Abstract
Intracellular protein delivery is a transformative tool for biologics research and medicine. Delivery into the cytosol allows proteins to diffuse throughout the cell and access subcellular organelles. Inefficient delivery caused by endosomal entrapment is often misidentified as cytosolic delivery. This inaccuracy muddles what should be a key checkpoint in assessing delivery efficiency. Green fluorescent protein (GFP) is a robust cargo small enough to passively diffuse from the cytosol into the nucleus. Fluorescence of GFP in the nucleus is a direct readout for cytosolic access and effective delivery. Here we highlight recent examples from literature for the accurate assessment of cytosolic protein delivery using GFP fluorescence in the cytosol and nucleus.
Introduction
Protein therapeutics are powerful tools for targeted manipulation of cellular processes.1 Proteins are responsible for modulating cellular activities like autophagy and homeostasis, and protein dysfunction can cause disease on a cellular or organismal level.2-4 Controlled introduction of functional proteins into the cell is a promising strategy to alleviate disease symptoms.5,6 Further, cytosolic delivery of exogenous proteins provides a key tool for the fundamental study of protein function in cells.7,8
Entry into the cytosol is a critical checkpoint for the assessment of most protein therapeutics.9,10 From the cytosol, proteins can access subcellular organelles, apoptotic machinery, and the nuclear membrane.11,12 Direct entry into the cytosol is generally barred to proteins by the selectively permeable cell membrane.13
A number of nanocarriers14,15 and cell-penetrating peptides16,17 have been engineered for delivery of proteins into the cell. However access to the cytosol is often limited in efficiency because these systems are generally uptaken by endocytosis18 through a range of mechanisms.19 Endocytic uptake entraps particles in vesicles (endosomes) that sequester them from the cytosol, ultimately leading to either endo/lysosomal degradation or exocytosis.20,21 Choice of carrier material, formulation, and treatment conditions are critical considerations for effective delivery, and all contribute to both cellular uptake and intracellular distribution.22-24 Engineered delivery vehicles can promote endosomal ‘escape’ through various methods of endosomal rupture, 25-28 but recent reports estimate that even utilizing these methods <10% of endosomally delivered cargo can be expected to enter the cytosol.29,30 Recently, vehicles capable of non-endosomal uptake through direct cytosolic entry have been reported with highly efficient protein delivery, providing a potential alternative pathway31,32 Distinguishing cytosolic access from endosomal entrapment however, is a critically important yet often overlooked checkpoint in the development of effective intracellular delivery systems.23,33
Fluorescent proteins provide a straightforward and effective readout to evaluating cytosolic access. The integral fluorescence of these proteins eliminates artifacts arising from intracellular separation of the fluorophore from the protein cargo.34,35 Green fluorescent protein (GFP) is a robust and versatile model cargo that can passively diffuse from the cytosol into the nucleus through pores in the nuclear membrane.36 Nuclear fluorescence of GFP thus provides definitive evidence of cytosolic access, making it a versatile tool for evaluating intracellular protein delivery (Figure 1). In this topical review we will discuss the use of both GFP and fluorophore-tagged small proteins as tools for validating cytosolic protein delivery, highlighting fundamental concepts and recent applications of this strategy.
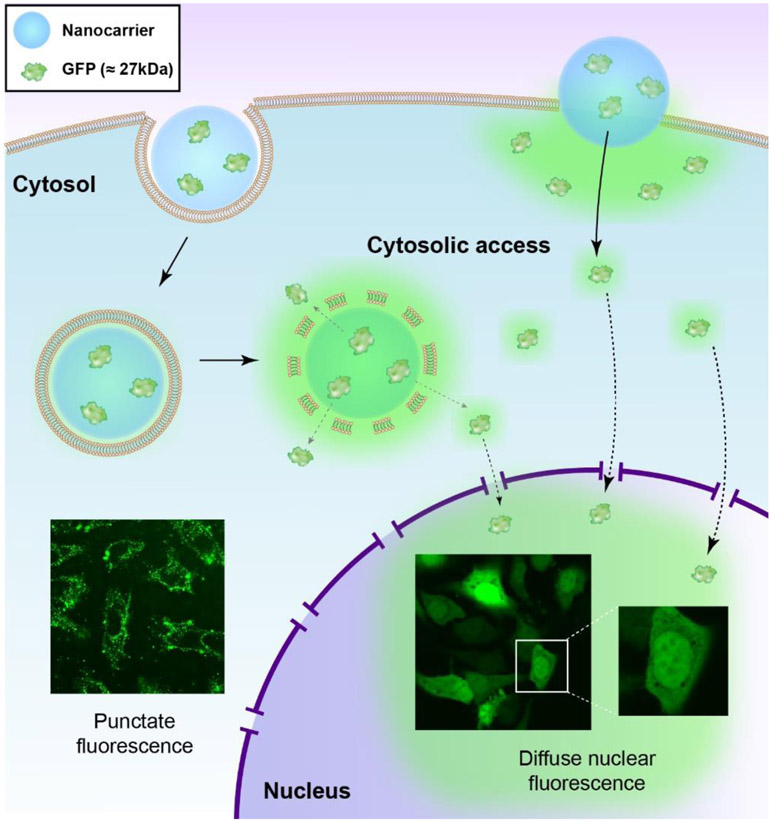
Figure 1.
Protein nanocarriers are uptaken by the cell through endocytosis (left) or direct cytosolic entry (right). Endocytosis presents as punctate fluorescence in the cytosol. Diffuse fluorescence suggests cytosolic access. Small proteins (< 60kDa) like GFP can passively diffuse from the cytosol into the nucleus, definitively establishing cytosolic access.
Diffuse Fluorescence Indicates Cytosolic Access
There are two main indicators of cytosolic access of small fluorescent (or fluorophore-tagged) proteins that are easily evaluated by fluorescence microscopy: punctate versus diffuse fluorescence signal and fluorescence signal in the nucleus. Diffuse fluorescence qualitatively defines endosomal entrapment from cytosolic access. Fluorescence in the nucleus provides a direct readout, as small proteins like GFP will diffuse from the cytosol through nuclear membrane pores and illuminate the nucleus.
Fluorescent proteins trapped in endosomes are visible as highly localized, punctate fluorescence signals. In contrast fluorescent proteins free to move about the cytosol display diffuse, evenly spread fluorescence, as seen in cells that constitutively express fluorescent protein. 37 Recently, Gao reported a method allowing small proteins to avoid endocytosis and pass into the cytosol through noncovalent tagging with Coomassie blue (CB)-cholesterol conjugates.38 Cytosolic access of four dye-labeled proteins was assessed by diffuse versus punctate fluorescence signal (Figure 2) as well as co-localization with LysoTracker. The two smaller proteins were delivered to the cytosol, as evidenced by diffuse fluorescence throughout the cytosol. Lysozyme displayed a mixture of diffuse and punctate fluorescence. Notably, the small size of these proteins also allowed them to diffuse across the nuclear pore and into the nuclei. The larger proteins showed only localized punctate fluorescence, indicating vesicular entrapment.
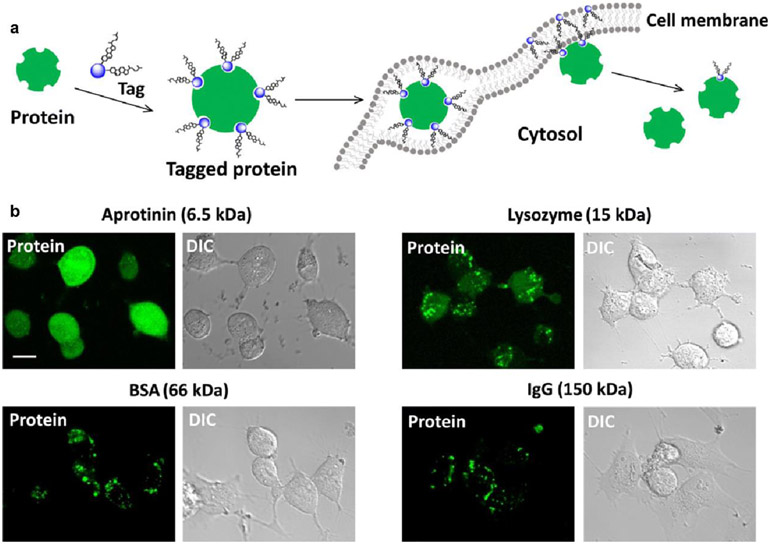
Figure 2.
Cytosolic delivery by protein tagging. (a) Proposed delivery mechanism. Proteins tagged with CB-cholesterol conjugates embedded within the lipid bilayer. The noncovalent nature of the tagging causes dissociation, after which untagged hydrophilic protein can ‘slip’ into the cytosol. Deliveries were performed in serum-free RPMI 1640 media for 3 hours. (b) Confocal fluorescence and bright-field micrographs of live HeLa cells treated with tagged proteins of different molecular weights. All proteins were labeled with a green fluorescent dye (AF488). Adapted with permission from [35].
Co-localization of protein fluorescence with an endosomal marker is a common method to validate endosomal entrapment.39 A common misconception is that protein cargo not co-localized with endosomal marker has avoided the endosome and entered the cytosol, even if fluorescence signal remains punctate. Endosomal markers like those of the LysoTracker series40 work through pH-responsiveness, making them specific for early (pH ~6.3) or late stage (pH 4.5 - 5) endo/lysosomes.33 When LysoTracker dyes aggregate, intracellular pH also rises considerably, potentially quenching the fluorophore of the delivery cargo.41,42 Lysotracker co-localization should therefore always be qualified by fluorescence signal to accurately assess endosomal escape. Flavell recently reported a method to quantify endosomal escape of fluorescently labelled saporin toxin in Daudi cells using flow cytometry. 43 Endosomal escape was induced by treatment with chloroquine, an inhibitor of endolysosomal acidification. Pulse width analysis was used to distinguish punctate signal from diffuse cytosolic fluorescence. A fluorescence shift from punctate to diffuse corresponded with protein escape from the endosome, providing a metric of cytosolic access.
GFP in the Cytosol will Enter the Nucleus
GFP is an ideal model cargo to visualize protein trafficking into and throughout the cell. The fluorophore of GFP is composed of amino acid residues within the polypeptide chain, providing robust fluorescence (quantum yield up to 80 percent)44 while avoiding the issues of photobleaching or enzymatic cleavage associated with fluorescent dyes. 45-47 Fluorescence is only lost upon denaturation, which can be used as a readout for retention of protein structure through the delivery process.
In addition to the diffuse fluorescence discussed above, GFP also provides a reliable readout of cytosolic access through nuclear fluorescence. 48 The nuclear membrane pores limit entry by passive diffusion to approximately 60kDa.49,50 Small proteins like GFP (238 residues, 27 kDa) will passively translocate from the cytosol into the nucleus through these size-restrictive pores. 51 For proteins larger than the nuclear pore, passive nuclear diffusion is dramatically decreased and generally requires the aid of active nuclear targeting elements.52, 53 Nuclear fluorescence therefore provides a direct readout of cytosolic access, as endosomally entrapped GFP can access neither the cytosol nor the nucleus. In one example of this effect, Raines modified GFP allowing it to pass through the cell membrane and into the cytosol by cloaking its carboxyl groups with a hydrophobic moiety.54 Modified GFP showed diffuse fluorescence throughout the cytosol and nucleus. Delivery was compared with a super-charged GFP variant, which displayed dark nuclei and punctate signal indicative of endocytosis (Figure 3). In this study deliveries were performed through incubation for 30, 120, or 240 min at 37 °C in serum-free culture media.
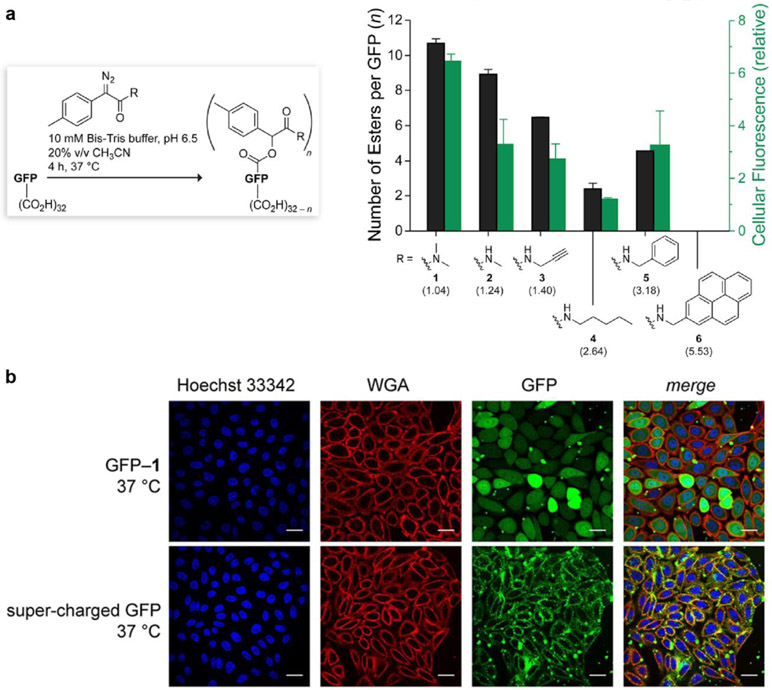
Figure 3.
(a) Bar graph showing the extent of esterification of the superfolder variant of GFP with diazo compounds 1–6 (black) with parenthetical log P values, and the internalization of the ensuing esterified GFPs into CHO-K1 cells (green). Values (±SD) were determined by mass spectrometry and flow cytometry, respectively. (b) Images of the cellular internalization in CHO-K1 cells of esterified GFP (top) and its supercharged variant (bottom). Cells were stained with Hoechst 33342 and wheat germ agglutinin (WGA)–AlexaFluor 647. Scale bars: 25 μm. Adapted with permission from [51].
We recently reported an imaging flow cytometric method that utilized the passive translocation of GFP into the nucleus as a quantitative indicator of cytosolic delivery. Cytosolic delivery efficiency was quantified through co-localization of GFP signal with nuclear stain. Supramolecular nanocomposite vehicles were generated between engineered oligo-glutamate, or E-tagged GFP and guanidinium-functionalized poly(oxanorbornene) imide (PONI) polymers. The charge ratio between guanidinium (polymer) and E-tag (protein) during formulation was found to be critical to nanocomposite generation. These vehicles efficiently delivered GFP directly to the cytosol, with diffuse cytosolic and nuclear fluorescence (Figure 4).55 Notably these vehicles demonstrated cytosolic delivery in the presence of serum (media supplemented with 10% fetal bovine serum). Under the most efficient delivery conditions, GFP was delivered to the cytosol in >90% of the cell population as indicated through GFP fluorescence in the nucleus. This simple and straightforward approach to quantifying cytosolic delivery is widely applicable to small fluorescent or dye-labeled protein delivery, regardless of carrier vehicle.56-59
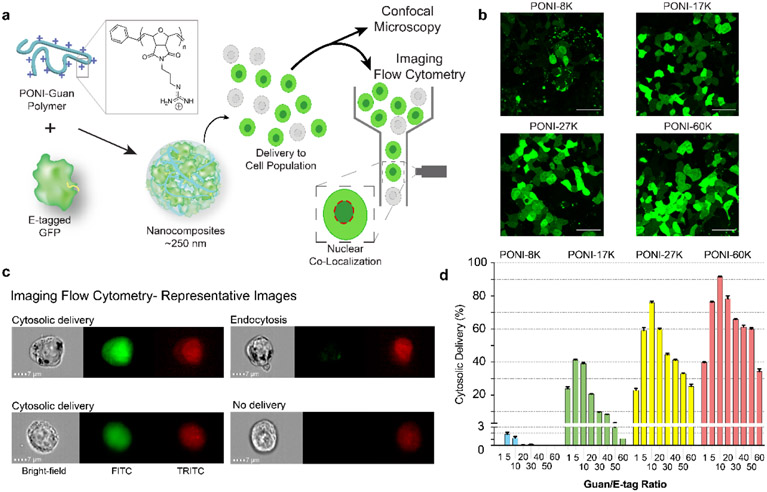
Figure 4.
Polymer mediated intracellular delivery of GFP, quantified by imaging flow cytometry. (a) PONI polymer electrostatically complexed with ‘E-tagged’ GFP to form supramolecular polymer-protein nanocomposites. Delivered cell population was analyzed by confocal microscopy or imaging flow cytometry, through co-localization with a nuclear stain. (b) Confocal images of GFPE20 showing cytosolic delivery and diffuse nuclear fluorescence in HEK-293T cells. Scale bars = 50 μm. (c) Representative imaging flow cytometry micrographs showing cytosolic delivery and nuclear localization of GFPE20 as compared to endosomally entrapped GFP and undelivered cells. Channels displayed are bright-field, FITC for GFP, and TRITC for nuclear stain (DRAQ5). (d) Percentage of cell population with cytosolic delivery of GFP using a family of different molecular weight PONI polymers. Adapted with permission from [52].
Conclusions
In sum, GFP and fluorophore-tagged proteins provide two key readouts for cytosolic access. Diffuse fluorescence provides a simple readout but is challenging to quantify in a cell population. Diffuse nuclear fluorescence definitively demonstrates cytosolic access of delivered GFP and can be quantified in cell populations using microscopy and imaging flow cytometry. These robust, versatile methods are suitable for fluorophore-tagged small proteins, and using fluorescent proteins like GFP further eliminates artifacts arising from cargo degradation and fluorophore release. Widespread utilization of these tactics provides rigorous assessment of cytosolic access that will promote the development of more efficient platforms for protein delivery.
Acknowledgements
This research was supported by the NIH (DK121351 and EB022641), National Research Service Award T32 GM008515 from the NIH and by a grant from the Korean Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (grant number: HI19C0753).
References
- (1).Scaletti F; Hardie J; Lee YW; Luther DC; Ray M; Rotello VM Protein Delivery into Cells Using Inorganic Nanoparticle-Protein Supramolecular Assemblies. Chem. Soc. Rev 2018, 47 (10), 3421–3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Chiti F; Dobson CM Protein Misfolding, Functional Amyloid, and Human Disease. Annu. Rev. Biochem 2006, 75 (1), 333–366. [DOI] [PubMed] [Google Scholar]
- (3).Gandhi J; Antonelli AC; Afridi A; Vatsia S; Joshi G; Romanov V; Murray IVJ; Khan SA Protein misfolding and aggregation in neurodegenerative diseases: a review of pathogeneses, novel detection strategies, and potential therapeutics. Rev. Neurosci 2019, 30, 4, 339–358. [DOI] [PubMed] [Google Scholar]
- (4).Dalle-Donne I; Aldini G; Carini M; Colombo R; Rossi R; Milzani A Protein carbonylation, cellular dysfunction, and disease progression. J. Cell. Mol. Med 2006, 10, (2), 389–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Mantej J; Polasik K; Piotrowska E; Tukaj S Autoantibodies to Heat Shock Proteins 60, 70, and 90 in Patients with Rheumatoid Arthritis. Cell Stress Chaperones 2019, 24 (1), 283–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Sands RW; Tabansky I; Verbeke CS; Keskin D; Michel S; Stern J; Mooney DJ Steroid-Peptide Immunoconjugates for Attenuating T Cell Responses in an Experimental Autoimmune Encephalomyelitis Murine Model of Multiple Sclerosis. Bioconjug. Chem 2020, 31 (12), 2779–2788. [DOI] [PubMed] [Google Scholar]
- (7).Darwish M; Shatz W; Leonard B; Loyet K; Barrett K; Wong JL; Li H; Abraham R; Lin M; Franke Y; Tam C; Mortara K; Zilberleyb I; Blanchette C Nanolipoprotein Particles as a Delivery Platform for Fab Based Therapeutics. Bioconjug. Chem 2020, 31 (8), 1995–2007. [DOI] [PubMed] [Google Scholar]
- (8).Lieser RM; Yur D; Sullivan MO; Chen W Site-Specific Bioconjugation Approaches for Enhanced Delivery of Protein Therapeutics and Protein Drug Carriers. Bioconjug. Chem 2020, 31 (10), 2272–2282. [DOI] [PubMed] [Google Scholar]
- (9).Deprey K; Becker L; Kritzer J; Pluckthun A Trapped! A critical evaluation of methods for measuring total cellular uptake versus cytosolic localization. Bioconjug. Chem 2019, 30 (4), 1006–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Liu C; Shen W; Li B; Li T; Chang H; Cheng Y Natural Polyphenols Augment Cytosolic Protein Delivery by a Functional Polymer. Chem. Mater 2019, 31 (6), 1956–1965. [Google Scholar]
- (11).Goswami R; Jeon T; Nagaraj H; Zhai S; Rotello VM Accessing Intracellular Targets through Nanocarrier-Mediated Cytosolic Protein Delivery. Trends in Pharmacol. Sci 2020, 41 (10), 743–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Lee YW; Luther DC; Kretzmann JA; Burden A; Jeon T; Zhai S; Rotello VM Protein Delivery into the Cell Cytosol Using Non-Viral Nanocarriers. Theranostics. 2019, 9 (11) 3280–3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Mitragotri S; Burke PA; Langer R Overcoming the Challenges in Administering Biopharmaceuticals: Formulation and Delivery Strategies. Nat. Rev. Drug Discov 2014, 13, 655–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Filipczak N; Pan J; Yalamarty SSK; Torchilin VP Recent Advancements in Liposome Technology. Adv. Drug Deliv. Rev 2020, 156, 4–22. [DOI] [PubMed] [Google Scholar]
- (15).Chiper M; Niederreither K; Zuber G Transduction Methods for Cytosolic Delivery of Proteins and Bioconjugates into Living Cells. Adv. Healthc. Mater 2018, 7 (6), e1701040. [DOI] [PubMed] [Google Scholar]
- (16).Schneider AFL; Wallabregue ALD; Franz L; Hackenberger CPR Targeted Subcellular Protein Delivery Using Cleavable Cyclic Cell-Penetrating Peptides. Bioconjug. Chem 2019, 30 (2), 400–404. [DOI] [PubMed] [Google Scholar]
- (17).Sumito N; Koeda S; Umezawa N; Inoue Y; Tsukiji S; Higuchi T; Mizuno T Development of Cell-Penetration PG-Surfactants and Its Application in External Peptide Delivery to Cytosol. Bioconjug. Chem 2020, 31 (3), 821–833. [DOI] [PubMed] [Google Scholar]
- (18).Fu A; Tang R; Hardie J; Farkas ME; Rotello VM Promises and Pitfalls of Intracellular Delivery of Proteins. Bioconjug. Chem 2014, 25 (9), 1602–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Sahay G; Alakhova DY; Kabanov AV Endocytosis of Nanomedicines. J. Control. Release 2010, 145 (3), 182–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Du S; Liew SS; Li L; Yao SQ Bypassing Endocytosis: Direct Cytosolic Delivery of Proteins. J. Am. Chem. Soc 2018, 140 (47), 15986–15996. [DOI] [PubMed] [Google Scholar]
- (21).Luzio JP; Rous BS; Bright NA; Pryor PR; Mullock BM; Piper RC Lysosome-Endosome Fusion and Lysosome Biogenesis. J. Cell Sci 2000, 113 (9), 1515–1524. [DOI] [PubMed] [Google Scholar]
- (22).Luther DC; Huang R; Jeon T; Zhang X; Lee YW; Nagaraj H; Rotello VM Delivery of Drugs, Proteins, and Nucleic Acids Using Inorganic Nanoparticles. Advanced Drug Delivery Reviews. 2020, 156, 188–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Geraldes DC; Beraldo-de-Araujo VL; Pardo BOP; Pessoa Junior A; Stephano MA; de Oliveira-Nascimento L Protein Drug Delivery: Current Dosage Form Profile and Formulation Strategies. Journal of Drug Targeting. 2020, 28 (4), 339–355. [DOI] [PubMed] [Google Scholar]
- (24).Patel SG; Sayers EJ; He L; Narayan R; Williams TL; Mills EM; Allemann RK; Luk LYP; Jones AT; Tsai YH Cell-Penetrating Peptide Sequence and Modification Dependent Uptake and Subcellular Distribution of Green Florescent Protein in Different Cell Lines. Sci. Rep 2019, 9 (1), 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Stewart MP; Langer R; Jensen KF Intracellular Delivery by Membrane Disruption: Mechanisms, Strategies, and Concepts. Chem. Rev 2018, 118 (16), 7409–7531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Brock DJ; Kondow-Mcconaghy HM; Hager EC; Pellois JP Endosomal Escape and Cytosolic Penetration of Macromolecules Mediated by Synthetic Delivery Agents. Bioconjug. Chem, 2019, 30 (2), 293–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Rangasamy L; Chelvam V; Kanduluru AK; Srinivasarao M; Bandara NA; You F; Orellana EA; Kasinski AL; Low PS New Mechanism for Release of Endosomal Contents: Osmotic Lysis via Nigericin-Mediated K + /H + Exchange. Bioconjug. Chem 2018, 29 (4), 1047–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Benjaminsen RV; Mattebjerg MA; Henriksen JR; Moghimi SM; Andresen TL The Possible "proton Sponge " Effect of Polyethylenimine (PEI) Does Not Include Change in Lysosomal PH. Mol. Ther 2013, 21 (1), 149–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Smith SA; Selby LI; Johnston APR; Such GK The Endosomal Escape of Nanoparticles: Toward More Efficient Cellular Delivery. Bioconjug. Chem 2019, 30 (2), 263–272. [DOI] [PubMed] [Google Scholar]
- (30).Teo SLY; Rennick JJ; Yuen D; Al-Wassiti H; Johnston APR; Pouton CW Unravelling Cytosolic Delivery of Endosomal Escape Peptides with a Quantitative Endosomal Escape Assay (SLEEQ). bioRxiv, 2020, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Mout R; Ray M; Yesilbag Tonga G; Lee YW; Tay T; Sasaki K; Rotello VM Direct Cytosolic Delivery of CRISPR/Cas9-Ribonucleoprotein for Efficient Gene Editing. ACS Nano 2017, 11 (3), 2452–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Brock R The Uptake of Arginine-Rich Cell-Penetrating Peptides: Putting the Puzzle Together. Bioconjug. Chem 2014, 25 (5), 863–868. [DOI] [PubMed] [Google Scholar]
- (33).Pei D; Buyanova M Overcoming Endosomal Entrapment in Drug Delivery. Bioconjug. Chem 2019, 30 (2) 273–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Poc P; Gutzeit VA; Ast J; Lee J; Jones BJ; D’Este E; Mathes B; Lehmann M; Hodson DJ; Levitz J; Broichhagen J Interrogating Surface: Versus Intracellular Transmembrane Receptor Populations Using Cell-Impermeable SNAP-Tag Substrates. Chem. Sci 2020, 11 (30), 7871–7883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Gallo E Fluorogen-Activating Proteins: Next-Generation Fluorescence Probes for Biological Research. Bioconjug. Chem 2020, 31 (1), 16–27. [DOI] [PubMed] [Google Scholar]
- (36).Cooper GM The Nuclear Envelope and Traffic between the Nucleus and Cytoplasm. 2000. The Cell: A Molecular Approach. 2nd edition. [Google Scholar]
- (37).Soboleski MR; Oaks J; Halford WP Green Fluorescent Protein Is a Quantitative Reporter of Gene Expression in Individual Eukaryotic Cells. FASEB J. 2005, 19 (3), 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Tai W; Zhao P; Gao X Cytosolic Delivery of Proteins by Cholesterol Tagging. Sci. Adv 2020, 6 (25), 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Liu BR; Lo S-Y; Liu C-C; Chyan C-L; Huang Y-W; Aronstam RS; Lee H-J Endocytic Trafficking of Nanoparticles Delivered by Cell-Penetrating Peptides Comprised of Nona-Arginine and a Penetration Accelerating Sequence. PLoS One 2013, 8 (6), e67100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Chazotte B Labeling Lysosomes in Live Cells with LysoTracker. Cold Spring Harb. Protoc 2011, (2), 5571. [DOI] [PubMed] [Google Scholar]
- (41).Pierzynska-Mach A; Janowski PA; Dobrucki JW Evaluation of Acridine Orange, LysoTracker Red, and Quinacrine as Fluorescent Probes for Long-Term Tracking of Acidic Vesicles: Tracking Acidic Vesicles. Cytometry A 2014, 85 (8), 729–737. [DOI] [PubMed] [Google Scholar]
- (42).Yapici NB; Bi Y; Li P; Chen X; Yan X; Mandalapu SR; Faucett M; Jockusch S; Ju J; Gibson KM; Pavan WJ; Bi L Highly Stable and Sensitive Fluorescent Probes (LysoProbes) for Lysosomal Labeling and Tracking. Sci. Rep 2015, 5 (1), 8576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Wensley HJ; Johnston DA; Smith WS; Holmes SE; Flavell SU; Flavell DJ A Flow Cytometric Method to Quantify the Endosomal Escape of a Protein Toxin to the Cytosol of Target Cells. Pharm. Res 2019, 37 (1), 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Ozbakir HF; Anderson NT; Fan K-C; Mukherjee A Beyond the Green Fluorescent Protein: Biomolecular Reporters for Anaerobic and Deep-Tissue Imaging. Bioconjug. Chem 2020, 31 (2), 293–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Remington SJ Green Fluorescent Protein: A Perspective. Protein Sci. 2011, 20 (9) 1509–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Liu J; Cui Z Fluorescent Labeling of Proteins of Interest in Live Cells: Beyond Fluorescent Proteins. Bioconjug. Chem 2020, 31 (6), 1587–1595. [DOI] [PubMed] [Google Scholar]
- (47).Toseland CP Fluorescent Labeling and Modification of Proteins. J. Chem. Biol 2013, 6 (3), 85–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Pan S; Jeon T; Luther DC; Duan X; Rotello VM Cytosolic Delivery of Functional Proteins in Vitro through Tunable Gigahertz Acoustics. ACS Appl. Mater. Interfaces 2020, 12 (13), 15823–15829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Timney BL; Raveh B; Mironska R; Trivedi JM; Kim SJ; Russel D; Wente SR; Sali A; Rout MP Simple Rules for Passive Diffusion through the Nuclear Pore Complex. J. Cell Biol 2016, 215 (1), 57–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Wang R; Brattain MG The Maximal Size of Protein to Diffuse through the Nuclear Pore is Larger than 60 KDa. FEBS Lett. 2007, 581 (17), 3164–3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Seibel NM; Eljouni J; Nalaskowski MM; Hampe W Nuclear Localization of Enhanced Green Fluorescent Protein Homomultimers. Anal. Biochem 2007, 368 (1), 95–99. [DOI] [PubMed] [Google Scholar]
- (52).Ray M; Tang R; Jiang Z; Rotello VM Quantitative Tracking of Protein Trafficking to the Nucleus Using Cytosolic Protein Delivery by Nanoparticle-Stabilized Nanocapsules. Bioconjug. Chem 2015, 26 (6), 1004–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Tang R; Wang M; Ray M; Jiang Y; Jiang Z; Xu Q; Rotello VM Active Targeting of the Nucleus Using Nonpeptidic Boronate Tags. J. Am. Chem. Soc 2017, 139 (25), 8547–8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Mix KA; Lomax JE; Raines RT Cytosolic Delivery of Proteins by Bioreversible Esterification. J. Am. Chem. Soc 2017, 139 (41), 14396–14398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Lee Y-W; Luther D; Goswami R; Jeon T; Clark V; Elia J; Gopalakrishnan S; Rotello V,M Direct Cytosolic Delivery of Proteins through Coengineering of Proteins and Polymeric Delivery Vehicles. J. Am. Chem. Soc 2020, 142 (9), 4349–4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Wadia JS; Stan RV; Dowdy SF Transducible TAT-HA Fusogenic Peptide Enhances Escape of TAT-Fusion Proteins after Lipid Raft Macropinocytosis. Nat. Med 2004, 10 (3), 310–315. [DOI] [PubMed] [Google Scholar]
- (57).Mandal S; Mann G; Satish G; Brik A Enhanced Live-Cell Delivery of Synthetic Proteins Assisted by Cell-Penetrating Peptides Fused to DABCYL. Angew. Chemie Int. Ed 2021, 60 (13), 7333–7343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Ngwa VM; Axford DS; Healey AN; Nowak SJ; Chrestensen CA; McMurry JL A Versatile Cell-Penetrating Peptide-Adaptor System for Efficient Delivery of Molecular Cargos to Subcellular Destinations. PLoS One 2017, 12 (5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Banga J; Srinivasan D; Sun CC; Thompson CD; Milletti F; Huang K.Sen; Hamilton S; Song S; Hoffman AF; Qin YG; Matta B; LaPan M; Guo Q; Lu G; Li D; Qian H; Bolin DR; Liang L; Wartchow C; Qiu J; Downing M; Narula S; Fotouhi N; DeMartino JA; Tan SL; Chen G; Barnes BJ Inhibition of IRF5 Cellular Activity with Cell-Penetrating Peptides That Target Homodimerization. Sci. Adv 2020, 6 (20). [DOI] [PMC free article] [PubMed] [Google Scholar]