Abstract
Cryptococcus neoformans is an opportunistic fungal pathogen with a defined sexual cycle. The gene encoding a heterotrimeric G-protein β subunit, GPB1, was cloned and disrupted. gpb1 mutant strains are sterile, indicating a role for this gene in mating. GPB1 plays an active role in mediating responses to pheromones in early mating steps (conjugation tube formation and cell fusion) and signals via a mitogen-activated protein (MAP) kinase cascade in both MATα and MATa cells. The functions of GPB1 are distinct from those of the Gα protein GPA1, which functions in a nutrient-sensing cyclic AMP (cAMP) pathway required for mating, virulence factor induction, and virulence. gpb1 mutant strains are also defective in monokaryotic fruiting in response to nitrogen starvation. We show that MATa cells stimulate monokaryotic fruiting of MATα cells, possibly in response to mating pheromone, which may serve to disperse cells and spores to locate mating partners. In summary, the Gβ subunit GPB1 and the Gα subunit GPA1 function in distinct signaling pathways: one (GPB1) senses pheromones and regulates mating and haploid fruiting via a MAP kinase cascade, and the other (GPA1) senses nutrients and regulates mating, virulence factors, and pathogenicity via a cAMP cascade.
Cryptococcus neoformans is an opportunistic fungal pathogen that infects the central nervous system to cause meningoencephalitis in individuals with compromised immune function (26, 39). Virulence is associated with mating type (28), production of melanin (29, 30, 48, 56) and a polysaccharide capsule (4, 16, 30), and growth at 37°C (30, 45).
The life cycle of this organism has been defined (25). Mating occurs between MATa and MATα cells and involves cell fusion, filamentation, nuclear migration and fusion, meiosis, and sporulation. Mating type is linked to physiology and virulence. MATα strains are more prevalent in the environment, and most clinical isolates are MATα (27); MATα strains are more virulent in mice than are congenic MATa strains (28). In response to nitrogen starvation, MATα cells differentiate to form filaments, basidia, and spores (haploid fruiting) (62). Thus, genes linked to the MATα locus regulate the physiology and virulence of C. neoformans. A homolog of the Saccharomyces cerevisiae and Candida albicans STE12 transcription factor is encoded by the C. neoformans MATα locus (61), and ste12 mutant strains have defects in haploid fruiting (67). Recent studies of a GTP-binding protein, GPA1, underscore the importance of signaling cascades in C. neoformans virulence (2, 54).
Heterotrimeric guanine nucleotide binding proteins interact with G-protein-coupled receptors to sense external signals and regulate cell growth and development (14). G-protein-mediated signals include responses to hormones and neurotransmitters, vision and olfaction, and pheromone-induced mating in S. cerevisiae and Schizosaccharomyces pombe. Heterotrimeric G proteins are comprised of alpha (α), beta (β), and gamma (γ) subunits. In response to binding of ligand to receptors, the Gαβγ complex is recruited, leading to GDP-GTP exchange on Gα and release of Gβγ.
In most examples, the Gα-GTP subunit actively transduces signals. However, Gβγ subunits can also signal (5, 21–23, 32, 33, 36, 52, 64). For example, the S. cerevisiae Gβγ complex Ste4-Ste18 is released from the Gα subunit Gpa1 by pheromone (6, 32, 33, 59). The Gβγ complex recruits the Ste5 scaffold, allowing activation of the Ste5-bound kinase Ste11 by membrane-localized Ste20 kinase (32–34, 47). The Gα subunit Gpa1 plays a negative role in S. cerevisiae mating (9, 40). In contrast, in Schizosaccharomyces pombe, the Gα subunit Gpa1 positively signals mating and, together with Ras1, activates a mitogen-activated protein (MAP) kinase cascade (8, 42, 44, 58, 65). In the chestnut blight fungus Cryphonectria parasitica, the Gβ subunit CPGB-1 regulates sporulation and virulence, likely with the Gα subunit CPG-1 (13, 21).
In C. neoformans, the Gα subunit GPA1 is required for mating and virulence (2). GPA1 regulates responses to nutritional starvation signals required for mating and induction of the virulence factors capsule and melanin. Cyclic AMP (cAMP) suppresses the mating and virulence defects of gpa1 mutant cells, suggesting that GPA1 activates adenylyl cyclase similarly to Gαs in mammals and Gpa2 during S. cerevisiae pseudohyphal growth (24, 37). In the present study, we investigated the roles of a G-protein β subunit, GPB1, in C. neoformans.
MATERIALS AND METHODS
Strains and media.
C. neoformans strains used in this study included H99 (serotype A, MATα) and the isogenic ade2 mutant M049. Strains JEC34 and JEC43 are isogenic ura5 mutant serotype D MATa and MATα strains, respectively (41). Strain BAC20 is a gpa1::ADE2 mutant of the MATa strain JEC20 (provided by B. Allen and A. Alspaugh). Yeast extract-peptone-dextrose (YPD) and yeast nitrogen base (YNB) media, synthetic medium, V8 agar, filament agar, niger seed agar for melanin production, and low-iron medium plus 56 μM ethylenediamine-di(o-hydroxyphenylacetic acid) (EDDHA) for induction of capsule formation were as described in references 2 and 55.
Isolation of the C. neoformans GPB1 gene.
Primers 5′-AT(ATC)TA(TC)GC(GATC)ATGCA(TCT)TGG and 5′-AA(AG)TC(AG)TA(GATC)CC(GATC)GC encompassed conserved residues IYAMHW and AGYDDF, respectively, of Gβ subunits. PCR parameters were as follows: 94°C for 40 s, 40°C for 1 min, and 72°C for 2 min (40 cycles). C. neoformans cDNA (strain B3501; 200 ng) served as the template. PCR products were excised, cloned, and used to clone the GPB1 gene. For size-selected libraries, DNA was cleaved with HindIII and electrophoresed, and 4.9-kb fragments were excised. DNA was recovered by using a QIAEX DNA extraction kit (Qiagen), ligated in HindIII-cleaved plasmid pUC18, and transformed into Escherichia coli, and bacterial colonies were screened with the GPB1 PCR product as a probe (49).
Nucleic acid manipulations.
DNA and RNA were extracted from cells that were lyophilized overnight and broken with glass beads (4 mm diameter) by the use of a Vortex mixer. Total DNA for Southern blot analysis was isolated as described in reference 46. Total DNA for PCRs was obtained as described in reference 18. Total RNA was extracted with a buffer containing 150 mM sodium acetate, 100 mM LiCl, 4% sodium dodecyl sulfate, 10 mM EDTA, 10 mM EGTA, and 20 mM β-mercaptoethanol, extracted with phenol (pH 4.0), and precipitated with LiCl.
The GPB1 cDNA clone was obtained in two steps. First, cDNA was synthesized from total RNA of strain H99 by using a reverse transcription-PCR kit (Stratagene) with random primers to generate cDNA from the 5′ region and oligo(dT) primers to obtain 3′ cDNA. Second, the two cDNA pools were used as templates for PCR with primers corresponding to the GPB1 gene based on the genomic sequence to amplify 5′- and 3′-proximal fragments of the gene which span an internal EcoRI site. The full-length GPB1 cDNA was obtained by ligating these two EcoRI fragments. Southern and Northern blot analyses and hybridizations were performed by standard procedures (49).
Two-hybrid assays.
For two-hybrid interactions, a GPB1 cDNA was cloned in plasmids pGBT9 and pGAD424 (Clontech) to yield plasmids pGBT9::GPB1 and pGAD424:GPB1, expressing GAL4(DB)-GPB1 and GAL4(AD)-GPB1 fusion proteins, respectively. DNA of plasmids pGAD424::GPB1 and pGBT::GPA1 (2) or of plasmids pGBT9::GPB1 and pGAD424::GPA1 (2) was used to transform the yeast strain PJ69-4A (20).
GPB1 gene disruption.
pCnGPB1 is a pUC18-derived clone containing a 4.9-kb HindIII fragment spanning the GPB1 gene from strain H99. For the gpb1::ADE2 gene disruption, pCnGPB1 was digested with ApaI (for which there is a unique cleavage site in the GPB1 gene), blunt ended with T4 DNA polymerase, and dephosphorylated with calf intestinal alkaline phosphatase. Two plasmids were constructed for the GPB1 gene disruption. Either a 2.4-kb XhoI or 2.9-kb KpnI-BamHI DNA fragment containing the ADE2 gene from C. neoformans serotype D strain B3501 (51, 53) was blunt ended and inserted at the blunted ApaI site in plasmid pCnGPB1 to yield the gpb1::ADE2 disruption alleles. The ade2 serotype A strain M049 was grown for 40 h in liquid YPD and transformed with the gpb1::ADE2 disruption allele by the use of a biolistic DNA delivery apparatus (Bio-Rad) as described elsewhere (53). Transformants were selected on synthetic medium lacking adenine but containing 1 M sorbitol. Primers used for PCRs to verify the presence of gpb1::ADE2 alleles were 5′-AGAGAGCTCAGCGCACAC-3′ and 5′-GTAGTCATCGTAGCCGGC-3′.
Mating assays.
Mating assays were conducted by coculturing MATα strains with the tester MATa strain JEC20 on V8 or filament agar medium containing 0.5% galactose (inducing) or 0.5% glucose (repressing) (62). Plates were incubated at 22°C, and resultant colonies were examined by using a Nikon Eclipse E400 microscope.
Expression of GPB1, GAL7-GPB1, GAL7-CPK1, GAL7-STE12α, MFα1, and Ras1-Q67L.
A ura5 derivative of the gpb1 mutant strain was obtained by plating cells on 5-fluoroorotic acid medium, and the ura5 mutation was then complemented by introducing plasmid pCnTel1 (10, 31). A 2.5-kb XbaI fragment containing the wild-type GPB1 gene was inserted into plasmid pCnTel1 (pCnTel1::GPB1) for complementation tests. The genomic clone of the C. neoformans Fus3/Kss1 MAP kinase homolog CPK1 (pCnTel1::CPK1) and the CPK1 cDNA clone (R. Davidson and J. Heitman, unpublished data) were cloned under the control of the C. neoformans GAL7 promoter (60), and the resulting plasmids (pCnTel1::GAL7-CPK1) were used for epistasis. The cDNA clone of the C. neoformans STE12α gene expressed from the GAL7 promoter in plasmid pCGS-1 (61) was also used. Plasmids were transformed in circular form by biolistic transformation into the gpb1 ura5 mutant. The GPB1 cDNA clone was placed under the control of the GAL7 promoter (pCnTel1Δ::GAL7-GPB1) and used to transform a ura5 strain of H99, the gpb1 ura5 mutant, and the ura5 serotype D MATa and MATα strains JEC34 and JEC43. pCnTel1Δ differs from pCnTel1 in that it lacks the NotI fragment containing telomeric sequences. The dominant active Ras1 Q67L mutant was expressed with the C. neoformans actin gene promoter and was introduced with the hygromycin B resistance or URA5 gene as a marker. The cloned MFα1 gene (plasmid pCnTel1::MFα1) (41) was expressed in the serotype D strain JEC34 (MATa ura5) and the isogenic gpa1 mutant strain BAC20 (MATa gpa1::ADE2 ura5).
Haploid fruiting assays.
For haploid fruiting assays, the Ras1 Q67L mutant protein was expressed by introducing linear DNA fragments containing the mutant RAS1 gene, linked to the hygromycin resistance or URA5 gene as a marker, by biolistic transformation. Isolates containing the mutant allele for Ras1 Q67L were identified by PCR of genomic DNA with primers flanking the RAS1 gene and by XbaI cleavage to detect the Q67L mutation. Haploid fruiting was assayed by incubating spotted suspensions of cells on filament agar at 24°C for up to 4 weeks.
For confrontation assays, isolated colonies were streaked on the surface of filament agar as lines with sterile toothpicks. In some cases, a sterile dialysis membrane was interposed between the cell types by inserting it with sterile forceps into an agar cut that was sealed with molten agar.
Virulence test.
Virulence was evaluated using a rabbit model of cryptococcal meningitis (2, 45). Cells of the isogenic wild-type strain (H99) and the gpb1 mutant strain were grown for 48 h in liquid YPD medium and resuspended in 15 mM phosphate-buffered saline. New Zealand White male rabbits (four in a group for each strain) weighing 2 to 3 kg were administered cortisone acetate (2.5 mg/kg of body weight) intramuscularly 1 day prior to inoculation of C. neoformans and then daily for 14 days. Twenty-four hours following initial steroid treatment, rabbits were anesthetized with xylazine and ketamine intramuscularly and inoculated intracisternally with 0.3 ml of cell suspension (3 × 108 cells/ml). Rabbits were sedated on days 4, 7, 10, and 14 postinoculation, and cerebrospinal fluid (CSF) was withdrawn. Cell cultures were performed by plating dilutions of CSF (in phosphate-buffered saline) on YPD medium, incubating the plates at 30°C for 3 days, and counting viable colonies.
Nucleotide sequence accession number. The GPB1 gene sequence has been submitted to GenBank under accession no. AF091120.
RESULTS
Identification of the C. neoformans G-protein β subunit GPB1.
Previous studies revealed that the Gα protein GPA1 (54) regulates mating and virulence in C. neoformans (2). To further address the role of G proteins in mating and physiology, we identified a heterotrimeric G-protein β subunit from C. neoformans.
Oligonucleotides were designed against conserved regions of Gβ subunits and used as primers in low-stringency PCRs with a C. neoformans cDNA library or C. neoformans genomic DNA as a template. Primers encompassing two conserved peptides, IYALHW and AGYDDY, amplified a partial Gβ cDNA homolog from the serotype D strain B-3501. This cDNA clone was sequenced and then used to probe a Southern blot of genomic DNA isolated from the serotype A MATα strain H99 and from the congenic serotype D strains JEC20 (MATa) and JEC21 (MATα). The GPB1 gene was present in a single copy in both mating types and serotypes (data not shown).
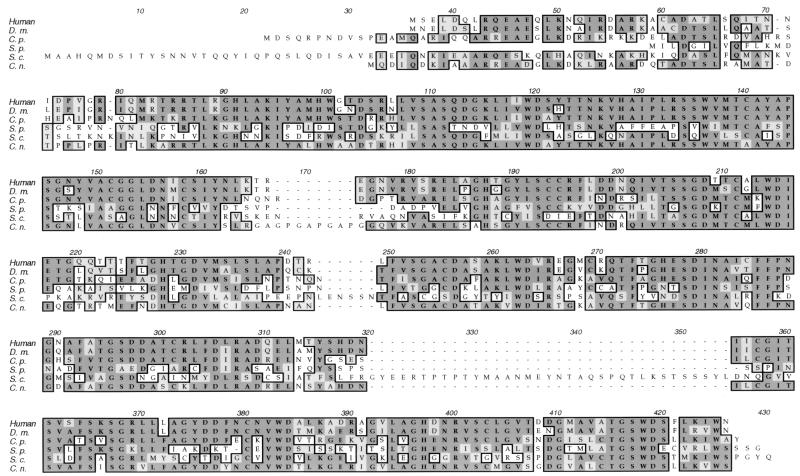
The complete GPB1 genomic locus was cloned from a size-selected genomic library. Sequence analysis revealed an open reading frame of 1,059 nucleotides encoding a 352-amino-acid protein (GenBank accession no. AF091120). Four introns were identified by sequence comparison with a cDNA clone from strain H99. The predicted GPB1 protein shares marked identity with G-protein β subunits from other organisms, including Gβ subunits from humans (68%), Drosophila melanogaster (67%), C. parasitica (70%), Schizosaccharomyces pombe (40%), and S. cerevisiae (38%) (Fig. 1).
FIG. 1.
C. neoformans GPB1 exhibits identity to G-protein β subunits. The sequences of Gβ subunits from humans (12), D. melanogaster (D.m.) (66), Cryphonectria parasitica (C.p.) (21), Schizosaccharomyces pombe (S.p.) (23), and S. cerevisiae (S.c.) (59) were aligned with that of the C. neoformans (C.n.) GPB1 protein. Identical amino acids are boxed and darkly shaded; conservative amino acid substitutions are boxed and lightly shaded.
Disruption of the C. neoformans GPB1 gene.
The GPB1 gene was disrupted by inserting the ADE2 gene into the GPB1 open reading frame, and the resulting gpb1::ADE2 disruption allele was introduced into the ade2 strain M049 by biolistic DNA transformation and homologous recombination. Genomic DNA was extracted from candidate gpb1::ADE2 strains (18). PCRs with primers flanking the ADE2 gene insertion were used to identify gpb1 mutations and generate a 550-bp product from the GPB1 allele and a 3,450-bp product from the gpb1::ADE2 allele.
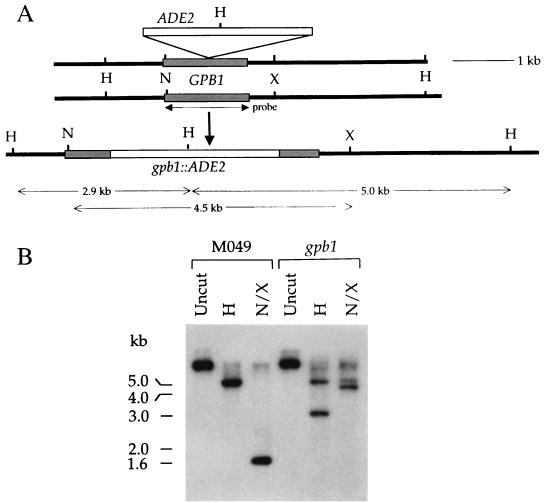
In total, six gpb1::ADE2 mutant strains were identified from 306 adenine-prototrophic transformants by PCR analysis. In subsequent analyses, independent gpb1 mutations conferred the same phenotypes. Southern blot analysis confirmed that the GPB1 gene had been replaced by the gpb1::ADE2 disruption allele by homologous recombination at the GPB1 locus in all six mutant strains (Fig. 2). The wild-type GPB1 gene is located on a 4.9-kb HindIII fragment and a 1.6-kb NotI-XbaI fragment. In the gpb1::ADE2 mutant, the wild-type 4.9-kb HindIII fragment is replaced by 2.9- and 5.0-kb HindIII fragments (Fig. 2). In addition, the 1.6-kb NotI-XbaI wild-type GPB1 locus is missing from the gpb1::ADE2 mutant, having been replaced by 4.5- and 5.1-kb NotI-XbaI fragments (Fig. 2).
FIG. 2.
Disruption of the C. neoformans GPB1 gene. (A) A schematic illustration of the GPB1 gene replacement; (B) Southern analysis of the wild type and the gpb1 mutant. The ADE2 gene was inserted at an ApaI site in the GPB1 coding domain, and the gpb1::ADE2 disruption allele was used to biolistically transform the Δade2 strain M049 to adenine prototrophy. Genomic DNAs from the isogenic GPB1 wild-type strain H99 and the gpb1::ADE2 disruption mutant were isolated, cleaved with HindIII (H) or with NotI (N) and XbaI (X), separated by 1% agarose gel electrophoresis, transferred to a nylon membrane, and probed with the 32P-labeled GPB1 open reading frame (indicated by an arrow labeled “probe”). Sizes of DNA fragments resulting from gene disruption are indicated by horizontal arrows. The positions of DNA molecular size standards are indicated on the left.
GPB1 is required for mating in C. neoformans.
We tested whether GPB1 regulates mating in C. neoformans. MATα and MATa strains of C. neoformans mate when cocultured on nutrient-limiting medium (25). Mating consists of conjugation tube formation, cell fusion, and filamentation (1). Subsequent nuclear migration results in the formation of dikaryotic filaments that differentiate to form terminal basidia, in which nuclear fusion, meiosis, and sporulation occur. When one or both parents are sterile, few or no filaments or spores are produced.
The wild-type GPB1 MATα serotype A strain (H99) yielded abundant filaments and basidiospores when crossed with the MATa serotype D strain JEC20 (Fig. 3). In contrast, no filaments or spores were ever observed when any of the independent gpb1 mutant MATα strains were mated with their MATa mating partners (Fig. 3). Reintroduction of the wild-type GPB1 gene into the gpb1 mutant strain restored filamentation and spore production to the wild-type level (Fig. 3).
FIG. 3.
The C. neoformans G-protein β subunit GPB1 is required for mating. The isogenic C. neoformans wild-type MATα strain H99 (GPB1 GPA1) and the gpb1::ADE2 (gpb1) and gpa1::ADE2 (gpa1) MATα mutant strains were mated with the MATa strain JEC20 on V8 agar medium (upper panels) and V8 agar medium supplemented with 2 or 10 mM cAMP as indicated (lower panels). The wild-type GPB1 gene was reintroduced into the gpb1 mutant strain as described in Materials and Methods (gpb1+GPB1). Mating was at 22°C for 7 days. Magnification, ×25.
GPB1 and the Gα subunit GPA1 play different roles in mating.
Several findings suggest that the Gα protein GPA1 and the Gβ protein GPB1 function in distinct pathways to regulate mating. First, the gpb1 mutation confers an absolute mating defect, whereas, following prolonged incubation, gpa1 mutants eventually mate to a limited extent with a wild-type mating partner, forming filaments, basidia, and recombinant basidiospores (Fig. 3) (2). Second, cAMP suppresses the mating defect of gpa1 mutants, but not that of gpb1 mutants (Fig. 3) (2). Third, no interaction between GPA1 and GPB1 was detected in the two-hybrid system (data not shown) (see Materials and Methods).
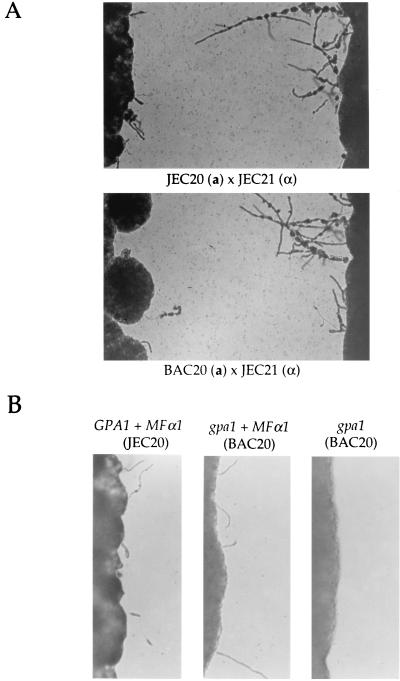
Several additional findings indicate the Gα subunit GPA1 is not required for pheromone sensing. First, in confrontation assays, the congenic MATα strain JEC21 and the MATa strain JEC20 both produced conjugation tubes in response to pheromone secreted by their mating partners (Fig. 4A). Most importantly, when the wild-type MATα strain JEC21 was grown in confrontation with a gpa1 MATa mutant strain (BAC20), both the gpa1 mutant and the wild-type strain produced conjugation tubes (Fig. 4A). Second, when a plasmid expressing the MFα1 pheromone was introduced into wild-type and gpa1 mutant MATa strains, both produced conjugation tubes (Fig. 4B). The response of gpa1 mutants to pheromones was somewhat reduced from that of the wild type, but taken together these findings indicate that GPA1 is not required for pheromone sensing. In an assay that detects cell fusion during mating (MATα ura5 strains were coincubated with MATa lys1 strain JEC30 on V8 agar, and prototrophic self-filamenting heterokaryons were detected by replica plating to YNB medium), the gpb1 mutation prevented cell fusion whereas the gpa1 mutation reduced but did not block fusion (data not shown). In a mating assay in which recombinant basidiospores were quantified (MATα prototrophic strains were mated with MATa ura5 lys1 strain JEC53 on V8 agar, and LYS1 ura5 recombinants were selected on 5-fluoroorotic acid–lysine medium), no recombinant basidiospores were produced by the gpb1 mutant whereas the gpa1 mutant produced a reduced number of basidiospores.
FIG. 4.
The Gα subunit GPA1 is not required for responses to pheromones. (A) Cells of the wild-type MATα serotype D strain JEC21 were grown in confrontation with the isogenic MATa GPA1 wild-type strain JEC20 (upper panel) or the gpa1 mutant strain BAC20 (lower panel), with incubation for 3 days at 24°C on filament agar, and conjugation tubes were photographed. Magnification, ×25. (B) A ura5 derivative of the GPA1 wild-type strain JEC20 (MATa ura5) and the isogenic gpa1 mutant strain BAC20 (MATa gpa1::ADE2 ura5) were transformed with plasmid pCnTel1 lacking or expressing the MFα1 pheromone gene, grown on filament agar for 2 days at 24°C, and photographed. Magnification, ×50.
GPB1 is not required for melanin or capsule production or virulence.
The Gα protein GPA1 regulates the production of the virulence factors melanin and capsule in response to nutrient limitation (2). To determine whether the functions of GPB1 and GPA1 are distinct, we tested whether the gpb1 mutation alters virulence factors or virulence.
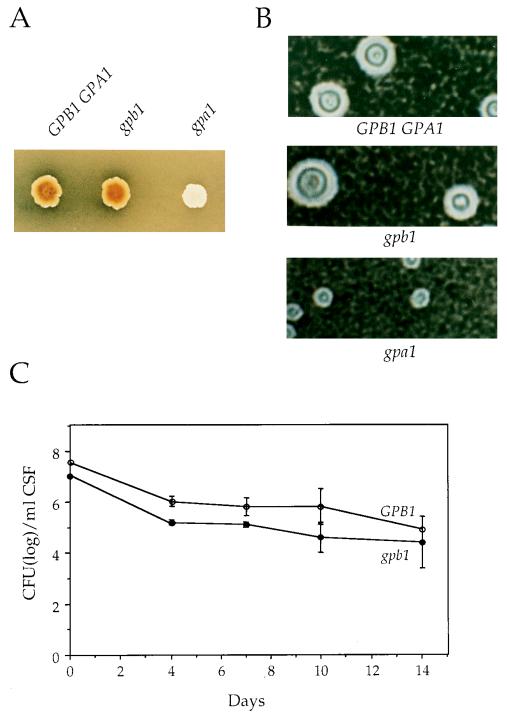
C. neoformans produces melanin when grown in the presence of diphenolic precursors under carbohydrate-limiting conditions. Melanin is required for virulence and may protect cells from nitrogen- and oxygen-derived radicals produced by host immune cells (56, 57). When cultured on a medium containing niger seed extract as a source of diphenolic compounds, gpa1 mutants did not produce melanin (Fig. 5A) (2). In contrast, gpb1 mutant strains produced melanin to the same extent as the GPB1 wild-type strain (Fig. 5A). By a quantitative spectrophotometric assay, it was determined that gpb1 mutant and GPB1 wild-type cells produced similar levels of laccase activity (data not shown) (63).
FIG. 5.
GPB1 is not required for virulence factors or virulence in C. neoformans. (A) The isogenic GPB1 GPA1 wild-type strain H99 and the gpb1::ADE2 (gpb1) and gpa1::ADE2 (gpa1) mutant strains were grown on niger seed agar for 72 h at 37°C. Strains that produce melanin (GPB1 GPA1, gpb1) form brown colonies on this medium, whereas strains that do not produce melanin (gpa1) are white. (B) Cells of the wild-type strain H99 (GPB1 GPA1) and the gpb1::ADE2 (gpb1) and gpa1::ADE2 (gpa1) mutant strains were grown in low-iron medium plus EDDHA at 30°C for 48 h to induce capsule synthesis. The polysaccharide capsule was identified by India ink staining and photographed. Magnification, ×200. (C) The GPB1 wild-type (H99) and gpb1 mutant strains were inoculated intracisternally into immunosuppressed rabbits. CSF was withdrawn on days 4, 7, 10, and 14 postinfection, and the numbers of surviving yeast cells were determined by plating serial dilutions of CSF on YPD medium. The mean cell count for each strain was plotted with the standard error of the mean.
C. neoformans is distinguished from many pathogenic yeast by its polysaccharide capsule, which inhibits phagocytosis by host cells and is required for virulence (3). Formation of the capsule is induced during infection or in response to low-iron or elevated-CO2 conditions in vitro (16, 55). To assess capsule production, the wild-type strain H99 and the gpa1 and gpb1 mutant strains were grown in liquid iron-limiting medium. Capsule production in wild-type cells was readily observed by staining with India ink, and the capsule size was decreased in gpa1 mutant cells (Fig. 5B) (2). In contrast, gpb1 mutant cells produced capsules similar to those of wild-type cells (Fig. 5B).
We next tested whether the gpb1 mutation alters virulence. An animal model of cryptococcal meningitis was employed in which glucocorticoid-immunosuppressed rabbits were inoculated intrathecally with C. neoformans strains and survival in the central nervous system was determined by removing CSF and quantifying yeast cells by serial dilution and culture (2, 45). As shown in Fig. 5C, virulence of the gpb1 mutant was similar to that of the GPB1 wild-type strain H99. Both wild-type and gpb1 mutant cells persisted for up to 14 days in the CSF, and they were recovered in similar quantities, although cell counts for the gpb1 mutant were slightly reduced on days 4 and 7. Similar results were obtained with a second gpb1 mutant, as well as when the inoculum size was reduced 10-fold. gpb1 mutant cells recovered from infected animals still exhibited a mating defect in vitro. In summary, in contrast to GPA1, GPB1 is not required for melanin or capsule production and is not a major virulence determinant.
GPB1 regulates mating upstream of a MAP kinase cascade.
Our findings suggested that the Gβ subunit GPB1 activates a signaling pathway that regulates mating in parallel with the GPA1-cAMP-regulated nutrient-sensing pathway. We tested whether the Gβ protein GPB1 regulates a MAP kinase cascade during mating in C. neoformans.
In addition to the G-protein β subunit, two other MAP kinase cascade components have been identified in C. neoformans: a MAP kinase homolog, CPK1 (R. Davidson and J. Heitman, unpublished data), and a homolog of the STE12 transcription factor (61, 67). We tested whether CPK1 or STE12α functions downstream of GPB1 by epistasis, using cloned genes under the control of the C. neoformans GAL7 promoter, which is induced by galactose and repressed by glucose (62).
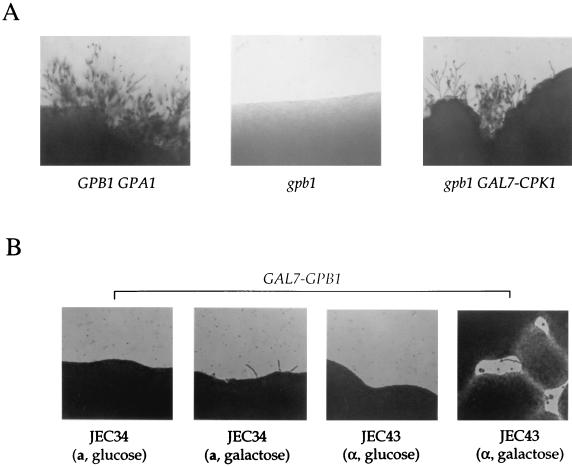
When the gpb1 mutant strain was transformed with the GAL7-CPK1 gene fusion, mating with a MATa strain was restored on galactose filament agar but not on glucose (Fig. 6A). Thus, expression of the CPK1 MAP kinase suppresses the gpb1 mating defect, providing evidence that GPB1 functions upstream of this MAP kinase. The GAL7-CPK1 gene fusion did not restore mating in gpa1 mutants (data not shown), indicating that CPK1 functions downstream of GPB1 but not of GPA1.
FIG. 6.
GPB1 activates a MAP kinase cascade involving the CPK1 kinase. (A) The CPK1 gene expressed from the C. neoformans GAL7 promoter in the URA5 plasmid pCnTel1 was introduced into a gpb1 ura5 mutant strain (see Materials and Methods) by biolistic transformation. The isogenic MATα wild-type strain H99 (GPB1 GPA1), the gpb1 mutant strain, and the gpb1 mutant strain transformed with the GAL7-CPK1 gene fusion (gpb1 GAL7-CPK1) were cocultured with a MATa mating partner (JEC20). Mating was for 21 days at 22°C on filament agar containing 0.5% galactose (shown here) or 0.5% glucose (data not shown). Magnification, ×25. (B) The congenic serotype D MATa ura5 strain JEC34 and the MATα ura5 strain JEC43 were transformed with the GAL7-GPB1 gene fusion linked to the URA5 gene and grown for 72 h at 24°C on filament agar with glucose or galactose. Conjugation tubes emanating from cell patches were photographed. Magnification, ×25.
In contrast to the effects of CPK1, the GAL7-STE12α gene fusion did not restore mating of the gpb1 mutant strain on glucose or galactose filament agar (data not shown). The functions of STE12α likely involve haploid fruiting and not mating, because STE12α overexpression stimulates haploid fruiting (61) whereas ste12α mutations block haploid fruiting but not mating (67).
GPB1 stimulates conjugation tube formation in MATα and MATa cells.
We next tested whether GPB1 plays an active signaling role upstream of the MAP kinase cascade, analogous to that of the Gβγ complex in S. cerevisiae (50). During mating in C. neoformans, the mating partners secrete pheromones that trigger the formation of conjugation tubes in the opposite cell type (1, 41; R. Davidson and J. Heitman, unpublished data). We tested whether GPB1 overexpression stimulates conjugation tube formation in cells not exposed to pheromones.
The GAL7 promoter was fused upstream of the GPB1 gene, and the GAL7-GPB1 gene fusion was introduced into congenic MATα and MATa serotype D strains. Growth on galactose filament agar induced the formation of conjugation tubes in both MATa and MATα strains (Fig. 6B). Conjugation tubes produced in response to GPB1 overexpression were similar to those observed in confrontation assays or in MATa cells in response to expressed or synthetic MFα1 pheromone (1, 41) (Fig. 4). MATa cells produced more conjugation tubes than did MATα cells, suggesting that the mating responses of the two cell types differ (Fig. 6B).
GPB1 and MATa cells regulate monokaryotic fruiting.
Mating of MATa and MATα cells of C. neoformans is regulated by both pheromones and nitrogen starvation. In contrast, in response to nitrogen starvation alone, MATα haploid strains differentiate, forming monokaryotic filaments, basidia, and spores by haploid fruiting (62). This filamentous differentiation shares some features with pseudohyphal growth in S. cerevisiae (15). Components of the mating pheromone response pathway are required for pseudohyphal growth, whereas mating pheromones, pheromone receptors, and the coupled heterotrimeric G protein are not (35). We therefore hypothesized that the Gβ protein GPB1 would not be required for haploid fruiting in C. neoformans.
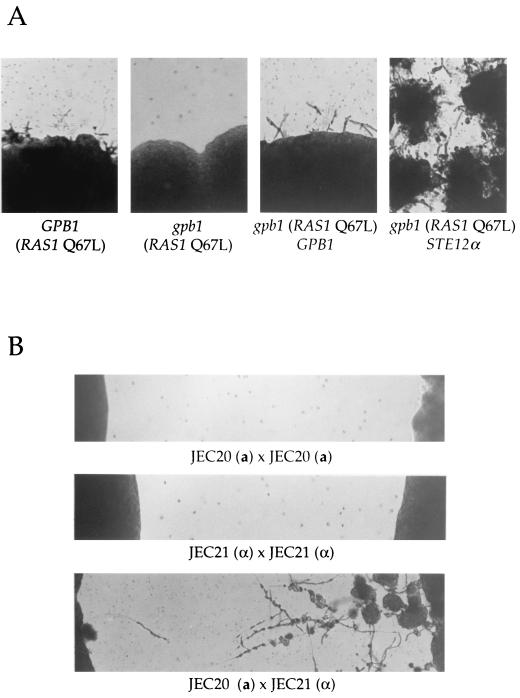
To our surprise, we found that GPB1 is required for haploid fruiting in C. neoformans. Similar to the many lab strains of S. cerevisiae which do not undergo pseudohyphal growth, C. neoformans strains also differ in their ability to form filaments in response to nitrogen starvation. The serotype A strain H99 does not exhibit haploid fruiting under a variety of conditions. Introduction of a dominant active RAS1 mutant (Ras1 Q67L) does stimulate haploid fruiting of strain H99 (Fig. 7A) (J. A. Alspaugh and J. Heitman, unpublished data). However, the dominant active Ras1 Q67L mutant protein did not stimulate haploid fruiting in the gpb1 mutant strain (Fig. 7A). Reintroduction of the wild-type GPB1 gene restored haploid fruiting of the gpb1 mutant (Fig. 7A). The GAL7-STE12α gene fusion (Fig. 7A) and the GAL7-CPK1 gene fusion (data not shown) suppressed the haploid fruiting defect of gpb1 mutants on galactose filament agar. Thus, GPB1 is required for monokaryotic fruiting and functions upstream of CPK1 and STE12α.
FIG. 7.
GPB1 and MATa cells regulate haploid fruiting. (A) The isogenic GPB1 wild-type strain H99 (far-left panel), the gpb1::ADE2 mutant strain (second panel from left), and the gpb1::ADE2 mutant strain reconstituted with the GPB1 wild-type gene (third panel from left) were transformed with the dominant active Ras1 Q67L mutant gene, grown on glucose filament agar medium for 7 days at 24°C, and photographed. The gpb1 mutant strain was also transformed with plasmid pCGS-1 expressing the GAL7-STE12 fusion gene and grown on galactose filament agar (far-right panel) for 7 days at 24°C. Magnification, ×25. (B) Cells of the serotype D MATα strain JEC21 were grown in confrontation with themselves (middle panel) or with congenic cells of the opposite (MATa) mating type (strain JEC20) (lower panel). As a control, the MATa strain JEC20 was grown in confrontation with itself (upper panel). Cells were incubated for 10 days at 24°C on filament agar and photographed. Magnification, ×25.
We next addressed why the pheromone-sensing Gβ protein is required for haploid fruiting if this process normally occurs in response to nitrogen limitation. We found that when MATα cells are grown in confrontation with MATa cells, monokaryotic fruiting of the MATα cells is dramatically stimulated and abundant filaments, basidia, and basidiospores are produced (Fig. 7B). In contrast, a much lower level of monokaryotic fruiting is observed when MATα cells are grown in isolation or when MATα cells are grown in confrontation with MATα cells (Fig. 7B). The response of MATα cells to confronting MATa cells does not require cell-cell or cell-filament contact, and it occurs before any of the projecting filaments touch the confronting cells. Moreover, monokaryotic fruiting was still observed when a dialysis membrane with a molecular mass cutoff of 3,800 Da was interposed between MATα and MATa cells (data not shown). The C. neoformans mating pheromones are predicted to diffuse through this membrane.
By microscopic observation and nuclear staining with the DNA-specific dye DAPI (4′,6′-diamidino-2-phenylindole), it was determined that the filament cells are linked by unfused clamp connections and are monokaryotic, hallmarks of monokaryotic fruiting. In addition, micromanipulation and mating type tests confirmed that basidiospores produced by MATα cells in response to confronting MATa cells are all MATα and are thus products of asexual monokaryotic fruiting (data not shown). Our findings indicate that monokaryotic fruiting of MATα cells is stimulated by MATa cells, possibly in response to MATa pheromones sensed by a receptor coupled to GPB1.
DISCUSSION
We have identified the gene encoding a heterotrimeric G-protein β subunit, GPB1, from C. neoformans. GPB1 is required for mating and plays a role in the pheromone response in both MATα and MATa cells by activating a MAP kinase cascade leading to conjugation tube formation and cell fusion. Two distinct signal transduction pathways regulate mating: one involves pheromone sensing and requires GPB1, and the second senses nutrients via the Gα protein GPA1-cAMP pathway and is also required for virulence factor production and pathogenicity. These signal transduction cascades coordinately regulate mating in C. neoformans, analogous to the role of the MAP kinase and Gα-cAMP signal transduction cascades in development in other organisms, including pseudohyphal growth in S. cerevisiae and mating in Schizosaccharomyces pombe. We found a novel role for the pheromone-sensing Gβ subunit GPB1 in haploid fruiting in C. neoformans. We have also discovered that monokaryotic fruiting of MATα cells is dramatically stimulated by MATa cells, suggesting that this differentiation cascade may function in mating. Finally, we have shown that gpb1 mutant strain virulence is similar to that of wild-type strains, indicating that this component of the mating pathway does not play a prominent role in virulence.
C. neoformans GPB1 Gβ and GPA1 Gα subunits have distinct functions.
Our studies support a model in the which the GPB1 Gβ subunit and the GPA1 Gα subunit function in two different signal transduction cascades that regulate different steps in mating. Several observations indicate that GPB1 functions in the pheromone response pathway and regulates early steps in mating involving conjugation tube formation and cell fusion. First, gpb1 mutants are completely sterile and exhibit a profound defect in a cell fusion assay. Second, GPB1 overexpression stimulates conjugation tube formation. Third, overexpression of the MAP kinase CPK1 suppresses the mating defect of gpb1 mutant strains, whereas cAMP does not. Later in mating, the pheromone response pathway likely also plays a second role involving the fusion of the clamp cells during filament formation.
Our findings also contribute to the understanding of the role of GPA1-cAMP signaling in mating. The mating defect of gpa1 mutant strains is suppressed by cAMP but not by the MAP kinase CPK1. In addition, gpa1 mutants can respond to pheromones in confrontation assays and in response to expression of the MFα1 pheromone gene (Fig. 4). In quantitative mating assays, the gpa1 mutation reduces but does not block cell fusion and also reduces filamentation and the production of recombinant basidiospores. The nutrient-sensing GPA1-cAMP cascade is required for melanin and capsule production and virulence, whereas GPB1 is not.
These findings support a model in which the GPA1 Gα and GPB1 Gβ subunits are components of two different signaling cascades. We propose that GPA1 and GPB1 are components of two different G proteins and function in distinct signaling cascades, one that senses nutrients via a cAMP pathway (GPA1) and another that senses mating pheromones and signals via a MAP kinase cascade (GPB1).
MAP kinase signaling in MATα and MATa cells.
Our findings reveal that the Gβ subunit GPB1 regulates conjugation tube formation in both MATa and MATα cells. The GPB1 gene is present in both MATα and MATa strains and is expressed in cells of both mating types. This is in contrast to other MAP kinase cascade components recently identified in C. neoformans, including STE11α, STE12α, and STE20α homologs, which are encoded by the MATα locus and are specific to MATα cells (61; B. Wickes and J. Edman, personal communication; P. Wang and J. Heitman, unpublished results). These findings raise the conundrum of how signaling occurs in MATa cells during mating if several components are present only in MATα cells. Our findings suggest that the MAP kinase cascade functions during mating in both MATα and MATa cells. We propose that mating in both cell types is regulated by GPB1 signaling via two divergent versions of a conserved MAP kinase cascade: one, containing components encoded by the MATα locus, that supports mating and can also function in haploid fruiting and virulence, and another, with components encoded by the MATa locus, that plays a more restricted role in mating, does not support haploid fruiting, and remains to be identified. Such a model may be related to the situation in the yeast S. cerevisiae, which expresses two related MAP kinases with divergent functions: Fus3, which is specialized for mating of haploid cells, and Kss1, which regulates pseudohyphal differentiation of diploid cells (7, 38).
G-protein signaling roles in other yeasts.
Our studies on G-protein function are relevant to previous studies of G proteins in other yeasts. In S. cerevisiae, two G proteins regulate responses to pheromone and nutrients (50). During pseudohyphal differentiation, nutrients regulate the Gα protein Gpa2, which signals via a cAMP cascade (24, 37). During mating, pheromone binding to the Ste2 or Ste3 receptors recruits the Gαβγ complex (Gpa1-Ste4-Ste18), and the released Ste4-Ste18 βγ complex activates the MAP kinase cascade by recruiting signaling components to the membrane (32–34, 47). The Gα subunit Gpa1 inhibits signaling by βγ. C. neoformans Gβ subunit GPB1 functions analogously to the Ste4 Gβ subunit in S. cerevisiae mating, whereas the functions of the C. neoformans Gα subunit GPA1 are analogous to nutrient sensing by S. cerevisiae GPA2.
In the fission yeast Schizosaccharomyces pombe, mating is also regulated by two G proteins, composed of the Gα subunit Gpa1 and the Gα and Gβ subunits Gpa2 and Gpb1 (65). Gpa1 is required for the pheromone response during mating and, in contrast to the situation for S. cerevisiae, plays a positive role in activating the MAP kinase cascade. Gpa2 plays a role analogous to that of the S. cerevisiae Gpa2 and C. neoformans GPA1 subunits, and it functions in a nutrient-sensing cAMP pathway regulating mating (19, 43). Mutants lacking the Gβ subunit Gpb1 exhibit a phenotype similar to that of gpa2 mutants and mate and sporulate under nutrient-rich conditions (23). The sensing of pheromones by the Gβ subunit GPB1 in C. neoformans appears to be distinct from the role for the Schizosaccharomyces pombe Gβ subunit in nutrient sensing.
GPB1 and MATa cells regulate haploid fruiting in C. neoformans.
We found that the Gβ subunit GPB1 is also required for haploid fruiting in C. neoformans. Haploid fruiting is a differentiation pathway whereby MATα cells can form filaments and sporulate in response to nitrogen starvation in the absence of a mating partner (62). Haploid fruiting shares features with pseudohyphal differentiation in the yeast S. cerevisiae, which is regulated by a MAP kinase cascade and induced by nitrogen limitation (15). However, the mating pheromones, receptors, and the coupled G protein are not required for filamentation in S. cerevisiae (35).
Although haploid fruiting occurs to a limited extent in some MATα strains in response to nitrogen starvation alone (61), we found that monokaryotic fruiting of C. neoformans MATα cells is markedly stimulated by confrontation with MATa cells. This stimulation does not require cell-cell or filament-filament contact. We propose that MATa cells stimulate haploid fruiting of adjacent MATα cells by secreting a peptide mating pheromone. Stimulation of monokaryotic fruiting by this pheromone may function to disperse MATα spores to locate uncommon MATa mating type cells at a distance. Thus, haploid fruiting could function as a prelude to mating in an organism in which MATα cells are more abundant than MATa cells.
We propose that haploid fruiting occurs in isolated C. neoformans MATα cells at a low level due to basal signaling of the pheromone-responsive MAP kinase pathway in the absence of pheromone and is then markedly stimulated by a pheromone produced by adjacent MATa cells. MATa cells do not undergo haploid fruiting, in part because they lack the MATα locus-encodes transcription factor STE12α required for haploid fruiting (67). The model in which haploid fruiting is regulated by pheromones makes the testable prediction that the MFa1 pheromone and its receptor are also required.
Although haploid invasive growth and diploid filamentous growth in S. cerevisiae are not normally regulated by the Gβ subunit Ste4, our findings may be related to studies of altered differentiation in mutant S. cerevisiae strains and the basal signaling state of the yeast pheromone response pathway. First, in fus3 mutant yeast strains, haploid invasive growth and expression of filamentous reporter genes are increased, and this requires the Gβ subunit Ste4 and results from inappropriate cross-talk between the pheromone-responsive MAP kinase cascade and the filamentous growth pathway (38). Second, even in the absence of pheromones, the S. cerevisiae pheromone response pathway is active at a basal level. The Gβ subunit Ste4, the kinases Ste11 and Ste7, the scaffold Ste5, and the Ste12 transcription factor are required for basal signaling (11, 17, 50). By analogy, we propose that signaling by the Gβ subunit GPB1 in the absence of pheromones supports a basal level of monokaryotic fruiting in C. neoformans that is then stimulated by pheromones.
Perspective.
The MATα locus has been linked to virulence in C. neoformans. Our finding that gpb1 mutant strains are defective in mating but not impaired in virulence indicates that mating is not required for virulence, and further studies are needed to establish the link between the MATα locus and virulence.
ACKNOWLEDGMENTS
We thank Cristl Arndt, Lora Cavallo, and Wiley Schell for assistance; Maria Cardenas for advice; Andy Alspaugh, Maria Cardenas, Cristina Cruz, Rob Davidson, Danny Lew, and Rey Sia for comments; and Andy Alspaugh, Rob Davidson, Don Nuss, and Brian Wickes for reagents and strains.
This work was supported by NIAID R01 grants AI39115 and AI42159 and program project grant P01 AI44975 from NIAID to the Duke University Mycology Research Unit. Joseph Heitman is an associate investigator of the Howard Hughes Medical Institute and a Burroughs Wellcome Scholar in Molecular Pathogenic Mycology.
REFERENCES
- 1.Alspaugh, J. A., R. C. Davidson, and J. Heitman. Morphogenesis of Cryptococcus neoformans. In J. F. Ernst and A. Schmidt (ed.), Dimorphism in human pathogenic and apathogenic yeasts, in press. S. Karger, Basel, Switzerland.
- 2.Alspaugh J A, Perfect J R, Heitman J. Cryptococcus neoformans mating and virulence are regulated by the G-protein α subunit GPA1 and cAMP. Genes Dev. 1997;11:3206–3217. doi: 10.1101/gad.11.23.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang Y C, Kwon-Chung K J. Complementation of a capsule-deficient mutation of Cryptococcus neoformans restores its virulence. Mol Cell Biol. 1994;14:4912–4919. doi: 10.1128/mcb.14.7.4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang Y C, Penoyer L A, Kwon-Chung K J. The second capsule gene of Cryptococcus neoformans, CAP64, is essential for virulence. Infect Immun. 1996;64:1977–1983. doi: 10.1128/iai.64.6.1977-1983.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clapham D E, Neer E J. New roles for G-protein βγ-dimers in transmembrane signalling. Nature. 1993;365:403–406. doi: 10.1038/365403a0. [DOI] [PubMed] [Google Scholar]
- 6.Cole G M, Reed S I. Pheromone-induced phosphorylation of a G protein β subunit in S. cerevisiae is associated with an adaptive response to mating pheromone. Cell. 1992;64:703–716. doi: 10.1016/0092-8674(91)90500-x. [DOI] [PubMed] [Google Scholar]
- 7.Cook J G, Bardwell L, Thorner J. Inhibitory and activating functions for MAPK Kss1 in the S. cerevisiae filamentous-growth signalling pathway. Nature. 1997;390:85–88. doi: 10.1038/36355. [DOI] [PubMed] [Google Scholar]
- 8.Crespo P, Xu N, Simonds W F, Gutkind J S. Ras-dependent activation of MAP kinase pathway mediated by G-protein βγ subunits. Nature. 1994;369:418–420. doi: 10.1038/369418a0. [DOI] [PubMed] [Google Scholar]
- 9.Dietzel C, Kurjan J. The yeast SCG1 gene: a Gα-like protein implicated in the α- and a-factor response pathway. Cell. 1987;50:1001–1010. doi: 10.1016/0092-8674(87)90166-8. [DOI] [PubMed] [Google Scholar]
- 10.Edman J C. Isolation of telomerelike sequences from Cryptococcus neoformans and their use in high-efficiency transformation. Mol Cell Biol. 1992;12:2777–2783. doi: 10.1128/mcb.12.6.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fields S, Chaleff D T, Sprague G F., Jr Yeast STE7, STE11, and STE12 genes are required for expression of cell-type-specific genes. Mol Cell Biol. 1988;8:551–556. doi: 10.1128/mcb.8.2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fong H K W, Hurley J B, Hopkins R S, Miake-Lye R, Johnson M S, Doolittle R F, Simon M I. Repetitive segmental structure of the transducin β subunit: homology with the CDC4 gene and identification of related mRNAs. Proc Natl Acad Sci USA. 1986;83:2162–2166. doi: 10.1073/pnas.83.7.2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao S, Nuss D L. Distinct roles for two G protein α subunits in fungal virulence, morphology, and reproduction revealed by targeted gene disruption. Proc Natl Acad Sci USA. 1996;93:14122–14127. doi: 10.1073/pnas.93.24.14122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilman A G. G-proteins: transducers of receptor-generated signals. Annu Rev Biochem. 1987;56:615–649. doi: 10.1146/annurev.bi.56.070187.003151. [DOI] [PubMed] [Google Scholar]
- 15.Gimeno C J, Ljungdahl P O, Styles C A, Fink G R. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell. 1992;68:1077–1090. doi: 10.1016/0092-8674(92)90079-r. [DOI] [PubMed] [Google Scholar]
- 16.Granger D L, Perfect J R, Durack D T. Virulence of Cryptococcus neoformans: regulation of capsule synthesis by carbon dioxide. J Clin Investig. 1985;76:508–516. doi: 10.1172/JCI112000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hagen D C, McCaffrey G, Sprague G F., Jr Pheromone response elements are necessary and sufficient for basal and pheromone-induced transcription of the FUS1 gene of Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:2952–2961. doi: 10.1128/mcb.11.6.2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoffman C S, Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57:267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- 19.Isshiki T, Mochizuki N, Maeda T, Yamamoto M. Characterization of a fission yeast gene, gpa2, that encodes a Gα subunit involved in the monitoring of nutrition. Genes Dev. 1992;6:2455–2462. doi: 10.1101/gad.6.12b.2455. [DOI] [PubMed] [Google Scholar]
- 20.James P, Halladay J, Craig E A. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kasahara S, Nuss D L. Targeted disruption of a fungal G-protein β subunit gene results in increased vegetative growth but reduced virulence. Mol Plant-Microbe Interact. 1997;10:984–993. doi: 10.1094/MPMI.1997.10.8.984. [DOI] [PubMed] [Google Scholar]
- 22.Katz A, Wu D, Simon M I. Subunits βγ of heterotrimeric G protein activate βγ isoform of phospholipase C. Nature. 1992;360:686–689. doi: 10.1038/360686a0. [DOI] [PubMed] [Google Scholar]
- 23.Kim D-U, Park S-K, Chung K-S, Choi M-U, Yoo H-S. The G protein β subunit Gpb1 of Schizosaccharomyces pombe is a negative regulator of sexual development. Mol Gen Genet. 1996;252:20–32. doi: 10.1007/BF02173201. [DOI] [PubMed] [Google Scholar]
- 24.Kübler E, Mösch H U, Rupp S, Lisanti M P. Gpa2p, a G-protein alpha-subunit, regulates growth and pseudohyphal development in Saccharomyces cerevisiae via a cAMP-dependent mechanism. J Biol Chem. 1997;272:20321–20323. doi: 10.1074/jbc.272.33.20321. [DOI] [PubMed] [Google Scholar]
- 25.Kwon-Chung K J. Morphogenesis of Filobasidiella neoformans, the sexual state of Cryptococcus neoformans. Mycologia. 1976;68:942–946. [PubMed] [Google Scholar]
- 26.Kwon-Chung K J, Bennett J E. Medical mycology. Baltimore, Md: Williams & Wilkins; 1992. pp. 397–446. [Google Scholar]
- 27.Kwon-Chung K J, Bennett J E. Distribution of α and a mating types of Cryptococcus neoformans among natural and clinical isolates. Am J Epidemiol. 1978;108:337–340. doi: 10.1093/oxfordjournals.aje.a112628. [DOI] [PubMed] [Google Scholar]
- 28.Kwon-Chung K J, Edman J C, Wickes B L. Genetic association of mating types and virulence in Cryptococcus neoformans. Infect Immun. 1992;60:602–605. doi: 10.1128/iai.60.2.602-605.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kwon-Chung K J, Polacheck I, Popkin T J. Melanin-lacking mutants of Cryptococcus neoformans and their virulence for mice. J Bacteriol. 1982;150:1414–1421. doi: 10.1128/jb.150.3.1414-1421.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kwon-Chung K J, Rhodes J C. Encapsulation and melanin formation as indicators of virulence in Cryptococcus neoformans. Infect Immun. 1986;51:218–223. doi: 10.1128/iai.51.1.218-223.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kwon-Chung K J, Varma A, Edman J C, Bennett J E. Selection of ura5 and ura3 mutants from the two varieties of Cryptococcus neoformans on 5-fluoroorotic acid medium. J Med Vet Mycol. 1992;30:61–69. [PubMed] [Google Scholar]
- 32.Leberer E, Dignard D, Harcus D, Thomas D Y, Whiteway M. The protein kinase homologue Ste20p is required to link the yeast pheromone response G-protein βγ subunits to downstream signalling components. EMBO J. 1992;11:4815–4824. doi: 10.1002/j.1460-2075.1992.tb05587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leberer E, Dignard D, Hougan L, Thomas D Y, Whiteway M. Dominant-negative mutants of a yeast G-protein β subunit identify two functional regions involved in pheromone signalling. EMBO J. 1992;11:4805–4813. doi: 10.1002/j.1460-2075.1992.tb05586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leeuw T, Wu C, Schrag J D, Whiteway M, Thomas D Y, Leberer E. Interaction of G-protein β-subunit with a conserved sequence in Ste20/PAK family protein kinases. Nature. 1998;391:191–195. doi: 10.1038/34448. [DOI] [PubMed] [Google Scholar]
- 35.Liu H, Styles C A, Fink G R. Elements of the yeast pheromone response pathway required for filamentous growth of diploids. Science. 1993;262:1741–1744. doi: 10.1126/science.8259520. [DOI] [PubMed] [Google Scholar]
- 36.Logothetis D E, Kurachi Y, Galper J, Neer E J, Clapham D E. The βγ subunits of GTP-binding proteins activate the muscarinic K+ channel in heart. Nature. 1987;325:321–326. doi: 10.1038/325321a0. [DOI] [PubMed] [Google Scholar]
- 37.Lorenz M C, Heitman J. Yeast pseudohyphal growth is regulated by GPA2, a G protein α homolog. EMBO J. 1997;16:7008–7018. doi: 10.1093/emboj/16.23.7008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Madhani H D, Styles C A, Fink G R. MAP kinases with distinct inhibitory functions impart signaling specificity during yeast differentiation. Cell. 1997;91:673–684. doi: 10.1016/s0092-8674(00)80454-7. [DOI] [PubMed] [Google Scholar]
- 39.Mitchell T G, Perfect J R. Cryptococcosis in the era of AIDS—100 years after the discovery of Cryptococcus neoformans. Clin Microbiol Rev. 1995;8:515–548. doi: 10.1128/cmr.8.4.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miyajima I, Nakafuku M, Nakayama N, Brenner C, Miyajima A, Kaibuchi K, Arai K, Kaziro Y, Matsumoto K. GPA1, a haploid-specific essential gene, encodes a yeast homolog of mammalian G protein which may be involved in mating factor signal transduction. Cell. 1987;50:1011–1019. doi: 10.1016/0092-8674(87)90167-x. [DOI] [PubMed] [Google Scholar]
- 41.Moore T D E, Edman J C. The α-mating type locus of Cryptococcus neoformans contains a peptide pheromone gene. Mol Cell Biol. 1993;13:1962–1970. doi: 10.1128/mcb.13.3.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Neiman A M, Stevenson B J, Xu H P, Sprague G F, Herskowitz I, Wigler M, Marcus S. Functional homology of protein kinase required for sexual differentiation in Schizosaccharomyces pombe and Saccharomyces cerevisiae suggests a conserved signal transduction module in eukaryotic organisms. Mol Biol Cell. 1993;4:107–120. doi: 10.1091/mbc.4.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nocero M, Isshiki T, Yamamoto M, Hoffman C S. Glucose repression of fbp1 transcription in Schizosaccharomyces pombe is partially regulated by adenylate cyclase activation by a G protein α subunit encoded by gpa2 (git8) Genetics. 1994;138:39–45. doi: 10.1093/genetics/138.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Obara T, Nakafuku M, Yamamoto M, Kaziro Y. Isolation and characterization of a gene encoding a G-protein alpha subunit from Schizosaccharomyces pombe: involvement in mating and sporulation pathways. Proc Natl Acad Sci USA. 1991;88:5877–5881. doi: 10.1073/pnas.88.13.5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Odom A, Muir S, Lim E, Toffaletti D L, Perfect J, Heitman J. Calcineurin is required for virulence of Cryptococcus neoformans. EMBO J. 1997;16:2576–2589. doi: 10.1093/emboj/16.10.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perfect J R, Toffaletti D L, Rude T H. The gene encoding phosphoribosylaminoimidazole carboxylase (ADE2) is essential for growth of Cryptococcus neoformans in cerebrospinal fluid. Infect Immun. 1993;61:4446–4451. doi: 10.1128/iai.61.10.4446-4451.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pryciak P M, Huntress F A. Membrane recruitment of the kinase cascade scaffold protein Ste5 by the Gβγ complex underlies activation of the yeast pheromone response pathway. Genes Dev. 1998;12:2684–2697. doi: 10.1101/gad.12.17.2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Salas S D, Bennett J E, Kwon-Chung K J, Perfect J R, Williamson P R. Effect of the laccase gene, CNLAC1, on virulence of Cryptococcus neoformans. J Exp Med. 1996;184:377–386. doi: 10.1084/jem.184.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 50.Sprague G F, Jr, Thorner J W. Pheromone response and signal transduction during the mating process of Saccharomyces cerevisiae. In: Jones E W, Pringle J R, Broach J R, editors. The molecular and cellular biology of the yeast Saccharomyces. 2. Gene expression. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 657–744. [Google Scholar]
- 51.Sudarshan S, Davidson R C, Heitman J, Alspaugh J A. Molecular analysis of the Cryptococcus neoformans ADE2 gene, a selectable marker for transformation and gene disruption. Fungal Genet Biol. 1999;27:36–48. doi: 10.1006/fgbi.1999.1126. [DOI] [PubMed] [Google Scholar]
- 52.Tang W-J, Gilman A G. Type-specific regulation of adenylyl cyclase by G protein βγ subunits. Science. 1991;254:1500–1503. doi: 10.1126/science.1962211. [DOI] [PubMed] [Google Scholar]
- 53.Toffaletti D L, Rude T H, Johnston S A, Durack D T, Perfect J R. Gene transfer in Cryptococcus neoformans by use of biolistic delivery of DNA. J Bacteriol. 1993;175:1405–1411. doi: 10.1128/jb.175.5.1405-1411.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tolkacheva T, McNamara P, Piekarz E, Courchesne W. Cloning of a Cryptococcus neoformans gene, GPA1, encoding a G-protein α-subunit homolog. Infect Immun. 1994;62:2849–2856. doi: 10.1128/iai.62.7.2849-2856.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vartivarian S E, Anaissie E J, Cowart R E, Sprigg H A, Tingler M J, Jacobson E S. Regulation of cryptococcal capsular polysaccharide by iron. J Infect Dis. 1993;167:186–190. doi: 10.1093/infdis/167.1.186. [DOI] [PubMed] [Google Scholar]
- 56.Wang Y, Aisen P, Casadevall A. Cryptococcus neoformans melanin and virulence: mechanism of action. Infect Immun. 1995;63:3131–3136. doi: 10.1128/iai.63.8.3131-3136.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang Y, Casadevall A. Susceptibility of melanized and nonmelanized Cryptococcus neoformans to nitrogen- and oxygen-derived oxidants. Infect Immun. 1994;62:3004–3007. doi: 10.1128/iai.62.7.3004-3007.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Y, Xu H-P, Riggs M, Rodgers L, Wigler M. byr2, a Schizosaccharomyces pombe gene encoding a protein kinase capable of partial suppression of the ras1 mutant phenotype. Mol Cell Biol. 1991;11:3554–3563. doi: 10.1128/mcb.11.7.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Whiteway M, Hougan L, Dignard D, Thomas D Y, Bell L, Saari G C, Grant F J, O'Hara P, MacKay V L. The STE4 and STE18 genes of yeast encode potential β and γ subunits of the mating factor receptor-coupled G protein. Cell. 1989;56:467–477. doi: 10.1016/0092-8674(89)90249-3. [DOI] [PubMed] [Google Scholar]
- 60.Wickes B L, Edman J C. The Cryptococcus neoformans GAL7 gene and its use as an inducible promoter. Mol Microbiol. 1995;16:1099–1109. doi: 10.1111/j.1365-2958.1995.tb02335.x. [DOI] [PubMed] [Google Scholar]
- 61.Wickes B L, Edman U, Edman J C. The Cryptococcus neoformans STE12α gene: a putative Saccharomyces cerevisiae STE12 homologue that is mating type specific. Mol Microbiol. 1997;26:951–960. doi: 10.1046/j.1365-2958.1997.6322001.x. [DOI] [PubMed] [Google Scholar]
- 62.Wickes B L, Mayorga M E, Edman U, Edman J C. Dimorphism and haploid fruiting in Cryptococcus neoformans: association with the α-mating type. Proc Natl Acad Sci USA. 1996;93:7327–7331. doi: 10.1073/pnas.93.14.7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Williamson P R. Biochemical and molecular characterization of the diphenol oxidase of Cryptococcus neoformans: identification as a laccase. J Bacteriol. 1994;176:656–664. doi: 10.1128/jb.176.3.656-664.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu L, Valkema R, Haastert P J M V, Devreotes P N. The G protein β subunit is essential for multiple responses to chemoattractants in Dictyostelium. J Cell Biol. 1995;129:1667–1675. doi: 10.1083/jcb.129.6.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yamamoto M, Imai Y, Watanabe Y. Mating and sporulation in Schizosaccharomyces pombe. In: Pringle J R, Broach J R, Jones E W, editors. The molecular and cellular biology of the yeast Saccharomyces. Vol. 3. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 1037–1106. [Google Scholar]
- 66.Yarfitz S, Provost N M, Hurley J B. Cloning of a Drosophila melanogaster guanine nucleotide regulatory protein β-subunit gene and characterization of its expression during development. Proc Natl Acad Sci USA. 1988;85:7134–7138. doi: 10.1073/pnas.85.19.7134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yue, C., L. M. Cavallo, J. A. Alspaugh, P. Wang, G. M. Cox, J. R. Perfect, and J. Heitman. The STE12α homolog is required for haploid filamentation but largely dispensable for mating and virulence in Cryptococcus neoformans. Genetics, in press. [DOI] [PMC free article] [PubMed]