Abstract
Adoptive cell therapy using tumor infiltrating lymphocytes (TIL) has shown activity in melanoma, but has not been previously evaluated in metastatic non-small cell lung cancer (NSCLC). We conducted a single-arm open-label phase 1 trial (NCT03215810) of TIL administered with nivolumab in 20 patients with advanced NSCLC following initial progression on nivolumab monotherapy. The primary endpoint was safety, and secondary endpoints included objective response rate, duration of response, and T-cell persistence. Autologous TIL was expanded ex vivo from minced tumors cultured with IL-2. Patients received cyclophosphamide and fludarabine lymphodepletion, TIL infusion, and interleukin-2, followed by maintenance nivolumab. The endpoint of safety was met according to the pre-specified criteria of ≤17% rate of severe toxicity (95% confidence interval, 3–29%). Of 13 evaluable patients, 3 had confirmed responses and 11 had reduction in tumor burden, with a median best change of 35%. Two patients achieved complete responses ongoing 1.5 years later. In exploratory analyses, we found T cells recognizing multiple types of cancer mutations were detected after TIL treatment, and were enriched in responding patients. Neoantigen-reactive T cell clonotypes increased and persisted in the peripheral blood after treatment. Cell therapy with autologous TIL is generally safe and clinically active, and may constitute a new treatment strategy in metastatic lung cancer.
Introduction
Despite progress with programmed death receptor 1 (PD-1) immune checkpoint inhibitors in metastatic lung cancer, the majority of patients fail to achieve an objective response. 1Even when combined with first-line platinum doublet chemotherapy, most patients suffer cancer progression within 12 months. 2 While substantial effort is currently dedicated toward identifying new immune checkpoint combination partners, the clinical results thus far have been incremental.3 One limitation of current combination approaches is that non-small cell lung cancers (NSCLC) are often immunologically ‘cold’ tumors, with a paucity of activated, tumor-specific T cells. 4 Therefore, more effective combination immunotherapy is needed for metastatic NSCLC.
To address this challenge, adoptive cell therapy (ACT) using either autologous or allogeneic T cells is a potent strategy. In ACT, patients receive an infusion of a large number of T cells with tumor specificity. ACT using tumor-infiltrating lymphocytes (TIL) cultured from a patient’s tumor has caused durable complete responses in a subset of patients with metastatic melanoma.5 A hallmark of this therapy is the possibility for durable remissions which can last for decades, due in part to the trans-differentiation potential and lifespan of memory T cells. Durable regressions with TIL have also previously been reported in a variety of epithelial malignancies, including cholangiocarcinoma6, cervical7, colorectal8, and breast cancer9. However, this approach has not been evaluated in metastatic lung cancer.
Ex vivo expansion of TIL can release T cells from a suppressive microenvironment and re-activate them to target the tumor. By this method, billions of activated T cells can be produced and infused back into a patient. In contrast, T cells transduced with recombinant monoclonal receptors such as chimeric antigen receptor (CAR) or recombinant T cell receptor (TCR) have met with problems in solid tumors, including absence of stable tumor antigen expression and the need for human leukocyte antigen (HLA) restriction. Toxicity can be severe and unpredictable, due to cross-reactivity or trace expression of tumor-associated antigens in normal epithelial and neural tissue.10,11 Even if a clinical response is observed, acquired resistance is frequently conferred within several months.12,13 This may be due to deletion or mutation of the requisite antigen, antigenic heterogeneity,14 or impaired trafficking.15 In contrast to recombinant monoclonal technology such as CAR, TIL is composed of polyclonal cells capable of targeting multiple tumor antigens. As TIL are derived from native genetically unmodified cells, complications due to engagement of normal host cells is uncommon.16 Since TIL has the potential to target ‘truncal’ neoantigens which are clonally expressed by a cancer cell, resistance due to deficient target antigen expression may be less common.17
We hypothesized that TIL would be feasible and have clinical activity in metastatic NSCLC. Like melanoma, non-small cell lung cancers (NSCLC) also contain T cells that recognize tumor antigens.18 Between 31– 40% of advanced NSCLC patients may have accessible metastatic supraclavicular adenopathy19,20,21, and up to 40% have pleural metastases accessible through video-assisted thoracotomy.22 Moreover, approximately 65% of advanced NSCLC have pulmonary CO diffusion capacity of ≥50% sufficient to tolerate the interstitial fluid shifts induced by interleukin-2 (IL-2) and lymphodepletion.23 We also hypothesized that the infused TIL could recognize antigens comprised from genomic alterations expressed by each individual patients’ tumor, and that the infused TIL clones may persist within the peripheral blood.
We performed a phase I pilot trial of nivolumab followed by TIL in metastatic NSCLC patients, with the primary endpoint of safety, and secondary endpoints of response rate and survival. We harvested tumors from patients naïve to PD(L)1 blockade, to reduce the proportion of terminally differentiated T cells in culture. Those with tumor enlargement or progression proceeded to receive TIL. We found that infusion of TIL in combination with lymphodepletion and IL-2 had manageable toxicity, and mediated tumor regressions in several patients, including complete responses. We retrospectively screened if expanded TIL were capable of reactivity to genomic alterations, and identified that the TIL was capable of recognizing a variety of types of cancer antigens.
Results
Patient Characteristics
Twenty patients were enrolled. Among them, 50% were current or former smokers. The median age was 54 years (range 38 – 75) and median PD-L1 proportion score was 6% (Supplementary Table 1). Forty percent were PD-L1 0% and 30% were PD-L1 >50%. Median non-synonymous tumor mutation burden (TMB) by whole-exome sequencing was 1.5 mutations per megabase of DNA (range 0.1 – 10.2). Four patients had EGFR mutation including two classical Exon 19 deletions, and two patients had EML4-ALK translocations. The predominant enrolled histology was lung adenocarcinoma. One half of patients had not received prior systemic therapy. Most patients had bulky disease, with a mean sum of target lesion diameters of 8.5 cm prior to TIL treatment.
Outcome after TIL Harvest and Initial Nivolumab
Ninety percent of patients were discharged within one day or less after the excisional biopsy for TIL harvest. Nivolumab treatment did not have any previously unreported adverse effects.24 After excisional biopsy of the metastasis, all patients were treated with at least 4 cycles of intravenous nivolumab at a dose of 240 mg every 2 weeks (Fig. 1a). If patients had evidence of clinical benefit after two sequential CT scans, then nivolumab was continued until progression. If there was evidence of progression, as defined by tumor enlargement or new lesions, then we proceeded to administer lymphodepletion and TIL treatment. At least two sequential CT scans were required to reduce the possibility of confounding error due to pseudo-progression.25 Sixteen patients had tumor enlargement or new lesions (Extended Data Fig. 1a). Three patients had eventual PR or CR with initial nivolumab, and therefore nivolumab was continued. Two of these patients with an initial nivolumab benefit eventually had biopsy-proven progression while on nivolumab (Extended Data Fig. 1b) and were subsequently treated with TIL.
Figure 1. Schematic representing study design and patient disposition.
a, Clinical trial schema. Day count is relative to TIL infusion. b, Patient disposition per Consolidated Standards of Reporting Trials (CONSORT) guidelines. Of the 16 patients treated with TIL, 2 of them had initial response to nivolumab, followed by progressive disease prior to receiving TIL therapy. Abbreviations: TKI denotes tyrosine kinase inhibitor; IL-2, interleukin-2; TIL, tumor-infiltrating lymphocyte treatment; PD, progressive disease; q w, every week; ID, patient number.
Feasibility of TIL Expansion and Infusion
TIL were successfully expanded for 95% of patients to a median dose of 95 billion CD3+ cells (range 4.3 – 175). Specific reactivity to autologous tumor cell suspensions was detected in oligoclonal TIL cultures within 13 of 18 patients, as determined by interferon-γ capture by enzyme-linked immunosorbent assay (ELISA). The autologous reactivity of individual patients is presented in Extended Data Fig. 2. Four patients did not receive TIL due to reasons outlined in Fig. 1b. In total, 16 patients received lymphodepletion chemotherapy with cyclophosphamide and fludarabine followed by TIL infusion and IL-2 for 5 days. All of these patients received full course lymphodepletion chemotherapy without dose modification. All treated patients initiated infusional IL-2, and the majority (56%) received all planned doses (Extended Data Fig. 3b). Six patients remained as inpatient after completing cyclophosphamide, and the median length of stay for the entire inpatient stay was 12 days with a range of 7 to 22 days (Extended Data Fig. 3c). Patients with smoking history appeared to have a longer inpatient recovery time.
Safety and Adverse Events
Adverse effects were primarily attributable to the lymphodepletion and interleukin-2 combination (Extended Data Fig. 4 and Table 1). Common non-hematologic adverse events included hypoalbuminemia, hypophosphatemia, nausea, hyponatremia, and diarrhea. Although manageable for the majority of patients, two patients died prior to a response assessment (ID 13, 15). Both patients had declined to Eastern Cooperative Oncology Group (ECOG) performance status 3 and were requiring supplemental oxygen prior to start of lymphodepletion. Patient 13 was an active smoker with severe right carotid stenosis, and had an ischemic right middle cerebral artery stroke at home at Day +12. Patient 15 was 75 year old limited to wheelchair with metastases replacing 70% of his pulmonary parenchyma. As he continued to decline post-TIL, he transitioned to comfort care without a CT scan. After these patients, more stringent pre-lymphodepletion performance status criteria were added. According to pre-specified criteria for the safety endpoint, these two events were defined as severe toxicities, with a final severe toxicity rate of 12.5%. Following lymphodepletion, patients recovered lymphoid and myeloid lineages (Extended Data Fig. 5), with neutrophil count recovering at median 7.5 days (range 4.7 to 20.6). The majority of treatment-emergent adverse events had resolved within one month after TIL. After six months of maintenance nivolumab, one patient had severe thrombocytopenia (ID 16). This event resolved with corticosteroid taper and cessation of nivolumab. The adverse events of each individual patient is listed separately in the Supplementary Appendix 1.
Table 1:
Treatment-emergent adverse events reported with cyclophosphamide, fludarabine, TIL, or interleukin-2.
| Grade | ||||||
|---|---|---|---|---|---|---|
|
|
||||||
| NCI CTCAE Preferred Term | 1 | 2 | 3 | 4 | 5 | Any |
| Lymphocyte count decreased | 0 (0%) | 0 (0%) | 0 0%) | 16 (100%) | 0 (0%) | 16 (100%) |
| White blood cell count decreased | 0 (0%) | 0 (0%) | 1 (6%) | 15 (94%) | 0 (0%) | 16 (100%) |
| Anemia | 1 (6%) | 2 13%) | 13 (81%) | 0 (0%) | 0 0%) | 16 (100%) |
| Platelet count decreased | 0 (0%) | 1 (6%) | 3 19%) | 11 (69%) | 0 (0%) | 15 (94%) |
| Neutrophil count decreased | 0 (0%) | 2 13%) | 1 6%) | 11 (69%) | 0 0%) | 14 (88%) |
| Hypoalbuminemia | 0 (0%) | 13 (81%) | 1 (6%) | 0 (0%) | 0 (0%) | 14 (88%) |
| Nausea | 7 (44%) | 7 44%) | 0 0%) | 0 (0%) | 0 0%) | 14 (88%) |
| Hypophosphatemia | 0 (0%) | 4 (25%) | 8 (50%) | 0 (0%) | 0 (0%) | 12 (75%) |
| Hyponatremia | 8 (50%) | 0 (0%) | 2 (13%) | 1 (6%) | 0 (0%) | 11 (69%) |
| Hypocalcemia | 6 (38%) | 2 (13%) | 2 (13%) | 0 (0%) | 0 (0%) | 10 (63%) |
| Aspartate aminotransferase (AST) increased | 6 (38%) | 1 (6%) | 2 (13%) | 0 (0%) | 0 (0%) | 9 (56%) |
| Fever | 3 (19%) | 5 (31%) | 1 (6%) | 0 (0%) | 0 (0%) | 9 (56%) |
| Diarrhea | 5 (31%) | 4 (25%) | 0 (0%) | 0 (0%) | 0 (0%) | 9 (56%) |
| Rash | 7 (44%) | 2 (13%) | 0 (0%) | 0 (0%) | 0 (0%) | 9 (56%) |
| Hypoxia | 1 (6%) | 5 (31%) | 2 (13%) | 0 (0%) | 0 (0%) | 8 (50%) |
| Dyspnea | 1 (6%) | 6 (38%) | 1 (6%) | 0 (0%) | 0 (0%) | 8 (50%) |
| Alkaline phosphatase increased | 5 (31%) | 2 (13%) | 1 (6%) | 0 (0%) | 0 (0%) | 8 (50%) |
| Sinus tachycardia | 3 (19%) | 5 (31%) | 0 (0%) | 0 (0%) | 0 (0%) | 8 (50%) |
| Alanine aminotransferase (ALT) increased | 5 (31%) | 3 (19%) | 0 (0%) | 0 (0%) | 0 (0%) | 8 (50%) |
| Hyperkalemia | 6 (38%) | 0 (0%) | 0 (0%) | 1 (6%) | 0 (0%) | 7 (44%) |
| Hypotension | 2 (13%) | 4 (25%) | 1 (6%) | 0 (0%) | 0 (0%) | 7 (44%) |
| Chills | 6 (38%) | 1 (6%) | 0 (0%) | 0 (0%) | 0 (0%) | 7 (44%) |
| Anorexia | 3 (19%) | 3 (19%) | 0 (0%) | 0 (0%) | 0 (0%) | 6 (38%) |
| Edema limbs | 3 (19%) | 3 (19%) | 0 (0%) | 0 (0%) | 0 (0%) | 6 (38%) |
| Creatinine increased | 3 (19%) | 1 (6%) | 1 (6%) | 0 (0%) | 0 (0%) | 5 (31%) |
| Prothrombin time (INR) increased | 4 (25%) | 1 (6%) | 0 (0%) | 0 (0%) | 0 (0%) | 5 (31%) |
| Vomiting | 1 (6%) | 4 (25%) | 0 (0%) | 0 (0%) | 0 (0%) | 5 (31%) |
| Hypermagnesemia | 5 (31%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 5 (31%) |
| Dysphagia | 3 (19%) | 1 (6%) | 0 (0%) | 0 (0%) | 0 (0%) | 4 (25%) |
| Dry mouth | 4 (25%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 4 (25%) |
| Hypokalemia | 4 (25%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 4 (25%) |
| Pulmonary edema | 0 (0%) | 2 (13%) | 0 (0%) | 1 (6%) | 0 (0%) | 3 (19%) |
| Hyperglycemia | 1 (6%) | 1 (6%) | 1 (6%) | 0 (0%) | 0 (0%) | 3 (19%) |
| Respiratory term, not otherwise specified | 0 (0%) | 3 (19%) | 0 (0%) | 0 (0%) | 0 (0%) | 3 (19%) |
| Weight loss | 0 (0%) | 3 (19%) | 0 (0%) | 0 (0%) | 0 (0%) | 3 (19%) |
| Dehydration | 2 (13%) | 1 (6%) | 0 (0%) | 0 (0%) | 0 (0%) | 3 (19%) |
| Dizziness | 2 (13%) | 1 (6%) | 0 (0%) | 0 (0%) | 0 (0%) | 3 (19%) |
| Mucositis oral | 2 (13%) | 1 (6%) | 0 (0%) | 0 (0%) | 0 (0%) | 3 (19%) |
| Febrile neutropenia | 3 (19%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 3 (19%) |
Footnotes: Shown are all adverse events (n, %) of the highest grade recorded for all TIL treated patients (n = 16) with possible, probable and definite attribution for any of the trial treatment drugs cyclophosphamide, fludarabine, tumor-infiltrating lymphocytes, or interleukin-2 occurring in >2 patients. National Cancer Institute Common Terminology Criteria for Adverse Events version 4.
Clinical Activity
Initial tumor regression occurred in the majority of patients (11 of 16) at the first CT scan performed one month after TIL adoptive cell therapy. Overall, the median best change in sum of target lesion diameters was −35.5% (range +20 to −100). Radiographic response, including unconfirmed response, occurred for 6 of 13 evaluable patients. These included two complete responses which remain ongoing 1.5 years later (Fig. 2a). Two patients had unconfirmed partial response due to subsequent new brain metastases (ID 02, 05), and another two patients maintained a clinical remission by local ablative therapy of a new ‘escape’ lesion (ID 03, 08) performed between 6 to 17 months post-TIL infusion (Fig. 2b). Another patient (ID 14) had enlargement of her only target lesion, and biopsy showed fibrosis tissue (Extended Data Fig. 6). Since this was a core biopsy, we could not rule out the presence of occult tumor cells elsewhere in the lesion. She remained without disease-related symptoms on trial for 1.5 years, before eventual progression with new lesions. Among the twenty patients enrolled, median overall survival was not reached on an intention-to-treat basis (Extended Data Fig. 7) or post-TIL (Fig. 2d).
Figure 2: Clinical activity of TIL and patient survival.
a, Features of patients who were treated with TIL and their clinical outcomes (n = 16). b, Change in sum of tumor diameters, relative to Week −1 prior to TIL. Non-evaluable patients (ID 03, 13, 15) are not shown. Pt 14, designated as radiographic “PD” post-TIL, had biopsy of only target lesion showing fibrosis with no tumor cells at the time of progression. c, Waterfall plot showing best overall change in sum of diameter of tumor lesions. Change in sum of tumor diameters by RECIST is compared to Week −1 prior to TIL. Pt 14, designated as radiographic “PD” post-TIL, had biopsy showing of only target lesion showing fibrosis with no tumor cells. All patients evaluable with a post-TIL CT scan are included (n =13). Non-evaluable patients (ID 03, 13, 15) are not shown. “*” denotes initial PR with nivolumab, followed by biopsy-proven unequivocal PD (ID 07, 09), ‡ Unconfirmed PR, subsequent non-target PD (ID 02, 05, 31); §§ Had 10 mm increase in target lesion on prior nivolumab. (ID 08).d, Overall survival of all treated patients from date of TIL infusion (n = 16 patients). Hashmarks denote 95% confidence intervals. Median follow-up of 1.6 years (range 0.5 – 2.9) by reverse Kaplan-Meier method. Abbreviations: TPS denotes tumor proportion score by 22C3 antibody immunohistochemistry; IL-2, interleukin-2 and TIL, tumor-infiltrating lymphocyte treatment. TMB, estimated tumor mutation burden; mut/MB, mutations per megabase. ‘Ex’ denotes exon, and Δ denotes exon deletion. Smoking pack-years represents self-reported cigarette packs per day multiplied by total years.
Presence of T cells Recognizing Cancer Mutations
We sequenced whole-exomes and transcriptomes of pretreatment tumors with paired germline and human leukocyte antigen sequencing (Supplementary Table 2). For each patient, we screened infused T cells for recognition of tumor non-synonymous alterations, including indels and translocations. If aberrant cancer testis antigen was detected, these proteins were also screened. We selected between 13–100% of somatic variants for screening, with a median of 54 mutations per patient and range of 8 to 117. If the number of predicted neoantigens was greater than 100, we limited the tested antigens to those with predicted pMHC < 500 nM, 26confirmed mutation coverage, and detectable RNA expression (FPKM >0). In patients who did not have a research apheresis performed at Day +30, we restricted testing to the highest priority neoantigens due to limited cell sample. A summary of how antigens were selected is included in Supplementary Table S3. Custom-synthesized peptide pools were pulsed with autologous antigen-presenting cells (APCs). For each ELISpot assay, co-culture of T cells and antigen presenting cells but without any peptides was used as negative control and a one-way ANOVA with Dunnett’s multiple comparison test was used to determine if the peptide elicited reactivity relative to control. We considered the peptides as TIL-recognized peptides if p < 0.05. If peptides displaying tumor genomic alterations were confirmed as eliciting T cell reactivity, then these peptides were co-cultured with autologous APCs and T cells over 10 days. A summary of all detected neoantigens is shown as a Supplementary Table S4.
One patient with EGFRΔEx19 lung adenocarcinoma (ID 25) had cancer refractory to nivolumab yet achieved a sustained complete response after TIL treatment (Fig. 3a). Her infused TIL contained not only T clonotypes capable of recognizing one somatic mutation, but also several melanoma associated gene (MAGE) cancer testis antigens discovered to be aberrantly expressed in her tumor (Fig. 3b). Using post-TIL peripheral blood T cells, we used an antigen-recognition method to evaluate in vitro expansion of T-cell clones after peptide stimulation.27 Nineteen T cell clonotypes exhibited antigen-specific stimulation (Extended Data Fig. 8a), with mean 140-fold expansion relative to controls. There was a significant increase in these T cell clonotypes circulating in peripheral blood at post-infusion time-points, which persisted over one year (Fig. 3c). Another patient (ID 09) had a complete response to TIL after progressing with a new biopsy-proven soft tissue metastasis while on initial nivolumab (Fig. 3d). Three mutations elicited reactivity from both CD4+ and CD8+ sorted TIL (Fig 4e) and peripheral blood T cells. Like Pt 25, a similar surge in antigen-linked T cells clonotypes occurred after TIL infusion within the peripheral blood (Fig. 3f and Extended Data Fig. 8b). This effect was not evident with nivolumab alone, despite an initial partial response. Another patient (ID 16) with a confirmed partial response had antigens targeting aberrant expression of MAGE-A4. Notably, clinical responses were associated with polyclonal T cell responses against neoantigens derived from a variety of genetic alterations, including single nucleotide variants, insertions/deletions and gene fusions, in addition to cancer testis antigens (Fig. 4). However, specific antigens could not be found in some patients. In an exploratory comparison, T cells functionally recognizing aberrant tumor antigens were identified in most patients who achieved uPR, PR, or CR after TIL compared to non-responders (p = 0.02, Fishers exact test).
Figure 3. Examples of complete responses mediated by TIL recognizing tumor antigens.
a, Patient (ID 25) with an EGFR ΔEx19 tumor had progressive metastases with nivolumab, followed by complete response to TIL. The sum of radiographic target lesions is shown over time with representative contrast-enhanced axial CT images. b, IFN-γ spot formation after co-culture of her post-TIL T cells with her autologous dendritic cells pulsed with synthesized long peptides displaying tumor antigens. Five proteins are shown, of 104 tested, mean ± SEM of 3 plates over 2 experiments. c, Absolute number of antigen-specific T cell receptor clonotypes within the peripheral blood after TIL. The absolute number and proportion increased after TIL, and then gradually decayed. Data is derived from a total of 10 serial blood samples. The proportion of antigen-specific clones in her infused TIL product is shown in the pie chart. d, Patient (ID 09) is a former smoker with lung adenocarcinoma. She had partial response to nivolumab followed by enlargement of metastatic lymph nodes and a new biopsy-proven rib soft tissue metastasis 10 months later. She was then treated with TIL and had a complete response with ongoing absence of radiographic target lesions. Representative coronal contrast-enhanced CT images are shown over time. e, Co-culture of either CD4+ or CD8+ T cells sorted from the final infused TIL with autologous dendritic cells and custom peptides corresponding to tumor neoantigens elicited reactivity. Three positive peptides are shown, of 85 tested. Mean ± SEM of 3 plates over two experiments. f, Absolute number of antigen-specific clonotypes in the peripheral blood increased and then gradually decayed after the TIL infusion. Data is derived from a total of 13 serial blood samples.
Abbreviations: TRVβ, T cell receptor variable beta chain; MAGE, melanoma associated gene; IFN, interferon; Pt, patient.
Figure 4. Summary of tumor-specific antigens tested for all patients.
Top panel shows the total sum of unique genomic alterations tested for each patient. Bottom panel shows the tumor-specific antigens which were detected by autologous T cells. Positive antigens were assessed for T cell reactivity by synthesis of the corresponding recombinant peptides, and identification of IFN-γ colony formation by ELISpot, relative to controls with autologous T cells and dendritic cells only. Mean spot forming units per positive Ag are shown, with n = 2 – 5 tests per antigen. Cultured TIL was used to assess antigen reactivity for all patients except 02, 25, 32, 33 due to high IFN-γ background of the cultured TIL. In these instances, autologous T cells isolated from post-TIL peripheral blood were used.
Abbreviations: IFN, interferon; Pt, patient; TIL, tumor-infiltrating lymphocyte; Ag, antigen.
Persistence and Phenotype of T cells
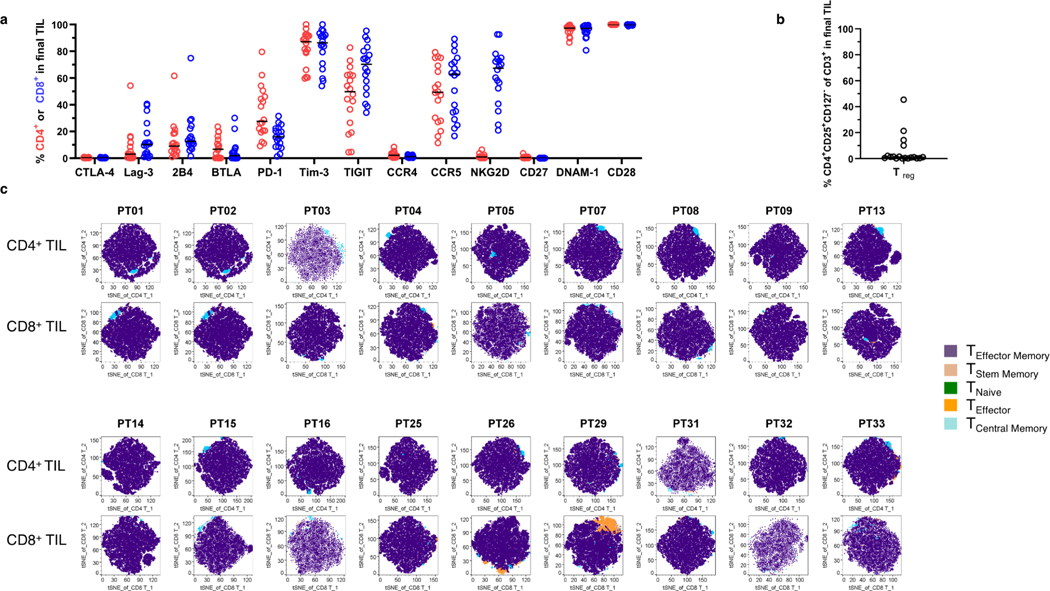
We characterized the infused T cells for markers of terminal differentiation, which can be an indirect sign of impaired function or persistence. We performed flow cytometry upon retained aliquots from the final infused TIL product, and detected high expression of several immune checkpoint ligands, as shown in Extended Data Fig. 9a. In particular, all TIL had high expression of T-cell immunoglobulin and mucin-domain containing-3 (Tim-3) and T cell immunoreceptor with Ig and ITIM domains (TIGIT). Cell surface expression of both Tim-3 and TIGIT receptors is known to inhibit Th1-mediated immune responses and promote tolerance. Moreover, several patients had a high proportion of cells classified as regulatory T cells (Extended Data Fig. 9b). We performed t-distributed stochastic neighbor embedding (t-SNE) of the memory differentiation markers based on lineage-specifying transcription factors, and the TIL differentiation status was primarily composed of CCR7-CD45RA- T effector memory cells28 (Extended Data Fig. 9c). This predominance of terminally differentiated cells were likely due to the ≥7 total weeks of accrued time in culture media under chronic IL-2 stimulation.
Patients achieved durable conversion of the phenotype of their peripheral T cells after TIL infusion. This effect was not evident from initial nivolumab alone. In particular, we compared the ability of T cells to secrete cytokine proteins, derived before and after TIL infusion. We assessed the polyfunctional strength index (PSI), which is an indicator of the number of cells capable of secreting multiple types of cytokines. Since PSI reflects the ability of a T cell to carry out multiple functions, it is recognized as a metric for the potency of cell therapy, and for the efficacy of vaccines. 29 We performed a single-cell assay of 12 cytokines, testing up to 1000 individual CD8+ and CD4+ cells after stimulation with CD28 antibody, relative to normalized background levels of each protein. PSI was increased at post-infusion timepoints after TIL (Fig. 5a). Likewise, peripheral T cells changed to an effector memory (TEM) lineage characteristic of the infused TIL product for CD8+ and CD4+ cells, followed by gradual recovery of naïve T cell types (Fig. 5b) Since this treatment effect upon the peripheral compartment was not limited to the lifespan of the infused T cell pool, it may have been related in part to immune reconstitution or persistence.
Figure 5. Change in phenotype and genotype of peripheral T cells after TIL infusion.
a, Increase in circulating subsets of polyfunctional CD8+ or CD4+ T cells at post-infusion timepoints. *Friedman test using mixed-effects model (REML) with stacked matching with Geisser-Greenhouse correction. Shown is the multiplicity adjusted, two-sided p value using Dunnett’s control for multiple comparisons, with family-wise alpha of 0.05. Exact p value 0.0000008. Stacked bars denote mean ± SEM of available dataset with 12 tested patients. b, Increase in peripheral CD45RA−CCR7− effector memory T cells at post-infusion timepoints for all patients with available timepoints (n = 15) with 8 serial timepoints collected per patient. Stacked area curves denote mean ± SEM percentage of CD8+ or CD4+ T cells. Lineage definitions: Naïve T cells, CD45RA+CCR7+CD95−; central memory, CCR7+CD45RA−; effector memory, CCR7-CD45RA−; stem cell-like memory, CCR7+CD45RA+CD95+; effector T cells, CCR7-CD45RA+”.
d, Overlap of TRVβ chain productive rearrangements between final infused TIL product and peripheral blood T cells at indicated time-points, using Morasita’s index. Bars denote mean ± SEM of all patients with available timepoints for analysis (n = 16). Abbreviations: TRVβ, T cell receptor variable beta chain; SEM, standard error; TIL, tumor-infiltrating lymphocyte infusion; nivo, nivolumab; MIP, macrophage inflammatory protein; IFN, interferon. REML, restricted (or residual, or reduced) maximum likelihood; CCR, chemokine receptor.
To address this question of persistence, we examined the complementarity-determining regions of the T cell receptor beta chain (TRVB) over time. Each receptor motif is unique to a clonal population of cells and reflects the total number of each clone in circulation. We performed TRVB sequencing on snap-frozen tumor and serial peripheral blood samples for patients. We found the infused TIL clonotypes largely partially replaced the patients’ baseline peripheral T cell repertoire, and then gradually decayed in proportion over the ensuing months (Fig. 5c). No apparent association between TIL clonotype systemic persistence and response was observed. Likewise, in an exploratory comparison, no clear association between radiographic response and either infused cell dose or autologous tumor reactivity of TIL was evident (Extended Data Fig. 10). There was also no discernable differences in the yield of TIL final cell count based on prior lines of treatment. TIL cultures did undergo change in clonotypic composition, with apparent enrichment of T cell clones in several patients during TIL culture compared to baseline tumor (Extended Data Fig. 10c). This enrichment may have been due in part to antigen-driven expansion of T cells in culture, although the specificity of the clonotypes was unknown. This was accompanied by turnover in the most prevalent T cell clonotypes, possibly related to either clonal selection or stochastic drift (Extended Data Fig. 10d). Overall, the TIL cultures were more clonal and less diverse than the baseline intratumoral T cells (median 6% vs. 31%, p < 0.0001), (Extended Data Fig. 10e). We concluded the final infused TIL composition was not simply a scaled up reproduction of the original T cells resident within the harvested tumor of each patient. Instead, the TIL underwent clonal contraction within culture, which may have impaired the polyvalence and ultimate efficacy of the cell product.
Discussion
We found that excisional tumor biopsy and nivolumab followed by administration of cyclophosphamide, fludarabine, IL-2 with TIL infusion in pretreated metastatic lung cancer was feasible in an academic center setting, and had manageable adverse effects. We observed that lymphocytes could be successfully expanded from most patients’ tumors, and were largely capable of autologous tumor recognition. In addition to two durable complete responses, we also observed some patients who derived clinical benefit in a variety of other ways, including local ablation of a single metachronous new lesion. One durable complete response occurred in a TMBlow, PD-L1negative never smoker, who was refractory to nivolumab. This may be particularly encouraging for the large subset of never-smoker patients, for whom immune checkpoint inhibitors have historically had limited efficacy.30 Although neoantigen load was predictive of clinical benefit in a small melanoma cohort treated with TIL, 31 it seems that infusion of T cells which target anti-tumor antigens is of particular importance. Together with results in breast cancer9, our data indicate that TIL can mediate effective responses in tumor subtypes which are not sensitive to traditional immune checkpoint targeted therapy. Therefore, therapy with TIL may extend the scope and impact of immunotherapy into wider populations.
Our report adds to growing evidence that TIL adoptive cell therapy may be active in a variety of epithelial malignancies. In contrast to prior experience in melanoma, we found a high proportion of CD4+ TIL in NSCLC. Despite this, CD4+ predominant TIL were capable of mediating responses, and antigen-specific CD4 clones recognizing immunogenic tumor mutations were identified (Fig. 3). Antigen-specific CD4 cells recognizing mutations such as BRAFV600E have previously been identified in melanoma,32 and have mediated tumor regressions in gastrointestinal malignancies.33 Therefore, an important requirement for TIL efficacy may be the presence of sufficient T cells which recognize immunogenic genomic alterations. In our case, we found autologous T cells reactive to against multiple types of tumor antigens (Fig. 4). Further study is needed to determine the threshold polyclonality, or diversity of antigens targeted by an infused TIL repertoire, required to achieve a durable clinical response.
Although infusion of mutation-reactive T cells appears to be important for a successful TIL response, the lineage differentiation status of the T cells may be paramount. CD8+ T cells with stem-like surface markers were associated with tumor cytolysis and durability of responses in a large cohort of melanoma patients treated with TIL. 34 In fact, the majority of mutation-reactive T cells were terminally differentiated, and not associated with clinical benefit. However, a small subset of mutation-reactive T cells were detected in the CD39-CD69- stem-like state, and these were associated durable remission. Therefore, both specificity and lineage appear to be critical factors in melanoma, and further study in NSCLC is warranted.
Lymphodepletion chemotherapy and IL-2 are prerequisite to ensure homeostatic expansion and engraftment of the infused TIL. This incumbent toxicity, together with the required production infrastructure, has historically limited experimentation with TIL to select centers. We had 2 patients with early deaths related to worsening performance status and inanition caused by rapidly progressive disease, combined with physiologic stress of lymphodepletion and IL-2. By the time TIL could be manufactured, these patients were infirm and requiring supplemental oxygen. Future strategies to expedite TIL manufacturing 35 and curtail the physiologic footprint of lymphodepletion may be needed to ensure the broader adoption of TIL in the lung cancer community. To this end, newer, less invasive approaches to isolate and expansion tumor neoepitope reactive T cells are being pioneered to expand to indications with limited tumor access. For example, expansion of TIL is possible from core needle biopsies, with final product counts in the 1 – 10 billion cell range across a variety of epithelial malignancies.36 This approach is currently being tested in registration trials.37 PD-1+ selection may enrich for neoantigen-reactive T cells from the peripheral blood of patients with melanoma and gastrointestinal cancer 38. In ovarian cancer, tumor-reactive TIL are reported primarily in the PD-1+ fraction of cells, and a T-cell-inflamed genetic signature or high TMB may also help predict for antitumor reactivity.39 Likewise, sorting for memory T cell markers may capture most of the mutated neoantigen-reactive T cells in the peripheral blood of epithelial cancer patients. 40,41
Our study was characterized by important limitations derived from its design. The completion of the trial within two academic cancer centers does not necessarily reflect feasibility of daily practice in a less specialized setting. Along these lines, we enrolled a highly selected group with a larger-than-normal proportion of treatment-naïve, never-smoker, and oncogene-addicted lung cancer patients. This may have diminished the external validity of our results, since severe adverse events may be more frequent in patients with smoking-related diseases who are encountered in common practice. Within this combination therapy, the individual component contributions of IL-2, cyclophosphamide, fludarabine, and nivolumab are difficult to assess. To the best of our knowledge, neither lymphodepletion nor IL-2 monotherapy have historically reported durable responses in NSCLC. In this trial, we gave initial nivolumab monotherapy in unselected NSCLC patients, as a trial alternative to a standard immune checkpoint inhibitor and applicable chemotherapy. Patients were informed that this nivolumab was not superior to standard chemotherapy.42 Although uncorroborated, many patients opted for nivolumab followed by the prospect of cell therapy, due in part to interest in avoiding platinum-doublet chemotherapy and the historical experience of TIL in other cancers. Our trial included post-TIL nivolumab for up to 1 year, with the intent of deterring subsequent relapse related to PD-L1 expression. Nonetheless, the necessity or optimal duration of maintenance therapy after TIL remains to be proven.
We chose to exclusively manufacture TIL from anti-PD(L)-1 treatment naïve tumors. Exhausted TIL clones appear to have limited ability for phenotype transition to an activated state after PD(L)-1 blockade.43 Therefore, we expected that pre-treatment tumor harvest followed by PD-1 blockade would represent the most ideal scenario to test TIL efficacy, while still controlling for the effect of nivolumab monotherapy. In particular, one patient (ID 09) still had a complete response to TIL after suffering relapse after 1 year on nivolumab. This suggests that anti-PD-1 treatment-naive TIL can still mediate specificity and cytotoxicity against PD-1-experienced tumor cells. Despite the observation that bulk TIL expressed multiple immune checkpoint proteins, we demonstrated that neoepitope-specific T cells retained functionally capable of interferon-γ secretion after antigen challenge. In a recent phase II trial of TIL for treatment refractory metastatic melanoma, 79% of responders had ipilimumab-refractory disease, and all had previous PD-1 blockade, demonstrating that TIL still has efficacy for some patients after progression on immune checkpoint inhibitors.44 Based on its activity and safety profile, TIL is a rational therapy to further investigate for fit, motivated metastatic NSCLC patients. Future TIL trials will ideally enroll either NSCLC patients who have progressed after combination chemotherapy and immunotherapy, or patients who are predicted to have poor chance of response to traditional immune checkpoint inhibitors. Larger trials are required to further define the optimal biomarkers of response, and the efficacy of TIL harvested from patients who have progressed on prior PD-(L)1 based treatments.
Online Methods
Data Availability
TRVβ sequencing of tumor and peripheral T cells as the project “TIL trial for NSCLC” are available in the immuneACCESS free public database: https://clients.adaptivebiotech.com/immuneaccess. Tumor whole exome sequencing and RNA-Seq raw data files (accession code: phs002486) is available in the public NIH dbGaP data browser, https://www.ncbi.nlm.nih.gov/projects/gapprev/gap/cgi-bin/preview1.cgi?GAP_phs_code=UgsrhvZ3dGC4uQSh. Raw data on expression, allelic frequency, and predicted MHC affinity for all identified neoantigens are provided in the excel spreadsheet uploaded as in Supplementary Table S4. The study protocol is uploaded as a supplementary file. In such circumstances that source data is not inferred from specified Tables/Figures, it is available upon request as a comma-delimited format from the study authors.
Patients
From October 2017 to January 2020, we enrolled patients in a dual center phase I investigator-initiated trial designed to test the feasibility of adoptive cell therapy with TIL (ClinicalTrials.gov NCT03215810). Eligible patients were adults with histologically proven stage 4 NSCLC who were not candidates for surgical resection for curative intent or a curative-intent radiation modality. Patients were required to have an accessible metastasis to procure for TIL with acceptable anticipated perioperative risk, and also at least one separate additional measurable tumor lesion on computed tomography (CT). All patients were required to have received fewer than six prior lines of systemic therapy. Prior therapy could not have included a programmed death 1 (PD-1) or PD-L1 inhibitor. Patients were required to have sufficient cardiopulmonary function, including: a cardiac stress test showing no reversible ischemia; adjusted diffusing capacity for carbon monoxide (DLCO) of greater than 50% of predicted as assessed by pulmonary function testing; and left ventricular ejection fraction of greater than 50% as assessed by multigated acquisition (MUGA) scan. Patients with active brain metastases, autoimmune conditions, or acquired immunodeficiency were excluded. Active brain metastases were excluded due to concern regarding symptomatic central nervous system progression during the ≥63 day TIL production to infusion time. Patients were required to have adequate normal organ and marrow function, defined as: hemoglobin ≥ 9 g/dL; absolute neutrophil count ≥ 1.0 × 109/L (≥1000 per mm3); platelet count ≥ 100 × 109/L (>100,000 per mm3); albumin ≥ 2.5 g/dl; prothrombin time, partial thromboplastin time, serum bilirubin, serum creatinine ≤ 1.5x of the institutional upper limit of normal; and aspartate aminotransferase and alanine aminotransferase ≤ 2.5x of the institutional upper limit of normal.
Patients with epidermal growth factor receptor (EGFR) mutation or anaplastic lymphoma kinase (ALK) translocation were allowed if they had progressed on ≥1 prior approved tyrosine kinase inhibitor (TKI). Comprehensive exome sequencing was performed on the resected tumor obtained after enrollment. This step revealed an uncommon EGFR L861Q in Patient 07 and a variant EML4-ALK in Patient 12 which was not detected on cell-free DNA assay prior to enrollment. Full details on eligibility are provided in the Supplementary Protocol and clinicaltrials.gov.
Study Conduct
This study was conducted at two academic medical centers in the United States. The primary endpoints were safety and feasibility. Key secondary endpoints were radiologic response rate and overall survival. Adverse events were recorded according to National Cancer Institute Common Terminology Criteria for Adverse Events, v4.0. Tumors were assessed per Response Evaluation Criteria in Solid Tumors (RECIST), version 1.1. Patients were deemed eligible and enrolled after passing all screening procedures, prior to tumor resection.
Study Design
After excisional biopsy of the metastasis, patients were treated with 4 cycles of intravenous nivolumab at a dose of 240 mg every 2 weeks (Fig. 1a). If patients had evidence of clinical benefit after two sequential CT scans, then nivolumab was continued until progression. If there was evidence of progression, as defined by tumor enlargement or new lesions, then we proceeded to administer lymphodepletion and TIL treatment. TIL infusion was defined as Day 0. Patients received lymphodepletion intravenously with cyclophosphamide at a dose of 60 mg per kg daily for Day −7 and −6 with fludarabine daily at a dose of 30 mg per m2 of body surface area for Days −7 to −3. Patients were then admitted to the hospital and received TIL infusion on Day 0, followed by continuous aldesleukin infusion beginning 12 hours later at a dose of 18 million international units (MIU) per m2 over 6, then 12, and then 24 hours followed by 4.5 MIU/m2 over 24 hours for 3 consecutive days.45 Patients were discharged once they had >1000 k/μL peripheral neutrophils. CT scans were performed every 6 weeks and leukapheresis at Week 4 to collect immune cells for functional studies. Then patients resumed nivolumab 480 mg every 4 weeks for up to 1 year. If patients had a single site of progressive tumor, this was formally recorded as progression. However, these patients were permitted to remain on trial after successful local ablative therapy using either resection or stereotactic radiation. Oral prophylaxis for pneumocystis and herpes simplex virus was given for 6 months after TIL treatment.
Study Supervision
This study was approved by Adverra Institutional Review Board (FWA 0023875) and underwent FDA safety review (IND 17489). Written informed consent was obtained, including germline DNA sequencing. The trial was designed and reported by the authors, who assure the accuracy of the data. Only the authors participated in manuscript preparation. The trial was primarily supported by Stand Up to Cancer Foundation. Nivolumab was supplied by E.R. Squibb & Sons LLC. Aldesleukin (IL-2) was supplied by Clinigen Group, Inc. Funding to the Moffitt Cell Therapies Facility was provided by Iovance Biotherapeutics, Inc. The companies played no other role in the study or report.
TIL Product Processing, Selection and Expansion
The majority of harvested metastases were pleural nodules or supraclavicular lymph nodes. Freshly resected patient tumors were dissected by the surgeon in a sterile back table of the operating room and adventitial tissue removed. Tumor was then placed in sterile RPMI media container and delivered to the Moffitt Cell Therapy Facility. Tumors were processed within 3 to 12 hours of receipt by a cell technician within an ISO 7 cGMP certified clean room. Tumor fragments were minced into 3-mm cubes. Each tumor was cut into 48 fragments. Tumor fragments were placed individually into two separate 24-well culture plates in media supplemented with 600 IU/mL of IL-2 and 10% human serum from type AB donors. Fragments were surveyed for proliferation every 2–3 days for up to 5 weeks. TIL were expanded to a new well of a 24-well plate when they became ≥80% confluent. TIL derived from each fragment was kept separate. Media containing 6,000-IU/mL IL-2 was supplemented every 3–4 days to maintain TIL growth.
T cells were screened for specificity to autologous tumor by incubation with a suspension of tumor cells and interferon-gamma (IFN-γ) quantification. For autologous reactivity testing, a portion of excess tumor was disaggregated into a single-cell suspension to create substrate tumor for enzyme linked immunoassays (ELISA). Tumor was minced and carefully mixed in digestion media with collagenase (type II and type IV), hyaluronidase, and DNAase (Fisher Scientific Co., Pittsburgh, PA, USA). Then undigested tumor was filtered from the cell suspension to generate a single-cell suspension. Intact cells were enriched with a Ficoll-Hypaque (GE Healthcare Bio-Sciences, Pittsburgh, PA, USA) gradient, and viable tumor cells were enumerated by trypan blue exclusion. Lymphocytes from fragment wells with the most pronounced proliferation were tested for autologous tumor reactivity by overnight co-culture with autologous tumor or allogeneic control tumor cell line in a 1:5 concentration. Allogeneic tumor cells were included as optional controls, based on predicted partial class I allele match to the patient. These included research resource identification (RRID): CVCL_0B68, CVCL_6789, CVCL_8058, CVCL_A164, CVCL_8051, CVCL_4632, CVCL_8054, and CVCL_8787. Two 96-well plates were used per patient. A standard curve between 62 to 1000 pg/mL was set up for each plate. IFN-γ within the supernatants was detected by ELISA, as shown in Extended Data Fig. 2, and analysed using SkanIt Software 4.1. Autologous reactivity was judged based on approximately twofold or more higher IFN-γ compared with culture media alone. These TIL pools containing the reactive TIL were selected, pooled and cryopreserved for later rapid expansion protocol (REP). If insufficient or no autologously reactive TIL were identified, then additional fragments were used based on their growth rate.
Positive cultures were pooled and cryopreserved for each patient. If patients had progressive disease on nivolumab, then TIL were thawed and expanded in CD3 antibody, irradiated allogeneic feeder cells, and IL2 over 14 days. For REP, TIL were cultured in in G-Rex 100 MCS flasks and incubated at 37°C and 5% CO2 at a 1:200 ratio with a layer of irradiated allogeneic donor feeder cells, and IL-2 at a concentration of 3,000 IU/mL and anti-CD3 (Ortho Biotech) at a concentration of 30 ng/mL were added to the flasks. Flasks were incubated at 37◦C and monitored for the next 7 days and split as needed to maintain the TIL concentration at 2 × 106 /mL. Then cells were collected, washed, and concentrated to 1.0 L or less. Cell viability was tested using acridine orange (AO) and propidium iodide (PI) dyes. Total live cells were counted using a Cellometer Auto 2000 (Nexcelom Bioscience, Lawrence, MA, USA). The final product was tested for sterility and then gravity infused into the patient at a rate of approximately 300 mL per hour. To characterize the final product, cells from were stained with 7-AAD, CD3-FITC, CD4-PE, CD45-V500, and CD8-APC. Flow cytometry data was collected using a FACSCanto (BD Biosciences, San Jose, CA, USA) and FlowJo Software (Treestar, Ashland, OR, USA). Upon harvest, TIL product underwent Quality Control testing and had to meet the following criteria prior to release for patient administration: ≥ 45% CD45+ by flow cytometry, <5EU/Kg of endotoxin, no detectable mycoplasma, negative gram stain, ≥70% viability and sterile blood culture from day −3 of REP. The TIL were also assessed by flow cytometry for CD8 (mean = 57.27 ± 33.77%) and CD4 (mean = 41.00 ± 32.86%) composition.
Flow Cytometry of Lymphocytes
Human peripheral blood samples were collected in 4 heparin tubes at baseline, following nivolumab infusion and multiple timepoints following TIL infusion. Peripheral blood mononuclear cells (PBMC) were collected using a Ficoll gradient and cryopreserved in 10% DMSO and FBS. Cells were thawed in media, and subsequently stained in PBS containing 5% FBS (vol/vol, FACS buffer) with: CD3 BUV496, CD56 BUV563, CD4 BUV737, CD197 BV421, CD28 BV480, CD14 BV605, CD19 BV605, CD95 BV711, CD195 BV786, CD127 PE, CD194 PE-Cy7, CD45RA Alexa488, CD25 PerCP-Cy5.5, NKG2D APC, Tim3 BV421, PD1 BV480, CD226 BV711, CTLA4 BV786, Lag3 PE, TIGIT PE-Cy7, CD244 Alexa488, CD27 PerCP-Cy5.5, BTLA APC from BD Biosciences. Dead cells were excluded using the Zombie NIR Fixable Viability Kit from Biolegend, incubated at 4 °C for 1 hr, then washed twice with FACS buffer, and finally fixed in PBS containing 1% paraformaldehyde before running flow cytometry. Cells were acquired on a BD FACSymphony™ A5, and data were analyzed with FlowJo Version 10.0 software. All cell gates were drawn uniformly for analysis across patients and time points, with gating strategy provided in Supplementary Appendix 2. Plots of t-SNE were generated by Flowjo Version 10.6.1 according to the expression of CD45RA, CCR7, CD28 and CD95. Different memory T cell subsets were shown using separate colors.
Polyfunctional Strength Index
Polyfunctional strength index (PSI) measures the ability of single CD3+ cells to secrete multiple different cytokines after stimulation. 29 CD8+ T cells were sorted from TIL and peripheral blood, and stimulated with CD28 and CD3 antibody as previously described. 46 Testing was performed using the Isoplexis IsoCode chip using the 12 human cytokine single-cell proteome panel. For PSI, data from empty chambers are used to measure the background level for each protein. These data were used to generate protein abundance histograms, which are fitted by normal distributions and nonparametric methods, judged by goodness of fit.
The mean of the histogram, identified by the best fit, is used as the background level. Single-cell data were then normalized by subtracting this background, so that different samples can be compared. The single-cell data were then fitted by finite mixture models, and the gate that separates the cytokine-producing and non–cytokine-producing cells was identified. Pre- and post- infusion timepoints were compared using a mixed-effects model (REML) with stacked matching with Geisser-Greenhouse correction, in an exploratory comparison. Multiplicity adjusted p value was performed using Dunnett’s control.
Generation of autologous dendritic cells
The plastic adherence method was used to generate monocyte-derived dendritic cells. Autologous PBMCs or apheresis samples were thawed and resuspended at 2–5·106/mL with AIM-V media (Life Technologies). The cells were incubated in a tissue culture flask of an appropriate size at 37°C, 5% CO2. After 90 minutes, non-adherent cells were collected and the flasks were washed with AIM-V media twice with an interval of 60 minutes, after which DC media were added. DC media were made from RPMI 1640 containing 5% human serum (Sigma), 100 U/ml penicillin and 100 μg/ml streptomycin, 2 mM L-glutamine (media supplements were from Life Technologies), 800 IU/ml GM-CSF and 800 U/ml IL-4 (cytokines were from Peprotech). On day 3, fresh DC media was supplemented. Dendritic cells were harvested on day 5–7 and frozen in 10% DMSO for co-culture experiments.
Screening T cells for reactivity to antigens using the ELISpot assay
Both effector T cells & DCs were pre-thawed 24 hours before co-culture in IL-2 free media. In the ELISpot assay, PVDF membrane plates (Millipore, MAIPSWU10) were pre-activated with 50 μl 70% ethanol per well for 2 min and washed thrice with PBS. Then 50 μl purified IFN-γ capture antibody (Mabtech, clone: 1-D1K) were added per well for incubation at 4°C overnight. Before co-culture, wells were washed thrice with PBS and incubated with 100 μl AIM-V media at room temperature for 1 hour. In co-culture, when using TIL as effector T cells, 1•104 to 3•104 T cells were placed per well in a 96-well flat bottom plate. When T cells isolated from apheresis samples (4 weeks post TIL infusion) were used as effector cells, 1•105 cells were used per well in a 96-well flat bottom plate. Effector T cells were co-cultured with 5•103 - 1•104 DCs cells loading 1–10 μg/mL tumor antigen peptides. Cytomegalovirus, Epstein-Barr, or influenza (CEF) viral peptides and/or plate-bound OKT3 (1 μg/mL) were used as positive controls. Effector T cells only and/or effector T cells co-cultured with unloaded DCs were used as negative controls.
After 24 hours of co-culture, the plate was washed 6x with PBS plus 0.05% Tween-20 and then incubated with 100 μl per well of 0.22 μm filtered 1 μg/mL biotinylated anti-IFN-γ detection antibody (Mabtech, clone: 7-B6–1) for 2 hours. Another 4x wash with PBS-T was performed and 100 μl per well of streptavidin-ALP (Mabtech, diluted 1:1000) were added for a 1-hour incubation. The plate was washed 6x with PBS and developed with 100 μl per well of 0.45 μm filtered BCIP/NBT substrate solution (KPL, Inc.) for 10 minutes. Finally, the plate was washed thoroughly with cold tap water and then scanned and auto-counted using an ImmunoSpot ELISpot plate reader (Cellular Technologies, Ltd).
Identification of tumor antigen specific TCR clonotypes (FEST)
Our protocol to identify tumor antigen specific TCR clonotypes followed the published MANAFEST assay 47 with modifications. Apheresis samples (4 weeks post TIL infusion) were thawed and T cells were isolated using EasySep Human T cell Enrichment Kit (Stemcell Technologies). Then T cells were washed and resuspended at 2•106 /mL in AIM-V media. Dendritic cells were thawed and resuspended at 5•104 /mL in AIM-V media. After identifying T cell-recognized tumor antigens, 125 μl T cells were co-cultured with 125 μl DCs in a 96-well round bottom plate and tumor antigen proteins/peptides were added at the concentration of 1–10 μg/mL. On day 3, supernatants were half replaced with fresh AIM-V media containing 100 IU/mL IL-2, 50 ng/mL IL-7 and 50 ng/mL IL-15. On day 7, supernatants were half replaced with fresh AIM-V media containing 200 IU/mL IL-2, 50 ng/mL IL-7 and 50 ng/mL IL-15. Cell cultures were harvested on day 10 for DNA extraction (Qiagen) and further TCRVβ sequencing. Analysis of TCRseq data to figure out tumor antigen specific expansion of TCR clonotypes was performed using the online tool http://www.statapps.onc.jhmi.edu/FEST47
The absolute number of T cell clonotypes in the peripheral blood was calculated by using the absolute lymphocyte count derived from automated cell differential provided by the medical laboratory, multiplied by the percentage of CD3+ gated of all lymphocytes by flow cytometry, multiplied by the frequency of this specific clone as a proportion of all productive rearrangements derived from immunoSEQ TRVβ DNA sequencing of that sample.
Analysis of Tumor Markers
Resected baseline tumors were stained for PD-L1 using 22C3 pharmDx immunohistochemistry antibody and scored using tumor proportion score (TPS) by a trained pathologist. Tumor mutation burden (TMB) was originally estimated using targeted exome sequencing from Foundation Medicine. After completion of WES, it was calculated by dividing the total number of non-synonymous mutations by the total length of exome regions. All tumor marker inferences were defined as exploratory in the study protocol.
Nucleic Acid Extraction
Portions of the same tumor resected for TIL were snap-frozen in liquid nitrogen and stored at −80 C. In batches, tumor regions of interest were selected based on H&E sections using laser-capture macrodissection (LCMD) and pulverized into aliquots of 15 mg. Aliquots were extracted for RNA and DNA using Allprep kits (Qiagen). DNA and RNA was quantified using Invitrogen Qubit Fluorometer, purity assessed using Thermo Scientific™ NanoDrop and integrity assessed with Agilent 4200 TapeStation system. Mononuclear cells were isolated from peripheral blood collected in EDTA tubes, using a Ficoll gradient. Cell pellets were snap-frozen, and later DNA was extracted in water using mini-prep kit (Qiagen). Quality metrics were assessed as described above.
Whole Exome Sequencing
Whole exome sequencing was performed in order to identify somatic mutations in DNA extracted from pre-treatment tumor tissue. Two hundred ng of DNA was used as input into the Agilent SureSelect XT Clinical Research Exome kit, which includes the exon targets of Agilent’s v5 whole-exome kit, with increased coverage at 5000 disease-associated targets. Briefly, for each tumor DNA sample and germline DNA sample, a genomic DNA library was constructed according to the manufacturer’s protocol and the size and quality of the library was evaluated using the Agilent BioAnalyzer. An equimolar amount of library DNA was used for a whole-exome enrichment using the Agilent capture baits and after quantitative PCR library quantitation and quality control analysis on the BioAnalyzer, 75-base paired-end sequences were generated using v2 chemistry on an Illumina NextSeq 500 sequencer.
Human Leukocyte Antigen (HLA) Class I and II locus sequencing was performed in the Moffitt HLA laboratory (ASHI accreditation 07–3-FL-18–1). DNA from nucleated peripheral blood cells underwent QC using Nanodrop followed by next generation sequencing using the MIA FORA FLEX HLA Typing Kit (Immucor, Norcross GA). A confirmatory sequence-specific oligonucleotide (SSO) DNA typing (LABType, One Lambda) was performed to resolve ambiguous alleles.
Transcriptome Sequencing
RNA-sequencing libraries were prepared from RNA extracted from pre-treatment tumor tissue using the NuGen FFPE RNA-Seq Multiplex System (Tecan US, Inc., Morrisville, NC). Briefly, 50 ng of DNase-treated RNA was used to generate cDNA and a strand-specific library following the manufacturer’s protocol. Library molecules containing ribosomal RNA sequences were depleted using the NuGen AnyDeplete probe-based enzymatic process. The final libraries were assessed for quality on the Agilent TapeStation (Agilent Technologies, Inc., Wilmington DE), and quantitative RT-PCR for library quantification was performed using the Kapa Library Quantification Kit (Roche Sequencing, Pleasanton, CA). The libraries were sequenced on the Illumina NextSeq 500 sequencer with a 75-base paired-end run in order to generate 80–100 million read pairs per sample.
T Cell Repertoire Sequencing
T-cell receptor repertoire analysis was performed using Adaptive Biotechnologies immunoSEQ v3 assay, which employs bias-controlled multiplex PCR amplification and high-throughput sequencing to target rearranged T-cell receptor genes. The manufacturer’s protocol was followed in order to utilize the immunoSEQ hsTCRB kit to amplify the complementarity determining region 3 (CDR3) locus from genomic DNA extracted from sorted T-cells or tissue. Following the confirmation of amplification and a successful final library preparation, sequencing was performed on the Illumina NextSeq 500 to a depth of 2 million pairs of sequencing reads per sample for survey-level analysis, or 5–6 million pairs per sample for deep-level analysis. The data were then analyzed using the Adaptive Biotechnologies immunoSEQ Analyzer software, which identifies and counts the V, D, and J genes, filters non-productive sequences, and reports and tracks T cell clonality.
Proteomics
Tissue samples were pulverized and denaturing buffer was used to extract the proteins, followed by protein reduction, alkylation and trypsin digestion. The tryptic peptides were acidified and desalted with C18 cartridge. After lyophilization, the peptides were fractionated off-line with basic pH reversed phase HPLC and 24 concatenated fractions were collected. LC-MS/MS analysis of each fraction was performed with a nanoflow ultra high performance liquid chromatograph (RSLC, Dionex, Sunnyvale, CA) coupled to an electrospray bench top orbitrap mass spectrometer (Q-Exactive plus, Thermo, San Jose, CA). The result was analyzed with MaxQuant software for protein identification and label free quantitation. 48
Identification of Somatic Mutations
Somatic mutations were identified from whole-exome sequence reads and aligned to the reference human genome (hg19) with the Burrows-Wheeler Aligner (BWA). 49 Insertion/deletion realignment and quality score recalibration were performed with the Genome Analysis ToolKit.50 Tumor specific mutations were identified with Strelka 51 and MuTect 52, and were annotated to determine genic context, including non-synonymous, missense, splicing calls, using ANNOVAR 53. Only somatic mutations predicted by both algorithms were included for subsequent neoantigen prediction. Additional contextual information was incorporated, including allele frequency was derived from available resources including 1000 Genomes, the NHLBI Exome Sequence Project, and the Exome Aggregation Consortium 54, in silico function impact predictions, and observed impacts from databases including ClinVar (http://www.ncbi.nlm.nih.gov/clinvar/) and the Collection Of Somatic Mutations In Cancer (COSMIC).
Gene Expression and Fusion Detection
Sequence reads were aligned to the human reference genome in a splice-aware fashion using STAR55, allowing for accurate alignments of sequences across introns. Aligned sequences were assigned to exons using the HTseq package. 56 DESeq2 was used for normalization of gene expression.57 Aberrant cancer testis antigen (MAGE) expression was identified based on gene expression level and verified using the sum of peptide intensities for the corresponding fasta protein proteomic sequencing described above. Gene fusions were identified from RNAseq data using FusionCatcher, https://www.biorxiv.org/content/10.1101/011650v1.
Neoantigen Prediction and Prioritization
Neoantigens will be identified by extracting altered peptides from mutation data with ANNOVAR or directly from FusionCatcher results, and then predicting MHC binding against patient-specific HLA type using NetMHC 58 NetMHCpan 26 and NetMHCIIpan 59 as implemented by the Immune Epitope Database 60. HLA types were derived from clinical HLA laboratory sequencing data. Neoantigens were prioritized by a combination of predicted MHC binding affinity, variant allele frequency, RNA expression of the genes where the mutation is potentially located, and evidence of reads carrying the mutated base.
Statistical Analysis
Adverse events were continuously monitored. Acceptable safety was prospectively defined as a dose-limiting toxicity rate of 17% or less, using a Pocock–type stopping boundary with continuous monitoring.61 Dose-limiting toxicity was defined as grade ≥4 toxicity with attribution to ACT, detailed with sample size justification in the Supplementary Protocol. Baseline tumor measurements were defined on Week −1 CT prior to TIL, with subsequent assessment every 6 weeks. Overall survival was calculated using the Kaplan–Meier method, and 95% confidence interval included for medians and curves. Pathological, genomic, and functional immunologic testing was performed and analyzed as described in the Supplementary Protocol. Reported p values were two-sided with a significance of 0.05, unless otherwise noted. Normality was not assumed, and non-parametric tests were performed where applicable. For functional expansion of specific T-cells (FEST) analysis, a threshold p-value of 0.05 was adjusted by Benjamini-Hochberg procedure to control for multiple comparisons, as previously published.47 Data were analyzed in R version 3.6.3 (2020–02-29), using Windows 10 ×64 operating system, ming w32, user interface “RTerm” language English, United States, and Prism 9.0 (121) October 22, 2020.
Extended Data
Extended Data Fig. 1.
Clinical follow-up after initial nivolumab. a, Sum of tumor diameters per RECIST v1.1 over time measured by serial CT scan. Patients with evidence of tumor enlargement or new lesions within 10 weeks of initial nivolumab were treated with TIL infusion. All enrolled patients (n = 20) are included. b, Clinical follow-up of patients after initial nivolumab with clinicopathologic features in a Swimmer’s plot. Pt 04 received bridging crizotinib due to rapid progression on nivolumab. Abbreviations: TPS denotes tumor proportion score using 22C3 PD-L1 antibody. TMB; estimated tumor mutation burden based upon exome sequencing. mut/MB mutations per mega-base. ‘Ex’ denotes exon, and ‘Δ’ denotes mutation conferring exon deletion. Smoking packyears represents self-reported cigarette packs per day multiplied by total years.
Extended Data Fig. 2.
Autologous reactivity screening of TIL cultured from individual tumor fragments in combination with baseline tumor cell suspensions. Shown are the interferon-γ enzyme-linked immunosorbent assays conducted without (a) and with (b) CD3 bead positive controls. Each graph is labeled with the trial patient ID. The x-axis represents the TIL fragment selected for testing. Each fragment number represents an autologous T cell line generated from a unique tumor morsel cultured in IL-2 and media between 3 to 5 weeks. Each culture is labelled by its original fragment identification number, from 48 total per patient. Post-decimal numerals indicate the tumor number, if multiple tumor sites were harvested for the same patient. An asterisk denotes whether the fragment was deemed autologously reactive. Shown is the mean ± SEM of triplicate wells from a single ELISA experiment. Each bar denotes one biologically independent culture sample (n = 1). “≤10” denotes a sample with absorbance reading below the lowest point of the standard curve performed for each assay. “TIL Ctrl” denotes a TIL culture derived from an unrelated patient to serve as a positive control for the CD3 beads and a negative control for autologous substrate. Allogeneic cancer cell lines were selected as an optional control based on partial HLA class I allele compatibility with the patient. Patient ID 12 did not have sufficient TIL growth for manufacture, and ID 26 did not have sufficient tumor for autologous reactivity testing.
Extended Data Fig. 3.
Feasibility of TIL harvest and intermediate-dose interleukin-2 infusion for all patients. a, Total hours of length of stay (LOS) for inpatient admissions after TIL harvest surgery. Two patients had postoperative airleak requiring multiple days of inpatient observation. All enrolled patients (n = 20) are included. b, The total duration of infusional IL-2 for all 16 patients after TIL treatment. Patients received TIL infusion on Day 0, followed by continuous IL-2 infusion beginning 12 hours later at a dose of 18 million international units (MIU) per m2 over 6, then 12, and then 24 hours followed by 4.5 MIU/m2 over 24 hours for 3 consecutive days. c, Total inpatient length of stay for all patients. Some patients were discharged to outpatient between cyclophosphamide admission and TIL admission.
Extended Data Fig. 4.
Treatment-emergent adverse events reported with cyclophosphamide, fludarabine, TIL, and IL-2. a, Adverse events grouped by National Cancer Institute preferred term occurring in 20% or more of patients attributable to cyclophosphamide, fludarabine, TIL, or IL-2. All TIL treated patients (n = 16) are included. b, All adverse events grouped by date of onset within 3 months of TIL. Patients with multiple events for a given term are counted once, using the maximum grade under each preferred term. Abbreviations: AE denotes adverse events; AST denotes aspartate aminotransferase; ALT denotes alanine aminotransferase; IL-2, interleukin-2.
Extended Data Fig. 5.
Change in lymphocytes and neutrophils with lymphodepletion, TIL, and IL2. a, Initial peak followed by recovery of peripheral blood absolute lymphocyte count. b, peripheral blood absolute neutrophil count recovered by median of 7.5 days (range 4.7 – 20.6). Shown is absolute cell count (1000 cells per mm3). TIL infusion is “Day 0”. All TIL treated patients are shown (n = 16)
Extended Data Fig. 6.
Radiographic enlargement of a target lesion in patient 14. Patient had biopsy-proven lung adenocarcinoma biopsy proven from a lymph node metastasis and pleural mass. She had increase in her only target lesion pleural metastasis post-TIL and biopsy revealed fibrosis. Whole exome sequencing conducted on the original tumor showed over four hundred somatic mutations. Only 3 mutations could be detected in the post-TIL biopsy despite 200x depth of sequencing. She had new metastases appear 1.4 years later. Shown is a representative hematoxylin and eosin (H&E) image from 15 high-powered field views, selected by diagnostic clinical pathologists. Bar denotes approximately 100 μm. Pre-TIL biopsy comprises 5 separate preserved blocks from 1 tumor from a single procedure performed on one day. Post-TIL biopsy comprises 3 core needle samples obtained from 4 total passes from a single procedure performed on one day. Each somatic mutation count represents 1 biologically independent sample.
Extended Data Fig. 7.
Overall survival from enrollment. a, Overall survival of all enrolled patients (n = 20) from date of enrollment. b, Overall survival of patients who had not received any anti-cancer systemic therapy prior to enrollment (n = 12), and c, of patients who had received at least one line of previous systemic therapy (n = 8). Hashmarks denote 95% confidence intervals for survival probability. All patients were assessed for survival through the data cut-off of 10 Sept. 2020.
Extended Data Fig. 8.
In vitro expansion of autologous T-cell clonotypes after stimulation with peptide antigens. Clonotypes with significant increase using autologous T and dendritic cells from a, Patient 25 and b, Patient 9. T cells were co-cultured with autologous dendritic cells for 10 days and compared to controls wells including T cells only and no peptides. TCR Vβ CDR3 AA denotes T-cell receptor Vβ complementaritydetermining region 3 amino acid. Fold expansion is relative to baseline control without peptide.
Extended Data Fig. 9.
Expression of immune checkpoint ligands and cell composition of final infused TIL product a, Expression of immune checkpoint and cell activation markers assessed by flow cytometry as a proportion of either CD4+ or CD8+ T cells (n = 18 patients with 1 biologically independent experiment per patient, bar denotes median). b, Proportion of regulatory T cells (CD4+ CD25+CD127dim). (n = 18 patients with 1 biologically independent experiment per patient). c, TIL cell population stratified by tdistributed stochastic neighbor embedding (t-SNE) mapping of memory differentiation subsets. Defined cell populations were assigned specific colors. Each patient is labeled by Pt ID. There are 18 patients presented in total, with 1 biologically independent experiment per patient.
Extended Data Fig. 10.
Phenotype and clonotype features of final infused TIL Phenotype and clonotype features of final infused TIL a, Best overall response (BORR) and proportion of tumor-specific fragments in the final product. All patients with sufficient TIL for autologous reactivity testing performance are shown (n = 18). Pt 26 had insufficient tumor digest for autologous reactivity testing, and Pt 12 had insufficient pre-REP TIL for testing. A proportion were tested for reactivity with autologous tumor suspension using ELISA for interferon-γ. Pink is the proportion of autologous-reactively cultures pooled for the final rapid expansion. Gray are either non-specific or negative cultures added to the pool, to ensure sufficient starting cell numbers. b, Patient BORR and dose of CD8+ or CD4+ cells in final expanded TIL product. All patients with final manufactured TIL are shown (n =18). Pt 30 pre-REP TIL is still cryopreserved, and Pt 12 had insufficient pre-REP TIL to manufacture. 48 minced tumor fragments were cultured in IL-2. c, Change in T cell clonotypic composition between original intratumor T cells and final infused TIL. Across patients, there were variable numbers of enriched clones in the TIL culture, with a range 13 to 1011 clones. A large proportion of clones in each patient were not detected in the baseline tumor. d, Comparison of the most prevalent one hundred clones in the manufactured TIL culture with baseline tumor, with total number shown on the x-axis and overlap in middle. High repertoire turnover during TIL culture was observed in most samples. Several cultures shared no high frequency clones with their baseline tumors. e, Productive clonality Simpson clonality increased from baseline tumor to final cultured TIL (Exact p = 0.000013, two-sided Wilcoxon signed rank test). Shown is the median and 95% confidence interval overlaying the individual values. All patients with manufactured TIL are shown (n =19). Abbreviations: NT; not treated. NE; not evaluable. uPR, unconfirmed partial response.
Supplementary Material
Acknowledgments
Supported by a Catalyst Award Clinical Trial grant through the Stand Up to Cancer Foundation (SU2C-AACR-CT04-17; EBH, SJA); Barbara Bauer Prelude to a Cure Foundation grant to BCC; E.R. Squibb L.L.C. for nivolumab supply; Iovance Biotherapeutics Inc as a research grant, Clinigen Inc for aldesleukin supply, a NCI Cancer Center Support Grant (P30-CA076292; EBH) and 2018 Young Investigator award from Adaptive Biotechnologies Inc. to BCC. Stand Up 2 Cancer is a program of the Entertainment Industry Foundation administered by the American Association for Cancer Research.
We thank the patients and their families who participated in this research; members of the Moffitt Cell Therapies Facility including L. Kelley and C. Cox for cell manufacturing; C. Yang, T Mesa, at the Moffitt Molecular Genomics Facility, M. Wloch, N. Smith, K. Fahrner at the Moffitt Tissue Core and P. Cano at the Moffitt Histocompatibility Leukocyte Antigen laboratory; T Boyle, G. Zhang for assistance with clinical specimens; C. Ulge for grant administration; M. Avedon for translational project management; and other members of our clinical and laboratory research teams. Cell sorting for this project was done on instruments in the Moffitt Flow Cytometry Core. Sequencing was performed by the Moffitt Molecular Genomics and Proteomics & Metabolomics Core Facilities (supported in part by NCI Cancer Center Support Grant P30-CA076292). Clinical care and trial coordination was provided by the Moffitt Immune and Cellular Therapies (ICE-T) department. We thank Dr B. Banbury for assistance with the graphs of T cell clonotypes.
Moffitt Cancer Center (PI: B.C.C.) was the sponsor of the trial. The trial was primarily supported by Stand Up to Cancer Foundation through the AACR Catalyst grant mechanism. Nivolumab was supplied by E.R. Squibb & Sons LLC. Aldesleukin (IL-2) was supplied by Clinigen Group Inc. Funding to the Moffitt Cell Therapies Facility was provided by Iovance Biotherapeutics. The companies played no other role in the study or report.
B.C.C. has received speaking fees from AstraZeneca plc, ARIAD Pharmaceuticals, Inc. Hoffmann-La Roche AG.and consultant fees; Xilio Inc, Achilles plc, ; E.R. Squibb LLC. ; F. Hoffmann-La Roche AG. ; AstraZeneca plc.; AbbVie, KSQ Therapeutics Inc. ; GlaxoSmithKline plc. ; Gilead Sciences inc. ; Celgene corp ; Research funding; NeoGenomics Laboratories; Adaptive Biotechnologies Corp. and patent application filed WO2020263919A1. A.A.C. has received speaking fees from Genentech, Merck, Celgene, Takeda, and consultant fees; Amgen, Jazz, AstraZeneca, Pfizer, Novartis, AbbVie, BMS, research funding Astra Zeneca, Novartis, BMS, outside the submitted work. J.R.C.G. consultant fees with Leidos, consultant and non-financial support Anixa Biosciences, consultant and and non-financial support from Compass Therapeutics; and patent WO2020033923A1 pending. E.B.B. has consultant fees with Amgen, Janssen, outside the submitted work. S.K. reports non-financial and research financial support from BMS and AstraZeneca plc. A.M.L. received financial support from by Iovance Biotherapeutics, Inc. J.E.M reports laboratory research support from Iovance Biotherapeutics, Inc., for research outside the submitted work; and a patent 62668246 licensed to Iovance Biotherapeutics, Inc. S.A.P.T. reports grants from Swim Across America Foundation Grant Recipient, grants from V Foundation Grant Recipient, grants from American Cancer Society Research Scholar Grant, grants from State of Florida Bankhead-Coley Grant, grants from Iovance Biotherapeutics, grants from Intellia Therapeutics, grants from Myst Therapeutics, grants from NIH-NCI: U54 CA193489-01A1, grants from NIH-NCI: R01 CA241559, grants from NIH-NCI: U01 CA244100-01, grants from NIH-NCI: R01 CA239219-01A1, grants from Provectus Biopharaceuticals, outside the submitted work; In addition, S.A.P.T. has a patent 61/973,006 61/978,112 with royalties paid to Iovance Biotherapeutics, and a patent Immunotherapy targeted to tumors expressing receptors with royalties paid to Tuhura Biopharma. A.N.S. received non-financial and research grants from Daiichi Sankyo, Novartis, Mersana, Genmab, Eli Lilly, outside the submitted work. A.A.S. reports grants from National Cancer Institute K23 receipient, grants from Ocala Royal Dames Grant Recipient, grants from Swim Across America Foundation Grant Recipient, personal fees from Guidepoint, Inc, personal fees from Gerson Lehrman Group, Inc, consultant fees from Defined health, Inc, Iovance Biotherapeutics, Inc, Physicians’ Educational Resource (PER), LLC, Medscape, outside the submitted work; In addition, A.A.S. has a patent Compositions and methods for improving tumor-infiltrating lymphocytes for adoptive cell therapy 61/973,002 with royalties paid to Iovance Biotherapeutics, a patent Rapid method for culture of tumor-infiltrating lymphocytes from core needle biopsies of solid tumors issued, and a patent Tumor-infiltrating lymphocytes and stapled peptoid peptide hybrid peptidomimetics, 16/157,174 issued. J.K.T. has grant support through P30-CA76292 and patent pending for Large Data Set Negative Information Storage Model. C.W. has patent application filed for Generation of T-cell receptors (TCRs) that react to neoantigens. S.J.A. has received advisor fees from Achilles plc, Amgen Inc., AstraZeneca plc, E.R. Squibb LLC, Caris Life Sciences Inc, Celsius Therapeutics Inc, G1 Therapeutics Inc, GlaxoSmithKline plc, Memgen LLC, Merck & Co. Inc, Nektar Therapeutics, RAPT Therapeutics Inc, Venn Therapeutics LLC, Glympse Inc, EMD Serano Inc, Samyang Biopharm USA, Inc., and research funding from Cellular Biomedicine Group Inc.
Footnotes
Author Contributions Statement
The trial protocol was designed and written by the authors (B.C.C., A.A.S., S.J.A.). S.A.A. and F.J.K. obtained grant funding. L.B.M., W.C. coordinated the trial initiation and study procedures. Z.J.T., D-T.C. analyzed the clinical data and statistical analysis. E.B, B.C.C, S.J.A. interpreted clinical data. C.W. and D.R.N performed the immunologic assays. S.K. and C.W. performed and supervised flow cytometry. J.K.T., J.Y. performed and interpreted bioinformatics analyses. J.T.S. performed immunohistochemical staining scoring. Work related to TIL production was conducted and supervised by A.M.L. RNA and DNA sequencing preparations were supervised by B.C.C and C.W. Genomic sequencing was performed and supervised by S.Y. Proteomic sequencing was performed and supervised by J.M.K. and B.Y. T.T., A.N.S., A.A.C., S.J.A., and B.C.C., informed patients and were responsible for patient care. E.M.T. and J.E.M performed TIL surgery. S.P.T. E.B.H., S.J.A., J.C.G. provided scientific input during protocol design and interpretation of the study. The manuscript was written by B.C.C. and C.W., together with all co-authors, who vouch for the accuracy of the data reported and adherence to the protocol. All authors edited and approved the manuscript.
Competing Interest Statement
The remaining authors have no relevant conflicts to disclose.
References
- 1.Man J, Millican J, Mulvey A, Gebski V. & Hui R. Response rate and survival at key timepoints with PD-1 blockade versus chemotherapy in PD-L1 subgroups: Meta-analysis of metastatic NSCLC trials. JNCI Cancer Spectrum (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Addeo A, Banna GL, Metro G. & Di Maio M. Chemotherapy in combination with immune checkpoint inhibitors for the first-line treatment of patients with advanced non-small cell lung cancer: a systematic review and literature-based meta-analysis. Frontiers in oncology 9, 264 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hellmann MD, et al. Nivolumab plus ipilimumab in advanced non–small-cell lung cancer. New England Journal of Medicine 381, 2020–2031 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Schoenfeld AJ & Hellmann MD Acquired resistance to immune checkpoint inhibitors. Cancer Cell 37, 443–455 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosenberg SA, et al. Durable complete responses in heavily pretreated patients with metastatic melanoma using T-cell transfer immunotherapy. Clinical Cancer Research 17, 4550–4557 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tran E, et al. Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science 344, 641–645 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stevanović S, et al. Complete regression of metastatic cervical cancer after treatment with human papillomavirus–targeted tumor-infiltrating T cells. Journal of Clinical Oncology 33, 1543 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tran E, et al. T-cell transfer therapy targeting mutant KRAS in cancer. New England Journal of Medicine 375, 2255–2262 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zacharakis N, et al. Immune recognition of somatic mutations leading to complete durable regression in metastatic breast cancer. Nature medicine 24, 724–730 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duinkerken CW, et al. Sensorineural Hearing Loss after Adoptive Cell Immunotherapy for Melanoma Using MART-1 Specific T Cells: A Case Report and Its Pathophysiology. Otology & Neurotology 40, e674–e678 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Blumenschein G, et al. 278 Phase I clinical trial evaluating the safety of ADP-A2M10 SPEAR T-cells in patients with MAGE-A10+ advanced non-small cell lung cancer. (BMJ Specialist Journals, 2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu Y-C, et al. Treatment of patients with metastatic cancer using a major histocompatibility complex class II–restricted T-cell receptor targeting the cancer germline antigen MAGE-A3. Journal of Clinical Oncology 35, 3322 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.D’Angelo SP, et al. Antitumor activity associated with prolonged persistence of adoptively transferred NY-ESO-1 c259T cells in synovial sarcoma. Cancer discovery 8, 944–957 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xia Y, et al. Treatment of metastatic non small cell lung cancer with NY ESO 1 specific TCR engineered T cells in a phase I clinical trial: A case report. Oncology letters 16, 6998–7007 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haas AR, et al. Phase I study of lentiviral-transduced chimeric antigen receptor-modified T cells recognizing mesothelin in advanced solid cancers. Molecular Therapy 27, 1919–1929 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yeh S, et al. Ocular and systemic autoimmunity after successful tumor-infiltrating lymphocyte immunotherapy for recurrent, metastatic melanoma. Ophthalmology 116, 981–989. e981 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kast F, Klein C, Umaña P, Gros A. & Gasser S. Advances in identification and selection of personalized neoantigen/T-cell pairs for autologous adoptive T cell therapies. OncoImmunology 10, 1869389 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Renkvist N, Castelli C, Robbins PF & Parmiani G. A listing of human tumor antigens recognized by T cells. Cancer Immunology, Immunotherapy 50, 3–15 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ahmed M, Flannery A, Daneshvar C. & Breen D. PET and neck ultrasound for the detection of cervical lymphadenopathy in patients with lung cancer and mediastinal lymphadenopathy. Respiration 96, 138–143 (2018). [DOI] [PubMed] [Google Scholar]
- 20.Fultz PJ, et al. Detection and diagnosis of nonpalpable supraclavicular lymph nodes in lung cancer at CT and US. Radiology 222, 245–251 (2002). [DOI] [PubMed] [Google Scholar]
- 21.Niwa H, et al. Patterns of mediastinal and supraclavicular metastases in apical invasive lung cancer--importance of supraclavicular lymph node dissection. [Zasshi][Journal]. Nihon Kyobu Geka Gakkai 42, 1142–1147 (1994). [PubMed] [Google Scholar]
- 22.Doebele RC, et al. Oncogene status predicts patterns of metastatic spread in treatment-naive nonsmall cell lung cancer. Cancer 118, 4502–4511 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guerra JLL, et al. Changes in pulmonary function after three-dimensional conformal radiotherapy, intensity-modulated radiotherapy, or proton beam therapy for non-small-cell lung cancer. International Journal of Radiation Oncology* Biology* Physics 83, e537–e543 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weber JS, et al. Safety profile of nivolumab monotherapy: a pooled analysis of patients with advanced melanoma. (2017). [DOI] [PubMed] [Google Scholar]
- 25.Katz SI, et al. Radiologic Pseudoprogression during Anti–PD-1 Therapy for Advanced Non–Small Cell Lung Cancer. Journal of Thoracic Oncology 13, 978–986 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Jurtz V, et al. NetMHCpan-4.0: improved peptide–MHC class I interaction predictions integrating eluted ligand and peptide binding affinity data. The Journal of Immunology 199, 3360–3368 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Forde PM, et al. Neoadjuvant PD-1 blockade in resectable lung cancer. New England Journal of Medicine 378, 1976–1986 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Golubovskaya V. & Wu L. Different subsets of T cells, memory, effector functions, and CAR-T immunotherapy. Cancers 8, 36 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Precopio ML, et al. Immunization with vaccinia virus induces polyfunctional and phenotypically distinctive CD8+ T cell responses. The Journal of experimental medicine 204, 1405–1416 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gainor JF, et al. EGFR mutations and ALK rearrangements are associated with low response rates to PD-1 pathway blockade in non–small cell lung cancer: a retrospective analysis. Clinical cancer research 22, 4585–4593 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lauss M, et al. Mutational and putative neoantigen load predict clinical benefit of adoptive T cell therapy in melanoma. Nature communications 8, 1–11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Veatch JR, et al. Tumor-infiltrating BRAF V600E-specific CD4+ T cells correlated with complete clinical response in melanoma. The Journal of clinical investigation 128, 1563–1568 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tran E, et al. Immunogenicity of somatic mutations in human gastrointestinal cancers. Science 350, 1387–1390 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krishna S, et al. Stem-like CD8 T cells mediate response of adoptive cell immunotherapy against human cancer. Science 370, 1328–1334 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simpson-Abelson M, D’Arigo K, Hilton F, Fardis M. & Chartier C. 1053P Iovance generation-2 tumour-infiltrating lymphocyte (TIL) product is reinvigorated during the manufacturing process. Annals of Oncology 31, S720 (2020). [Google Scholar]
- 36.Abelson M, D’Arigo K, Hilton F, Fardis M. & Chartier C. Expanding Iovance’s tumor infiltrating lymphocytes (TIL) from core biopsies for adoptive T cell therapy using a 22-day manufacturing process. in JOURNAL FOR IMMUNOTHERAPY OF CANCER, Vol. 7 (BMC CAMPUS, 4 CRINAN ST, LONDON N1 9XW, ENGLAND, 2019). [Google Scholar]
- 37.NCT04614103. Autologous LN-145 in Patients With Metastatic Non-Small-Cell Lung Cancer. in U.S. National Library of Medicine (Clinicaltrials.gov, 2021). [Google Scholar]
- 38.Gros A, et al. Recognition of human gastrointestinal cancer neoantigens by circulating PD-1+ lymphocytes. The Journal of clinical investigation 129(2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salas-Benito D, et al. The mutational load and a T-cell inflamed tumour phenotype identify ovarian cancer patients rendering tumour-reactive T cells from PD-1+ tumour-infiltrating lymphocytes. British Journal of Cancer, 1–12 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Malekzadeh P, et al. Antigen experienced T cells from peripheral blood recognize p53 neoantigens. Clinical Cancer Research 26, 1267–1276 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cafri G, et al. Memory T cells targeting oncogenic mutations detected in peripheral blood of epithelial cancer patients. Nature communications 10, 1–9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carbone DP, et al. First-line nivolumab in stage IV or recurrent non–small-cell lung cancer. New England Journal of Medicine 376, 2415–2426 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yost KE, et al. Clonal replacement of tumor-specific T cells following PD-1 blockade. Nature medicine 25, 1251–1259 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sarnaik A, et al. Long-term follow up of lifileucel (LN-144) cryopreserved autologous tumor infiltrating lymphocyte therapy in patients with advanced melanoma progressed on multiple prior therapies. J Clin Oncol 38, 10006 (2020). [Google Scholar]
References for Online Methods
- 45.Andersen R, et al. Long-lasting complete responses in patients with metastatic melanoma after adoptive cell therapy with tumor-infiltrating lymphocytes and an attenuated IL2 regimen. Clinical Cancer Research 22, 3734–3745 (2016). [DOI] [PubMed] [Google Scholar]
- 46.Xue Q, et al. Single-cell multiplexed cytokine profiling of CD19 CAR-T cells reveals a diverse landscape of polyfunctional antigen-specific response. Journal for immunotherapy of cancer 5, 85 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Danilova L, et al. The Mutation-Associated Neoantigen Functional Expansion of Specific T Cells (MANAFEST) Assay: A Sensitive Platform for Monitoring Antitumor Immunity. Cancer Immunol Res 6, 888–899 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cox J. & Mann M. MaxQuant enables high peptide identification rates, individualized ppb-range mass accuracies and proteome-wide protein quantification. Nature biotechnology 26, 1367–1372 (2008). [DOI] [PubMed] [Google Scholar]
- 49.Li H. & Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature genetics 43, 491 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saunders CT, et al. Strelka: accurate somatic small-variant calling from sequenced tumor–normal sample pairs. Bioinformatics 28, 1811–1817 (2012). [DOI] [PubMed] [Google Scholar]
- 52.Cibulskis K, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nature biotechnology 31, 213–219 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang K, Li M. & Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic acids research 38, e164–e164 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lek M, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Anders S, Pyl PT & Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Anders S. & Huber W. Differential expression analysis for sequence count data. Nature Precedings, 1–1 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nielsen M, et al. Reliable prediction of T-cell epitopes using neural networks with novel sequence representations. Protein Science 12, 1007–1017 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reynisson B, et al. Improved prediction of MHC II antigen presentation through integration and motif deconvolution of mass spectrometry MHC eluted ligand data. Journal of Proteome Research (2020). [DOI] [PubMed] [Google Scholar]
- 60.Vita R, et al. The immune epitope database (IEDB): 2018 update. Nucleic acids research 47, D339–D343 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ivanova A, Qaqish BF & Schell MJ Continuous toxicity monitoring in phase II trials in oncology. Biometrics 61, 540–545 (2005). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
TRVβ sequencing of tumor and peripheral T cells as the project “TIL trial for NSCLC” are available in the immuneACCESS free public database: https://clients.adaptivebiotech.com/immuneaccess. Tumor whole exome sequencing and RNA-Seq raw data files (accession code: phs002486) is available in the public NIH dbGaP data browser, https://www.ncbi.nlm.nih.gov/projects/gapprev/gap/cgi-bin/preview1.cgi?GAP_phs_code=UgsrhvZ3dGC4uQSh. Raw data on expression, allelic frequency, and predicted MHC affinity for all identified neoantigens are provided in the excel spreadsheet uploaded as in Supplementary Table S4. The study protocol is uploaded as a supplementary file. In such circumstances that source data is not inferred from specified Tables/Figures, it is available upon request as a comma-delimited format from the study authors.