2015;362:fnu027. doi: 10.1093/femsle/fnu027. [DOI] [PubMed] [Google Scholar]
- 67.Luo E., Aylward F.O., Mende D.R., DeLong E.F. Bacteriophage Distributions and Temporal Variability in the Ocean’s Interior. MBio. 2017;8:e01903-17. doi: 10.1128/mBio.01903-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pietilä M.K., Demina T.A., Atanasova N.S., Oksanen H.M., Bamford D.H. Archaeal Viruses and Bacteriophages: Comparisons and Contrasts. Trends Microbiol. 2014;22:334–344. doi: 10.1016/j.tim.2014.02.007. [DOI] [PubMed] [Google Scholar]
- 69.Ackermann H.-W. Classification of Bacteriophages. Bacteriophages. 2006;2:8–16. [Google Scholar]
- 70.Ackermann H.-W. 5500 Phages Examined in the Electron Microscope. Arch. Virol. 2007;152:227–243. doi: 10.1007/s00705-006-0849-1. [DOI] [PubMed] [Google Scholar]
- 71.Ackermann H.-W. Bacteriophages. Springer; Cham, Switzerland: 2009. Phage classification and characterization; pp. 127–140. [DOI] [PubMed] [Google Scholar]
- 72.Adriaenssens E.M., Edwards R., Nash J.H., Mahadevan P., Seto D., Ackermann H.-W., Lavigne R., Kropinski A.M. Integration of Genomic and Proteomic Analyses in the Classification of the Siphoviridae Family. Virology. 2015;477:144–154. doi: 10.1016/j.virol.2014.10.016. [DOI] [PubMed] [Google Scholar]
- 73.Lavigne R., Darius P., Summer E.J., Seto D., Mahadevan P., Nilsson A.S., Ackermann H.W., Kropinski A.M. Classification of Myoviridae Bacteriophages Using Protein Sequence Similarity. BMC Microbiol. 2009;9:1–16. doi: 10.1186/1471-2180-9-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lavigne R., Seto D., Mahadevan P., Ackermann H.-W., Kropinski A.M. Unifying Classical and Molecular Taxonomic Classification: Analysis of the Podoviridae Using BLASTP-Based Tools. Res. Microbiol. 2008;159:406–414. doi: 10.1016/j.resmic.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 75.Paterson W., Douglas R., Grinyer I., McDermott L. Isolation and Preliminary Characterization of Some Aeromonas salmonicida Bacteriophages. J. Fish. Board Can. 1969;26:629–632. doi: 10.1139/f69-056. [DOI] [Google Scholar]
- 76.Carlson K. Working with Bacteriophages: Common Techniques and Methodological Approaches. Volume 1 CRC Press; Boca Raton, FL, USA: 2005. [Google Scholar]
- 77.Dubrovin E.V., Voloshin A.G., Kraevsky S.V., Ignatyuk T.E., Abramchuk S.S., Yaminsky I.V., Ignatov S.G. Atomic Force Microscopy Investigation of Phage Infection of Bacteria. Langmuir. 2008;24:13068–13074. doi: 10.1021/la8022612. [DOI] [PubMed] [Google Scholar]
- 78.Zago M., Scaltriti E., Fornasari M.E., Rivetti C., Grolli S., Giraffa G., Ramoni R., Carminati D. Epifluorescence and Atomic Force Microscopy: Two Innovative Applications for Studying Phage–Host Interactions in Lactobacillus helveticus. J. Microbiol. Methods. 2012;88:41–46. doi: 10.1016/j.mimet.2011.10.006. [DOI] [PubMed] [Google Scholar]
- 79.Dubrovin E.V., Popova A.V., Kraevskiy S.V., Ignatov S.G., Ignatyuk T.E., Yaminsky I.V., Volozhantsev N.V. Atomic Force Microscopy Analysis of the Acinetobacter baumannii Bacteriophage AP22 Lytic Cycle. PLoS ONE. 2012;7:e47348. doi: 10.1371/journal.pone.0047348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Remuzgo-Martínez S., Lázaro-Díez M., Padilla D., Vega B., El Aamri F., Icardo J.M., Acosta F., Ramos-Vivas J. New Aspects in the Biology of Photobacterium damselae subsp. piscicida: Pili, Motility and Adherence to Solid Surfaces. Vet. Microbiol. 2014;174:247–254. doi: 10.1016/j.vetmic.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 81.Vivas J., Padilla D., Real F., Bravo J., Grasso V., Acosta F. Influence of Environmental Conditions on Biofilm Formation by Hafnia alvei Strains. Vet. Microbiol. 2008;129:150–155. doi: 10.1016/j.vetmic.2007.11.007. [DOI] [PubMed] [Google Scholar]
- 82.Cai W., Arias C.R. Biofilm Formation on Aquaculture Substrates by Selected Bacterial Fish Pathogens. J. Aquat. Anim. Health. 2017;29:95–104. doi: 10.1080/08997659.2017.1290711. [DOI] [PubMed] [Google Scholar]
- 83.Hall-Stoodley L., Costerton J.W., Stoodley P. Bacterial Biofilms: From the Natural Environment to Infectious Diseases. Nat. Rev. Microbiol. 2004;2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 84.Acosta F., Montero D., Izquierdo M., Galindo-Villegas J. High-Level Biocidal Products Effectively Eradicate Pathogenic γ-Proteobacteria Biofilms from Aquaculture Facilities. Aquaculture. 2021;532:736004. doi: 10.1016/j.aquaculture.2020.736004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Koo H., Allan R.N., Howlin R.P., Stoodley P., Hall-Stoodley L. Targeting Microbial Biofilms: Current and Prospective Therapeutic Strategies. Nat. Rev. Microbiol. 2017;15:740–755. doi: 10.1038/nrmicro.2017.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Clutterbuck A., Woods E., Knottenbelt D., Clegg P., Cochrane C., Percival S. Biofilms and Their Relevance to Veterinary Medicine. Vet. Microbiol. 2007;121:1–17. doi: 10.1016/j.vetmic.2006.12.029. [DOI] [PubMed] [Google Scholar]
- 87.MacKenzie K.D., Palmer M.B., Köster W.L., White A.P. Examining the Link between Biofilm Formation and the Ability of Pathogenic Salmonella Strains to Colonize Multiple Host Species. Front. Vet. Sci. 2017;4:138. doi: 10.3389/fvets.2017.00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Curtin J.J., Donlan R.M. Using Bacteriophages to Reduce Formation of Catheter-Associated Biofilms by Staphylococcus epidermidis. Antimicrob. Agents Chemother. 2006;50:1268–1275. doi: 10.1128/AAC.50.4.1268-1275.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Melo L.D., Ferreira R., Costa A.R., Oliveira H., Azeredo J. Efficacy and Safety Assessment of Two Enterococci Phages in an in vitro Biofilm Wound Model. Sci. Rep. 2019;9:1–12. doi: 10.1038/s41598-019-43115-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shlezinger M., Friedman M., Houri-Haddad Y., Hazan R., Beyth N. Phages in a Thermoreversible Sustained-Release Formulation Targeting E. faecalis in vitro and in vivo. PLoS ONE. 2019;14:e0219599. doi: 10.1371/journal.pone.0219599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gutierrez D., Ruas-Madiedo P., Martínez B., Rodríguez A., García P. Effective Removal of Staphylococcal Biofilms by the Endolysin LysH5. PLoS ONE. 2014;9:e107307. doi: 10.1371/journal.pone.0107307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gutiérrez D., Fernández L., Martínez B., Ruas-Madiedo P., García P., Rodríguez A. Real-Time Assessment of Staphylococcus aureus Biofilm Disruption by Phage-Derived Proteins. Front. Microbiol. 2017;8:1632. doi: 10.3389/fmicb.2017.01632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fu W., Forster T., Mayer O., Curtin J.J., Lehman S.M., Donlan R.M. Bacteriophage Cocktail for the Prevention of Biofilm Formation by Pseudomonas aeruginosa on Catheters in an in vitro Model System. Antimicrob. Agents Chemother. 2010;54:397–404. doi: 10.1128/AAC.00669-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Magin V., Garrec N., Andrés Y. Selection of Bacteriophages to Control in vitro 24 h Old Biofilm of Pseudomonas aeruginosa Isolated from Drinking and Thermal Water. Viruses. 2019;11:749. doi: 10.3390/v11080749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cano E.J., Caflisch K.M., Bollyky P.L., Van Belleghem J.D., Patel R., Fackler J., Brownstein M.J., Horne B., Biswas B., Henry M. Phage Therapy for Limb-Threatening Prosthetic Knee Klebsiella pneumoniae Infection: Case Report and in vitro Characterization of Anti-Biofilm Activity. Clin. Infect. Dis. 2021;73:e144–e151. doi: 10.1093/cid/ciaa705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fong S.A., Drilling A., Morales S., Cornet M.E., Woodworth B.A., Fokkens W.J., Psaltis A.J., Vreugde S., Wormald P.-J. Activity of Bacteriophages in Removing Biofilms of Pseudomonas aeruginosa Isolates from Chronic Rhinosinusitis Patients. Front. Cell. Infect. Microbiol. 2017;7:418. doi: 10.3389/fcimb.2017.00418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Oliveira A., Sousa J.C., Silva A.C., Melo L.D., Sillankorva S. Chestnut Honey and Bacteriophage Application to Control Pseudomonas Aeruginosa and Escherichia coli Biofilms: Evaluation in an ex vivo Wound Model. Front. Microbiol. 2018;9:1725. doi: 10.3389/fmicb.2018.01725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Oliveira A., Ribeiro H.G., Silva A.C., Silva M.D., Sousa J.C., Rodrigues C.F., Melo L.D., Henriques A.F., Sillankorva S. Synergistic Antimicrobial Interaction between Honey and Phage against Escherichia coli Biofilms. Front. Microbiol. 2017;8:2407. doi: 10.3389/fmicb.2017.02407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Papadopoulou A., Dalsgaard I., Wiklund T. Inhibition Activity of Compounds and Bacteriophages against Flavobacterium psychrophilum Biofilms in vitro. J. Aquat. Anim. Health. 2019;31:225–238. doi: 10.1002/aah.10069. [DOI] [PubMed] [Google Scholar]
- 100.Le T.S., Nguyen T.H., Vo H.P., Doan V.C., Nguyen H.L., Tran M.T., Tran T.T., Southgate P.C., Kurtböke D.İ. Protective Effects of Bacteriophages against Aeromonas hydrophila Causing Motile Aeromonas septicemia (MAS) in Striped Catfish. Antibiotics. 2018;7:16. doi: 10.3390/antibiotics7010016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Liu J., Gao S., Dong Y., Lu C., Liu Y. Isolation and Characterization of Bacteriophages against Virulent Aeromonas hydrophila. BMC Microbiol. 2020;20:1–13. doi: 10.1186/s12866-020-01811-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Easwaran M., Dananjaya S., Park S.C., Lee J., Shin H., De Zoysa M. Characterization of Bacteriophage PAh-1 and Its Protective Effects on Experimental Infection of Aeromonas hydrophila in Zebrafish (Danio Rerio) J. Fish Dis. 2017;40:841–846. doi: 10.1111/jfd.12536. [DOI] [PubMed] [Google Scholar]
- 103.Akmal M., Rahimi-Midani A., Hafeez-ur-Rehman M., Hussain A., Choi T.-J. Isolation, Characterization, and Application of a Bacteriophage Infecting the Fish Pathogen Aeromonas hydrophila. Pathogens. 2020;9:215. doi: 10.3390/pathogens9030215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chandrarathna H., Nikapitiya C., Dananjaya S., De Silva B., Heo G.-J., De Zoysa M., Lee J. Isolation and Characterization of Phage AHP-1 and Its Combined Effect with Chloramphenicol to Control Aeromonas hydrophila. Braz. J. Microbiol. 2020;51:409–416. doi: 10.1007/s42770-019-00178-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cheng Y., Gao D., Xia Y., Wang Z., Bai M., Luo K., Cui X., Wang Y., Zhang S., Xiao W. Characterization of Novel Bacteriophage AhyVDH1 and Its Lytic Activity Against Aeromonas hydrophila. Curr. Microbiol. 2021;78:329–337. doi: 10.1007/s00284-020-02279-7. [DOI] [PubMed] [Google Scholar]
- 106.Cao Y., Li S., Wang D., Zhao J., Xu L., Liu H., Lu T., Mou Z. Genomic Characterization of a Novel Virulent Phage Infecting the Aeromonas hydrophila Isolated from Rainbow Trout (Oncorhynchus mykiss) Virus Res. 2019;273:197764. doi: 10.1016/j.virusres.2019.197764. [DOI] [PubMed] [Google Scholar]
- 107.Wu J.-L., Lin H.-M., Jan L., Hsu Y.-L., CHANG L.-H. Biological Control of Fish Bacterial Pathogen, Aeromonas hydrophila, by Bacteriophage AH 1. Fish Pathol. 1981;15:271–276. doi: 10.3147/jsfp.15.271. [DOI] [Google Scholar]
- 108.Tu V.Q., Nguyen T.-T., Tran X.T., Millard A.D., Phan H.T., Le N.P., Dang O.T., Hoang H.A. Complete Genome Sequence of a Novel Lytic Phage Infecting Aeromonas hydrophila, an Infectious Agent in Striped Catfish (Pangasianodon hypophthalmus) Arch. Virol. 2020;165:2973–2977. doi: 10.1007/s00705-020-04793-2. [DOI] [PubMed] [Google Scholar]
- 109.Hoang Hoang A., Xuan Tran T.T., Nga L.E.P., Oanh Dang T.H. Selection of Phages to Control Aeromonas hydrophila–an Infectious Agent in Striped Catfish. Biocontrol Sci. 2019;24:23–28. doi: 10.4265/bio.24.23. [DOI] [PubMed] [Google Scholar]
- 110.Jun J.W., Kim J.H., Shin S.P., Han J.E., Chai J.Y., Park S.C. Protective Effects of the Aeromonas Phages PAh1-C and PAh6-C against Mass Mortality of the Cyprinid Loach (Misgurnus anguillicaudatus) Caused by Aeromonas hydrophila. Aquaculture. 2013;416:289–295. doi: 10.1016/j.aquaculture.2013.09.045. [DOI] [Google Scholar]
- 111.Wang J.-B., Lin N.-T., Tseng Y.-H., Weng S.-F. Genomic Characterization of the Novel Aeromonas hydrophila Phage Ahp1 Suggests the Derivation of a New Subgroup from PhiKMV-like Family. PLoS ONE. 2016;11:e0162060. doi: 10.1371/journal.pone.0162060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Haq I.U., Chaudhry W.N., Andleeb S., Qadri I. Isolation and Partial Characterization of a Virulent Bacteriophage IHQ1 Specific for Aeromonas punctata from Stream Water. Microb. Ecol. 2012;63:954–963. doi: 10.1007/s00248-011-9944-2. [DOI] [PubMed] [Google Scholar]
- 113.Vincent A.T., Paquet V.E., Bernatchez A., Tremblay D.M., Moineau S., Charette S.J. Characterization and Diversity of Phages Infecting Aeromonas salmonicida subsp. salmonicida. Sci. Rep. 2017;7:1–10. doi: 10.1038/s41598-017-07401-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kim J.H., Son J.S., Choi Y.J., Choresca C.H., Jr., Shin S.P., Han J.E., Jun J.W., Park S.C. Complete Genome Sequence and Characterization of a Broad-Host Range T4-like Bacteriophage PhiAS5 Infecting Aeromonas salmonicida subsp. salmonicida. Vet. Microbiol. 2012;157:164–171. doi: 10.1016/j.vetmic.2011.12.016. [DOI] [PubMed] [Google Scholar]
- 115.Kim J., Son J., Choi Y., Choresca C., Shin S., Han J., Jun J., Kang D., Oh C., Heo S. Isolation and Characterization of a Lytic Myoviridae Bacteriophage PAS-1 with Broad Infectivity in Aeromonas salmonicida. Curr. Microbiol. 2012;64:418–426. doi: 10.1007/s00284-012-0091-x. [DOI] [PubMed] [Google Scholar]
- 116.Yang Z., Yuan S., Chen L., Liu Q., Zhang H., Ma Y., Wei T., Huang S. Complete Genome Analysis of Bacteriophage AsXd-1, a New Member of the Genus Hk97virus, Family Siphoviridae. Arch. Virol. 2018;163:3195–3197. doi: 10.1007/s00705-018-3977-5. [DOI] [PubMed] [Google Scholar]
- 117.Silva Y.J., Moreirinha C., Pereira C., Costa L., Rocha R.J., Cunha Â., Gomes N.C., Calado R., Almeida A. Biological Control of Aeromonas salmonicida Infection in Juvenile Senegalese Sole (Solea senegalensis) with Phage AS-A. Aquaculture. 2016;450:225–233. doi: 10.1016/j.aquaculture.2015.07.025. [DOI] [Google Scholar]
- 118.Duarte J., Pereira C., Costa P., Almeida A. Bacteriophages with Potential to Inactivate Aeromonas hydrophila in Cockles: In vitro and in vivo Preliminary Studies. Antibiotics. 2021;10:710. doi: 10.3390/antibiotics10060710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Imbeault S., Parent S., Lagacé M., Uhland C.F., Blais J.-F. Using Bacteriophages to Prevent Furunculosis Caused by Aeromonas salmonicida in Farmed Brook Trout. J. Aquat. Anim. Health. 2006;18:203–214. doi: 10.1577/H06-019.1. [DOI] [Google Scholar]
- 120.Petrov V., Karam J. Diversity of Structure and Function of DNA Polymerase (Gp43) of T4-Related Bacteriophages. Biochemistry (Moscow) 2004;69:1213–1218. doi: 10.1007/s10541-005-0066-7. [DOI] [PubMed] [Google Scholar]
- 121.He Y., Huang Z., Zhang X., Zhang Z., Gong M., Pan X., Wei D., Yang H. Characterization of a Novel Lytic Myophage, PhiA8-29, Infecting Aeromonas Strains. Arch. Virol. 2019;164:893–896. doi: 10.1007/s00705-018-4109-y. [DOI] [PubMed] [Google Scholar]
- 122.Jia K., Yang N., Zhang X., Cai R., Zhang Y., Tian J., Raza S.H.A., Kang Y., Qian A., Li Y. Genomic, Morphological and Functional Characterization of Virulent Bacteriophage IME-JL8 Targeting Citrobacter freundii. Front. Microbiol. 2020;11:2967. doi: 10.3389/fmicb.2020.585261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Walakira J., Carrias A., Hossain M., Jones E., Terhune J., Liles M. Identification and Characterization of Bacteriophages Specific to the Catfish Pathogen, Edwardsiella ictaluri. J. Appl. Microbiol. 2008;105:2133–2142. doi: 10.1111/j.1365-2672.2008.03933.x. [DOI] [PubMed] [Google Scholar]
- 124.Carrias A., Welch T.J., Waldbieser G.C., Mead D.A., Terhune J.S., Liles M.R. Comparative Genomic Analysis of Bacteriophages Specific to the Channel Catfish Pathogen Edwardsiella ictaluri. Virol. J. 2011;8:1–12. doi: 10.1186/1743-422X-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Yasuike M., Kai W., Nakamura Y., Fujiwara A., Kawato Y., Hassan E.S., Mahmoud M.M., Nagai S., Kobayashi T., Ototake M. Complete Genome Sequence of the Edwardsiella ictaluri-Specific Bacteriophage PEi21, Isolated from River Water in Japan. Genome Announc. 2014;2:e00228-14. doi: 10.1128/genomeA.00228-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hassan E.S., Mahmoud M.M., Kawato Y., Nagai T., Kawaguchi O., Iida Y., Yuasa K., Nakai T. Subclinical Edwardsiella ictaluri Infection of Wild Ayu Plecoglossus Altivelis. Fish Pathol. 2012;47:64–73. doi: 10.3147/jsfp.47.64. [DOI] [Google Scholar]
- 127.Hoang H.A., Yen M.H., Ngoan V.T., Nga L.P., Oanh D.T. Virulent Bacteriophage of Edwardsiella ictaluri Isolated from Kidney and Liver of Striped Catfish Pangasianodon Hypophthalmus in Vietnam. Dis. Aquat. Org. 2018;132:49–56. doi: 10.3354/dao03302. [DOI] [PubMed] [Google Scholar]
- 128.Kim S.G., Giri S.S., Yun S., Kim H.J., Kim S.W., Kang J.W., Han S.J., Kwon J., Jun J.W., Oh W.T. Genomic Characterization of Bacteriophage PEt-SU, a Novel PhiKZ-Related Virus Infecting Edwardsiella tarda. Arch. Virol. 2020;165:219–222. doi: 10.1007/s00705-019-04432-5. [DOI] [PubMed] [Google Scholar]
- 129.Cui H., Zhang J., Cong C., Wang L., Li X., Murtaza B., Xu Y. Complete Genome Analysis of the Novel Edwardsiella tarda Phage VB_EtaM_ET-ABTNL-9. Arch. Virol. 2020;165:1241–1244. doi: 10.1007/s00705-020-04600-y. [DOI] [PubMed] [Google Scholar]
- 130.Yasuike M., Nishiki I., Iwasaki Y., Nakamura Y., Fujiwara A., Sugaya E., Kawato Y., Nagai S., Kobayashi T., Ototake M. Full-Genome Sequence of a Novel Myovirus, GF-2, Infecting Edwardsiella tarda: Comparison with Other Edwardsiella myoviral Genomes. Arch. Virol. 2015;160:2129–2133. doi: 10.1007/s00705-015-2472-5. [DOI] [PubMed] [Google Scholar]
- 131.Laanto E., Bamford J.K., Ravantti J.J., Sundberg L.-R. The Use of Phage FCL-2 as an Alternative to Chemotherapy against Columnaris Disease in Aquaculture. Front. Microbiol. 2015;6:829. doi: 10.3389/fmicb.2015.00829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Laanto E., Sundberg L.-R., Bamford J.K. Phage Specificity of the Freshwater Fish Pathogen Flavobacterium columnare. Appl. Environ. Microbiol. 2011;77:7868–7872. doi: 10.1128/AEM.05574-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Prasad Y., Kumar D., Sharma A. Lytic Bacteriophages Specific to Flavobacterium columnare Rescue Catfish, Clarias batrachus (Linn.) from Columnaris Disease. J. Environ. Biol. 2011;32:161–168. [PubMed] [Google Scholar]
- 134.Christiansen R.H., Dalsgaard I., Middelboe M., Lauritsen A.H., Madsen L. Detection and Quantification of Flavobacterium psychrophilum-Specific Bacteriophages in vivo in Rainbow Trout upon Oral Administration: Implications for Disease Control in Aquaculture. Appl. Environ. Microbiol. 2014;80:7683–7693. doi: 10.1128/AEM.02386-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Stenholm A.R., Dalsgaard I., Middelboe M. Isolation and Characterization of Bacteriophages Infecting the Fish Pathogen Flavobacterium psychrophilum. Appl. Environ. Microbiol. 2008;74:4070–4078. doi: 10.1128/AEM.00428-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kim J.H., Gomez D.K., Nakai T., Park S.C. Isolation and Identification of Bacteriophages Infecting Ayu Plecoglossus altivelis Altivelis Specific Flavobacterium psychrophilum. Vet. Microbiol. 2010;140:109–115. doi: 10.1016/j.vetmic.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 137.Yamaki S., Kawai Y., Yamazaki K. Characterization of a Novel Bacteriophage, Phda1, Infecting the Histamine-producing Photobacterium damselae subsp. damselae. J. Appl. Microbiol. 2015;118:1541–1550. doi: 10.1111/jam.12809. [DOI] [PubMed] [Google Scholar]
- 138.Veyrand-Quirós B., Gómez-Gil B., Lomeli-Ortega C.O., Escobedo-Fregoso C., Millard A.D., Tovar-Ramírez D., Balcázar J.L., Quiroz-Guzmán E. Use of Bacteriophage VB_Pd_PDCC-1 as Biological Control Agent of Photobacterium Damselae Subsp. Damselae during Hatching of Longfin Yellowtail (Seriola rivoliana) Eggs. J. Appl. Microbiol. 2020;129:1497–1510. doi: 10.1111/jam.14744. [DOI] [PubMed] [Google Scholar]
- 139.Kawato Y., Yasuike M., Nakamura Y., Shigenobu Y., Fujiwara A., Sano M., Nakai T. Complete Genome Sequence Analysis of Two Pseudomonas plecoglossicida Phages, Potential Therapeutic Agents. Appl. Environ. Microbiol. 2015;81:874–881. doi: 10.1128/AEM.03038-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Park S.C., Shimamura I., Fukunaga M., Mori K.-I., Nakai T. Isolation of Bacteriophages Specific to a Fish Pathogen, Pseudomonas plecoglossicida, as a Candidate for Disease Control. Appl. Environ. Microbiol. 2000;66:1416–1422. doi: 10.1128/AEM.66.4.1416-1422.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Khairnar K., Raut M.P., Chandekar R.H., Sanmukh S.G., Paunikar W.N. Novel Bacteriophage Therapy for Controlling Metallo-Beta-Lactamase Producing Pseudomonas aeruginosa Infection in Catfish. BMC Vet. Res. 2013;9:1–9. doi: 10.1186/1746-6148-9-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Yang Z., Tao X., Zhang H., Rao S., Gao L., Pan Z., Jiao X. Isolation and Characterization of Virulent Phages Infecting Shewanella baltica and Shewanella putrefaciens, and Their Application for Biopreservation of Chilled Channel Catfish (Ictalurus punctatus) Int. J. Food Microbiol. 2019;292:107–117. doi: 10.1016/j.ijfoodmicro.2018.12.020. [DOI] [PubMed] [Google Scholar]
- 143.Kawato Y., Istiqomah I., Gaafar A.Y., Hanaoka M., Ishimaru K., Yasuike M., Nishiki I., Nakamura Y., Fujiwara A., Nakai T. A Novel Jumbo Tenacibaculum maritimum Lytic Phage with Head-Fiber-like Appendages. Arch. Virol. 2020;165:303–311. doi: 10.1007/s00705-019-04485-6. [DOI] [PubMed] [Google Scholar]
- 144.Kokkari C., Sarropoulou E., Bastias R., Mandalakis M., Katharios P. Isolation and Characterization of a Novel Bacteriophage Infecting Vibrio alginolyticus. Arch. Microbiol. 2018;200:707–718. doi: 10.1007/s00203-018-1480-8. [DOI] [PubMed] [Google Scholar]
- 145.Lal T.M., Sano M., Hatai K., Ransangan J. Complete Genome Sequence of a Giant Vibrio Phage ValKK3 Infecting Vibrio alginolyticus. Genom. Data. 2016;8:37–38. doi: 10.1016/j.gdata.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kalatzis P.G., Bastías R., Kokkari C., Katharios P. Isolation and Characterization of Two Lytic Bacteriophages, ΦSt2 and ΦGrn1; Phage Therapy Application for Biological Control of Vibrio alginolyticus in Aquaculture Live Feeds. PLoS ONE. 2016;11:e0151101. doi: 10.1371/journal.pone.0151101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Higuera G., Bastías R., Tsertsvadze G., Romero J., Espejo R.T. Recently Discovered Vibrio anguillarum Phages Can Protect against Experimentally Induced Vibriosis in Atlantic Salmon, Salmo salar. Aquaculture. 2013;392:128–133. doi: 10.1016/j.aquaculture.2013.02.013. [DOI] [Google Scholar]
- 148.Silva Y.J., Costa L., Pereira C., Mateus C., Cunha A., Calado R., Gomes N.C., Pardo M.A., Hernandez I., Almeida A. Phage Therapy as an Approach to Prevent Vibrio anguillarum Infections in Fish Larvae Production. PLoS ONE. 2014;9:e114197. doi: 10.1371/journal.pone.0114197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Tan D., Gram L., Middelboe M. Vibriophages and Their Interactions with the Fish Pathogen Vibrio anguillarum. Appl. Environ. Microbiol. 2014;80:3128–3140. doi: 10.1128/AEM.03544-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Nuidate T., Kuaphiriyakul A., Surachat K., Mittraparp-Arthorn P. Induction and Genome Analysis of HY01, a Newly Reported Prophage from an Emerging Shrimp Pathogen Vibrio campbellii. Microorganisms. 2021;9:400. doi: 10.3390/microorganisms9020400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lomelí-Ortega C.O., Martínez-Sández A., Barajas-Sandoval D.R., Reyes A.G., Magallón-Barajas F., Veyrand-Quíros B., Gannon L., Harrison C., Michniewski S., Millard A. Isolation and Characterization of Vibriophage VB_Vc_SrVc9: An Effective Agent in Preventing Vibrio campbellii Infections in Brine Shrimp Nauplii (Artemia franciscana) J. Appl. Microbiol. 2021;131:36–49. doi: 10.1111/jam.14937. [DOI] [PubMed] [Google Scholar]
- 152.Vinod M., Shivu M., Umesha K., Rajeeva B., Krohne G., Karunasagar I., Karunasagar I. Isolation of Vibrio harveyi Bacteriophage with a Potential for Biocontrol of Luminous Vibriosis in Hatchery Environments. Aquaculture. 2006;255:117–124. doi: 10.1016/j.aquaculture.2005.12.003. [DOI] [Google Scholar]
- 153.Oakey H., Owens L. A New Bacteriophage, VHML, Isolated from a Toxin-producing Strain of Vibrio harveyi in Tropical Australia. J. Appl. Microbiol. 2000;89:702–709. doi: 10.1046/j.1365-2672.2000.01169.x. [DOI] [PubMed] [Google Scholar]
- 154.Phumkhachorn P., Rattanachaikunsopon P. Isolation and Partial Characterization of a Bacteriophage Infecting the Shrimp Pathogen Vibrio harveyi. Afr. J. Microbiol. Res. 2010;4:1794–1800. [Google Scholar]
- 155.Stalin N., Srinivasan P. Efficacy of Potential Phage Cocktails against Vibrio harveyi and Closely Related Vibrio Species Isolated from Shrimp Aquaculture Environment in the South East Coast of India. Vet. Microbiol. 2017;207:83–96. doi: 10.1016/j.vetmic.2017.06.006. [DOI] [PubMed] [Google Scholar]
- 156.Wang Y., Barton M., Elliott L., Li X., Abraham S., O’Dea M., Munro J. Bacteriophage Therapy for the Control of Vibrio harveyi in Greenlip Abalone (Haliotis laevigata) Aquaculture. 2017;473:251–258. doi: 10.1016/j.aquaculture.2017.01.003. [DOI] [Google Scholar]
- 157.Crothers-Stomps C., Høj L., Bourne D., Hall M., Owens L. Isolation of Lytic Bacteriophage against Vibrio harveyi. J. Appl. Microbiol. 2010;108:1744–1750. doi: 10.1111/j.1365-2672.2009.04578.x. [DOI] [PubMed] [Google Scholar]
- 158.Patil J.R., Desai S.N., Roy P., Durgaiah M., Saravanan R.S., Vipra A. Simulated Hatchery System to Assess Bacteriophage Efficacy against Vibrio harveyi. Dis. Aquat. Org. 2014;112:113–119. doi: 10.3354/dao02806. [DOI] [PubMed] [Google Scholar]
- 159.Karunasagar I., Shivu M., Girisha S., Krohne G., Karunasagar I. Biocontrol of Pathogens in Shrimp Hatcheries Using Bacteriophages. Aquaculture. 2007;268:288–292. doi: 10.1016/j.aquaculture.2007.04.049. [DOI] [Google Scholar]
- 160.Shivu M.M., Rajeeva B.C., Girisha S.K., Karunasagar I., Krohne G., Karunasagar I. Molecular Characterization of Vibrio harveyi Bacteriophages Isolated from Aquaculture Environments along the Coast of India. Environ. Microbiol. 2007;9:322–331. doi: 10.1111/j.1462-2920.2006.01140.x. [DOI] [PubMed] [Google Scholar]
- 161.Lal T.M., Sano M., Ransangan J. Isolation and Characterization of Large Marine Bacteriophage (Myoviridae), VhKM4 Infecting Vibrio harveyi. J. Aquat. Anim. Health. 2017;29:26–30. doi: 10.1080/08997659.2016.1249578. [DOI] [PubMed] [Google Scholar]
- 162.Echeverría-Vega A., Morales-Vicencio P., Saez-Saavedra C., Ceh J., Araya R. The Complete Genome Sequence and Analysis of VB_VorS-PVo5, a Vibrio Phage Infectious to the Pathogenic Bacterium Vibrio Ordalii ATCC-33509. Stand. Genom. Sci. 2016;11:1–8. doi: 10.1186/s40793-016-0166-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Peng Y., Ding Y., Lin H., Wang J. Isolation, Identification and Lysis Properties Analysis of a Vibrio parahaemolyticus Phage VPp1. Mar. Sci. 2013;37:96–101. [Google Scholar]
- 164.Matsuzaki S., Inoue T., Tanaka S., Koga T., Kuroda M., Kimura S., Imai S. Characterization of a Novel Vibrio parahaemolyticus Phage, KVP241, and Its Relatives Frequently Isolated from Seawater. Microbiol. Immunol. 2000;44:953–956. doi: 10.1111/j.1348-0421.2000.tb02589.x. [DOI] [PubMed] [Google Scholar]
- 165.Matsuzaki S., Tanaka S., Koga T., Kawata T. A Broad-host-range Vibriophage, KVP40, Isolated from Sea Water. Microbiol. Immunol. 1992;36:93–97. doi: 10.1111/j.1348-0421.1992.tb01645.x. [DOI] [PubMed] [Google Scholar]
- 166.Onarinde B.A., Dixon R.A. Prospects for Biocontrol of Vibrio parahaemolyticus Contamination in Blue Mussels (Mytilus edulus)—A Year-Long Study. Front. Microbiol. 2018;9:1043. doi: 10.3389/fmicb.2018.01043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kim J.H., Jun J.W., Choresca C.H., Shin S.P., Han J.E., Park S.C. Complete Genome Sequence of a Novel Marine Siphovirus, PVp-1, Infecting Vibrio parahaemolyticus. J. Virol. 2012;86:7013–7014. doi: 10.1128/JVI.00742-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Jun J.W., Kim H.J., Yun S.K., Chai J.Y., Park S.C. Eating Oysters without Risk of Vibriosis: Application of a Bacteriophage against Vibrio parahaemolyticus in Oysters. Int. J. Food Microbiol. 2014;188:31–35. doi: 10.1016/j.ijfoodmicro.2014.07.007. [DOI] [PubMed] [Google Scholar]
- 169.Yang M., Liang Y., Huang S., Zhang J., Wang J., Chen H., Ye Y., Gao X., Wu Q., Tan Z. Isolation and Characterization of the Novel Phages VB_VpS_BA3 and VB_VpS_CA8 for Lysing Vibrio parahaemolyticus. Front. Microbiol. 2020;11:259. doi: 10.3389/fmicb.2020.00259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Matamp N., Bhat S.G. Genome Characterization of Novel Lytic Myoviridae Bacteriophage ΦVP-1 Enhances Its Applicability against MDR-Biofilm-Forming Vibrio parahaemolyticus. Arch. Virol. 2020;165:387–396. doi: 10.1007/s00705-019-04493-6. [DOI] [PubMed] [Google Scholar]
- 171.Lal T.M., Sano M., Ransangan J. Genome Characterization of a Novel Vibriophage VpKK5 (Siphoviridae) Specific to Fish Pathogenic Strain of Vibrio parahaemolyticus. J. Basic Microbiol. 2016;56:872–888. doi: 10.1002/jobm.201500611. [DOI] [PubMed] [Google Scholar]
- 172.Lal T.M., Ransangan J. Complete Genome Sequence of VpKK5, a Novel Vibrio parahaemolyticus Lytic Siphophage. Genome Announc. 2015;3:e01381-14. doi: 10.1128/genomeA.01381-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Li Z., Li X., Zhang J., Wang X., Wang L., Cao Z., Xu Y. Use of Phages to Control Vibrio splendidus Infection in the Juvenile Sea Cucumber Apostichopus japonicus. Fish Shellfish. Immunol. 2016;54:302–311. doi: 10.1016/j.fsi.2016.04.026. [DOI] [PubMed] [Google Scholar]
- 174.Katharios P., Kalatzis P.G., Kokkari C., Sarropoulou E., Middelboe M. Isolation and Characterization of a N4-like Lytic Bacteriophage Infecting Vibrio splendidus, a Pathogen of Fish and Bivalves. PLoS ONE. 2017;12:e0190083. doi: 10.1371/journal.pone.0190083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Kim H.J., Jun J.W., Giri S.S., Chi C., Yun S., Kim S.G., Kim S.W., Kang J.W., Han S.J., Kwon J. Application of the Bacteriophage PVco-14 to Prevent Vibrio coralliilyticus Infection in Pacific Oyster (Crassostrea gigas) Larvae. J. Invertebr. Pathol. 2019;167:107244. doi: 10.1016/j.jip.2019.107244. [DOI] [PubMed] [Google Scholar]
- 176.Lee H.S., Choi S., Choi S.H. Complete Genome Sequence of Vibrio vulnificus Bacteriophage SSP002. J. Virol. 2012;86:7711. doi: 10.1128/JVI.00972-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Lee H.S., Choi S., Shin H., Lee J.-H., Choi S.H. Vibrio vulnificus Bacteriophage SSP002 as a Possible Biocontrol Agent. Appl. Environ. Microbiol. 2014;80:515–524. doi: 10.1128/AEM.02675-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Kim H.-J., Kim Y.-T., Kim H.B., Choi S.H., Lee J.-H. Characterization of Bacteriophage VVP001 and Its Application for the Inhibition of Vibrio Vulnificus Causing Seafood-Borne Diseases. Food Microbiol. 2021;94:103630. doi: 10.1016/j.fm.2020.103630. [DOI] [PubMed] [Google Scholar]
- 179.Srinivasan P., Ramasamy P. Morphological Characterization and Biocontrol Effects of Vibrio vulnificus Phages against Vibriosis in the Shrimp Aquaculture Environment. Microb. Pathog. 2017;111:472–480. doi: 10.1016/j.micpath.2017.09.024. [DOI] [PubMed] [Google Scholar]
- 180.Chen L., Fan J., Yan T., Liu Q., Yuan S., Zhang H., Yang J., Deng D., Huang S., Ma Y. Isolation and Characterization of Specific Phages to Prepare a Cocktail Preventing Vibrio Sp. Va-F3 Infections in Shrimp (Litopenaeus vannamei) Front. Microbiol. 2019;10:2337. doi: 10.3389/fmicb.2019.02337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Welch T.J. Characterization of a Novel Yersinia ruckeri Serotype O1-specific Bacteriophage with Virulence-neutralizing Activity. J. Fish Dis. 2020;43:285–293. doi: 10.1111/jfd.13124. [DOI] [PubMed] [Google Scholar]
- 182.Stevenson R., Airdrie D. Isolation of Yersinia ruckeri Bacteriophages. Appl. Environ. Microbiol. 1984;47:1201–1205. doi: 10.1128/aem.47.6.1201-1205.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Kiljunen S., Hakala K., Pinta E., Huttunen S., Pluta P., Gador A., Lönnberg H., Skurnik M. Yersiniophage ΦR1-37 Is a Tailed Bacteriophage Having a 270 Kb DNA Genome with Thymidine Replaced by Deoxyuridine. Microbiology. 2005;151:4093–4102. doi: 10.1099/mic.0.28265-0. [DOI] [PubMed] [Google Scholar]
- 184.Leskinen K., Pajunen M.I., Vilanova M.V.G.-R., Kiljunen S., Nelson A., Smith D., Skurnik M. YerA41, a Yersinia ruckeri Bacteriophage: Determination of a Non-Sequencable DNA Bacteriophage Genome via RNA-Sequencing. Viruses. 2020;12:620. doi: 10.3390/v12060620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Park K., Matsuoka S., Nakai T., Muroga K. A Virulent Bacteriophage of Lactococcus garvieae (Formerly Enterococcus Seriolicida) Isolated from Yellowtail Seriola quinqueradiata. Dis. Aquat. Org. 1997;29:145–149. doi: 10.3354/dao029145. [DOI] [Google Scholar]
- 186.Park K.H., Kato H., Nakai T., Muroga K. Phage Typing of Lactococcus garvieae (Formerly Enterococcus Seriolicida) a Pathogen of Cultured Yellowtail. Fish. Sci. 1998;64:62–64. doi: 10.2331/fishsci.64.62. [DOI] [Google Scholar]
- 187.Nakai T., Sugimoto R., Park K.-H., Matsuoka S., Mori K., Nishioka T., Maruyama K. Protective Effects of Bacteriophage on Experimental Lactococcus garvieae Infection in Yellowtail. Dis. Aquat. Org. 1999;37:33–41. doi: 10.3354/dao037033. [DOI] [PubMed] [Google Scholar]
- 188.Ooyama T., Hirokawa Y., Minami T., Yasuda H., Nakai T., Endo M., Ruangpan L., Yoshida T. Cell-Surface Properties of Lactococcus Garvieae Strains and Their Immunogenicity in the Yellowtail Seriola quinqueradiata. Dis. Aquat. Org. 2002;51:169–177. doi: 10.3354/dao051169. [DOI] [PubMed] [Google Scholar]
- 189.Eraclio G., Tremblay D.M., Lacelle-Côté A., Labrie S.J., Fortina M.G., Moineau S. A Virulent Phage Infecting Lactococcus garvieae, with Homology to Lactococcus lactis Phages. Appl. Environ. Microbiol. 2015;81:8358–8365. doi: 10.1128/AEM.02603-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Hoai T.D., Nishiki I., Yoshida T. Properties and Genomic Analysis of Lactococcus garvieae Lysogenic Bacteriophage PLgT-1, a New Member of Siphoviridae, with Homology to Lactococcus lactis Phages. Virus Res. 2016;222:13–23. doi: 10.1016/j.virusres.2016.05.021. [DOI] [PubMed] [Google Scholar]
- 191.Hoai T., Yoshida T. Induction and Characterization of a Lysogenic Bacteriophage of Lactococcus garvieae Isolated from Marine Fish Species. J. Fish Dis. 2016;39:799–808. doi: 10.1111/jfd.12410. [DOI] [PubMed] [Google Scholar]
- 192.Hoai T.D., Nishiki I., Fujiwara A., Yoshida T., Nakai T. Comparative Genomic Analysis of Three Lytic Lactococcus garvieae Phages, Novel Phages with Genome Architecture Linking the 936 Phage Species of Lactococcus lactis. Mar. Genom. 2019;48:100696. doi: 10.1016/j.margen.2019.100696. [DOI] [PubMed] [Google Scholar]
- 193.Ghasemi S.M., Bouzari M., Yoon B.H., Chang H.-I. Comparative Genomic Analysis of Lactococcus garvieae Phage WP-2, a New Member of Picovirinae Subfamily of Podoviridae. Gene. 2014;551:222–229. doi: 10.1016/j.gene.2014.08.060. [DOI] [PubMed] [Google Scholar]
- 194.Luo X., Liao G., Liu C., Jiang X., Lin M., Zhao C., Tao J., Huang Z. Characterization of Bacteriophage HN 48 and Its Protective Effects in Nile Tilapia Oreochromis niloticus against Streptococcus agalactiae Infections. J. Fish Dis. 2018;41:1477–1484. doi: 10.1111/jfd.12838. [DOI] [PubMed] [Google Scholar]
- 195.Wright E., Elliman J., Owens L. Induction and Characterization of Lysogenic Bacteriophages from Streptococcus iniae. J. Appl. Microbiol. 2013;114:1616–1624. doi: 10.1111/jam.12192. [DOI] [PubMed] [Google Scholar]
- 196.Hoai T.D., Mitomi K., Nishiki I., Yoshida T. A Lytic Bacteriophage of the Newly Emerging Rainbow Trout Pathogen Weissella Ceti. Virus Res. 2018;247:34–39. doi: 10.1016/j.virusres.2018.01.016. [DOI] [PubMed] [Google Scholar]
- 197.Schulz P., Pajdak-Czaus J., Robak S., Dastych J., Siwicki A.K. Bacteriophage-based Cocktail Modulates Selected Immunological Parameters and Post-challenge Survival of Rainbow Trout (Oncorhynchus mykiss) J. Fish Dis. 2019;42:1151–1160. doi: 10.1111/jfd.13026. [DOI] [PubMed] [Google Scholar]
- 198.Moye Z.D., Woolston J., Abbeele P.v.d., Duysburgh C., Verstrepen L., Das C.R., Marzorati M., Sulakvelidze A. A Bacteriophage Cocktail Eliminates Salmonella Typhimurium from the Human Colonic Microbiome While Preserving Cytokine Signaling and Preventing Attachment to and Invasion of Human Cells by Salmonella in vitro. J. Food Prot. 2019;82:1336–1349. doi: 10.4315/0362-028X.JFP-18-587. [DOI] [PubMed] [Google Scholar]
- 199.Moye Z.D., Woolston J., Sulakvelidze A. Bacteriophage Applications for Food Production and Processing. Viruses. 2018;10:205. doi: 10.3390/v10040205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Fischer S., Kittler S., Klein G., Glünder G. Impact of a Single Phage and a Phage Cocktail Application in Broilers on Reduction of Campylobacter Jejuni and Development of Resistance. PLoS ONE. 2013;8:e78543. doi: 10.1371/journal.pone.0078543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Sulakvelidze A. Using Lytic Bacteriophages to Eliminate or Significantly Reduce Contamination of Food by Foodborne Bacterial Pathogens. J. Sci. Food Agric. 2013;93:3137–3146. doi: 10.1002/jsfa.6222. [DOI] [PubMed] [Google Scholar]
- 202.Hsu C., Lo C., Liu J., Lin C. Control of the Eel (Anguilla Japonica) Pathogens, Aeromonas Hydrophila and Edwardsiella tarda, by Bacteriophages. J. Fish. Soc. Taiwan. 2000;27:21–31. [Google Scholar]
- 203.Blanco-Picazo P., Roscales G., Toribio-Avedillo D., Gómez-Gómez C., Avila C., Ballesté E., Muniesa M., Rodríguez-Rubio L. Antibiotic Resistance Genes in Phage Particles from Antarctic and Mediterranean Seawater Ecosystems. Microorganisms. 2020;8:1293. doi: 10.3390/microorganisms8091293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Castillo D., Kauffman K., Hussain F., Kalatzis P., Rørbo N., Polz M.F., Middelboe M. Widespread Distribution of Prophage-Encoded Virulence Factors in Marine Vibrio Communities. Sci. Rep. 2018;8:1–9. doi: 10.1038/s41598-018-28326-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Christiansen R.H., Madsen L., Dalsgaard I., Castillo D., Kalatzis P.G., Middelboe M. Effect of Bacteriophages on the Growth of Flavobacterium psychrophilum and Development of Phage-Resistant Strains. Microb. Ecol. 2016;71:845–859. doi: 10.1007/s00248-016-0737-5. [DOI] [PubMed] [Google Scholar]
- 206.Castillo D., Christiansen R.H., Dalsgaard I., Madsen L., Middelboe M. Bacteriophage Resistance Mechanisms in the Fish Pathogen Flavobacterium psychrophilum: Linking Genomic Mutations to Changes in Bacterial Virulence Factors. Appl. Environ. Microbiol. 2015;81:1157–1167. doi: 10.1128/AEM.03699-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Middelboe M., Holmfeldt K., Riemann L., Nybroe O., Haaber J. Bacteriophages Drive Strain Diversification in a Marine Flavobacterium: Implications for Phage Resistance and Physiological Properties. Environ. Microbiol. 2009;11:1971–1982. doi: 10.1111/j.1462-2920.2009.01920.x. [DOI] [PubMed] [Google Scholar]
- 208.Verner–Jeffreys D.W., Algoet M., Pond M.J., Virdee H.K., Bagwell N.J., Roberts E.G. Furunculosis in Atlantic Salmon (Salmo salar L.) Is Not Readily Controllable by Bacteriophage Therapy. Aquaculture. 2007;270:475–484. doi: 10.1016/j.aquaculture.2007.05.023. [DOI] [Google Scholar]
- 209.Carrizo J.C., Griboff J., Bonansea R.I., Nimptsch J., Valdés M.E., Wunderlin D.A., Amé M.V. Different Antibiotic Profiles in Wild and Farmed Chilean Salmonids. Which Is the Main Source for Antibiotic in Fish? Sci. Total. Environ. 2021;800:149516. doi: 10.1016/j.scitotenv.2021.149516. [DOI] [PubMed] [Google Scholar]
- 210.Firmino J.P., Galindo-Villegas J., Reyes-López F.E., Gisbert E. Phytogenic bioactive compounds shape fish mucosal immunity. Front. Immunol. 2021;12:695973. doi: 10.3389/fimmu.2021.695973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Siriyappagouder P., Galindo-Villegas J., Dhanasiri A.K., Zhang Q., Mulero V., Kiron V., Fernandes J.M. Pseudozyma Priming Influences Expression of Genes Involved in Metabolic Pathways and Immunity in Zebrafish Larvae. Front. Immunol. 2020;11:978. doi: 10.3389/fimmu.2020.00978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Monzón-Atienza L., Bravo J., Torrecillas S., Montero D., González-de Canales A.F., de la Banda I.G., Galindo-Villegas J., Ramos-Vivas J., Acosta F. Isolation and Characterization of a Bacillus velezensis D-18 Strain, as a Potential Probiotic in European Seabass Aquaculture. Probiotics Antimicrob. Proteins. 2021:1–9. doi: 10.1007/s12602-021-09782-8. [DOI] [PubMed] [Google Scholar]
- 213.Montalban-Arques A., De Schryver P., Bossier P., Gorkiewicz G., Mulero V., Gatlin III D.M., Galindo-Villegas J. Selective Manipulation of the Gut Microbiota Improves Immune Status in Vertebrates. Front. Immunol. 2015;6:512. doi: 10.3389/fimmu.2015.00512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Sanchez M., Sepahi A., Casadei E., Salinas I. Symbiont-derived sphingolipids regulate inflammatory responses in rainbow trout (Oncorhynchus mykiss) Aquaculture. 2018;495:932–939. doi: 10.1016/j.aquaculture.2018.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Galindo-Villegas J., Masumoto T., Hosokawa H. Effect of Continuous and Interval Administration of Peptidoglycan on Innate Immune Response and Disease Resistance in Japanese Flounder Paralichthys olivaceus. Aquac. Sci. 2006;54:163–170. [Google Scholar]
- 216.Rodrigues M.V., Zanuzzo F.S., Koch J.F.A., de Oliveira C.A.F., Sima P., Vetvicka V. Development of Fish Immunity and the Role of β-Glucan in Immune Responses. Molecules. 2020;25:5378. doi: 10.3390/molecules25225378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Żaczek M., Weber-Dąbrowska B., Górski A. Phages as a Cohesive Prophylactic and Therapeutic Approach in Aquaculture Systems. Antibiotics. 2020;9:564. doi: 10.3390/antibiotics9090564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Borin J.M., Avrani S., Barrick J.E., Petrie K.L., Meyer J.R. Coevolutionary phage training leads to greater bacterial suppression and delays the evolution of phage resistance. Proc. Natl. Acad. Sci. USA. 2021;118:e2104592118. doi: 10.1073/pnas.2104592118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Richards G.P. Bacteriophage Remediation of Bacterial Pathogens in Aquaculture: A Review of the Technology. Bacteriophage. 2014;4:e975540. doi: 10.4161/21597081.2014.975540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Sieiro C., Areal-Hermida L., Pichardo-Gallardo Á., Almuiña-González R., de Miguel T., Sánchez S., Sánchez-Pérez Á., Villa T.G. A Hundred Years of Bacteriophages: Can Phages Replace Antibiotics in Agriculture and Aquaculture? Antibiotics. 2020;9:493. doi: 10.3390/antibiotics9080493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Park S.Y., Han J.E., Kwon H., Park S.C., Kim J.H. Recent Insights into Aeromonas salmonicida and Its Bacteriophages in Aquaculture: A Comprehensive Review. J. Microbiol. Biotechnol. 2020;30:1443–1457. doi: 10.4014/jmb.2005.05040. [DOI] [PMC free article] [PubMed] [Google Scholar]